Abstract
Soybean stem canker (SSC) caused by the fungal pathogen Diaporthe caulivora is an important disease affecting soybean production worldwide. However, limited information related to the molecular mechanisms underlying soybean resistance to Diaporthe species is available. In the present work, we analyzed the defense responses to D. caulivora in the soybean genotypes Williams and Génesis 5601. The results showed that compared to Williams, Génesis 5601 is more resistant to fungal infection evidenced by significantly smaller lesion length, reduced disease severity and pathogen biomass. Transcriptional profiling was performed in untreated plants and in D. caulivora-inoculated and control-treated tissues at 8 and 48 h post inoculation (hpi). In total, 2.322 and 1.855 genes were differentially expressed in Génesis 5601 and Williams, respectively. Interestingly, Génesis 5601 exhibited a significantly higher number of upregulated genes compared to Williams at 8 hpi, 1.028 versus 434 genes. Resistance to D. caulivora was associated with defense activation through transcriptional reprogramming mediating perception of the pathogen by receptors, biosynthesis of phenylpropanoids, hormone signaling, small heat shock proteins and pathogenesis related (PR) genes. These findings provide novel insights into soybean defense mechanisms leading to host resistance against D. caulivora, and generate a foundation for the development of resistant SSC varieties within soybean breeding programs.
Subject terms: Molecular biology, Plant sciences
Introduction
Soybean (Glycine max L.) is a major global crop affected by biotic stress caused by microbial pathogens, nematodes and insects, as well as abiotic stress such as drought, nutrient deficiency, salt and cold1. Soybean stem canker (SSC) caused by fungal Diaporthe species is an important soybean disease worldwide. D. aspalathi (E. Jansen, Castl. & Crous), D. caulivora (Athow & Caldwell) and D. longicolla (Hobbs) are the principal agents causing SSC in different countries2–5. Disease symptoms appear on the stem as 1–2 mm spots that expand as elongated brown lesions associated to withered brown leaves3. SSC control is based on integrating management practices such as crop rotation and fungicides application. However, the most effective way to control SSC is to develop and use resistant cultivars. Five Rdm loci confer resistance to D. aspalathi3,6, although these resistance loci are not effective against D. caulivora3. Recently, an Rdc1 locus of G. max was identified as a resistance source for D. caulivora7, although the molecular identity of Rdc1 is currently unknown.
Plants perceive pathogens and trigger cellular and molecular modifications associated with defense responses such as signaling, transcriptional activation, synthesis of defense molecules, and their transport to specific sites in the plant8. Recognition occurs at the plasma membrane by pattern-recognition receptors (PRRs), and in the cytoplasm by nucleotide-binding domain leucine-rich repeat containing receptors (NLRs)9,10. PRRs perceive conserved microbe- or damage-associated molecular patterns (MAMPs or DAMPs) at the surface of the plant cells to activate pattern-triggered immunity (PTI). PRRs include receptor-like kinases (RLKs) or receptor-like proteins (RLP) with different extracellular domains. NLRs perceive pathogen effectors delivered inside the plant cell leading to effector-triggered immunity (ETI). Both PTI and ETI activate overlapping events such as mitogen-activated protein kinases (MAPKs) cascades, Ca2+ flux, hormonal signaling, reactive oxygen species (ROS) burst, callose deposition, and transcriptional reprogramming11,12.
During soybean-D. caulivora interaction, plant cells activate the expression of genes encoding pathogenesis-related proteins (PR-1, PR-2, PR-3, PR-4, PR-10), and enzymes involved in phenylpropanoid and oxylipin synthesis such as phenylalanine-ammonia lyase (PAL), chalcone synthase (CHS), and lipoxygenase (LOX)2. Most of these defense genes were also induced in soybean tissues infected with D. aspalathi13. The phenylpropanoid and oxylipin pathways participate in plant defenses against pathogens by producing important compounds with antimicrobial activities, and contribute to reinforcement of the plant cell walls and defense signaling14,15. Recently, we sequenced the genome of D. caulivora (isolate D57) and performed transcriptional profiling during stem colonization to reveal the molecular basis of fungal pathogenesis16. In this analysis, we identified a high number of fungal upregulated genes that encode enzymes involved in plant cell wall degradation and modification such as polygalacturonases, endoglucanases, exoglucanases, pectate lyases, pectin lyases, and glycoside hydrolases, among others16. D. caulivora infection strategy also relies on detoxification of plant compounds, transporter activities, and toxin production that could kill plant cells, enabling nutrient uptake and mycelium growth16. Moreover, D. caulivora genes encoding secreted effector candidates are induced during soybean infection, suggesting that plant defense evasion contributes to the plant colonization process16. However, the molecular mechanisms employed by plant cells to recognize D. caulivora and activate an effective defense response leading to resistance are still unknown.
Transcriptomic studies have allowed to understand complex gene regulatory networks operating in soybean plants infected with different pathogens, including Phytophthora sojae (Kaussmann & Gerdemann)17, Phakopsora pachyrhizi (Sydow & P. Sydow)18,19, Fusarium oxysporum (Schltdl.)20, and soybean mosaic virus (SMV)21. To unravel the mechanisms involved in defense activation and resistance against D. caulivora, we performed RNAseq profiling in two contrasting soybean genotypes, the susceptible cultivar Williams and the resistant cultivar Génesis 5601. The results revealed a complex and differential gene expression network during the activation of plant defense responses between cultivars upon pathogen infection.
Results
D. caulivora infection of soybean genotypes
Williams and Génesis 5601 cultivars were inoculated with D. caulivora and stem canker progress was monitored during 14 days post inoculation (dpi). The first symptoms of SSC were observed at 3 dpi, and lesions were more evident at 5 dpi showing typical stem browning (Supplementary Fig. S1). Lesions expanded in both directions of the stem and disease symptoms were clearly visible in Williams, while Génesis 5601 exhibited smaller stem lesions (Fig. 1a, Supplementary Fig. S1). Moreover, withered leaves above de canker lesion were only observed in Williams (Fig. 1a). From 5 to 14 dpi, lesion length was significantly higher in Williams compared to Génesis 5601, varying between 36 and 50% (Fig. 1a, b). Disease severity index and area under the disease progress curve (AUDPC) were evaluated in soybean stems until 14 dpi. According to a disease severity scale2, D. caulivora infection resulted in more symptom development in Williams compared to Génesis 5601 throughout time (Fig. 1c, d).
Figure 1.
Soybean stem canker disease progress after D. caulivora inoculation. (a) Symptoms in susceptible and resistant soybean plants following D. caulivora inoculation at 7 days post-inoculation (dpi), (b) Lesion length in susceptible and resistant soybeans at 3, 5, 7, 11 and 14 dpi, (c) Disease severity index in susceptible and resistant soybeans at 3, 5, 7, 11 and 14 dpi, (d) AUDPC in susceptible and resistant soybeans at 3, 5, 7, 11 and 14 dpi. D. caulivora biomass in susceptible and resistant soybeans at 8, 24, 48, 72 and 96 hpi. Asterisk indicates a significant difference between the soybean genotypes at p-value < 0.05 (one-way ANOVA).
We previously showed that D. caulivora biomass starts to increase at 8 hpi in Williams stems, and at 96 hpi fungal DNA became predominant in the host tissues2. Here, we measured by quantitative PCR (qPCR) D. caulivora biomass in stems of both genotypes at 8, 24, 48, 72 and 96 h post inoculation (hpi). The results show that pathogen biomass was significantly higher in Williams compared to Génesis 5601 at 24 to 96 hpi (2–4 fold) (Fig. 1e). Taken together, these results indicate that Génesis 5601 is more resistant to D. caulivora infection than Williams.
Transcriptome profiles of contrasting soybean genotypes infected with D. caulivora
To identify molecular components involved in soybean defense responses against D. caulivora, we first compared the transcriptomes of Williams and Génesis 5601 under normal conditions in untreated plants. A second comparison included Williams and Génesis 5601 tissues inoculated with D. caulivora versus control treatment. Two time points, 8 and 48 hpi were selected based on previous expression patterns of PRs, PAL, CHS and LOX in D. caulivora-infected soybean plants2. In total, 819 million reads were generated after removing adapter sequences and low-quality reads. Reads mapped uniquely to the G. max nuclear reference genome (approximately 737 million reads) were considered for further analyses (Supplementary Table S1). The principal component analysis of the different treatments showed a clear separation, and variability among biological replicates was very low as indicated (Supplementary Figure S2).
