Abstract
Background:
At diagnosis, up to one-third of patients with Crohn’s disease (CD) have a complicated phenotype with stricturing (B2) or penetrating (B3) behavior or require early surgery. We evaluated protein biomarkers and anti-microbial antibodies in serum archived years before CD diagnosis to assess whether complicated diagnoses were associated with a specific serological signature.
Methods:
Pre-diagnosis serum was obtained from 201 patients with CD and 201 healthy controls (HC). Samples were evaluated with a comprehensive panel of 1129 proteomic markers (SomaLogic®) and anti-microbial antibodies. CD diagnosis and complications were defined by ICD-9 and CPT codes. Cox regression models were utilized to assess the association between markers and the subsequent risk of being diagnosed with complicated CD. In addition, biological pathway and network analyses were performed.
Results:
Forty-seven CD subjects (24%) had a B2 (n=36) or B3 (n=9) phenotype or CD-related surgery (n=2) at diagnosis. Subjects presenting with complicated CD at diagnosis had higher levels of anti-microbial antibodies 6 years before diagnosis as compared to those diagnosed with non-complicated CD. Twenty-two protein biomarkers (reflecting inflammatory, fibrosis, and tissue protection markers) were found associated with CD complication. Pathway analysis of the altered protein biomarkers identified higher activation of the innate immune system and complement/coagulation cascades up to six years before diagnosis in complicated CD.
Conclusions:
Proteins and anti-microbial antibodies associated with dysregulated innate immunity, excessive adaptive response to microbial antigens, and fibrosis, precede and predict a complicated phenotype at the time of diagnosis in CD patients.
Keywords: Preclinical period, Crohn’s disease, Complications, Serologic biomarkers
Introduction
Crohn’s disease (CD) is a chronic inflammatory condition of the gastrointestinal tract with peak onset between the ages of 15 and 30 years.1, 2 CD is a progressive disease that can lead to complications including strictures, fistulae, or abscesses that require aggressive medical and/or surgical treatment.3, 4 Up to one-third of patients present with a complicated phenotype at disease onset.5
Mounting evidence suggests that the diagnosis of CD is preceded by a lengthy asymptomatic pre-clinical period.6–10 Gaining insight into this phase may allow a better understanding of the primary events that lead to its development and offer potential strategies to predict and prevent the disease including its complications. The PREDICTS (Proteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects) cohort was initiated to identify biomarkers and altered biologic pathways that precede inflammatory bowel disease (IBD) onset using pre-clinical sera from the United States Department of Defense Serum Repository (DoDSR).11 In our prior studies, we have observed that antibodies against several microbial antigens and specific proteins can be detected years before diagnosis and are highly predictive of CD.9, 10 We have also demonstrated that a higher prevalence of anti-microbial antibodies in preclinical CD serum samples has shown to be associated with complications at diagnosis.9 We thus postulated that aberrant innate and adaptive immunity against gut microbiota which occurs in the pre-clinical stage of CD is further amplified and distinct in those patients with complications at diagnosis. Using the PREDICTS cohort, we evaluated anti-microbial antibodies and protein biomarkers in longitudinal serum samples before diagnosis to assess whether CD patients with complications at diagnosis had a specific preclinical anti-microbial or proteomic profile.
Methods
Study Design and Study Population
We conducted a nested case-control study using the previously described PREDICTS study.9–11 Briefly, patients with an incident diagnosis of CD were identified between 1998 and 2013 in the Department of Defense Medical surveillance system (DMSS).11, 12 Linked serum samples from each subject were obtained through the DoDSR.11, 12
Incident CD was defined based on procedural and ICD-9 (International Classification of Diseases, Ninth Revision) codes. The date of CD diagnosis was based on the first ICD-9 code for CD. For each subject, 4 serum samples were obtained from the DoDSR: sample A was the closest sample available to the date of CD diagnosis, and sample D was the earliest serum sample available in the repository before clinical diagnosis; samples B and C were approximately 2 and 4 years before diagnosis, respectively. For sample D, SomaLogic proteomic panel and anti-microbial antibodies were tested for 201 CD, while for sample B and sample C, the number of samples was 116 and 166, respectively. For this reason, sample B and sample C groups were aggregated into one group. Samples of healthy controls (HC) were also obtained from the DoDSR. As previously described,10 controls were matched on age, gender, race and timing of the diagnostic sample, and required to have no medical encounter with evidence of IBD, rheumatoid arthritis, celiac disease or colorectal cancer (based on ICD9 codes). From each subject, 3 to 4 serum samples were retrieved. Sample A from HC subjects was matched to Sample A from IBD cases based on the year of collection (± 1 year)., whose serum samples were available and stored from the three-preceding biennial (e.g., every two years) HIV test.
Phenotype Classification
Disease phenotype (i.e., behavior) was categorized according to the Montreal classification.22 Complicated CD was defined by the presence of penetrating (P) (B3 behavior), stricturing (S) (B2 behavior), or surgical history of intestinal resection using ICD-9 and CPT codes from time of diagnosis (index ICD-9 code). The detailed information for phenotype classification is provided in Supplemental Table 1.
