Abstract
BALB/c mice sensitized to vaccinia virus expressed G protein of respiratory syncytial virus (RSV) develop a Th2-type cytokine response and pulmonary eosinophilia when challenged with live RSV. In this study, BALB/c mice were immunized or challenged with an RSV mutant lacking the G and SH proteins or with DNA vaccines coding for RSV G or F protein. F or G protein DNA vaccines were capable of sensitizing for pulmonary eosinophilia. The absence of the G and/or SH protein in the infecting virus resulted in a consistent increase both in pulmonary natural killer cells and in gamma interferon and tumor necrosis factor expression, as well as, with primary infection, a variable increase in neutrophils and CD11b+ cells. The development of pulmonary eosinophilia in formalin-inactivated RSV-vaccinated mice required the presence of the G and/or SH protein in the challenge virus. These data show that G and/or SH protein has a marked impact on the inflammatory and innate immune response to RSV infection.
Respiratory syncytial virus (RSV) is the primary agent of serious lower respiratory tract disease in young children and is also associated with serious lower respiratory tract disease throughout life (4, 10, 20). For these reasons, it is important to develop a safe and efficacious vaccine. Early studies of children given a formalin-inactivated RSV (FI-RSV) vaccine had the disastrous consequences of enhanced pulmonary disease upon reexposure to live RSV (5, 35). The gravity of the results from this early vaccination attempt widened the vaccine search to include subunit vaccines as well as live, attenuated virus vaccines (8, 9, 17, 21, 24, 33). The use of a live, attenuated virus vaccine is a promising approach, since attenuated vaccines mimic natural infection, which is not associated with enhanced disease following reinfection. For development of both live, attenuated and subunit vaccines, an understanding of the pathogenesis of disease is likely to be important. In the case of subunit vaccines, it is critical to understand the pathogenesis of the enhanced disease associated with FI-RSV vaccination to ensure that new vaccines will not have similar deleterious consequences. In the case of live, attenuated vaccines, it is important to understand the pathogenesis of natural disease, to improve our ability to design a safe and effective vaccine from an RSV infectious clone.
The G and SH proteins of RSV are unusual compared to most other respiratory viruses. Relatively little is known about the SH protein, but the G protein contributes to both protective immunity and immune-mediated disease. The G protein has both a secreted and a membrane-bound form. Recent studies with BALB/c mice suggest that it is the secreted form that is most effective in inducing pulmonary eosinophilia and enhanced disease (12, 16, 22). The G protein has also been linked to increased expression of Th2 cytokines (13, 14, 23, 28). Recent evidence suggests that there is a T-cell epitope on the G protein that by itself is capable of inducing eosinophilia (25); in addition, immunization of BALB/c mice with native or vaccinia virus-expressed G glycoprotein has previously been shown to induce eosinophilia (12, 16, 28).
A cold-adapted temperature-sensitive strain of RSV, termed CP52, was evaluated in animals and humans as an RSV vaccine candidate (7). CP52 was shown to have a deletion of both the SH and G genes, resulting in a loss of expression of the SH and G proteins during infection (18). Despite the lack of SH and G genes, CP52 replicates and forms syncytia in tissue culture and has restricted replication in mice, nonhuman primates, and humans (18, 37). The deletion of the SH and G proteins provided us with an opportunity to examine the impact of these two proteins on the RSV immune response.
In this report, we describe studies of CP52 compared to its parent strain, B1. These studies provide important insights into the effect that the G and/or SH protein has on the type of inflammatory cells, cytokines expressed by immune cells, and the cytotoxic T lymphocytes (CTL) seen in response to RSV infection.
MATERIALS AND METHODS
Viruses.
The B1 strain of RSV, B1 derivative strain CP52, and the JS strain of human parainfluenza virus 3 (PIV3) were propagated in Vero cells (African green monkey kidney fibroblasts [ATCC CCL 81]) maintained in RPMI 1640 (GIBCO Laboratories, Grand Island, N.Y.) supplemented with 2% heat-inactivated (56°C) fetal bovine serum (FBS; HyClone Laboratories, Salt Lake City, Utah), 1% l-glutamine, and 1% antibiotic-antimycotic (all from GIBCO) (TCM). Upon detectable cytopathic effect, the TCM was decanted and replaced with a minimal volume of Dulbecco’s modified phosphate-buffered saline (D-PBS) and frozen at −70°C. The flask was thawed, and the loosely adherent cell monolayer was scraped off with a cell scraper (Costar, Cambridge, Mass.) and collected. The cells and supernatant were frozen at −70°C, thawed, and then centrifuged at 2,000 × g for 15 min at 4°C. The titer was determined by methylcellulose plaque assay on Vero cells.
Virus titer in lungs.
The quantities of infectious virus present in individual lung homogenates at various days after infection with either B1 or CP52 were determined. Identical weights of individual lung isolates were homogenized in PBS and assayed by plaque assay on monolayers of Vero cells (18, 34).
Vaccines.
Formalin-inactivated virus was prepared as described previously (30). Briefly, 1 part formalin (Sigma, St. Louis, Mo.) was incubated with 4,000 parts clarified virus lysate (B1 or PIV3) for 3 days at 37°C and pelleted by centrifugation for 1 h at 50,000 × g. The volume of virus was adjusted to a 1:25 dilution of the original volume in minimal essential medium (MEM; GIBCO) and subsequently precipitated with aluminum hydroxide (4 mg/ml; Sigma), resuspended in 1:100 the original volume in serum-free MEM, and stored at 4°C. DNA vaccines were prepared by using pcDNA3 plasmids (InVitrogen Corp., San Diego, Calif.) constructed with G or F protein cDNA from RSV strain A2 as described previously (29, 36). Plasmid-purified G protein DNA vaccine (pcDNA-G), F protein DNA vaccine (pcDNA-F), and control (pcDNA only) were diluted in PBS containing 25% sucrose. Mice were anesthetized and then immunized in each tibialis anterior muscle with 100 μl of DNA vaccine (100 μg/ml). Mice were boosted with a total of 200 μl of DNA vaccine (100 μg/ml) every week for 4 weeks. To confirm plasmid expression in vivo, sera from eye bleeds was collected weekly and analyzed by indirect enzyme-linked immunosorbent assay for antibodies against RSV-infected and uninfected Vero cells. Prior to live virus challenge, RSV-specific antibody titers for G-immunized mice ranged from 1:64 to 1:256, and those for F-immunized mice ranged from 1:128 to 1:256 by enzyme-linked immunosorbent assay.