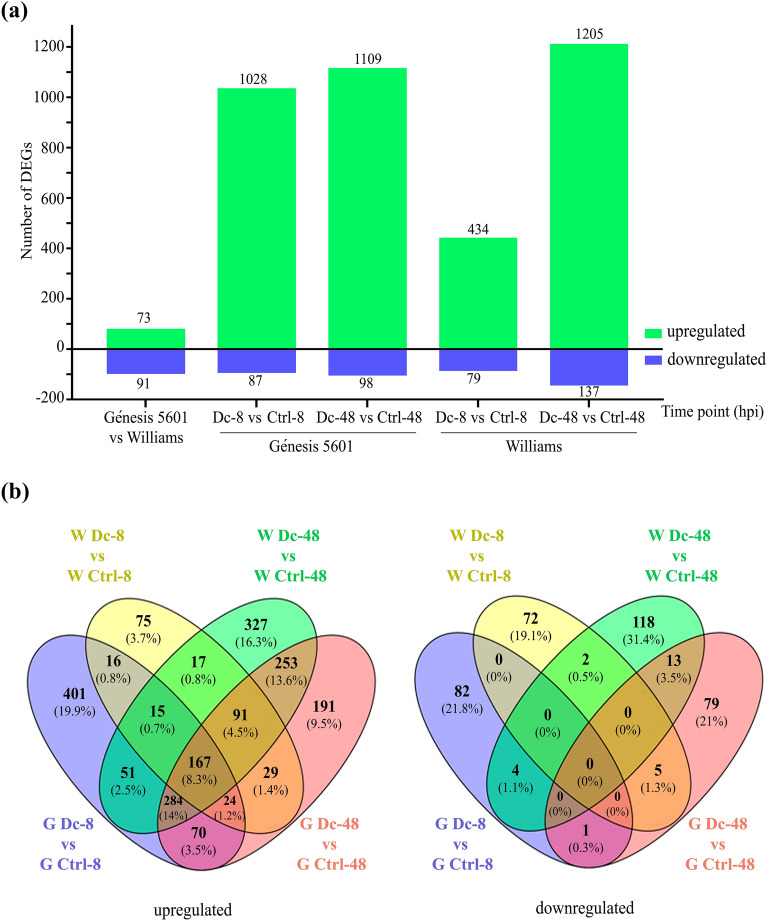
Soybean expression profile analysis identified 2.384 differentially expressed genes (DEGs) between soybean genotypes (Supplementary Table S2). A significant transcriptional shift towards upregulation was observed at 8 and 48 hpi after D. caulivora inoculation compared to control treatment in both cultivars (from now on named as Génesis-8 and -48 and Williams-8 and -48) (Fig. 2a). Untreated Génesis 5601 plants exhibited 164 DEGs (73 up- and 91 downregulated) compared to untreated Williams plants. During infection, Williams-48 has more DEGs than Williams-8, 1.342 and 513, respectively (Fig. 2a, Supplementary Table S2). However, the number of DEGs in Génesis-8 and Génesis-48 were similar, 1.115 and 1.207, respectively, indicating that Génesis-8 has significantly more DEGs than Williams-8. Among DEGs, 167 were upregulated in both genotypes at both time points, while some genes were uniquely expressed in one condition (Fig. 2b). Interestingly, 335 DEGs (32,6%) that were upregulated in Génesis-8 and not in Williams-8, were upregulated in Williams-48, indicating that these genes were later expressed in the susceptible genotype. In total, 930 DEGs were commonly upregulated in both genotypes, representing 46% of the upregulated DEGs (2.011 genes). In contrast, 22 DEGs were downregulated in both genotypes, representing 5.9% of the downregulated DEGs (376 genes). Furthermore, 75 and 327 upregulated DEGs were only found in Williams-8 and Williams-48, and 401 and 191 upregulated DEGs were only present in Génesis-8 and Génesis-48, respectively (Fig. 2b). Thus, the presence of a significantly higher number of upregulated DEGs in Génesis-8 compared to Williams-8 could be related to the molecular mechanisms involved in resistance against D. caulivora.
Figure 2.
Differentially expressed genes (DEGs) identification in susceptible (Williams) and resistant (Génesis 5601) soybean plants without treatment and after D. caulivora inoculation. (a) Number of upregulated and downregulated DEGs for each treatment in both genotypes. Log2 FC ≥ 2.0 or ≤ 2.0 and false discovery rate (FDR) ≤ 0.05 were considered for DEGs identification, (b) Venn diagram showing the number of upregulated and downregulated soybean genes at 8 and 48 h post inoculation (hpi) with D. caulivora in susceptible and resistant soybean plants. Overlap of expressed soybean genes are indicated by bold numbers. See Supplementary Table S2 for complete information.
Functional enrichment of differentially expressed genes
In order to identify biological processes (BP) and molecular functions (MF) mostly affected by D. caulivora infection, we performed Gene Ontology (GO) term enrichment analysis of the upregulated DEGs. Enriched GO were not found for untreated Williams or Génesis 5601 tissues. Most of the top 25 significantly enriched GO terms in D. caulivora-inoculated versus control soybean plants were similar at 8 and 48 hpi (Fig. 3a, Supplementary Table S2). Génesis 5601 showed a significantly higher number of genes per category respect to Williams, principally at 8 hpi. The most represented BP in both genotypes at 8 and 48 hpi were protein phosphorylation, regulation of transcription, defense response, and transmembrane transport, among others. Other top GO terms enrichment in BP at 8 hpi included ethylene-activated signaling pathway, response to oxidative stress and hydrogen peroxide, response to heat and salt stress, ABA activated signaling pathway, response to auxin and cell wall modification. Most of these enriched BP related to defense were also present at 48 hpi, and additional upregulated defense-related GO terms at 48 hpi included cellular oxidant detoxification, flavonoid biosynthetic process, and response to biotic stimulus, bacterium, chitin and fungus. In general, biotic related process, abiotic related process, hormones and secondary metabolites represented 60% of the BP terms identified in the upregulated genes. In both genotypes, MF terms at 8 and 48 were represented by ATP binding, DNA-binding transcription factor activity, heme- , protein- , DNA- and iron ion-binding, sequence-specific DNA binding, protein kinase- , oxidoreductase- , monooxygenase- , peroxidase- , glycosyltransferase- and protein serine threonine kinase-activity.
Figure 3.
Enriched gene ontology (GO) and KEGG pathways analysis of upregulated genes in soybean plants inoculated with D. caulivora. (a) Top 25 enrichment GO biological process and molecular function terms (p < 0.05) of upregulated genes in Williams (green bars) and Génesis 5601 (blue bars) at 8 and 48 h post inoculation (hpi) with D. caulivora. See Supplementary Table S2 for complete information. (b) Upregulated genes KEGG pathway analysis in Williams (green bars) and Génesis 5601 (blue bars) at 8 and 48 h post inoculation (hpi) with D. caulivora. See Supplementary Table S3 for complete information.
To study the host pathways altered during D. caulivora infection, we performed a KEGG enrichment analysis (Fig. 3b, Supplementary Table S3). The most enriched KEGG pathways for upregulated DEGs were related to phenylpropanoid, flavonoid and isoflavonoid biosynthesis, plant hormone signal transduction, mitogen-activated protein kinase (MAPK) signaling, plant-pathogen interaction, and steroid hormone biosynthesis. Both genotypes shared most of the KEGG pathways, although there were more genes within pathways associated to Génesis 5601 compared to Williams.
