Abstract
To better understand the virological aspect of the expanding AIDS epidemic in southern Africa, a set of 23 near-full-length clones of human immunodeficiency virus type 1 (HIV-1) representing eight AIDS patients from Botswana were sequenced and analyzed phylogenetically. All study viruses from Botswana belonged to HIV-1 subtype C. The interpatient diversity of the clones from Botswana was higher than among full-length isolates of subtype B or among a set of full-length HIV-1 genomes of subtype C from India (mean value of 9.1% versus 6.5 and 4.3%, respectively; P < 0.0001 for both comparisons). Similar results were observed in all genes across the entire viral genome. We suggest that the high level of HIV-1 diversity might be a typical feature of the subtype C epidemic in southern Africa. The reason or reasons for this diversity are unclear, but may include an altered replication efficiency of HIV-1 subtype C and/or the multiple introduction of different subtype C viruses.
The majority of new human immunodeficiency virus (HIV) infections in the global AIDS epidemic are appearing in sub-Saharan Africa and Southeast Asia. Compared with the situation a decade ago, the main AIDS epidemics have shifted from central and eastern Africa to the southern regions. The most severe HIV epidemics have recently afflicted such southern African countries as Zimbabwe, Zambia, Namibia, South Africa, and Botswana (43). HIV-1 subtype C has been estimated to account for 48% of HIV-1 infections worldwide and 51.5% of HIV-1 infections in Africa (4, 7, 14–16, 21, 31), where the main mode of transmission is heterosexual (43, 44, 47).
A rapid expansion of the HIV-1 epidemic in Botswana has occurred since the early to mid 1990s. According to the UNAIDS and World Health Organization (WHO) Global HIV/AIDS & STD Surveillance data, HIV prevalence among antenatal clinic attendees tested in the major urban areas of Botswana (Gaborone, Francistown, and Selebi-Phikwe) increased from 6% in 1990 to 39% in 1997 (range of 34 to 43%) (42). Among women 20 to 29 years of age, 43 to 44% tested HIV positive. Outside of the major urban areas, median HIV prevalence increased from no evidence of infection in 1985 to 1987 to 34% in 1997. In 1997, HIV prevalence in Botswana ranged from 28 to 38%. As such, locally circulating HIV-1 needs to be characterized thoroughly, and vital information about the nature of the epidemic should be extended (2, 4, 7, 21, 31, 37, 45–47). Moreover, Botswana’s central geographic position makes a comprehensive HIV-1 molecular epidemiological study that much more urgent, because it may serve as an example of the burgeoning epidemic in southern Africa.
In this study, we report the molecular cloning and phylogenetic analysis of 23 near-full-length clones from Botswana. All of them were identified as belonging to HIV-1 subtype C and demonstrated high levels of intersample diversity across the entire viral genome. By providing new genetic information regarding locally circulating viruses, this study may contribute to AIDS vaccine design for the southern Africa region countries and, in particular, for Botswana.
Specimens for this study were selected from HIV-seropositive patients in Gaborone, Botswana. All HIV-1 infections in this study were likely to be heterosexually acquired. The times of infection were not known. The HIV-1-seropositive status of patients was confirmed by enzyme-linked immunosorbent assay and Western blot analysis. Clinical classification was performed by using the 1987 Centers for Disease Control and Prevention (CDC) revised classification (9) (data not shown).
Genomic DNA was obtained directly from the patients’ peripheral blood mononuclear cells (PBMCs)—buffy coats—without passage through cell culture or donor PBMCs. All clones in this study were amplified in heminested PCR with three primers from the LA set (18) or their modifications. The Expand Long Template PCR system (Boehringer Mannheim, Indianapolis, Ind.) was used according to the manufacturer’s instructions. Gel purification of the first-round PCR product was essential for direct amplification of 9.0-kb fragments from uncultured PBMCs. Estimation of the expanded PCR sensitivity (based on 8E5/LAV) revealed a successful amplification of the 9.0-kb fragment in the first round when at least 8 × 102 to 4 × 103 proviral copies were present in the reaction (data not shown). These results were consistent with those from other studies (10, 39). The TA pCR2.1 TOPO system (Invitrogen, Carlsbad, Calif.) and JM109 competent cells (Promega Corporation, Madison, Wis.) were used for cloning. Positive colonies were screened by PCR. To obtain sufficient plasmids for sequence analysis, we amplified the constructs under the previously described conditions with some modifications (41). Purified plasmid DNA served as a template for sequencing. Both-strand sequencing was combined with a strategy involving overlapping sequences. Dye terminator sequencing on an automated DNA Sequenator (model 373A; Applied Biosystems, Inc., Foster City, Calif.) was used.
