Abstract
The Graffi murine leukemia virus (MuLV) is a nondefective retrovirus that induces granulocytic leukemia in BALB/c and NFS mice. To identify genes involved in Graffi MuLV-induced granulocytic leukemia, tumor cell DNAs were examined for genetic alterations at loci described as common proviral integration sites in MuLV-induced myeloid, lymphoid, and erythroid leukemias. Southern blot analysis revealed rearrangements in c-myc, Fli-1, Pim-1, and Spi-1/PU.1 genes in 20, 10, 3.3, and 3.3% of the tumors tested, respectively. These results demonstrate for the first time the involvement of those genes in granulocytic leukemia.
Murine retroviruses have been widely used to understand the mechanisms by which they perturb normal hematopoiesis and to identify genes involved in the process of leukemogenesis. Retroviral insertional mutagenesis is the major mechanism by which these nondefective viruses cause oncogenesis, and it is responsible for the alterations found in several genes implicated in leukemic transformation (2, 23, 47). Graffi murine leukemia virus (MuLV) was originally a retroviral mixture that predominantly induced myeloid leukemia in some strains of mice, although a large spectrum of leukemia could be observed after repeated passages (18, 19). Two molecular clones (GV-1.2 and GV-1.4) have been derived and characterized (34). They both induced granulocytic leukemias in BALB/c and NFS mice (34). They have very similar structures, but clone GV-1.2 induces pathology with shorter latency period and shows a perfect 60-bp duplication in the U3 region of the long terminal repeat (34). Most mice develop thymic and lymph node enlargements, and in peripheral blood smears, the granulocytic leukemia can take two distinct appearances: a juvenile type characterized by myeloblastic to promyelocytic stages or a more mature one with metamyelocytic to segmented granulocytic stages. Approximately 15 to 20% of the blasts showed positive staining for myeloperoxidase, and 60 to 70% showed positive staining for naphthol AS-D chloroacetate esterase (34). These tumors, although clearly myeloid demonstrate DNA rearrangements in the immunoglobulin heavy-chain and T-cell receptor β genes (34). This phenomenon is also observed in acute and chronic human myeloid leukemias (35). Therefore, the Graffi MuLV could be an excellent model to study the mechanisms of myeloid leukemia induction and progression.
In an attempt to investigate the possible contribution of Graffi MuLV in the process of leukemic disease, we examined 30 granulocytic tumors for DNA structure integrity and expression of cellular genes found to be activated by proviral insertion in myeloid and other types of leukemias. Tumors were generated by inoculating newborn NFS and BALB/c mice intraperitoneally with Graffi MuLV molecular clones and parental mixture as described previously (34). DNA from thymic, splenic, and lymph node tumors was extracted and analyzed by Southern blot hybridization for gene rearrangement. Table 1 summarizes the probes used for the analysis and the results obtained. Six tumors had rearrangements in the c-myc gene, one tumor had rearrangements in Spi-1/PU.1, three tumors had rearrangements in Fli-1, and one had rearrangements in Pim-1 (also in c-myc) (Fig. 1). No rearrangements could be detected for the other oncogenes listed in Table 1, although we cannot exclude the possibility of locus alterations beyond the region covered.
TABLE 1.