Differential expression of genes involved in plant defense during D. caulivora infection
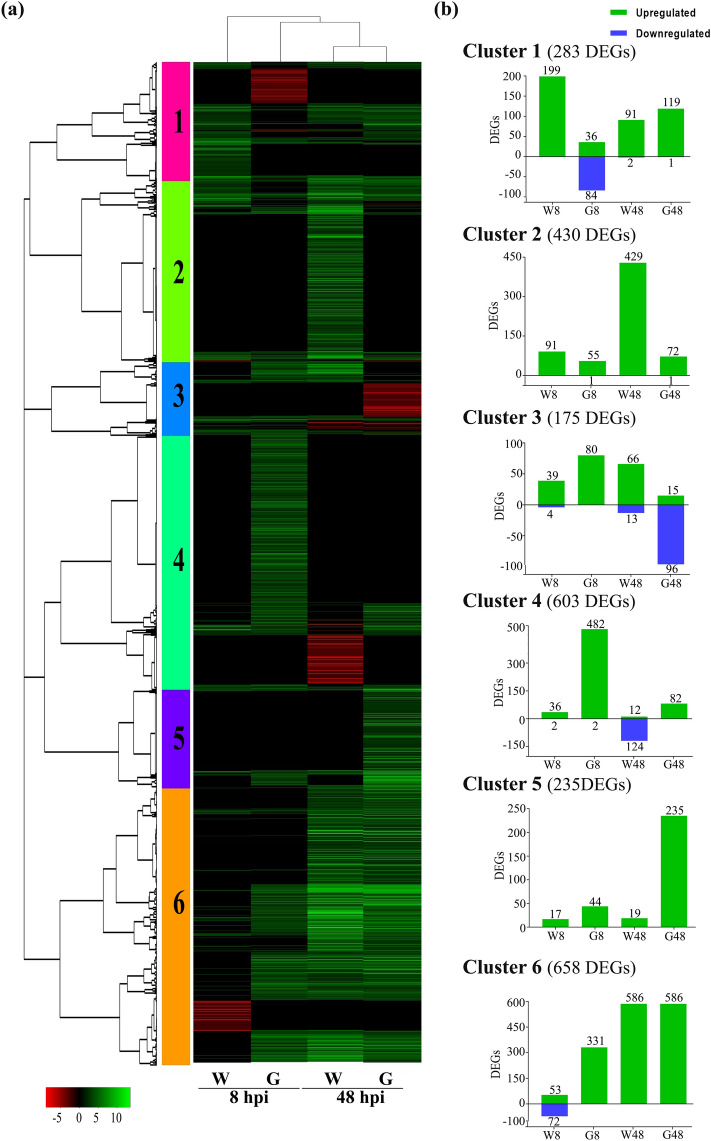
Hierarchical clustering was performed to group similar expression patterns across genotypes and treatments. This analysis grouped Williams-48 with Génesis-48, and Génesis-8 was more related to this group than to Williams-8, which is consistent with a significantly higher number of upregulated DEGs in Génesis-8 (Fig. 4, Supplementary Table S4). The analysis of total DEGs identified six clusters with different gene numbers and expression patterns (Fig. 4a, b). Clusters 4, 5, and 6 were strongly associated with increased expression of genes in Génesis-8 and Génesis-48 compared to the same time points in Williams. Génesis-8 contained 482 and 331 upregulated genes in cluster 4 and 6 respectively, while Génesis-48 comprises 235 upregulated DEGs in cluster 5. The corresponding numbers of upregulated DEGs in the Williams genotype were 36, 53 and 19 DEGS (Fig. 4b). To explore the BP associated with Génesis 5601 resistance, GO terms enrichment analysis was performed for DEGs present in the different clusters (Supplementary Table S5). Common GO terms identified in most clusters included regulation of DNA-templated transcription, protein phosphorylation, defense response, transmembrane transport, flavonoid or isoflavonoid biosynthetic process, among others. Other major GO terms associated with Génesis-8 in cluster 4, included response to hydrogen peroxide, response to oxidative stress, hydrogen peroxide catabolic process, glutathione metabolic process, response to salt stress and response to heat stress. Response to oxidative stress and cellular oxidant detoxification were also GO terms enriched in cluster 5 and 6, and the latter comprises other GO terms related to hormones. By performing a deeper inspection of genes within GO terms among genotypes, we found that the term response to heat stress in cluster 4 included DEGs that encode small heat shock proteins (sHSPs) that were only upregulated in Génesis 5601, while they did not show differential expression in Williams. In total, 17 sHSPs and one sHSP were upregulated in Génesis-8 and Génesis-48, respectively. Taken together, these results suggest that regulation of transcription, signaling, phenylpropanoid and flavonoid pathways, ROS detoxification and sHSPs play important functions in plant resistance against D. caulivora.
Figure 4.
Differentially expressed genes (DEGs) in soybean plants inoculated with D. caulivora. (a) Heat map of hierarchical clustering of all DEGs. Green represents upregulated DEGs and red downregulated DEGs. (b) Numbers of total, upregulated and downregulated DEGs for each cluster. See Supplementary Table S4 for complete information.
Induced expression of genes involved in pathogen perception, signaling and transcription during soybean infection by D. caulivora
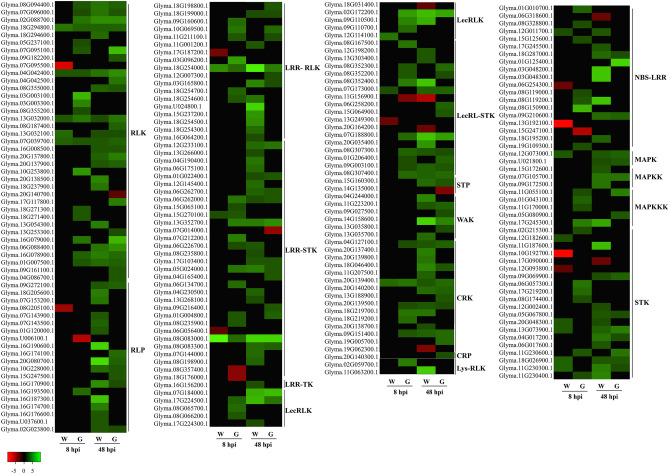
Pathogen recognition, signaling and transcriptional reprogramming are important steps in the activation of plant defense mechanism against pathogens. We identified 159 DEGs encoding PRRs, most of which were upregulated during D. caulivora infection, including leucine-rich repeat receptor-like protein kinase (LRR- RLK), RLKs, RLPs and lectin domain containing receptor kinase (LecRLKs), among others (Fig. 5, Supplementary Table S6). While only 11 receptor genes were upregulated in Williams-8, this number increased to 59 in Génesis-8, and included LRR-RLKs, RLPs, cysteine-rich receptor-like protein kinase (CRKs) and LecRLKs. At 48 hpi, 76 and 94 receptor genes were upregulated in Génesis-48 and Williams-48, respectively. Furthermore, 15 upregulated DEGs encoded NLR; seven in Génesis-8, one in Williams-8, three in Génesis-48 and eight in Williams-48. In untreated plants, a higher number of PRRs and NLR genes showed increased expression levels in Génesis 5601 compared to Williams.
Figure 5.
Heat map of differentially expressed genes (DEGs) encoding for proteins with roles in perception and signaling. Individual genes are listed and colors represent the log2 fold change value based on the comparison of the transcript levels between D. caulivora-inoculated and control treatment for both genotypes (Williams and Génesis 5601). Green represents upregulated DEGs and red downregulated DEGs. RLK receptor-like protein kinase, RLP receptor-like protein, LRR-RLK leucine-rich repeat receptor-like protein kinase, LecRLK lectin domain containing receptor kinase, WAK wall-associated receptor kinase, CRK cysteine-rich receptor-like protein kinase, CRP cysteine-rich protein, LysM-RLK LysM domain receptor-like kinase, NLR nucleotide-binding site leucine-rich repeat, MAPK mitogen-activated protein kinase, MAPKK mitogen-activated protein kinase kinase, MAPKKK mitogen-activated protein kinase kinase kinase, and STK serine/threonine-protein kinase. See Supplementary Table S6 for complete information.
Thirty DEGs encoded members of MAPK cascades, including MAPK, MAPKK and MAPKKK and serine/threonine-protein kinase (STKs) (Fig. 5, Supplementary Table S6). Four MAPKKK were only induced in Génesis-8 and different STKs were upregulated in Génesis-8 and Williams-8. Moreover, 219 DEGs encoded TFs related to plant defenses to biotic stress, including 48 WRKYs, 71 Apetala2/ethylene responsive factor (AP2/ERFs), 28 myeloblastosis-related (MYB), 17 basic helix–loop–helix (bHLH), 15 no apical meristem, ATAF1/2, and cup-shaped cotyledon (NAC), and eight Gibberellin-insensitive, repressor of GA1–3, and Scarecrow (GRAS) (Fig. 6, Supplementary Table S7). Most TFs were upregulated during D. caulivora infection and the number of upregulated TFs increased significantly at 48 hpi respect to 8 hpi in both genotypes. Génesis-8 exhibited more upregulated ERFs, MYBs and bHLHs compared to Williams-8.
Figure 6.
Heatmap of differentially expressed genes (DEGs) encoding for transcription factors. Individual genes are listed and colors represent the log2 fold change value based on the comparison of the transcript levels between D. caulivora-inoculated and control treatment for both genotypes (Williams and Génesis 5601). Green represents upregulated DEGs and red downregulated DEGs. ERF ethylene responsive transcription factor, MYB myeloblastosis-related transcription factor, bHLH basic helix–loop–helix transcription factor, NAC no apical meristem, ATAF1/2, and cup shaped cotyledon, GRAS gibberellin-insensitive, repressor of GA1-3 and Scarecrow, basic region/leucine zipper motif (bZIP). See Supplementary Table S7 for complete information.
Activation of pathogenesis-related genes and the phenylpropanoid pathway during D. caulivora infection
Pathogenesis-related proteins play important functions in plant immune responses22. During D. caulivora infection, soybean induced the expression of 139 PR genes encoding PR-1, β-1,3-glucanases (PR-2), chitinases (PR-3, PR-4, PR-8), thaumatins (PR-5), proteinase inhibitors (PR-6), endoproteinasse (PR-7), peroxidases (PR-9), PR-10 (ribonuclease-like protein), defensins (PR-12), lipid-transfer proteins (PR-14) and germin-like proteins (PR-16) (Supplementary Fig. S3, Supplementary Table S8). A higher number of genes encoding PRs were upregulated in Génesis-8 compared to Williams-8, while at 48 hpi the number of upregulated genes were similar among genotypes. Four PR-2 were only induced in Génesis-8 and not in Williams-8, while six PR-14 were induced in Génesis-8 and only one in Williams-8. Similarly, 27 PR-9 and 10 PR-10 were upregulated in Génesis-8, while this number decreased to seven and four in Williams-8.
