Abstract
RNA vaccines possess significant clinical promise in counteracting human diseases caused by infectious or cancerous threats. Self-amplifying replicon RNA (repRNA) has been thought to offer the potential for enhanced potency and dose sparing. However, repRNA is a potent trigger of innate immune responses in vivo, which can cause reduced transgene expression and dose-limiting reactogenicity, as highlighted by recent clinical trials. Here, we report that multivalent repRNA vaccination, necessitating higher doses of total RNA, could be safely achieved in mice by delivering multiple repRNAs with a localizing cationic nanocarrier formulation (LION). Intramuscular delivery of multivalent repRNA by LION resulted in localized biodistribution accompanied by significantly upregulated local innate immune responses and the induction of antigen-specific adaptive immune responses in the absence of systemic inflammatory responses. In contrast, repRNA delivered by lipid nanoparticles (LNPs) showed generalized biodistribution, a systemic inflammatory state, an increased body weight loss, and failed to induce neutralizing antibody responses in a multivalent composition. These findings suggest that in vivo delivery of repRNA by LION is a platform technology for safe and effective multivalent vaccination through mechanisms distinct from LNP-formulated repRNA vaccines.
Keywords: replicating RNA, lipid nanoparticles, RNA vaccines, infectious diseases, innate immunity, LION formulation, nanoparticle emulsion, reactogenicity, side-effect, multivalent vaccines
Graphical abstract
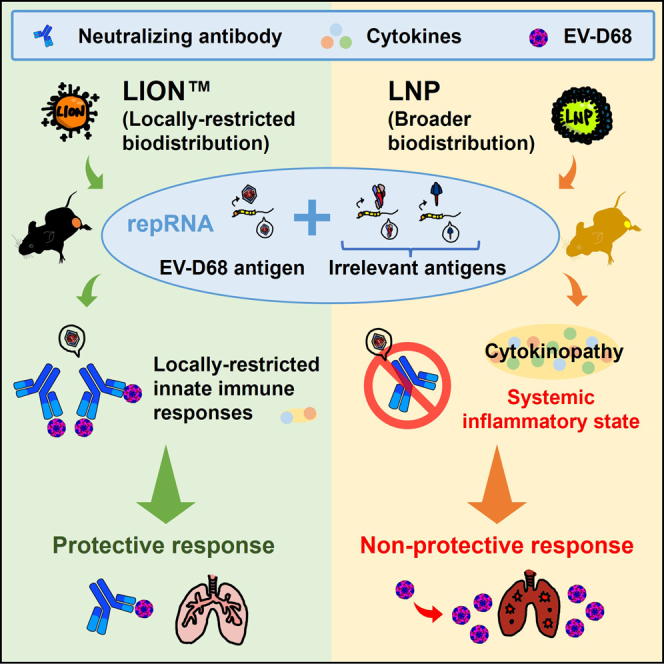
Kimura, Erasmus, and colleagues demonstrate that the delivery of multivalent self-amplifying replicon RNA (repRNA) vaccines via LION, a cationic nanocarrier formulation, prompts robust localized immune responses without systemic inflammation. This approach overcomes challenges faced by lipid nanoparticle (LNP) formulations, suggesting LION as a promising platform for safe, efficient multivalent vaccination.
Introduction
Non-viral in vivo RNA delivery is an appealing modality for therapeutic and prophylactic medicines.1,2 Two types of RNA have been mainly used for protein expression in vivo to date: conventional mRNA and replicon RNA (repRNA). The latter is a type of RNA that encodes a viral replicase to amplify protein-encoding mRNA intracellularly, thus typically resulting in the expression of large amounts of protein from fewer copies of RNA for an extended period.3,4 To achieve efficient delivery to cells, various RNA delivery formulations have been developed over the past 30 years.1,2 Lipid nanoparticles (LNPs) are widely used for mRNA delivery; phase I studies conducted with current COVID vaccines ranging from 25 to 250 μg have demonstrated that conventional mRNA modified in their nucleosides and purified by columns can be efficiently and safely delivered up to 100 μg (clinicaltrial.gov NCT04470427 and NCT04368728).5 Nevertheless, LNP-formulated repRNA has shown some limitations. Imperial College London recently reported that vaccination with repRNA/LNP induced only 61% seroconversion as defined by enzyme-linked immunosorbent assay (ELISA) in a phase I clinical trial.6 In addition, two clinical studies published by Imperial College London and Arcturus Therapeutics (ARCT-021) with dose ranging from 1 to 10 μg reported some cases of severe systemic adverse events in a dose-dependent manner,6,7 where over 70% of participants in a cohort receiving a single dose of 10 μg of ARCT-021 experienced fever, a measurable parameter of reactogenicity.7 Due to this dose-limiting reactogenicity, Arcturus advanced a lower 5-μg dose into a phase III efficacy trial where they demonstrated equivalent efficacy (clinicaltrial.gov NCT04480957) (Link) to Moderna’s 100-μg dose of conventional mRNA vaccine illustrating the potency and dose-sparing features of repRNA,8 consistent with that previously published in a preclinical setting.9 However, this apparent dose ceiling of LNP-delivered repRNA may significantly limit the utility of this next-generation approach to RNA medicine and vaccines that may require increased dose levels, such as multivalent vaccines and cancer vaccines.10 We have developed a family of oil-in-water emulsion-based cationic nanocarriers named LION. Unlike LNPs, which shield RNA molecules from RNases by encapsulation, LION nanoparticles and RNA molecules form a complex where the RNA is electrostatically attached to the surface of the nanoparticle, which results in complete protection from RNase-mediated degradation. We previously reported that repRNA/LION vaccination elicits robust immune responses against encoded viral antigens, including protective immunity, high neutralizing antibody (Ab) titers, and T cell responses against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Crimean-Congo hemorrhagic fever virus, and enterovirus D68 (EV-D68) in mice and nonhuman primates11,12,13,14,15 (Warner et al., manuscript submitted). Moreover, a repRNA/LION coronavirus disease 19 (COVID-19) vaccine based on repRNA/LION technology recently received regulatory approval for emergency use in India (Link); the first repRNA-based product to achieve this milestone since repRNA was first described in 198916 and demonstrated as a tool for in vivo expression and immunization studies in 1994.17 Furthermore, other products with the same indication are now being evaluated in clinical studies in Brazil (clinicaltrial.gov NCT04844268), South Korea, and the United States (clinicaltrial.gov NCT05132907). Importantly, the COVID-19 vaccine platform was tolerable at all doses evaluated (5–25 μg), where the maximum dose of 25 μg was 5-fold higher than the maximum tolerable dose previously reported for repRNA/LNP: The group receiving a 10-μg dose in phase II/III clinical trial (clinical trial identifier: CTRI/2021/09/036379) in India demonstrated 94.1% seroconversion and non-inferior immunogenicity compared to the COVISHIELD (a ChAdOx1-based COVID-19 vaccine of Serum Institute of India) control group. Less than 30% of subjects reported any systemic solicited adverse event, only 15.6%–23.7% reported fever, 1.3%–6.3% reported nausea, and no cases of myocarditis/pericarditis were reported (Link). Factors such as RNA, formulation, or a combination of the two are thought to play a role in reactogenicity. While multiple RNA sensors such as Toll-like receptors and RIG-like receptors may be involved in the inflammatory responses that could contribute to the reactogenicity,18,19 the observed differences in reactogenicity among repRNA-based vaccines described above suggest an important role of the formulation. Preclinical studies of repRNA/LNP have shown systemic biodistribution of RNA9 and dose-dependent induction of systemic cytokines.19,20 Similar information on repRNA/LION is so far limited.
The present study was undertaken to better understand innate and adaptive immune responses to repRNA/LION vaccination in mice by comparing them with those induced by repRNA formulated with LNPs sharing a composition similar to that used in approved mRNA/LNP vaccines (Link). Our data show that repRNA elicits antigen-specific immunity following high-dose multivalent immunization only when delivered by LION but not LNP. In addition, our data show that intramuscular (i.m.) administration of repRNA/LNP is highly reactogenic in mice, providing mechanistic insight into the differential frequency of systemic adverse events observed between LION and LNP clinical trials and demonstrating that repRNA formulated with cationic nanocarriers is an effective platform for protective multivalent RNA vaccination.
Results
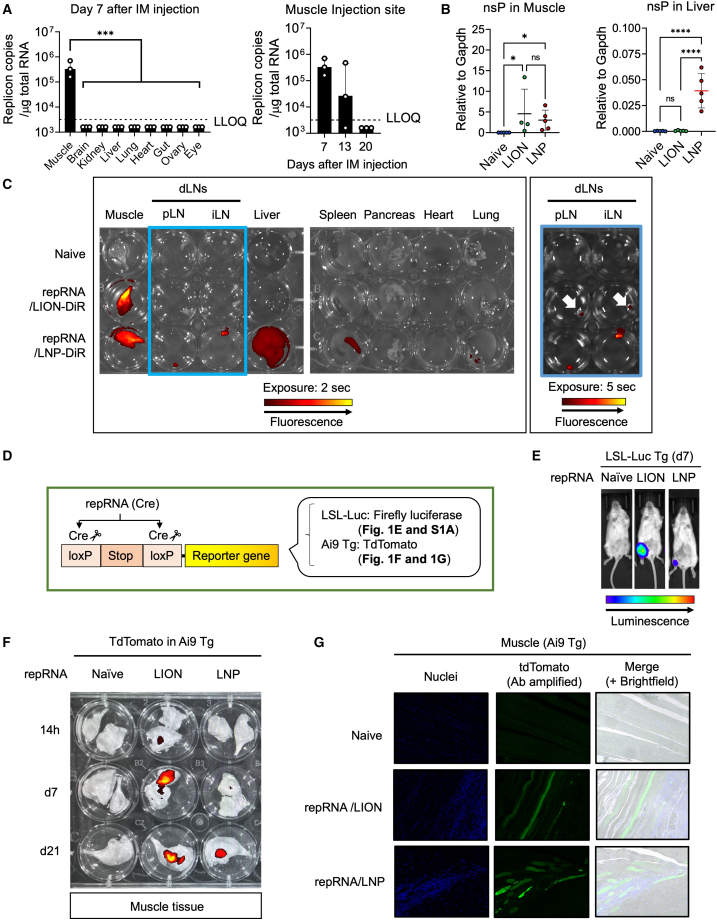
Intramuscular injection of repRNA/LION restricts transgene expression to the muscle in mice
Others have recently shown that LNP-formulated repRNA disseminates throughout the body after i.m. injection in Sprague-Dawley rats.9 To examine the biodistribution of repRNA delivered by LION, we injected C57BL/6 mice with repRNA/LION through an i.m. route, then, 7 days later, measured the RNA levels of transgene in multiple tissues. Unlike LNP-formulated RNA reported by others, LION-formulated repRNA was detectable only in the muscle over 13 days after i.m. injection, which was no longer detectable at day 20 (Figure 1A). To directly compare the biodistribution of the repRNA delivered by LION and repRNA delivered by LNP, we next injected C57BL/6 mice with repRNA/LION and repRNA/LNP, then, 14 h later, quantified repRNA copy number in the muscle and the liver. RepRNA delivered by both LION and LNP was detected in the muscle, while only that delivered by LNP was detected in the liver (Figure 1B). To corroborate this finding further, we formulated repRNA with a fluorescent dye (XenoLight DiR)-labeled LION (LION-DiR) or co-encapsulated in LNP with the same dye (LNP-DiR) and injected them into C57BL/6 mice through the i.m. route. Twenty-four hours later, their muscle, draining popliteal, and inguinal lymph nodes (pLN and iLN, respectively), liver, spleen, pancreas, heart, and lung were harvested and analyzed by an in vivo imaging system (IVIS). A higher level of LION-DiR signal was detected in the muscle and a relatively weaker level in the draining lymph nodes (dLNs) 24 h after injection, while the fluorescent signal was not detected in any of the other sampled organs (Figure 1C). Conversely, the LNP-DiR signal was detected beyond the injected muscle and dLNs, with fluorescence detectable in the liver, spleen, and, to a lesser degree the lungs, consistent with others’ observation of broad biodistribution of RNA delivered by LNP.9 Together, these data indicate that LION remains local to the injection site, whereas LNP disseminates throughout the body and can be found in multiple major organs.
