Abstract
The Us9 protein is a phosphorylated membrane protein present in the lipid envelope of pseudorabies virus (PRV) particles in a unique tail-anchored type II membrane topology. In this report, we demonstrate that the steady-state residence of the Us9 protein is in a cellular compartment in or near the trans-Golgi network (TGN). Through internalization assays with an enhanced green fluorescent protein epitope-tagged Us9 protein, we demonstrate that the maintenance of Us9 to the TGN region is a dynamic process involving retrieval of molecules from the cell surface. Deletion analysis of the cytoplasmic tail reveals that an acidic cluster containing putative phosphorylation sites is necessary for the recycling of Us9 from the plasma membrane. The absence of this cluster results in the relocalization of Us9 to the plasma membrane due to a defect in endocytosis. The acidic motif, however, does not contain signals needed to direct the incorporation of Us9 into viral envelopes. In this study, we also investigate the role of a dileucine endocytosis signal in the Us9 cytoplasmic tail in the recycling and retention of Us9 to the TGN region. Site-directed mutagenesis of the dileucine motif results in an increase in Us9 plasma membrane staining and a partial internalization defect.
The unique short (Us) region of most alphaherpesvirus genomes contains an open reading frame called Us9. The Us9 gene was first described and the protein product was first characterized in herpes simplex virus type 1 (HSV-1) (18, 42). Sequences homologous to HSV-1 Us9 have been found in the Us regions of HSV-2 (16) and varicella-zoster virus (VZV) (15), as well as in the animal pathogens pseudorabies virus (PRV) (51, 61), bovine herpesvirus 1 (35), equine herpesvirus 1 (EHV-1) (12, 17, 57), feline herpesvirus 1 (68), canine herpesvirus (21, 60), and simian herpesvirus B (29). The only alphaherpesviruses sequenced to date that do not contain a Us9 gene are the oncogenic avian Marek’s disease herpesvirus (7) and herpesvirus of turkeys (69). Although the Us9 gene product has not yet been assigned a function in vitro or in vivo (47), the fact that the gene is conserved among the alphaherpesviruses, nonessential in tissue culture for HSV-1 and PRV, and absent from several attenuated strains of PRV (37, 44, 45, 50) implies that the Us9 protein plays a role in virus-host interactions.
We have recently shown that the 98-amino-acid PRV Us9 protein is a type II membrane protein present abundantly in the virion envelope (6). We also demonstrated that the protein is expressed as multiple phosphorylated polypeptides ranging from 17 to 20 kDa late in infection (6). A striking finding was that the PRV Us9 protein is localized in a compartment reminiscent of the Golgi apparatus in both infected and transfected cells. The protein was also visible in small cytoplasmic vesicles and on the plasma membrane, but it was difficult to detect in the endoplasmic reticulum (ER) or the nuclear membranes (6). The Us9 protein is predicted to be anchored in membranes by a 26-amino-acid stretch of hydrophobic amino acids that is bracketed by several charged amino acids (6). The topology of the protein in a type II orientation predicts that there is a carboxy-terminal extension of only 3 amino acids on the outside of the viral particle or plasma membrane, 26 amino acids spanning the lipid bilayer, and 68 amino acids in the tegument region or cellular cytosol. The PRV Us9 protein falls into a unique subclass of type II membrane proteins called tail-anchored membrane proteins (31, 32, 36, 66). Tail-anchored membrane proteins have no obvious signal sequence, an amino terminus exposed to the cytosol, and a carboxy-terminal membrane anchor. Unlike type I membrane proteins, tail-anchored type II membrane proteins are posttranslationally inserted into membranes (31, 32, 36, 66).
Given this unusual topology, it was of interest to determine the protein sequences responsible for the intracellular trafficking and steady-state localization of the PRV Us9 protein to the Golgi apparatus and virion envelopes. Immunogold electron microscopy of PK15 cells stably expressing a Us9-enhanced green fluorescent protein (EGFP) fusion protein was used to show that the fusion protein is highly concentrated both in cytoplasmic vesicles and Golgi-associated membranes. Through double indirect immunofluorescence microscopy with known Golgi and trans-Golgi network (TGN) markers, we have been able to define the intracellular localization of the Us9-EGFP fusion protein to the TGN region. We then used indirect immunofluorescence with confocal microscopy to show that the intracellular distribution of Us9 is dynamic resulting, in part, from the retrieval of molecules from the plasma membrane. We introduced defined mutations in the Us9 gene and found that the amino-terminal cytoplasmic tail contains at least two motifs important in the maintenance of Us9 in the TGN region and recycling from the plasma membrane. These motifs include an acidic cluster containing putative tyrosine and casein kinase I and II phosphorylation sites as well as a dileucine endocytosis signal. Deletion of the acidic region resulted in a protein that was redistributed from the TGN region to the plasma membrane. This mislocalized protein was still fully competent to be incorporated efficiently into virion envelopes. Site-directed mutagenesis of the dileucine endocytosis signal resulted in an increase of Us9 molecules in the plasma membrane and a partial defect in internalization.
MATERIALS AND METHODS
Virus strains and cells.
The PRV Becker (Be) strain used in this study was propagated on PK15 cells as described previously (65). PK15-BB14 cells expressing a Us9-EGFP fusion protein (6) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1 mg of G418 (Gibco/BRL) per ml.
PK15-AB35 cells and PK15-AB37 cells express Us9(d46-55)-EGFP and Us9(L30-31A)-EGFP fusion proteins, respectively (see below for descriptions of Us9 mutants). To construct these cell lines, 40% confluent dishes of PK15 cells were transfected by the calcium phosphate method (19) with 10 μg of plasmid pAB35 or pAB37. Two days after transfection, the cells were split 1:20 and replated into selection medium containing 1 mg of G418 per ml. G418-positive cells were pooled and grown to confluence. A uniform population of EGFP-expressing cells were sorted from nonexpressing, low-expressing, and high-expressing cells by using a FACscan cell sorter (Becton Dickinson).
Antisera.
The polyvalent rabbit Us9 antiserum was described previously (6). Rabbit polyvalent and mouse monoclonal antisera directed against GFP were purchased from Clontech. The gE monoclonal antibody pool (M133, M138, and M156) was received from T. Ben-Porat. The rabbit polyvalent mannosidase II antiserum was provided by K. Moremen (46). 115-2, a monoclonal antiserum raised against p115, was provided by M. G. Waters (64). The rabbit polyvalent TGN38 antiserum was provided by G. Banting (67).
Mouse monoclonal antiserum 2A2 was raised against the PRV Us9 protein. A glutathione S-transferase–Us9 fusion protein was expressed in Escherichia coli DH5α cells and isolated as inclusion bodies essentially as described previously (6). The fusion protein was dialyzed against phosphate-buffered saline (PBS) and used to immunize BALB/c mice according to standard procedures.
Construction of PRV 162.
PRV 162 is an isogenic strain of PRV Be encoding Us9(d46-55), a mutant Us9 protein in which the nucleotide sequence encoding amino acids 46 to 55 has been removed. To construct PRV 162, a transfer vector was first engineered by overlap extension PCR mutagenesis (23) in which 30 bp encoding amino acids 46 to 55 of the Us9 protein were deleted. Specifically, two fragments flanking the 30-bp region to be deleted were PCR amplified with Taq DNA polymerase (Gibco/BRL) and plasmid pALM94 as the template (8). The two amplified fragments containing an overlapping region of homology were mixed together, denatured, and subjected to another round of PCR amplification with outside flanking primers. The final PCR product containing the 30-bp deletion was directly ligated to the T-cloning vector pT7Blue (Novagen) to yield plasmid pAB30. Plasmid pAB30 was digested with EagI and BbsI to release a 404-bp fragment containing the 30-bp deletion from the Us9 gene. This fragment was used to replace the EagI-BbsI fragment from plasmid pGS166, and the resulting plasmid was named pAB31. The nucleotide sequence of pAB31 was then verified by DNA sequencing with Sequenase (United States Biochemical). A transfer vector, pAB33, was then constructed by replacing the SphI-MluI fragment of pPH2 with the SphI-MluI fragment of pAB31. pPH2 contains the SalI-MluI region of the PRV Be BamHI 7 fragment. Plasmid pAB33 was cotransfected by the calcium phosphate method (19) with PRV 91 viral DNA, in which the gE sequences are deleted (65). After complete cytopathic effect was observed, the infected cells were harvested and replated onto PK15 cells. Recombinant viruses were screened for gE expression by an immunoreactivity assay with a gE monoclonal antibody pool. Recombinant viruses were picked and plaque purified three times, and the deletion of the 30-bp region encoding amino acids 46 to 55 from the Us9 open reading frame was confirmed by PCR and Southern blot analysis.
Plasmids.
Plasmid pBB14 contains a hybrid gene in which the EGFP open reading frame was fused to sequences encoding the carboxy terminus of Us9 (6). pAB7 contains the Us9 gene under the control of the cytomegalovirus (CMV) immediate-early promoter (6).