Virus infection and sampling.
Four- to six-week-old, specific-pathogen-free, female BALB/c mice were purchased from Harlan Sprague Dawley Laboratories (Indianapolis, Ind.). The mice were housed in microisolator cages and fed sterilized water and food ad libitum. Mice were anesthetized with Avertin (2,2,2-tribromoethanol), then intranasally (i.n.) infected with 104 PFU of B1 or CP52 diluted in PBS (GIBCO). Mice were immunized intraperitoneally (i.p.) with 104 PFU equivalents of FI-B1 or FI-PIV3 in the superficial gluteal muscle. All immunized animals were rested for >3 weeks prior to challenge. Mice were i.n. challenged with 104 PFU of either B1 or CP52. At various time points postinfection p.i., mice were anesthetized and exsanguinated by severing the right caudal artery, and lymphoid organs were removed. All organs were collected on ice in Hanks balanced salt solution. To collect bronchoalveolar lavage (BAL) cells, the lung was lavaged three times with Hanks balanced salt solution containing 1% bovine serum albumin (BSA; Sigma).
Flow cytometry.
Single-cell suspensions of BAL cells were blocked with 10% normal mouse serum (Jackson Laboratories, Bar Harbor, Maine) in D-PBS, and then stained with the appropriate combinations of fluorescein thiocyanate (FITC)- or phycoerythrin (PE)-labeled anti-CD3ɛ (145-2C11), anti-CD45R/B220 (RA3-6B2), anti-pan natural killer (NK) cell (DX5), antineutrophil (RB6-8C5), anti-adhesion molecule (CD11b), and mouse isotype antibody controls (all from PharMingen, San Diego, Calif.). A lymphocyte gate was used to select 10,000 events for CD3+ and B220+ lymphocytes, and 10,000 ungated events were used for analysis of DX5+, RB6-8C5+, and CD11b+ cells. The distribution of cell surface markers was determined in two-color mode on a FACScan with CellQUEST software (Becton Dickinson, Mountain View, Calif.). The procedure used for intracellular cytokine (IC cytokine) staining was modified for microculture staining from the protocol described by PharMingen. Briefly, the intracellular transport of cytokines was inhibited by culturing cells in PBS containing GolgiStop (PharMingen) for 3 h at 37°C, thereby allowing for accumulation of cytokines in the Golgi complex of the cells. The cells were washed in PBS (GIBCO), stained with an appropriate dilution of FITC anti-CD3 for 30 min on ice, washed, and resuspended in Cytofix/Cytoperm (PharMingen) for 15 min on ice. The cells were washed in Cytofix/Cytoperm and resuspended in the appropriate dilution of PE-labeled anti-interleukin-2 (IL-2) (JES6-5H4), anti-IL-4 (BVD4-1D11), anti-IL-5 (TRFK5), anti-IL-6 (MP5-20F3), anti-IL-12 (C15.6), anti-gamma interferon (IFN-γ) (XMG1.2), or anti-tumor necrosis factor alpha (TNF-α) (MP6-XT22) antibody (all from PharMingen) diluted in D-PBS containing 1% BSA and 0.1% saponin. The cells were stained on ice for 30 min, washed, resuspended in D-PBS containing 1% BSA, and analyzed on the FACScan.
H&E staining of BAL cells.
BAL cells were washed from the lungs of anesthetized mice with PBS (GIBCO) containing 1% BSA (Sigma) by using a 1-ml syringe and 18-gauge cannula (Baxter, Deerfield, Ill.) as previously described (31, 32). Cells were kept at 4°C, and portions were cytospun onto glass microscope slides, fixed, and stained in hematoxylin and eosin (H&E).
MHC class I- and II-restricted CTLp assays.
Major histocompatibility complex (MHC) class I-restricted target cells used were the mouse mastocytoma line P815 (ATCC TIB 64). Class II-restricted target cells used were a low-expressing MHC class I subclone of the B-cell lymphoma line A20 (ATCC TIB 208). Both cell lines were maintained in RPMI 1640 (GIBCO) containing 10% FBS (HyClone) plus 1% antibiotic-antimycotic (GIBCO). The target cells were prepared by suspending 106 cells in 1.0 ml of serum-free MEM (GIBCO) containing 104 PFU of B1 (or comparable dilution of uninfected cell control lysate) for 18 h at 37°C followed by addition of 1.0 ml of MEM containing 10% FBS and 200 μCi of 51Cr (Na2CrO4; Amersham, Arlington Heights, Ill.) and incubation for an additional 2 h at 37°C. The cells were then washed and resuspended to an appropriate concentration in TCM comprised of S-MEM (GIBCO) containing 10% FBS (HyClone), 1% essential amino acids, 2% nonessential amino acids, 2% sodium pyruvate, 2% l-glutamine, 1% antibiotic-antimycotic (all from GIBCO), and 50 μM 2-mercaptoethanol (Sigma). Virus-specific CTL precursor (CTLp) prevalence was determined by using a modification of a well-established limiting-dilution assay (30).
RESULTS
BAL Cell infiltrate during primary infection, immunization, and challenge.