Figure 1.
Intramuscular injection of repRNA/LION restricts transgene expression to the muscle in mice
(A) C57BL/6 mice received i.m. injection of 2.5 μg of repRNA/LION encoding SARS-CoV-2 Spike protein, and SARS-CoV-2 Spike RNA in the indicated tissues 7 days after i.m. injection was determined by qRT-PCR. Values for individual mice are plotted along with the geometric mean (n = 3). LLOQ, lower limit of quantification. Samples below the assay’s lower level of quantitation were plotted as ½ the LLOQ. Statistical comparison of mean values was performed by ordinary one-way ANOVA and Dunnett’s multiple comparisons test between the values in the muscle group vs. the values in other individual groups. (B) C57BL/6 mice received i.m. injection of repRNA encoding SEAP (10 μg). RNA transcripts for non-structural protein (NSP) in the muscle and the liver 14 h after i.m. injection were determined by qRT-PCR. Values were normalized to the values of Gapdh. Means ± SD are shown, and each symbol indicates individual values (n = 4–5). Statistical comparison of mean values was performed by Kruskal-Wallis test and Dunn’s multiple comparisons test between the values in individual groups (muscle) or ordinary one-way ANOVA and Tukey’s multiple comparisons test between the values in individual groups (liver). (C) C57BL/6 mice received i.m. injection of repRNA (10 μg) formulated with LION-DiR or LNP-DiR. Lipid biodistribution was analyzed by IVIS using XenoLight-DiR-conjugated formulations (representative data from two independent experiments are shown). (D) Schematic representation of reporter mouse models used in (E–G and Figure S1A). (E) LSL-Luc Tg mice received i.m. injection of 10 μg of repRNA encoding Cre recombinase formulated with LION or LNP. At day 7, luciferase expression was analyzed in vivo by IVIS. Data from one representative mouse in each group are shown, and other data are included in Figure S1A. (F and G) Ai9 Tg mice received i.m. injection of repRNA encoding Cre recombinase (10 μg) formulated with LION or LNP. (F) At the indicated time points, muscle tissues were isolated, and tdTomato expression was analyzed by IVIS (representative data from two independent experiments are shown). (G) Seven days later, their muscles were isolated, sectioned, and analyzed by immunofluorescence. The nuclei and tdTomato were stained with Hoechst (blue) and anti-RFP Ab (green), respectively. Statistically significant results are shown as asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
To determine whether in vivo transgene expression patterns would follow the vehicle biodistribution patterns, we employed two reporter mice (LSL-Luc Tg and Ai9 Tg) in which Cre/loxP-mediated genomic recombination drives stable expression of reporter genes (luciferase and tdTomato, respectively). As such, these strains allow for the detection of cells/tissues that received repRNA-expressing Cre recombinase at some point in time before harvest (Figure 1D). Seven days after i.m. injection of repRNA-expressing Cre recombinase, the luciferase-expressing area was restricted to the injected leg muscle of both repRNA/LION- and repRNA/LNP-injected LSL-Luc Tg mice (Figures 1E and S1A). LNP formulation mediated efficient transgene expression of repRNA in multiple cell types in vitro, including muscle, liver, and monocyte cell lines (Figure S1B), suggesting that the lack of reporter gene detection in extra muscular tissues is not due to an impaired ability to express the transgene in those cells in vivo. Notably, the total transfected area was more extensive in repRNA/LION-injected mice compared with repRNA/LNP-injected mice (Figures 1E and S1A). A similar observation was made in the extracted tissue of Ai9 Tg mice after repRNA delivery (Figure 1F). In the muscle of repRNA/LION-injected Ai9 Tg mice, the transfected region was detected as early as 14 h, and peaked at day 7, after i.m. injection. In the muscle of repRNA/LNP-injected mice, the total transfected area was barely detectable by day 7 but was evident at day 21. To further analyze what cells are transfected by repRNA/LION and repRNA/LNP in the muscle, we analyzed the muscle sections of Ai9 mice that received repRNA. Immunofluorescence assay revealed that both LION and LNP predominantly deliver repRNA to myocytes (Figure 1G). In addition, a mononuclear cellular infiltration with some transgene expression could be observed in the proximity of these transfected myocytes. Taken together, these data indicate that, following i.m. delivery, formulation and transgene expression have differing biodistribution profiles. While LION remains in the muscle, LNP disseminates throughout the body, but in both cases the repRNA expression appears to be restricted to the muscle.
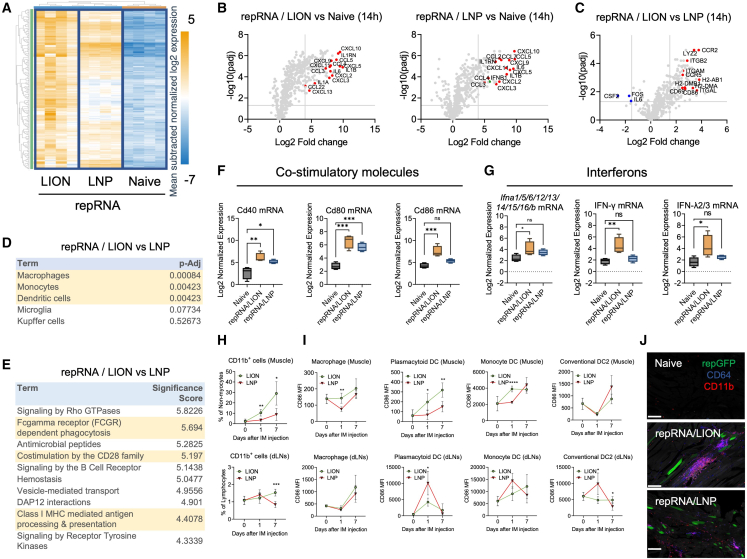
Intramuscular injection of repRNA/LION activates the local muscle innate immune response in mice
The mononuclear cellular infiltration to the proximity of transfected myocytes prompted us to analyze the early local innate immune response in the muscle of repRNA/LION and repRNA/LNP-injected mice. We performed a NanoString analysis (mouse host response 773-gene panel) of RNA extracted from muscle tissues of naive mice or mice receiving LNP- or LION-formulated repRNA 14 h after i.m. injection. Many gene transcripts that were upregulated by repRNA delivered by LION and LNP compared with naive samples were genes for chemokines and pro-inflammatory cytokines (Figures 2A, 2B, and S2A), suggesting that repRNA-transfected muscle is in a pro-inflammatory state. Of the 192 gene transcripts that were differentially expressed between repRNA/LION- and repRNA/LNP-injected mice, the vast majority (188 genes) were preferentially induced by repRNA/LION (Figure 2C). These preferentially induced genes in repRNA/LION-injected mice included characteristic gene transcripts of antigen-presenting cells (APCs) such as macrophages, monocytes, and dendritic cells (DCs) (e.g., H2-ab1/I-Ab, Itgal/Cd11a, and Itgam/Cd11b) (Figure 2D). At the molecular pathway levels, the preferentially induced genes in repRNA/LION-injected mice included gene transcripts for Fcγ receptor-dependent phagocytosis, co-stimulation by the CD28 family, and class I MHC-mediated antigen processing and presentation pathway (Figure 2E). Individual genes of interest for co-stimulatory molecules (Figure 2F) and multiple types of interferons (Figures 2G and S2B) were preferentially induced by repRNA/LION over repRNA/LNP, except for IFN-β (Ifnb1) showing an opposite trend (Figure S2B).
Figure 2.