Plasmid pAB35 contains a hybrid Us9 gene encoding a product in which EGFP was fused to the carboxy terminus of Us9(d46-55). pAB35 was created by PCR mutagenesis with Taq DNA polymerase and plasmid pAB30 (see above) as the template. The forward PCR primer corresponded to the 5′ end of the Us9 gene, beginning 24 nucleotides upstream and including the first ATG. The forward PCR primer also introduced an EcoRI site upstream of the first ATG. The reverse primer replaced the Us9 stop codon (TAG) with the codon for an arginine residue. The reverse primer also introduced a BamHI restriction site after the mutated stop codon to allow an in-frame fusion with EGFP. The 328-bp PCR product was digested with EcoRI and BamHI and ligated to pEGFP-N1 (Clontech).
Plasmid pAB15 contains the Us9(d46-55)-encoding gene under the control of the CMV immediate-early promoter. PCR was used to amplify the Us9(d46-55) open reading frame from plasmid pAB30. The forward primer introduced an EcoRI site upstream of the Us9 starting methionine residue. The reverse primer corresponded to the 3′ end of the Us9 gene including the stop codon and 12 downstream nucleotides. The 335-bp PCR product was ligated to the T-cloning vector pT7Blue (Novagen) and subsequently cloned into the mammalian expression vector pcDNA1/Amp (Invitrogen).
pAB37 contains a hybrid Us9 gene in which the EGFP open reading frame was fused to the carboxy terminus of Us9(L30-31A), a mutant Us9 protein in which the leucine residues at positions 30 and 31 were substituted with alanine residues. Site-directed mutagenesis of the nucleotide sequence encoding Us9 amino acids 30 and 31 was performed with an Altered Sites kit (Promega). Specifically, amino acids 30 and 31 of the Us9 open reading frame were changed to alanine residues by oligonucleotide mutagenesis on plasmid pRS3 containing the SphI-MluI fragment of PRV BamHI 7 fragment. The oligonucleotide used to introduce the alanine residues at amino acid positions 30 and 31 also introduced a unique SacII restriction enzyme site to facilitate screening of mutated clones. Potential positive clones were selected by restriction digest analysis and verified by DNA sequencing to contain the leucine-to-alanine mutations. One positive clone was chosen for further manipulation and designated pAB36. Us9(L30-31A) was then fused to EGFP by PCR mutagenesis with pAB36 as the template as described above for the construction of pAB35.
Cycloheximide treatment.
PK15-BB14 monolayers grown on glass coverslips were treated with cycloheximide (50 μg/ml; Sigma) for 0, 2, 4, and 6 h. At the indicated times, the cells were fixed and the localization of Us9-EGFP was detected by fluorescence microscopy.
Electron microscopy.
PK15-BB14 cells grown in DMEM containing 10% FBS and 1 mg of G418 per ml were fixed with increasing amounts of paraformaldehyde (2 to 8%) and prepared for cryosectioning as previously described (5). Ultrathin frozen sections were cut with a Leica/Reichert FCS ultracryomicrotome. Thawed sections were incubated with a rabbit polyvalent Us9 antiserum and then with 10-nm colloidal gold-conjugated protein A (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands) diluted in 1% bovine serum albumin (BSA) in 0.1 M phosphate buffer (pH 7.4). Sections were then stained with 0.3% uranyl acetate in 1.8% methylcellulose and viewed with a Philips CM100 electron microscope.
Transient transfections.
PK15 cells grown to 40 to 50% confluence on glass coverslips were transfected with 10 μg of plasmid pBB14, pAB35, or pAB37 by the calcium phosphate coprecipitation method (19). At 72 h after transfection, the intracellular localization of the EGFP fusion proteins was detected by confocal microscopy. The relative level of Us9-EGFP, Us9(d46-55)-EGFP, and Us9(L30-31A)-EGFP on the cell surface was visualized by fluorescence microscopy.
Indirect colocalization immunofluorescence microscopy.
PK15 cells grown on glass coverslips to approximately 40 to 50% confluency were transfected with plasmid pBB14 or pAB7. At 36 h posttransfection, the coverslips were rinsed with PBS and fixed with 2% paraformaldehyde. The pBB14-transfected coverslips were incubated with p115 mouse monoclonal antiserum (diluted 1:100 in PBS containing 3% BSA and 0.5% saponin) or with mannosidase II rabbit polyvalent antiserum (1:500) for 30 min in a 37°C humidified chamber. The coverslips were rinsed three times with PBS containing 3% BSA and 0.5% saponin and incubated with Alexa 568-conjugated secondary antibodies (1:400; Molecular Probes) for 30 min at 37°C. The pAB7-transfected cells were incubated with a mixture of Us9 monoclonal antiserum 2A2 (1:2) and TGN38 rabbit polyvalent antiserum (1:500). Us9 staining was detected with a fluorescein isothiocyanate-conjugated secondary antibody (Pierce), and TGN38 staining was detected with an Alexa 568-conjugated secondary antibody. The cells were rinsed three times with PBS plus 3% BSA and 0.5% saponin, rinsed once with distilled water, and then mounted onto microscope slides with VectaShield (Vector Laboratories).
Indirect immunofluorescence internalization assay.
The assay was performed on PK15 cells stably expressing Us9-EGFP (PK15-BB14 cells), Us9(d46-55)-EGFP (PK15-AB35 cells), or Us9(L30-31A)-EGFP (PK15-AB37 cells). All cells were grown on glass coverslips and cooled on ice by being washed two times with cold PBS. Incubation on ice inhibited internalization of cell surface molecules. The EGFP fusion protein molecules on the cell surface were then labeled by the addition of polyvalent GFP antiserum (diluted 1:75 in PBS–3% BSA) for 30 min on ice. The cells were then washed three times with PBS to remove any unbound antiserum and either fixed immediately or shifted to 37°C by the addition of pre-warmed DMEM–10% FBS and placement in a 37°C incubator. In one case (see figure legends), the cells were placed in a 32°C incubator rather than a 37°C incubator in order to slow down the process of endocytosis (14). At various times after temperature shift, the cells were fixed with 2% paraformaldehyde and the localization of the GFP antiserum was detected by indirect immunofluorescence microscopy with an Alexa 568-conjugated goat anti-rabbit secondary antibody as described above. Single optical sections were taken through the center of the cells by using a Nikon MRC600 confocal microscope.
Biotinylation internalization assay.
A 90% confluent monolayer of PK15-BB14 cells growing in a 100-mm-diameter dish was cooled to 4°C by three rinses with Hanks’ balanced salt solution (HBSS). The cells were then biotinylated on ice with 0.5 mg/ml Sulfo-NHS-SS-biotin (Pierce) in HBSS for 15 min. The excess biotin was removed by washing the cells three times with HBSS-5 mM Tris (pH 7.4). The cells were then lysed immediately (representing total amount of biotinylated proteins), cleaved immediately (control for cleavage reaction), or shifted to 37°C to allow internalization by addition of prewarmed DMEM–10% FBS and placement in a 37°C incubator. At various times after temperature shift, the cells were chilled on ice and the biotin was cleaved from the cell surface by three 20-min incubations in glutathione cleavage solution (15.5 mg of reduced glutathione [Sigma] per ml, 75 mM NaCl, 1 mM EDTA [pH 8.0], 75 mM NaOH, 10% FBS). The cells were then washed three times in HBSS, and free glutathione was quenched by the addition of PBS-CM (PBS, 0.1 mM CaCl2, 1 mM MgCl2) containing 1% FBS and 5 mg of iodoacetamide (Sigma) per ml. Biotinylated proteins were affinity purified from cell lysates with streptavidin-agarose (Gibco/BRL) and loaded onto a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel. Western blot analysis with GFP monoclonal antiserum was then performed as described previously (6).
Isolation of virions.
Viral particles from the medium of PRV Be-infected or PRV 162-infected cells were isolated, electrophoresed on a SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and subjected to Western blot analysis with Us9 antiserum as described previously (6).
RESULTS
Steady-state localization of Us9.
