Abstract
The Epstein-Barr virus (EBV) genome can persist in dividing human B cells as multicopy circular episomes. Viral episomes replicate in synchrony with host cell DNA and are maintained at a relatively constant copy number for a long time. Only two viral elements, the replication origin OriP and the EBNA-1 protein, are required for the persistence of viral genomes during latency. EBNA-1 activates OriP during the S phase and may also contribute to the partition and/or retention of viral genomes during mitosis. Indeed, EBNA-1 has been shown to interact with mitotic chromatin. Moreover, viral genomes are noncovalently associated with metaphase chromosomes. This suggests that EBNA-1 may facilitate the anchorage of viral genomes on cellular chromosomes, thus ensuring proper partition and retention. In the present paper, we have investigated the chromosome-binding activity of EBV EBNA-1, herpesvirus papio (HVP) EBNA-1, and various derivatives of EBV EBNA-1, fused to a variant of the green fluorescent protein. The results show that binding to metaphase chromosomes is a common property of EBV and HVP EBNA-1. Further studies indicated that at least three independent domains (CBS-1, -2, and -3) mediate EBNA-1 binding to metaphase chromosomes. In agreement with the anchorage model, two of these domains mapped to a region that has been previously demonstrated to be required for the long-term persistence of OriP-containing plasmids.
Epstein-Barr virus (EBV) can establish a latent infection in dividing human B cells. During latency, viral genomes replicate in synchrony with host cell DNA and only once per cell cycle (1, 10, 41). The genomes are nearly always maintained as multicopy, circular episomes (15, 20, 26) although they may sometimes integrate (8, 13).
Strikingly, EBV-infected B-cell lines maintain relatively constant numbers of episomal copies for long periods (reviewed in reference 24), which reflects both the replication of the viral genomes and their efficient partition in daughter cells during cell division. The replication and stable maintenance of latent genomes require only two viral elements: the nuclear protein, EBNA-1 (12, 22), and the latent replication origin, OriP (34, 37, 38). OriP contains multiple copies of an 18-bp EBNA-1 recognition site clustered in two functional elements, namely, the family repeat and the dyad symmetry element (31). In addition to its well-known function in DNA replication, several studies have suggested that EBNA-1 might also facilitate the retention of viral genomes. Indeed, EBNA-1 interaction with the family of repeats contributes to the persistence of nonreplicating plasmids in human and hamster cells and leads to a phenomenon known as transient drug resistance (16, 25, 30, 31). However, EBNA-1 nuclear localization and specific binding to OriP are not sufficient to support efficient retention, indicating that other, as yet unknown, functions are involved (25). In this respect, it is important to note that (i) EBNA-1 is the only EBNA protein that associates with mitotic chromatin in human and mouse cell lines (9, 21, 27–29) and (ii) there is a close, but not covalent, association of extrachromosomal EBV DNA with metaphase chromosomes in various infected B cells (11). In addition, circularized yeast artificial chromosomes that contained OriP have been shown to associate with human chromosomes in the presence of EBNA-1 (32). Thus, EBNA-1 may play a role in episome maintenance both by activating their replication and by controlling their anchorage on cellular chromosomes and their subsequent partition into daughter cells.
To date, no EBNA-1 domains required for its binding to metaphase chromosomes have been identified. Here we took advantage of the bioluminescence properties of a variant of the green fluorescent protein (EGFP) in mammalian cells to investigate the interaction of EBNA-1 with human metaphase chromosomes (4). In agreement with previous studies, it has been shown that EBV EBNA-1 interacts specifically with human chromosomes during mitosis. Moreover, this property is also shared by the EBNA-1 homolog from herpesvirus papio (HVP). By two different strategies, i.e., the observation of chromosomes spread on slides and low-light-level fluorescence microscopy of mitotic living cells, three independent chromosome binding sites have been identified, namely, CBS-1, -2, and -3. Interestingly, CBS-1 and -2 mapped to a region that has previously been shown to be required for long-term maintenance of OriP-containing plasmids. Taken together, these data reinforce the hypothesis that EBNA-1 interaction with mitotic chromosomes is a key determinant of viral episome retention and/or segregation during mitosis.
MATERIALS AND METHODS
Cell lines.
The human epithelial cell line HeLa (ATCC CCL2) was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, streptomycin (105 U/liter), vancomycin (0.1 g/liter), and glutamine (2 mM). 26 CB1 is a nonproducing lymphoblastoid cell line carrying HVP (ATCC CRL 1495). 26 CB1 cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, streptomycin (105 U/liter), vancomycin (0.1 g/liter), and glutamine (2 mM).
Plasmids.