A multiple alignment procedure for the full-length HIV genome was performed by using the hidden Markov model. Constructed through the HIV-1 HMMER computer program of the Los Alamos National Laboratory, the model has been previously shown to provide the best description of the true nucleotide substitution pattern of HIV-1 gag and env genes (26). The HIV-1 HMMER model (11, 12) constructed at Los Alamos National Laboratory for the full-length HIV-1 genomes (24) was employed. Sixty full-length reference sequences were included in the alignment from the GenBank data bank (5). The 3′ end of the alignment, which included the nef coding region and 3′ long terminal repeat (LTR), was adjusted manually. The pairwise evolutionary distances from nucleotide sequences were computed by the DNADIST program under Kimura’s two-parameter model (17). All alignments were globally gap stripped for the generation of the trees. The transition/transversion ratio parameters were set at 3.0 for the gag gene, 1.5 for the env gene, 1.42 for the V1-V2 and V3 fragments, and 2.0 for the other viral loci (25). A tree was drawn by the Njplot (33) and TreeView (32) programs. To analyze patterns of variability along the HIV-1 genomes, the program SWAN, which utilizes a “sliding window” approach was used (34). Positions with gaps either were or were not excluded from the analysis. The variability distribution was estimated as an entropy function of the nucleotide variation observed at a particular position. The Recombinant Identification Program (RIP) (40) and HIV-1 Subtyping Basic BLAST (3) were used in searching for recombination among the clones studied.
Sequence analysis of the Botswana HIV-1 revealed that 10 of 23 clones had an intact genomic organization with open reading frames. The other clones had point mutations and/or insertions and deletions resulting in frameshifts, disabled start codons, or premature stop codons. No major deletions or rearrangements were observed. Determined length polymorphism among studied sequences was limited to the vpu (15- to 18-nucleotide [nt] insertion at the 5′ end), env (from 3- to 9-nt deletions to 48-nt insertions, GIGRGQ motif in the BW17 V3 loop), 2nd exon of rev (a 13-amino-acid truncation at the 3′ end), nef (6- to 15-nt deletions in some clones), and regulatory regions of the LTR (three or four NF-κB sites with GGGACTTTCT as a potential fourth NF-κB in two clones of BW05).
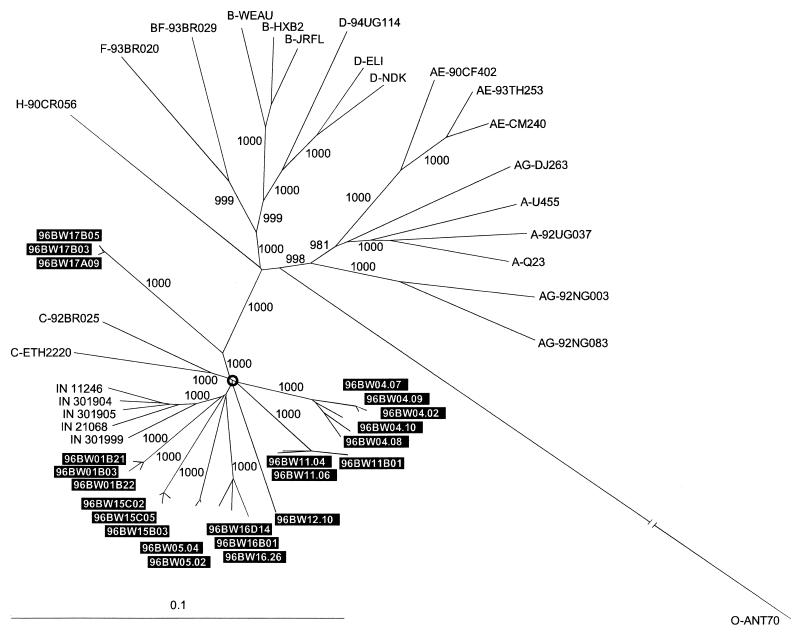
An evolutionary tree in Fig. 1 shows the phylogenetic relationship of the full-length Botswana clones to other representative full-length HIV-1 sequences. All Botswana sequences, a set from India (27), and two subtype C reference sequences (C-ETH2220 [38] and C-92BR025 [19]) clustered together, forming a compelling subtype C outcropping on the phylogenetic tree. This cluster was separated from other HIV-1 sequences by the extremely high bootstrap value of 1,000 (out of 1,000 resampling). Phylogenetic relationships within the subtype C bush were also noteworthy. Assuming that the circled node at the center of the bush could represent the potential ancestral subtype C node, we observed the following. (i) The star-like phylogeny of the subtype C bush together with its branching order may demonstrate the relatively high diversity of the Botswana samples. (ii) Four Botswana sample clones (BW01, BW05, BW15, and