Abstract
Background
Cardiac hypertrophy, which develops in middle-aged and older individuals as a consequence of hypertension and obesity, is an established risk factor for sudden cardiac death (SCD). However, it is sometimes difficult to differentiate SCD with acquired cardiac hypertrophy (SCH) from compensated cardiac hypertrophy (CCH), at autopsy. We aimed to elucidate the proteomic alteration in SCH, which can be a guideline for future postmortem diagnosis.
Methods
Cardiac tissues were sampled at autopsy. SCH group consisted of ischemic heart failure, hypertensive heart failure, and aortic stenosis. CCH group included cases of non-cardiac death with cardiac hypertrophy. The control group comprised cases of non-cardiac death without cardiac hypertrophy. All patients were aged > 40 years, and hypertrophic cardiomyopathy was not included in this study. We performed histological examination and shotgun proteomic analysis, followed by quantitative polymerase chain reaction analysis.
Results
Significant obesity and myocardial hypertrophy, and mild myocardial fibrosis were comparable in SCH and CCH cases compared to control cases. The proteomic profile of SCH cases was distinguishable from those of CCH and control cases, and many sarcomere proteins were increased in SCH cases. Especially, the protein and mRNA levels of MYH7 and MYL3 were significantly increased in SCH cases.
Conclusion
This is the first report of cardiac proteomic analysis in SCH and CCH cases. The stepwise upregulation of sarcomere proteins may increase the risk for SCD in acquired cardiac hypertrophy before cardiac fibrosis progresses significantly. These findings can possibly aid in the postmortem diagnosis of SCH in middle-aged and older individuals.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00414-023-03038-6.
Keywords: Sudden cardiac death, Cardiac hypertrophy, Fibrosis, Proteomics, Sarcomere proteins, MYH7, MYL3
Introduction
Sudden cardiac death (SCD) refers to unexpected death not attributable to an extracardiac cause, occurring usually within 1 h of symptom onset (or within 24 h of last being seen in good health if the death is unwitnessed) [1]. Lethal arrhythmia due to coronary artery disease is considered the major cause of SCD in middle-aged and older individuals [2, 3]. High levels of coronary atherosclerotic plaques are often observed at autopsy in middle-aged and older individuals who die of non-cardiac cause. Thus, together with coronary plaque, myocardial molecular remodeling can also be a substrate for lethal arrythmia which can contribute to SCD in the middle-aged and older population.
Cardiac hypertrophy is an established risk factor for SCD, known to increase the risk of SCD with an odds ratio of 2.5–4.2 [4–6]. Among patients with cardiac hypertrophy, hypertrophic cardiomyopathy (HCM) has been cited as a leading cause of SCD in younger populations, whereas hypertension/obesity-related cardiac hypertrophy is a more common finding in SCD in middle-aged and older populations [6–8]. Physiologically, cardiac hypertrophy occurs in response to pressure and volume overload, progressing through the compensated phase without symptoms until heart failure in the decompensated phase [9, 10]. Although years or decades may pass between the first episode of heart failure and cardiac death in the decompensated phase, some patients die suddenly before symptoms of heart failure appear. Given accelerated population aging in recent years, the clinical impact of SCD with acquired cardiac hypertrophy (SCH) remains substantial and is likely to increase.
At autopsy, it is sometimes difficult to grossly differentiate SCH from compensated cardiac hypertrophy (CCH), which does not contribute to the cause of death in middle-aged and older generations. Histological findings such as coronary atherosclerosis and cardiac hypertrophy are not sufficient to diagnose SCD; circumstantial information of sudden death and negative results of toxic screening are often necessary for the final diagnosis of SCD. Therefore, further information of cardiac molecular alterations that contribute to SCD is essential to differentiate SCH from CCH cases at postmortem examination.
Recently, genetic variants have gained attention as an explanation of the variable risk of SCD in patients with acquired cardiac diseases. Postmortem genetic testing has revealed that 3–10% of SCH cases are likely caused by pathogenic variants, where HCM is histologically denied [8, 11–13]. These results indicate that commonly acquired cases of cardiac hypertrophy may interact with genetic variants. However, the broad genetic panel mostly detects variants of unknown significance; therefore, genetic screening confers less diagnostic utility in cases of acquired cardiac hypertrophy than in cases of inherited cardiomyopathy, as it cannot further evaluate the pathogenicity of variants.
In this study, we aimed to elucidate the proteomic changes in middle-aged and older human heart, which can directly contribute to the pathology of SCH and may be used for the postmortem diagnosis.
Materials and methods
Study design
Forensic cases in which patients were discovered within 24 h after death were enrolled in this study (Table 1 and Online Resource 1). All patients were > 40 years of age. To focus on the acquired cardiac hypertrophy, HCM cases were not included. As well, to focus on the early proteomic changes, cases after cardiac operation or grossly presenting cardiac fibrosis were not included. This study was approved by the institutional ethical committee (21R177), and informed consent was obtained from the relatives of all patients.