pEGFP-CI (Clontech) encoded a variant of the GFP with enhanced fluorescence (EGFP). It was used to express EBV EBNA-1, HVP EBNA-1, and the different EBV EBNA-1 derivatives fused to the carboxy terminus of EGFP. The EBV EBNA-1 coding sequence (amino acids 8 to 641) was obtained from pCEP4 (Invitrogen) following digestion with RsaI and MluNI. After addition of EcoRI (5′) and BamHI (3′) linkers, the EBNA coding sequence was cloned into pEGFP-CI. The resulting plasmid (pEGFP-EBV-EBNA1) was used to generate the following deletion mutants by PCR with the indicated primers: M1 (E619, GGTCGTGGACCTCGAGAAAAG; EBamHI, ATCTAGATCCGGTGGATCCAG), M5 (E234, ATGGACGAGCTGTACAAGTCC; E339, TGCTCCTGCTCGAGTTCCACCG), M6 (E234; E318, CTCCGGACTCGAGCTCTATG), M7 (E234; E305, CGCCCGGGGCTCGAGGTCTTC), M9 (E306, GGCGGAAGACCTCGAGCCCCG; E339), M10 (E312, CATAGAGCTCGAGTCCGGAG; E339), M15 (E619; E852, CCAGGGATCCAAATCTACTCC), M16 (E360, CACGGTGGAACTCGAGCAGGA; E702, GCGCCTGGATCCATCACCCTG), M17 (E360; E656, TTCAAAATAATCGGGATCCCC), M18 (E600, GGAGCAGGAGCTCGAGGCCGG; E702), M19 (E600; E656), M20 (E600; E391, GCGCGGTGGGGATCCGGATG), M21 (E600; E361, TCTTTCACGGGATCCCCCCCT), M22 (E619; E702), M23 (E619; E656), and M24 (E372, GAAAGAGCTCGAGGGAGAG; E391). M6 dimer contained two M6 inserts cloned in frame which resulted in the duplication of the region encoding amino acids 8 to 67. M2 was generated by inserting the SacI-SacI fragment from pEGFP-EBV-EBNA1 into pEGFP-CI. M3 was generated by cloning the SmaI-SmaI fragment from pEGFP-EBV-EBNA1 into pEGFP-CI. The mutant M4 contained a large, in-frame deletion, which arose spontaneously during PCR with primers E234 and E656. M8 was generated by subcloning the SmaI-SmaI fragment from M6 into pEGFP-CI. M12 was obtained by replacing the internal XhoI/BstXI EBNA-1 fragment from pEGFP-EBV-EBNA1 with the XhoI/BstXI fragment from M3. M14 was obtained by replacing the internal XhoI/BstXI EBNA-1 fragment from pEGFP-EBV-EBNA1 with the XhoI/BstXI fragment from M16. Inserting the XhoI-XhoI fragment of M6 into M14 generated M13. Mutant M11, Rand1, and Rand2 were generated by inserting the following synthetic linkers into pEGFP-CI: M11.S, TCGAGGGAGACCCCAAAAACGTCCAAGTTGCATTGGCTGCAAAG; M11.AS, GATCCTTTGCAGCCAATGCAACTTGGACGTTTTTGGGGTCTCCC; Rand1.S, TCGAGGGGGCAGATGCATTTGCAAACCCCAACGTAAAAGTCCAG; Rand1.AS, GATCCTGGACTTTTACGTTGGGGTTTGCAAATGCATCTGCCCCC; Rand2.S, TCGAGGGATTTGCAAAAGTGGCCCATGCCCAAGAAAACAAAGAG; and Rand2.AS, GATCCTCTTTGTTTTCTTGGGCATGGGCCACTTTTGCAAATCCC.
The HVP EBNA-1 coding sequence (amino acids 9 to 476) (HVP EBNA-1) was generated by PCR from 26 CB1 DNA with primers Pap1 (GGGCCTCGAGCGAACAACG) and Pap2 (GGCGGATCCTAACAAGTTAC) and subcloned into pEGFP-CI. The region encoding amino acids 67 to 93 of HVP EBNA-1 was amplified from 26 CB1 DNA with primers Pap3 (CTGGGATCCGGTCCCCGCC) and Pap4 (ACCGGATCCTCCTGATGTAC) and subcloned into pEGFP-CI.
Plasmid DNA was purified with the Qiagen plasmid Maxi kit (Qiagen). DNA sequencing was performed by automated sequencing by the dideoxynucleotide chain termination method according to the manufacturer’s recommendations (ABI Prism dRhodamine terminator cycle sequencing ready mix; Applied Biosystems).
Transfections.
HeLa cells were grown in 24-well plates until they reached approximately 80% confluence. Purified plasmid DNA (0.5 μg) and Lipofectamine (2 μl; Life Technologies) were used for each transfection according to the manufacturer’s recommendations. The DNA-Lipofectamine complex was overlaid on the cells and incubated at 37°C for 5 h in serum and antibiotic-free medium. Fetal calf serum was added to a 10% final volume, and the cells were further incubated for 12 to 16 h. To evaluate the transfection efficiency and intracellular localization of the fusion proteins, an aliquot of the cells was stained for 15 min with Hoechst 33342 (1 μg/ml) at 37°C, washed once, and observed with an inverted fluorescence microscope, either at 365 nm (Hoechst) or at 488 nm (EGFP). A second aliquot was washed in serum-free medium, trypsinized, washed in sterile 0.9% NaCl, and submitted to flow cytometry analysis (Epics XL; Coulter).
Western blot analysis.
Soluble extracts were prepared from transiently transfected HeLa cells 48 h following transfection. Briefly, the cells were scraped into culture medium, washed once in ice-cold phosphate-buffered saline (PBS), and resuspended in 10 packed cell volumes of denaturing lysis buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol). The cell lysate was pushed 10 times through a 25-gauge needle, incubated for 5 min in boiling water, and then centrifuged for 15 min at 20,000 × g. Total proteins (20 μg per lane) were separated on SDS–10% polyacrylamide gels as previously described (17) and transferred on nitrocellulose membranes (Hybond ECL; Amersham Pharmacia Biotech). EGFP fused proteins were detected with a 1:1,000 dilution of a rabbit serum directed against the GFP (Clontech) and a 1:1,000 dilution of a peroxidase-conjugated anti-rabbit immunoglobulin G polyclonal antibody (Amersham Pharmacia Biotech). Detection was performed by chemiluminescence according to the manufacturer’s recommendations (ECL Western blotting detection reagents; Amersham Pharmacia Biotech).