Intramuscular injection of repRNA/LION activates the local muscle innate immune response in mice
(A–G) C57BL/6 mice received i.m. injection of repRNA/LION and repRNA/LNP encoding SEAP (10 μg) (n = 4). Fourteen hours later, the animals were sacrificed, and total RNA was isolated from the muscle of these animals. Transcripts were analyzed by NanoString. (A) The expression of muscle genes included in the host response panel is represented by heatmaps. The color gradient of blue to orange indicates mean subtracted normalized log2 expression values. (B and C) repRNA-induced genes are shown as volcano plots. Genes of special interest are indicated in red. Differential expression in muscle genes between repRNA/LION and naive mice, repRNA/LNP and naive mice (B), and between repRNA/LION and repRNA/LNP (C) are shown as a volcano plot. (D) PanglaoDB cell type analysis for the differentially expressed genes in the muscle of repRNA/LION vs. repRNA/LNP. (E) NanoString annotation analysis for the differentially expressed genes in the muscle of repRNA/LION vs. repRNA/LNP. (F) Normalized expression of the genes for CD40, CD80, and CD86 in the muscle of indicated mice is shown as boxplots. (G) Normalized expression of the genes for interferons in the muscle of indicated mice is shown as boxplots. (F and G) All the boxplots show min to max values. Statistical comparison of mean values among groups was performed by ordinary one-way ANOVA and Dunnett’s multiple comparison test between values in the naive group vs. values in each repRNA-delivered group. (H–J) C57BL/6 mice received i.m. injection of repRNA/LION or repRNA/LNP expressing GFP (10 μg) (n = 5). (H and I) At indicated time points, mice were sacrificed, and their muscles and dLNs were processed using a skeletal muscle dissociation kit and collagenase-based cell isolation, respectively. The frequencies of CD11b+ cells in the muscle and dLNs (H) and surface CD86 expression levels on the indicated cell types (I) were determined by flow cytometry. Statistical comparison of mean values among groups was performed by multiple unpaired t tests between values in the LION group vs. values in the LNP group at each time point. (J) Mice were sacrificed at day 7 after i.m. injection of repRNA/LION or repRNA/LNP expressing GFP. Their muscle tissues were isolated, sectioned, and analyzed by multiplex immunofluorescence. CD11b and CD64 were stained with antibodies against each protein. Scale bar, 200 μm. Statistically significant results are shown as asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
We next analyzed the innate immune cell composition in the muscle of repRNA-delivered C57BL/6 mice at days 1 and 7. C57BL/6 mice received i.m. injection of repRNA formulated with either LION or LNP, then non-myocytes were analyzed by enzymatically removing myocytes from dissected muscle. Similar to the previous observations using a squalene emulsion-based adjuvant MF59 and a cationic nanoemulsion (CNE),21,22 CD11b+ cells were significantly expanded in the muscle and, by day 7, in the dLNs of repRNA/LION-delivered mice compared with those of repRNA/LNP-delivered mice (Figure 2H. The gating strategy is included in Figure S2C). At the cell-type level, repRNA/LION induced more infiltration of many cell types, including macrophages (CD11b+ CD64− CD11c−), monocyte-derived DCs (MoDC) (CD64+ CD11c+ MHC-II+), conventional DC2 (CD11c+ MHC-II+ XCR1− CD11b+), neutrophils (CD11b+ Ly6Ghi Ly6Clo), and NK cells (Ly6G− CD64− CD3− NK1.1+), in the muscle than repRNA/LNP (Figure S2D). MoDCs were also rapidly expanded in the dLNs of repRNA/LION-injected mice compared with repRNA/LNP-injected mice (Figure S2E). Non-myocytes, in particular, macrophages, pDCs, and MoDCs were all activated to a greater extent following injection in the muscle of repRNA/LION relative to repRNA/LNP, as determined by qPCR and flow cytometry of an early activation marker, CD86 expression (Figures 2I and S2F). Confocal microscopic analysis confirmed the infiltration of CD64+ cells and CD11b+ cells in the proximity of repRNA/LION-transfected myocytes by day 7, while the infiltration of CD64+ cells and CD11b+ cells were relatively minor in the muscles of repRNA/LNP-injected mice (Figures 2J and S2G). Of note, repRNA/LNP induced substantial activation of all the analyzed cell types in dLNs as early as 1 day after injection, and the activation levels had reverted to background levels by day 7 except for macrophages (Figure 2I). RepRNA/LION, on the other hand, did so only to a minor extent. These data indicate differential impacts on the location and kinetics of innate immune responses between the two formulations; repRNA/LION mainly activates the innate immune system in the injected muscle, while repRNA/LNP does so in the dLNs. Taken together, these data indicate that i.m. injection of repRNA/LION robustly induces the local innate immune response by rapidly upregulating genes for cytokines and chemokines, and subsequent innate immune cell activation in the injected muscle.
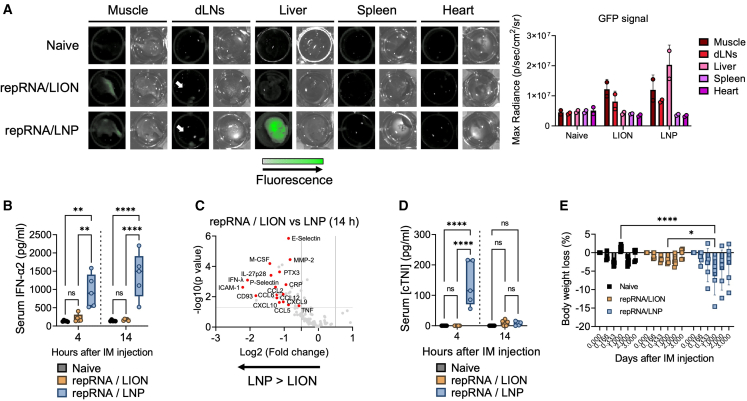
Intramuscular injection of repRNA/LION induces minimal systemic cytokine response in mice
Many studies reported that repRNA/LNP induces systemic innate responses.19,20 Therefore, we next compared the systemic innate immune response to repRNA/LION and repRNA/LNP. We first utilized IFN stimulation response element reporter mice (Mx1-GFP Tg) that express GFP in response to type I and type III IFN signaling, two central mediators of the innate immune system.23 IVIS analysis of the extracted tissues from repRNA-delivered mice detected a repRNA-induced GFP reporter signal in the injected muscle and dLNs in both repRNA/LION- and repRNA/LNP-injected mice at 24 h after i.m. injection (Figure 3A). When more distal organs were examined, the strong GFP signal was detected in the liver of repRNA/LNP-injected mice. However, no signal was detected in the liver of repRNA/LION-injected mice 24 h after i.m. injection. Neither group of animals had a detectable GFP signal in the heart and spleen 24 h after i.m. injection.
Figure 3.
Intramuscular injection of repRNA/LION induces minimal systemic cytokine response in mice
(A) Mx1-GFP Tg mice received i.m. injection of repRNA/LION or repRNA/LNP encoding SEAP (10 μg). Twenty-four hours later, mice were sacrificed, and the indicated tissues were isolated. GFP signaling (reporting type I/III IFN responses) in the extracted tissues was analyzed by IVIS (representative data of two biological replicates are shown). The bar graphs indicate means +SD (n = 2) with individual values. (B) C57BL/6 mice received i.m. injection of repRNA/LION or repRNA/LNP encoding SEAP (10 μg). Serum IFN-α2 levels at the indicated time points were determined by ELISA and shown as boxplots (n = 5). (C) Cytokine profiling of sera isolated from 10 μg of repRNA/LION- and repRNA/LNP-injected mice at 14 h was determined by cytokine array. The results from the calculation of the values of repRNA/LION divided by the values of repRNA/LNP are shown (n = 3). (D) C57BL/6 mice were i.m. injected with 10 μg of repRNA/LION or repRNA/LNP encoding SEAP. Serum levels of cardiac troponin-I (cTNI) at indicated time points were determined by ELISA and shown as boxplots (n = 5). (E) C57BL/6 mice were i.m. injected with 10 μg total of repRNA/LION or repRNA/LNP encoding mixed antigens derived from EV-D68 and RSV. Their body weight changes were monitored over 3 days after the immunization, and the percentages of the body weight loss are shown as bar graphs (mean) ± SD (error bars) with individual values (square dots). All the boxplots show min to max values with all data points. Statistical comparison of mean values among groups was performed by ordinary two-way ANOVA and Tukey’s multiple comparisons test between each group. Statistically significant results are shown as asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
We next measured systemic (circulating) type I IFN levels in mice injected with repRNA delivered by either LION or LNP via the i.m. route. Consistent with previous reports,19,20 repRNA/LNP induced abundant serum type I IFN levels within 1 day after i.m. injection (Figure 3B). In contrast, the serum levels of type I IFNs in mice injected with repRNA/LION were not statistically significant compared with mice in the naive group. Similar observations, with extended time points, were made in experiments using repRNA expressing a different transgene (Cre recombinase) (Figure S3A), non-replicating 5′-triphosphate RNA (Figure S3B), and non-replicating 5′-capped mRNA (Figure S3C). In addition to type I IFN, several chemokines (e.g., CCL2 and CXCL10), selective cytokines (IFN-λ, IL-27p28, and TNF), and C-reactive protein (CRP), a protein linked to reactogenic response to vaccination in humans,24 and some markers associated with cardiovascular disease (PTX5,25 selectin families,26,27 and ICAM-128) were also detected at significantly lower levels in sera of repRNA/LION-injected mice compared with the high levels circulating in repRNA/LNP-injected mice (Figures 3C, S3D, and S3E). These data indicate that i.m. injection of repRNA/LION does not induce detectable systemic inflammatory responses.
Since increased risks of myocarditis and/or pericarditis have been associated with mRNA/LNP COVID-19 vaccination,29,30,31 albeit lower than the risk associated with COVID-19, we measured a serological marker for cardiac damage, cardiac troponin-I (cTNI), in the repRNA-injected mice. At 4 h after i.m. injection, repRNA/LNP-injected mice had significantly elevated cTNI levels, whereas repRNA/LION-injected mice showed only a minor elevation that was statistically insignificant compared with naive mice (Figures 3D and S3F). Despite the observed increases in cTNI, none of the repRNA/LNP-injected mice in our study showed ongoing inflammation in heart tissue (Figure S3G), suggesting that cardiac damage caused by repRNA/LNP is transient and not sufficient to induce overt myocarditis in C57BL/6 mice.
We next monitored the signs of systemic reactogenicity in the immunized mice. Overt changes in body condition of the immunized mice were not observed. But monitoring of body weight after immunization revealed a very minor weight loss in repRNA/LION-injected mice, whereas more and significant body weight loss was revealed in repRNA/LNP-injected mice (Figure 3E). Taken together, our data demonstrate that i.m.-administered repRNA/LION restricts innate immune signaling locally in the injection site but does not trigger systemic inflammatory responses. In contrast, repRNA/LNP triggers both local and systemic innate immune/inflammatory responses, and significant body weight loss.