Table 1.
Clinical characteristics of included patients
| SCH (n = 10) | CCH (n = 10) | Control (n = 10) | |
|---|---|---|---|
| Cause of death |
SCD: IHF (6), HHF (3), AS (1) |
Accident (8), Cerebral hemorrhage (2) |
Accident (8), Pneumonia (2) |
| Age, years | 63 (41–82) | 59 (43–74) | 58 (42–76) |
| Sex | 10 men | 10 men | 8 men, 2 women |
| BMI (kg/m2) | 25.0 (17.5–34.5) * | 25.4 (19.4–28.4) * | 20.6 (14.0–27.5) |
| BNP (pg/mL) | 30.0 (2.0–56.6) | 16.4 (2.0–81.5) | 6.0 (2.0–21.8) |
SCD sudden cardiac death; SCH SCD with acquired cardiac hypertrophy; CCH compensated cardiac hypertrophy; IHF ischemic heart failure; HHF hypertensive heart failure; AS aortic stenosis; BMI body mass index; BNP B-type natriuretic peptide whose lowest detectable level is 2.0 pg/mL in our assay. * p < 0.05; ** p < 0.001, compared with control cases. Individual characteristics are detailed in Online Resource 1
SCD was defined as death within 1 h of symptom onset or unwitnessed death with the individual having normal health within 24 h before death [1]. Cardiac hypertrophy was diagnosed as heart weight/body height (g/cm) > 2.38 in men and > 2.15 in women, respectively [6]. SCH was defined as SCD with acquired cardiac hypertrophy, consisting of ischemic heart failure, hypertensive heart failure, and aortic stenosis. Ischemic heart failure was diagnosed based on a high level of coronary atherosclerosis or coronary thrombus, and exclusion of other causes of death. Hypertensive heart failure was diagnosed based on a clinical history of hypertension and hypertension-related changes such as nephrosclerosis in those without a high level of coronary atherosclerotic plaques. Because there are some cases in which cardiac hypertrophy contributes relatively less to the cause of death, we set CCH as the subcontrol group in this study. The CCH group included cases of non-cardiac death in patients with cardiac hypertrophy without a clinical history of heart failure. The control group included cases of non-cardiac death in patients without cardiac hypertrophy or a clinical history of heart failure.
Tissue sampling and histological analysis
The body was stored at 4 ℃ until autopsy, and the cardiac tissue was extracted during autopsy. At the middle level between the base and apex, small tissue samples from the middle layer of the left ventricular free wall were removed and immediately frozen at -80 ℃ and stored until protein isolation. The other cardiac tissues were preserved in 10% formalin, and 4-μm sections were stained with hematoxylin and eosin (H&E), and picrosirius red dyes. Microscopic measurements were performed in the sections from the tissues obtained from the middle layer of the left ventricular free wall. The endocardium, epicardium, and trabeculae carneae were excluded. The cardiomyocyte diameter was measured at the nuclear point with H&E staining at × 200 magnification, and the values were averaged from 10 fields per case. The myocardial fibrosis was quantitatively assessed with picrosirius red at × 40 magnification using Image J software (https://imagej.net/), and the percentage to the total image were averaged in 10 fields per case [14, 15].
Proteomic analysis
Cardiac tissue samples were homogenized using a Shake Master Neo system (Bio Medical Science, Tokyo, Japan) in an equal volume of phase-transfer surfactant-based lysis buffer containing 12 mM deoxycholic acid, 12 mM sodium N-lauroylsarcosine, 50 mM ammonium bicarbonate, and protease inhibitors [16]. After centrifugation at 15,000 × g for 5 min, the collected supernatant was subjected to protein quantification using a DC protein assay (Bio-Rad, Hercules, CA, USA).
For quantitative proteomic analysis, 5 μg protein from the supernatant samples were used. Protein samples were enzymatically digested with mass spectrometry (MS)-grade trypsin (Promega, Madison, WI, USA). Digested samples were dried using a SpeedVac vacuum concentrator (Thermo Fisher Scientific, Waltham, MA, USA) and dissolved in 50 mM ammonium bicarbonate buffer containing 0.1% trifluoroacetic acid (TFA). Samples were loaded onto C18 StageTips [17] and subsequently eluted with 30 and 60% acetonitrile in 50 mM ammonium bicarbonate. The eluted samples were dried using a SpeedVac vacuum concentrator and dissolved in 13 μL of 2% acetonitrile in 0.1% formic acid, of which 6 μL was applied for liquid chromatography–MS (LC–MS). LC–MS-grade ultrapure water and acetonitrile were purchased from Merck (Darmstadt, Germany). TFA was purchased from Wako Pure Chemical Industries (Osaka, Japan). LC–MS grade formic acid was obtained from Thermo Fischer Scientific.