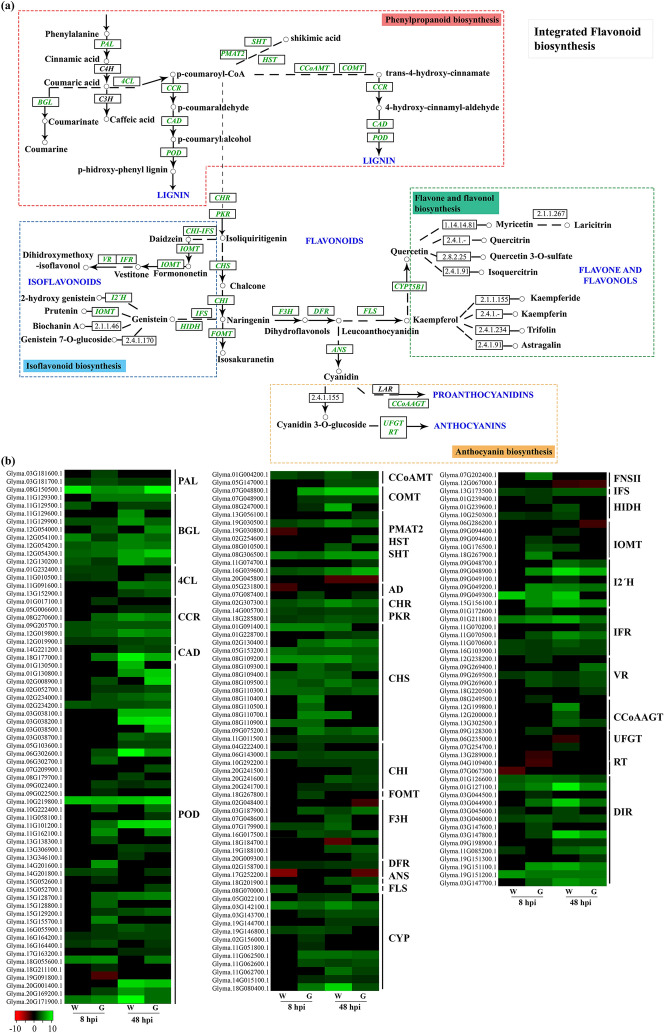
The phenylpropanoid pathway produces multiple compounds involved in defense mechanisms against biotic stress14. In total, 169 DEGs related to this pathway were identified during D. caulivora infection. A high number of genes encoding enzymes involved in flavonoids, isoflavonoids, flavonone, flavonols, flavones and anthocyanins biosynthesis were upregulated in both genotypes at 48 hpi (Fig. 7, Supplementary Table S9). The most remarkable differences between genotypes were observed in Génesis-8, which has significantly more upregulated DEGs than Williams-8 (117 versus 50). For example, while 10 cytochrome P450 (CYP) and 14 CHS encoding genes were induced in Génesis-8, this number decreased to two CYPs and eight CHSs in Williams-8. Similarly, Génesis-8 showed increased expression of genes encoding for three isoflavone 7-O-methyltransferases (IOMT) and four isoflavone 2'-hydroxylases (I2¨H), while Williams-8 showed only increased expression of one I2¨H and differential expression of IOMT genes could not be observed. Other genes encoding dirigent proteins, involved in the synthesis of lignans and lignin, were also upregulated during D. caulivora infection in both genotypes.
Figure 7.
Activation of the phenylpropanoid and flavonoids pathways in response to D. caulivora. (a) Integrated and simplified scheme of phenylpropanoid, flavonoid, isoflavonoid, anthocyanin, flavone and flavonol biosynthesis KEGG pathways. Upregulated genes encoding enzymes of this pathway are highlighted in green. (b) Heatmap of DEGs encoding genes of the phenylpropanoid and flavonoids biosynthetic pathway. Individual genes are listed and colors represent the log2 fold change value based on the comparison of the transcript levels between D. caulivora-inoculated and control treatment for both genotypes (Williams and Génesis 5601). Green represents upregulated DEGs and red downregulated DEGs. PAL phenylalanine ammonia-lyase, BGL beta-glucosidase, 4CL 4-coumarate–CoA ligase, CCR cinnamoyl-CoA reductase, CAD cinnamyl alcohol dehydrogenase, POD peroxidase, CCoAMT caffeoyl-CoA O-methyltransferase, COMT caffeic acid 3-O-methyltransferase, PMAT2 phenolic glucoside malonyltransferase 1-like, HST spermidine hydroxycinnamoyl transferase, SHT shikimate O-hydroxycinnamoyl transferase, AD aldehyde dehydrogenase, CHR chalcone reductase, CHS chalcone synthase, CHI chalcone isomerase, FOMT isoflavone 7-O-methyltransferase, F3H flavanone 3-hydroxylase, DFR dihydroflavonol-4-reductase, ANS 2-oxoglutarate-dependent dioxygenase, FLS DMR6-like oxygenase 2, CYP cytochrome P450, FNSII flavone synthase II, IFS isoflavone synthase 2, HIDH 2-hydroxyisoflavanone dehydratase, IOMT isoflavone 7-O-methyltransferase, I2'H isoflavone 2'-hydroxylase, IFR isoflavone reductase, VR vestitone reductase, CCoAAGT coumaroyl-CoA: anthocyanidin 3-O-glucoside-6''-O-coumaroyltransferase, UFGT UDP-glycosyltransferase, RT UDP-rhamnose: rhamnosyltransferase, DIR Dirigent protein. See Supplementary Table S9 for complete information.
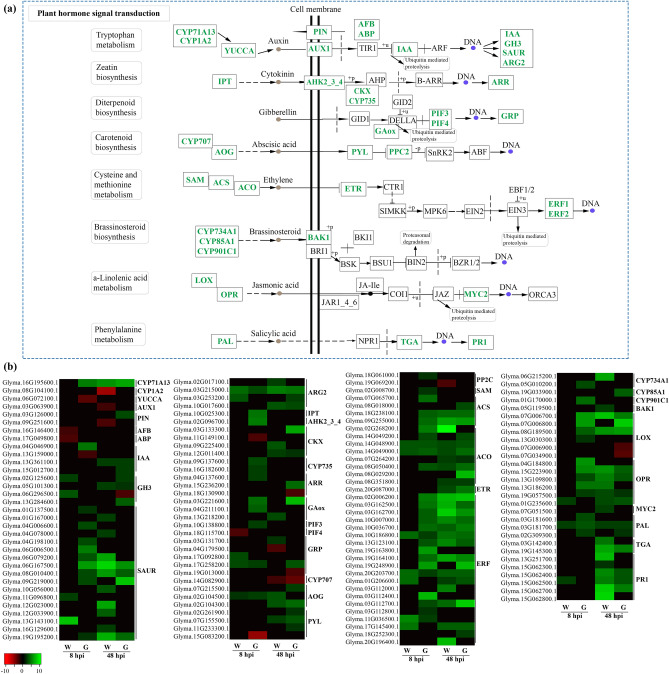
Plant hormones involved in soybean defense against D. caulivora
GO enrichment analysis and overrepresented DEGs showed that salicylic acid (SA), auxin, ET, jasmonic acid (JA), abscisic acid (ABA), cytokinins (CK) and brassinosteroids (BR) probably participate in defense responses against D. caulivora. In total, 131 DEGs were involved in phytohormone pathways. Three PAL-encoding genes with possible roles in SA synthesis were upregulated during D. caulivora infection, and two of these PAL genes were only upregulated in Génesis 5601. However, isochorismate synthase genes (ICSs) were not induced after D. caulivora inoculation (Fig. 8, Supplementary Table S10). Auxin-related DEGs included 37 genes involved in biosynthesis, signaling and response to indole-3-acetic acid (IAA), and most of them were upregulated. A higher number of small auxin-up RNA (SAUR) genes were induced in Génesis-8 and in Williams-48 (nine and 10), compared to two SAUR genes in Williams-8 and four in Génesis-48 (Fig. 8, Supplementary Table S10). Furthermore, significantly more upregulated DEGs involved in CK synthesis and signaling were observed in Génesis-8 compared to Williams-8. We found three genes encoding abscisate beta-glucosyltransferases, and three PYL4 ABA receptors that were upregulated with D. caulivora. Moreover, one ET receptor (ETR) gene was upregulated in both genotypes at 48 hpi, and 34 DEGs were related to ET synthesis and signaling; 24 were upregulated in Génesis-8 and only seven in Williams-8. Most of these DEGs encode 1-aminocyclopropane-1-carboxylate synthases (ACC), 1-aminocyclopropane-1-carboxylate oxidases (ACO) and ERF TFs. Nine upregulated DEGs encoded proteins involved in JA biosynthesis and signaling in Génesis-8 such as LOX, 12-oxophytodienoate reductase (OPR) and MYC2 TF, while in Williams-8 only one OPR was upregulated. At 48 hpi, the number of DEGs related to ET and JA pathways were similar among soybean genotypes. Taken together, these results suggest that several hormones, in particular IAA, ET, CK and JA are involved in resistance mechanisms against D. caulivora.