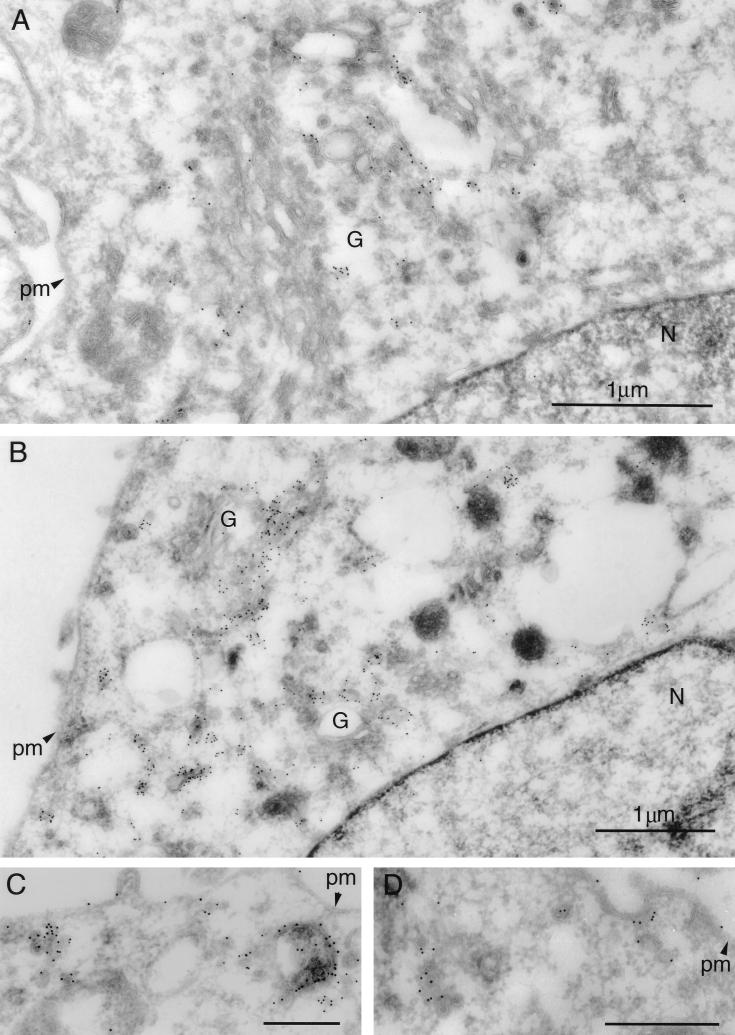
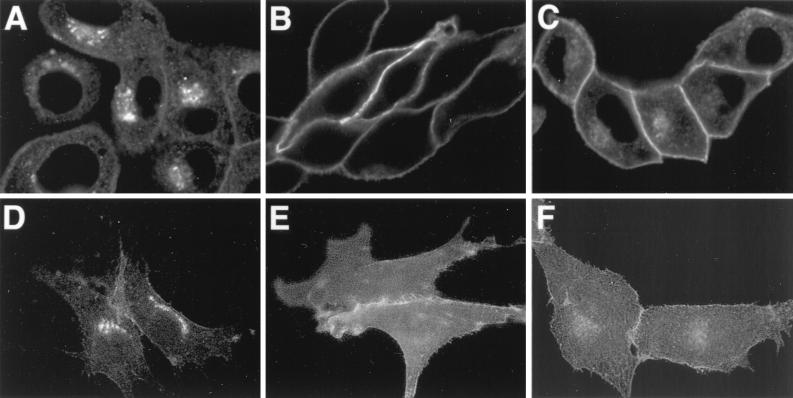
We recently reported that the PRV Us9 protein is a tail-anchored type II membrane protein highly localized to the Golgi complex in both infected and transfected cells (6). In that same study, we showed that the addition of the jellyfish EGFP to the carboxy terminus of Us9 did not interfere with either the targeting of Us9 to the Golgi region or its ability to be incorporated into viral particles (6). We have also shown that the Us9-EGFP protein has no effect on cell cycle progression or cell growth (28). In addition to a predominant Golgi complex localization pattern, both the wild-type Us9 protein and the Us9-EGFP fusion protein can be detected in cytoplasmic vesicles and along the plasma membrane by fluorescence microscopy (6). To extend the observations made on the intracellular localization of Us9 as determined by fluorescence microscopy, we performed immunogold electron microscopy with Us9 polyvalent antiserum on PK15 cells stably expressing the Us9-EGFP fusion protein. The electron microscopy images revealed a predominant intracellular distribution of Us9-EGFP to cytoplasmic vesicles and Golgi-associated structures (Fig. 1). Figures 1A and B are low-magnification views clearly showing the concentration of Us9-EGFP in vesicular structures; they also demonstrate that the staining observed in the Golgi region of these cell appears to be predominantly on one side of the complex, indicating that perhaps Us9-EGFP localization is restricted and not evenly distributed throughout the Golgi apparatus. Vesicles containing immunoreactive Us9-EGFP were also seen singularly throughout the cytoplasm, in clusters, or in structures reminiscent of endosomes (Fig. 1C). In some cases, the Us9-EGFP containing vesicles appeared closely juxtaposed to the cell surface as if in the process of fusing with the plasma membrane (Fig. 1D). A small amount of Us9-EGFP staining was observed on the plasma membrane, while the nuclear membrane appears to be devoid of immunoreactive Us9-EGFP.
FIG. 1.
Immunoelectron microscopy of Us9-EGFP. Thawed ultrathin sections of PK15 cells stably expressing a Us9-EGFP fusion protein were stained with a polyvalent rabbit Us9 antiserum followed by protein A conjugated to colloidal gold (10 nm). Low-magnification views illustrate the predominant staining of Golgi-associated membranes and vesicular structures (A and B). No staining of the nuclear membrane is observed. Low levels of Us9-EGFP can be detected on the plasma membrane (C and D). Vesicular structures throughout the cytoplasm were seen singularly, in clusters, or in structures reminiscent of endosomes (C). In some instances, Us9-EGFP-containing vesicles are observed close to the plasma membrane and seem to be in the process of fusing with the plasma membrane (D). Magnification bars in panels C and D represent 500 nm. pm, plasma membrane; G, Golgi apparatus; N, nucleus.
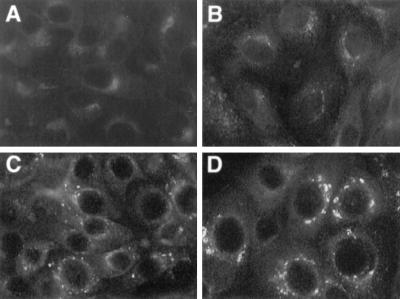
The localization of Us9 predominantly to Golgi-associated membranes and vesicles is distinct from that of other PRV envelope proteins, which are easily detected in all aspects of the secretory system, including the nuclear membrane and the ER. Consequently, it was of interest to determine if Us9 encoded signals involved in maintaining the protein in the Golgi region or whether the protein accumulated there transiently as it passed through the secretory system. To address this, we treated PK15-BB14 cells stably expressing the Us9-EGFP fusion protein with the protein synthesis inhibitor cycloheximide (50 μg/ml) and at various times after treatment examined the steady-state localization of Us9-EGFP by fluorescence microscopy. If the steady-state localization of Us9 is in the Golgi region, then the fusion protein should not relocalize to the plasma membrane even after several hours of cycloheximide treatment as was observed for the Punta Toro virus Golgi-resident G1 and G2 proteins (40). As seen in Fig. 2, cycloheximide treatment did not significantly alter the localization of the Us9-EGFP fusion protein. Even after 6 h of cycloheximide treatment (Fig. 2D), Us9-EGFP was still detected in a perinuclear staining pattern reminiscent of the Golgi apparatus. However, unlike the uniform Golgi-like staining of Us9-EGFP observed in untreated cells (Fig. 2A), the Us9-EGFP staining pattern became very vesicular in appearance beginning at approximately 4 h of cycloheximide treatment (Fig. 2C). The appearance of Us9-EGFP-containing vesicles during cycloheximide treatment was most apparent at the 6-h time point (Fig. 2D). As a control for this experiment, the Golgi apparatus itself was visualized with the dye Bodipy-FL-C5 ceramide (data not shown). In these experiments, the Golgi apparatus remained unaltered throughout cycloheximide treatment, thereby indicating that this effect observed with cycloheximide treatment is specific to Us9-EGFP and does not represent a general vesiculation of the Golgi apparatus. However, the appearance of perinuclear Us9-EGFP-containing vesicles following cycloheximide treatment suggests that Us9-EGFP maintains its steady-state localization in the Golgi region by a mechanism involving the endocytic pathway.
FIG. 2.
Steady-state localization of Us9-EGFP. Monolayers of Us9-EGFP-expressing cells grown on glass coverslips were treated with cycloheximide (50 μg/ml) for 0 h (A), 2 h (B), 4 h (C), and 6 h (D). At the indicated time points, the cells were fixed with paraformaldehyde and the localization of Us9-EGFP was visualized by fluorescence microscopy under UV illumination.
Us9 partially colocalizes with TGN38.