Quantitative proteomic analysis was performed as previously described [18], with minor modifications. A Q-Exactive mass spectrometer coupled with an UltiMate 3000 High Performance Liquid Chromatography (HPLC) system (Thermo Fisher Scientific) was used. The injected peptides were separated using a nano-LC capillary column (75 μm × 180 mm, C18, 3 μm; Nikkyo Technos, Tokyo, Japan) at a flow rate of 300 nL/min. The aqueous mobile phase (eluent A) was 0.1% formic acid in 2% acetonitrile and the organic mobile phase (eluent B) was 0.1% formic acid in 95% ACN. The elution gradient was 2–33% B for 120 min. Data were acquired using a data-dependent acquisition protocol in which an MS1 scan ranging from 350 to 1,500 m/z was selected, followed by higher-energy C-trap dissociation (HCD)-based MS/MS fragmentation against the 20 highest precursor peaks. Raw data files were subjected to protein identification analysis using Mascot server ver.2.7 (Matrix Science, London, UK). Protein searches were conducted against the SwissProt human database (January 2020). The mass tolerances for the precursor and product ions were 5 ppm and 0.05 Da, respectively. Two trypsin missed cleavages and variable modifications of N-terminal acetylation, N-terminal carbamylation, cysteine carbamidomethylation, and methionine oxidation were allowed. Proteins were identified with FDR 1% and peptide ion score > 30.
Label-free quantitative analysis of the identified proteins was performed using the Progenesis QI for proteomics v2 software (Nonlinear, Durham, NC, USA). Protein levels were normalized using the ratiometric method in log space, and a p-value < 0.05 in an analysis of variance among the groups was considered significant. Principal component analysis and differential analysis were performed using SIMCA (Infocom, Tokyo, Japan). Gene ontology analysis was performed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/), and a p-value < 0.05 obtained using the Benjamini–Hochberg method was considered significant.
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from the cardiac tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed into cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA). qPCR was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems). Triplicate Ct values obtained for tenfold diluted cDNA were averaged, and the relative expression of target mRNA was determined via the ΔΔCt method [19], using GAPDH as an internal control. The relative levels were normalized to the average values of the control cases. The primers for MYH7 (Hs01110632_m1), MYL3 (Hs00264820_m1), ACTC1(Hs01109515_m1), and GAPDH (4333764F) were obtained using TaqMan Assay (Applied Biosystems).
Statistical analysis
Multiple comparisons were performed using the Steel–Dwass test for clinical value, qPCR, and histological results in Excel Statistics 2015 (SSRI, Tokyo, Japan). Differences with p values < 0.05 were considered statistically significant.
Results
Histological analysis
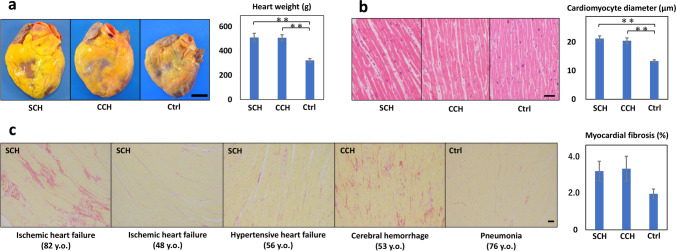
The heart weights of SCH (511 ± 34 g) and CCH (508 ± 25 g) were significantly heavier than the control cases (322 ± 15 g), but there was no significant difference between SCH and CCH cases (Fig. 1a). As well, the cardiomyocyte diameter was significantly increased in SCH (21.2 ± 0.9 µm) and CCH (20.5 ± 1.0 µm) compared with the control cases (13.4 ± 0.4 µm), but there was no significant difference between SCH and CCH cases (Fig. 1b). No significant accumulation of inflammatory cells was observed in any case.
Fig. 1.
Histological characteristics of the specimens obtained from the cases. a Representative macroscopic images of the heart. There was comparable increase in heart weights in SCH and CCH cases when compared with that in control cases. Scale bar = 3 cm. b Representative microscopic images of cardiomyocytes with hematoxylin and eosin staining. The cardiomyocytes diameters are increased in equal proportions in SCH and CCH cases when compared with that in control cases. Scale bar = 50 μm. c Representative microscopic images with picrosirius red staining. The myocardial fibrosis fractions are moderately visible in SCH and CCH cases when compared with that in control cases. Scale bar = 200 μm. Graphical error bars represent mean ± SE. *p < 0.05, **p < 0.001. SCH, sudden cardiac death with acquired cardiac hypertrophy; CCH, compensated cardiac hypertrophy
The myocardial fibrosis level was moderately increased in SCH (3.1 ± 0.5%) and CCH (3.2 ± 0.7%) compared with the control cases (1.9 ± 0.3%), but no significant substantial variation was observed (Fig. 1c). Elderly SCH cases with ischemic heart failure or aortic stenosis presented the most prominent patchy fibrosis, while the middle-aged SCH cases presented a little to moderate levels of myocardial fibrosis. In contrast, CCH cases with cerebral hemorrhage presented more prominent diffuse fibrosis than SCH cases with hypertensive heart failure. In the control cases, a little to moderate levels of myocardial fibrosis was observed elderly cases.