Serum Testing
To evaluate protein abundance, serum was tested using the SomaLogic® (Boulder, CO, USA) assay, a multiplex platform profiling 1129 protein biomarkers, representing a range of biological functions including innate immune response and inflammatory signals. Samples were also tested for a panel of anti-microbial antibodies, including anti-ASCA-IgA and -IgG, anti-CBir1, anti-OmpC, anti-Flagellin 2, and anti-Flagellin X. These antibodies were measured by a standardized enzyme-linked immunosorbent assay (ELISA) using a Freedom EVO 200 liquid-handling robot (Tecan) at Prometheus® Laboratories (San Diego, CA, USA).13 Abbreviations of serologic and protein biomarkers are in the Supplemental Table 2.
Statistical Analyses
The association between each marker (SomaLogic® proteomic biomarker and serum antibodies against microbiota) and complication was assessed via Cox regression after adjusting by age and gender. P-values were adjusted for multiple comparisons via Benjamini-Hochberg14 and markers passing a false-discovery rate of 10% were reported as significant. Cox-regression models were estimated for different times before diagnosis (i.e., 2–4 and 6 years before diagnosis). We repeated this analysis adjusting by disease location in Cox-regression model considering a subset of 167 patients for which disease location was measured. We also performed differential analysis via Wilcoxon rank sum test between patients with ileal involvement and colonic involvement (L1/L3 vs. L2). P-values were adjusted for multiple comparison via Benjamini-Hochberg14 adjustment and only markers with adjusted p-value less than 10% were reported as significant. To visualize the association between each marker abundance and complication, Kaplan–Meier (KM) survival curves were utilized. To display the association of a set of markers and time of complication, the estimated mean of the Cox regression model was utilized in order to stratify patients into high and low risk groups and derive KM curves. Different multivariate cox-regression models were compared using the Akaike’s information criterion (AIC).15 This index is used to determine how well the model fits the data while accounting for the total number of parameters in the model. Although models having more markers result in a better fit, they are usually less suitable to predict other datasets. For this reason, finding a good balance between model fit and parsimony is essential to select the best model. One of the ways to compare the goodness of fit among models with different number of parameters is AIC, which is a function of the log-likelihood of the estimated model and the number of parameters utilized. The model with lowest AIC is the preferred model. The log-likelihood of the Cox-regression was computed via the logLik function available in the Survival R package16,17.
Biological pathways enriched in the set of proteins associated with complication were identified at each time point via Fisher Exact test. For the analysis, pathways are from Kyoto Encyclopedia of Genes and Genomes (KEGG)18 and Reactome databases.19 were utilized. For each patient, pathway scores were computed as the average abundance of protein mapping to each pathway after z-score them across patients (mean 0 and standard deviation 1). Beside pathway analysis, we performed co-expression network analysis to identify the association across proteomic markers and anti-microbial antibodies for patients with CD and HC using joint random Forest (JRF).20 Permutation-based techniques were used to find association significant at 10% FDR.20 Network modules based on the CD network were identified based on the cluster-edge-betweenness function available in the iGraph R package.21 All analyses were performed by using R statistical software (version 3.6.3, R Foundation for Statistical Computing, Vienna, Austria).
