Abstract
In sub-Saharan Africa, where the effects of human immunodeficiency virus type 1 (HIV-1) have been most devastating, there are multiple subtypes of this virus. The distribution of different subtypes within African populations is generally not linked to particular risk behaviors. Thus, Africa is an ideal setting in which to examine the diversity and mixing of viruses from different subtypes on a population basis. In this setting, it is also possible to address whether infection with a particular subtype is associated with differences in disease stage. To address these questions, we analyzed the HIV-1 subtype, plasma viral loads, and CD4 lymphocyte levels in 320 women from Nairobi, Kenya. Subtype was determined by a combination of heteroduplex mobility assays and sequence analyses of envelope genes, using geographically diverse subtype reference sequences as well as envelope sequences of known subtype from Kenya. The distribution of subtypes in this population was as follows: subtype A, 225 (70.3%); subtype D, 65 (20.5%); subtype C, 22 (6.9%); and subtype G, 1 (0.3%). Intersubtype recombinant envelope genes were detected in 2.2% of the sequences analyzed. Given that the sequences analyzed represented only a small fraction of the proviral genome, this suggests that intersubtype recombinant viral genomes may be very common in Kenya and in other parts of Africa where there are multiple subtypes. The plasma viral RNA levels were highest in women infected with subtype C virus, and women infected with subtype C virus had significantly lower CD4 lymphocyte levels than women infected with the other subtypes. Together, these data suggest that women in Kenya who are infected with subtype C viruses are at more advanced stages of immunosuppression than women infected with subtype A or D. There are at least two models to explain the data from this cross-sectional study; one is that infection with subtype C is associated with a more rapid disease progression, and the second is that subtype C represents an older epidemic in Kenya. Discriminating between these possibilities in a longitudinal study will be important for increasing our understanding of the role of specific subtypes in the transmission and pathogenesis of HIV-1.
The genetic diversity and rapid variation of human immunodeficiency virus type 1 (HIV-1) continues to complicate the development of effective vaccines to limit the AIDS pandemic. While there are regional clusters of more closely related HIV-1 variants, viral strains that originate from different continents show more significant genetic differences. Group M represents the main group of HIV-1 strains; in addition, there is an outgroup of HIV-1 variants (group O) whose envelope sequences have diverged from those of the group M viruses by approximately 50% (52). The group M viruses have been further subdivided into subtypes, or clades, A through J, whose envelope gene sequences differ from each other by as much as 30 to 35% (52). Intersubtype recombinant genomes have also been identified (3, 4, 11, 12, 23, 28, 47, 48, 50), and this has complicated the analysis of HIV-1 genetic diversity.
Many of the first HIV-1 genomes to be analyzed at both the molecular and phenotypic levels were subtype B variants from the United States and Europe. Thus, much of our understanding of the biology and pathogenesis of HIV-1 comes from analyses of clade B viruses (2). It remains unclear whether the subtypes, which are defined purely on the basis of sequence similarity, also define groups with any biological or immunological differences. As a result, it is also unclear whether the development of effective vaccines will require the use of geographically and/or subtype-specific vaccine strains. Over the past few years, more and more viral strains from throughout the world have been characterized (35). However, the majority of these studies have focused on a relatively small number of viral isolates, in some cases repetitive isolates from a single individual. Thus, the true extent of viral diversity within a particular HIV-1-infected endemic population has rarely been examined in detail.
Representatives of most of the group M HIV-1 subtypes have been found in sub-Saharan Africa (2, 15), where the global impact of HIV-1 infection and disease has been most apparent (55). It is estimated that 70% of individuals infected with HIV-1 reside in sub-Saharan Africa and that new infections are occurring there at a rate of 4 million per year, including 0.5 million new infections per year in infants (55). The distribution of different subtypes within African populations is usually not linked to particular risk behaviors. In contrast, different subtypes are clustered within distinct risk groups in some Asian countries (8, 22, 42, 56). Thus, Africa is an ideal setting in which to examine in more detail the diversity and mixing of viruses of different subtypes on a population basis.
In Kenya, HIV-1 infection was first reported in the mid-1980s in a group of Nairobi sex workers (21, 38). Of the African countries, Kenya has one of the highest prevalences of HIV-1 (32), and the epidemic has included the spread of at least two group M HIV-1 subtypes (17, 26, 45, 57). To gain a comprehensive picture of HIV-1 diversity in Kenya, we have determined HIV-1 subtypes by using proviral envelope gene sequences from 320 seropositive Nairobi women who were participants in a breast-feeding transmission study of HIV-1 (19, 36). Subtype A, C, D, and G genomes were identified, as well as several recombinant envelope gene sequences that are distinct from previously described HIV-1 group M or O genomes. Among these subjects, plasma RNA levels and CD4 lymphocyte counts were measured at the same time to determine whether there were differences in disease stage among persons infected with different subtypes.
MATERIALS AND METHODS
Study population.
The 347 women who provided blood samples for this study were participants in a randomized clinical trial of breast and formula feeding to determine the frequency of breast milk transmission of HIV-1 in Nairobi, Kenya (19, 36). After giving informed consent, the women were tested for HIV-1-specific antibodies, typically during their third trimester. Sera were tested by the use of a peptide enzyme-linked immunosorbent assay (Behring, Ausgabe, Germany), and samples that were reactive were confirmed by using a second enzyme-linked immunosorbent assay (Cambridge Biotech, Rockville, Md.). Each HIV-1-seropositive woman who enrolled in the trial underwent a baseline clinical evaluation, including determination of CD4 and CD8 lymphocyte counts by flow-cytometric analysis using specific monoclonal antibodies (Becton Dickinson). The blood samples were separated into cellular and plasma fractions by standard methods, and the samples were stored in liquid nitrogen and a −70°C freezer, respectively, for subsequent virological analyses.
PCR amplification.
Subjects’ peripheral blood mononuclear cell (PBMC) samples, which were isolated from approximately 5 to 10 ml of blood, were lysed in the presence of proteinase K as described previously (34). Typically, 2 μl of the total 100-μl volume of lysate was used in a PCR. Primers and reaction conditions that have been used extensively in our previous studies to amplify envelope sequences from Kenyan subjects were used in the first-round PCR (44, 46). These primers (env13 [5′-CCA CTC TAT TTT GTG CAT CAG A-3′] and env12 [5′-CCT GGT GGG TGC TAC TCC TA-3′]) amplify a 1.2-kb fragment encoding V1 through V5 of the extracellular envelope glycoprotein. This first-round product was used for all subsequent second-round PCRs.
Two different primer pairs were used to amplify a fragment spanning V1 through C3 in a second-round PCR. For the first 35 samples analyzed, primers designed by Delwart et al. (5, 6) ED5 (5′-ATG GGA TCA AAG CCT AAA GCC ATG TG-3′) and ED33 (5′-TTA CAG TAG AAA AAT TCC CCT C-3′), were used. These primers would be predicted to amplify a 824-bp fragment from an African-derived clade A proviral clone (U455 [41]). However, the amount of product obtained from Kenyan samples with these primers was frequently suboptimal. Therefore, we designed other primers for amplification of V1 through C3 sequences and examined their ability to amplify HIV-1 sequences from Kenyan samples. A primer, designated env22 (5′-GTG TTG TAA TTT CTA GAT CCC CTC CTG-3′), which binds 37 bases 5′ to ED33, was chosen for use with ED5 to amplify the remaining samples. The predicted product from this amplification is 787 bp (on a U455 proviral template). Two microliters of round one product was used as a template for the round two amplification. Amplification conditions for both primer pairs were as follows: 1 cycle of 94°C for 5 min; 3 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and 32 cycles of 94°C for 15 s, 55°C for 45 s, and 72°C for 1 min.