Probes used for DNA analysis in this study
| Probe | Murine restriction fragment(s) used as probe | No. of tumors with rearrange-ment in gene | Refer-ence |
|---|---|---|---|
| Evi-1 | 1.0-kbp HindIII-PstI | 0 | 28 |
| Evi-2 | 0.5-kbp PstI (probe B) | 0 | 11 |
| c-myb | 4.2-kbp EcoRI, 0.5-kbp EcoRI | 0 | 24 |
| Fim-1 | 1.5-kbp PvuII | 0 | 42 |
| Fim-2 | 1.0-kbp PvuII | 0 | 42 |
| Fim-3 | 0.7-kbp HindIII, 0.7-kbp EcoRI | 0 | 10 |
| His-1 | 1.5-kbp EcoRI | 0 | 1 |
| Spi-1 | 1.0-kbp PstI, 2.1-kbp EcoRI-SaII | 1 | 26 |
| Fli-1 | 1.4-kbp EcoRI | 3 | 9 |
| Pim-1 | 0.7-kbp BamHI | 1 | 15 |
| c-myc | 5.6-kbp BamHI | 6 | 40 |
| Fis-1 | 1.8-kbp EcoRI-BamHI | 0 | 41 |
| Meis 1 | Probe p19-24 | 0 | 30 |
| p53 | 0.9-kbp PstI-BglII cDNA | 0 | 22 |
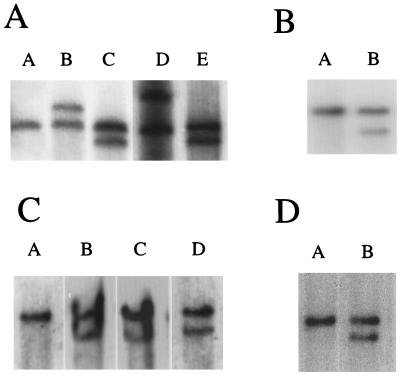
FIG. 1.
Southern blot analysis of Graffi MuLV-induced leukemias (A) Eco-RI-digested DNAs probed with c-myc. Lanes: A, spleen; B, tumor F9; C, tumor F14; D, tumor B30; E, tumor B32. (B) EcoRV-digested DNAs probed with Pim-1. Lanes: A, spleen; B, tumor F9. (C) EcoRV-digested DNAs probed with Fli-1. Lanes: A, spleen; B, tumor B9; C, tumor F11; D, tumor F17. (D) EcoRV-digested DNAs probed with Spi-1/PU.1. Lanes: A, spleen; B, tumor B38.
The c-myc proto-oncogene is the most commonly activated gene in retrovirus-induced tumors. It is frequently activated in both B- and T-cell lymphomas induced by Moloney or Friend MuLV (13, 39). Rearrangements of the c-myc gene were also described in Friend helper virus-induced erythroleukemias (16). Our results from Southern blots of EcoRI- and KpnI-digested DNAs suggest that all c-myc rearrangements in these myeloid tumors are due to proviral insertions upstream of the first exon of c-myc (Fig. 2). Analysis with other enzymes did not unambiguously allow the determination of the proviral orientation in those tumors. Northern blot analysis performed on total RNA revealed a high level of expression of the normal-size c-myc transcript for the tumors with rearrangements in c-myc as well as for many other tumors with no rearrangements in any known oncogene tested (not shown). These results are not surprising, since c-myc is expressed in almost all proliferating normal cells and downregulated in many types of cells when induced to terminally differentiate (17, 20). High expression of c-myc has also been observed in leukemic cells of acute myeloid leukemia patients (43). The constitutive expression of c-myc appears to be an important leukemogenic event occurring in a variety of MuLV-induced leukemias.
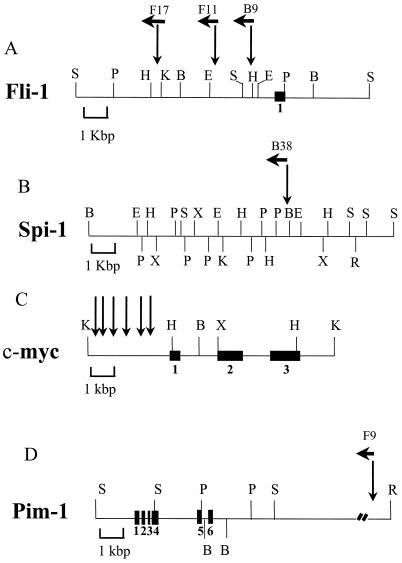
FIG. 2.