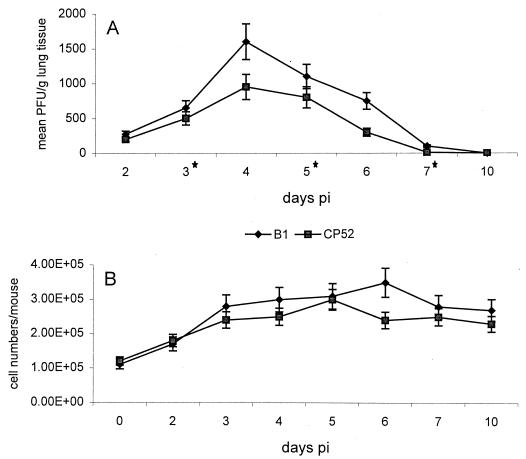
Effects of the G and/or SH protein on the types of cells responding to pulmonary infection were examined at days 3 and 6 after B1 or CP52 primary infection (Table 1) or challenge (Table 2). Loss in body weight following infection was used as one indicator of the severity of illness. There was a decrease in body weight following infection with both B1 and CP52, but no significant differences in weight loss were detected between B1- and CP52-infected mice (data not shown). Determination of titers of infectious virus in the lungs of B1- and CP52-infected mice revealed that significantly more infectious virus was present at days 4 to 6 p.i. in B1-infected mice than in CP52-infected mice (Fig. 1A). The numbers of cells responding to pulmonary infection were similar except for day 6 p.i. (Fig. 1B). BAL infiltrate (hereafter referred to simply as BAL) was examined for inflammatory cells by microscopic examination of H&E-stained cells and by flow cytometry to broadly characterize the cellular components in the BAL. For H&E studies, at least 100 cells/BAL were scored as either macrophage (MAC), polymorphonuclear cell (PMN), eosinophil (EOS), or lymphocyte (LYM) based on morphologic and staining features of the cells. In three separate experiments (three mice per experiment), the cell types resulting from either B1 or CP52 primary infection were similar at days 3 and 6 p.i. (Table 1). At days 3 and 6 p.i., MAC was the predominant cell type in the BAL, the second most common cell type being LYM. Small numbers of both PMN and EOS were observed. By flow cytometry, however, we did see significant differences in some cell types. At both day 3 and day 6 p.i., the CP52-infected mice had considerably higher numbers of DX5+ and RB6-8C5+ cells. There were significantly more CD11b+ cells and significantly fewer B220+ cells in CP52-infected mice at day 6 p.i. (Table 1). Taken together, these experiments strongly suggest that the G and/or SH protein decreases the NK cell (DX5+ cells) response to RSV infection and can alter expression of the adhesion molecule CD11b as well as the presence of RB6-8C5+ cells. In B1-immunized mice challenge with CP52, compared to challenge with B1, extensive differences in the types of some infiltrating BAL cells were evident (Table 2). There was a small increase in the percentage of MAC at both day 3 and day 6 p.i. and a decrease in EOS at day 6 p.i. (Table 2). The most striking difference, however, was in the increase in the percentage of DX5+ cells (Table 2). Similar to primary infection with CP52 (Table 1), B1-immune mice challenged with CP52 had a dramatically higher percentage of DX5+ cells in the BAL at both day 3 and day 6 p.i. These results support the primary infection data and show that the G and/or SH protein suppresses the NK response to RSV infection. By day 6 p.i., the percentage of RB6-8C5+ cells in the BAL was no longer increased in CP52-challenged mice (Table 2). The results for Vero-immunized mice challenged with B1 and CP52 suggest that the cellular antigens did not have a substantial role in the pulmonary response to RSV challenge, as the distribution of cell types was similar to that seen after primary infection (Table 3). Additionally, two controls in the B1 and CP52 challenge experiments (Vero-immunized mice [Table 3] and pcDNA-immunized mice [Table 6]) demonstrate the consistent increase in DX5+ cells, RB6-8C5+ cells, and CD11b expression at day 3 p.i.
TABLE 1.
BAL cell types by H&E staining and flow cytometry at days 3 and 6 after B1 or -CP52 primary infection
| Infectiona | H&E staining
|
Flow cytometry
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell typeb | Day 3 p.i.
|
Day 6 p.i.
|
Cell phenotyped | Day 3 p.i.
|
Day 6 p.i.
|
|||||
| Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | |||
| B1 | MAC | 80–88 | 84 (>0.10) | 78–90 | 86 (>0.10) | CD3 | 38–44 | 42 (>0.05) | 30–44 | 36 (>0.05) |
| PMN | 2–6 | 4 (>0.10) | 0–4 | 1 (>0.10) | B220 | 4–10 | 6 (>0.10) | 9–16 | 12 (<0.05) | |
| EOS | 1–5 | 2 (>0.10) | 0–5 | 3 (>0.10) | DX5 | 2–8 | 5 (<0.01) | 8–12 | 8 (<0.01) | |
| LYM | 4–15 | 10 (>0.10) | 2–12 | 10 (>0.05) | RB6-8C5 | 0–4 | 2 (<0.01) | 8–12 | 8 (<0.01) | |
| CD11b | 5–12 | 8 (<0.01) | 6–10 | 7 (<0.01) | ||||||
| CP52 | MAC | 75–90 | 85 | 75–88 | 87 | CD3 | 27–32 | 30 | 19–25 | 23 |
| PMN | 2–8 | 4 | 2–11 | 4 | B220 | 1–4 | 2 | 2–5 | 4 | |
| EOS | 0–3 | 1 | 1–8 | 4 | DX5 | 18–25 | 20 | 24–35 | 28 | |
| LYM | 6–16 | 10 | 2–5 | 5 | RB6-8C5 | 14–18 | 15 | 18–25 | 22 | |
| CD11b | 20–24 | 22 | 15–24 | 21 | ||||||
Mice were i.n. infected with 104 PFU of virus.
BAL was collected and analyzed by H&E staining for percent MAC, PMN, EOS, and LYM.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1- and CP52-infected mice ranged from 4 × 105 to 4.5 × 105/ml and from 3.8 × 105 to 5.5 × 105/ml, respectively.
BAL was collected and analyzed by flow cytometry for percent CD3+ T cells, B220+ B cells, DX5+ NK cells, RB6-8C5 neutrophils, and CD11b+ PMN.
TABLE 2.