In the round two PCR, a smaller fragment, spanning C2 through C3, was amplified from the env12-env13 first-round product with four primers—two that have been used previously (5, 6) for subtyping by heteroduplex mobility analysis (HMA) (ED31 [5′-CCT CAG CCA TTA CAC AGG CCT GTC CAA AG-3′] and ED33), and two derivatives of these primers that included several single-base differences (in boldface) more commonly found in the subtype consensus sequences (35) (env69 [5′-CCT CAG CCA TTA CAC AGG CTT GTC CAA AG-3′] and env70 [5′-TTG CAA TAG AAA AAT TCT CCT C-3′]). This combination of primers resulted in a more uniform amplification of the C2-C3 product from the samples analyzed in this study. The product size was predicted to be 564 bases on a U455 clade A sequence template.
For all round two PCRs, 5 μl of product was subjected to agarose gel electrophoresis. Sample lysates that did not yield a product were tested with both larger (up to 10 μl) and smaller amounts of template in the round one reaction so as to increase the proviral copy number and decrease potentially inhibitory blood contaminants, respectively. Samples that were PCR negative were tested with all possible primer combinations to determine if envelope sequences could be amplified. HIV-1 gag-specific primers (34), which amplify a 142-bp product, were also used to detect HIV-1 sequences in samples that tested negative with envelope-specific primers.
HMA.
The protocol for subtype analysis was based on an HIV-1 envelope gene HMA method described previously (5–7). However, in order to maximize and streamline our ability to determine Kenyan HIV-1 subtypes, modifications were made to this protocol as we accumulated subtype information from our cohort. Specifically, envelope sequences from known Kenyan clade A and D sequences were added to the reference strain panel when they became available (45), as described below.
HMA was performed as described elsewhere (6). To determine which bands in the HMA were the result of intrapatient viral duplexes, envelope diversity within the subject’s sample was also examined, in parallel, by carrying out all of the HMA steps in the absence of a reference strain. In cases in which there was extensive heterogeneity in this control reaction, the first-round PCR was repeated with smaller amounts of template so that the HMA pattern of the subject’s viral quasispecies did not complicate the interpretation of heteroduplexes in the presence of reference strains.
In the primary screen, standard subtype reference strains were used, and plasmids encoding these envelope sequences as well as detailed protocols for HMA were generously provided by the National Institutes of Health AIDS Research and Reference Research Program (6). Because of the expected subtypes in this population (45), samples were first subjected to a primary screen using two subtype A (SF170 [Rwanda] and IC144 [Ivory Coast]), two subtype D (UG21 [Uganda] and UG46 [Uganda]), and two subtype C (ZM18 [Zambia] and MA959 [Malawi]) references (35). A subject’s viral envelope sequence was assigned to a particular subtype if the heteroduplex that it formed with each of the two reference strains for that subtype migrated with more rapid mobility than all other heteroduplexes and at least as far as the single-stranded DNA form.
For samples that could not be characterized by the use of the subtype reference sequences, a secondary screen, using Kenyan reference sequences that had previously been analyzed by nucleotide sequence and phylogenetic methods, was performed. The Kenyan envelope sequences included two subtype A (Q23 and Q06) and two subtype D (T16 and Y61) clones (45) (clade C variants from Kenya were not available). Because of the encouraging results obtained when the Kenyan sequences were used as reference strains, 59 samples were tested directly with these sequences, bypassing the primary screen. This approach also allowed us to determine the efficacy of using these Kenyan strains as a primary screen to predict subtype.
If the subtype could not be clearly assigned after testing V1 through C3 sequences of samples with both subtype reference and Kenyan reference sequences, a smaller fragment, excluding the V1 and V2 regions, was analyzed. This C2-C3 product was tested against the six subtype reference sequences used in the primary screenings (SF170, IC144, UG21, UG46, ZM18, and MA959). This third assay had a tendency to exaggerate migration trends seen in previous tests, and lane-by-lane heteroduplex band positions were compared with data from the first two assays to define subtypes. If the results of this third, C2-C3 HMA analysis were either inconsistent with those of the analysis of V1 through C3 or otherwise inconclusive, a segment of the envelope gene of this viral isolate was sequenced.
Cloning and sequence analyses.
The 1.2-kb envelope fragment spanning V1 through V5 was amplified and cloned into M13 by methods described previously (44, 46). For each subject, two to four clones were obtained from at least two independent first-round PCRs. The sequences of both strands were determined by using an ABI 377 automated sequencer (Applied Biosystems).
Phylogenetic and recombinant analyses.
A multiple alignment of the 16 new envelope sequences from Kenya with several reference HIV-1 sequences of each subtype was generated. For 15 of the 16 Kenyan sequences, sequences encompassing C2 through V5 were analyzed. For one envelope gene (MM2227), the clone included only V1 through V3 sequences, and so this sequence was analyzed in a separate phylogenetic analysis. Gaps which had to be introduced to create the alignment were eliminated in the analysis. Reference isolates of subtype A (Q23 from Kenya, UG037 from Uganda, DJ263 from Djibouti, and IbNG from Nigeria), subtype B (MN and SF2 from the United States), subtype C (C2220 from Ethiopia, BR025 from Brazil, UG268 from Uganda, and SM145 from Somalia), subtype D (MB2059 and TK1316 from Kenya, UG274 and UG114 from Uganda, and NDK and ELI from Zaire), subtype E (CM240 from Thailand and CAR402 from Central African Republic), subtype F (F9363 from Zaire and BZ163 from Brazil), G (HH8793 from Kenya, NG083 from Nigeria, and SE6165 from Congo), H (V1991 from Zaire and CF056 from Central African Republic), and J (SE9280 and SE9173 from Zaire) were used (35). The multiple alignment was then broken into overlapping segments of equal length, and each segment was analyzed separately. Briefly, phylogenetic trees were constructed and the consistency of branching order was evaluated by using the SEQBOOT, DNADIST, NEIGHBOR, CONSENSE, and DNAPARS modules of the Phylip package (V3.52c) (10) and TREETOOL (27). Bootscanning, an analytical approach that tracks the bootstrap value of the node joining an unknown sequence with known sequences progressively across the genome, was used to identify putative recombination breakpoints (51). A bootstrap value equal to or greater than 70% was considered definitive (14). In addition, distance scanning (3) was performed by computing the genetic distances between isolates, using the technique of maximum likelihood with a transition-transversion ratio of 2.0, and the distances between the unknown isolate and isolates of known subtypes were evaluated for each segment. These distances were normalized within each fragment in order to aid comparability across the genome. Using these two scanning techniques, breakpoints were identified, and the segments were then used in separate phylogenetic analyses to confirm the subtype origin of the segment. Each segment was analyzed by building a phylogenetic tree via the neighbor-joining method (49), and the stability of the nodes was assessed by using maximum parsimony (10, 53) with the bootstrap value (9).
Plasma RNA methods.