Proteomic analysis
Proteomic profiles were obtained from three patients with SCH (all hypertensive heart failure), three with CCH, and four control patients. A total of 9,581 peptides and 1,551 proteins were identified, and the abundance of 140 proteins significantly differed between the groups (Online Resources 2 and 3). Among the top 1,000 abundant proteins, 19 exhibited significant differences with fold changes > 5 among the groups, and most of them were upregulated in the SCH group (Table 2).
Table 2.
Major cardiac proteins differentially expressed based on proteomic analysis
| Gene ID | Description | p-value | Fold change | Highest group | Lowest group | Average abundance | ||
|---|---|---|---|---|---|---|---|---|
| A: SCH | B: CCH | C: Control | ||||||
| MYH7 | Myosin-7 (β-Myosin heavy chain) | 0.00001 | 31.7 | A | C | 4,398,718,463 | 240,223,132 | 138,631,218 |
| MYL3 | Myosin light chain 3 | 0.00515 | 5.4 | A | C | 2,278,403,475 | 774,826,120 | 419,683,634 |
| ACTC1 | Actin, alpha cardiac muscle 1 | 0.00011 | 6.4 | A | C | 661,847,239 | 111,049,697 | 102,836,296 |
| ACTA1 | Actin, alpha skeletal muscle | 0.01325 | 5.0 | A | C | 237,838,909 | 79,671,590 | 47,534,401 |
| H4C1 | Histone H4 | 0.02068 | 6.1 | A | C | 126,050,127 | 63,734,082 | 20,503,322 |
| BLVRA | Biliverdin reductase A | 0.00001 | 7.4 | A | B | 11,870,823 | 1,594,859 | 1,906,043 |
| RBP4 | Retinol-binding protein 4 | 0.00050 | 6.1 | A | C | 6,842,534 | 2,062,599 | 1,124,422 |
| MYH9 | Myosin-9 | 0.00014 | 46.4 | A | B | 5,151,286 | 111,106 | 130,214 |
| MYOM3 | Myomesin-3 | 0.00319 | 6.9 | A | C | 4,905,559 | 2,122,198 | 714,203 |
| MT-CO1 | Cytochrome c oxidase subunit 1 | 0.00390 | 6.4 | A | C | 3,188,028 | 1,204,247 | 501,341 |
| MT-ATP6 | ATP synthase subunit a | 0.00442 | 5.4 | C | A | 2,604,003 | 11,058,033 | 14,132,003 |
| PSMA5 | Proteasome subunit alpha type-5 | 0.00006 | 9.1 | A | C | 1,780,039 | 355,736 | 196,202 |
| MACROH2A1 | Core histone macro-H2A.1 | 0.04699 | 5.2 | A | C | 1,695,102 | 813,034 | 328,393 |
| MYH11 | Myosin-11 | 0.00118 | 33.0 | A | C | 1,229,860 | 43,050 | 37,230 |
| C1QB | Complement C1q subcomponent subunit B | 0.02465 | 5.5 | A | B | 1,165,830 | 210,848 | 535,773 |
| PSMB1 | Proteasome subunit beta type-1 | 0.00014 | 13.7 | A | C | 1,072,370 | 194,629 | 78,070 |
| RHOC | Rho-related GTP-binding protein RhoC | 0.03362 | 8.4 | C | A | 232,937 | 834,397 | 1,948,366 |
| SLFN12L | Schlafen family member 12-like | 0.01401 | 8.0 | C | A | 135,208 | 498,154 | 1,075,500 |
| THBS4 | Thrombospondin-4 | 0.00455 | 14.9 | B | A | 69,740 | 1,035,738 | 691,891 |
ATP adenosine triphosphate; GTP guanosine-5'-triphosphate; SCD sudden cardiac death; SCH SCD with acquired cardiac hypertrophy; CCH compensated cardiac hypertrophy
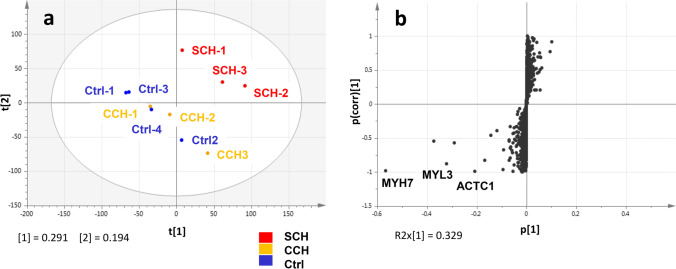
Principal component analysis of the total peptides indicated that SCH cases had profiles distinguishable from those of CCH and control cases, with the latter two exhibiting relatively similar profiles (Fig. 2a). Differential analysis of the total proteins revealed some sarcomeric proteins, including MYH7, MYL3, and ACTC1, that characterized SCH (vs. CCH) (Fig. 2b).