Figure 8.
Activation of plant hormone signal transduction pathways after D. caulivora inoculation. (a) Simplified scheme of KEGG hormones signaling. Upregulated genes are represented in green. (b) Heatmap of DEGs encoding enzymes involved in defense hormone signaling. Individual genes are listed and colors represented the log2 fold change value based on the comparison of the transcript levels between D. caulivora inoculated and control treatment for both genotypes (Williams and Génesis 5601). Green represents upregulated DEGs and red downregulated DEGs. CYP71A13, CYP1A2 cytochrome P450, YUCCA indole-3-pyruvate monooxygenase YUCCA, AUX1 auxin influx carrier, PIN auxin efflux carrier, AFB auxin signaling F-box, ABP auxin-binding protein, IAA auxin responsive protein, GH3 auxin responsive GH3 gene family, SAUR SAUR family protein, ARG2 indole-3-acetic acid-induced protein ARG2, IPT adenylate isopentenyltransferase, AHK2_3_4 Arabidopsis histidine kinase 2/3/4, cytokinin receptors, CKX cytokinin dehydrogenase, CYP735 cytokinin hydroxylase, ARR two-component response regulator ARR family, GAox gibberellin oxidase, PIF phytochrome-interactin factor, GRP gibberellin-regulated protein, CYP707 abscisic acid 8'-hydroxylase, AOG abscisate beta-glucosyltransferase, PYL abscisic acid receptor PYR/PYL family, PP2C protein phosphatase 2C, SAM S-adenosylmethionine synthase, ACS 1-aminocyclopropane-1-carboxylate synthase, ACO 1-aminocyclopropane-1-carboxylate oxidase, ETR ethylene receptor, ERF ethylene-responsive transcription factor, CYP734A1, CYP85A1, CYP901C1 cytochrome P450, BAK1 brassinosteroid insensitive 1-associated receptor 1, LOX linoleate lipoxygenase, OPR 12-oxophytodienoate reductase, MYC2 transcription factor MYC2, PAL phenylalanine ammonia-lyase, TGA transcription factor TGA, PR-1 pathogenesis-related protein 1. See Supplementary Table S10 for complete information.
Expression analysis of selected candidate genes by RT-qPCR
Twenty-four candidate genes were selected for qRT-PCR validation of the differential response observed in the transcriptomic analysis (Supplementary Fig. S4). Genes encoding proteins with important functions in plant defense against pathogens were included; five RLKs, two NLR, five TFs, one HSP70, three sHSP, four PRs, a CHS, one dirigent, one Bcl-2-associated athanogene (BAG) cochaperone, and a beta-glucosidase. Relative transcript accumulation observed by qPCR showed a strong correlation with transcriptomic expression profiles (R2 = 0.9502), validating the RNA-seq data (Supplementary Table S11).
Discussion
Host responses to biotic stress rely on the timely recognition of the pathogen and the efficient activation of a defense response that involves transcriptional reprogramming. The first stages of the interaction are decisive for the outcome of the disease. Hence, we focused on two early stages of D. caulivora infection, 8 and 48 hpi, according to stem colonization process and induction of PR gene expression2. Comparative transcriptional profiles between Génesis 5601 and Williams detected more than 2.000 DEGs. From these, 46% were commonly upregulated in both genotypes during D. caulivora infection, indicating overlapping responses. Interestingly, Génesis-8, showed 2.4 fold more upregulated DEGs than Williams-8 (1.028 versus 434 DEGs), while at 48 hpi the number of upregulated DEGs did not differ significantly between genotypes (1109 in Génesis-48 and 1205 in Williams-48). Comparative GO and KEGG pathway enrichment analysis resulted in similar terms and pathways in both infected genotypes, although Génesis-8 exhibited a greater number of genes in each category. Most enriched terms and pathways were related to plant defense, including defense response, response to oxidative stress and oxidant detoxification, phenylpropanoid and flavonoid biosynthesis, plant hormone signal transduction, plant-pathogen interaction, as well as protein phosphorylation and regulation of transcription. These findings suggest that an adequate recognition of the pathogen and activation of defense mechanisms may underlie the observed resistance in Génesis 5601 to D. caulivora.
The interplay between pathogen perception and defense activation has a profound effect on plant resistance. Our results revealed that 15 PRRs exhibited higher transcript levels in untreated Génesis 5601 compared to untreated Williams, including LRR-STKs, RLK and RLPs. This number increased to 59 upregulated PRRs in Génesis-8 versus 11 in William-8, including RLKs, LRR-RLKs, LecRLKs, RLPs and CRKs. Thus, basal PRRs expression levels as well as an early induction of PRRs upon D. caulivora inoculation might play important roles in MAMPs recognition and PTI activation. In accordance with these results, narrow-leafed lupin RLKs, LRR-RLKs, LecRLKs and RLP encoding genes were earlier induced in resistant compared to susceptible genotypes in response to D. toxica (Will., Highet, Gams, and Sivasith)23. Similarly, several PRR genes such as CRKs and LRR-RLKs were higher expressed in resistant compared to susceptible soybean plants in response to Phakopsora pachyrhizi24, soybean mosaic virus (SMV)25, and soybean cyst nematode (Heterodera glycines Ichinohe)26. Moreover, RLKs and RLPs are candidate soybean resistance genes against the fungus Phialophora gregata (sin. Cadophora gregata) (Allington & D.W. Chamb.) W. Gams 27.
Suppression of PTI by pathogen effectors activates ETI through the action of NLRs10. D. caulivora genome analysis evidenced the presence of 133 secreted effector candidates, and several of these effector genes were induced during soybean colonization, suggesting that they could interfere with soybean defense16. Here, we show that seven NLR genes exhibited higher expression levels in untreated Génesis 5601 versus Williams. Similarly, seven and only one NLR were upregulated in Génesis-8 and Williams-8, respectively. The induction of NLR genes could lead to earlier activation of downstream immune events in Génesis 5601. Soybean NLR genes co-localize with disease-resistance quantitative-trait loci (QTL)28, supporting their role in plant resistance against different pathogens. NLR were identified as candidate resistance genes for several soybean diseases caused by bacterial, fungal, oomycete and virus29–36. Some of these NLR genes have been functionally validated31,37,38. Furthermore, genomic regions identified by genome-wide association studies (GWAS) and related to resistance against several soybean diseases were enriched in LRR-RLK and NLR39.
MAPKs, MAPKKs, MAPKKKs and STKs activate downstream signaling after pathogen recognition40. D. caulivora inoculation increased expression of genes encoding for these type of kinases in both cultivars. Several MAPKKKs were only upregulated in Génesis-8 and STKs were differentially expressed in Génesis-8 and Williams-8. Interestingly, STKs are candidate genes for resistance to soybean mosaic virus and several STKs were higher expressed in resistant compared to susceptible plants25.
WRKY, AP2/ERF, MYB, bHLH, NAC, GRAS, and other TFs play important roles in defense responses to biotic and abiotic stress41–43. Our study found that 219 of these TFs were upregulated during D. caulivora colonization. Génesis-8 showed significantly more upregulated ERFs, MYB and bHLH compared to Williams-8, which could trigger coordinated induction of target genes involved in plant immunity. Similarly, higher expression levels of bHLHs were observed in P. sojae resistant compared to susceptible soybean cultivars44. Interestingly, GmMYBs and GmNAC regulate biosynthesis of flavonoids leading to the production of phytoalexins that increase resistance against pathogen45–47. Moreover, GmERF5- and GmERF113-overexpressing soybean plants showed enhanced resistance to P. sojae and positively regulated expression of PR-10 and PR-148,49.