BW16), together with all five sequences from India, formed a potential subcluster, although the bootstrap value was not high. (iii) All Indian samples were separated by bootstrap values of 1,000, possibly reflecting a “founder effect” among these sequences. (iv) Three Botswana sample clones (BW04, BW11, and BW12) may represent individualized groups of sequences inherited from a common subtype C ancestor. (v) Two reference sequences ETH2220 and 92BR025 deviated together with a high bootstrap value (1,000), possibly reflecting another subtype C subcluster differing significantly from Botswana or Indian samples. (vi) One of the Botswana samples (BW17) strayed rather far off the main subtype C bush and may be the least representative of Botswana HIV-1 samples. (vii) The topology of the Botswana clones confirms that clones of the same samples are closely related to each other based on full-length genome sequences. A multilocus analysis was congruent with full-genome phylogenetic analysis and confirmed clustering of newly derived Botswana clones within subtype C across the entire HIV-1 genome (data not shown).
FIG. 1.
Phylogenetic relationship of the newly characterized full-length clones from Botswana (boxed in black) to other representative full-length HIV-1 sequences of subtypes A, B, C, D, F, and H and recombinant subtypes AE, AG, and BF. Full-length subtype C sequences from India were also included in the analysis. A neighbor-joining tree was constructed on the basis of the hidden Markov model nucleotide alignment of full-length HIV-1 genomes. Subtype O ANT70 sequence was used as an outgroup. Values along the branches indicate the bootstrap values that support branching (out of a 1,000 resampling).
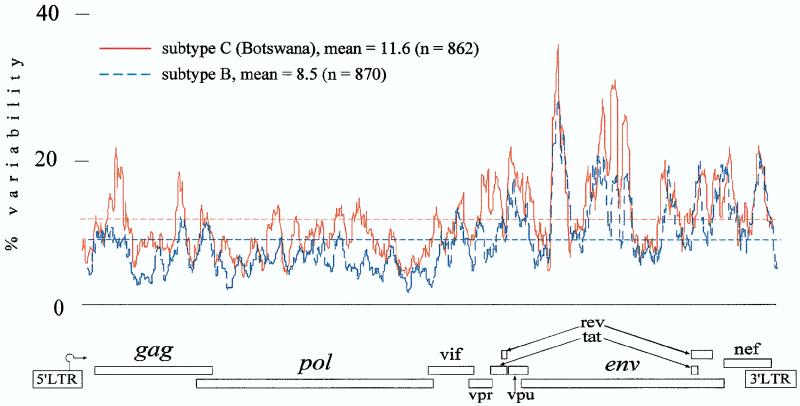
To characterize the level of variability among Botswana clones across the entire HIV-1 genome, we performed SWAN program analysis as an entropy function of the nucleotide variation. The Botswana set had greater variability than subtype B samples (Fig. 2) (mean values of 11.6 and 8.5%, respectively, for gap-stripped analysis). The profiles of viral variability across the HIV-1 genome were similar among subtype B and C viruses. Comparison of gap-stripped and gap-nonstripped plots revealed that the differences in mean values between the two methods of computing and the shape of variability plot profiles were not significant (data not shown). Gap stripping slightly decreased the mean value and the number of sliding window sites across the genome. It also hid the extreme regions with the highest level of variability. Both variability when measured as an entropy function in this study and when described before diversity as a pairwise comparison of the sequence (19) exhibited similar profiles of variable and conservative genomic regions. Variability plots (especially non-gap stripped) revealed higher peaks in variable regions than diversity plots.
FIG. 2.
Variability plots comparing subtype B and C (Botswana) sequences across the entire HIV-1 genome. The variability distribution was estimated as an entropy function of the nucleotide variation in the SWAN program based on the hidden Markov model alignment of the complete HIV-1 genome. The subtype B sequences used in the analysis were DH123, 89.6, RF, WEAU, OYI, HXB2, JRFL, and YU2. n, number of sliding window sites across the HIV-1 genome with gap stripping.