Fig. 2.
Multivariate analysis of the proteomic data. a Principal component analysis of total peptides. b S-plot loading diagram for the differentiation of the total proteins between SCH and CCH. SCH, sudden cardiac death with acquired cardiac hypertrophy; CCH, compensated cardiac hypertrophy
Gene ontology analysis of the 85 proteins that were significantly upregulated in SCH revealed their significant contributions to proteasomes, cardiac muscle contraction, and tight junction function (Table 3). Similarly, analysis of the 41 proteins that were significantly downregulated in SCH revealed their moderate contributions to oxidative phosphorylation and ribosome function (Table 4).
Table 3.
Functional annotation summary of highly abundant proteins in SCH cases
| Term | Count | % | Gene ID | p-value |
|---|---|---|---|---|
| Proteasome | 11 | 10.9 | PSMA4, PSMA5, PSMB4, PSMB5, PSMC1, PSMC4, PSMC5, PSMC6, PSMB1, PSMD1PS, MD12 | 6.30E-10 |
| Cardiac muscle contraction | 9 | 8.9 | MYH7, ACTC1, MYL3, TPM1, TPM2, MT-CO1, MT-CO2, COX5A, COX7C | 2.29E-05 |
| Tight junction | 9 | 8.9 | MYH4, MYH3, MYH9, MYH11, MYH13, MYH14, MYH15, ACTB, MYL12B | 4.81E-05 |
| Ribosome | 8 | 7.9 | RPS2, RPS9, RPS27A, RPLP2, MRPS7, RPL17, RPL27, RPL34 | 0.00704 |
| Adrenergic signaling in cardiomyocytes | 7 | 6.9 | MYH7, ACTC1, TPM1, TPM2, MYL3, PPP1CB, PPP1CC | 0.03677 |
SCH sudden cardiac death with acquired cardiac hypertrophy
Table 4.
Functional annotation summary of weakly abundant proteins in SCH cases
| Term | Count | % | Gene ID | p-value |
|---|---|---|---|---|
| Oxidative phosphorylation | 5 | 9.6 | COX15, MT-ATP6, NDUFB1, NDUFV1, NDUFA5 | 0.034 |
| Ribosome | 5 | 9.6 | RPS5, RPL15, RPLP1, MRPL23, MRPL27 | 0.034 |
SCH sudden cardiac death with acquired cardiac hypertrophy
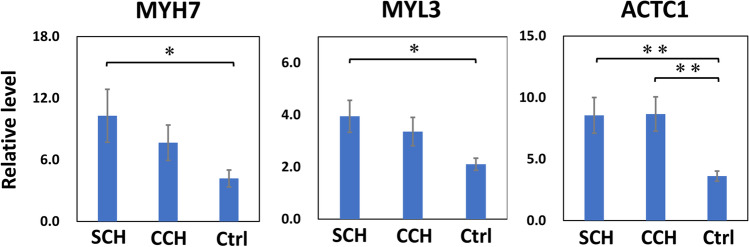
qPCR
Levels of MYH7 and MYL3 mRNA were significantly increased in SCH and moderately increased in CCH, when compared with levels observed in control cases (Fig. 3). The level of ACTC1 expression was also significantly higher in SCH and CCH cases than that in control cases.
Fig. 3.
qPCR for MYH7, ACTC1, and MYL3. Quantitative PCR was performed using GAPDH as an endogenous control in human hearts. Error bars represent mean ± SE. *p < 0.05, **p < 0.001. SCH, sudden cardiac death with cardiac hypertrophy (n = 10); CCH, compensated cardiac hypertrophy (n = 10); Ctrl, control (n = 10)
Discussion
Our study focused on acquired cardiac hypertrophy which accompanies obesity, hypertension, coronary arteriosclerosis, or aortic stenosis, which are common diseases of middle-aged and older adults. To the best of our knowledge, this study is the first to reveal proteomic changes in human cardiac tissue samples of patients with acquired cardiac hypertrophy resulting in SCD. Interestingly, protein and mRNA levels of MYH7, MYL3, and ACTC1, the major sarcomere genes whose variants can cause inherited cardiomyopathies such as HCM [20–22], and dilated cardiomyopathy [23, 24] were significantly increased in SCH cases. Studies have reported that some types of genetic mutations in these sarcomere proteins may increase the risk of SCD in patients with inherited cardiomyopathy [25–28]. The proteomic analysis in this study indicates that both qualitative and quantitative changes in these sarcomere proteins can influence cardiac function and increase the risk of SCD. Moreover, the proteomic changes here preceded the histological changes: the levels of sarcomere proteins were increased stepwise from CCH to SCH, while the levels of myocardial hypertrophy and fibrosis were unchanged. Thus, the elevation of sarcomere proteins may differentiate SCH from CCH in the early phase acquired cardiac hypertrophy before the myocardial fibrosis, a classical pathological finding of heart failure, progresses significantly.