To determine the viral loads of plasma samples, we used a quantitative assay for HIV-1 RNA which is under development at Gen-Probe Incorporated. This method utilizes an integrated approach in which sample preparation, amplification, and detection are performed in a single tube. The assay protocol includes three hybridization-based procedures: (i) target capture and magnetic microparticle-based sample preparation, (ii) amplification of viral sequences by transcription-mediated amplification (TMA), and (iii) detection of the amplicon by use of the hybridization protection assay. This integrated approach is highly effective and allows processing of 200 samples in less than 6 h (13). The specimen processing method releases and stabilizes the viral RNA, which is then captured with oligonucleotides that contain sequences complementary to the viral RNA and poly(dA) tails complementary to poly(T) tails on a magnetic particle. The use of magnetic racks allows washing of the magnetic particles and elimination of unwanted clinical specimen. Amplification of HIV-1-specific viral sequences is performed by TMA (29). TMA is an exponential isothermal reaction that utilizes reverse transcriptase and T7 RNA polymerase. Briefly, the reaction is initiated by the annealing of a chimeric primer that contains a T7 polymerase promoter coupled to an HIV-1-specific primer, which serves to prime DNA synthesis via reverse transcriptase. The T7 primer is extended by reverse transcriptase to form an RNA-DNA duplex, and the RNase H activity of the reverse transcriptase degrades the RNA in this duplex. A second HIV-1-specific primer is used to create a double-stranded DNA copy of the target RNA. The DNA, which is engineered to include 5′ promoter sequences, then serves as a template for RNA synthesis by T7 polymerase. The RNA product is then subjected to the same cycle of DNA synthesis and RNA amplification, leading to a greater than 109-fold amplification of the specific target nucleic acid. The amplified product is detected by using chemiluminescence-labeled oligonucleotide probes in a homogeneous hybridization assay, the hybridization protection assay (37). Results (in relative light units) from each sample are converted to copies per milliliter of HIV-1 virus by interpolation against an external standard curve run at the same time as the samples. The current Gen-Probe quantitative assay has a dynamic range of 50 to 100,000 copies/ml and between-run coefficients of variation of less than 20% down to 500 copies/ml. The use of capture oligonucleotides, primers, and detection probes targeted to conserved regions of the pol gene of HIV-1 allows detection and quantification of all HIV-1 viral subtypes, including group O strains, with similar efficiencies.
Plasma samples were obtained from heparinized blood and stored at −70°C. Two hundred microliters of plasma was diluted with 800 μl of a negative plasma matrix (Boston Biomedica, Inc.), and 500 μl of the diluted plasma was tested in duplicate by using the Gen-Probe HIV-1 viral load assay. The viral load was calculated from the duplicate tests if they met the following criteria: the values agreed within threefold, and the average of the two values fell between 100 and 100,000 copies. Samples that were below or above the limits of detection were retested undiluted or at a 1:50 dilution, respectively, and the same criteria were applied to the results of duplicate tests at these dilutions. Although a threefold (half-log) difference between duplicate tests was allowed, 99% of duplicate tests of the same sample agreed within twofold.
Statistical methods.
Median CD4 cell counts and plasma viral loads of subtypes were compared by nonparametric statistical tests. Comparisons were conducted between each subtype and other subtypes, with the latter being considered both as a grouped and as a separate variable (e.g., for subtype A, A versus non-A, A versus D, and A versus C). Plasma viral load was divided into quartiles (<10,000, 10,000 to 42,000, 42,000 to 162,000, and >162,000). Women with viral loads in the highest and lowest quartiles were compared with the remainder of the cohort by the use of nonparametric statistical tests for continuous variables (CD4 cell counts) and chi-square tests for dichotomous variables (subtypes).
Nucleotide sequence accession numbers.
The 16 Kenyan envelope sequences described here are available under GenBank accession no. AF101456 to AF101471. The GenBank accession numbers for Q23, TK1316, and 2059 are AF004885, AF133822, and AF133821, respectively.
RESULTS
PCR amplification of sequences from PBMC samples.
A total of 347 PBMC samples from pregnant women participating in a breast feeding transmission study of HIV-1 in Nairobi were analyzed. The envelope sequences of 23 samples from subjects identified serologically as being HIV-1 positive could not be amplified. In 19 of these cases, a 142-bp HIV-1 gag sequence was amplified, which suggested that the sample contained HIV-1 proviruses. We were unable to determine why envelope sequences, from these samples could not be amplified, but possible explanations include partially degraded or very small amounts of PBMC sample, a very low proviral copy number, and inefficient amplification due to extensive sequence divergence from the primers. For four additional samples, the amount of envelope PCR product was not sufficient for HMA analysis. For the purposes of this report, these 4 samples, along with the 23 amplification-negative samples, were not considered further, and the analyses include only the 320 samples whose subtypes were determined.
Characterization of viral subtype by HMA of envelope sequences.
The envelope sequences amplified from the 320 PBMC samples were analyzed by HMA against reference strains of known subtype, using a sequential approach described in Materials and Methods. In the primary screen, a fragment encompassing V1 through C3 was analyzed against clade A, D, and C subtype reference strains, and the subtypes of 167 (64%) of 261 samples, including 123 subtype A, 32 subtype D, and 12 subtype C, were determined by this approach. However, the subtypes of the remaining 94 samples could not be assigned with these six reference strains by using V1 through C3 sequences.
The HIV-1 subtype reference sequences used in the original HMAs provide a broad representation of HIV-1 envelope sequences, but reference sequences from the geographic location of interest are more ideal for subtype analysis by HMA (6, 25). A total of 144 uncharacterized samples were tested, using V1 through C3 sequences, against four Kenyan sequences that had previously been characterized as clade A (Q23 and Q06) or clade D (T16 and T61) by phylogenetic methods (45). The samples tested included 94 envelope genes which could not be assigned to specific subtypes in the primary screen, and of these 78 (83%), including 57 clade A and 11 clade D sequences, were assigned subtypes by using the Kenyan clade A and D sequences. In this secondary screen, there may have been some sample bias toward those sequences that were refractory to HMA because of size variation or unusual sequence structure, since these 94 viruses had already failed one HMA. To examine whether initial screening of a Kenyan virus of unknown subtype with the Kenyan subtype A and D sequences would be more productive than screening with the reference panel, we analyzed an additional 59 samples directly with the Kenyan panel, bypassing the primary screen. To validate this approach, we first performed a secondary screen with 10 samples that yielded subtype information in the primary screen. In all cases (seven A and three D), concordant results were obtained. The subtypes of 49 (83%) of the 59 samples analyzed by initially screening with the four Kenyan strains were determined.
For the remaining 39 samples that could not be typed using V1 through C3 sequences, a smaller second-round PCR product was generated, spanning sequences from C2 through C3, and this was analyzed by HMA against the subtype reference strains. Twenty-three (14 subtype A, 6 subtype D, and 3 subtype C) envelope genes were characterized by using this smaller fragment.