BAL cell types by H&E staining and flow cytometry at days 3 and 6 after challenge with B1 or CP52 in previously infected mice
| Treatmenta
|
H&E staining
|
Flow cytometry
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Infection | Challenge | Cell typeb | Day 3 p.i.
|
Day 6 p.i.
|
Cell phenotyped | Day 3 p.i.
|
Day 6 p.i.
|
||||
| Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | ||||
| B1 | B1 | MAC | 80–88 | 82 (>0.10) | 78–88 | 86 (>0.10) | CD3 | 30–40 | 38 (>0.10) | 28–35 | 32 (>0.10) |
| PMN | 0–4 | 2 (>0.10) | 0–5 | 2 (>0.05) | B220 | 0–1 | 1 (>0.10) | 2–5 | 2 (>0.10) | ||
| EOS | 0–1 | 1 (>0.05) | 4–9 | 5 (>0.05) | DX5 | 5–8 | 5 (<0.01) | 4–9 | 8 (<0.01) | ||
| LYM | 12–20 | 15 (>0.10) | 5–10 | 7 (>0.10) | RB6-8C5 | 2–8 | 5 (<0.05) | 5–10 | 6 (>0.10) | ||
| CD11b | 2–8 | 6 (<0.01) | 2–8 | 5 (>0.10) | |||||||
| B1 | CP52 | MAC | 76–80 | 78 | 62–78 | 72 | CD3 | 36–45 | 40 | 26–38 | 32 |
| PMN | 0–6 | 2 | 5–13 | 8 | B220 | 2–6 | 4 | 1–6 | 2 | ||
| EOS | 2–8 | 6 | 9–20 | 12 | DX5 | 28–35 | 30 | 22–30 | 26 | ||
| LYM | 10–17 | 14 | 2–9 | 8 | RB6-8C5 | 10–15 | 12 | 5–8 | 5 | ||
| CD11b | 22–36 | 24 | 4–6 | 4 | |||||||
Mice were i.n. infected and challenged with 104 PFU of virus.
BAL was collected and analyzed by H&E staining for percent MAC, PMN, EOS, and LYM.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1- and CP52-infected mice ranged from 3 × 105 to 4.5 × 105/ml and from 3.5 × 105 to 5.5 × 105/ml, respectively.
BAL was collected and analyzed by flow cytometry for percent CD3+ T cells, B220+ B cells, DX5+ NK cells, RB6-8C5 neutrophils, and CD11b+ PMN.
FIG. 1.
(A) Mean PFU per gram of lung tissue ± standard error of the mean determined following infection with B1 or CP52 in BALB/c mice. Six to nine mice were examined at each time point except for days 3, 5, and 7 p.i., when 14 mice per time point were examined (∗). (B) Six to nine mice were i.n. infected with B1 or CP52, and total cell numbers in the BAL were determined by staining with 1% crystal violet dye and microscopic examination.
TABLE 3.
BAL cell types by H&E staining and flow cytometry at days 3 and 6 after challenge with B1 or CP52 in control mice
| Treatmenta
|
H&E staining
|
Flow cytometry
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell typeb | Day 3 p.i.
|
Day 6 p.i.
|
Cell phenotyped | Day 3 p.i.
|
Day 6 p.i.
|
||||||
| Immunization | Challenge | Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | ||
| Vero | B1 | MAC | 88–95 | 90 (>0.10) | 76–90 | 87 (>0.10) | CD3 | 35–40 | 38 (>0.10) | 30–44 | 35 (>0.10) |
| PMN | 0–2 | 1 (>0.10) | 1–4 | 1 (>0.05) | B220 | 0–6 | 4 (>0.10) | 1–3 | 1 (>0.10) | ||
| EOS | 0 | 0 (>0.10) | 0–2 | 1 (>0.10) | DX5 | 2–6 | 4 (<0.01) | 6–15 | 12 (<0.05) | ||
| LYM | 5–15 | 9 (>0.10) | 10–20 | 11 (>0.10) | RB6-8C5 | 0–2 | 2 (<0.01) | 1–7 | 3 (<0.05) | ||
| CD11b | 8–12 | 10 (<0.05) | 1–5 | 5 (<0.01) | |||||||
| Vero | CP52 | MAC | 80–92 | 86 | 72–80 | 80 | CD3 | 30–40 | 35 | 34–45 | 36 |
| PMN | 2–10 | 5 | 8–16 | 8 | B220 | 2–8 | 4 | 3–6 | 5 | ||
| EOS | 0–4 | 2 | 1–4 | 2 | DX5 | 18–30 | 25 | 16–25 | 23 | ||
| LYM | 2–10 | 7 | 8–12 | 10 | RB6-8C5 | 15–20 | 20 | 8–12 | 10 | ||
| CD11b | 25–30 | 28 | 10–15 | 15 | |||||||
,b,d See Table 2, footnotes a, b, and d.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1- and CP52-infected mice ranged from 3 × 105 to 5 × 105/ml and from 3 × 105 to 5.5 × 105/ml, respectively.
TABLE 6.
BAL cell types at day 3 and 6 after challenge of control mice with B1 or CP52
| Treatmenta
|
H&E staining
|
Flow cytometry
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell typeb | Day 3 p.i.
|
Day 6 p.i.
|
Cell phenotyped | Day 3 p.i.
|
Day 6 p.i.
|
||||||
| Immunization | Challenge | Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | ||
| pcDNA | B1 | MAC | 85–92 | 88 (>0.10) | 78–96 | 94 (>0.10) | CD3 | 25–36 | 0 (>0.10) | 33–42 | 35 (<0.01) |
| PMN | 0–5 | 2 (>0.10) | 1–5 | 3 (>0.10) | B220 | 1–10 | 5 (>0.10) | 1–16 | 8 (>0.10) | ||
| EOS | 0–2 | 2 (>0.10) | 0–2 | 1 (>0.10) | DX5 | 0–8 | 5 (<0.01) | 3–12 | 7 (<0.01) | ||
| LYM | 5–11 | 8 (>0.10) | 0–2 | 2 (>0.10) | RB6-8C5 | 2–10 | 5 (<0.05) | 1–9 | 6 (<0.01) | ||
| CD11b | 8–15 | 10 (<0.01) | 2–10 | 6 (>0.10) | |||||||
| pcDNA | CP52 | MAC | 80–95 | 87 | 70–94 | 92 | CD3 | 30–40 | 34 | 12–17 | 14 |
| PMN | 1–5 | 3 | 0–12 | 3 | B220 | 0–4 | 2 | 4–10 | 6 | ||
| EOS | 0 | 0 | 1–6 | 1 | DX5 | 30–38 | 35 | 37–54 | 48 | ||
| LYM | 2–12 | 10 | 1–5 | 4 | RB6-8C5 | 8–28 | 22 | 19–25 | 22 | ||
| CD11b | 25–40 | 38 | 3–10 | 6 | |||||||
Mice were i.n. infected with 104 PFU of virus or with pcDNA as described in Materials and Methods.