A high number of PR genes were induced during D. caulivora colonization in both cultivars, suggesting their involvement in soybean defense against this fungal pathogen. Likewise, PR-1, β-1,3-glucanase (PR-2), chitinases (PR-3 and PR-4), PR-10 and defensin were induced in a susceptible cultivar infected with D. aspalathi13. Expression profiles highlighted β-1,3-glucanase, peroxidases class III (PR-9) and PR-10 function in the early response of Génesis 5601. Similarly, peroxidases encoding genes were upregulated in a resistant genotype of narrow-leafed lupin infected with D. toxica23. As observed for other pathogens, these enzymes could protect plant cells against D. caulivora by degrading fungal cell wall polysaccharides (β-1,3-glucanases), and probably by inhibiting hyphal growth through RNase activity (PR-10)50,51. Peroxidases could increase soybean defenses by reinforcing plant cell walls, synthesis of phytoalexins, or participate in ROS metabolism as has been observed in other pathogen-infected plants52. In addition to peroxidases, we found increased expression of other oxidative stress-related genes encoding glutathione S-transferase, thioredoxin, ferredoxin oxidoreductases in Génesis-8, supporting the importance of cellular homeostasis to maintain a redox balance in soybean tissues to resist further infection. ROS accumulation can lead to a hypersensitive response (HR), a programmed localized cell death that occurs at the site of infection and is associated with restriction of the pathogen and disease resistance53. Interestingly, the induced expression of three genes encoding BAG cochaperone (Glyma.18G284900, Glyma.18G285100 and Glyma.07G061500) in Génesis-8, suggest a possible involvement of programmed cell death in D. caulivora resistance. This type of proteins trigger autophagy in the host to limit fungal colonization and confer resistance to fungal pathogens54.
Plants respond to pathogen infection by activating the phenylpropanoid pathway14,55. Here, we show that a high proportion of genes required for phenylpropanoid synthesis and lignin production were induced upon D. caulivora infection, suggesting that reinforcement of the cell wall through lignin and phenolic compounds, and synthesis of antimicrobial compounds such as flavonoids, isoflavonoids, coumarins and lignans are important defense mechanisms against this fungal pathogen. Interestingly, a significant number of upregulated DEGs were only present in Génesis-8 and not in Williams-8, suggesting that some of the metabolites produced by the phenylpropanoid pathway could be involved in resistance mechanisms against D. caulivora. Phytoalexin production in response to pathogens is regulated by the enzymes PAL, CHS, and chalcone isomerase (CHI), among others14. The number of upregulated DEGs encoding these enzymes were significantly higher in Génesis-8 compared to Williams-8. Interestingly, phenylpropanoids such as isoflavones daidzein, genistein and glyceollins are produced in soybean resistant plants after treatment with D. aspalathi elicitors56. Moreover, GmMYB29 regulates isoflavonoid biosynthesis in soybean through the activation of isoflavone synthase and CHS encoding genes45. Functional analysis demonstrated that GmMYB29A2 is crucial for the accumulation of glyceollin I and expression of P. sojae resistance47. Likewise, R2R3-MYB involved in lignin synthesis and genes responsive to chitin were significantly induced in P. pachyrhizi resistant genotypes46. Furthermore, activation of cell wall reinforcement by incorporation of phenolic compounds was previously observed in D. caulivora infected tissues2. Thus, these findings suggest that several metabolites of the phenylpropanoid could be produced during D. caulivora colonization, although further studies are needed to decipher the involvement of these metabolites in soybean resistance against this fungal pathogen.
Plant hormones conform a complex network that regulate plant resistance against pathogens57. Soybean plants activate hormonal pathways during D. caulivora infection in both genotypes. The significantly higher number of upregulated DEGs involved in IAA, ET, CK and JA pathways in Génesis-8 compared to Williams-8 suggests that these hormones could be involved in resistance responses against D. caulivora. Interestingly, induction of ACS and ACO genes involved in ET synthesis has been associated with resistance of soybean plants to vascular disease caused by Fusarium virguliforme (Link ex Grey)58. In addition, genes encoding enzymes involved in the production of JA, including LOXs and OPR, were earlier induced in D. caulivora-inoculated Génesis 5601 plants compared to Williams plants. Consistently, expression levels of LOXs increased in resistant genotypes of narrow-leafed lupin compared to susceptible genotypes in response to D. toxica23. Oxylipins produced by the LOX pathway play different roles during defense responses against biotic stress through antimicrobial activities, contribution to HR, and production of signaling molecules such as JA and related compounds that lead to induced expression of genes with multiple roles in defense15,59.
Small HSP are chaperones that play important roles in immunity by protecting cells from stress-induced protein aggregation and misfolding60. Remarkably, sHSPs encoding genes were only upregulated in Génesis 5601, and generally at 8 hpi, suggesting their important contribution in the early stages of plant resistance. HSPs are involved in stability and accumulation of PRRs and NLRs and further defense signaling60,61. In Nicotiana tabacum, a sHSP was shown to be involved in disease resistance against Ralstonia solanacearum (Smith)62. Furthermore, GmHsp22.4 was highly induced in a nematode resistant soybean genotype and its overexpression in Arabidopsis plants renders lower nematode multiplication63. Interestingly, some pathogen effectors interact with sHSPs to suppress chaperone activity and promote virulence64, highlighting the role of these plant chaperones in disease resistance.
Conclusions
This comparative transcriptomic approach between contrasting soybean genotypes revealed that resistance of Génesis 5601 to D. caulivora infection could be related to induced expression of a higher number of genes encoding proteins involved in perception through PRR and NLR, as well as TFs, PRs, biosynthesis of phenylpropanoid derived metabolites, hormones, sHSPs and genes with different roles in defense. Future studies comprising functional characterization of soybean candidate genes and target genes of D. caulivora effectors will contribute to a valuable comprehension of soybean-Diaporthe interactions. These findings provide novel molecular insights into soybean defense mechanisms used to control this pathogen, and establish a foundation for improving resistance in breeding programs.
Methods
Plant materials and D. caulivora inoculation
For this study, we used the D. caulivora isolate D57, collected from canker lesions of soybean plants grown in Uruguay during 20152. Two soybean genotypes were used for all plant assays: SSC-susceptible Williams PI548631 obtained from USDA ARS Soybean germplasm collection (seed source 13U-9280, order 253444, 2014), and the SSC-resistant Génesis-5601 from the Instituto Nacional de Investigación Agropecuaria (INIA) breeding program (Stewart S, personal communication). Three soybean seeds of each genotype were individually planted in a 10-cm-diameter pot filled with a mix of soil and vermiculite at a rate of 3:1. Soybean seedlings were grown in a growth room under a 16 h light/8 h dark photoperiod regime at 24°C. For all experiments, 3-week-old plants at V2 were used. D. caulivora D57 isolate was inoculated using the stem wounding method where an agar plug containing mycelium was applied to the wounded stem2. As a control an agar plug without mycelium was used. All experiments were performed with accordance to relevant regulations and guidelines.
Development of SSC symptoms were compared between both soybean genotypes. Ten plants were used per treatment and the experiment was repeated three times. Lesion length (mm) and disease severity (Scale 1–7), was determined at various time points (3, 5, 7, 11, and 14 dpi]. Disease severity index and area under disease progress curve (AUDPC) was calculated according to Mena et al.2. Differences between treatments were determined by non-parametric Kruskal–Wallis and Mann– Whitney tests using SPSS Statistics v. 21.0. P values of < 0.01 were considered as significant.
Quantitative PCR
After soybean stem inoculation with D. caulivora, fungal DNA was quantified at 8, 24, 48, 72, and 96 hpi. Three plants per treatment were used as biological replicates and samples were frozen in liquid nitrogen. DNA was extracted from stem tissues (stem section of 1.5 cm including the wounded area) using the DNeasy kit (Qiagen, Germany). DNA concentration and quality were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). Quantitative PCR (qPCR) was performed using primers designed for the elongation factor gene Ef1a of soybean (5′-GATTTCATGTAGCCGTAGCC-3′ and 5′-ATTTAAGACATCCCTCCTCAG-3′) and the β-tubulin gene of D. caulivora (5′-CCGTGGAAAGGTCTCTATGAAG-3′ and 5′-TCTGGACGTTGTTGGGAATC-3′). qPCR was performed using the QuantiNova Probe SYBR Green PCR Kit (Qiagen, Germany) in a 96-well thermocycler (New Applied Biosystems QuantStudio 3). Each reaction consisted in 20 μl containing 10 μl of SYBR Green PCR Master mix (2×), 0.7 mM primers mix, and DNA (~ 25 ng). The thermocycler was programmed to run for 2 min at 95°C, followed by 40 cycles of 15 s at 94°C and 20 s at 60°C. Water was used as negative control. As a standard, a serial dilution of genomic DNA from D. caulivora with known concentrations (60 ng, 6 ng, 600 pg, 60 pg, and 6 pg) were analyzed to determine the sensitivity and linear range of the assay. Pathogen standard curve was generated by plotting the CT values of a tenfold dilution series of D. caulivora DNA stock solution versus the logarithm of the concentration. The resulting regression equations were used to calculate fungal DNA in stem samples. Similarly, a standard curve was generated to estimate the amount of soybean DNA present in each sample. Pathogen β-tubulin estimated was expressed relative to soybean elongation factor. Each data point is the mean value of three biological replicates. Results were expressed as ng of D. caulivora/ng of soybean tissue. Student’s t-test was applied to all qPCR data, and values of p ≤ 0.01 were considered statistically significant.