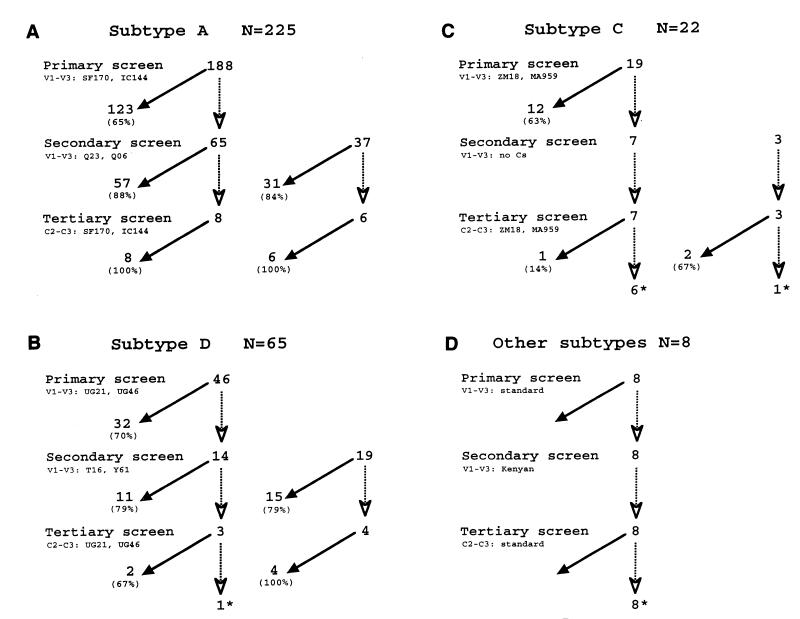
Figure 1 summarizes the detailed results of this approach from the perspective of the actual subtype of the virus in the sample. Analyses using the Kenyan reference strains were superior in predicting subtype, even if the samples were first tested, and found to be ambiguous, using the standard subtype reference strains. For example, only 65% of the subtype A viruses in this study could be identified by using the standard subtype references, which were from Rwanda and Ivory Coast, while 88% of the remaining samples were characterized with the Kenyan panel (Fig. 1A). In 84% of the cases in which subtype A samples were first tested with the Kenyan strains, the subtype was defined. The Kenyan subtype D reference strain also performed better overall than the standard subtype D reference sequences, which were both from Uganda (Fig. 1B). Only 15 of 22 subtype C variants were characterized by HMA with envelope sequences from Malawi and Zambia (Fig. 1C). It is likely that our ability to assign subtype C variants in our cohort by HMA was hampered by the lack of Kenyan subtype C reference strains. Eight viruses representing other subtypes (see below) could not be characterized by our HMA approach (Fig. 1D). Overall, using this sequential HMA approach, the subtypes of 304 of 320 samples were assigned. Two hundred and twenty-five samples were classified as subtype A, 64 were determined to be subtype D, and 15 were found to be subtype C. The remaining 16 could not be assigned a subtype with an acceptable degree of confidence by this method.
FIG. 1.
Summary flowchart of subtype analysis based on the eventual subtype identified in the sample. The different steps in this analyses included a primary screen using V1 through V3 sequences versus standard reference strains from the AIDS Research and Reference Reagent Program, a secondary screen using V1 through V3 sequences against Kenyan reference strains, and a tertiary screen using a C2-C3 fragment against the standard reference strains, as described in Materials and Methods. The envelope sequences used for HMA are indicated for each subtype, and the number of samples tested against these reference sequences is shown in the flowchart. Samples that were successfully typed at each stage are indicated at the base of a closed arrow, and samples that could not be typed with the indicated sequences are indicated following a dotted line at the base of an open arrowhead. The latter were then tested with the next screen, and the results of this screen are similarly displayed. In cases in which samples were not analyzed with the primary screen, the flowchart begins at the secondary screen, and these samples are shown separately to the right. The percentages that were typed or not typed at each screen are indicated in parentheses. Viral envelope genes that could not be assigned subtypes based on the HMA sequential screening method and required sequence analysis are indicated with asterisks. The results are for subtypes A (A), D (B), and C (C) and recombinants or subtype G (D).
Sequence analysis of unusual envelope genes.
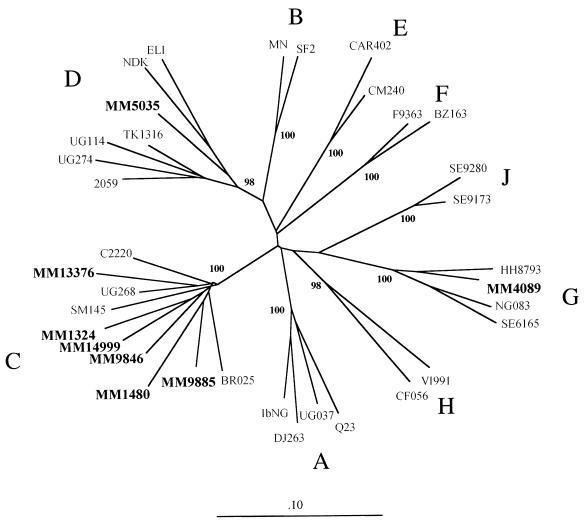
The envelope gene fragments of 16 proviral genomes that could not be assigned a subtype by the HMA approach were amplified and cloned. Two to four envelope clones from at least two PCRs were obtained from the PBMCs from each subject. The sequences spanning V1 through V5 from each of the 16 subjects were compared to a panel of HIV-1 subtype sequences by phylogenetic methods (Fig. 2). The sequences from an individual clustered closely together in the analysis (data not shown), indicating that the ambiguity in the HMA was not likely to be the result of extensive intrapatient viral sequence heterogeneity. For convenience, only one subject’s sequence is represented in Fig. 2. The known-subtype reference sequences included in this analysis are representative of subtypes A through J (sequences for subtype I were not available). Seven of the Kenyan envelope genes (MM13376, MM9846, MM14999, MM1324, MM1480, MM9885, and MM2227 grouped with the clade C viruses (Fig. 2 and data not shown). Interestingly, one envelope gene (MM4089) clearly grouped with type G HIV-1; this is the only example of a subtype G variant in our cohort. One sequence (MM5035) grouped with clade D. MM5035 was unusual relative to other subtype D viruses from Kenya that we have examined because it grouped midway between the Zairian variants and the East African viruses. A more detailed analysis of this envelope sequence was performed, but the precise structure could not be resolved (data not shown). Seven sequences were outliers that showed weak associations with either subtype A, C, D, or G but were clearly distinct (data not shown).
FIG. 2.
Phylogenetic tree of HIV-1 envelope sequences. Phylogenetic analysis was performed as described in Materials and Methods, using C2 through V5 sequences and the subtype references sequences indicated. The number at each node indicates the percentage of bootstrap support as determined from 100 bootstrap resamplings and maximum-parsimony analysis. The phylogenetic tree was built by neighbor-joining analysis. The sequences from this cohort are shown in boldface.
Detection of subtype recombinants.