Preparation of chromosome spreadings.
At 12 to 16 h following transfection, HeLa cells were washed once in prewarmed culture medium, incubated for an additional 16 h in the presence of 0.1 μg of colcemid (Sigma) per ml, and then stained for 15 min with Hoechst 33342 (1 μg/ml) at 37°C. Mitotic cells were then collected by gentle pipetting, washed once, counted, and resuspended in 0.075 M KCl. After a 15-min incubation at room temperature, swelling cells were cytocentrifuged on slides (500 rpm, 3 min [Cytospin 3; Shandon]). The slides were then briefly air dried and immediately observed by epifluorescence microscopy in the presence of PBS.
Fluorescence microscopy at low light level.
HeLa cells were transfected as described above, trypsinized, and grown on cover slides (diameter, 32 mm; Bachofer, Reutlingen, Germany) for an additional 12-h period. The cover slides were then mounted on a thermostat-regulated holder for direct microscopic observation and epifluorescence measurements, with an objective with a high numerical aperture, i.e., 100× (numerical aperture = 1.3) ultrafluor objective of an inverted microscope (Leica; DMIRBE). The cells were then incubated with Hoechst 33342 (0.1 μg/ml) for 15 min at 37°C, washed once, and placed in the presence of prewarmed phenol red-free medium. Fluorescent images were acquired with low excitation light levels (a neutral density filter of optical density = 1 was placed on the excitation path) for 20 s. The excitation source was a 50-W pressure mercury lamp. The detector was a digital, cooled, slow-scan charge-coupled device camera (S1-8M; Silar Ltd, Electronic Imaging, St. Petersburg, Russia). The fluorescent image was focused on the charge-coupled device sensor (1,040 by 1,160 pixels of 16 by 16 μm2) and was digitalized on a dynamic range of 4,096 grey levels. The digitalized image was further processed as described previously (6) with Khoros software (Khoral Research, Albuquerque, N.Mex.) running on a Sun workstation. The images were displayed over 256 grey levels in false color (blue for Hoechst fluorescence image and green for EGFP fluorescent image). For Hoechst fluorescence, the excitation wavelength was 365 nm, and the emission wavelengths were >400 nm. For EGFP fluorescence, the excitation wavelengths were between 430 and 480 nm and the emission wavelengths were >500 nm (dichroic filter set L4 from Leica).
RESULTS
EBV and HVP EGFP EBNA-1 are specifically associated with metaphase chromosomes.
Previous studies have suggested that EBNA associates with human and mouse chromosomes during mitosis. However, this was demonstrated mainly by anticomplement immunofluorescence with sera from EBV-seropositive patients. In addition, these experiments were performed on chemically fixed cells, an approach that may alter intracellular structures and induce a mislocation of the protein of interest. To circumvent these drawbacks, an experimental procedure that did not require any chemical fixative or immunological detection was designed. Briefly, a fragment encoding amino acids 8 to 641 of EBV EBNA-1 was cloned in frame with the 3′ end of the enhanced GFP gene. It should be underlined that deletion of amino acids 1 to 8 does not alter any known function of EBNA-1 (39). The resulting plasmid, pEGFP-EBV-EBNA1, was transfected into HeLa cells, and the cellular localization of EGFP EBV EBNA-1 was determined by fluorescence microscopy on living cells 24 to 36 h following the transfection. As confirmed by Hoechst counterstaining, EGFP EBV EBNA-1 was strictly localized to the nucleus and exhibited a rather homogeneous staining, compatible with the localization of the native EBNA-1 protein. Due to its small size and lack of nuclear localization signal, EGFP alone diffused freely, both in the cytoplasm and in the nucleus (5) (Fig. 1). To further investigate EGFP EBV EBNA-1 association with metaphase chromosomes, transiently transfected HeLa cells were grown for an additional 16 h in the presence of colcemid before staining with Hoechst 33342. Mitotic cells were then selectively collected by gentle pipetting, and their chromosomes were subsequently spread on slides and observed by epifluorescence microscopy in the presence of PBS. As shown in Fig. 2, the EGFP was localized in the nucleoplasm and was sometimes excluded from the chromosomes. In most cases, the protein quickly diffused in the mounting medium, suggesting that the procedure employed altered the plasma membrane. In contrast, EGFP EBV EBNA-1 was closely associated with metaphase chromosomes in this assay. It was thus confirmed that EBV EBNA-1 specifically binds to mitotic chromatin in nonfixed human cells.
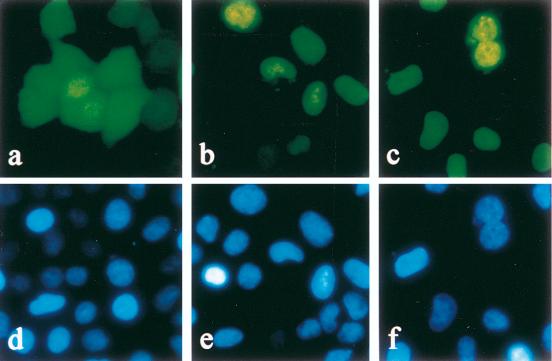
FIG. 1.