Positions of viral integration in the Fli-1 (A), Spi-1/PU.1 (B), c-myc (C), and Pim-1 (D) regions. The black boxes represent the exons. The Spi-1/PU.1 exon 1 is located 10 kbp downstream of the integration site. Arrows above the maps indicate the positions and orientations of the Graffi MuLV integrations. The integration in the Pim-1 locus is located approximately 13 kbp downstream of the last exon (exon 6). Restriction site abbreviations: S, SacI; P, PstI; H, HindIII; K, KpnI; B, BamHI; E, EcoRI; X, XbaI; R, EcoRV.
One of the tumors with rearrangements in c-myc also had rearrangements in the Pim-1 proto-oncogene, which is also activated by proviral insertions in erythroleukemias and T-cell lymphomas in mice (14, 16, 36, 38). It was demonstrated that Pim-1 is one of the most efficient collaborators of c-myc in the induction of lymphomagenesis in mice (29, 45, 46). Our results suggest that the association of c-myc and Pim-1 activation may also play a role in myeloid leukemogenesis.
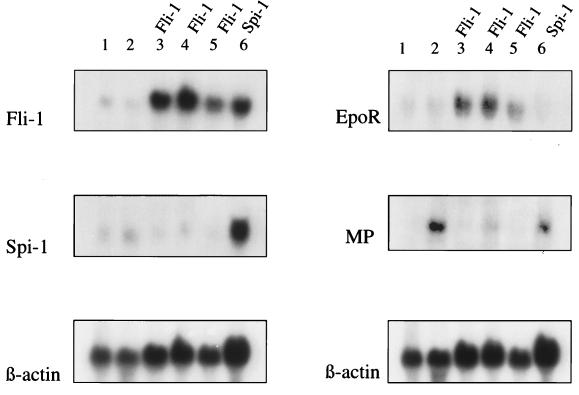
Spi-1/PU.1 and Fli-1 are both members of the ets family of transcription factors and were identified as a consequence of their activation in Friend virus-induced erythroleukemia (6, 7, 26, 32). For both genes, our results show that the retrovirus is integrated in the same region and in an orientation opposite to transcriptional orientation of the gene as does the Friend MuLV (Fig. 2). The Fli-1 gene is activated in 72% of the non-T, non-b lymphomas induced by Cas-Br-E MuLV in NIH/Swiss mice (9). The 10A1 MuLV was also associated with Fli-1 activation in tumors similar to those induced by Cas-Br-E (31). However, these integrations of Cas-Br-E and 10A1 are all clustered in exon 1 within 35 nucleotides directly upstream of the Fli-1 ATG start codon in the same transcriptional orientation as the gene (4, 9, 31). This could indicate the necessity of a promoter insertion mechanism to activate Fli-1 in this case due to a weak activating potential of Cas-Br-E long terminal repeat enhancer (3). Spi-1/PU.1 is found rearranged in Friend spleen focus-forming virus-induced erythroleukemia. Friend spleen focus-forming virus integrations are located 10 kbp upstream of the first exon of Spi-1/PU.1 (26). In no other cases were both Fli-1 and Spi-1/PU.1 genes reported to be activated in myeloid leukemias. These data confirm the involvement of those two genes in myeloid leukemogenesis in addition to erythroid leukemogenesis and suggest their importance in normal regulation of myeloid hematopoiesis. To further characterize the tumors with rearrangements in Fli-1 and Spi-1/PU.1 genes, we analyzed the expression of Fli-1, Spi-1/PU.1, and EpoR on Northern blots performed on total RNA from tumors with rearranged genes. Results depicted in Fig. 3 clearly demonstrate overexpression of the Fli-1 gene in the three tumors with rearrangement in that locus compared to the levels from healthy spleen or from a tumor with nonrearranged genes. In those three same tumors, a high level of EpoR is also observed (Fig. 3). High levels of EpoR expression were also observed in Cas-Br-E-induced non-T, non-B lymphomas that harbored a proviral integration in Fli-1 (8). However, it is possible that the high level of EpoR observed is correlated with the overexpression of Fli-1, suggesting that Fli-1 might be involved in the regulation of EpoR expression. Hybridization with a myeloperoxidase cDNA probe did not reveal high levels of transcription in the three tumors with rearrangements in Fli-1 gene. A control tumor and the tumor with rearrangement in the Spi-1/PU.1 gene both demonstrate a high level of expression of myeloperoxidase (Fig. 3), but analysis of several granulocytic tumors revealed different levels of myeloperoxidase expression (not shown). This nonuniformity of