Results
A total of 201 patients with CD and 201 healthy controls were included in the study. Among CD patients, 23% presented with complications at diagnosis with 36 B2, 9 B3, and two CD-related surgery (one colectomy and one small bowel resection). The detailed study population characteristics are provided in Supplemental Table 3.
Anti-Microbial Antibody Markers Associated with Complicated CD at Diagnosis
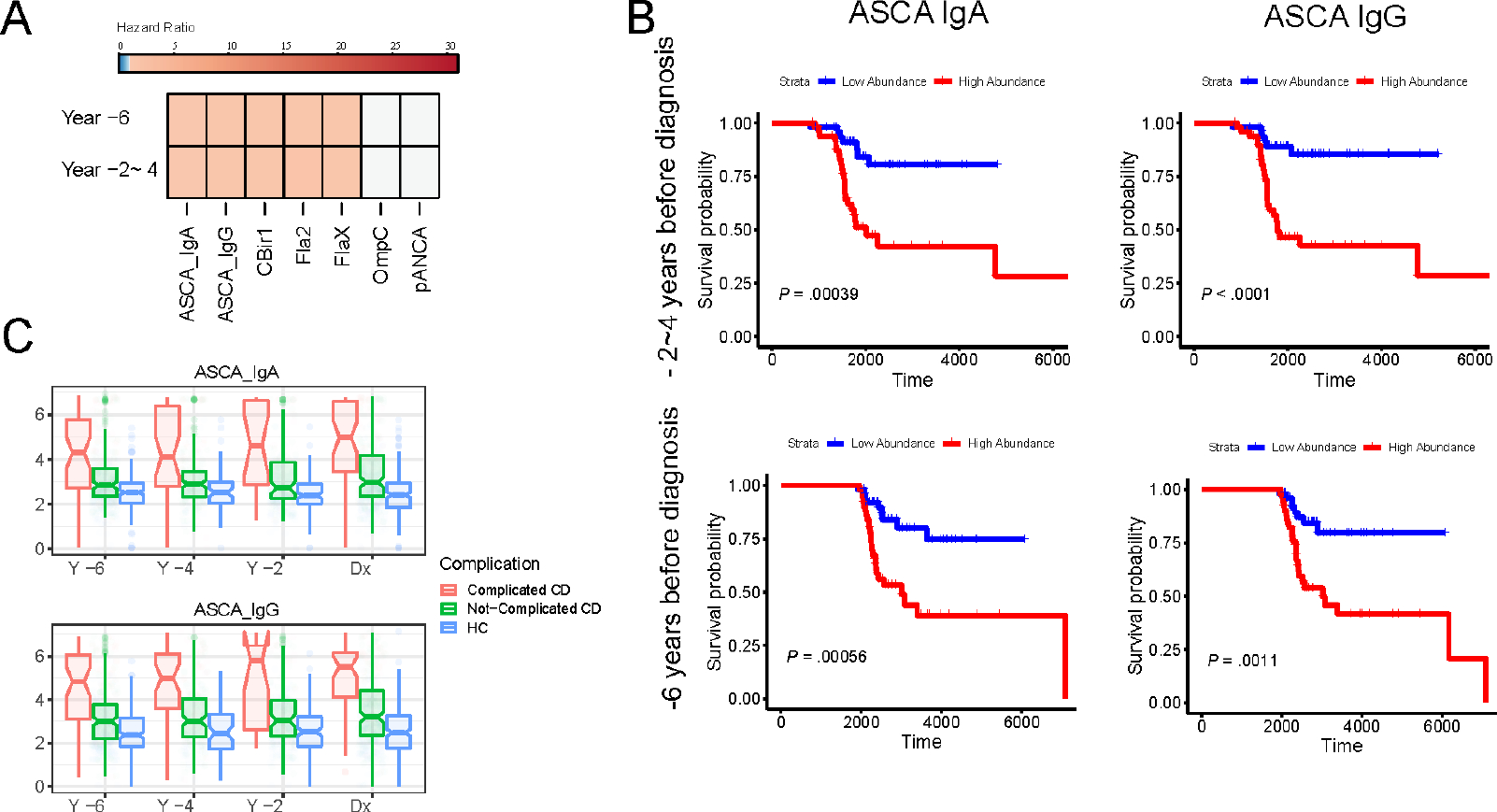
The mean concentration of all anti-microbial antibodies (except for OmpC) at all serum sample time-points before diagnosis was significantly higher in those diagnosed with complicated CD than in those diagnosed with non-complicated CD or HC (Figure 1A & Supplemental Figure 1). High anti-microbial antibodies (>75th percentile) such as ASCA-IgA were associated with an increased risk of developing complications (HR: 1.33; 95% CI: 1.13– 1.55 at 2–4 years before diagnosis and HR: 1.30; 95% CI: 1.10– 1.51 at 6 years before diagnosis) as compared to those with a low abundance of anti-microbial antibodies (<25th percentile) (Figure 1B). Anti-microbial antibodies were significantly higher in complicated cases, compared to non-complicated cases or HC (Figure 1C).
Figure 1.

(A) Heatmap of hazard ratios for serologic markers significantly associated with complications for different years before diagnosis.
(B) Kaplan-Meier curve of ASCA IgA and ASCA IgG for year 2–4 and year 6 from diagnosis.
(C) Boxplot of marker abundance for complicated Crohn’s, uncomplicated Crohn’s, and Healthy Controls for different years before diagnosis.
Proteomic Biomarkers Associated with Complicated CD at Diagnosis
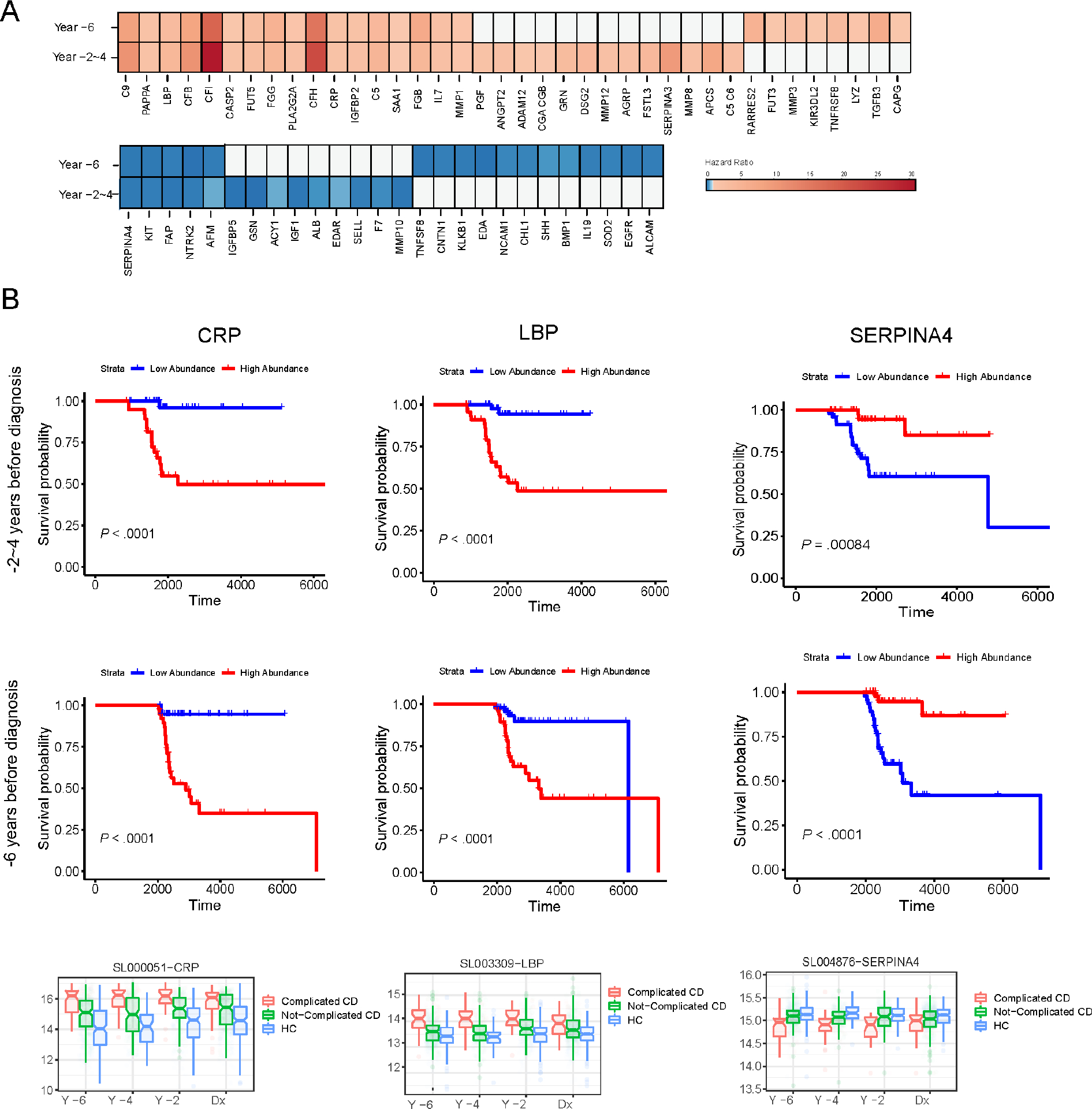
Overall, after adjusting for age and sex, 38 protein biomarkers (measured 6 or 2–4 years before diagnosis) were positively associated with the risk of developing CD complications, while 26 protein biomarkers were negatively associated with the risk of developing complications (Adjusted p-value < 10%) (Figure 2). Among these 38 protein biomarkers with a positive association, 17 were found significant at different times before diagnosis (i.e., 6 years and 2–4 years before diagnosis) (Figure 2A). Conversely, among the 26 negatively associated protein biomarkers, 5 biomarkers were found significant different times before diagnosis (i.e., 6 years and 2–4 years before diagnosis) (Figure 2A). An additional analysis was performed adjusting for disease location (L1/L3 vs. L2) in addition to sex and age, considering a subset of 167 Crohn’s disease patients (i.e., L1/L3: 128 samples, L2 disease location: 39 samples) for which disease location was available. After adjusting by age, gender, and disease location, 18 biomarkers (out of 38 protein biomarkers) were significantly associated with complicated CD (Supplemental Table 4). Association analysis results can be found in Supplemental Tables 4 & 5. The 95% confidence intervals of estimated coefficients from Cox proportional hazards regression for protein biomarkers, which remained significantly associated with complications, are provided in Supplemental Figures 4 & 5. In addition, we also