Table 1 depicts the high degree of intersample diversity across the entire HIV-1 genome among Botswana clones compared with subtype B and C sequences from India (27). Because AIDS patients might be expected to have higher variability, we made the same comparison, limiting the subtype B reference to eight sequences selected from confirmed AIDS patients (column 2). The intersample diversity among Botswana clones was significantly higher than that among subtype B references or Indian samples on the level of the full-length HIV-1 genome. Across the viral genome, the mean diversity among Botswana samples was congruent with the full-length genome analysis. The intersample diversity analysis statistically confirmed the phylogenetic study observations (Fig. 1 and 2) that the newly characterized Botswana clones were highly diversified.
TABLE 1.
Intersample HIV-1 diversity in this studya
| Gene or region | Mean % (range) HIV-1 diversityb
|
Subtype C, India | ||
|---|---|---|---|---|
| Subtype C, Botswana | Subtype B
|
|||
| AIDS patients (8 sequences) | 23 isolates | |||
| gag | 7.9 (5.9–9.2) | 5.6* (3.6–9.6) | 4.9* (1.0–8.6) | 3.4* (2.4–4.6) |
| pol | 5.9 (4.3–7.8) | 4.1* (2.9–5.4) | 3.9* (0.8–6.2) | 2.6* (1.5–3.7) |
| vif | 7.5 (4.3–12.8) | 6.3† (3.7–9.3) | 6.4‡ (0–9.5) | 3.2* (1.8–4.3) |
| vpr | 9.8 (5.3–14.9) | 6.2* (4.3–8.4) | 5.9* (1.7–10.8) | 4.8* (2.1–8.1) |
| tat | 9.9 (6.9–14.9) | 7.5* (4.6–10.7) | 7.1* (3.4–10.7) | 3.7* (2.7–5.1) |
| rev | 10.4 (7.7–16.6) | 9.4§ (4.2–14.2) | 8.2* (3.3–14.2) | 4.7* (2.8–5.7) |
| vpu | 13.9 (10.1–18.8) | 10.8* (6.1–14.3) | 9.5* (4.2–15.8) | 5.1* (2.1–8.1) |
| env | 12.4 (10.4–14.5) | 9.5* (6.2–11.9) | 9.5* (5.9–12.7) | 6.9* (5.1–9.3) |
| V1-V2 | 25.7 (15.9–36.7) | 18.0* (8.6–26.1) | 19.0* (8.6–30.8) | 25.8 (14.2–34.8) |
| V3 | 14.4 (11.5–18.9) | 11.6* (6.7–17.3) | 12.1* (6.0–18.2) | 6.4* (5.1–7.9) |
| nef | 11.3 (8.0–15.1) | 9.8∥ (6.3–15.7) | 9.2* (3.5–16.3) | 5.2* (4.4–6.4) |
| 3′ LTR | 9.7 (6.6–13.4) | 8.3# (5.1–11.6) | 7.3* (1.0–11.6) | 4.0* (3.2–5.4) |
| Full-length genome | 9.1 (7.7–10.7) | 6.6* (4.5–8.0) | 6.5* (3.5–9.6) | 4.3* (3.2–5.7) |
Fifty-six full-length HIV-1 sequences were used in the analysis, based on the hidden Markov model alignment of the entire HIV-1 genome. The following 28 sequences of subtype B were used: AUMBCC54, C18MBC, DH123, 89.6, RF, WEAU, HAN, HIVMN, BCSG3, OYI, CAM1, NY5, pNL43, LAI, HXB2, JRFL, JRCSF, AUMBC925, AUMBC200, YU2, YU10, ACH320A, ACH320B, SF2, HIV1AD8, D31, MANC, and WR27. Sequences AUMBCC54, C18MBC, and NY5 were excluded from the nef and 3′ LTR analysis because of deletions or the absence of sequences for these regions. Pairwise distances in four groups of sequences (pNL43, LAV, and HXB2; JRFL and JRCSF; YU2 and YU10; and ACH320A and ACH320B) were excluded from the analysis. The eight subtype B sequences from AIDS patients were JRFL, YU2, 89.6, RF, HAN, MN, BCSG3, and WR27. Thirteen sequences of subtype C were included in the analysis: 8 clones from Botswana (1 from each patient) and 5 sequences from India (301999, 21068, 301905, 301904, and 11246). All distances were calculated by DNADIST program from the PHYLIP v. 3.572 package based on hidden Markov model alignment. The transition/transversion ratios were set to 3.0 for gag, 1.5 for env, 1.42 for V3 and V1-V2, and 2.0 for all other HIV-1 genes.