Intramuscular injection of repRNA/LION induces a comparable binding Ab response to repRNA/LNP despite lacking systemic innate responses in mice
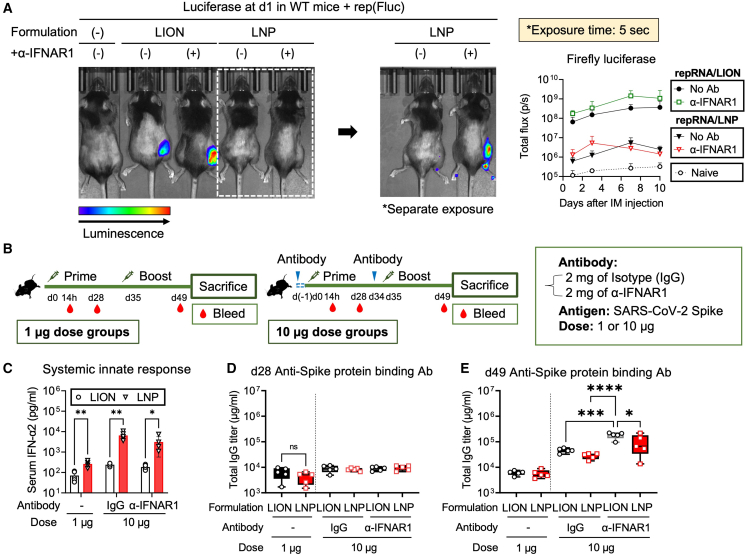
Next, we analyzed the impact of systemic and local innate responses on transgene expression and adaptive immune responses after i.m. vaccination with repRNA/LION and repRNA/LNP. We previously observed the inhibitory role of the type I IFN system on transgene expression from repRNA delivered by a cationic nanocarrier.32 The inhibitory effect of type I IFN on transgene expression from repRNA was confirmed in vitro with LION (Figure S4A). Similarly, systemic blockade of type I IFN signaling in repRNA/LION- and repRNA/LNP-injected mice showed greater reporter gene expression in the muscle-injected site at day 1, but the extent was much greater with LION (Figure 4A). Reporter protein expression was still restricted to the local injection site even in the absence of type I IFN signaling in vivo (Figure S4B), indicating that type I IFN signaling does not alter biodistribution, but controls protein expression locally.
Figure 4.
Intramuscular injection of repRNA/LION induces a comparable binding antibody response to repRNA/LNP despite lacking systemic innate responses in mice
(A) C57BL/6 mice received an intraperitoneal injection of anti-IFNAR1 Ab (2 mg) or were left untreated. Twenty-four hours later, mice were i.m. injected with 10 μg of repRNA encoding firefly luciferase (Fluc) delivered by LION and LNP. At 24 h after the i.m. injection, luciferase expression was analyzed in vivo by IVIS. The total flux data over the course of in vivo transfection of repRNA are shown as means ± SD (data are the pool of two independent experiments; n = 4). Statistical comparison of mean values between groups at each time point was performed by ordinary two-way ANOVA and Tukey’s multiple comparisons test. The complete list of the results of the statistical test is included in Table S3. (B) The experimental design for analyzing the antigen-specific binding Ab response to SARS-CoV-2 Spike protein. C57BL/6 mice received i.m. injection of repRNA encoding SARS-CoV-2 Spike protein (1 or 10 μg) formulated with LION or LNP at days 0 and 35 (n = 5). The left panel shows the regimen for a 1-μg dose. The right panel shows the regimen for a 10-μg dose. For the 10-μg dose regimen, the mice also received an intraperitoneal injection of isotype IgG or α-IFNAR1 (2 mg) 1 day before immunization. (C) At 14 h, serum IFN-α2 levels were measured by ELISA. Statistical comparison of mean values among groups was performed by multiple unpaired t tests between values in the LION group vs. values in the LNP group at each time point. (D and E) Anti-SARS-CoV-2/Spike protein binding Ab response at day 28 (post-prime) (D) and day 49 (post-boost) (E) determined by ELISA. Statistical comparison of mean values among groups was performed by ordinary two-way ANOVA and Tukey’s multiple comparisons test between the values in each group. Unless otherwise noted, only statistically significant results are shown as asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
The inhibitory effect of type I IFN signaling on vaccine immunogenicity is also reported by others using alternative local administration-based RNA vaccine modalities33 and viral vaccines,34 and it has been previously shown that targeted shutoff type I IFN production and action is effective for repRNA/LNP vaccination.3,35,36,37,38 To compare the immunogenicity of repRNA/LION and repRNA/LNP vaccinations with or without short-term blockade of type I IFN signaling, we immunized C57BL/6 mice with 1 or 10 μg of repRNA encoding SARS-CoV-2 Spike protein12 delivered via LION or LNP in the presence or absence of intraperitoneal injection of isotype control IgG or α-IFNAR1 (2 mg) at 1 day before the immunizations (Figure 4B). At 14 h after the prime dose, a dose-dependent systemic innate response was observed in all conditions with and without Ab treatment, with significantly lower levels in repRNA/LION-injected mice (Figure 4C). At day 28 (before the boost), repRNA/LION and repRNA/LNP induced comparable Ab response at both 1- and 10-μg doses (Figure 4D). At day 49 (2 weeks after the boost), repRNA delivered by both formulations induced comparable levels of spike-binding Ab response at the 1-μg dose (Figure 4E). Others have recently shown that systemic innate responses were correlated with Ab responses of repRNA/LNP vaccination.19 Nevertheless, at the 10-μg dose, where the difference in the systemic innate responses between LION and LNP are greater than those at the 1-μg dose (Figure 4C), repRNA/LION-injected mice still induced comparable, if anything slightly better, responses than repRNA/LNP-injected mice (Figure 4E). Furthermore, systemic blockade of type I IFN signaling increased the spike-binding Ab response (Figure 4E) and an antigen-specific IFN-γ-producing T cell response (Figure S4C) in repRNA/LION-vaccinated mice significantly, and only modestly and more variably in repRNA/LNP-vaccinated mice. Such variability in the effect of type I IFN signaling on the immunogenicity of repRNA/LNP vaccine is most likely due to its systemic biodistribution and interference by type I IFN response at different levels from a wide variety of cells. These data suggest that the local type I IFN response (Figure 2) may play a key role in regulating immunogenicity after i.m. vaccination of repRNA/LION. Thus, LION-mediated muscle-localization of repRNA and the subsequent upregulation of a robust local innate response induces antigen-specific binding Ab and T cell responses at comparable levels to repRNA/LNP, even in the absence of a systemic innate response. However, the adaptive immune response to repRNA is, to some extent, subdued by the type I IFN system.
Intramuscular injection of multivalent repRNA/LION but not repRNA/LNP induces a neutralizing Ab response in mice
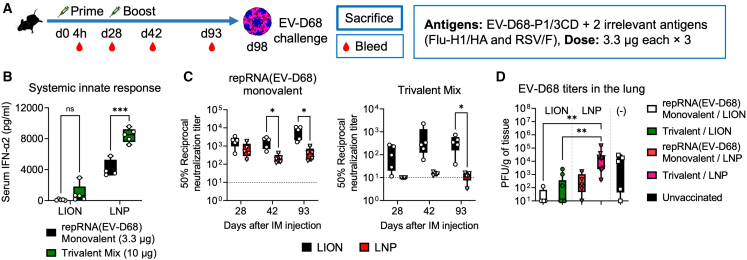
Multivalent RNA vaccines are expected to induce a broad immunity against multiple antigenic targets such as different virus species.10 However, the greater the number of target antigens, the larger the overall dose. Therefore, multivalent repRNA/LNP vaccines may cause dose-limiting reactogenicity (Figures 3 and 4C), considering the maximum tolerable dose observed in ongoing clinical trials for this modality is 5 μg. To examine this in mice in the context of the EV-D68 infection model, we immunized mice twice, 28 days apart, with 10 μg (total dose) of a trivalent vaccine formulated with LION or LNP and composed of 3.3 μg each of repRNA vaccines against EV-D68 P1/3CD (Warner et al., manuscript submitted) and two irrelevant antigens, influenza (Flu) hemagglutinin (HA), and respiratory syncytial virus (RSV) fusion (F) protein along with monovalent LION and LNP control groups (Figure 5A). The 10-μg trivalent repRNA/LNP induced higher levels of systemic type I IFN than the 3.3-μg monovalent control (Figures 5B and S5A), confirming a dose-dependent systemic innate response in the context of a multivalent composition. Similar to the observation following SARS-CoV-2 Spike vaccination (Figures 4B–4E), monovalent repRNA-expressing Flu/HA and RSV/F delivered by LION and LNP induced comparable levels of binding Ab responses (Figure S5B). In addition, the binding Ab responses were almost equally induced by both monovalent and trivalent repRNA vaccinations, indicating that the observed suboptimal innate response to the 10-μg dose of mixed repRNAs does not appear to negatively impact the binding Ab response. Since protection from enterovirus infection is thought to rely most heavily on neutralizing Ab response,39 we next measured the neutralizing Ab titers in sera from the vaccinated mice. Multivalent repRNA vaccination induced neutralizing Ab responses against EV-D68 when delivered by LION but not LNP, as measured at all time points post-prime and -boost (Figure 5C). To test whether these observed differential immune responses would lead to differential protective outcomes, all mice, including an unvaccinated group, were then challenged with EV-D68 via the intranasal route, and lungs were harvested 24 h later for measurement of viral loads by plaque assay. While no differences in viral loads in the lung were observed between mono- and trivalent LION-formulated repRNAs, as well as monovalent LNP-formulated repRNA, the trivalent LNP-formulated repRNA group exhibited a lack of viral load reduction (Figure 5D), confirming that the LNP multivalent composition was suboptimal. These data suggest that the suboptimal and elevated innate response to the total repRNA dose delivered by LNP impaired the neutralizing Ab response to EV-D68 P1/3CD repRNA, while repRNA/LION was only modestly affected by this. Overall, our data in mice demonstrate that multivalent repRNA/LION induces neutralizing Ab responses against a vaccine target without causing systemic innate responses.
Figure 5.