performed differential analysis between L1/L3 and L2 to find markers associated with disease location. No markers were found significantly associated with disease location at 6 years before diagnosis (Adjusted p-value < 10%). At 2–4 years before diagnosis, only 6 protein biomarkers (Complement factor I, CRP, C9, Complement factor B, Serum amyloid P, Stromal cell-derived factor 1) were significantly different between patients with ileal involvement and colonic involvement (L1/L3 vs. L2). Figure 2B shows the Kaplan–Meier curves of the risk of complicated CD with high and low abundance of representative protein biomarkers (CRP, LBP, and SERPINA4). CRP abundance was associated with an increased risk of developing complications at 2–4 years and 6 years before diagnosis (Hazard ratio, 2.15 [1.50–3.08] and Hazard ratio, 2.04 [1.46–2.88], respectively); while the abundance of SERPINA4 was associated with decreased risk of complications (Hazard ratio, 0.16 [0.08,0.35] and 0.16 [0.08, 0.34], respectively). All other Kaplan-Meier curves of each protein biomarker showing the cumulative incidence of complications are shown in Supplemental Figure 2. Figure 2C shows the box plots of CRP, LBP, and SERPINA4 as examples of protein biomarkers, significantly distinct in patients with complications at diagnosis, compared to patients without complications or HC. Additional box plots of each protein biomarker are presented in Supplemental Figure 3.
Figure 2.

(A) Heatmap of hazard ratios from Cox regression model for proteomic biomarkers significantly associated with CD complications (Adj. P-value < 10%) at different years before diagnosis.
(B) Kaplan-Meier curve of CRP, LBP, and CFB for year 2–4 and year 6 from diagnosis.
(C) Boxplot of marker abundance for complicated Crohn’s, uncomplicated Crohn’s, and Healthy Controls for different years before diagnosis. Among 38 protein biomarkers positively associated with complications (Adjusted p-value < 10%), 17 biomarkers were significantly higher in serum samples pre-diagnosis (both at 2–4 years and 6 years before diagnosis) in patients with complicated CD (A). Among 26 protein biomarkers negatively associated with complications, five biomarkers were significantly and consistently lower before diagnosis in patients diagnosed with complicated CD, compared to those diagnosed with non-complicated CD. Kaplan–Meier curves or the box plots (B or C, respectively) on the risk of complicated CD among CD cases with high and low abundance of CRP, LBP, SERPINA4.
Combination of Protein Biomarkers and Anti-Microbial Antibodies Associated with Complicated CD at Diagnosis
In this section, we present results based on multivariate analysis of anti-microbial antibodies and 22 protein biomarkers, which were found associated with the development of complications at diagnosis for all time points. Figure 3 shows the Kaplan-Meier curves based on a multivariate Cox regression model considering (i) serum anti-microbial antibodies, (ii) 22 protein biomarkers, and (iii) the integration of the two sets of markers. As shown in Figure 3, proteomic markers result in better separation between KM curves than anti-microbial antibodies. A comparison of the three models based on AIC revealed that the best explanatory model of CD complications was the model based on protein biomarkers alone (Supplemental Table 6). Note that although the serologic model is the most parsimonious, it does not result in the lowest AIC since it fails to adequately model the data.