Statistical significance versus Botswana HIV-1 clones is shown as follows: *, P < 0.0001; #, P = 0.005; ∥, P = 0.013. ‡, P = 0.028; †, P = 0.033; and §, P = 0.13.
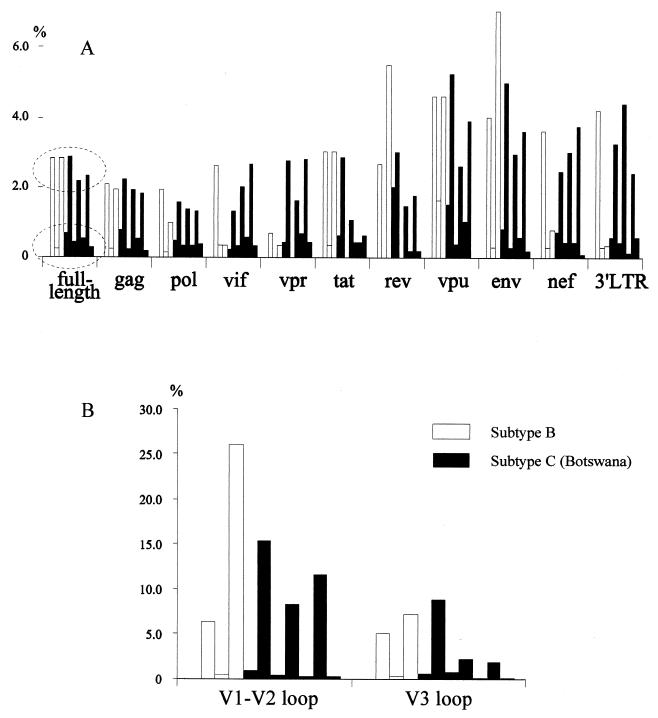
The results of intrasample diversity analysis were limited by the methods used (PCR amplification and cloning) and by the available number of multiple subtype B full-length clones. Seven Botswana samples (except BW12) and three subtype B sets (JRFL, YU, and ACH320) were compared across the HIV-1 genome. The range of full-length diversity among Botswana samples was 0.3% (BW17 clones) to 2.9% (BW04 clones), with an average mean value of 1.4%. Intrasample diversity showed no significant difference between subtype B and C sequences (Fig. 3). However, two concentrations of diversity (low and high) were revealed among both subtype B and C sets (Fig. 3). These concentrations of low and high diversity were distributed across the entire genome and were found to be more consistent in the structural genes (gag, pol, and env).
FIG. 3.
Intrasample diversity among subtype B (open bars) and newly derived subtype C (solid bars) clones from Botswana. Pairwise distances were computed by the DNADIST program from the PHYLIP package based on hidden Markov model alignment of the complete genome or individual genes or subgenomic regions (B) retrieved from the same alignment. Three sets of subtype B sequences were included: JRFL and FRCSF, YU2 and YU10, and ACH320A and ACH320B. Seven sets of subtype C Botswana clones were included: BW01, BW04, BW05, BW11, BW15, BW16, and BW17. The order of bars for each gene or region corresponds to the order in which the sets are mentioned. Dashed ovals depict low- and high-diversity groups.
All Botswana sequences were checked for potential recombination sites by the HIV-1 Subtyping Basic BLAST (3) and by RIP (40), the results consistently showing no evidence of recombination.
Clustering with HIV-1 subtype C and the high intersample diversity were the most exceptional attributes of the 23-clone set from Botswana. A star-like shape of the subtype C cluster in the phylogenetic tree was accompanied by extremely high bootstrap values across tree branches. The topology of the phylogenetic tree suggested that a common ancestor for the Botswana sequences might have existed before the common ancestor for the Indian sequences analyzed or before the strains C-92BR025 (Brazil) and C-ETH2220 (Ethiopia) diverged.
Intersample diversity within subtype C has been previously found to vary from 5 to 11.5% (1, 7, 38). Higher levels of diversity were found among Botswana clones in this study, in spite of the fact that samples were taken from one place and at one time point. Both full-genome sequences and multiple subgenomic loci demonstrated the same patterns, with a higher mean value of variability among the Botswana samples.