Localization of EGFP (a), EGFP EBV EBNA-1 (b), and EGFP HVP EBNA-1 (c) in HeLa cells. HeLa cells were transfected with plasmids encoding a variant of the GFP (EGFP) or encoding EBNA-1 from EBV (EGFP EBV EBNA-1) or from HVP (EGFP HVP EBNA-1) fused to EGFP. Following transfections, cells were grown for 16 h and stained with Hoechst 33342. Cells were observed by fluorescence microscopy in the presence of phenol red-free culture medium either at 488 nm (EGFP) (a to c) or at 365 nm (Hoechst) (d to f).
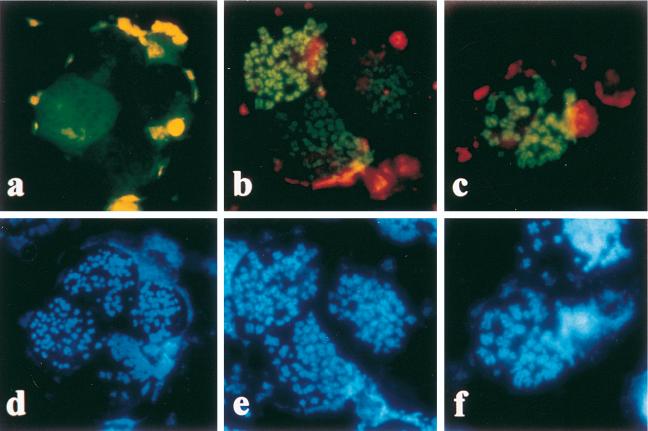
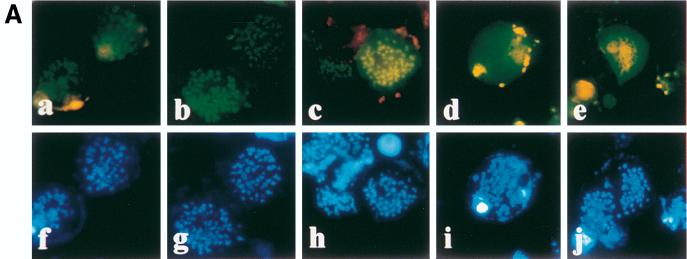
FIG. 2.
EBV and HVP EBNA-1 bind to human metaphase chromosomes. HeLa cells transfected with plasmids encoding EGFP (a), EGFP EBV EBNA-1 (b), or EGFP HVP EBNA-1 (c) were treated with colcemid, stained with Hoechst 33342, swollen in hypotonic buffer, centrifuged onto microscope slides, and observed in the presence of PBS either at 488 nm (EGFP) (a to c) or at 365 nm (Hoechst) (d to f). Whereas EGFP was localized in the nucleoplasm with frequent chromosome exclusion, EGFP EBV EBNA-1 and EGFP HVP EBNA-1 specifically bound to metaphase chromosomes.
If EBV EBNA-1 binding to metaphase chromosomes is important for its function, it might be expected that this activity is also shared by close homologs, such as the recently cloned and characterized HVP EBNA-1 (40), from HVP, a simian virus related to the EBV. To verify this, a PCR product encoding amino acids 9 to 476 of HVP EBNA-1 was introduced into pEGFP-CI. The resulting plasmid (pEGFP-HVP-EBNA1) was transfected in HeLa cells, and the localization of the fusion protein was determined as described above. In these assays, EGFP HVP EBNA-1 was localized in the nucleus but exhibited a less homogeneous staining than that observed for EGFP EBV EBNA-1 (Fig. 1). Depending on the cells, the protein was either concentrated in or excluded from Hoechst-negative regions that were found to be nucleoli (data not shown). Following chromosome spreading, EGFP HVP EBNA-1 was found exclusively in association with human chromosomes (Fig. 2). These results clearly demonstrate that binding to metaphase chromosomes is a common property of EBV EBNA-1 and its homolog in HVP. Consequently, EBNA-1 binding to mitotic chromatin is likely to be important for its function. Similar results were obtained following transfection of EGFP EBV EBNA-1- or EGFP HVP EBNA-1-encoding plasmids in mouse BALB/C 3T3 fibroblasts (data not shown), suggesting that the EBNA-1 target is also conserved between human and mouse cells.
Mapping the EBNA-1 chromosome binding site following chromosome spreading.
Several deletion mutants of EBV EBNA-1 fused to the EGFP were constructed and tested for their ability to associate with mitotic chromatin in HeLa cells (Fig. 3 and Table 1). All fusion proteins localized either in the nucleus or in both the nucleus and the cytoplasm. Thus, lack of binding could not be attributed to a specific retention of the proteins in the cytoplasm. Prior to chromosome spreading, transfection efficiencies were measured both by flow cytometry and by conventional fluorescence microscopy and were found to range reproductively from 20 to 50%, depending on the plasmid. A Western blot analysis was performed with a serum directed against EGFP. In this assay, the relative mobility of the fusion proteins following SDS-polyacrylamide gel electrophoresis was compatible with the expected mobility of the full-length proteins, although some minor breakdown products could also be observed in some cases (Fig. 4). Following transfection and treatment with colcemid, metaphase chromosomes were spread on slides, briefly air dried, and mounted in PBS as described above. The binding activity was assessed on an average of 25,000 cells per slide, which corresponded to between 5,000 and 12,000 transfected cells. All assays were carried out at least three and up to six times for each mutant.