Figure 3.

Kaplan-Meier curves and corresponding 95% confidence intervals for anti-microbial antibodies-based model, SomaLogic protein biomarker-based model, and the model integrating anti-microbial antibodies and protein biomarkers. For each model, the estimated mean from Cox regression was utilized to stratify patients into high and low risk individual considering the median of the fitted mean as threshold.
Pre-Diagnostic Pathways and Network-Based Approach
Complement and coagulation cascades and innate immune system pathways were significantly enriched in the set of proteins associated with complicated CD at different time points before diagnosis (Figure 4A). Figure 4B shows the KM for the group of samples with high-pathway activity and low-pathway activity (p-value from log-rank test 0.0066 and 0.014 at year 6 before diagnosis for Innate Immune Response and Complement and Coagulation Cascade, respectively). Next, we employed a network-based approach to study the co-expression pattern across different proteins in CD patients and HC considering samples collected at the furthest year from diagnosis. For this network analysis, we integrated both SomaLogic® and anti-microbial antibodies to identify associations between the two sets of markers. Figure 4C shows the Pearson’s correlation of markers in this network module for CD patients and HC. As shown, this network module contains proteins that are positively associated with the development of complications such as CFB, C9, and CRP; and negatively associated with the development of complications such as SERPINA4. ASCA IgA and IgG were strongly correlated with protein biomarkers in patients with CD. This correlation structure was not captured in the HC (Figure 4C)
Figure 4.
(A) Pathway score for “Innate Immune Response” and “Complement and Coagulation Cascades” pathways stratified by different groups of patients corresponding to complicated Crohn’s, non-complicated Crohn’s, and healthy controls.
(B) Kaplan-Meier for samples at the furthest year of diagnosis based on the score of “Complement and coagulation” pathway and “innate immune response”. (C) Heatmap of Pearson’s correlation of proteins and serum antibodies contained in the network cluster identified based on co-expression network analysis of Crohn’s disease patients. Pearson’s correlation of protein and serum antibodies markers is shown for different groups of patients corresponding to complicated Crohn’s, non-complicated Crohn’s, and healthy controls.
Discussion
In this study, we demonstrate that complicated CD at diagnosis is associated with a distinct serologic profile compared to uncomplicated CD years before diagnosis. This profile is characterized by increased levels of ASCA-IgA and IgG, anti-flagellin antibodies, a high abundance of protein biomarkers associated with innate immunity, fibrosis, and adaptive immunity, and a low abundance of protein biomarkers related to protection against tissue damage or fibrosis (SERPINA4, FAP, KIT, NTRK2, and afamin). Moreover, using network-based analysis, we found a significant correlation of ASCA IgA/IgG with protein biomarkers related to innate immunity and lack of tissue-protective factors.
Consistent with previous findings, we confirmed the presence of higher levels of anti-microbial antibodies years before diagnosis in patients with complicated CD at diagnosis as compared with uncomplicated.9 Alexander et al.,22 also showed that patients with CD displayed a strong adaptive immune response to flagellin antigens with a subset of CD patients having multi-flagellin reactivity, which was related to a high frequency of CD complications. Together with these findings, our data suggest a preclinical aberrant adaptive immune response against gut microbiota many years before CD diagnosis which is amplified in patients with a complicated phenotype at diagnosis.
Using the SomaLogic® platform, we identified a unique set of 22 protein biomarkers associated with complicated CD at diagnosis. Among these 22 protein biomarkers associated with disease complication, 15 protein biomarkers were not found associated with disease onset in our previous study10 (Supplemental Figure 6). Additional analysis confirmed that the association between proteomic markers with CD complications was for the most part not confounded with disease location. These biomarkers have biological plausibility. Twelve out of the 22 biomarkers, like CRP, LBP, and complement proteins, are associated with the innate immune response and inflammation. Another two biomarkers (IL7 and FUT5) were also significantly increased in complicated CD. IL-7 plays a central role in B and T cell development and modulates T cell homeostasis.23 Few studies have shown that the overexpression of the IL-7 or IL-7R signaling pathway in IBD patients with an aggressive course.24, 25 FUT5 is involved in host-commensal interactions with certain bacteria, causing the upregulation of fucosylation in the intestine.26 Among the 22 protein biomarkers, 6 were related to fibrosis (3 markers [MMP1, PAPPA, IGFBP2] increased and another 3 markers [KIT, FAP, & NTRK2] decreased). MMP1 has shown to be upregulated in the areas of intestinal stenosis in patients with CD.27 PAPPA is a metalloproteinase, working as an interactive cellular mechanism promoting pulmonary fibrosis.28 In addition, IGFBP2, which is a transport protein for insulin-like growth factors, is also increased in patients with pulmonary fibrosis or systemic sclerosis.29 Regarding FAP, Corsi et al. also showed that circulating FAP (cFAP) concentration was reduced in patients with IBD, especially those undergoing surgery.30 The decreased levels of circulating FAP have also been shown to be related to organ damage and fibrosis in other diseases.31, 32 Serum protein KIT is significantly reduced in patients with hypertrophic cardiomyopathy, which is another fibrosis-related condition.33 NTRK2 is a receptor brain-derived neurotrophic factor (BDNF), and BDNF/TrKB axis activation has shown an association with lung fibrosis.34
Two biomarkers (SERPINA4 and afamin), known to be linked to protection from tissue injury or anti-inflammatory properties,35–37 were significantly decreased in the preclinical serum of patients with complications at diagnosis, compared to uncomplicated CD or HC. SERPINA4 is a unique protein, showing protective roles against tissue damage by preventing apoptosis, oxidative stress, and inflammation in several conditions like sepsis and cardiovascular diseases.35, 38 Stadnicki et al. showed that intestinal tissue SERPINA4 was significantly decreased in the inflamed intestine in patients with active IBD.39 Afamin, a vitamin E-binding protein, also plays a protective role in conditions of oxidative and inflammatory stress.36, 37
Finally, we identified two pathways linked to innate immunity and coagulation and complement cascade to be significantly up-regulated in the preclinical serum samples of patients with complications at diagnosis, compared to non-complicated CD. The network analysis showed the clustering of anti-microbial antibodies and protein biomarkers together in serum samples collected 6 years before the diagnosis of CD. Overall, our findings may suggest that perturbations in innate immune response against gut microbiota may induce the overproduction of inflammatory proteins and stimulate adaptive immunity, leading to the production of anti-microbial antibodies in complicated phenotypes. Figure 5 summarizes the potential mechanism of complications in the preclinical stage of the disease.