The increased genetic diversity of subtype C viruses in Botswana might have different underlying causes, including a variety of host and viral factors. Among the latter factors, a combination of the genetic flexibility of subtype C virus and its multiple introductions might be the most important. A number of recent findings argue that one possible cause of the high viral diversity in the Botswana epidemic could be higher flexibility of subtype C virus and its altered ability to diversify. These arguments include, but are not limited to the following. (i) Subtype C is predominant in most recent HIV-1 epidemics worldwide (1, 7, 27, 35, 36, 38, 45, 47). (ii) The highest prevalence of HIV-1 infection in various epidemics is caused mainly by subtype C virus. (iii) Subtype C virus may have a faster disease progression (20), and patients infected with HIV-1 subtype C developed AIDS earlier than patients with subtype A virus (23). (iv) Three or four NF-κB sites (instead of two) might lead to more efficient viral transcription (13, 29, 30). (v) The TNF-α response to subtype C virus is significantly higher than to HIV-1 subtype B (28, 29), suggesting the possibility of increased viral transcription and replication in correlation with NF-κB copy number (28, 29). (vi) The viral load of subtype C infections may be higher in different compartments that might cause an increased level of viral transmission (22). On the other hand, a scenario that suggests independent diversification of the virus in other regions and delayed entry of the epidemic in Botswana, followed by multiple introductions of the subtype C virus from adjacent countries cannot be excluded (42–44).
Botswana is geographically located at the center of the AIDS epidemic in Southern Africa. UNAIDS and World Health Organization surveillance data suggest that the widespread rise of the HIV-1 epidemic in Botswana started in the early to mid-1990s and reached one of the highest prevalence rates in Africa (42–44). For more recent HIV-1 epidemics, such as those described in Thailand and India, one might expect to find a highly homogeneous pool of local viruses that formed a monophyletic phylogenetic subcluster with relatively short and aggregated branches. However the findings in this study contradict the established trend.
Extremely high interpatient diversity across the genome was supported by long branch lengths in the phylogenetic trees throughout the Botswana viruses within the genetic subtype C. No multiple subtypes or recombination were found in this study. Because it currently has the highest incidence rates of HIV-1, the region of southern Africa, including Botswana and Zimbabwe, is an important site for the design and development of an anti-HIV vaccine. Recent studies of cross-clade cytotoxic T-lymphocyte recognition (6, 8, 48) have made a significant contribution to AIDS vaccine development. However, the extent of cross-clade immune responses in a highly diverse HIV-1 environment and the mechanism of vaccination throughout different HLA profiles in the population are still open questions. HIV-1 strains circulating locally (or appropriate combinations of local strains) should provide a more effective prototype or candidate for a vaccine than a distinct virus circulating elsewhere. As such, a comprehensive molecular epidemiology study of locally circulating HIV-1 strains would facilitate the design of an effective AIDS vaccine for a particular population, country, or geographic region.
Nucleotide sequence accession number.
The 23 full-length HIV-1 sequences from Botswana are available under GenBank accession no. AF110959 to AF110981.
Acknowledgments
This study was supported in part by OIG grant R35 CA39805-13 from the National Institutes of Health and grant D43 TW00004 from the Fogarty International Center, National Institutes of Health.
We thank Bette T. Korber for critical analysis, Suranjana Choudhury for HIV-1 culturing, Peter Gilbert for help with statistical analysis, and Rajiv Rawat for editorial assistance.
REFERENCES
- 1.Abebe A, Kuiken C L, Goudsmit J, Valk M, Messele T, Sahlu T, Yeneneh H, Fontanet A, De Wolf F, Rinke De Wit T F. HIV type 1 subtype C in Addis Ababa, Ethiopia. AIDS Res Hum Retroviruses. 1997;13:1071–1075. doi: 10.1089/aid.1997.13.1071. [DOI] [PubMed] [Google Scholar]
- 2.Alaeus A, Leitner T, Lidman K, Albert J. Most HIV-1 genetic subtypes have entered Sweden. AIDS. 1997;11:199–202. doi: 10.1097/00002030-199702000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipmann D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker M L B, de Jager G, Becker W B. Analysis of partial gag and env gene sequences of HIV type 1 strains from Southern Africa. AIDS Res Hum Retroviruses. 1995;11:1265–1267. doi: 10.1089/aid.1995.11.1265. [DOI] [PubMed] [Google Scholar]
- 5.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C, N’Gandu N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredell H, Williamson C, Sonnenberg P, Martin D J, Morris L. Genetic characterization of HIV type 1 from migrant workers in three South African gold mines. AIDS Res Hum Retroviruses. 1998;14:677–684. doi: 10.1089/aid.1998.14.677. [DOI] [PubMed] [Google Scholar]
- 8.Cao H, Kanki P, Sankalé J-L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker B D. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Centers for Disease Control and Prevention: revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. Morbid Mortal Weekly RP. 1987;36(Suppl 1):S1–S15. [Google Scholar]
- 10.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy S R. Hidden Markov models. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 12.Eddy, S. R. April 1995, posting date. HMMER. [Online.] http://hmmer.wustl.edu/. [15 October 1998, last date accessed.]