RNA extraction, cDNA library preparation and sequencing
Samples of both genotypes were taken from untreated plants, and from plants inoculated with plugs containing D. caulivora mycelium and their respective controls (plugs without mycelium) at 8 and 48 hpi. Total RNA was extracted and purified from 100 mg soybean stems, 10 mm above the inoculation point of each sample with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and Invitrogen PureLink RNA Extraction Mini kit (Invitrogen, USA), including an on-column digestion with RNase-Free DNase I, according to the manufacturer’s instructions. RNA quality was checked by running samples on 1.2% formaldehyde agarose gel. RNA concentration was measured using a NanoDrop 2000c (Thermo Scientific, Wilmington, USA). RNA quality control, library preparation, and sequencing were performed at Macrogen Inc. (Seoul, Korea). Three biological replicates were included per treatment. Libraries for each biological replicate were prepared for paired-end sequencing by TruSeq Stranded Total RNA LT Sample Prep Kit (Plant) with 1 μg input RNA, following the TruSeq Stranded Total RNA Sample Prep Guide, Part # 15031048 Rev. E. Sequencing was performed on Illumina platform (Illumina, CA, USA) to generate paired-end 101 bp reads, obtaining 41.6 to 65.2 M reads per sample with Q20 > 98% and Q30 > 95%.
Pre-processing of raw data, mapping of reads and annotation
RNA-seq processing steps were done through Galaxy platform (https://usegalaxy.org/)65 and according to Reboledo et al.66. Raw reads quality was subjected to a quality control check using FastQC software ver. 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequences were trimmed, and the adapters removed using Trimmomatic Version 0.38.0 software67. Additionally, to the default options, the following parameters were adjusted: adapter sequence TruSeq3 (paired-ended (PE), for MiSeq and HiSeq), always kept both PE reads, and SLIDINGWINDOW: 4:15 HEADCROP: 13 MINLEN:50. Trimmed reads were mapped to reference genome of Glycine max Gmax_275_Wm82.a2.v1.fa68 as the reference genome file and Gmax_275_Wm82.a2.v1.gene.gff3 as a reference file for annotation gene models from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) using Hisat2 software69. The BAM files were obtained with Samtools View software ver.1.9 and then sorted by name with Samtools Sort software ver. 2.0.370, for further analysis.
Differential gene expression and functional analysis
Cluster analysis of replicates from each time point and control samples were performed by Principal Component Analysis (PCA) using log2 fold changes for all datasets. Reads were counted using FeatureCounts software ver. 1.6.471. Additionally, to default options, the following parameters were set: Stranded (Reverse), Count fragments instead of reads -p, Allow read to map to multiple features True, Count multi-mapping reads/fragments -M and Reference sequence file Glycine max Wm82.a2.v1. Differential expression analyses were performed using EdgeR software ver. 3.24.172, with p-value adjusted method of Benjamini and Hochberg adjusted threshold 0.0573, and Minimum log2 Fold Change 2. Counts were normalized to counts per million (cpm) with the TMM method and low expressed genes filtered for count values ≥ 3 in all samples. In this study, a false discovery rate (FDR) ≤ 0.05 was used to determine significant differentially expressed genes (DEGs) between D. caulivora inoculated plants and mock; and expression values were represented by log2 ratio. Heat maps were generated using the Heatmapper server (https://www.heatmapper.ca/expression). Hierarchical clustering analysis of expressed genes were performed on log2 Fold-Change expression values using the “hclust” tool from R package “stats” ver. 3.6.0. To visualize the obtained data, heatmap plots were performed using the “heatmap.2” tool from R package “gplots” ver. 3.1.0.
Gene ontology (GO) and functional annotations were assigned with the Blast2GO, through Omicbox software (https://www.biobam.com/omicsbox)74. Gene models were compared with several databases (NCBI nonredundant protein database, GO, and InterpoScan) with BlastP finding single hit at an e-value threshold of e-value ≤ 1.0E−3 using taxIds for Viridiplantae. InterproScan analysis was used to identify domains in the genome75 DEG functional enrichment analysis was performed using OmicBox software. GO terms with a FDR ≤ 0.05 were considered for the analysis. DEGs of each dataset were divided into upregulated and downregulated subsets. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of all DEGs were obtained through Omicbox software for D. caulivora inoculated vs. control samples in both soybean genotypes. All heat maps were generated using the Heatmapper server (https://www.heatmapper.ca/expression).
Quantitative real-time PCR
The expression level of twenty-four selected candidate genes was analyzed via quantitative reverse transcription PCR (RT-qPCR). cDNA was generated from 1 μg of RNA using RevertAid Reverse transcriptase (Thermo Scientific) and oligo (dT) according to the manufacturer’s protocol. RT-qPCR was performed in an Applied Biosystems QuantStudio 3 thermocycler using the QuantiNova Probe SYBR Green PCR Kit (Qiagen, Germany); mix proportions and cycling parameters were used as described in manufacturer’s instructions. Relative expression of each gene was normalized to the quantity of constitutively expressed elongation factor 1-alpha gene, using the 2−ΔΔCt method76. Gene expression of soybean-inoculated tissues was expressed relative to the corresponding control samples at the indicated time points, with its expression level set to one. Each data point is the mean value of three biological replicates. Student’s t-test was performed to determine the significance for quantitative gene expression analysis using GraphPad Prism software ver. 8.0.1. P-values < 0.01 were considered statistically significant. Primer pairs used for qPCR analyses are provided in Supplementary Table S12. In all cases, amplification efficiencies were between 95 and 110%.
Supplementary Information
Acknowledgements
Authors thank Ricardo Larraya for technical assistance. This work was supported by “Agencia Nacional de Investigación e Innovación (ANII) (graduate fellowship and grant FCE_3_2022_1_172688)” Uruguay, “Programa de Desarrollo de las Ciencias Básicas (PEDECIBA)” Uruguay, and “Programa Grupo de I+D Comisión Sectorial de Investigación Científica, Universidad de la República”, Uruguay.
Author contributions
E.M. performed all the experiments, interpreted the data, contributed to discussions, and helped to write the manuscript. G.R. performed the hierarchical clustering analysis. S.S. contributed to the selection of the soybean genotypes and discussions. M.M. contributed to discussions and helped to write the manuscript. I.P. designed and supervised the study, interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
Data availability
RNA sequencing data were deposited at the National Center for Biotechnology Information (NCBI) in the Sequence Read Archive (SRA) under the PRJNA878492 Bioproject accession.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-39695-1.
References
- 1.Miransari, M. Abiotic and biotic stresses in soybean production in soybean production. in Soybean Production and Heavy Metal Stress (Ed. Miransari, M.). 197–216 (Academic Press, 2015).
- 2.Mena E, Stewart S, Montesano M, Ponce de León I. Soybean stem canker caused by Diaporthe caulivora; pathogen diversity colonization process and plant defense activation. Front. Plant Sci. 2020;10:1733. doi: 10.3389/fpls.2019.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández, F. A., Philips, D. V., Russin, J. S.,& Rupe, J. C. Diaporthe–Phomopsis complex. in Compendium of Soybean Diseases (Eds. Hartman, G. L., Sinclair, J. B. & Rupe, J. C.). 4th ed. 33–35 (Minnesota, 1999).