Figure 5.
Overview of immune dysregulation and tissue destruction in Crohn’s disease (CD). After infections or dysbiosis of gut microbiota trigger disease development, induced acute and chronic inflammation could cause epithelial damage, which consequently activates the innate immune response and complement system. Then, several cytokines are released by intestinal epithelium and innate immune cells, which subsequently activate the adaptive immune response against gut microbiota. Lack of protective cytokines against tissue damage enhances dysregulated tissue repair. Finally, chronic inflammatory reactions due to persistent interactions between host and environment could result in developing fibrosis in Crohn’s disease.
Our study has several strengths. We used preclinical samples from a well-characterized cohort, which allowed us to explore the early changes of the innate and adaptive immune response against microbiota and to discover protein biomarkers of the early host response. The unique availability of preclinical samples collected at multiple time points allowed us to examine the sequence of immunological changes and protein biomarkers that occurred before diagnosis. We also evaluated a wide array of protein biomarkers, utilizing a novel proteomic platform, and applied novel rigorous statistical approaches, which allowed us to discover the potential biomarkers and biologic pathways for the complicated phenotypes even before diagnosis.
Our study also has limitations. First, the study’s findings may not be generalizable since the study population was mainly white and male. Second, the cases and complicated phenotypes were identified based on the diagnostic and procedural codes, and not validated using medical records review. However, the case ascertainment for IBD, utilizing the diagnostic codes, has shown a high level of accuracy in similar populations.11 Third, it is possible that our findings are confounded by a diagnostic delay in CD. This is however unlikely since significant changes in biomarkers were observed 6 or more years before diagnosis.5 Finally, data on other potentially important risk factors for complications (e.g., smoking history, genetics) were not available in this cohort.
In conclusion, a complicated CD phenotype at diagnosis is associated with a specific serological profile years before diagnosis including biomarkers of innate and adaptive immunity, fibrosis, tissue damage, and amplified antibody response to commensal microorganisms. Altogether, the hypothesis can be proposed that the combination of increasing levels of inflammatory cytokines, loss of anti-inflammatory proteins, and production of anti-microbial antibodies could accelerate and magnify tissue destruction and fibrosis driving complications at diagnosis in CD. These data support the concept that complicated CD may not always be the result of progression of an uncontrolled inflammatory disease but may also be the consequence of a unique pathophysiological process. The serological signature that we identified could help to further select subjects at risk of developing complicated CD who could be preferential candidates for preventative strategies.
Supplementary Material
What You Need to Know.
Background:
Crohn’s disease (CD) has a preclinical period, similar to other autoimmune diseases like type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. However, it is unclear whether early pathophysiologic changes in preclinical serum distinguish patients with complicated versus uncomplicated CD.
Findings:
A specific preclinical serological signature, including biomarkers of innate and adaptive immunity, fibrosis, tissue damage, and amplified antibody response to commensal microorganisms, is strongly associated with complicated (stricturing or penetrating) behavior at diagnosis.
Implications for patient care:
The findings of the current study suggest that specific preclinical signatures predict the development of CD-related complications years before diagnosis, which could result in improved prevention strategies and uncover pathways important in disease progression.
Acknowledgments:
This work was prepared as part of the official duties of U.S. Government personnel and service members (Chad Porter & Mark Riddle). Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties. The opinions and assertions expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Uniformed Services University, US Navy, or the Department of Defense. The study protocols were approved by the Naval Medical Research Center Institutional Review Board in compliance with all applicable federal regulations governing the protection of human subjects.
Funding:
Support for this study was provided by Janssen Pharmaceuticals and Prometheus Laboratories under a Cooperative Research and Development Agreement entitled, “Antimicrobial Antibodies as Predictors of Inflammatory Bowel Diseases,” (CRADA number NMR-11-3920). Joseph A. Murray and Rok Seon Choung received a grant from the Crohn’s & Colitis Foundation (BMRP Grant No. 342367), and Joana Torres received support from the Sandford J. Grossman Charitable Trust. RCU supported by an NIH K23 Career Development Award (5K23DK111995). Francesca Petralia and Jean Frederic Colombel were supported by the Kenneth-Rainin Foundation (Grant #20210021).