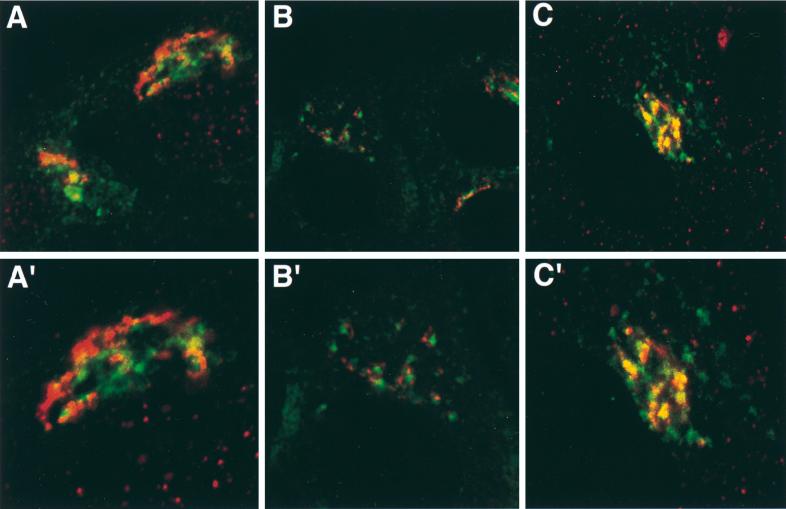
The immunogold electron microscopy images in Fig. 1 clearly show that Us9 is highly localized to both cytoplasmic vesicles and Golgi-associated membranes. Although staining appears to be concentrated on one side of the Golgi complex, we are unable to determine from these images if the Us9-EGFP containing membranes represent cis-, medial-, or trans-Golgi stacks or the TGN. Consequently, we performed double indirect immunofluorescence microscopy with Golgi and TGN markers in order to more precisely define the nature of this Golgi compartment in which Us9 resides. Specifically, we determined if Us9 colocalized with the medial-Golgi marker mannosidase II (Fig. 3A), the peripheral Golgi protein p115 (Fig. 3B), or the TGN marker TGN38 (Fig. 3C). While the staining pattern of Us9 is similar to those of both mannosidase II (Fig. 3A) and p115 (Fig. 3B), the intracellular distribution of Us9 did not overlap with either of the Golgi markers. However, double indirect immunofluorescence microscopy of Us9 with TGN38 (Fig. 3C) revealed partial colocalization of these two membrane proteins. These results suggest that Us9 resides in a cellular compartment in or near the TGN.
FIG. 3.
Colocalization of Us9 with Golgi and TGN markers. PK15 cells grown on glass coverslips were transfected with plasmids pBB14 (A and B) and pAB7 (C). After 36 h of transfection, the cells were fixed and stained for Us9 and mannosidase II (A), for Us9 and p115 (B), and for Us9 and TGN38 (C). Us9 was detected either by GFP fluorescence (A and B) or with a fluorescein isothiocyanate-conjugated secondary antibody (C) which fluoresces green. Mannosidase II (A), p115 (B), and TGN38 (C) were visualized with an Alexa 568-conjugated secondary antibody which fluoresces red. High-magnification views are shown in panels A′, B′, and C′.
Internalization of Us9.
The targeting of both the TGN protein TGN38 and the endoprotease furin to the TGN has been demonstrated to be due, in part, to the retrieval of molecules from the plasma membrane via endocytic vesicles (4, 9, 38). To test the hypothesis that Us9 also maintains its steady-state localization in the TGN region by a recycling mechanism, indirect immunofluorescence internalization assays were performed on PK15 Us9-EGFP cells. An EGFP epitope-tagged Us9 protein was used in these experiments because the three carboxy-terminal amino acids of the wild-type Us9 protein predicted to extend from the plasma membrane are not detectable by Us9 antiserum on nonpermeabilized cells (unpublished observations). Fusion of EGFP to the carboxy terminus of Us9 exposes the EGFP moiety to the extracellular environment, where it can be recognized by GFP-specific antiserum on intact cells. In this experiment, PK15 Us9-EGFP cells were cooled to 4°C on ice to inhibit internalization of cell surface molecules. These cells were then incubated with an antiserum specific for the GFP portion of the fusion protein. Since this incubation was performed on ice, only Us9-EGFP molecules on the cell surface were labeled with the GFP antiserum. The cells were then shifted to 32°C to allow internalization of cell surface molecules. At various times after the temperature shift, the cells were fixed and permeabilized, and the localization of the cell surface Us9-EGFP molecules was examined by indirect immunofluorescence with an Alexa 568-conjugated secondary antibody. The Alexa 568-conjugated secondary antibody fluoresces red and can therefore be easily distinguished from GFP fluorescence. The images shown in Fig. 4 are confocal microscopy sections taken through the center of the cell where the red fluorescence staining represents the localization of the GFP antiserum and the green fluorescence staining represents the total pool of Us9-EGFP molecules. When PK15 Us9-EGFP cells were incubated with GFP antiserum on ice (Fig. 4A), only those molecules on the plasma membrane were labeled red. However, when these cells were shifted to 32°C to allow internalization to occur, Us9-EGFP molecules began to internalize into the interior of the cell in a series of fine cytoplasmic vesicles. The internalization of Us9-EGFP began as early as 30 min after temperature shift (Fig. 4C). These Us9-EGFP-containing vesicles continued to internalize into the interior of the cell with time (Fig. 4D) and eventually accumulated in a perinuclear region staining pattern (Fig. 4E). It was evident by 90 min after temperature shift that the internalized Us9-EGFP molecules traveled to an intracellular compartment in or near the TGN, where they were closely associated with the total Us9-EGFP pool. These internalized molecules, however, appeared to remain in vesicles and did not fuse with the total Us9-EGFP pool. Figure 4F is a confocal image showing only the red fluorescence of internalized Us9-EGFP molecules after a 90 min shift to 37°C. It is clear from this image that the Us9-EGFP molecules on the plasma membrane are efficiently internalized into the interior of the cell to a perinuclear compartment reminiscent of the TGN. Moreover, it is also evident that Us9 internalization is faster at 37°C than at 32°C (compare Fig. 4E and F) as has been previously observed (14). We interpret these findings to mean that Us9 maintains its steady-state localization in a TGN-like compartment by means of a recycling mechanism similar to that of TGN38 and furin.
FIG. 4.
Us9-EGFP is maintained in the TGN region by retrieval of molecules from the plasma membrane. Glass coverslips plated with PK15 cells stably expressing Us9-EGFP were cooled and incubated with GFP-specific antiserum on ice for 30 min. The cells were then shifted to 32°C by the addition of prewarmed medium and placement in a 32°C incubator. At 0 min (A), 15 min (B), 30 min (C), 60 min (D), or 90 min (E), the cells were fixed with paraformaldehyde and the localization of the GFP antiserum was detected with an Alexa 568-conjugated goat anti-rabbit IgG secondary antibody (red). The red fluorescence represents the localization of internalized Us9-EGFP molecules, and the green fluorescence represents the total pool of Us9-EGFP molecules. Panel F shows the localization of internalized Us9-EGFP after 90 min of temperature shift to 37°C. All images are confocal sections taken through the center of the cell.
Internalization of Us9 is not antibody dependent.
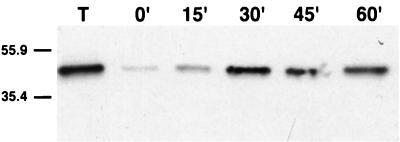
To confirm that the ability of Us9-EGFP to internalize into the interior of the cell is not an artifact of GFP antiserum binding, we performed an internalization assay which is not dependent on the binding of antibodies. In this experiment, Us9-EGFP molecules on the cell surface were labeled with a cleavable form of biotin. The biotinylated cells were then shifted to 37°C to allow internalization, and at various times after temperature shift, the extracellular biotin was cleaved by the addition of glutathione. Only those Us9-EGFP molecules able to internalize into the interior of the cell were protected from cleavage. Lane T in Fig. 5 represents the total amount of cell surface Us9-EGFP molecules biotinylated. When the extracellular biotin was cleaved from these cells before endocytosis was allowed to occur, the majority of the biotinylated Us9-EGFP molecules were sensitive to glutathione cleavage (0 min). However, when these cells were shifted to 37°C to allow internalization before cleavage, a portion of the biotinylated Us9-EGFP molecules became protected from cleavage as early as 15 min after temperature shift. The amount of biotinylated Us9-EGFP molecules protected from proteolysis increased with time, and the maximum amount protected appeared to be after 30 min of shift to 37°C. The residual amount of biotinylated Us9-EGFP observed in the 0-min lane is most likely the result of inefficient cleavage. These results suggest that the internalization of Us9-EGFP from the cell surface is not dependent on the binding of antibodies.
FIG. 5.
Us9-EGFP internalization is not an antibody dependent process. PK15 cells stably expressing a Us9-EGFP fusion protein were incubated for 15 min (15′) on ice with Sulfo-NHS-SS-biotin. The cells were then lysed immediately to show the total amount biotinylated (lane T) or shifted to 37°C to allow internalization by the addition of prewarmed medium and placement in a 37°C incubator. At 0 min, 15 min, 30 min, 45 min, or 60 min after the temperature shift, biotin was cleaved from the cell surface with glutathione, cell lysates were prepared, and biotinylated proteins were affinity purified with streptavidin-agarose. The affinity-purified molecules were separated on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and Western blotted with monoclonal GFP antiserum. The migration of molecular mass markers is indicated on the left in kilodaltons.
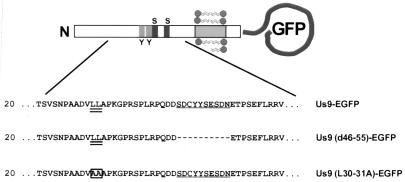
Targeting determinants in the Us9 cytoplasmic tail.