Intramuscular injection of multivalent repRNA/LION but not repRNA/LNP induces a neutralizing antibody response in mice
(A) The experimental design for analyzing the adaptive immune response to the multivalent antigens encoded in repRNA in mice. C57BL/6 mice received i.m. injection of repRNA encoding three indicated antigens (EV-D68 P1/3CD, Flu/H1-HA, and RSV/F) (3.3 μg each) formulated with LION or LNP at days 0 and 28. (B) Serum IFN-α2 levels at 4 h after the prime dose were determined by ELISA and shown as boxplots. Min to max values with all data points are shown (n = 4–5). (C) EV-D68 neutralizing activity of sera of vaccinated mice was measured by neutralization assay. Geometric means with each individual value are shown as bars and symbols (n = 4–5). (B and C) Statistical comparison of mean values among groups was performed by multiple unpaired t tests between values in the monovalent group vs. values in the trivalent mix group (B) for each formulation, or values in the LION group vs. values in the LNP group at each time point (C). (D) EV-D68 titers in the lung of infected mice were determined by plaque assays (n = 5–10). Min to max values with all data points are shown. Statistical comparison of mean values among groups was performed by Kruskal-Wallis test and Dunn’s multiple comparisons test between the values in each group. Unless otherwise noted, only statistically significant results are shown as asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
Discussion
Rapid innate immune responses to in-vivo-administered mRNA vaccines hamper adaptive immunogenicity.18,24,40 Accordingly, innovation around the production and processing of this molecule has been the major focus in the field of RNA medicine.38,41 The advances by Karikó and co-workers in reducing the host’s detection of such RNA molecules by modifying nucleotides and removing dsRNA are considered key developments that have enabled mRNA vaccine technology.42,43,44 However, next-generation repRNA approaches, which have recently delivered on their promise of dose-sparing capacity, are not amenable to nucleotide modifications since (1) we previously observed that pseudouridine-modified repRNA failed to produce transgene,32 (2) the impact of the modified nucleoside would no longer be present after the RNA amplification,45 and (3) dsRNA intermediates are always produced as a byproduct of replication.3 Therefore, alternative approaches are needed to enable the safe and effective administration of repRNA to realize the potential these next-generation RNA medicines offer. The recent emergency use approval of repRNA/LION in India, a milestone for the repRNA field in general, with demonstrated safety at a 5-fold higher dose than could be achieved with an LNP formulation, suggested that cationic nanocarriers may play a role in achieving these goals. Our present data showed that repRNA/LION elicits an effective adaptive immune response in the absence of systemic innate responses, including type I IFN and CRP, both of which have been implicated in reactogenic outcomes in humans.24,46 repRNA/LION-injected mice also showed only minor body weight loss compared with repRNA/LNP-injected mice. This may account for the low frequency of systemic adverse events observed in clinical trials of a repRNA/LION-based vaccine platform in multiple countries (Link) (India: CTRI/2021/09/036379; Brazil: clinicaltrial.gov NCT04844268; USA: clinicaltrials.gov NCT05132907; data not shown and personal communication). Of note, our data suggest that in vivo delivery of repRNA could be cardiotoxic when delivered by LNP, but not LION. We do, however, note that neither LNP biodistribution nor IFN response (Mx1-GFP) was detected in the hearts of mice after repRNA/LNP injection. Therefore, the cardiotoxic effect of repRNA/LNP in mice is likely attributed to extracardiac factors. Importantly, repRNA/LNP delivery itself is likely insufficient to trigger myocarditis. Others reported that IL-1RA antibodies are detectable in some patients with myocarditis after mRNA/LNP vaccination47,48 (Thurner et al., 2021 bioRxiv, doi: https://doi.org/10.1101/2021.04.23.441188). In addition, elevated circulating free spike protein and inflammatory cytokine profiling were reported in the blood of adolescents and young adults who developed myocarditis compared with those who had no vaccine-related complications.49 As such, we expect that i.m. injection of repRNA/LNP may not induce myocarditis unless other factors (e.g., comorbidities, genetic factors, and repeat dosing) coexist. However, a very recent study by Barmada and Klein et al. revealed elevations in circulating cytokines, chemokines, matrix metalloproteases, and interferon-stimulated genes in peripheral blood cells, in a cohort of patients who developed rare myocarditis and pericarditis after mRNA/LNP vaccination, in most cases after the second dose.50 Of note, they detected neither immune targeting of cardiac and IL-1RA autoantigens nor enhanced clonal expansion of B and T lymphocytes. We detected significantly elevated serum levels of similar protein homologs (e.g., CRP, cTNI, CXCL10, MMP, and IFN-⍺) specifically in the blood of the repRNA/LNP-injected mice, which parallels the cytokinopathy observations by Barmada and Klein et al.50 Thus, our present data suggest that more careful investigation, including measurement of transient cardiac damage, is warranted for several RNA vaccine modalities that have been previously reported to increase serum cytokine levels after administration. Indeed, recommendations for increased monitoring diligence for cardiac events has been communicated by the FDA to developers of RNA vaccines (Link), including revisions to patient and provider fact sheets for the currently licensed mRNA COVID-19 vaccines (Link) (Link) (Link), indicating the associated increased risks of myocarditis and pericarditis. Nevertheless, it is worth noting that repRNA/LION did not induce significant elevation in serum cardiac troponin and other cytokines compared with naive mice.
Our present data also link these safety features of repRNA/LION to biodistribution. We hypothesize that the differential biodistribution between repRNA/LION, which remains localized at the injection site, and repRNA/LNP, which shows a broad biodistribution, is due to structural differences between the nanoparticles. While we are still elucidating all the structural attributes of cationic nanocarriers that can affect biodistribution, surface charge, measured as zeta potential, is a distinguishing feature between the two formulations. The LNPs we used in these studies are similar in composition to LNPs currently approved in mRNA vaccines, characterized by a relatively neutral surface charge, and further shielded by polyethylene glycol (PEG), allowing the formulation to drain out from the injection site and actively being taken up by phagocytic cells and hepatocytes.18,51 In contrast, repRNA/LION has a net positive surface charge that may enable active transfection of muscle cells in the injection site, and could explain the larger transfected area and elevated levels of expressed protein we observed in mouse muscle compared with LNP/repRNA.
Although many researchers have examined repRNA in RNA medicines, the only platform authorized for human use to date is based on LION. Unlike repRNA/LNP-induced adaptive immunity, which was recently shown to correlate positively with systemic innate responses,19 our findings show that repRNA/LION-induced adaptive responses are likely shaped by early local, but not systemic, innate responses. Therefore, repRNA/LION and repRNA/LNP are likely to induce adaptive immune responses via distinct mechanisms due to their differential biodistribution. While further investigation is required to characterize the functional impact of muscle infiltrating immune cells on the overall immune response, it is noteworthy that we observed intensive expansion of MoDCs in the muscle and dLNs of repRNA/LION-injected mice, and that CD64+ MoDCs are reported to be the most efficient inducers of IFN-γ-producing T cells in alum-injected muscle.52 The advantage of the locally restricted innate response induced by repRNA/LION not only efficiently induces innate and adaptive immunity but also mitigates unwanted systemic reactogenicity. By limiting systemic innate responses, we aim to safely achieve effective vaccination against multiple antigens in the context of multivalent drug products or immunotherapy against poorly immunogenic self-antigens, in the context of oncology, approaches that are currently impeded by the dose-limiting reactogenicity of repRNA. Importantly, our present data demonstrated that LION allows us to achieve immunogenic multivalent vaccination and protective efficacy in mice, without triggering systemic cytokine production that could not be achieved with repRNA/LNP. Although our present study focused on repRNA, Arevalo et al. recently showed the successful protection of mice and ferrets from Influenza virus with 50 and 60 μg total dose, respectively, of conventional mRNA/LNP encoding HA antigens from all 20 known influenza A virus subtypes and influenza B virus lineages.10 However, Moderna reported that, in their interim results for their phase III clinical trial of mRNA-1010, which evaluates their quadrivalent vaccines of influenza antigens (A/H3N2, A/H1N1, B/Victoria, B/Yamagata) encoded in their mRNA/LNP vaccine mRNA-1010 at 50 μg total dose, achieved superiority in seroconversion rates for A/H3N2 and A/H1N1, as well as superiority in geometric mean titer ratios for A/H3N2 and non-inferiority in geometric mean titer ratios for A/H1N1 but non-inferiority was not met for an either endpoint for the influenza B/Victoria- and B/Yamagata-lineage strains, highlighting the challenges that multivalent mRNA vaccination continues to face. Given their reported 2- to 3-fold increase in rates of systemic reactogenicity compared with the standard influenza vaccine, increasing the dose to overcome this problem is likely not an option. Therefore, the 20-valent approach described by Arevalo et al., will need to overcome dose-limiting reactogenicity challenges. Finally, while many of the innate-evading advances that enabled high-dose mRNA vaccines, such as modified nucleotides or sequence optimization, cannot be applied to repRNA to raise the dose ceiling, this study highlights that advances in formulation innovation can provide alternative solutions.
Materials and methods
Study design
The primary objective of this study was to analyze the innate response to LION/repRNA vaccine in mice. Endpoints were selected before the start of each study on the basis of the primary objective of characterizing the safety and immune responses to vaccination with a LION/repRNA vaccine. Group sizes were based on power analyses using data from previous experiments using a similar repRNA platform and, although no blinding was used, mice were randomly distributed between groups. Replication of experiments and the number of biological and technical replicates varied between experiments, as described in the figure legends.
Animal studies
All animal studies were approved by the University of Washington Department of Comparative Medicine (assurance no. A3381-01) Institutional Animal Care and Use Committee. The facility where animal studies were conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International, and follows guidelines set forth by the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
In vivo animal treatments
Age-matched 6- to 12-week-old female C57BL/6, Mx1-GFP Tg (The Jackson Laboratory, no. 033219), Ai9 Tg (The Jackson Laboratory, no. 007909), and LSL-Luc Tg (The Jackson Laboratory, no. 034320) mice were used for in vivo studies. For in vivo neutralization of IFNAR1 signaling, mice were injected with 2 mg of α-IFNAR1 polyclonal antibody (clone MAR1-5A3, BioXcell, no. BE0241) intraperitoneally. For repRNA vaccination, the vaccine was prepared as described below and consistent with previous reports12 using a LION complexed with repRNA encoding the full-length spike of SARS-CoV-2 (repRNA-CoV-2S), Influenza H1/HA protein, RSV-F protein, and EV-D68 P1/3CD protein (manuscript in preparation). The vaccines were delivered by 50-μL i.m. injection into the quadriceps. Animals received two doses of a 1-, 3.3-, or 10-μg dose of repRNA/LION or remained unvaccinated (naive).
Cells
Huh7.5 and RD cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% FBS, 100 units (U)/mL penicillin, 100 mg/mL streptomycin, and 2 mM L-glutamine (Life Technologies) at 37°C/5% CO2. THP-1 cells were maintained in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, and 2 mM L-glutamine (Life Technologies) at 37°C/5% CO2. Primary non-myocytes were isolated from the muscles using Skeletal Muscle Dissociation Kit (Miltenyi Biotech, no. 130-098-305) according to the manufacturer’s instructions. Popliteal and inguinal lymph nodes were isolated as the muscle-dLNs, and single-cell suspensions were prepared by the collagenase digestion method.