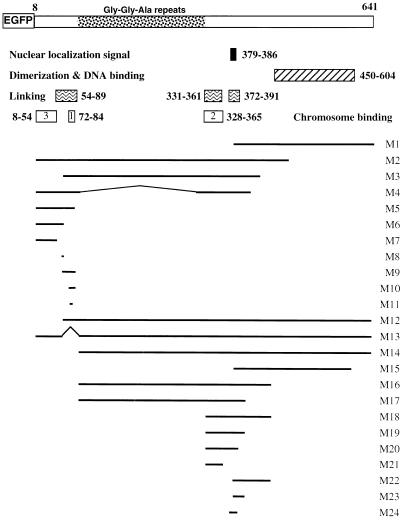
FIG. 3.
Functional map of EBV EBNA-1 and structure of EBNA-1 derivatives fused to EGFP. Several functional domains of EBV EBNA-1 are indicated, including the nuclear localization signal, dimerization and DNA binding domains (2), DNA linking regions (23), and the chromosome binding sites described herein. All EBNA-1 derivatives were fused to EGFP and tested for their ability to associate with metaphase chromosomes. Their precise sequences as well as their relative chromosome binding activities are indicated in Table 1.
TABLE 1.
Chromosome binding activity of EBNA-1 derivatives following chromosome spreading and in living cells
| Mutant | Amino acids | Binding to chromosome spreadingsa | Binding in mitotic living cellsb | % Positive mitotic living cellsc |
|---|---|---|---|---|
| EBNA-1 | 8–641 | + | ++ | 100 |
| M1 | 378–641 | − | − | 0 |
| M2 | 8–487 | + | ++ | 100 |
| M3 | 57–432 | + | ++ | 100 |
| M4 | 8–410Δ95–314 | + | ++ | 100 |
| M5 | 8–89 | + | + | 100 |
| M6 | 8–67 | − | ++ | 100 |
| M7 | 8–54 | − | + | <10 |
| M8 | 57–67 | − | − | 0 |
| M9 | 56–89 | + | + | 100 |
| M10 | 70–89 | + | ++ | 100 |
| M11 | 72–84 | + | + | 100 |
| M12 | 57–641 | + | ++ | 100 |
| M13 | 8–641Δ68–90 | − | ++ | 100 |
| M14 | 91–641 | − | + | <10 |
| M15 | 378–606 | − | − | 0 |
| M16 | 91–456 | − | + | <10 |
| M17 | 91–410 | − | + | <10 |
| M18 | 328–456 | − | ++ | 100 |
| M19 | 328–410 | − | ++ | 100 |
| M20 | 328–393 | − | ++ | 100 |
| M21 | 328–365 | − | ++ | 100 |
| M22 | 378–456 | − | − | 0 |
| M23 | 378–410 | − | − | 0 |
| M24 | 370–393 | − | − | 0 |
Binding was observed by fluorescence microscopy in the presence of PBS following centrifugation of transfected mitotic cells onto microscope slides.
Transfected HeLa cells were grown on coverslips and observed by fluorescence microscopy at a low light level. When binding was observed, the protein was localized mainly on the chromosomes (++) or localized both on the chromosomes and in the nucleoplasm (+).
For some mutants, binding to metaphase chromosomes could be observed in only a fraction of transfected mitotic cells.
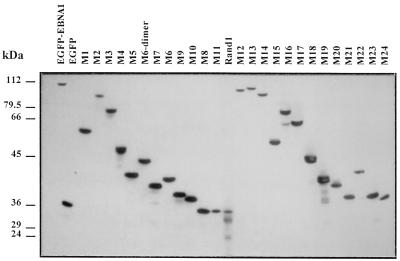
FIG. 4.
Immunoblot analysis of various EBNA-1 mutants fused to EGFP. Expression of EBNA-1 derivatives fused to EGFP was analyzed by Western blotting in transiently transfected HeLa cells. In this experiment, HeLa cells were 24 to 28% transfected, as shown by fluorescence-activated cell sorter analysis. Total proteins were separated by SDS-polyacrylamide gel electrophoresis (20 μg per lane), transferred on nitrocellulose, and incubated with a rabbit serum directed against GFP. Detection was performed by chemiluminescence. The relative mobility of the fusion proteins and the absence of free GFP were compatible with the absence of major proteolytic degradation, except for mutants M19 and Rand1, which showed moderate degradation. Mutant Rand2 (data not shown) and mutant M11, which contained the same amino acids as Rand1 in a different order, did not show any significant proteolysis. Note that the average expression level of the various EGFP EBNA-1 derivatives is unrelated to their ability to associate with metaphase chromosomes.
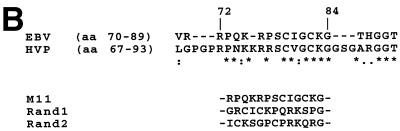
The results are summarized in Table 1. The N-terminally truncated mutant, M1, did not exhibit any binding activity, indicating that the DNA binding-dimerization domain was not involved in this interaction. In contrast, mutant M2, which comprised amino acids 8 to 487, bound mitotic chromatin with the same efficiency as the full-length EBNA-1. Further deletions delineated a chromosome binding site between amino acids 70 and 89 (M10) (Fig. 5A). Since a deletion spanning amino acids 68 to 90 (M13) totally abrogated EGFP EBV EBNA-1 binding activity in these assays, it was initially concluded that EBV EBNA-1 contains a unique chromosome binding site. In addition, as HVP EBNA-1 also binds to metaphase chromosomes, it was assumed that the region in HVP homologous to amino acids 70 to 89 of EBV EBNA-1 would also be responsible for chromosome binding. A construct encoding EGFP fused to amino acids 67 to 93 of HVP (EGFP-HVP.67-93) was therefore transfected in HeLa cells, and the fusion protein was tested for its ability to bind mitotic chromatin. As shown in Fig. 5A, EGFP-HVP.67-93 was able to interact with human metaphase chromosomes. This raised the question of whether a conserved set of amino acids was responsible for EBV and HVP EBNA-1 binding activity. A search for sequence homologies revealed a short stretch of highly conserved amino acids, encompassing amino acids 72 to 84 of EBV EBNA-1 (M11) (Fig. 5B). When fused to EGFP and expressed in HeLa cells, this 13-amino-acid-long region was sufficient to associate with cellular chromosomes. The binding was specific, since two mutants containing a randomized combination of the same amino acids (Rand1 and Rand2) did not show any binding activity (Fig. 5). Whereas Rand1 partial proteolysis may account for lack of binding, no proteolysis was observed for Rand2 in HeLa cells (Fig. 4 and data not shown). Moreover, a frameshift mutation, which preserved only amino acids 72 to 78 and amino acid 83, totally abrogated the binding (data not shown). In this assay, we could thus identify a unique chromosome binding site localized between amino acids 72 and 84 of EBV EBNA-1 that specifically associated with metaphase chromosomes.