Abbreviations used in this paper:
- AFM
Afamin
- ASCA
anti-Saccharomyces cerevisiae antibodies
- OmpC
anti-outer membrane protein C precursor
- CASP2
Caspase-2
- CRP
C-reactive protein
- CD
Crohn’s disease
- CFB
Complement factor B
- CFI
Complement factor I
- CFH
Complement factor H
- C5
Complement C5a
- C9
Complement 9
- DoDSR
Department of Defense Serum Repository
- DMSS
Department of Defense Medical surveillance system
- ELISA
Enzyme-linked immunosorbent assay
- FAP
Fibroblast activation protein α
- FGG
Fibrinogen gamma chain dimer
- FGB
d-dimer
- FUT5
Fucosyltransferase 5
- IGFBP2
Insulin like growth factor binding protein 2
- IL-7
Interleukin-7
- ICD-9
International Classification of Diseases, Ninth Revision
- IBD
Inflammatory Bowel Disease
- HC
Healthy controls
- KM
Kaplan–Meier
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KIT
Stem cell factor receptor/CD117/c-Kit
- LBP
Lipopolysaccharide-binding protein
- MMP1
Matrix metalloproteinase-1
- NTRK2
Neurotrophic tyrosine kinase receptor type 2
- PLA2G2A
Phospholipase A2 Group IIA
- PREDICTS
Proteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects
- PAPPA
Pregnancy-associated plasma protein-A
- SERPINA4
Serpina family A member 4
- SAA1
Serum amyloid A
Footnotes
Conflict of Interest:
These authors disclose the following: Joana Torres has received speaker fees from Janssen, Abbvie, Galapagos and Pfizer, and grants support from Abbvie and Janssen. RCU has served as a consultant for AbbVie, Bristol Myers Squibb, Janssen, Pfizer, and Takeda. Takahiro Sato, Shannon Telesco, and Richard Strauss are employees of Janssen Research and Development. Fred Princen is an employee of Prometheus. Scott Plevy is an employee of Protagonist Therapeutics, a past employee of Janssen, and a shareholder of Johnson & Johnson. Joseph A. Murray has received research grants from Nexpep/ImmusanT, National Institutes of Health, Immunogenix, Takeda Pharmaceutical, Allakos, ProventionBio, Oberkotter Foundation, and 9Meters, Inc.; contract (to institution) from Kanyos Bio (a wholly owned subsidiary of Anokion); and consultancy fees from Johnson and Johnson, Bristol Myers Squibb, Intrexon Corporation, Dren Bio, Neoleukin, Reistone pharma, Immunic Therapeutics, Senda Biosciences, Brightseed Bio, Chugai Pharma, Alimentiv, Equillium, Ukko, Vial Health Technologies and has received royalties from Torax Medical and Evelo. Jean Frédéric Colombel reports receiving research grants from AbbVie, Janssen Pharmaceuticals, and Takeda; receiving payment for lectures from AbbVie, Amgen, Allergan Inc, Ferring Pharmaceuticals, Shire, and Takeda; receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Celgene Corporation, Celltrion, Eli Lilly, Enterome, Ferring Pharmaceuticals, Genentech, Janssen Pharmaceuticals, Landos, Ipsen, Medimmune, Merck, Novartis, Pfizer, Shire, Takeda, and TiGenix; and holding stock options in Intestinal Biotech Development and Genfit. A patent application named, “A panel of biomarkers for inflammatory bowel disease and uses thereof,” has been filed between NRMC, Mount Sinai, Janssen Research and Development, and Prometheus (JBI5154USPSP). The remaining authors disclose no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loftus EV Jr., Silverstein MD, Sandborn WJ, et al. Crohn’s disease in Olmsted County, Minnesota, 1940–1993: incidence, prevalence, and survival. Gastroenterology 1998;114:1161–8. [DOI] [PubMed] [Google Scholar]
- 2.Aniwan S, Park SH, Loftus EV Jr. Epidemiology, Natural History, and Risk Stratification of Crohn’s Disease. Gastroenterol Clin North Am 2017;46:463–480. [DOI] [PubMed] [Google Scholar]
- 3.Peyrin-Biroulet L, Loftus EV Jr., Colombel JF, et al. The natural history of adult Crohn’s disease in population-based cohorts. Am J Gastroenterol 2010;105:289–97. [DOI] [PubMed] [Google Scholar]
- 4.Thia KT, Sandborn WJ, Harmsen WS, et al. Risk factors associated with progression to intestinal complications of Crohn’s disease in a population-based cohort. Gastroenterology 2010;139:1147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Lago I, Zabana Y, Barreiro-de Acosta M. Diagnosis and natural history of preclinical and early inflammatory bowel disease. Ann Gastroenterol 2020;33:443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Israeli E, Grotto I, Gilburd B, et al. Anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic antibodies as predictors of inflammatory bowel disease. Gut 2005;54:1232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Schaik FD, Oldenburg B, Hart AR, et al. Serological markers predict inflammatory bowel disease years before the diagnosis. Gut 2013;62:683–8. [DOI] [PubMed] [Google Scholar]
- 8.Lochhead P, Khalili H, Ananthakrishnan AN, et al. Association Between Circulating Levels of C-Reactive Protein and Interleukin-6 and Risk of Inflammatory Bowel Disease. Clin Gastroenterol Hepatol 2016;14:818–824 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choung RS, Princen F, Stockfisch TP, et al. Serologic microbial associated markers can predict Crohn’s disease behaviour years before disease diagnosis. Aliment Pharmacol Ther 2016;43:1300–10. [DOI] [PubMed] [Google Scholar]
- 10.Torres J, Petralia F, Sato T, et al. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology 2020;159:96–104. [DOI] [PubMed] [Google Scholar]