Virus
One representative isolate of EV-D68 (US/IL/14–18952 [NR-49131]) was obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI resources). Virus stocks were grown in RD cells at 33°C with 5% CO2. Confluent flasks of RD cells were infected at MOI of 0.1 and incubated until cytopathic effect was observed in the cell monolayer after ∼2 days. Cells and supernatants were harvested, sonicated, and clarified supernatants titered by plaque assay.
Accession numbers for the antigens
Accession numbers for the antigens encoded in repRNA we used in this study are summarized in Table S1.
In vitro synthesis of RNA
repRNA and mRNA were enzymatically generated by T7 polymerase as described previously.53 Following in vitro transcription with T7 polymerase, RNA was treated with DNase I, and a 7-methylguanylate (Cap-0) structure was added with vaccinia capping enzyme. All RNA was precipitated with lithium chloride, washed with 70% ethanol, resuspended in nuclease-free water, and stored in small aliquots at −80°C. Quality control metrics included integrity by capillary gel electrophoresis (Agilent 5200 Fragment Analyzer System) and purity and concentration by UV-vis spectroscopy (NanoDrop 2000). For the synthesis of nucleotide-modified mRNA, pseudouridine replaced uridine in the in vitro transcription reaction.
Formulations
-
(1)
LION was prepared as described previously.11,54 In brief, the oil phase (squalene, Span 60, and DOTAP) was sonicated for 30 min in a 65°C water bath. Separately, the aqueous phase, containing Tween 80 and sodium citrate dihydrate solution in water, was prepared with continuous stirring until all components were dissolved. The oil and aqueous phases were then mixed and emulsified using a high-shear mixer (Silverson L5M-A), and the crude colloid was subsequently processed by passaging through a microfluidizer at 30,000 psi with an M110-P microfluidizer (Microfluidics) equipped with a 75-μm Y-geometry diamond interaction chamber and an auxiliary H30Z 200-μm ceramic interaction chamber until the Z-average hydrodynamic diameter, measured by dynamic light scattering (Malvern Zetasizer Nano S), was approximately 60 nm with a polydispersity index of 0.2 or lower prior to complexing. The microfluidized LION was terminally filtered with a 200-nm pore size polyethersulfone filter and stored at 2°–8°C.
-
(2)
LNP:RNA was encapsulated into LNP’s using methods described previously.55 In brief, lipid components were dissolved in ethanol at a ratio of 50:10:38:2 (ionizable lipid [SM-102]/helper lipid [DSPC]/cholesterol/DMG-PEG 2000) and mixed with RNA buffer at pH 4.5 at an N:P 5.5 using a glass micromixer chip. After mixing, the formulations were dialyzed against PBS (pH 7.4) for 16–24 h. Formulated LNPs were concentrated using Amicon Ultra centrifugal filter devices (EMD Millipore, Billerica, MA) and stored at 5°C. RNA encapsulation was quantified using a Ribogreen assay using Triton to disrupt formulated LNPs, all LNPs had 92% ± 9% (n = 20) encapsulation. Particle size (average Z-average diameter = 87 ± 18 nm, n = 20), PDI (average = 0.19 ± 0.08, n = 20), and zeta potential (average = 8 ± 5 mV, n = 20) were measured using a Malvern Zetasizer Ultra.
ELISA
-
(1)
IFN-α2 and TNF ELISA
Serum IFN-α2 and TNF levels were measured by using Lumikine Xpress IFN-α 2.0 ELISA kit (InvivoGen, no. luex-mifnav2) and a Mouse TNF alpha Uncoated ELISA kit (Invitrogen, no. 88–7324), respectively, according to the manufacturer’s instructions.
-
(2)
Serum cTNI ELISA
Serum cTNI levels were measured by using cTNI ELISA kit (MyBiosource, no. MBS766175) according to the manufacturer’s instructions.
-
(3)
Binding Ab ELISA
Antigen-specific IgG responses were evaluated by ELISA as described previously.11 In brief, ELISA plates were coated with 1 μg/mL of recombinant SARS-CoV-2 Spike protein,56 1 μg/mL of influenza virus H1 HA protein (Sino Biological, no. 11052-V08H), and 1 μg/mL of human RSV A2 F protein (Sino Biological, no. 11049-V08B). Before serially diluted serum samples were added and detected via anti-mouse IgG-HRP (Southern Biotech, no. 1031-05). Plates were developed using a TMB substrate (Seracare, no. 5120-0083) and absorbance was measured at 450 nm (ELX808, Bio-Tek Instruments). Mouse total IgG concentrations were determined from a standard curve using purified mouse IgG.
EV-D68 neutralization assay
To measure the neutralizing Ab activity of polyclonal sera from immunized mice, the Agilent xCELLigence real-time cell analysis (RTCA) multiple plate (MP) instrument was used to quantify cell monolayer integrity. RD cells were plated on 96-well electronic microplates (E-Plate 96) at 60,000 cells/well. Cell impedance was measured every 15 min and recorded on the RTCA-MP instrument. The following day, individual mouse serum was diluted 4-fold starting at 1:10, in infection medium containing DMEM high glucose, 2% FBS, and 1% Pen/Strep. One hundred microliters of 1 × 106 PFU/mL of EV-D68 was added to diluted serum. Serum-virus mixture was incubated at 37°C for 30 min to allow Ab-virus interaction. Plates were then removed from the RTCA, and medium was removed from each well and replaced with serum-virus mixtures. Plates were put back on the RTCA and impedance was measured every 15 min for 160 h.
To calculate the 50% neutralization titer (1) area under the normalized cell index curve over time was determined using the RTCA software for each serum dilution as well as control wells containing virus alone or no virus, (2) percent neutralization at each serum dilution was then calculated, normalized between virus-alone and no-virus conditions, and plotted for regression analysis in Prism, and (3) serum dilution interpolated at the 50% neutralization level.
Cytokine array analysis
Sera from mice that received i.m. injection of 10 μg of repRNA/LION or repRNA/LNP were subjected to cytokine array. The array was performed using Proteome Profiler Mouse XL Cytokine Array (R&D Systems no. ARY028) according to the manufacturer’s instructions. In brief, membranes were blocked for 1 h at room temperature. Meanwhile, samples were incubated with detection antibodies for 1 h at room temperature. Then, the blocked membranes were incubated in the sample/detection Ab mixtures overnight at 4°C. Next day, the membranes were washed three times and incubated with diluted Streptavidin-HRP for 30 min at room temperature. After washing three times, the membranes were incubated with Chemi Reagent Mix, and each spot was visualized on ChemiDoc touch imaging system (Bio-Rad). The signal intensities of each spot were calculated using ImageJ software, and each value was normalized against the mean values of reference spots (pre-defined by the manufacturer).
IVIS
For ex vivo IVIS analysis, mice were intraperitoneally injected with D-luciferin. Mice were sacrificed 5 min later, and tissues were removed. Extracted tissues were soaked in PBS and analyzed by IVIS. Fluorescent images were obtained by using specific filters, and bioluminescent images were obtained by the open filter.
For in vivo IVIS analysis, mice were anesthetized, their hair was clipped, and intraperitoneally injected with 150 μg/mL of D-luciferin. After 5 min of the injection, ventral and dorsal photon emissions were analyzed during 5 s exposure, and the average total flux values of the region of interest were used for analyses.
Isolation of RNA from tissues
For muscle RNA isolation, muscle tissues were collected from mice, and RNA was isolated using the Fibrous RNeasy Mini Kit (QIAGEN). In brief, the injected muscles were homogenized in RLT buffer (a component of RNeasy Mini Kit) using TissueLyzer LT (QIAGEN). Cell debris was removed by centrifugation, and the supernatant was transferred to the RNeasy Mini column. After a centrifuge, followed by a one-time wash of the column with RW buffer, DNA was digested on the column using DNase II by incubation for 30 min at 55°C. Columns were washed with RW buffer once, then with RPE buffer twice, and the final elution was done with nuclease-free water. For repRNA (SARS-CoV-2 Spike protein) biodistribution, organs were collected from mice, and RNA was isolated using RNeasy Mini Plus Universal kit (QIAGEN). In brief, tissues were homogenized in QIAzol reagent using preset RNA purification protocol on gentleMACS Octo Dissociator (Miltenyi Biotech). Cell debris was removed by centrifugation, and the supernatant was transferred to the RNeasy Mini column.
NanoString analysis
Purified RNA (100 ng) was assessed for gene expression using the nCounter Mouse Host Response Panel cartridge (NanoString Technologies, Seattle, WA) on the nCounter MAX instrument.
Data were analyzed with NanoString nSolver software and ROSALIND.
Real-time RT-PCR
For the analyses of muscle and liver gene expression and NSP gene detection, up to 1 μg of RNA was reverse transcribed using an Invitrogen SuperScript First-Strand Synthesis System (Invitrogen, no. 11904018) or using an iScript cDNA Synthesis Kit (Bio-Rad, no. 1708891) for RT-PCR according to the manufacturer’s instruction. Transcribed cDNA libraries were diluted at 1:10 in nuclease-free water and used for quantitative real-time RT-PCR using SYBR Green Master Mix (Applied Biosystems, no. 4367659) with specific primer sets (primer information is included in Table S2).
For SARS-CoV-2 Spike RNA biodistribution, 0.5 μg RNA from each tissue, as well as a standard curve of SARS-CoV-2S repRNA was converted to cDNA in a reverse transcription reaction. Spike RNA levels in each tissue were quantified compared with the SARS-CoV-2S standard curve, and replicon copies per microgram of total RNA from each tissue were plotted below. The SARS-CoV-2S standard curve was used to establish the Lower Limit of Quantification, defined as the replicon copy number in the last dilution of RNA falling within the linear range of the assay. A positive reaction was considered any real-time RT-PCR amplification with a signal above the cycle threshold at the LLOQ. Samples that did not reach this threshold are plotted as half the LLOQ below.