FIG. 5.
Mapping of CBS-1 following metaphase spreadings. (A) Various derivatives of EBV and HVP EBNA-1 were tested for their ability to interact with metaphase chromosomes following transfection and chromosome spreading on slides. Mutants M10 (a) and M11 (c) associated with metaphase chromosomes, whereas Rand1 (d) and Rand2 (e), two mutants containing random combinations of M11 amino acids, did not. EGFP-HVP.67-97 (b) encoded the HVP region homologous to amino acids 70 to 89 of EBV EBNA-1. The same cells were observed at 365 nm (Hoechst) (f to j). (B) Amino acids (aa) 70 to 89 of EBV EBNA-1 and amino acids 67 to 93 of HVP EBNA-1 contain a short stretch of highly homologous amino acids (amino acids 72 to 84). Three mutants containing amino acids 72 to 84 (M11) or random combinations of these amino acids (Rand1 and Rand2) were constructed and tested for their ability to interact with metaphase chromosomes.
Other domains contribute to EBNA-1 binding in living cells.
The initial identification of a unique N-terminal chromosome binding site between amino acids 72 and 84 was based on an experimental approach that may be detrimental to the detection of weak binding activities, i.e., the analysis of chromosomes spread on a slide in the presence of PBS. In particular, as already discussed, EGFP alone or EBNA-1 derivatives that did not bind to chromosomes quickly diffused in the mounting medium. To gain further insight into the modalities of EBV EBNA-1 interactions with metaphase chromosomes, the localization of EBV EBNA-1 and truncated mutants was investigated in living mitotic cells. For this purpose, HeLa cells were transfected as described above, grown on cover slides, and observed in phenol red-free culture medium by fluorescence microscopy at a low light level. For each mutant, the analysis was performed on at least 30 transfected mitotic cells, and the assays were repeated at least three times for each construct. In some cases, binding to chromosomes could be detected in only a fraction of transfected mitotic cells, as indicated in Table 1. In any cases, we systematically verified that the binding occurred in cells expressing both low and high levels of EGFP and thus did not depend on the absolute amount of the fusion protein in a given cell. In addition, the immunoblot analysis demonstrated that the binding efficiency was not related to the average expression level of the fusion protein in transiently transfected HeLa cells.
As expected, EBV EBNA-1 and HVP EBNA-1 specifically bound to metaphase chromosomes in living cells (Fig. 6). M10 and M11 also bound to chromosomes, although binding appeared to be weaker with the latter mutant. In contrast, no binding could be observed with Rand1 and Rand2 (data not shown). Surprisingly, the CBS-1 deletion mutant (M13) also bound to metaphase chromosomes in this assay, suggesting that other domains may contribute to EBV EBNA-1 binding in vivo.
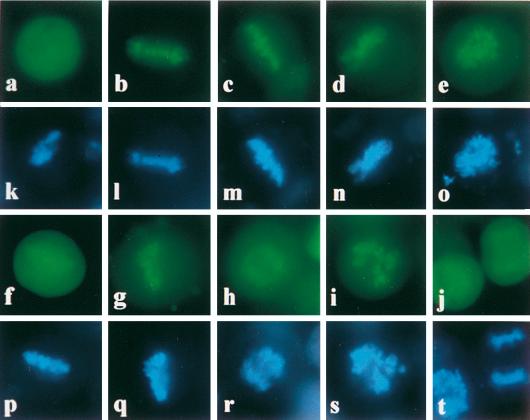
FIG. 6.
Mapping EBNA-1 chromosome binding sites in living cells. HeLa cells were transfected with plasmids encoding EGFP or various EBNA-1 derivatives fused to the EGFP protein: EGFP (a), EBV EBNA-1 (b), HVP EBNA-1 (c), M13 (d), M21 (e), M9 (f), M10 (g), M11 (h), M6 (i), and M7 (j). Cells were grown on cover slides, stained with Hoechst 33342, and observed in phenol red-free medium by low-light fluorescence microscopy either at 430 to 480 nm (EGFP) (a to j) or at 365 nm (Hoechst) (k to t).
The results indicated that a second chromosome binding site (CBS-2) could be localized between amino acids 328 and 365 (M21). In addition, whereas M18 and M19 binding was detected in 100% of transfected mitotic cells analyzed, M16 and M17 binding was weaker and could be observed in less than 10% of the cells. This suggested that the Gly-Gly-Ala repeats might exert a negative effect on CBS-2 activity, at least in the context of the fusion protein.