- 4.Pioli RN, et al. Morphologic, molecular, and pathogenic characterization of Diaporthe phaseolorum variability in the core soybean-producing area of Argentina. Phytopathology. 2003;93:136–146. doi: 10.1094/PHYTO.2003.93.2.136. [DOI] [PubMed] [Google Scholar]
- 5.Santos JM, Correia VG, Phillips AJL. Primers for mating-type diagnosis in Diaporthe and Phomopsis: their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010;114:255–270. doi: 10.1016/j.funbio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Chiesa MA, Pioli RN, Morandi EN. Specific resistance to soybean stem canker conferred by theRdm4locus. Plant Pathol. 2009;58:1032–1038. doi: 10.1111/j.1365-3059.2009.02145.x. [DOI] [Google Scholar]
- 7.Peruzzo AM, Hernández FE, Pratta GR, Ploper LD, Pioli RN. Identification and inheritance of an Rdc gene resistance to soybean stem canker (Diaporthe phaseolorum var. caulivora) Eur. J. Plant Pathol. 2019;154:1179–1184. doi: 10.1007/s10658-019-01716-z. [DOI] [Google Scholar]
- 8.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 9.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol. Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Yuan M, Ngou BPM, Ding P, Xin XF. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021;62:102030. doi: 10.1016/j.pbi.2021.102030. [DOI] [PubMed] [Google Scholar]
- 12.Ngou BPM, Ding P, Jones JDG. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell. 2022;34:1447–1478. doi: 10.1093/plcell/koac041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upchurch RG, Ramirez ME. Defense-related gene expression in soybean leaves and seeds inoculated with Cercospora kikuchii and Diaporthe phaseolorum var. meridionalis. Physiol. Mol. Plant Pathol. 2010;75:64–70. doi: 10.1016/j.pmpp.2010.08.007. [DOI] [Google Scholar]
- 14.Dixon RA, et al. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002;5:371–390. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]
- 15.Blée E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002;7:315–322. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- 16.Mena E, Garaycochea S, Stewart S, Montesano M, Ponce de León I. Comparative genomics of plant pathogenic Diaporthe species and transcriptomics of Diaporthe caulivora during host infection reveal insights into pathogenic strategies of the genus. BMC Genomics. 2022;23:175. doi: 10.1186/s12864-022-08413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin F, et al. Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genomics. 2014;15:18. doi: 10.1186/1471-2164-15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tremblay A, Hosseini P, Alkharouf NW, Li S, Matthews BF. Gene expression in leaves of susceptible Glycine max during infection with Phakopsora pachyrhizi using next generation sequencing. Sequencing. 2011;2011:827250. doi: 10.1155/2011/827250. [DOI] [Google Scholar]
- 19.Bencke-Malato M, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of gene involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 2014;14:236. doi: 10.1186/s12870-014-0236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanubile A, Muppirala UK, Severi AJ, Marocco A, Munkvold GP. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genomics. 2015;16:1089. doi: 10.1186/s12864-015-2318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Comparison of transcriptome differences in soybean response to soybean mosaic virus under normal light and in the shade. Viruses. 2019;11:793. doi: 10.3390/v11090793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Loon LC, Rep M, Pietersen CMJ. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 23.Książkiewicz M, et al. The resistance of narrow-leafed lupin to Diaporthe toxica Is based on the rapid activation of defense response genes. Int. J. Mol. Sci. 2021;22(574):2021. doi: 10.3390/ijms22020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delgado-Cerrone L, Alvarez A, Mena E, Ponce de León I, Montesano M. Genome wide analysis of the soybean CRK-family and transcriptional regulation by biotic stress signals triggering plant immunity. PLoS ONE. 2018;13:e0207438. doi: 10.1371/journal.pone.0207438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rui R, et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to soybean mosaic virus. Theor. Appl. Genet. 2017;130:2395–2410. doi: 10.1007/s00122-017-2966-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Kjemtrup-Lovelace S, Li C, Luo Y, Chen LP, Song BH. Comparative RNA-seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja) Sci. Rep. 2017;7:9699. doi: 10.1038/s41598-017-09945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCabe CE, Cianzio SR, Örourke JA, Graham MA. Leveraging RNA-seq to characterize resistance to brown stem rot and the Rbs3 locus in soybean. Mol. Plant Microb. Interact. 2018;31:1083–1094. doi: 10.1094/MPMI-01-18-009-R. [DOI] [PubMed] [Google Scholar]
- 28.Lin F, et al. Breeding for disease resistance in soybean: A global perspective. Theor. Appl. Genet. 2022;2022:35790543. doi: 10.1007/s00122-022-04101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang YJ, et al. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 2012;12:139. doi: 10.1186/1471-2229-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashfield T, et al. Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol. 2012;159:336–354. doi: 10.1104/pp.112.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H, Narayanan NN, Ellison L, Bhattacharyya MK. Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microb. Interact. 2005;18:1035–1045. doi: 10.1094/MPMI-18-1035. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya MK, et al. Identification of a large cluster of coiled coil-nucleotide binding site-leucine rich repeat-type genes from the Rps1 region containing Phytophthora resistance genes in soybean. Theor. Appl. Genet. 2005;111:75–86. doi: 10.1007/s00122-005-1993-9. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, et al. A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat. Commun. 2021;12:6263. doi: 10.1038/s41467-021-26554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer JDF, et al. Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol. 2009;150:295–307. doi: 10.1104/pp.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedley KF, et al. Rpp1 encodes a ULP1-NBS-LRR protein that controls immunity to Phakopsora pachyrhizi in soybean. Mol. Plant Microbe Interact. 2019;32:120–133. doi: 10.1094/MPMI-07-18-0198-FI. [DOI] [PubMed] [Google Scholar]
- 36.Su JS, et al. The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome. 2011;4:55–64. doi: 10.3835/plantgenome2010.11.0024. [DOI] [Google Scholar]
- 37.Zhou L, et al. A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. Crop J. 2022 doi: 10.1016/j.cj.2022.03.003. [DOI] [Google Scholar]
- 38.Xun H, et al. Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019;99:95–111. doi: 10.1007/s11103-018-0804-z. [DOI] [PubMed] [Google Scholar]
- 39.Chang HX, Lipka AE, Domier LL, Hartman GL. Characterization of disease resistance loci in the USDA soybean germoplasm collection using genome-wide associations. Phytopathology. 2016;106:1139–1151. doi: 10.1094/PHYTO-01-16-0042-FI. [DOI] [PubMed] [Google Scholar]
- 40.Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Jin J, et al. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erpen L, Devi HS, Grosser JW, Dutt M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 2018;132:1–25. doi: 10.1007/s11240-017-1320-6. [DOI] [Google Scholar]
- 43.Baillo EH, Kimotho RN, Zhang Z, Xu P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 2019;10:771. doi: 10.3390/genes10100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Q, et al. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018;69:2527–2541. doi: 10.1093/jxb/ery103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu S, et al. An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet. 2017;10:e1006770. doi: 10.1371/journal.pgen.1006770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoyagi LN, et al. Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci. 2014;229:32–42. doi: 10.1016/j.plantsci.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Jahan MA, et al. Glyceollin transcription factor GmMYB29A2 regulates soybean resistance to Phytophthora sojae. Plant Physiol. 2020;183:530–546. doi: 10.1104/pp.19.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong L, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015;66:2635–2647. doi: 10.1093/jxb/erv078. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y, et al. A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant Sci. 2017;8:299. doi: 10.3389/fpls.2017.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balasubramanian V, Vashisht D, Cletus J, Sakthivel N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012;34:1983–1990. doi: 10.1007/s10529-012-1012-6. [DOI] [PubMed] [Google Scholar]
- 51.Xu TF, et al. A pathogenesis related protein, VpPR-10.1, from Vitis pseudoreticulata: An insight of its mode of antifungal activity. PLoS ONE. 2014;9:e95102. doi: 10.1371/journal.pone.0095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Almagro L, et al. Class III peroxidases in plant defense reactions. J. Exp. Bot. 2009;60:377–390. doi: 10.1093/jxb/ern277. [DOI] [PubMed] [Google Scholar]
- 53.Lamb, C. & Dixon, R. A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol.48, 251–275. 10.1146/annurev.arplant.48.1.251 (1997). [DOI] [PubMed]
- 54.Li Y, Kabbage M, Liu W, Dickman MB. Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell. 2016;28:233–247. doi: 10.1105/tpc.15.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 56.Modolo LV, Cunha FQ, Braga MR, Salgado I. Nitric oxide synthase-mediated phytoalexin accumulation in soybean cotyledons in response to the Diaporthe phaseolorum f. sp. meridionalis elicitor. Plant Physiol. 2002;130:1288–1297. doi: 10.1104/pp.005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandey, P. & Senthil-Kumar, M. Plant responses to combined drought and pathogen infection: Current understanding on the role of phytohormones. in Plant Tolerance to Individual and Concurrent Stresses (ed. Senthil-Kumar, M). 133–149 (Springer India, 2017).
- 58.Abdelsamad NA, MacIntosh GC, Leandro LFS. Induction of ethylene inhibits development of soybean sudden death syndrome by inducing defense-related genes and reducing Fusarium virguliforme growth. PLoS ONE. 2019;14:e0215653. doi: 10.1371/journal.pone.0215653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park CJ, Seo YS. Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015;31:323–333. doi: 10.5423/PPJ.RW.08.2015.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 62.Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007;145:1588–1599. doi: 10.1104/pp.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hishinuma-Silva, S. M. et al. The soybean gene GmHsp22.4 is involved in the resistance response to Meloidogyne javanica in Arabidopsis thaliana. BMC Plant Biol.20, 535. 10.1186/s12870-020-02736-2 (2020). [DOI] [PMC free article] [PubMed]
- 64.Ahmed AA, et al. The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 2015;168:321–333. doi: 10.1104/pp.15.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:537–544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reboledo G, et al. Transcriptional profiling reveals conserved and species-specific plant defense responses during the interaction of Physcomitrium patens with Botrytis cinerea. Plant Mol. Biol. 2021;107:365–385. doi: 10.1007/s11103-021-01116-0. [DOI] [PubMed] [Google Scholar]
- 67.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 69.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H. et al. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics15, 2078–2079. 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed]
- 71.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Ser. Stat. Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 74.Conesa A, Götz S. Blast2gGO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008 doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zdobnov EM, Apweiler R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 76.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data were deposited at the National Center for Biotechnology Information (NCBI) in the Sequence Read Archive (SRA) under the PRJNA878492 Bioproject accession.