expression could be linked to the different stages of differentiation of the leukemic cells present in each tumor. Indeed, myeloperoxidase mRNA is detectable by Northern blot analysis solely in late myeloblastic and promyelocytic stages (44). Histochemical examination of the three tumors with rearrangements in the Fli-1 gene clearly revealed their granulocytic origin (not shown). The tumor with rearrangements in Spi-1 shows a higher expression of the Spi-1 transcript, suggesting an activation by transcriptional enhancement since the proviral integration is located 10 kbp upstream of the first exon (Fig. 2). Although only one Graffi MuLV-induced tumor had rearrangements in the Spi-1/PU.1 locus, involvement of Spi-1/PU.1 in granulocytic leukemia is not surprising, since this proto-oncogene has been shown to take an important part in myeloid cell development (5, 12, 25, 37). These data strongly suggest that proviral activation of the Fli-1 and Spi-1 genes resulting in their deregulated expression plays an important role in the development of granulocytic leukemia.
FIG. 3.
Northern blot analysis of Graffi MuLV-induced tumors. Lanes: 1, control spleen; 2, control tumor; 3, tumor B9; 4, tumor F11; 5, tumor F17; 6, tumor B32. DNA rearrangements observed in the tumors are indicated over the lanes. RNAs have been hybridized with probes indicated to the left of the blots. MP, myeloperoxidase.
In conclusion, we have observed rearrangements in the c-myc, Pim-1, Fli-1, and Spi-1 genes in 20, 3.3, 10, and 3.3%, respectively, of the Graffi MuLV-induced myeloblastic leukemias. These relatively low percentages of tumors with rearrangements in known oncogenes indicate that other genes could be involved in Graffi MuLV-induced leukemias.
Acknowledgments
We are grateful to Corinne Barat for helpful discussions and critical review of the manuscript.
This work was supported in part by grant 007072 from the National Cancer Institute of Canada. C.D. is a recipient of a Cancer Research Society Inc. studentship.
REFERENCES
- 1.Askew D S, Bartholomew C, Buchberg A M, Valentine M B, Jenkins N A, Copeland N G, Ihle J N. His-1 and His-2: identification and chromosomal mapping of two commonly rearranged sites of viral integration in a myeloid leukemia. Oncogene. 1991;6:2041–2047. [PubMed] [Google Scholar]
- 2.Askew D S, Bartholomew C, Ihle J N. Insertional mutagenesis and the transformation of hematopoietic stem cells. Hematol Pathol. 1993;7:1–22. [PubMed] [Google Scholar]
- 3.Barat C, Rassart E. Nuclear factors that bind to the U3 region of two myeloid leukemia-inducing retroviruses, Cas-Br-E and Graffi. Virology. 1999;252:82–95. doi: 10.1006/viro.1998.9435. [DOI] [PubMed] [Google Scholar]
- 4.Barbeau B, Bergeron D, Beaulieu M, Nadjem Z, Rassart E. Characterization of the human and mouse Fli-1 promoter regions. Biochim Biophys Acta. 1996;1307:220–232. doi: 10.1016/0167-4781(96)00060-7. [DOI] [PubMed] [Google Scholar]
- 5.Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood. 1997;90:1828–1839. [PubMed] [Google Scholar]
- 6.Ben-David Y, Giddens E B, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus—insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 7.Ben-David Y, Giddens E, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend. Proc Natl Acad Sci USA. 1990;87:1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergeron D, Houde J, Poliquin L, Barbeau B, Rassart E. Expression and DNA rearrangement of proto-oncogenes in Cas-Br-E induced non-T, non-B cell leukemias. Leukemia. 1993;7:954–962. [PubMed] [Google Scholar]