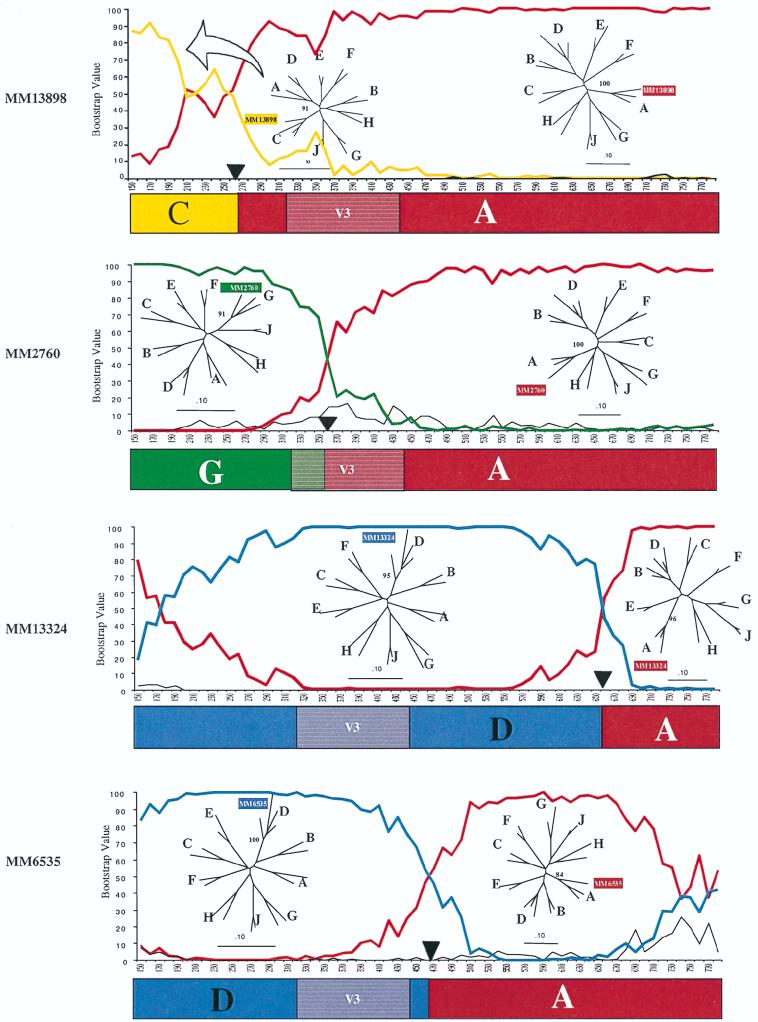
The sequences of the seven outlier envelope genes were subjected to additional analyses, using a bootscan method designed to characterize subtype recombinants of HIV-1 (51). By using this method, segments of sequence that were highly related to known subtypes (A, C, D, and G) were defined, although other sequences were not clearly derived from any HIV-1 subtype A to J group. The bootscan analysis results for the four sequences derived from known subtypes are presented in Fig. 3. All four of these recombinants had subtype A-derived V4-V5 sequences, and two of these also had subtype A V3 sequences. Each viral envelope gene is highly distinct, with different apparent sites of recombination. In particular, there are examples among these clones of 5′ sequences that originated from each of the other subtypes found in the Nairobi cohort (clades C, D, and G). Thus, each of these recombinants has a unique origin.
FIG. 3.
Recombination analysis: bootscans. Bootscans were performed as described in Materials and Methods, using subtype A (Q23), subtype C (SM145), subtype D (UG274), and subtype G (HH8793) HIV-1. The analysis included C2 through V5 sequences. Analysis of MM13898 included A (red), C (gold), and D (black). Analysis of MM2760 used A (red), G (green), and D (black). Analyses of MM13324 and MM6535 used A (red), D (blue), and C (black). A black triangle marks the spot where the sequences were separated. The phylogenetic trees are neighbor-joining trees with parsimony bootstrap values at the nodes containing the unknown. Schematics showing the deduced subtype structures are shown below the bootscans. V3, the location of the V3 loop (shown for orientation).
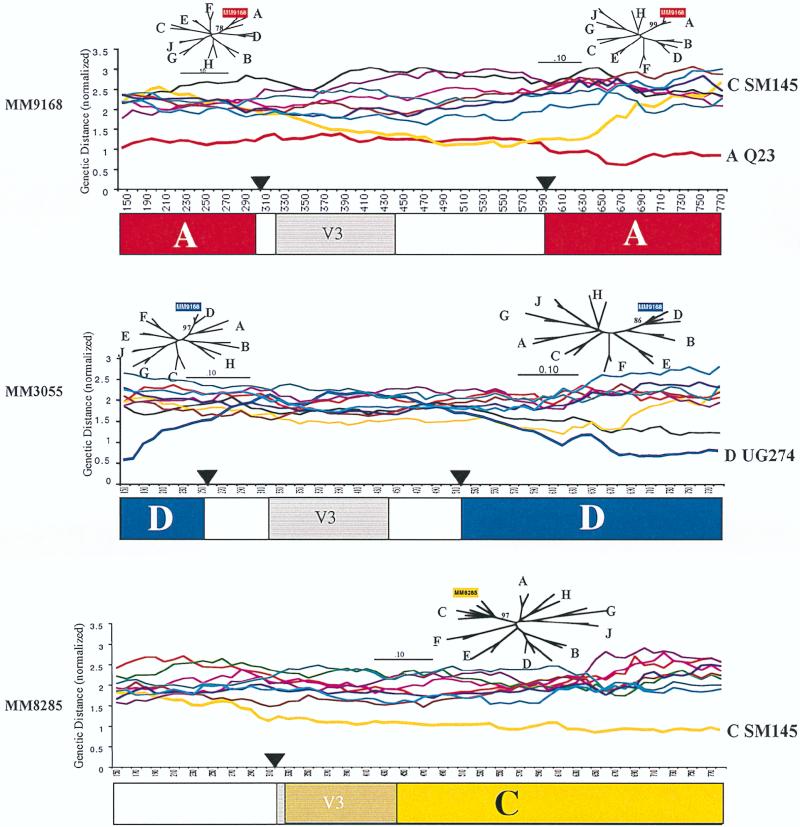
Three of the recombinants (MM9168, MM3055, and MM8285) encoded portions of the envelope gene that could not be readily assigned to any currently defined subtype; the results of a distance scan analysis of these envelope sequences are shown in Fig. 4. In the case of MM9168, which has two segments that were derived from subtype A, the subtype origin of V3 through C3 could not be determined. The distance scan of MM9168 suggests that these sequences have the highest degree of similarity to subtype A and C viruses. In a second example (MM3055), which involves a subtype D recombinant envelope gene, the origin of sequences within V2 and C3 also cannot be assigned to any known subtype. In the case of MM8285, which has mainly clade C-derived sequences, the subtype origin of the 5′ sequence encompassing C2 is not apparent. We cannot rule out the possibility that the sequences which do not appear to represent known subtypes were derived by multiple recombination events that cannot be detected by our methods. In all three cases, the distance scan shows that the distances between the unknown and known subtypes is in the usual range for group M HIV-1 (data not shown), and this rules out the possibility that they are from a less highly related virus group, such as O.
FIG. 4.
Recombination analysis: distance scans. Distance scanning was performed as described in Materials and Methods. The analysis included C2 through V5 sequences. Distances between the unknown and subtypes A (red), B (light blue), C (gold), D (dark blue), E (brown), F (magenta), G (green), H (black), and J (purple) were plotted. The black triangles represent locations where the sequences were broken to run phylogenetic trees. The phylogenetic trees were computed by the neighbor-joining method with parsimony bootstrap values at the nodes with the unknown. The deduced subtype structure of the each virus is shown below the distance scan. Regions in white could not be assigned to any known subtype. V3, the location of the V3 loop (shown for orientation).