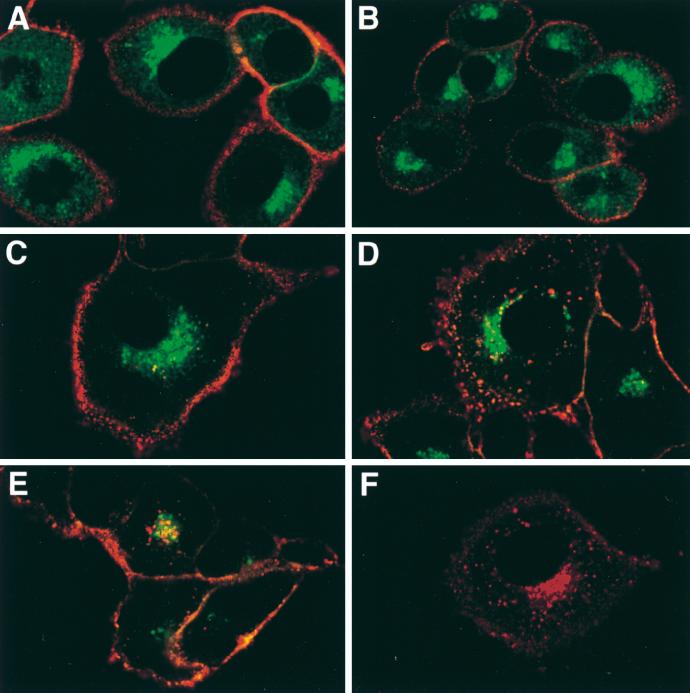
To map the domains involved in the maintenance of PRV Us9 to the TGN region, we introduced defined mutations in the Us9 cytoplasmic domain. Figure 6 shows the relative localization of these mutations in the Us9 protein. The intracellular trafficking of both the endoprotease furin (27, 55, 63) and the VZV gE membrane protein (1, 2) has been demonstrated to be dependent on an acidic segment containing casein kinase II-phosphorylatable residues. As a similar acidic domain containing putative tyrosine and casein kinase I and II sites is present in the Us9 cytoplasmic tail, we examined the effect of deleting this region on Us9 localization to the TGN region. Specifically, amino acids 46 to 55 were deleted from both the wild-type Us9 and Us9-EGFP proteins (encoded by plasmids pAB15 and pAB35, respectively). pAB15 and pAB35 were transiently transfected into PK15 cells as described in Materials and Methods, and the localization of Us9 was detected either by indirect immunofluorescence microscopy with Us9 polyvalent rabbit antiserum (for pAB15) or by GFP fluorescence (for pAB35) at 72 h posttransfection. As the localization of Us9 in PK15 cells transfected with either plasmid was identical, only the localization of Us9(d46-55) fused to EGFP (encoded by pAB35) is shown (Fig. 7B and E). When amino acids 46 to 55 were deleted from the Us9 protein, Us9-EGFP was no longer localized to the TGN region and cytoplasmic vesicles but was now found predominantly on the plasma membrane (compare Fig. 7A and D with Fig. 7B and E). Although at early times after transfection Us9(d46-55)-EGFP molecules could be detected in the TGN region as they transited through the secretory system (data not shown), very little Us9(d46-55)-EGFP protein could be detected in this region after approximately 60 h of transfection.
FIG. 6.
Amino acid sequence of the amino-terminal cytoplasmic tail of Us9 mutants. The dileucine motif at amino acids 30 and 31 is underlined twice, and the acidic domain from amino acids 46 to 55 is underlined once. Deletions are indicated by dashes, and amino acid substitutions are boxed. The relative locations of the tyrosine (Y) and casein kinase I and II sites (S) in the Us9 cytoplasmic tail are indicated in the diagram.
FIG. 7.
Transfection of Us9 constructs. PK15 cells grown on glass coverslips were transfected by the calcium phosphate method with pBB14 (A and D), pAB35 (B and E), or pAB37 (C and F). At 72 h posttransfection, the intracellular localization of the various Us9-EGFP fusion proteins was detected by confocal microscopy (A to C). The cell surface of the transfected cells was visualized by fluorescence microscopy (D to F).
In addition to the acidic peptide previously shown to be involved in the recycling and maintenance of furin to the TGN, trafficking of the endoprotease to endosomes requires a tyrosine-containing internalization signal (YKGL) (27, 55, 63). Although the cytoplasmic tail of PRV Us9 does not contain any previously described tyrosine-based internalization motifs, it does have a potential dileucine endocytosis signal (LL) (Fig. 6). Accordingly, we determined the significance of the dileucine motif in the trafficking of Us9 by mutating the leucine residues at positions 30 and 31 in the Us9-EGFP fusion protein to alanine residues. Olson and Grose demonstrated that a similar leucine-to-alanine substitution was able to disrupt the ability of a dileucine-type endocytosis motif (ML) to direct the internalization of the VZV gI protein in transfected cells (49). pAB37, containing a Us9(L30-31A)-EGFP gene fusion, was transfected into PK15 cells, and localization of the mutant Us9 protein was detected by confocal microscopy 72 h after transfection. As observed for wild-type Us9-EGFP (Fig. 7A), confocal images of the intracellular staining of pAB37-transfected cells revealed that Us9(L30-31A)-EGFP was found in the TGN region and on the plasma membrane (Fig. 7C). Moreover, the confocal sections showed that the plasma membrane of cells transfected with the Us9(L30-31A)-EGFP (Fig. 7C) construct appeared to stain more intensely than that of cells transfected with wild-type Us9-EGFP (Fig. 7A). The difference in plasma membrane staining intensity, however, is more evident when the cell surface fluorescence microscopy images of Us9-EGFP (Fig. 7D)- and Us9(L30-31A)-EGFP (Fig. 7F)-transfected cells are compared. The results presented above indicate that an acidic cluster from residues 46 to 55 and, to a lesser extent, a dileucine motif at residues 30 and 31 may both play a role in the localization of Us9-EGFP to the TGN region.
Role of the acidic cluster and dileucine motif in Us9 internalization.
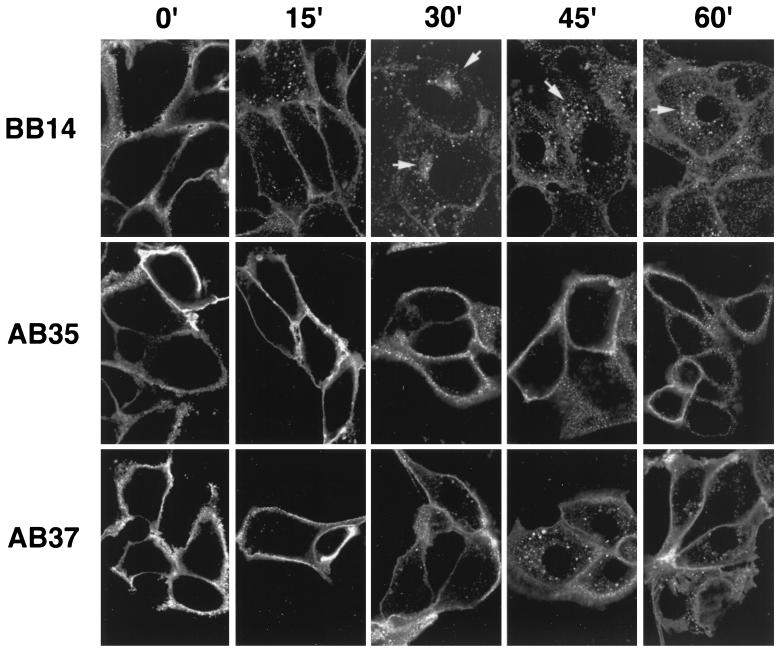
To determine if Us9(d46-55)-EGFP and Us9(L30-31A)-EGFP display increased plasma membrane staining compared to Us9-EGFP due to an internalization defect rather than a loss of TGN retention signals, indirect immunofluorescence internalization assays were performed on PK15 cells stably expressing the Us9-EGFP, Us9(d46-55)-EGFP, and Us9(L30-31A)-EGFP fusion proteins (PK15-BB14, PK15-AB35, and PK15-AB37 cells, respectively). Figure 8 compares the internalization of Us9 in the three cell lines, showing only the red fluorescence of the GFP antiserum. For all cell lines tested, the plasma membrane stained brightly at the 0-min time point. As early as 15 min after the shift to 37°C, wild-type Us9-EGFP molecules on the plasma membrane of PK15-BB14 cells began to internalize in the form of cytoplasmic vesicles. By 30 min, the majority of the wild-type molecules moved from the cell surface to the interior of the cell, where they started to collect in a TGN-like staining pattern. Although most of the wild-type plasma membrane Us9-EGFP molecules internalized by 30 min after temperature shift, there did appear to be a slight increase in the size and number of internalized vesicles at the 45- and 60-min time points. These results are consistent with the results of the antibody independent internalization assay described above (Fig. 5). In contrast to wild-type Us9-EGFP molecules, Us9(d46-55)-EGFP molecules (PK15-AB35 cells) did not internalize into the interior of the cell even 60 min after the shift to 37°C. Although a few small vesicles could be seen in the interior of some cells, this was by no means equivalent to the degree of endocytosis observed for wild-type Us9-EGFP in PK15-BB14 cells. Moreover, when the internalization assay was performed on PK15-AB37 cells, which express the Us9(L30-31A)-EGFP fusion protein, only a few internalized vesicles could be detected in the interior of the cell by 15 min after the shift to 37°C. An increase in Us9(L30-31A)-EGFP molecules in the interior of the cell was detected at the 30-, 45-, and 60-min time points, but the amount internalized was less than that observed for wild-type Us9-EGFP molecules in PK15-BB14 cells at the same time points. Examination of the 45-min time point indicated an accumulation of internalized Us9(L30-31A)-EGFP molecules in the TGN region, but not as common or as prevalent as that observed for wild-type Us9-EGFP (PK15-BB14 cells, 30-min time point). These experiments suggest that both the Us9(d46-55)-EGFP and Us9(L30-31A)-EGFP fusion proteins are defective in the ability to internalize from the plasma membrane. Whereas Us9(d46-55)-EGFP appears to be unable to internalize into the interior of the cell, the internalization defect observed for Us9(L30-31A)-EGFP seems to be in the overall rate of internalization. Us9(L30-31A)-EGFP molecules are capable of internalizing from the cell surface but more slowly than wild-type Us9-EGFP molecules. This difference in the internalization defect between Us9(d46-55)-EGFP and Us9(L30-31A)-EGFP correlates with the steady-state amounts of these fusion proteins on the plasma membrane.