- 11.Porter CK, Riddle MS, Gutierrez RL, et al. Cohort profile of the PRoteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects (PREDICTS) study: Rationale, organization, design, and baseline characteristics. Contemp Clin Trials Commun 2019;14:100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubertone MV, Brundage JF. The Defense Medical Surveillance System and the Department of Defense serum repository: glimpses of the future of public health surveillance. Am J Public Health 2002;92:1900–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology 2002;123:689–99. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira JA, Zwinderman AH. On the Benjamini–Hochberg method. The Annals of Statistics 2006;34:1827–1849, 23. [Google Scholar]
- 15.Bozdogan H. Akaike’s Information Criterion and Recent Developments in Information Complexity. J Math Psychol 2000;44:62–91. [DOI] [PubMed] [Google Scholar]
- 16.Therneau TM. A Package for Survival Analysis in R. R package version 3.5–0 https://CRAN.R-project.org/package=survival, 2023. [Google Scholar]
- 17.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, NY: Springer, 2000. [Google Scholar]
- 18.Kanehisa M. The KEGG database. Novartis Found Symp 2002;247:91–101; discussion 101–3, 119–28, 244–52. [PubMed] [Google Scholar]
- 19.Fabregat A, Jupe S, Matthews L, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018;46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petralia F, Song WM, Tu Z, et al. New Method for Joint Network Analysis Reveals Common and Different Coexpression Patterns among Genes and Proteins in Breast Cancer. J Proteome Res 2016;15:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csárdi G, Nepusz T. The igraph software package for complex network research, 2006.
- 22.Alexander KL, Zhao Q, Reif M, et al. Human Microbiota Flagellins Drive Adaptive Immune Responses in Crohn’s Disease. Gastroenterology 2021;161:522–535 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooms H. Interleukin-7: Fuel for the autoimmune attack. J Autoimmun 2013;45:40–8. [DOI] [PubMed] [Google Scholar]
- 24.Belarif L, Danger R, Kermarrec L, et al. IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J Clin Invest 2019;129:1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Hsu HC, Mountz JD, et al. Unmasking Fucosylation: from Cell Adhesion to Immune System Regulation and Diseases. Cell Chem Biol 2018;25:499–512. [DOI] [PubMed] [Google Scholar]
- 27.Warnaar N, Hofker HS, Maathuis MH, et al. Matrix metalloproteinases as profibrotic factors in terminal ileum in Crohn’s disease. Inflamm Bowel Dis 2006;12:863–9. [DOI] [PubMed] [Google Scholar]
- 28.Bale LK, Schafer MJ, Atkinson EJ, et al. Pregnancy-associated plasma protein-A (PAPP-A) is a key component of an interactive cellular mechanism promoting pulmonary fibrosis. J Cell Physiol 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guiot J, Bondue B, Henket M, et al. Raised serum levels of IGFBP-1 and IGFBP-2 in idiopathic pulmonary fibrosis. BMC Pulm Med 2016;16:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corsi F, Sorrentino L, Albasini S, et al. Circulating Fibroblast Activation Protein as Potential Biomarker in Patients With Inflammatory Bowel Disease. Frontiers in Medicine 2021;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uitte de Willige S, Keane FM, Bowen DG, et al. Circulating fibroblast activation protein activity and antigen levels correlate strongly when measured in liver disease and coronary heart disease. PLoS One 2017;12:e0178987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillmanns J, Fraccarollo D, Galuppo P, et al. Changes in concentrations of circulating fibroblast activation protein alpha are associated with myocardial damage in patients with acute ST-elevation MI. Int J Cardiol 2017;232:155–159. [DOI] [PubMed] [Google Scholar]
- 33.Sonnenschein K, Fiedler J, de Gonzalo-Calvo D, et al. Blood-based protein profiling identifies serum protein c-KIT as a novel biomarker for hypertrophic cardiomyopathy. Sci Rep 2021;11:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherubini E, Mariotta S, Scozzi D, et al. BDNF/TrkB axis activation promotes epithelial-mesenchymal transition in idiopathic pulmonary fibrosis. J Transl Med 2017;15:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin WC, Chen CW, Huang YW, et al. Kallistatin protects against sepsis-related acute lung injury via inhibiting inflammation and apoptosis. Sci Rep 2015;5:12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koninger A, Mathan A, Mach P, et al. Is Afamin a novel biomarker for gestational diabetes mellitus? A pilot study. Reprod Biol Endocrinol 2018;16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polkowska A, Pasierowska IE, Paslawska M, et al. Assessment of Serum Concentrations of Adropin, Afamin, and Neudesin in Children with Type 1 Diabetes. Biomed Res Int 2019;2019:6128410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li PF, Bledsoe G, Yang ZR, et al. Human kallistatin administration reduces organ injury and improves survival in a mouse model of polymicrobial sepsis. Immunology 2014;142:216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadnicki A, Mazurek U, Plewka D, et al. Intestinal tissue kallikrein-kallistatin profile in inflammatory bowel disease. Int Immunopharmacol 2003;3:939–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