- 13.Englund G, Hoggan M D, Theodore T S, Martin M A. A novel HIV-1 isolate containing alterations affecting the NF-κB element. Virology. 1991;181:150–157. doi: 10.1016/0042-6822(91)90479-u. [DOI] [PubMed] [Google Scholar]
- 14.Esparza, J. 1998. Personal communication.
- 15.Essex M. State of the HIV pandemic. J Hum Virol. 1998;1:427–429. [PubMed] [Google Scholar]
- 16.Essex, M. Human immunodeficiency viruses of the developing world. Semin. Virol., in press. [DOI] [PubMed]
- 17.Felsenstein J. PHYLIP: phylogeny inference package, 3.52c ed. Seattle: University of Washington; 1993. [Google Scholar]
- 18.Fujii S, Obaru K, Matsushita S, Morikita T, Higuchi H, Fujimoto K, Takatsuki K. Characterization of proviral DNA from an individual with long-term, nonprogressive infection with HIV-1 and nonrecoverable virus. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;15:247–256. doi: 10.1097/00042560-199708010-00001. [DOI] [PubMed] [Google Scholar]
- 19.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barré-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoelscher M, Eimannsberger D, Cheingsong-Popov R, Jordan-Harder B, Mwakagile D, Nkwera H, Nukulila T, Gurtler L, von Sonnenburg F. Program of the 5th Annual HIV Dynamics & Evolution Conference, Santa Fe, N.Mex. 1997. A shift in age specific subtype distribution gives evidence for different pathogenicity of subtypes A and C. [Google Scholar]
- 21.Janssens W, Buve A, Nkengasong J N. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 22.John G, Neilson J, Lewis P, Overbaugh J, Nduati R W, Mbori-Ngacha D A, Kreiss J. Program of the 12th World AIDS Conference, Geneva, Switzerland. 1988. HIV-1 subtype and shedding of cervicovaginal HIV-1 DNA during pregnancy, abstr. 13117. [Google Scholar]
- 23.Kanki P J, Hamel D J, Sankale J-L, Hsieh C-C, Thior I, Barin F, Woodcock S A, Gueye-Ndiaye A, Zhang E, Montano M, Siby T, Marlink R, NDoye I, Essex M E, MBoup S. HIV-1 subtypes differ in disease progression. J Infect Dis. 1999;179:68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- 24.Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuiken C, editors. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Group T-10; 1997. pp. I-1–I-127. [Google Scholar]
- 25.Leitner T, Korber B, Robertson D, Gao F, Hahn B. Updated proposal of reference sequences of HIV-1 genetic subtypes, p. III-19–III-24. In: Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuiken C, editors. Human retroviruses and AIDS. Theoretical Biology and Biophysics, Group T-10. N.Mex: Los Alamos; 1997. [Google Scholar]
- 26.Leitner T, Kumar S, Albert J. Tempo and mode of nucleotide substitutions in gag and env gene fragments in human immunodeficiency virus type 1 populations with a known transmission history. J Virol. 1997;71:4761–4770. doi: 10.1128/jvi.71.6.4761-4770.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montano M A, Nixon C P, Essex M. Dysregulation through the NF-κB enhancer and TATA box of the human immunodeficiency virus type 1 subtype E promoter. J Virol. 1998;72:8446–8452. doi: 10.1128/jvi.72.10.8446-8452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montano, M. A., C. P. Nixon, T. P. Ndung’u, A. V. A. Novitsky, H. Bussman, D. Dickman, S. M. Gaolekwe, and M. Essex. Transcriptional gain-of-function in TNF-α mediated activation of HIV-1 subtype C. Submitted for publication.