The myosin heavy chain (MHC) is a functional myosin motor molecule, and two isoforms, α-MHC encoded by MYH6 and β-MHC encoded by MYH7, are expressed in the mammalian heart. The contractile velocity and energy consumption are two- to three-fold higher for α-MHC than those for β-MHC [29, 30]. In the rodent ventricle, with > 90% of the total MHC proteins, α-MHC predominates and contributes to rapid contraction [31, 32]. Meanwhile, β-MHC predominates with > 95% of the total MHC proteins in the human ventricle and results in a lower resting heart rate [33, 34]. Ventricular MYH6 significantly decreases and MYH7 significantly increases in the rodent heart with pressure or volume overload [34–36]. In contrast, studies suggest that ventricular MYH7 does not increase as significantly in human heart failure as in rodent models of heart failure [37–39], and decreased levels of MYH7 have been observed at the end of heart failure in some clinical studies [40, 41]. Thus, direct extrapolation from model animals, such as rats and mice, to humans is often challenging in cardiac research [42, 43]. Molecular analysis of human cardiac tissue samples can produce valuable data for understanding human cardiac pathology. Moreover, SCD samples can be collected and studied in detail in forensics rather than general clinical field, because of the accessibility to the SCD cases outside the hospital. Therefore, analyzing autopsy tissue samples to reveal the molecular pathology of SCD can be an important objective for the forensic pathologists.
This study has some practical limitations. First, the sample size was limited as we selected relatively fresh cases of autopsy. Although postmortem changes have often prevented the large-scale molecular studies to date, we believe that accrual of SCD cases in small-size studies can help to elucidate the pathology and contribute to accurate postmortem diagnosis in the future. Second, there were some discrepancies between the quantitative results obtained using proteomics and qPCR in this study, which may have been caused by the heterogeneous distribution of the target proteins in the heart. Moreover, gene ontology analysis revealed that altered proteasome activity in SCH may decrease protein clearance, resulting in the accumulation of sarcomere proteins without dramatic alterations in their mRNA levels. Future studies including microscopic sampling with microdissection and analyses at the single-cell level, may reveal the more precise pathology of SCH in the human heart.
Conclusions
This is the first report of proteomic analysis in SCH and CCH using human cardiac tissue samples. Histologically SCH and CCH cases presented equal levels of significant myocardial hypertrophy and mild fibrosis. Meanwhile, the proteomic profiling demonstrated the significant upregulation of sarcomere proteins such as MYH7 and MYL3 in SCH cases. The stepwise upregulation of the sarcomere proteins from CCH to SCH may increase the risk for SCD in the acquired cardiac hypertrophy, and these findings can possibly be useful in the postmortem diagnosis of SCH in middle-aged and older individuals.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to the technical staff at the Forensic Laboratory and the Support Center for Medical Research and Education at Tokai University. We would also like to thank Editage for English language editing.
Authors’ contribution
Conceptualization: Yu Kakimoto; Methodology: Masatoshi Ito and Masayuki Tanaka; Formal analysis and investigation: Atsushi Ueda and Tomoko Kubota; Writing—original draft preparation: Yu Kakimoto and Masatoshi Ito; Writing—review and editing: Motoki Osawa; Funding acquisition: Yu Kakimoto; Resources: Yu Kakimoto and Motoki Osawa. Supervision: Shotaro Isozaki.
Funding
This work was supported by JSPS KAKEHNHI (grant number: 20K10565 and 23H03178).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval
This study was approved by the institutional ethical committee (21R177). The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments.
Consent
Informed consent was obtained from the relatives of all patients.
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/15/2023
Correcting the layout by following the citation first before image and ammending layout of the Tables.