BAL was collected and analyzed by H&E staining for percent positive MAC, PMN, EOS, and LYM.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1- and CP52-infected mice ranged from 2.0 × 105 to 3.5 × 105/ml and from 2.2 × 105 to 3.5 × 105/ml, respectively.
BAL was collected and analyzed by flow cytometry for percent CD3+ T cells, B220+ B cells, DX5+ NK cells, RB6-8C5 neutrophils, and CD11b+ PMN.
The characteristics of the inflammatory response in FI-B1-immunized mice were investigated (Table 4). In these experiments, the effects of the G and/or SH protein were quite remarkable. At day 3 p.i., there was a sixfold increase in percent EOS and a twofold decrease in percent MAC in the B1-compared to CP52-challenged mice (Table 4). Similar differences were observed at day 6 p.i., i.e., a fivefold decrease in percent MAC and an eightfold increase in EOS in B1- compared to CP52-challenged mice. These results were virus specific since they were not observed for mice similarly challenged after FI-PIV3 immunization (Table 4). The small increase in the numbers of EOS in the BAL of FI-PIV3-immunized mice challenged with CP52 (13%) compared to the numbers of EOS following primary CP52 infection (4% [Table 1]) suggests that some of the EOS response may have been induced by cellular antigens or adjuvant.
TABLE 4.
BAL cell types at days 3 and 6 after challenge with B1 or CP52 in mice vaccinated with formalin-inactivated virus
| Treatmenta
|
Cell typeb | H&E staining
|
||||
|---|---|---|---|---|---|---|
| Day 3 p.i.
|
Day 6 p.i.
|
|||||
| Immunization | Challenge | Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | |
| FI-B1 | B1 | MAC | 30–42 | 35 (<0.01) | 6–15 | 14 (<0.01) |
| PMN | 2–8 | 6 (>0.01) | 3–8 | 6 (>0.10) | ||
| EOS | 30–40 | 32 (<0.01) | 50–62 | 50 (<0.01) | ||
| LYM | 25–30 | 27 (>0.05) | 25–38 | 30 (<0.05) | ||
| FI-B1 | CP52 | MAC | 68–75 | 72 | 68–75 | 75 |
| PMN | 2–9 | 5 | 5–11 | 6 | ||
| EOS | 2–10 | 5 | 1–7 | 6 | ||
| LYM | 15–22 | 18 | 10–18 | 13 | ||
| FI-CP52 | B1 | MAC | 80–88 | 83 (>0.10) | 85–92 | 89 (>0.10) |
| PMN | 0–5 | 2 (>0.10) | 1–6 | 4 (>0.10) | ||
| EOS | 2–8 | 6 (>0.10) | 0–4 | 2 (>0.10) | ||
| LYM | 5–11 | 9 (>0.10) | 3–10 | 5 (>0.10) | ||
| FI-CP52 | CP52 | MAC | 78–85 | 81 | 87–96 | 94 |
| PMN | 3–9 | 5 | 1–6 | 2 | ||
| EOS | 0–6 | 2 | 0 | 0 | ||
| LYM | 10–18 | 12 | 2–8 | 4 | ||
| FI-PIV3 | B1 | MAC | 82–90 | 87 (>0.10) | 58–72 | 70 (>0.10) |
| PMN | 1–6 | 3 (>0.10) | 11–18 | 11 (>0.10) | ||
| EOS | 0–4 | 2 (>0.10) | 8–14 | 8 (>0.10) | ||
| LYM | 6–12 | 8 (>0.10) | 6–12 | 11 (>0.10) | ||
| FI-PIV3 | CP52 | MAC | 76–88 | 80 | 52–76 | 72 |
| PMN | 2–6 | 6 | 6–11 | 10 | ||
| EOS | 2–10 | 5 | 6–19 | 13 | ||
| LYM | 6–14 | 9 | 6–18 | 5 | ||
Mice were i.n. infected with 104 PFU of virus or i.p. administered 104 PFU equivalents of formalin-inactivated virus.
BAL was collected and analyzed by H&E staining for percent MAC, PMN, EOS, and LYM.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1- and CP52-infected mice ranged from 3 × 105 to 6 × 105/ml and from 3 × 105 to 6 × 105/ml, respectively.
To address the effect of sensitization with the G protein on the pulmonary inflammatory response, the cellular components in the BAL from mice immunized with cDNA designed to express the F protein (pcDNA-F), the G protein (pcDNA-G), or nothing (pcDNA) were examined (Table 5). In these experiments, prior immunization with both pcDNA-G and pcDNA-F was associated with similar high numbers of EOS in the BAL at days 3 and 6 after challenge with B1 (Table 5). In addition, challenge of pcDNA-F-sensitized mice with either B1 or CP52 resulted in high numbers of EOS at both day 3 and day 6 p.i. These data suggest that the F protein can sensitize for EOS in the absence of the G and/or SH protein (CP52) during challenge. Flow cytometry revealed that there were some differences in cell types between pcDNA-F- and pcDNA-G-immunized mice (Table 5). As seen in all other experiments, levels of DX5+ cells were higher in CP52-challenged mice than in B1-challenged mice. At day 3 p.i., pcDNA-F-immune mice challenged with B1 had slightly higher levels of LYM, RB6-8C5+ cells, and CD11b+ cells than similarly challenged pcDNA-G-immune mice; however, these differences were absent at day 6 p.i. (Table 5). In addition, both pcDNA-F- and pcDNA-G-immune mice challenged with CP52 had higher levels of CD11b+ cells than did B1-challenged mice at both day 3 and day 6 p.i. In control studies, mice immunized with the empty pcDNA vector and challenged with either B1 or CP52 did not show any difference in cell type at either day 3 or day 6 p.i. by H&E staining. However, at both day 3 and day 6 p.i., higher numbers of DX5+ and RB6-8C5+ cells were found in the BAL of CP52-challenged mice (Table 6), a result similar to that for primary infection (Table 1). At day 3 p.i., CP52-challenged mice had higher levels of CD11b expression (38%) than B1-challenged (10%) mice. By day 6 p.i., there were no significant differences in CD11b expression between groups.