FIG. 8.
Two determinants in the Us9 cytoplasmic tail are required for efficient internalization of Us9 from the plasma membrane. An indirect immunofluorescence assay was performed on PK15 cells stably expressing Us9-EGFP (PK15-BB14), Us9(d46-55)-EGFP (PK15-AB35), or Us9(L30-31A)-EGFP (PK15-AB37) essentially as described in the legend to Fig. 4. The temperature shift in this experiment was to 37°C. The composite shows only the red fluorescence of the Alexa 568-conjugated goat anti-rabbit IgG secondary antibody. Times are indicated in minutes.
Acidic cluster containing putative phosphorylation sites is not required for incorporation of Us9 into viral particles.
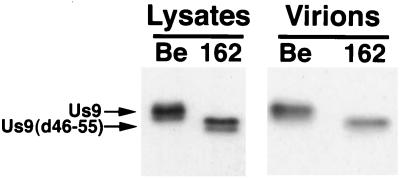
We also investigated if the acidic residues important in localization of Us9 to the TGN region were important for the incorporation of Us9 into viral particles. To address this question, Us9(d46-55) was crossed into the background of the wild-type Be strain of PRV by homologous recombination to create strain PRV 162. After the deletion of amino acids 46 to 55 from the Us9 open reading frame was confirmed by Southern blotting and PCR analysis, cell lysates were prepared and virions were isolated from the medium of PK15 cells infected with either PRV Be or PRV 162 (multiplicity of infection of 10). Both the cell lysates and the purified virions were separated on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and Western blotted with Us9 antiserum. As anticipated, the Us9 protein present in PRV 162-infected cells migrated slightly faster than wild-type Us9 present in Be-infected cells due to the deletion of amino acids 46 to 55 (Fig. 9). Similar to wild-type Us9, Us9(d46-55) was present as multiple polypeptides in PRV 162-infected cells. 33P labeling experiments indicated that the multiple Us9(d46-55) polypeptides present in PRV 162-infected cells are in fact phosphorylated despite the deletion of both putative tyrosine and casein kinase I and II phosphorylation sites (data not shown). This finding indicates that Us9(d46-55) is modified by the addition of phosphates to residues other than those previously predicted to be phosphorylated. Immunoreactive Us9 proteins present in virions isolated from Be- and 162-infected cells are also shown in Fig. 9. Both the wild-type Us9 protein and the Us9(d46-55) protein were incorporated into viral particles at approximately equivalent amounts, thereby indicating that the deletion of amino acids 46 to 55 from the Us9 protein did not affect the ability of Us9 to be incorporated into viral particles. Although it appears that only one of the two forms of Us9(d46-55) present in PRV 162-infected cells was incorporated into viral particles, we cannot conclude at this point which, if any, form is excluded.
FIG. 9.
Us9(d46-55) is efficiently incorporated into viral particles. Monolayers of PK15 cells were infected at a multiplicity of infection of 10 with either PRV Be or PRV 162 for 15 h. Cellular extracts were prepared, and virions were isolated from the medium by centrifugation through a 30% sucrose cushion. Both the cellular extracts and virions were fractionated on an SDS–12.5% polyacrylamide gel and analyzed by Western blotting with Us9 antiserum.
DISCUSSION
The lipid envelope of PRV virions contains at least 11 virally encoded membrane glycoproteins (26). Recently, we have characterized a novel PRV membrane protein designated Us9 (6). In the course of our studies, we discovered several characteristics of the Us9 protein that distinguish it from the other PRV membrane proteins. One striking difference is that Us9 is inserted into the viral envelope in a novel type II tail-anchored topology. In this topology, the majority of the Us9 protein (68 amino acids) is not presented on the outside of the viral particle but is buried within the tegument region of the viral particle, where it may interact with capsid proteins, tegument proteins, or the tails of other envelope proteins. As the Us9 protein is only 98 amino acids in length and 26 amino acids are predicted to span the lipid bilayer, Us9 appears to have only a 3-amino-acid ectodomain extending from the surface of the virion available for potential host cell interactions. This is the smallest ectodomain of any alphaherpesvirus membrane protein characterized thus far.
Us9 also differs from other characterized PRV envelope proteins in its steady-state intracellular localization during viral infection. In general, PRV envelope glycoproteins are detected in large quantities in all aspects of the secretory system, including the nuclear membrane and the ER. In contrast, the Us9 protein is localized predominantly to the TGN region and cytoplasmic vesicles. Localization of the protein to the plasma membrane is transient, as Us9 is rapidly internalized in small vesicles to a TGN-like compartment. By standard immunofluorescence microscopy, we are unable to detect any staining of Us9 in either the nuclear membrane or ER in infected cells, in transfected cells, or in a cell line constitutively expressing a Us9-EGFP fusion protein. In all these experiments, we are able to detect Us9 or Us9-EGFP on the plasma membrane. The electron microscopy images of immunogold-labeled Us9-EGFP presented in this report confirm the observations made by fluorescence microscopy that the fusion protein is concentrated mainly in cytoplasmic vesicles and Golgi-associated membranes. Moreover, we observed no labeling of Us9-EGFP in the nuclear membrane. Further analysis is necessary before we can conclude that the ER does not contain any Us9-EGFP molecules. In contrast to our observations with fluorescence microscopy, these electron microscope images show very little immunogold-labeled Us9-EGFP on the plasma membrane. We believe that the low level of Us9-EGFP on the plasma membrane in the micrographs reflects the transient state of the protein in this membrane and the resulting low concentration of immunoreactive molecules compared to cytoplasmic vesicles and Golgi-associated membranes. Indeed, in some cases we can observe Us9-EGFP-containing vesicles closely juxtaposed to the cell surface and apparently fusing with the plasma membrane (Fig. 1D). Therefore, these results indicate the difficulty of proving that there is absolutely no Us9 protein in nuclear membranes because it may be present in very low concentrations or for a very short period of time. Suffice it to say that the steady-state localization of Us9 protein is markedly different from that of the standard virion type I membrane glycoprotein.
Over the past years, several classes of internalization motifs have been identified in the cytoplasmic tails of membrane proteins (reviewed in references 30 and 39). These include the tyrosine-based motifs involved in low-density lipoprotein (10) and transferrin receptor (3, 25, 43) endocytosis and the dileucine motifs directing internalization of the GLUT4 glucose transporter (11, 62) and the insulin receptor (22, 52). A third motif identified is based on an acidic domain which often contains a casein kinase II phosphorylation site. Acidic domains containing a casein kinase II phosphorylation site have been demonstrated to be important in the intracellular trafficking of both the endoprotease furin (27, 55, 63) and the mannose-6-phosphate receptor (33, 41). Virus proteins have been shown to have similar endocytosis signals as cellular receptors. For example, the internalization of the human immunodeficiency virus type 1 envelope protein (53), the simian immunodeficiency virus envelope protein (34, 54), and the VZV gE protein (1, 2, 48) is dependent on a tyrosine-based internalization motif. In addition, a dileucine-like (ML) endocytosis motif has been shown to be important in the intracellular trafficking of the VZV gI protein (49). Moreover, the trafficking of the VZV gE protein (1, 2, 70) and a human immunodeficiency virus Nef-major histocompatibility complex class I complex (20) to the TGN has been demonstrated to be dependent on an acidic cluster.