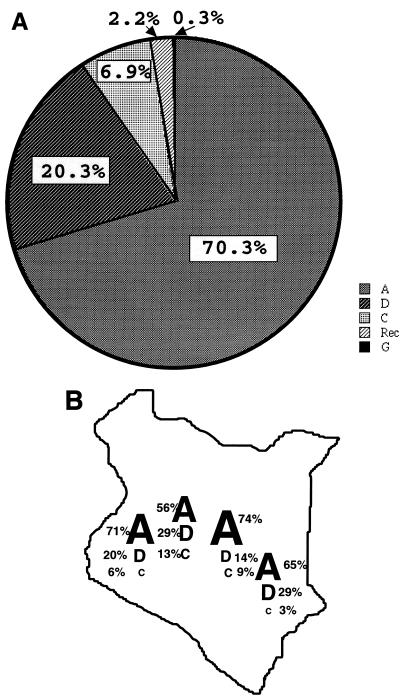
In summary, of the 320 envelope gene sequences evaluated here, 225 (70.3%) were subtype A, 65 (20.3%) were subtype D, 22 (6.9%) were subtype C, 1 (0.3%) was subtype G, and 7 (2.2%) were intersubtype recombinants (Fig. 5A). The geographic distribution of the subtypes was fairly similar throughout western, central, and eastern Kenya (Fig. 5B). There were no significant differences in the ages of women infected with different subtypes of virus.
FIG. 5.
Summary of the subtype distribution of viruses from the 320 women analyzed. (A) Pie chart of subtype distribution in the overall cohort. The percentage of viruses that belong to each subtype is indicated in the relevant pie section or above it. Rec, recombinant. (B) Geographic distribution of subtypes. The regions of origin were defined on the basis of the ethnicities of the women. This cohort represented ethnic groups from throughout Kenya.
Analysis of markers for disease in relation to viral subtype.
Both CD4 lymphocyte counts and plasma viral loads are markers for the stage of disease and the level of immunosuppression (30, 31, 39, 40). To address whether infection with a particular subtype was associated with a more advanced disease stage, plasma RNA levels were measured by a quantitative HIV-1 viral load assay (Gen-Probe) that was designed to detect all HIV-1 subtypes with the same efficiency (1, 13). Additional pilot studies were performed with Kenyan viral isolates and plasma samples, using the Gen-Probe quantitative assay versus a quantitative RNA PCR method (24), and these studies further validated the use of this approach for quantitative analysis of subtype A, C, and D HIV-1 (data not shown). The median viral load for this cohort was 41,400 copies per ml (range, 112 to 1,228,480). The highest median viral load was seen in women infected with subtype C HIV-1 (71,800 copies/ml), although this was not significantly different from the median plasma RNA levels in women infected with clade A or D virus (39,040 and 36,400 copies/ml, respectively) (Table 1). Women infected with clade C viruses were the most likely to have viral loads in the upper quartile and the least likely to have viral loads in the lowest quartile. Most striking was the fact that women infected with subtype C had significantly lower CD4 cell counts than women infected with subtype A and D (P = 0.01 for C versus non-C). Although women infected with subtype D had only a very slightly lower median viral load than the population as a whole, they were significantly less likely to have a viral load in the upper quartile.
TABLE 1.
Subtype versus demographic and clinical profiles
| Subtype | n | No. of CD4 cells/μl | Plasma viral RNA (copies/ml)
|
||
|---|---|---|---|---|---|
| Median | Highest quartile (>162,000), % | Lowest quartile (<10,000), % | |||
| A | 201 | 432 | 39,040 | 27 | 26 |
| D | 61 | 410 | 36,400 | 15a | 25 |
| C | 19 | 333b | 71,800 | 37 | 16 |
| Recc | 7 | 431 | 18,310 | 40 | 60 |
| G | 1 | 77 | 552,120 | ||
P = 0.03 (chi-square test).
P = 0.01 (Mann-Whitney U test).
Rec, recombinant.
DISCUSSION
This study presents a detailed analysis of the envelope subtypes that are currently circulating in Kenya, which has one of the highest prevalences of HIV-1 infection among countries in sub-Saharan Africa (33). In that country, particular subtypes are not preferentially circulating within particular risk groups, allowing us to generate a representative picture of both the pure subtypes and the intersubtype envelope recombinants in Kenya. The majority of women were infected with clade A virus (70.3%); a significant fraction were infected with clade D (20.3%) or clade C (6.9%) HIV-1. One (0.3%) subtype G virus and seven (2.2%) intersubtype recombinants were also detected. The cohort analyzed in this study included women from western, central, and eastern Kenya, suggesting a very similar distribution of subtypes throughout that country.
HMA provides a relatively rapid tool for determining HIV-1 subtypes (7). While the existing reference strains provide a reasonable starting point for subtype assessment in a new population, our analyses suggest that when using these strains it is difficult to assign a significant fraction of Kenyan viruses (about 30%) by HMA of envelope sequences. This is similar to what was seen in a French study with participants from various parts of the world, in which HMA did not predict all viral subtypes (25). The use of geographically specific viruses significantly enhanced our power to determine subtypes in this cohort. In fact, we were successful at assigning the subtype for each of the subtype A and all but one of the subtype D viruses by using a combination of Kenyan and standard reference strains. Because we lacked Kenyan subtype C reference strains, we were not able to achieve the same success in typing subtype C envelope sequences. Conversely, we were not able to assign a subtype to all of the envelope genes examined by using only Kenyan reference sequences. Thus, although local reference strains perform better than subtype reference strains from different geographic locations, a combination of both appears to be optimal when using HMA to define HIV-1 subtypes.
By surveying such a large cohort, intersubtype recombinants representing viruses that had exchanged sequences within the coding region for the extracellular envelope glycoprotein were also identified. Proviral genomes that are mosaic over the length of the genome as well as recombinant forms of individual HIV-1 genes have been described (3, 4, 11, 12, 23, 28, 47, 48, 50). Although some of these recombinants were characterized directly from patients’ samples (4, 50), many were identified in viruses amplified in culture. In the latter case, it is unclear whether the recombinant was present in the individual or it arose in culture from two pure subtypes in the individual. Moreover, there have been limited population-based studies of the prevalence of recombinant viruses. A survey of 320 subjects who came from throughout Kenya to work and live in Nairobi provided us with an opportunity to estimate the frequency of intersubtype recombinant genomes in the population. We detected seven (2.2%) intersubtype recombinants whose exchanges occurred within a segment of the gene coding for the extracellular envelope glycoprotein. This is likely to be a minimal estimate of the envelope recombinants in this population, since it is certainly possible that additional envelope recombinants were not identified in the HMA screen because either the majority of the sequence represented one subtype, leading it to behave like a pure subtype in the HMA, or recombination occurred outside of the V1 through C3 region analyzed in the HMA. Approximately 10% of the viral genome was analyzed. If we extrapolate from the frequency of recombinant envelope genes observed here and assume that there is not a strong bias for or against protein function of a recombinant envelope gene in comparison to other viral genes, then >20% of Kenyan HIV-1 proviral genomes would be predicted to be intersubtype recombinants. These estimates are likely to be relevant for most of sub-Saharan Africa because there are similar mixtures of multiple subtypes in many countries in this region (2).
Among the recombinant envelope genes analyzed here, sequences that did not closely associate with any known subtype were identified. Distance scan analyses demonstrated that these sequences originated from viruses that are within the group M cluster rather than from a more divergent HIV-1 group. These sequences may represent multiple small fragments of known subtype or novel group M subtypes not yet described.