TABLE 5.
BAL cell types at days 3 and 6 after challenge with B1 or CP52 in mice immunized with G or F DNA vaccine
| Treatmenta
|
H&E staining
|
Flow cytometry
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell typeb | Day 3 p.i.
|
Day 6 p.i.
|
Cell phenotyped | Day 3 p.i.
|
Day 6 p.i.
|
||||||
| Immunization | Challenge | Range, % positive | Median % (Pc) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | Range, % positive | Median % (P) | ||
| pcDNA-G | B1 | MAC | 48–60 | 54 (<0.01) | 40–52 | 49 (<0.01) | CD3 | 22–40 | 34 (>0.10) | 20–42 | 25 (>0.10) |
| PMN | 2–8 | 6 (>0.10) | 2–10 | 9 (>0.05) | B220 | 1–10 | 5 (>0.10) | 2–9 | 6 (>0.10) | ||
| EOS | 25–40 | 32 (<0.01) | 22–38 | 35 (<0.01) | DX5 | 16–28 | 18 (<0.01) | 22–38 | 25 (<0.05) | ||
| LYM | 5–12 | 8 (>0.10) | 3–20 | 7 (>0.10) | RB6-8C5 | 6–15 | 8 (<0.05) | 6–18 | 8 (>0.10) | ||
| CD11b | 14–25 | 16 (<0.01) | 3–7 | 7 (<0.01) | |||||||
| pcDNA-G | CP52 | MAC | 80–92 | 84 | 80–96 | 89 | CD3 | 25–34 | 28 | 10–25 | 17 |
| PMN | 4–12 | 6 | 0–3 | 1 | B220 | 1–8 | 4 | 0–12 | 5 | ||
| EOS | 0–4 | 2 | 1–4 | 4 | DX5 | 30–44 | 36 | 35–46 | 40 | ||
| LYM | 6–15 | 8 | 5–12 | 6 | RB6-8C5 | 10–20 | 15 | 3–14 | 12 | ||
| CD11b | 28–36 | 34 | 20–35 | 26 | |||||||
| pcDNA-F | B1 | MAC | 40–55 | 47 (>0.05) | 20–55 | 40 (>0.05) | CD3 | 25–32 | 30 (>0.10) | 20–28 | 24 (>0.10) |
| PMN | 6–14 | 10 (>0.05) | 18–30 | 18 (>0.05) | B220 | 5–12 | 10 (>0.05) | 5–12 | 8 (>0.10) | ||
| EOS | 20–34 | 28 (>0.10) | 38–47 | 40 (<0.01) | DX5 | 20–30 | 22 (<0.01) | 22–35 | 28 (<0.01) | ||
| LYM | 12–20 | 15 (>0.10) | 0–3 | 2 (>0.10) | RB6-8C5 | 12–20 | 18 (>0.10) | 10–16 | 12 (<0.01) | ||
| CD11b | 30–38 | 34 (>0.10) | 2–12 | 9 (<0.01) | |||||||
| pcDNA-F | CP52 | MAC | 58–72 | 68 | 54–80 | 61 | CD3 | 30–38 | 32 | 18–24 | 20 |
| PMN | 0–5 | 2 | 5–14 | 10 | B220 | 0–5 | 2 | 4–14 | 8 | ||
| EOS | 18–25 | 22 | 22–30 | 26 | DX5 | 36–48 | 44 | 48–59 | 55 | ||
| LYM | 5–16 | 8 | 2–8 | 3 | RB6-8C5 | 18–28 | 25 | 20–25 | 23 | ||
| CD11b | 34–40 | 36 | 22–30 | 25 | |||||||
Mice were i.n. infected with 104 PFU of virus or i.p. immunized with a DNA vaccine as described in Materials and Methods.
BAL was collected and analyzed by H&E staining for percent positive MAC, PMN, EOS, and LYM.
The nonparametric Mann-Whitney analysis was used to test for significant differences in percent cells between B1- and CP52-infected mice within each grouping. Total cell numbers in BAL of B1-challenged mice ranged from 2.2 × 105 to 5.5 × 105/ml, and those in CP52-infected mice ranged from 2.0 × 105 to 5.5 × 105/ml.
BAL was collected and analyzed by flow cytometry for percent positive CD3+ T cells, B220+ B cells, DX5+ NK cells, RB6-8C5 neutrophils, and CD11b+ PMN.
IC cytokine expression.
IC cytokine expression provided additional information on possible effects of the G-SH protein complex on the immune response to RSV (Table 7). A major difference between B1- and CP52-infected mice is that B1 mice exhibited more of a Th2-type cytokine profile than did CP52-infected mice. This was evident in the primary infection at days 3 and 6 p.i., when higher levels of IL-5 and IL-6 were expressed by B1-infected mice (Table 7). In contrast, CP52-immune mice expressed higher IFN-γ at both day 3 and day 6 p.i. A similar Th2 cytokine profile, manifested by an increase in IL-4 and IL-6 in B1-immune mice, was evident at day 6 after challenge with B1 but not CP52 (Table 7). An increase in IL-12, possibly associated with the increase of NK cells during CP52 challenge (Table 2), occurred in CP52-immune mice challenged with CP52, suggesting that expression of the G and/or SH protein may also induce a Th2-type response during secondary infection.
TABLE 7.