Immunofluorescence and histological analysis
Mice were sacrificed at the endpoint, and tissues were harvested and fixed using buffered zinc formalin at room temperature overnight. For histological analyses, formalin-fixed tissues were paraffin embedded, and 3-μm sections were cut. For standard histological analyses, sections were stained with hematoxylin and eosin. For immunofluorescence, sections were deparaffinized with Histo-Clear II (Electron Microscopy Sciences, no. 6411101) for 10 min, 100% ethanol for 10 min, 90% ethanol for 3 min, 70% ethanol for 3 min, then washed with PBS for 5 min. The sections were then washed with PBS for 5 min three times and permeabilized with 0.5% Triton/PBS for 30 min at room temperature, followed by blocking with 1% normal goat serum for 30 min at room temperature. The blocked sections were incubated with RFP polyclonal to amplify tdTomato signal overnight at 4°C. The next day, the sections were washed with 0.3% Triton/PBS and treated with goat anti-rabbit for at least 2 h at room temperature. After the staining, sections were washed with 0.3% Triton/PBS and treated with TrueView Auto fluorescence kit (Vector Laboratories, no. SP-8400) according to the manufacturer’s instructions. Before mounting with the reagent provided by the kit, sections were counterstained with Hoechst 33342 (1:10,000 dilution in PBS) (Thermo Fisher Scientific, no. H3570).
Multiplex confocal microscopy
For confocal imaging, isolated muscle tissue was fixed using Cytofix (BD Biosciences) buffer diluted 1:3 with PBS for 12 h at 4°C and then dehydrated with 30% sucrose for 24 h at 4°C. Tissues were then embedded in O.C.T. compound (Tissue-Tek) and stored at −80°C. Tissues were sectioned on a Thermo Scientific Microm HM550 cryostat into 20-μm sections and were then prepared and imaged as described previously.57 A Leica SP8 tiling confocal microscope equipped with a 20× 0.7 NA oil objective was used for confocal image acquisition. All acquired raw imaging data were processed and analyzed in Imaris (Bitplane).
Cell isolation and flow cytometry
For analysis of immune cell populations infiltrating the muscle injection site, single-cell suspensions were acquired by first cutting off the mouse leg below the hip and above the ankle and then separating the skeletal muscle tissue from the bone using surgical scissors. Muscle tissue was then minced into small pieces and placed into C tubes (Miltenyi Biotec) containing digestion reagent according to the Skeletal Muscle Dissociation Kit protocol (Miltenyi Biotec). In brief, tissues were then incubated at 37°C and subjected to the Muscle program of the OctoMACS automatic cell separator (Miltenyi Biotec). For analysis of immune cell populations in DLNs, single-cell suspensions were acquired by manually disrupting the tissue with forceps and then treating with 400 U/mL collagenase D (Roche Applied Science) solution consisting of one part complete RPMI and nine parts PBS for 30 min at 37°C. Single cells were then stained for 30 min at 4°C. Data were acquired on a FACSymphony A3 flow cytometer (BD). The acquired data were analyzed using FlowJo software (BD).
Antibodies and staining reagents
Antibodies used for flow cytometry and fluorescent microscopy include the following: anti-CD11c (1:200 dilution, BV510, clone N418, BioLegend), anti-Ly6C (1:200 dilution, BV605, clone HK1.4, BioLegend), anti-Ly6G (1:200 dilution, BV785, clone 1A8, BioLegend), anti-CD64 (1:200 dilution, BV421, clone X54-5/7.1, BioLegend), anti-MHC-II (1:400 dilution, AF700, clone M5/114.15.2, BioLegend), anti-CD11b (1:200 dilution, APC-Fire750, clone M1/70, BioLegend), anti-XCR1 (1:200 dilution, BV650, clone ZET, BioLegend), anti-CD3 (1:200 dilution, BUV737, clone 17A2, BD), anti-CD86 (1:200 dilution, PE, GL1 clone, BD), anti-CD4 (1:200 dilution, BUV395, clone RM4.5, BD), anti-PDCA-1 (1:200 dilution, BV711, clone 927, BioLegend), anti-CD8 (1:200 dilution, BUV805, clone 53–6.7, BD), anti-NK1.1 (1:200 dilution, PerCP-Cy5.5, clone S17016D, BioLegend), anti-CD11b (clone M1/70, eBioscience), anti-CD64 (X54–57.1, BioLegend), anti-Ki67 (clone B56, BD Biosciences), RFP Polyclonal Ab (Thermo Fisher Scientific, no. 600-401-379-RTU), and goat anti-rabbit (Alexa Fluor 488 conjugated).
Secreted enzyme alkaline phosphatase assays
Secreted enzyme alkaline phosphatase (SEAP) activity was measured using a NovaBright Phospha-Light EXP Assay Kit for the SEAP reporter gene detection kit (Invitrogen, no. N10578) according to the manufacturer’s instructions.
EV-D68 challenge
Mice were injected intraperitoneally with 150 μL of a 13 mg/mL ketamine and 0.88 mg/mL xylazine stock solution. Once animals were not responsive to a toe pinch, mice were then challenged with 4.2 × 106 PFU of EV-D68 (MA) intranasally. Viral stocks were diluted in PBS as necessary to meet appropriate concentrations for administration. Twenty-four hours after intranasal infection, mice were euthanized using a CO2 chamber, and nasal cavity cartilage and lungs were collected.
Plaque assays
For the titration of virus stocks and tissue homogenates, RD cells were seeded in 12-well tissue culture plates at 500,000 cells per well the day before using. Virus stocks were diluted in infection medium containing 2% FBS, DMEM with high glucose, and 1% Pen/Strep. Complete cell culture medium was removed from monolayers of RD cells, and 100 μL of 10-fold dilutions of virus stock or tissue homogenate was added to each well. Plates were rocked every 15 min for 1 h at 33°C. After 1 h, 1 mL of a 1% agarose overlay in infection medium was placed in each well. Plates were incubated at 33°C for 3 days. Plates were fixed with a 10% formaldehyde solution for 20 min. Agarose and formaldehyde were removed and 200 μL of crystal violet was added to each well for 2–5 min. Plates were washed in water and plaques counted.
Mouse IFN-γ ELISpot assay
Splenocytes were isolated from mice 14 days after the second vaccination. Multiscreen plates (Millipore, no. MAIPS4510) were coated with rat anti-mouse IFN-γ capture Ab (BD, no. 551216) in PBS and incubated overnight at 4°C. The plates were washed in PBS and then blocked (2 h, room temperature) with RPMI (Invitrogen) containing 10% heat-inactivated fetal calf serum (Gibco). Splenocytes were plated at 5 × 105 cells per well and stimulated with the 10 μg/mL of PepMix SARS-CoV-2 (Spike B.1.351/Beta) (JPT, no. PM-SARS2-SMUT02-1) or PMA (0.25 μg/mL) and ionomycin (5 μg/mL), and cultured for 20 h (37°C, 5% CO2). Biotinylated rat anti-mouse IFN-γ Ab (BD, no. 554410) and streptavidin-alkaline phosphatase substrate (BioLegend, no. 170–3554) were used to detect IFN-γ-secreting cells. Spot-forming cells were enumerated using an immunospot analyzer from CTL ImmunoSpot profession software (Cellular Technology).
Statistics
The indicated statistical comparisons, as described in the figure legends, were performed using GraphPad Prism (GraphPad). p values less than 0.05 were considered significant and are indicated in figures as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Kaitlyn LaCourse and the POPS team at NanoString for supporting NanoString analyses. This work was funded by NIH/NIAID grant 1R43AI165100-01, contract 75N93020C00028 (to J.H.E.). Additional support was from NIH/NIAID contract 75N93019C00037 (to M.S.D.), NIH/NIAID grant R61AI161811, and contract 75N93019C00008 (to A.P.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Author contributions
T.K., J.M.L., A.S., N.L.W., B.J.B., M.S.D., A.K., and J.H.E. conceived and designed the experiments. T.K., J.M.L., N.L.W., B.J.B., S.P., R.K., T.H., and S.S. performed the experiments. T.K., J.M.L., N.L.W., B.J.B., T.H., and J.H.E. analyzed the data. A.S., N.L.W., J.F.A., K.N., A.K., and J.H.E. contributed materials/analysis tools. T.K., B.J.B., M.S.D., P.B., S.G.R., A.K., and J.H.E. wrote the paper.
Declaration of interests
T.K., J.M.L., A.S., N.L.W., S.P., R.K., J.F.A., B.J.B., T.H., K.N., S.S., A.K., M.S.D., P.B., S.G.R., and J.H.E. have equity interest in HDT Bio. J.H.E. is a consultant for InBios. P.B. is a consultant for Karkinox, and Chimeron. P.B. and J.H.E. are co-inventors on US patent application no. 63/011,860 “Nucleic acid vaccines against COVID-19” pertaining to repRNA-CoV-2S. J.H.E., A.K., and S.G.R. are co-inventors on US patent application no. 62/993,307 “Compositions and methods for delivery of RNA” pertaining to the LION formulation.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.06.017.
Supplemental information
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information.