- 30.Montano M A, Novitsky V A, Blackard J T, Cho N L, Katzenstein D A, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orloff G M, Kalish M, Chiphangwi L J, Potts K E, Ou C-Y, Schochetman G, Dallabetta G, Saah A I, Miotti P G. V3 loops of HIV-1 specimens from pregnant women in Malawi uniformly lack a potential N-linked glycosylation site. AIDS Res Hum Retroviruses. 1993;9:705–706. doi: 10.1089/aid.1993.9.705. [DOI] [PubMed] [Google Scholar]
- 32.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 33.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 34.Proutski V, Holmes E. SWAN: sliding window analysis of nucleotide sequence variability. Bioinformatics. 1998;14:467–468. doi: 10.1093/bioinformatics/14.5.467. [DOI] [PubMed] [Google Scholar]
- 35.Ray S, Lole K S, Paranjape R S, Kulkarni S, Gadkari D, Sheppard H, Bollinger R C. Program of the 12th World AIDS Conference, Geneva, Switzerland. 1998. Cloning of full-length HIV-1 genomes from recent infections in India: recombination and CTL epitope, abstr. 11194. [Google Scholar]
- 36.Renjifo B, Chaplin B, Mwakagile D, Shah P, Vannberg F, Msamanga G, Hunter D, Fawzi W, Essex M. Epidemic expansion of HIV type 1 subtype C and recombinant genotypes in Tanzania. AIDS Res Hum Retroviruses. 1998;14:635–638. doi: 10.1089/aid.1998.14.635. [DOI] [PubMed] [Google Scholar]
- 37.Salminen M O, Carr J K, Robertson D L, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P M, Hahn B H, Burke D S, McCutchan F E. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salminen M O, Johansson B, Sonnerborg A, Ayehunie S, Gotte D, Leinikki P, Burke D S, McCutchan F E. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res Hum Retroviruses. 1996;12:1329–1339. doi: 10.1089/aid.1996.12.1329. [DOI] [PubMed] [Google Scholar]
- 39.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 40.Siepel A C, Halpern A L, Macken C, Korber B T. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 41.Trinh T, Jesse J, Bloom F R, Hirsch V. STBL2TM: an Escherichia coli strain for the stable propagation of retroviral clones and direct repeat sequences. Focus. 1994;16:78–80. [Google Scholar]
- 42.UNAIDS, and WHO. 22 June 1998, posting date. Global HIV/AIDS & STD Surveillance. Epidemiological fact sheets by country. [Online.] http://www.us.unaids.org/highband/document/epidemio/june98/fact_sheets/index.html. [15 October 1998, last date accessed.]
- 43.UNAIDS, WHO . Report on the Global HIV/AIDS Epidemic—June 1998. 1998. UNAIDS and WHO, Geneva, Switzerland. [Google Scholar]
- 44.UNAIDS, and WHO. 29 June 1998, posting date. The status and trends of the HIV/AIDS epidemic in the world. [Online.] http://www.us.unaids.org/highband/document/epidemio/map.pdf,Geneva. [15 October 1998, last date accessed.]
- 45.van Harmelen J, Bredell H, Morris L, van der Ryst E, Lyons S, York D, Williamson C. Program of the 12th World AIDS Conference, Geneva, Switzerland. 1998. Determination of prevalent HIV-1 subtypes in South Africa and the construction of a gp120 DNA vaccine, abstr. 33218. [Google Scholar]
- 46.van Harmelen J, Wood R, Lambrick M, Rybicki E P, Williamson A L, Williamson C. An association between HIV-1 subtypes and mode of transmission in Cape Town, South Africa. AIDS. 1997;11:81–87. doi: 10.1097/00002030-199701000-00012. [DOI] [PubMed] [Google Scholar]
- 47.Williamson C, Engelbrecht S, Lambrick M, van Rensburg E, Wood R, Bredell W, Williamson A-L. HIV-1 subtypes in different risk groups in South Africa. Lancet. 1995;346:782. doi: 10.1016/s0140-6736(95)91543-5. [DOI] [PubMed] [Google Scholar]
- 48.Wilson S E, Pedersen S L, Kunich J C, Wilkins V L, Mann D L, Mazzara G P, Tartaglia J, Celum C L, Sheppard H W. Cross-clade envelope glycoprotein 160-specific CD8+ cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res Hum Retroviruses. 1998;14:925–937. doi: 10.1089/aid.1998.14.925. [DOI] [PubMed] [Google Scholar]