The region encompassing amino acids 8 to 67 (M6) was also shown to contain a third chromosome binding site (CBS-3). The binding was significantly increased for M6 dimer, a mutant which encoded EGFP fused to two consecutive CBS-3 domains (data not shown). Deletion of amino acids 55 to 67 dramatically decreased but did not abolish CBS-3 activity (M7). Given that amino acids 57 to 67 (M8) did not bind to chromosomes, CBS-3 probably lies between amino acids 8 and 54 but requires amino acids 55 to 67 for maximal activity. CBS-2- and CBS-3-containing mutants bound to chromosomes in a specific manner, since their association with chromosomes was observed in transfected cells expressing low to high levels of the fusion proteins.
Finally, we noticed that M9 binding activity was much weaker than that of M10, although it could still be observed in 100% of transfected mitotic cells. This suggested that the region encompassing amino acids 56 to 69 negatively modulated CBS-1 activity, whereas it positively contributed to CBS-2 binding in vivo. However, since the relative binding activity of EBNA-1 derivatives was analyzed by using a fusion protein, it is not certain that these regions regulate the binding activity of the native EBNA-1.
CBS-1 does not have any significant amino acid homology with CBS-2 and CBS-3. In contrast, CBS-2 and CBS-3 have striking homologies, with amino acids 8 to 54 having more than 53% sequence identity with amino acids 325 to 367. Similarly, the corresponding regions in HVP EBNA-1 (amino acids 7 to 48 and 144 to 186) have 50% sequence identity. Both CBS-2 and CBS-3 mapped to a basic region that is rich in arginine and glycine residues. Taken together, these data suggest that CBS-2 and CBS-3 binding activity is related to the presence of a common arginine-glycine (RGG)-rich domain, whose mode of interaction with mitotic chromatin may differ from that of CBS-1.
DISCUSSION
In the present work, we took advantage of the fluorescence properties of a variant of the GFP to investigate the chromosome binding activity of EBV EBNA-1, HVP EBNA-1, and various truncated mutants of EBV EBNA-1. Using two distinct experimental procedures, we have demonstrated that both EBV and HVP EBNA-1 associate with human mitotic chromosomes. Three independent domains (CBS-1, -2, and -3) that mediate binding to chromosomes have been identified. CBS-1 activity (amino acids 72 to 84) was detected both in living cells and following chromosome spreading on slides. CBS-1 was also shown to associate with chromosomes in a sequence-specific manner. To our knowledge, CBS-1 is the shortest peptide known to be able to mediate a specific interaction with cellular chromosomes during mitosis. In addition to CBS-1, two other binding sites, namely, CBS-2 and CBS-3, contributed to EBNA-1 interaction with mitotic chromosomes in vivo. However, their activity was not detected when chromosomes were spread on slides and observed in the presence of PBS. This apparent discrepancy probably reflects a weaker affinity of CBS-2 and CBS-3 for metaphase chromosomes, compared to that of CBS-1. In line with this, transient CBS-2 and CBS-3 binding was observed when the mounting medium was complemented by up to 70% glycerol (data not shown). Glycerol is likely to prevent or limit diffusion in the mounting medium, and it has also been shown to stabilize protein structure (3) and to reinforce weak protein-protein interactions (14).
CBS-2 mapped to the central part of EBNA-1, between amino acids 328 and 365. CBS-3 was localized between amino acids 8 and 54 but required amino acids 55 to 67 for maximal activity. CBS-1, -2, and -3 are all rich in basic residues. Notably, CBS-2 and CBS-3 contain RGG repeats and are highly homologous. This is of particular interest, since similar RGG motifs were recently shown to be required for EBNA-2 interaction with histone H1 (36). However, although EBNA-2 has been shown to interact with cellular chromatin, it has not previously been found in association with mitotic chromosomes (28).
Several other viral proteins, including papillomavirus E1 and E2 proteins and simian virus 40 large T antigen, are known to interact with histones and/or with cellular chromosomes (7, 18, 33, 35). These proteins share several functional properties, including their specific requirement for activating viral replication origins. One tempting hypothesis is that they may facilitate the initiation of viral DNA replication by displacing histones from the replication origin, as suggested by several studies (19, 35). Alternatively, their simultaneous interaction with cellular chromosomes and viral genomes during mitosis would provide an efficient way to control the segregation and/or retention of viral episomes, as recently shown for the bovine papillomavirus (18). Several lines of evidence suggest that this may be the case for EBNA-1. Indeed it is known that viral episomes are noncovalently associated with chromosomes during the metaphase (11), and circularized yeast artificial chromosomes containing OriP have been shown to interact with chromosomes in human cells expressing EBNA-1 (32). In addition, given that EBNA-1 chromosome binding sites are distinct from the DNA binding-dimerization domain, EBNA-1 could mediate viral episome anchorage on the cellular chromosomes during mitosis, thus ensuring proper transmission to daughter cells. In this model, deleting one or several chromosome binding sites may severely impair the partition of viral episomes and consequently abrogate their long-term persistence. In agreement with this model, Yates and Camiolo showed that a large deletion, spanning amino acids 72 to 338 and including CBS-1 and most of CBS-2, resulted in an EBNA-1 protein that was still able to transiently activate replication but was unable to ensure long-term maintenance of OriP-containing plasmids in human cells (39). However, it is possible that this deletion also affected other functional domains of EBNA-1. Indeed, CBS-1 and -2 map within two regions that are involved in DNA linking, a function of EBNA-1 that is thought to be essential for OriP activation (23). Whether DNA linking and chromosome binding involve similar functional domains on EBNA-1 has still to be determined, although our data indicate that the DNA-linking region that has been localized between amino acids 372 and 391 is not involved in chromosome binding. It has generally been assumed that DNA linking is required for early activation of genome replication and would thus be mainly required during the S phase of the cell cycle. In contrast, our work and that of others have proved that EBNA-1 interacts with cellular chromatin during the M phase. This would suggest that DNA linking and chromosome binding are not mutually competitive. A more complete study of EBNA-1 interaction with cellular chromatin and regulation during the cell cycle is required in order to validate this model.