Recombination between the diploid RNA genomes during DNA synthesis is a common mechanisms for generating diversity in retroviruses (20). To generate a recombinant during reverse transcription, a single cell must be coinfected with two different viruses so that copackaging of heterologous RNAs can occur. Thus, detection of intersubtype recombinants implies that at some previous time a person was coinfected with HIV-1 of two different subtypes. This may have occurred as a result of simultaneous transmission of multiple variants, a common occurrence in women (43, 46), or by a second infection of the individual, with a different subtype, at a later time. At some point, dual infection must have occurred to initiate the cascade of events leading to the detection of recombinant genomes. Because recombination is predicted to occur quite commonly when there is coinfection of a cell by two highly related retroviruses (16, 20), the limiting event in the generation of intersubtype recombinants is likely to be dual infection. At this point, little is known about the factors that determine the frequency of dual infection in a population. As discussed herein, the predicted frequency of intersubtype recombinant genomes in this cohort is estimated to be approximately 20%, using fairly conservative assumptions. This implies either that reinfection occurs very commonly in HIV-1-infected individuals or that recombinant viruses are favored for transmission. In the latter case, the viruses could originate from a true genetic recombinant provirus, or they could represent particles assembled from viral proteins expressed from distinct viral genomes in a dually infected individual. Certainly it will be important to distinguish between these possibilities in considering the design of vaccines for African populations.
The subtypes define broad categories of viruses with similar sequence characteristics. In the viruses examined here, subtype was defined on the basis of sequences coding for the extracellular envelope protein, which is the protein that determines cell tropism as well as the replication patterns of the virus. Thus, it is possible that viruses that have different envelope subtypes will replicate with different efficiencies in the host, leading to different clinical outcomes. To address this possibility, we analyzed plasma viral loads and CD4 lymphocyte counts as markers for disease stage in this cohort. Women infected with clade C viruses tended to have higher viral loads, and they had significantly lower CD4 cell counts than women infected with clade A or D HIV-1. From these data, we cannot discriminate between the possibility that women infected with clade C virus have been infected for a longer period of time and the possibility that clade C infection is associated with a more rapid disease progression and, as a consequence, a more rapid CD4 cell count decline and higher viral loads. Recent studies suggest that viruses which can use the CXCR4 coreceptor are underrepresented among subtype C HIV-1, including viruses that were derived from individuals at later stages of infection and disease (54). This finding may suggest that subtype C variants use novel coreceptor proteins for entry into the cell. Interestingly, women infected with clade C viruses in the cohort described here were also significantly more likely to shed HIV-1-infected cells in their vaginal secretions (18). Together, these data suggest that viruses with clade C-derived envelope sequences may have unique replication properties which may affect transmission and/or progression. This can only be directly addressed by longitudinal studies of disease progression in relation to viral subtype, particularly in cohorts in which clade C virus infection represents a more significant fraction of all HIV-1 infections. Certainly, such information on differences in the biological properties or pathogenesis of different subtypes will be important in designing strategies to limit the spread of the most virulent HIV-1 variants.
ACKNOWLEDGMENTS
We thank all of the members of the Nairobi HIV/STD Research Project.
This work was supported by NIH National Institutes of Health grant AI38518 (J.O.); National Institute of Child Health and Human Development grants 23412, D43 TW00007, and T22 TW00001 (J.K.K.); a New Investigator Award from the University of Washington Center for AIDS Research (AI 27757) and an NIH Clinical Scientist Award (K08 HD01160-01) to G.C.J., and a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense (J.K.C.).
REFERENCES
- 1.Bodrug S, Domingo R, Holloway J, Sanders M, Nunomura K, Sloan C, Billyard B. Gen-Probe single-tube quantitative HIV assay. J Clin Microbiol Infect. 1997;3:1050. [Google Scholar]
- 2.Burke D S, McCutchan F E. Global distribution of human immunodeficiency virus-1 clades. In: Vincent S H, DeVita T Jr, Rosenberg Steven A, editors. AIDS: biology, diagnosis, treatment and prevention. Vol. 4. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 119–126. [Google Scholar]
- 3.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St. Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J the UNAIDS Network for HIV Isolation and Characterization. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 6.Delwart E L, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl. 1995;4:S202–S216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 7.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 8.Essex M E. Origin of acquired immunodeficiency syndrome. In: Vincent S H, DeVita T Jr, Rosenberg Steven A, editors. AIDS: biology, diagnosis, treatment and prevention. 4th ed. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 3–14. [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogenetic inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giachetti C, Kolk D, Dockter J, Knowlton J, Wang R, Hotaling H, McDonough S. Proceedings of the 12th World AIDS Conference. Bologna, Italy: Monduzzi Editore S. p. A.; 1998. High throughput assay for sensitive detection of HIV-1 RNA of diverse origins, including type O strains; pp. 151–155. [Google Scholar]
- 14.Hills D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic trees. Syst Biol. 1993;42:182–192. [Google Scholar]
- 15.Hu D J, Dondero T J, Rayfield M A, George J R, Schochetman G, Jaffe H W, Luo C C, Kalish M L, Weniger B G, Pau C P, Schable C A, Curran J W. The emerging genetic diversity of HIV. The importance of global surveillance for diagnostics, research, and prevention. JAMA. 1996;275:210–216. [PubMed] [Google Scholar]
- 16.Hu W S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssens W, Heyndrickx L, Fransen K, Temmerman M, Leonaers A, Ivens T, Motte J, Piot P, van der Groen G. Genetic variability of HIV type 1 in Kenya. AIDS Res Hum Retroviruses. 1994;10:1577–1579. doi: 10.1089/aid.1994.10.1577. [DOI] [PubMed] [Google Scholar]
- 18.John, G., J. Neilson, D. Panteleeff, R. Nduati, D. Mbori-Ngacha, J. Achola, J. Bwago, J. Overbaugh, and J. K. Kreiss. Unpublished data.