References
- 1.Fishman GI, Chugh SS, Dimarco JP, Albert CM, Anderson ME, Bonow RO, et al. Sudden cardiac death prediction and prevention: Report from a National Heart, Lung, and Blood Institute and Heart Rhythm Society Workshop. Circulation. 2010;122:2335–2348. doi: 10.1161/CIRCULATIONAHA.110.976092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isbister J, Semsarian C. Sudden cardiac death: an update. Intern Med J. 2019;49:826–833. doi: 10.1111/imj.14359. [DOI] [PubMed] [Google Scholar]
- 3.Luqman N, Sung RJ, Wang CL, Kuo CT. Myocardial ischemia and ventricular fibrillation: pathophysiology and clinical implications. Int J Cardiol. 2007;119:283–290. doi: 10.1016/j.ijcard.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Narayanan K, Reinier K, Teodorescu C, Uy-Evanado A, Aleong R, Chugh H, et al. Left ventricular diameter and risk stratification for sudden cardiac death. J Am Heart Assoc. 2014;3:e001193. doi: 10.1161/JAHA.114.001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayanan K, Reinier K, Teodorescu C, Uy-Evanado A, Chugh H, Gunson K, et al. Electrocardiographic versus echocardiographic left ventricular hypertrophy and sudden cardiac arrest in the community. Heart Rhythm. 2014;11:1040–1046. doi: 10.1016/j.hrthm.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakimoto Y, Asakura K, Osawa M. Cutoff value for hypertrophic heart weight in the Japanese population. Leg Med (Tokyo) 2021;48:101831. doi: 10.1016/j.legalmed.2020.101831. [DOI] [PubMed] [Google Scholar]
- 7.Hookana E, Junttila MJ, Puurunen VP, Tikkanen JT, Kaikkonen KS, Kortelainen ML, et al. Causes of nonischemic sudden cardiac death in the current era. Heart Rhythm. 2011;8:1570–1575. doi: 10.1016/j.hrthm.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Holmström L, Pylkäs K, Tervasmäki A, Vähätalo J, Porvari K, Pakanen L, et al. Genetic contributions to the expression of acquired causes of cardiac hypertrophy in non-ischemic sudden cardiac death victims. Sci Rep. 2021;11:11171. doi: 10.1038/s41598-021-90693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitoulis FG, Terracciano CM. Heart plasticity in response to pressure- and volume-overload: A review of findings in compensated and decompensated phenotypes. Front Physiol. 2020;11:92. doi: 10.3389/fphys.2020.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel ED, Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc Res. 2011;90:234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahrouchi N, Raju H, Lodder EM, Papatheodorou S, Miles C, Ware JS, et al. The yield of postmortem genetic testing in sudden death cases with structural findings at autopsy. Eur J Hum Genet. 2020;28:17–22. doi: 10.1038/s41431-019-0500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglesias M, Ripoll-Vera T, Perez-Luengo C, García AB, Moyano S, Canos JC et al (2021) Diagnostic yield of genetic testing in sudden cardiac death with autopsy findings of uncertain significance. J Clin Med 10. 10.3390/jcm10091806 [DOI] [PMC free article] [PubMed]
- 13.Larsen MK, Christiansen SL, Hertz CL, Frank-Hansen R, Jensen HK, Banner J, Morling N. Targeted molecular genetic testing in young sudden cardiac death victims from Western Denmark. Int J Legal Med. 2020;134:111–121. doi: 10.1007/s00414-019-02179-x. [DOI] [PubMed] [Google Scholar]
- 14.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leader CJ, Moharram M, Coffey S, Sammut IA, Wilkins GW, Walker RJ. Myocardial global longitudinal strain: An early indicator of cardiac interstitial fibrosis modified by spironolactone, in a unique hypertensive rat model. PLOS ONE. 2019;14:e0220837. doi: 10.1371/journal.pone.0220837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda T, Tomita M, Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 17.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 18.Kimura Y, Nakai Y, Shin J, Hara M, Takeda Y, Kubo S, et al. Identification of serum prognostic biomarkers of severe COVID-19 using a quantitative proteomic approach. Sci Rep. 2021;11:20638. doi: 10.1038/s41598-021-98253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Cirino AL, Ho C (1993) Hypertrophic cardiomyopathy overview. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Gripp KW et al (eds), GeneReviews. edn. University of Washington, Seattle. Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved 2022
- 21.Shahzadi SK, Naidoo N, Alsheikh-Ali A, Rizzo M, Rizvi AA, Santos RD, Banerjee Y (2021) Reconnoitering the role of long-noncoding RNAs in hypertrophic cardiomyopathy: A descriptive review. Int J Mol Sci 22. 10.3390/ijms22179378 [DOI] [PMC free article] [PubMed]