ACKNOWLEDGMENTS
We appreciate the technical assistance provided by Corinne Dutreuil for sequencing and Bakoli Rajoely for flow cytometry analysis. We thank Ann Beaumont for correcting the manuscript.
This work was supported by the D.R.E.D. (UPRES EA 2391) and a grant from the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M A, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler S L, Falke J J. Effects of protein stabilizing agents on thermal backbone motions: a disulfide trapping study. Biochemistry. 1996;35:10595–10600. doi: 10.1021/bi961107v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee S, Stochaj U. Monitoring nuclear transport in HeLa cells using the green fluorescent protein. BioTechniques. 1996;21:62–63. doi: 10.2144/96211bm13. [DOI] [PubMed] [Google Scholar]
- 6.Coppey-Moisan M, Delic J, Magdelenat H, Coppey J. Principle of digital imaging microscopy. Methods Mol Biol. 1994;33:359–393. doi: 10.1385/0-89603-280-9:359. [DOI] [PubMed] [Google Scholar]
- 7.D’Alisa R, Gershey E. Simian virus 40 T antigen binds to host cell chromosomes. Nature. 1978;274:164–166. doi: 10.1038/274164a0. [DOI] [PubMed] [Google Scholar]
- 8.Delecluse H J, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm G W. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J Virol. 1993;67:1292–1299. doi: 10.1128/jvi.67.3.1292-1299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grogan E A, Summers W P, Dowling S, Shedd D, Gradoville L, Miller G. Two Epstein-Barr viral neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampar B, Tanaka A, Nonoyama M, Derge J. Replication of the resident repressed Epstein-Barr virus genomes during the early S-phase (S-1 period) of non-producer Raji cells. Proc Natl Acad Sci USA. 1974;71:631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt’s lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hearing J C, Nicolas J C, Levine A J. Identification of Epstein-Barr virus sequences that encode a nuclear antigen expressed in latently infected lymphocytes. Proc Natl Acad Sci USA. 1984;81:4374–4377. doi: 10.1073/pnas.81.14.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurley E A, Agger S, McNeil J A, Lawrence J B, Calendar A, Lenoir G, Thorley-Lawson D A. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J Virol. 1991;65:1245–1254. doi: 10.1128/jvi.65.3.1245-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelesarov I, Bosshard H. Thermodynamics of ferredoxin binding to ferredoxin: NADP+ reductase and the role of water at the complex interface. Biochemistry. 1994;33:13321–13328. doi: 10.1021/bi00249a019. [DOI] [PubMed] [Google Scholar]
- 15.Kashka-Dierich C, Falk L, Bjursell G, Adams A, Lindahl T. Human lymphoid cell lines derived from individuals without lymphoproliferative disease contain the same latent forms of Epstein-Barr virus DNA as those found in tumor cells. Int J Cancer. 1977;20:173–179. doi: 10.1002/ijc.2910200203. [DOI] [PubMed] [Google Scholar]
- 16.Krysan P, Calos M. Epstein-Barr virus-based vectors that replicate in rodent cells. Gene. 1993;136:137–143. doi: 10.1016/0378-1119(93)90457-e. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lehman C W, Botchan M R. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindahl T, Adams A, Bjursell G, Bornkamm G W, Kaschaka-Dierich C, Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- 21.Luka J, Siegert W, Klein G. Solubilization of the Epstein-Barr virus-determined nuclear antigen and its characterization as a DNA-binding protein. J Virol. 1977;22:1–8. doi: 10.1128/jvi.22.1.1-8.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mecsas J, Sugden B. Replication of plasmids derived from bovine papilloma virus type 1 and Epstein-Barr virus in cells in culture. Annu Rev Cell Biol. 1987;3:87–108. doi: 10.1146/annurev.cb.03.110187.000511. [DOI] [PubMed] [Google Scholar]
- 25.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonoyama M, Pagano J S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972;238:169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- 27.Ohno S, Wiener F, Klein G. Histochemical studies on the EBV-determined nuclear antigen (EBNA) Biomedicine. 1977;26:268–275. [PubMed] [Google Scholar]
- 28.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 29.Reedman B, Klein G. Cellular localization of an Epstein-Barr virus-associated complement-fixing antigen in producer and nonproducer lymphoblastoid cell lines. Int J Cancer. 1973;11:499–520. doi: 10.1002/ijc.2910110302. [DOI] [PubMed] [Google Scholar]
- 30.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skiadopoulos M H, McBride A A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swindle C S, Engler J A. Association of the human papillomavirus type 11 E1 protein with histone H1. J Virol. 1998;72:1994–2001. doi: 10.1128/jvi.72.3.1994-2001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong X, Yalamanchili R, Harada S, Kieff E. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68:6188–6197. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yates J, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 39.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- 40.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 41.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]