References
- 1.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollock K.M., Cheeseman H.M., Szubert A.J., Libri V., Boffito M., Owen D., Bern H., O'Hara J., McFarlane L.R., Lemm N.M., et al. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. EClinicalMedicine. 2022;44:101262. doi: 10.1016/j.eclinm.2021.101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low J.G., de Alwis R., Chen S., Kalimuddin S., Leong Y.S., Mah T.K.L., Yuen N., Tan H.C., Zhang S.L., Sim J.X.Y., et al. A phase I/II randomized, double-blinded, placebo-controlled trial of a self-amplifying Covid-19 mRNA vaccine. NPJ Vaccin. 2022;7:161. doi: 10.1038/s41541-022-00590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and Delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruggi G., Mallett C.P., Westerbeck J.W., Chen T., Lofano G., Friedrich K., Qu L., Sun J.T., McAuliffe J., Kanitkar A., et al. A self-amplifying mRNA SARS-CoV-2 vaccine candidate induces safe and robust protective immunity in preclinical models. Mol. Ther. 2022;30:1897–1912. doi: 10.1016/j.ymthe.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arevalo C.P., Bolton M.J., Le Sage V., Ye N., Furey C., Muramatsu H., Alameh M.G., Pardi N., Drapeau E.M., Parkhouse K., et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science. 2022;378:899–904. doi: 10.1126/science.abm0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erasmus J.H., Khandhar A.P., O'Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., et al. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawman D.W., Meade-White K., Archer J., Leventhal S.S., Wilson D., Shaia C., Randall S., Khandhar A.P., Krieger K., Hsiang T.Y., et al. SARS-CoV2 variant-specific replicating RNA vaccines protect from disease following challenge with heterologous variants of concern. Elife. 2022;11:e75537. doi: 10.7554/eLife.75537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawman D.W., Meade-White K., Clancy C., Archer J., Hinkley T., Leventhal S.S., Rao D., Stamper A., Lewis M., Rosenke R., et al. Replicating RNA platform enables rapid response to the SARS-CoV-2 Omicron variant and elicits enhanced protection in naive hamsters compared to ancestral vaccine. EBioMedicine. 2022;83:104196. doi: 10.1016/j.ebiom.2022.104196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leventhal S.S., Meade-White K., Rao D., Haddock E., Leung J., Scott D., Archer J., Randall S., Erasmus J.H., Feldmann H., Hawman D.W. Replicating RNA vaccination elicits an unexpected immune response that efficiently protects mice against lethal Crimean-Congo hemorrhagic fever virus challenge. EBioMedicine. 2022;82:104188. doi: 10.1016/j.ebiom.2022.104188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor M.A., Hawman D.W., Meade-White K., Leventhal S., Song W., Randall S., Archer J., Lewis T.B., Brown B., Fredericks M.N., et al. A replicon RNA vaccine can induce durable protective immunity from SARS-CoV-2 in nonhuman primates after neutralizing antibodies have waned. PLoS Pathog. 2023;19:e1011298. doi: 10.1371/journal.ppat.1011298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong C., Levis R., Shen P., Schlesinger S., Rice C.M., Huang H.V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X., Berglund P., Rhodes G., Parker S.E., Jondal M., Liljeström P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–1514. doi: 10.1016/0264-410x(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 18.Li C., Lee A., Grigoryan L., Arunachalam P.S., Scott M.K.D., Trisal M., Wimmers F., Sanyal M., Weidenbacher P.A., Feng Y., et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 2022;23:543–555. doi: 10.1038/s41590-022-01163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tregoning J.S., Stirling D.C., Wang Z., Flight K.E., Brown J.C., Blakney A.K., McKay P.F., Cunliffe R.F., Murugaiah V., Fox C.B., et al. Formulation, inflammation, and RNA sensing impact the immunogenicity of self-amplifying RNA vaccines. Mol. Ther. Nucleic Acids. 2023;31:29–42. doi: 10.1016/j.omtn.2022.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosca F., Tritto E., Muzzi A., Monaci E., Bagnoli F., Iavarone C., O'Hagan D., Rappuoli R., De Gregorio E. Molecular and cellular signatures of human vaccine adjuvants. Proc. Natl. Acad. Sci. USA. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uccellini M.B., García-Sastre A. ISRE-Reporter Mouse Reveals High Basal and Induced Type I IFN Responses in Inflammatory Monocytes. Cell Rep. 2018;25:2784–2796.e3. doi: 10.1016/j.celrep.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hervé C., Laupèze B., Del Giudice G., Didierlaurent A.M., Tavares Da Silva F. The how's and what's of vaccine reactogenicity. NPJ Vaccin. 2019;4:39. doi: 10.1038/s41541-019-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peri G., Introna M., Corradi D., Iacuitti G., Signorini S., Avanzini F., Pizzetti F., Maggioni A.P., Moccetti T., Metra M., et al. PTX3, A prototypical long pentraxin, is an early indicator of acute myocardial infarction in humans. Circulation. 2000;102:636–641. doi: 10.1161/01.cir.102.6.636. [DOI] [PubMed] [Google Scholar]
- 26.Blann A.D., Nadar S.K., Lip G.Y.H. The adhesion molecule P-selectin and cardiovascular disease. Eur. Heart J. 2003;24:2166–2179. doi: 10.1016/j.ehj.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 27.McEver R.P. Selectins: initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc. Res. 2015;107:331–339. doi: 10.1093/cvr/cvv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang S.J., Ballantyne C.M., Sharrett A.R., Smith L.C., Davis C.E., Gotto A.M., Jr., Boerwinkle E. Circulating adhesion molecules VCAM-1, ICAM-1, and E-selectin in carotid atherosclerosis and incident coronary heart disease cases: the Atherosclerosis Risk In Communities (ARIC) study. Circulation. 1997;96:4219–4225. doi: 10.1161/01.cir.96.12.4219. [DOI] [PubMed] [Google Scholar]
- 29.Goddard K., Hanson K.E., Lewis N., Weintraub E., Fireman B., Klein N.P. Incidence of Myocarditis/Pericarditis Following mRNA COVID-19 Vaccination Among Children and Younger Adults in the United States. Ann. Intern. Med. 2022;175:1169–1771. doi: 10.7326/M22-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patone M., Mei X.W., Handunnetthi L., Dixon S., Zaccardi F., Shankar-Hari M., Watkinson P., Khunti K., Harnden A., Coupland C.A.C., et al. Risk of Myocarditis After Sequential Doses of COVID-19 Vaccine and SARS-CoV-2 Infection by Age and Sex. Circulation. 2022;146:743–754. doi: 10.1161/CIRCULATIONAHA.122.059970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weintraub E.S., Oster M.E., Klein N.P. Myocarditis or Pericarditis Following mRNA COVID-19 Vaccination. JAMA Netw. Open. 2022;5:e2218512. doi: 10.1001/jamanetworkopen.2022.18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erasmus J.H., Archer J., Fuerte-Stone J., Khandhar A.P., Voigt E., Granger B., Bombardi R.G., Govero J., Tan Q., Durnell L.A., et al. Intramuscular Delivery of Replicon RNA Encoding ZIKV-117 Human Monoclonal Antibody Protects against Zika Virus Infection. Mol. Ther. Methods Clin. Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hoecke L., Roose K., Ballegeer M., Zhong Z., Sanders N.N., De Koker S., Saelens X., Van Lint S. The Opposing Effect of Type I IFN on the T Cell Response by Non-modified mRNA-Lipoplex Vaccines Is Determined by the Route of Administration. Mol. Ther. Nucleic Acids. 2020;22:373–381. doi: 10.1016/j.omtn.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacio N., Dangi T., Chung Y.R., Wang Y., Loredo-Varela J.L., Zhang Z., Penaloza-MacMaster P. Early type I IFN blockade improves the efficacy of viral vaccines. J. Exp. Med. 2020;217:e20191220. doi: 10.1084/jem.20191220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Low J.G.H., Kalimuddin S., Matsuda D., et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021;29:1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blakney A.K., McKay P.F., Bouton C.R., Hu K., Samnuan K., Shattock R.J. Innate Inhibiting Proteins Enhance Expression and Immunogenicity of Self-Amplifying RNA. Mol. Ther. 2021;29:1174–1185. doi: 10.1016/j.ymthe.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepini T., Pulichino A.M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J., et al. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbeke R., Hogan M.J., Loré K., Pardi N. Innate immune mechanisms of mRNA vaccines. Immunity. 2022;55:1993–2005. doi: 10.1016/j.immuni.2022.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vermillion M.S., Dearing J., Zhang Y., Adney D.R., Scheuermann R.H., Pekosz A., Tarbet E.B. Animal Models of Enterovirus D68 Infection and Disease. J. Virol. 2022;96:e0083322. doi: 10.1128/jvi.00833-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahtinen S., Tong A.J., Himmels P., Oh J., Paler-Martinez A., Kim L., Wichner S., Oei Y., McCarron M.J., Freund E.C., et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2022;23:532–542. doi: 10.1038/s41590-022-01160-y. [DOI] [PubMed] [Google Scholar]
- 41.Minnaert A.K., Vanluchene H., Verbeke R., Lentacker I., De Smedt S.C., Raemdonck K., Sanders N.N., Remaut K. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: Getting the message across. Adv. Drug Deliv. Rev. 2021;176:113900. doi: 10.1016/j.addr.2021.113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y.X., Xu W.G., Sun X.J., Chen Y.Z., Liu X.Y., Tang H., Jiang C.L. Fever of recombinant human interferon-alpha is mediated by opioid domain interaction with opioid receptor inducing prostaglandin E2. J. Neuroimmunol. 2004;156:107–112. doi: 10.1016/j.jneuroim.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Thurner L., Kessel C., Fadle N., Regitz E., Seidel F., Kindermann I., Lohse S., Kos I., Tschöpe C., Kheiroddin P., et al. IL-1RA Antibodies in Myocarditis after SARS-CoV-2 Vaccination. N. Engl. J. Med. 2022;387:1524–1527. doi: 10.1056/NEJMc2205667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeifer J., Thurner B., Kessel C., Fadle N., Kheiroddin P., Regitz E., Hoffmann M.C., Kos I.A., Preuss K.D., Fischer Y., et al. Autoantibodies against interleukin-1 receptor antagonist in multisystem inflammatory syndrome in children: a multicentre, retrospective, cohort study. Lancet Rheumatol. 2022;4:e329–e337. doi: 10.1016/S2665-9913(22)00064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yonker L.M., Swank Z., Bartsch Y.C., Burns M.D., Kane A., Boribong B.P., Davis J.P., Loiselle M., Novak T., Senussi Y., et al. Circulating Spike Protein Detected in Post-COVID-19 mRNA Vaccine Myocarditis. Circulation. 2023;147:867–876. doi: 10.1161/CIRCULATIONAHA.122.061025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barmada A., Klein J., Ramaswamy A., Brodsky N.N., Jaycox J.R., Sheikha H., Jones K.M., Habet V., Campbell M., Sumida T.S., et al. Cytokinopathy with aberrant cytotoxic lymphocytes and profibrotic myeloid response in SARS-CoV-2 mRNA vaccine-associated myocarditis. Sci. Immunol. 2023;8:eadh3455. doi: 10.1126/sciimmunol.adh3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langlet C., Tamoutounour S., Henri S., Luche H., Ardouin L., Grégoire C., Malissen B., Guilliams M. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 53.Erasmus J.H., Khandhar A.P., Guderian J., Granger B., Archer J., Archer M., Gage E., Fuerte-Stone J., Larson E., Lin S., et al. A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika. Mol. Ther. 2018;26:2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khandhar A.P., Landon C.D., Archer J., Krieger K., Warner N.L., Randall S., Berube B.J., Erasmus J.H., Sather D.N., Staats H.F. Evaluation of repRNA vaccine for induction and in utero transfer of maternal antibodies in a pregnant rabbit model. Mol. Ther. 2023;31:1046–1058. doi: 10.1016/j.ymthe.2023.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerner M.Y., Kastenmuller W., Ifrim I., Kabat J., Germain R.N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information.