- 19.John G C, Nduati R W, Mbori-Ngacha D, Overbaugh J, Welch M, Richardson B A, Ndinya-Achola J, Bwayo J, Krieger J, Onyango F, Kreiss J K. Genital shedding of human immunodeficiency virus type 1 DNA during pregnancy: association with immunosuppression, abnormal cervical or vaginal discharge, and severe vitamin A deficiency. J Infect Dis. 1997;175:57–62. doi: 10.1093/infdis/175.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 21.Kreiss J K, Koech D, Plummer F A, Holmes K K, Lightfoote M, Piot P, Ronald A R, Ndinya-Achola J O, D’Costa L J, Roberts P. AIDS virus infection in Nairobi prostitutes. Spread of the epidemic to East Africa. N Engl J Med. 1986;314:414–418. doi: 10.1056/NEJM198602133140704. [DOI] [PubMed] [Google Scholar]
- 22.Kunanusont C, Foy H M, Kreiss J K, Rerks-Ngarm S, Phanuphak P, Raktham S, Pau C P, Young N L. HIV-1 subtypes and male-to-female transmission in Thailand. Lancet. 1995;345:1078–1083. doi: 10.1016/s0140-6736(95)90818-8. [DOI] [PubMed] [Google Scholar]
- 23.Leitner T, Escanilla D, Marquina S, Wahlberg J, Brostrom C, Hansson H B, Uhlen M, Albert J. Biological and molecular characterization of subtype D, G and A/D recombinant HIV-1 transmission in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 24.Lewis P, Nduati R, Kreiss J K, John G C, Richardson B, Mbori-Ngacha D, Ndinya-Achola J, Overbaugh J. Cell-free HIV-1 in breast milk. J Infect Dis. 1998;177:34–39. doi: 10.1086/513816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loussert-Ajaka I, Menu E, Apetrei C, Peeters M, Damond F, Mauclere P, Eberle J, Brengues C, Saragosti S, Barre-Sinoussi F, Brun-Vezinet F, Simon F. HIV type 1 diversity and the reliability of the heteroduplex mobility assay. AIDS Res Hum Retroviruses. 1998;14:877–883. doi: 10.1089/aid.1998.14.877. [DOI] [PubMed] [Google Scholar]
- 26.Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan F E, Burke D S. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciukenas S. Ribosomal RNA Database Project: TREETOOL. www.cme.msu.edu/RDP. 1994. [Google Scholar]
- 28.McCutchan F E, Salminen M O, Carr J K, Burke D S. HIV-1 genetic diversity. AIDS. 1996;10(Suppl. 3):S13–S20. [PubMed] [Google Scholar]
- 29.McDonough S, Bott M, Giachetti C. Application of transcription-mediated amplification to detection of nucleic acids from clinically relevant organisms. In: Lee H, Morse S, Olsvik O, editors. Nucleic acid amplification technologies: application to disease diagnosis. Cambridge, Mass: Eaton Publishing; 1997. pp. 113–123. [Google Scholar]
- 30.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 31.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 32.Mertens T E, Burton A. Estimates and trends of the HIV/AIDS epidemic. AIDS. 1996;10(Suppl. 1):S221–S228. doi: 10.1097/00002030-199601001-00031. [DOI] [PubMed] [Google Scholar]
- 33.Mertens T E, Burton A, Stoneburner R, Sato P, Beer D L, Caraël M, Belsey E. Global estimates and epidemiology of HIV-1 infections and AIDS. AIDS. 1994;8(Suppl. 1):S361–S372. [PubMed] [Google Scholar]
- 34.Moss G B, Overbaugh J, Welch M, Reilly M, Bwayo J, Plummer F A, Ndinya-Achola J O, Malisa M A, Kreiss J K. Human immunodeficiency virus DNA in urethral secretions in men: association with gonococcal urethritis and CD4 cell depletion. J Infect Dis. 1995;172:1469–1474. doi: 10.1093/infdis/172.6.1469. [DOI] [PubMed] [Google Scholar]
- 35.Myers G, Korber B, Berzofsky J A, Smith R F, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 36.Nduati R W, John G C, Richardson B A, Overbaugh J, Welch M, Ndinya-Achola J, Moses S, Holmes K, Onyango F, Kreiss J K. Human immunodeficiency virus type 1-infected cells in breast milk: association with immunodeficiency and vitamin A deficiency. J Infect Dis. 1995;172:1461–1468. doi: 10.1093/infdis/172.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson N C, Reynolds M A, Arnold L J. Detection of acridinium esters by chemiluminescence. In: Kricka L, editor. Nonisotopic probing, blotting, and sequencing. San Diego, Calif: Academic Press; 1995. pp. 391–428. [Google Scholar]
- 38.Obel A O, Sharif S K, McLigeyo S O, Gitonga E, Shah M V, Gitau W. Acquired immunodeficiency syndrome in an African. East Afr Med J. 1984;61:724–726. [PubMed] [Google Scholar]
- 39.O’Brien T R, Blattner W A, Waters D, Eyster E, Hilgartner M W, Cohen A R, Luban N, Hatzakis A, Aledort L M, Rosenberg P S, Miley W J, Kroner B L, Goedert J J. Serum HIV-1 RNA levels and time to development of AIDS in the Multicenter Hemophilia Cohort Study. JAMA. 1996;276:105–110. [PubMed] [Google Scholar]
- 40.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 41.Oram J D, Downing R G, Roff M, Clegg J C S, Serwadda D, Carswell J W. Nucleotide sequence of a Ugandan HIV-1 provirus reveals genetic diversity from other HIV-1 isolates. AIDS Res Hum Retroviruses. 1990;6:1073–1078. doi: 10.1089/aid.1990.6.1073. [DOI] [PubMed] [Google Scholar]
- 42.Ou C Y, Takebe Y, Weniger B G, Luo C C, Kalish M L, Auwanit W, Yamazaki S, Gayle H D, Young N L, Schochetman G. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341:1171–1174. doi: 10.1016/0140-6736(93)91001-3. [DOI] [PubMed] [Google Scholar]
- 43.Overbaugh, J., J. Kreiss, M. Poss, P. Lewis, S. Mostad, G. John, R. Nduati, D. Mbori-Ngacha, H. Martin, Jr., B. Richardson, S. Jackson, J. Neilson, E. M. Long, D. Panteleeff, M. Welch, J. Rakwar, D. Jackson, B. Chohan, L. Larvreys, K. Mandaliya, and J. Ndinya-Achola. Studies of HIV-1 mucosal viral shedding and transmission in Kenya. J. Infect. Dis., in press. [DOI] [PubMed]
- 44.Overbaugh J, Anderson R J, Ndinya-Achola J O, Kreiss J K. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- 45.Poss M, Gosink J, Thomas E, Kreiss J K, Ndinya-Achola J, Mandaliya K, Bwayo J, Overbaugh J. Phylogenetic evaluation of Kenyan human immunodeficiency virus type 1 isolates. AIDS Res Hum Retroviruses. 1997;13:493–499. doi: 10.1089/aid.1997.13.493. [DOI] [PubMed] [Google Scholar]
- 46.Poss M, Martin H L, Kreiss J K, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 48.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Salminen M O, Carr J K, Robertson D L, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P M, Hahn B H, Burke D S, McCutchan F E. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 52.Subbarao S, Schochetman G. Genetic variability of HIV-1. AIDS. 1996;10(Suppl. A):S13–S23. doi: 10.1097/00002030-199601001-00003. [DOI] [PubMed] [Google Scholar]
- 53.Swofford D. PAUP: phylogenetic analysis using parsimony. 3rd ed. Champaign: Illinois Natural History Survey; 1991. [Google Scholar]
- 54.Tscheming C, Alaeus A, Fredericksson R, Bjorndal A, Deng H, Littman D R, Fenyo E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 55.UNAIDS/World Health Organization. Global HIV/AIDS and STD surveillance 1998. Geneva, Switzerland: UNAIDS/World Health Organization; 1998. HIV/AIDS: regional statistics and features, December 1997. [Google Scholar]
- 56.Weniger B G, Takebe Y, Ou C-Y, Yamazaki S. The molecular epidemiology of HIV in Asia. AIDS. 1994;8(Suppl. 2):S13–S28. [PubMed] [Google Scholar]
- 57.Zachar V, Goustin A S, Zacharova V, Hager H, Koppelhus U, Wombel D D, Liu X, Bambra C, Nyongo A, Ebbesen P. Genetic polymorphism of envelope V3 region of HIV type 1 subtypes A, C, and D from Nairobi, Kenya. AIDS Res Hum Retroviruses. 1996;12:75–78. doi: 10.1089/aid.1996.12.75. [DOI] [PubMed] [Google Scholar]