- 9.Bergeron D, Poliquin L, Kozak C A, Rassart E. Identification of a common viral integration region in Cas-Br-E murine leukemia virus-induced non-T, non-B-cell lymphomas. J Virol. 1991;65:7–15. doi: 10.1128/jvi.65.1.7-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bordereaux D, Fichelson S, Sola B, Tambourin P E, Gisselbrecht S. Frequent involvement of the fim-3 region in Friend murine leukemia virus-induced mouse myeloblastic leukemias. J Virol. 1987;61:4043–4045. doi: 10.1128/jvi.61.12.4043-4045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchberg A M, Bedigian H G, Jenkins N A, Copeland N G. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol Cell Biol. 1990;10:4658–4666. doi: 10.1128/mcb.10.9.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H M, Zhang P, Voso M T, Hohaus S, Gonzalea D A, Glass C K, Zhang D E, Tenen D G. Neutrophils and monocytes express high levels of PU.1 (Spi.1) but not Spi-B. Blood. 1995;85:2918–2928. [PubMed] [Google Scholar]
- 13.Corcoran L M, Adams J M, Dunn A R, Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984;37:113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 14.Cuypers H T, Selten G, Quint W, Zijlstra M, Maandag E R, Boelens W, van Wezenbeek P, Melief C, Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 15.Cuypers H T M, Selten G C, Zijlstra M C, De Goede R E, Melief C J, Berns A J. Tumor progression in murine leukemia virus-induced T-cell lymphomas: monitoring clonal selections with viral and cellular probes. J Virol. 1986;60:230–241. doi: 10.1128/jvi.60.1.230-241.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreyfus F, Sola B, Fichelson S, Varlet P, Charron M, Tambourin P, Wendling F, Gisselbrecht S. Rearrangements of the Pim-1, c-myc, and p53 genes in Friend helper virus-induced mouse erythroleukemias. Leukemia. 1990;4:590–594. [PubMed] [Google Scholar]
- 17.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 18.Graffi A. Chloroleukemia of mice. Ann N Y Acad Sci. 1957;68:540–558. doi: 10.1111/j.1749-6632.1957.tb56107.x. [DOI] [PubMed] [Google Scholar]
- 19.Graffi A, Fey F, Schramm T. Experiments on the hematologic diversification of viral mouse leukemias. Natl Cancer Inst Monogr. 1996;22:21–31. [PubMed] [Google Scholar]
- 20.Hoffman-Liebermann B, Liebermann D A. Interleukin-6- and leukemia inhibitory factor-induced terminal differentiation of myeloid leukemia cells is blocked at an intermediate stage by constitutive c-myc. Mol Cell Biol. 1991;11:2375–2381. doi: 10.1128/mcb.11.5.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard J C, Bani M R, Hawley R G, Ben-David Y. Activation of the erythropoietin gene in the majority of F-MuLV-induced erythroleukemia results in growth factor independence and enhanced tumorigenicity. Oncogene. 1996;12:1405–1415. [PubMed] [Google Scholar]
- 22.Jenkins J R, Rudge K, Redmond S, Wade-Evans A. Cloning and expression analysis of full mouse c-DNA sequences encoding the transformation associated protein p53. Nucleic Acids Res. 1984;12:5609–5626. doi: 10.1093/nar/12.14.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonker J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 24.Lavu S, Reddy P. Structural organization and nucleotide sequence of the mouse c-myb oncogene. Nucleic Acids Res. 1986;14:5309–5320. doi: 10.1093/nar/14.13.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mckercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 26.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 27.Moreau-Gachelin F, Ray D, Mattei M G, Tambourin P, Taviatan A. The putative oncogene Spi-1: murine chromosomal localization and transcriptional activation in murine acute erythroleukemias. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- 28.Morishita K, Parker D S, Mucenski M L, Jenkins N A, Copeland N G, Ihle J C. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 29.Möröy T, Verbeek S, Ma A, Achacoso P, Berns A, Alt F. E(−N-myc and L-myc cooperate with Eμ-pim-1 to generate lymphoid tumors at high frequency in double transgenic mice. Oncogene. 1991;6:1941–1948. [PubMed] [Google Scholar]