IC cytokine expression by CD3+ T cells in BAL at days 3 and 6 after a primary or secondary infection with B1 or CP52
| Infectiona | Infection | Challenge | Mean % positive IC cytokine expression ± SEMb
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 3 p.i.
|
Day 6 p.i.
|
|||||||||||||||
| IL-2 | IL-4 | IL-5 | IL-6 | IL-12 | IFN-γ | TNF-α | IL-2 | IL-4 | IL-5 | IL-6 | IL-12 | IFN-γ | TNF-α | |||
| Primary | B1 | None | 7 ± 3 | 5 ± 2 | 20 ± 6 | 24 ± 6 | 7 ± 2 | 16 ± 9 | 12 ± 3 | 7 ± 2 | 17 ± 4 | 25 ± 5 | 28 ± 7 | 5 ± 2 | 19 ± 5 | 16 ± 6 |
| CP52 | None | 2 ± 2 | 11 ± 4 | 6 ± 3 | 10 ± 5 | 5 ± 2 | 36 ± 5 | 35 ± 11 | 13 ± 4 | 17 ± 5 | 11 ± 3 | 17 ± 3 | 12 ± 6 | 41 ± 9 | 18 ± 7 | |
| Secondary | B1 | B1 | 5 ± 3 | 19 ± 7 | 19 ± 6 | 9 ± 4 | 11 ± 4 | 10 ± 3 | 43 ± 12 | 6 ± 2 | 15 ± 4 | 8 ± 2 | 13 ± 2 | 8 ± 2 | 6 ± 2 | 28 ± 4 |
| B1 | CP52 | 10 ± 2 | 12 ± 2 | 14 ± 4 | 12 ± 4 | 9 ± 3 | 9 ± 4 | 45 ± 8 | 16 ± 8 | 8 ± 2 | 6 ± 3 | 6 ± 3 | 10 ± 3 | 10 ± 4 | 33 ± 4 | |
| CP52 | B1 | 8 ± 3 | 10 ± 4 | 16 ± 9 | 5 ± 2 | 7 ± 3 | 7 ± 3 | 32 ± 10 | 4 ± 2 | 8 ± 2 | 9 ± 3 | 4 ± 2 | 6 ± 3 | 6 ± 3 | 36 ± 6 | |
| CP52 | CP52 | 12 ± 3 | 6 ± 2 | 9 ± 3 | 9 ± 3 | 13 ± 3 | 10 ± 3 | 32 ± 9 | 10 ± 4 | 6 ± 2 | 4 ± 2 | 8 ± 2 | 10 ± 4 | 8 ± 2 | 28 ± 8 | |
Mice were i.n. immunized with 104 PFU of B1 or CP52, rested, and then challenged with 104 PFU of either B1 or CP52.
For three individual experiments, three mice per experiment.
MHC class I- and class II-restricted RSV-specific CTLp frequencies.
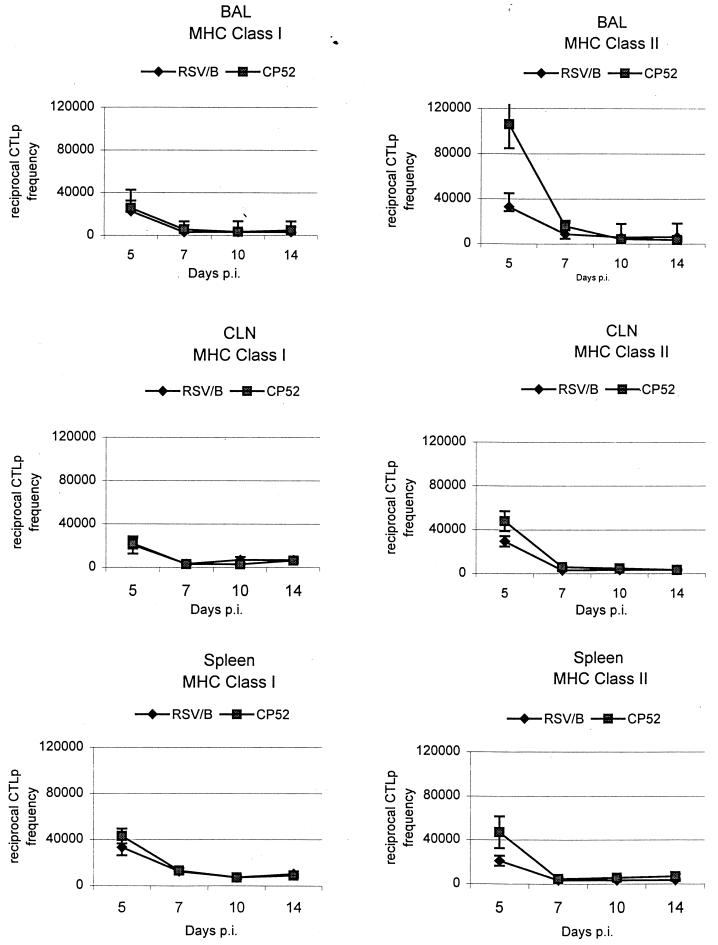
The CTLp frequencies were determined in the BAL, cervical lymph nodes (CLN), and spleen during primary (Fig. 2) and secondary (Table 8) infection with B1 or CP52. The MHC class I-restricted CTLp frequencies for BAL, CLN, and spleen were similar throughout the primary response following either B1 or CP52 infection (Fig. 2). However, a significant difference between CP52 and B1 was detected for all cell types at day 5 p.i., when MHC class II-restricted frequencies following CP52 infection were delayed compared to B1. By day 7 p.i., class II-restricted CTLp frequencies were similar and remained comparable throughout the period of study (Fig. 2). To determine if the G and/or SH protein affected CTLp frequencies in the secondary response, mice were immunized with B1, CP52, or Vero cell lysate control and then challenged with the same and reciprocal viruses (Table 8). The most prominent difference between B1 and CP52 challenge occurred in the spleen at day 3 p.i., when class I and II CTLp frequencies for CP52-immune mice challenged with CP52 were significantly lower (Table 8). The other difference in frequency occurred at day 5 p.i., in the BAL, when a significant decrease in class II CTLp frequency was detected in CP52-challenged B1-immune mice (Table 8).