In this report, we show that the Us9 protein internalizes into the interior of the cell from the plasma membrane and that internalization is critical for the intracellular localization of Us9 to the TGN region. Examination of the 68-amino-acid Us9 cytoplasmic tail revealed that the protein contains two of the internalization motifs described above: an acidic domain from amino acids 46 to 55 containing a casein kinase II phosphorylation site and a dileucine motif at amino acids 30 and 31. Our results indicate that both the acidic domain and the dileucine motif play a role in the intracellular localization of Us9 and the recycling of the protein from the plasma membrane. Deletion of both motifs individually resulted in an increase in plasma membrane staining compared to the amount of wild-type Us9 molecules on the cell surface. This increase in plasma membrane staining was shown to be due to a defect in internalization. Mutants deleted for the acidic domain were completely defective in Us9 internalization, while dileucine motif mutants were partially defective. Overall, these results indicate that Us9 localization and intracellular trafficking is dependent on at least two separate motifs. Additional experiments, however, must be conducted before we can conclude that these are the only trafficking motifs in the Us9 protein and that neither of these mutations had an effect on the overall structure of Us9. As observed with Us9, the TGN localization and endosome trafficking of furin have been demonstrated to rely on both a casein kinase II-containing acidic peptide and a tyrosine-based internalization signal (27, 55, 63). In this case, the authors have defined the role of the acidic domain in the retention of furin to the TGN and the role of the YKGL motif in retrieval of molecules from the cell surface (55). At this point, the specific roles of the acidic and dileucine motifs in Us9 localization and trafficking remain to be determined. As the Us9 intracellular trafficking results presented in this study were all performed on cell lines stably expressing EGFP epitope-tagged Us9 fusion proteins, the trafficking of these fusion proteins in the context of viral infection needs to be investigated.
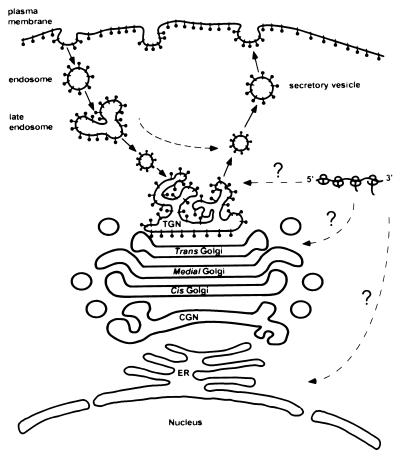
A model based on the conclusions reached in this study is shown in Fig. 10. We predict that the Us9 protein is synthesized in the cytoplasm and posttranslationally inserted into membranes. This hypothesis is based on the fact that the Us9 protein lacks an active signal sequence promoting cotranslational membrane insertion and that most of Us9’s carboxy-terminal membrane anchor will remain buried within the translating ribosome when the termination codon is reached. Although most tail-anchored membrane proteins are posttranslationally inserted into the ER before transport to their final destination, it is believed that some tail-anchored proteins may be incorporated directly into their target organelle (31, 32). In support of this concept, synaptobrevin (32), cytochrome b5 (13, 56), and Bcl-2 (24) have all been demonstrated to be inserted into a variety of membranes in vitro. As no staining of Us9 in the ER can be detected by immunofluorescence microscopy, it is conceivable that Us9 is posttranslationally inserted directly into the vesicles of either the Golgi or TGN. Experiments are being designed to test this hypothesis. The data presented in this report indicate that once Us9 is inserted into the secretory system, it establishes a steady-state residence in a late Golgi compartment such as the TGN. Although double immunofluorescence microscopy experiments show that there is partial colocalization between Us9 and TGN38, a detailed biochemical examination must be performed before we can conclude that these two proteins reside within the same compartment. Us9 molecules are then able to leave the TGN region and travel in cytoplasmic vesicles to the plasma membrane. In support of this concept, immunogold electron micrographs of Us9-EGFP-expressing cells clearly show Us9-EGFP-containing vesicles in the cytoplasm and in close approximation to the cell surface. In some cases, these vesicles appear to be in the process of fusing with the plasma membrane. Furthermore, Us9 molecules that have reached the cell surface are subsequently internalized into the interior of the cell in endocytic-like vesicles. The internalized Us9 molecules then travel to an intracellular compartment that is closely associated with the total pool of Us9 molecules. Our data suggest that the vesicles containing internalized Us9 molecules do not fuse with the membranes containing the total Us9 pool but rather remain as separate entities. The data presented in this report indicate that both an acidic domain containing putative phosphorylation sites and a dileucine motif in the Us9 cytoplasmic tail are needed for the appropriate localization of Us9 to the TGN region. Both of these determinants play a role in the ability of Us9 to be efficiently endocytosed into the interior of the cell and consequently retained in or near the TGN.
FIG. 10.
Model of intracellular Us9 trafficking (see Discussion for detailed description). The Us9 protein is represented by black lollipop-shaped structures. CGN, cis-Golgi network.
Although deletion of the acidic domain completely disrupted the ability of Us9 to localize to the TGN region and to recycle from the cell surface, the mislocalized protein was still incorporated into viral envelopes. Accordingly, the maintenance of Us9 to the TGN region and endocytic vesicles, as well as the capacity of the protein to be phosphorylated at either tyrosine or casein kinase I and II sites, is not necessary for the insertion of Us9 into viral envelopes. It is important to note that although the steady-state localization of Us9(d46-55) has been changed from the TGN region to the plasma membrane, this molecule can be found in a perinuclear staining pattern at early times posttransfection and postinfection (data not shown). Thus, we conclude that the maintenance of Us9 to the TGN region itself and the signals involved in this maintenance are not necessary for Us9 virion incorporation. These results do indicate, however, that the endocytosis of Us9 is not necessary for its incorporation in viral membranes. This is in agreement with the inability to detect internalized PRV gE protein in viral envelopes (58).
All Us9-homologous proteins contain either a tyrosine- or dileucine-based putative internalization motif. For instance, the Us9 proteins in HSV-1, HSV-2, and bovine herpesvirus 1 all share a common YXXφ motif (X represents any amino acid, and φ represents a bulky hydrophobic amino acid), whereas a dileucine-endocytosis motif is present in the PRV, equine herpesvirus 1, simian herpesvirus B, and VZV Us9 proteins. Based on the conservation of both the acidic domain and dileucine (or tyrosine)-based endocytosis motifs in the alphaherpesvirus Us9 homologues, a fundamental question remains as to the role of Us9 internalization in the viral replication cycle. One possibility is that the recycling of Us9 from the cell surface is required for maintaining Us9 in the TGN region, and the localization of Us9 to this cellular compartment is necessary for the function of the protein. We demonstrated in this report that at least in tissue culture, the maintenance of Us9 to a TGN-like compartment is not critical for the envelopment of Us9. Moreover, the maintenance of Us9 to the TGN region is not required for the general processes of PRV envelopment and egress, as a virus lacking the Us9 acidic domain (PRV 162) has no observable defects in tissue culture. For instance, initial examination of PRV 162 has shown that the virus grows to equivalent titers as the wild-type strain Be and has no defect in cell-to-cell spread as determined by plaque size in cultured epithelial cells (data not shown). Furthermore, the deletion of the acidic domain has no effect on the production or localization of other PRV envelope glycoproteins (data not shown). It is likely that Us9 internalization functions during in vivo infection. We are now examining this Us9 mutant virus as well as a virus containing a mutation in the dileucine motif for any spread or virulence defects in the rat nervous system. Another possibility is that the recycling property of Us9 functions during intracellular transport of the virus in vivo in polarized cells such as neurons or epithelial cells. Perhaps through interactions with its cytoplasmic domain, Us9 transports tegument proteins, other viral envelope proteins, or even cellular proteins to specific areas in the neuron such as the plasma membrane, endocytic compartment, or even the axon terminal. This type of transport must not be necessary within the confines of nonpolarized cells in tissue culture. However, if the speculation has merit, Us9 mutants should have marked phenotypes in vivo. For example, the inability of Us9 to direct the transport of proteins complexed to its cytoplasmic tail to the axon terminals may result in the transneuronal anterograde spread defect observed for gE and/or gI null viruses (65). Experiments are in progress to test this hypothesis. Last, it is conceivable that Us9 internalization is an early event during entry of a virus particle into the host cell. Recent reports have suggested that the inner part of the simian CMV viral tegument is an ordered structure and that some tegument proteins interact with the viral capsid itself (59). We speculate that in addition to those tegument proteins complexed to the viral capsid, there is a population of tegument proteins bound to the cytoplasmic tails of viral membrane proteins. In this hypothesis, only that population of tegument proteins bound to the capsid are transported along microtubules into the cell during viral entry and that the other population of tegument proteins enter the cell attached to the tail of internalized envelope proteins. The facts that Us9 is able to internalize into cells in endocytic-like vesicles and that the majority of the protein resides within the tegument region make Us9 a likely candidate to transport tegument proteins during viral entry.
ACKNOWLEDGMENTS
We acknowledge M. G. Waters (Princeton University), G. Banting (University of Bristol, Bristol, England), and K. Moremen (University of Georgia) for generously providing the p115, TGN38, and mannosidase II antisera, respectively. We gratefully acknowledge the expert technical assistance of J. Goodhouse, A. Beavis, and M. Marlow-Fonseca. Thanks also go to J. Schwarzbauer for use of her fluorescence microscope and imaging system and to members of the Enquist lab for advice and helpful comments.
A.D.B. and T.D.R. are supported by NIH training grant 5T32GM07388. This work was supported by NINDS grant 1RO133506 to L.W.E.