- 30.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott D, Keller J, Rein A. 10A1 MuLV induces a murine leukemia that expresses hematopoietic stem cell markers by a mechanism that induces Fli-1 integration. Virology. 1994;205:563–568. doi: 10.1006/viro.1994.1680. [DOI] [PubMed] [Google Scholar]
- 32.Paul R, Schuetze S, Kozak S L, Kozak C A, Kabat D. The Sfpi-1 proviral integration site of Friend erythroleukemia virus encodes the ets-related transcription factor Pu.1. J Virol. 1991;65:464–467. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rassart E, Houde J, Denicourt C, Ru M, Barat C, Edouard E, Poliquin L, Bergeron D. Molecular analysis and characterization of two myeloid leukemia inducing murine retroviruses. Curr Top Microbiol Immunol. 1995;211:201–210. doi: 10.1007/978-3-642-85232-9_20. [DOI] [PubMed] [Google Scholar]
- 34.Ru M, Shustik C, Rassart E. Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J Virol. 1993;67:4722–4731. doi: 10.1128/jvi.67.8.4722-4731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saikevych I A, Kerrigan D P, McConnell T S, Head D R, Appelbaum F R, Willman C L. Multiparameter analysis of acute mixed lineage leukemia: correlation of a B/myeloid immunophenotype and immunoglobulin and T-cell receptor gene rearrangements with the presence of the Philadelphia chromosome translocation in acute leukemias with myeloid morphology. Leukemia. 1991;5:373–382. [PubMed] [Google Scholar]
- 36.Saris C J, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 38.Selten G, Cuypers H T, Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T cell lymphomas. EMBO J. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selten G, Cuypers H T, Zijlstra M, Melief C, Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984;3:3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng-Ong G L C, Keath E J, Piccoli S P, Cole M D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell. 1982;1:443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- 41.Silver J, Kozak C. Common proviral integration region on mouse chromosome 7 in lymphomas and myelogenous leukemias induced by Friend murine leukemia virus. J Virol. 1986;57:526–533. doi: 10.1128/jvi.57.2.526-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sola B, Fichelson S, Bordereaux D, Tambourin P E, Gisselbrecht S. Fim-1 and Fim-2: two new integration regions of Friend murine leukemia virus in myeloblastic leukemias. J Virol. 1986;60:718–725. doi: 10.1128/jvi.60.2.718-725.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strassburg C P, Neubauer V, Poliwoda H, Benter T. Regulation of the proto-oncogenes c-cis, c-fos, c-myc and c-myb in acute myeloid leukemia. Neoplasma. 1992;39:343–347. [PubMed] [Google Scholar]
- 44.Tobler A, Miller C W, Johnson K R, Selsted M E, Rovera G, Koeffler H P. Regulation of gene expression of myeloperoxidase during myeloid differentiation. J Cell Physiol. 1988;136:215–225. doi: 10.1002/jcp.1041360203. [DOI] [PubMed] [Google Scholar]
- 45.van Lohuisen M, Verbeek J, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A. Predisposition to lyphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 46.Verbeek S, van Lohuisen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the Eμ-myc and Eμ-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol. 1991;11:1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolff I. Contribution of oncogenes and tumor suppressor genes to myeloid leukemia. Biochim Biophys Acta. 1997;1332:f67–f104. doi: 10.1016/s0304-419x(97)00006-1. [DOI] [PubMed] [Google Scholar]
- 48.Youssoufian H, Zon L I, Orkin S H, D’Andrea A D, Lodish H F. Structure and transcription of the mouse erythropoietin receptor gene. Mol Cell Biol. 1990;10:3675–3682. doi: 10.1128/mcb.10.7.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]