- 22.Marian AJ, Braunwald E. Hypertrophic cardiomyopathy: Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res. 2017;121:749–770. doi: 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Feng Y, Zhang YM, Ding XX, Song YZ, Zhang AM, et al. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med. 2015;36:1479–1486. doi: 10.3892/ijmm.2015.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Cao H, Song Y, Feng Y, Ding X, Pang M, et al. Identification of novel mutations including a double mutation in patients with inherited cardiomyopathy by a targeted sequencing approach using the Ion Torrent PGM system. Int J Mol Med. 2016;37:1511–1520. doi: 10.3892/ijmm.2016.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedaghat-Hamedani F, Kayvanpour E, Tugrul OF, Lai A, Amr A, Haas J, et al. Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: A meta-analysis on 7675 individuals. Clin Res Cardiol. 2018;107:30–41. doi: 10.1007/s00392-017-1155-5. [DOI] [PubMed] [Google Scholar]
- 26.Osborn DPS, Emrahi L, Clayton J, Tabrizi MT, Wan AYB, Maroofian R, et al. Autosomal recessive cardiomyopathy and sudden cardiac death associated with variants in MYL3. Genet Med. 2021;23:787–792. doi: 10.1038/s41436-020-01028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavilakandy A, Ahamed H. Mutation of the MYL3 gene in a patient with mid-ventricular obstructive hypertrophic cardiomyopathy. BMJ Case Rep. 2022;15:e244573. doi: 10.1136/bcr-2021-244573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JGW, Owen T, Bhagwan JR, Mosqueira D, Scott E, Mannhardt I, et al. Isogenic pairs of hiPSC-CMs with hypertrophic cardiomyopathy/LVNC-associated ACTC1 E99K mutation unveil differential functional deficits. Stem Cell Rep. 2018;11:1226–1243. doi: 10.1016/j.stemcr.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deacon JC, Bloemink MJ, Rezavandi H, Geeves MA, Leinwand LA. Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell Mol Life Sci. 2012;69:2261–2277. doi: 10.1007/s00018-012-0927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmiter KA, Tyska MJ, Dupuis DE, Alpert NR, Warshaw DM. Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms. J Physiol. 1999;519:669–678. doi: 10.1111/j.1469-7793.1999.0669n.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reiser PJ, Kline WO. Electrophoretic separation and quantitation of cardiac myosin heavy chain isoforms in eight mammalian species. Am J Physiol. 1998;274:H1048–H1053. doi: 10.1152/ajpheart.1998.274.3.H1048. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton N, Ianuzzo CD. Contractile and calcium regulating capacities of myocardia of different sized mammals scale with resting heart rate. Mol Cell Biochem. 1991;106:133–141. doi: 10.1007/BF00230179. [DOI] [PubMed] [Google Scholar]
- 33.Gorza L, Mercadier JJ, Schwartz K, Thornell LE, Sartore S, Schiaffino S. Myosin types in the human heart. An immunofluorescence study of normal and hypertrophied atrial and ventricular myocardium. Circ Res. 1984;54:694–702. doi: 10.1161/01.res.54.6.694. [DOI] [PubMed] [Google Scholar]
- 34.Reiser PJ, Portman MA, Ning XH, Schomisch Moravec C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol. 2001;280:H1814–H1820. doi: 10.1152/ajpheart.2001.280.4.H1814. [DOI] [PubMed] [Google Scholar]
- 35.Alpert NR, Mulieri LA. Functional consequences of altered cardiac myosin isoenzymes. Med Sci Sports Exerc. 1986;18:309–313. doi: 10.1249/00005768-198606000-00009. [DOI] [PubMed] [Google Scholar]
- 36.Satoh M, Nomura S, Harada M, Yamaguchi T, Ko T, Sumida T, et al. High-throughput single-molecule RNA imaging analysis reveals heterogeneous responses of cardiomyocytes to hemodynamic overload. J Mol Cell Cardiol. 2019;128:77–89. doi: 10.1016/j.yjmcc.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Narolska NA, Eiras S, van Loon RB, Boontje NM, Zaremba R, Spiegelen Berg SR, et al. Myosin heavy chain composition and the economy of contraction in healthy and diseased human myocardium. J Muscle Res Cell Motil. 2005;26:39–48. doi: 10.1007/s10974-005-9005-x. [DOI] [PubMed] [Google Scholar]
- 38.Lowes BD, Minobe W, Abraham WT, Rizeq MN, Bohlmeyer TJ, Quaife RA, et al. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. J Clin Invest. 1997;100:2315–2324. doi: 10.1172/JCI119770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 40.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 41.Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W, et al. Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol. 2008;51:129–136. doi: 10.1016/j.jacc.2007.07.087. [DOI] [PubMed] [Google Scholar]
- 42.Prodanovic M, Geeves MA, Poggesi C, Regnier M, Mijailovich SM. Effect of myosin isoforms on cardiac muscle twitch of mice, rats and humans. Int J Mol Sci. 2022;23:1135. doi: 10.3390/ijms23031135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milani-Nejad N, Janssen PM. Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol Ther. 2014;141:235–249. doi: 10.1016/j.pharmthera.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.