REFERENCES
- 1.Alconada A, Bauer U, Baudoux L, Piette J, Hoflack B. Intracellular transport of the glycoproteins gE and gI of the varicella-zoster virus. J Biol Chem. 1998;273:13430–13436. doi: 10.1074/jbc.273.22.13430. [DOI] [PubMed] [Google Scholar]
- 2.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein gI, a protein, localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez E, Girones N, Davis R J. A point mutation in the cytoplasmic tail domain of the transferrin receptor inhibits endocytosis. Biochem J. 1990;267:31–35. doi: 10.1042/bj2670031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos K, Wraight C, Stanley K K. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters P J, Bonifacino J. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunovskis P, Velicer L F. The Marek’s disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 8.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman R E, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W J, Goldstein J L, Brown M S. NPXY, a sequence often found in cytoplasmic tails is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 11.Corvera S, Chawla A, Chakrabarti R, Joly M, Buxton J, Czech M P. A double leucine within the GLUT4 glucose transporter COOH-terminal domain functions as an endocytosis signal. J Cell Biol. 1994;126:979–989. doi: 10.1083/jcb.126.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullinane A A, Rixon F J, Davison A J. Characterization of the genome of equine herpesvirus 1 subtype 2. J Gen Virol. 1988;69:1575–1590. doi: 10.1099/0022-1317-69-7-1575. [DOI] [PubMed] [Google Scholar]
- 13.Daily H A, Strittmatter P. Structural and functional properties of the membrane binding segment of cytochrome b5. J Biol Chem. 1978;253:8203–8209. [PubMed] [Google Scholar]
- 14.Damke H, Baba T, Warnock D E, Schmid S L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 16.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers C C, O’Callaghan D J. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type I Us9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology. 1992;190:307–315. doi: 10.1016/0042-6822(92)91217-i. [DOI] [PubMed] [Google Scholar]
- 18.Frame M C, McGeoch D J, Rixon F J, Orr A C, Marsden H S. The 10K virion phosphoprotein encoded by gene Us9 from herpes simplex virus type I. Virology. 1986;150:321–332. doi: 10.1016/0042-6822(86)90297-7. [DOI] [PubMed] [Google Scholar]
- 19.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haanes E J, Tomlinson C C. Genomic organization of the canine herpesvirus US region. Virus Res. 1998;53:151–162. doi: 10.1016/s0168-1702(97)00152-4. [DOI] [PubMed] [Google Scholar]
- 22.Hamer I, Renfrew Haft C, Paccaud J-P, Maeder C, Taylor S, Carpentier J-L. Dual role of a dileucine motif in insulin receptor endocytosis. J Biol Chem. 1997;272:21685–21691. doi: 10.1074/jbc.272.35.21685. [DOI] [PubMed] [Google Scholar]
- 23.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 24.Janiak F, Leber B, Andrews D W. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J Biol Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 25.Jing S Q, Spencer T, Miller K, Hopkins C, Trowbridge I S. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J Cell Biol. 1990;110:283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joens A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones B G, Thomas L, Molloy S S, Thulin C D, Fry M D, Walsh K A, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalejta, R. F., A. D. Brideau, B. W. Banfield, and A. J. Beavis. An integral membrane green fluorescent protein marker, US9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. J. Exp. Cell Biol., in press. [DOI] [PubMed]
- 29.Killeen A M, Harrington L, Wall L V M, Kelly D C. Nucleotide sequence analysis of a homologue of herpes simplex virus type I gene Us9 found in the genome of simian herpes B virus. J Gen Virol. 1992;73:195–199. doi: 10.1099/0022-1317-73-1-195. [DOI] [PubMed] [Google Scholar]
- 30.Kirchhausen T, Bonifacino J S, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 31.Kutay U, Hartmann E, Rapoport T A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 32.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport T A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Borgne R, Schmidt A, Mauxion F, Griffiths G, Hoflack B. Binding of AP-1 Golgi adaptors to membranes requires phosphorylated cytoplasmic domains of the mannose 6-phosphate/insulin-like growth factor II receptor. J Biol Chem. 1993;268:22552–22556. [PubMed] [Google Scholar]
- 34.LeBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung-Tack P, Audonnet J, Riviere M. The complete DNA sequence and the genetic organization of the short unique region (Us) of the bovine herpesvirus type 1 (ST strain) Virology. 1994;199:409–421. doi: 10.1006/viro.1994.1139. [DOI] [PubMed] [Google Scholar]
- 36.Linstedt A D, Foguet M, Renz M, Seelig H P, Glick B S, Hauri H P. A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golgi apparatus. Proc Natl Acad Sci USA. 1995;92:5102–5105. doi: 10.1073/pnas.92.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomniczi B, Blankenship M L, Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the gene. J Virol. 1984;49:970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marks M S, Ohno H, Kirchhausen T, Bonifacino J S. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 40.Matsuoka Y, Ihara T, Bishop D H L, Compans R W. Intracellular accumulation of Punta Toro virus glycoproteins expressed from cloned cDNA. Virology. 1988;167:251–260. doi: 10.1016/0042-6822(88)90075-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauxion F, Le Borgne R, Munier-Lehmann H, Hoflack B. A casein kinase II phosphorylation site in the cytoplasmic domain of the cation-dependent mannose 6-phosphate receptor determines the high affinity interaction of the AP-1 Golgi assembly proteins with membranes. J Biol Chem. 1996;271:2171–2178. doi: 10.1074/jbc.271.4.2171. [DOI] [PubMed] [Google Scholar]
- 42.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type I. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 43.McGraw T E, Maxfield F R. Human transferrin receptor internalization is partially dependent upon an aromatic amino acid on the cytoplasmic domain. Cell Regul. 1990;1:369–377. doi: 10.1091/mbc.1.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mettenleiter T C, Lukacs N, Rziha H J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J Virol. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moremen K W, Touster O, Robbins P W. Novel purification of the catalytic domain of Golgi α-mannosidase II: characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- 47.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The Us 9, 10, 11, and 12 genes of herpes simplex virus type I are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 48.Olson J, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson J K, Grose C. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor complex. J Virol. 1998;72:1542–1551. doi: 10.1128/jvi.72.2.1542-1551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrovskis E A, Timmins J, Gierman T, Post L. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrovskis E A, Post L E. A small open reading frame in pseudorabies virus and implications for evolutionary relationships between herpesviruses. Virology. 1987;159:193–195. doi: 10.1016/0042-6822(87)90368-0. [DOI] [PubMed] [Google Scholar]
- 52.Renfrew Haft C, Klausner R D, Taylor S I. Involvement of dileucine motifs in the internalization and degradation of the insulin receptor. J Biol Chem. 1994;269:26286–26294. [PubMed] [Google Scholar]
- 53.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 54.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer W, Stroh A, Berghoefer S, Seiler J, Vey M, Kruse M, Kern H F, Klenk H, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takagaki Y, Radhakrishnan R, Wirtz K W, Khorana H G. The membrane-embedded segment of cytochrome b5 as studied by cross-linking with photoactivatable phospholipids. II. The nontransferable form. J Biol Chem. 1983;258:9136–9142. [PubMed] [Google Scholar]
- 57.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 58.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trus B L, Gibson W, Cheng N, Steven A C. Tegument-capsid interactions of cytomegalovirus. Presented at the 23rd International Herpesvirus Workshop. 1998. York, England. [Google Scholar]
- 60.Tyack S G, Studdert M J, Johnson M A. Nucleotide sequence of canine herpesvirus homologues of herpes simplex virus type 1 US2, US3, glycoproteins I and E, US8.5 and US9 genes. DNA Seq. 1997;7:365–368. doi: 10.3109/10425179709034057. [DOI] [PubMed] [Google Scholar]
- 61.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. Identification of two genes in the unique short region of pseudorabies virus: comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]
- 62.Verhey K J, Birnbaum M J. A Leu-Leu sequence is essential for COOH-terminal targeting signal of GLUT4 glucose transporter in fibroblasts. J Biol Chem. 1994;269:2353–2356. [PubMed] [Google Scholar]
- 63.Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks M S, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waters M G, Clary D O, Rothman J E. A novel 115-kd peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 67.Wilde A, Reaves B, Banting G. Epitope mapping of two isoforms of a trans-Golgi network specific integral membrane protein TGN 38/41. FEBS Lett. 1992;313:235–238. doi: 10.1016/0014-5793(92)81199-v. [DOI] [PubMed] [Google Scholar]
- 68.Willemse M J, Strijdveen I G L, van Schooneveld S H B, van Den Berg M C, Sondermeijer P J A. Transcriptional analysis of the short segment of the feline herpesvirus type 1 genome and insertional mutagenesis of a unique reading frame. Virology. 1995;208:704–711. doi: 10.1006/viro.1995.1202. [DOI] [PubMed] [Google Scholar]
- 69.Zelnik V, Darteil R, Audonnet J C, Smith G D, Riviere M, Pastorek J, Ross L J N. The complete sequence and gene organization of the short unique region of herpesvirus of turkeys. J Gen Virol. 1993;74:2151–2162. doi: 10.1099/0022-1317-74-10-2151. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Z, Hao Y, Gershon M D, Ambron R T, Gershon A A. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J Virol. 1996;70:6563–6575. doi: 10.1128/jvi.70.10.6563-6575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]