FIG. 2.
Frequencies of virus-specific, MHC class I and II-restricted CTLp in the BAL, CLN, and spleen determined following immunization with B1 and CP52. CTLp frequencies were estimated by using linear regression and 95% confidence intervals about the slope of the regression line, plotting the number of cells versus the number nonresponding cultures. The 95% confidence intervals were used to determine significance, which is indicated by P < 0.05%.
TABLE 8.
Frequencies of virus-specific, MHC class I- and II-restricted CTLp in BAL and spleen
| Day post-challenge | Treatmenta
|
Reciprocal CTLp frequencyb
|
|||
|---|---|---|---|---|---|
| Immunization | Challenge | Source | Class I | Class II | |
| 3 | B1 | B1 | SPLc | 53,800 | 85,300 |
| B1 | CP52 | SPL | 33,500 | 56,300 | |
| CP52 | B1 | SPL | 33,200 | 68,200 | |
| CP52 | CP52 | SPL | 88,200d | >106d | |
| Vero | B1 | SPL | 55,500 | >106 | |
| Vero | CP52 | SPL | 79,900d | >106d | |
| B1 | B1 | BAL | 42,200 | 78,400 | |
| B1 | CP52 | BAL | 41,800 | 52,200 | |
| CP52 | B1 | BAL | 21,500 | 72,600 | |
| CP52 | CP52 | BAL | 77,500 | 81,900 | |
| Vero | B1 | BAL | 45,400 | 454,600 | |
| Vero | CP52 | BAL | 112,300d | >106d | |
| 5 | B1 | B1 | SPL | 9,400 | 20,300 |
| B1 | CP52 | SPL | 6,500 | 33,400 | |
| CP52 | B1 | SPL | 15,500 | 22,700 | |
| CP52 | CP52 | SPL | 13,000 | 14,500 | |
| Vero | B1 | SPL | 7,900 | 62,100 | |
| Vero | CP52 | SPL | 13,000 | 73,800d | |
| B1 | B1 | BAL | 8,200 | 7,700 | |
| B1 | CP52 | BAL | 11,400 | 28,900d | |
| CP52 | B1 | BAL | 6,400 | 19,300 | |
| CP52 | CP52 | BAL | 13,300 | 5,600 | |
| Vero | B1 | BAL | 10,500 | 21,700 | |
| Vero | CP52 | BAL | 45,400d | 42,200d | |
Mice were immunized and/or challenged with 104 PFU of B1 or CP52.
Estimated by using linear regression and 95% confidence intervals about the slope of the regression line, plotting the number of cells versus the number nonresponding cultures. The 95% confidence intervals were used to determine significance, which is indicated by P < 0.05%.
SPL, spleen.
Indicates a statistically significant difference (P < 0.05%) of CTLp frequencies between groups for the same MHC-restricted target cell.
DISCUSSION
With RSV infection, Th2-type cytokine expression and eosinophilia are associated with increased pulmonary disease, and the G protein of RSV has been shown to prime for both. Our studies, which contrast the response to CP52, which lacks the G and SH genes, with that of the B1 parent strain containing both genes, provide some new insights into the impact of the G and/or SH protein on the immune response to RSV infection. Most notably, the presence of the G and/or SH protein after both primary and secondary infections appears to markedly decrease NK cells and increase Th2 cytokines in the pulmonary response to infection. Additionally, the marked pulmonary eosinophilia observed in FI-RSV-immunized mice challenged with RSV appears to require the G and/or SH protein in the challenge virus.
We did not expect the pronounced effect that the G and/or SH protein had on DX5+ cells. DX5 expression is unique to NK cells and belongs to the Ly-49 gene family of class I-recognizing receptors (19). NK cells serve as an early defense against certain intracellular infections, including viral infections, and are recruited to the lung during the initial phase of primary RSV infection (15). When the G and/or SH protein was absent in a primary infection, a consistent increase in DX5+ cells and often a corresponding increase in neutrophils (RB6-8C5+ cells) and CD11b+ cells was observed. The increase in NK cells and neutrophils in the BAL when the G and/or SH protein was absent was also observed following secondary infection. We suspect that the G and/or SH protein inhibits trafficking and/or activation of these cells, possibly in a fashion similar to the inhibition of neutrophil induction shown to occur for Ebola virus (38). Interestingly, the Ebola G protein has many similarities to the RSV G protein, including high levels of glycosylation and production of a secreted form.
The dramatic pulmonary eosinophilia resulting from RSV challenge of mice immunized with FI-RSV, pcDNA-G, or pcDNA-F highlights the potential for priming, or vaccination, to impart a detrimental inflammatory response to subsequent RSV infection. The apparent critical role of the G and/or SH protein in the pulmonary eosinophilic response in FI-RSV-immunized mice is consistent with other studies demonstrating that the G protein can sensitize for eosinophilia (1–3, 12, 16). We did not, however, expect to see significant pulmonary eosinophilia in pcDNA-F-immunized mice nor, given the results for FI-RSV-immunized mice, to see eosinophilia in mice challenged with CP52. These results clearly show that the G and/or SH protein is not essential for eosinophilia and, contrary to some reports (1, 12, 26, 28), show that the F protein can prime for an eosinophilic response under appropriate conditions (11, 27). We suspect that the difference in the eosinophilic response between our studies and other studies probably results from differences in route of administration or expression vector (vaccinia virus versus DNA vaccine). Others have noted that the route and form of immunization can determine the outcome of the eosinophilic response (3). In these studies, mice immunized i.p. with a recombinant vaccinia virus expressing either soluble or membrane-anchored G glycoprotein did not generate an eosinophilic response compared to the strong eosinophilic response induced by scarification. One explanation for the apparent G and/or SH protein requirement for eosinophilia after FI-RSV immunization, but not after pcDNA-F immunization of mice follows from recent studies suggesting that CD8+ T cells play an important regulatory role in the type of response to RSV infection (15, 27). In these studies, mice were primed with a vaccinia virus construct that generates a strong CD8+ T-cell response; when challenged with live RSV, the mice developed a Th1-type T-cell response without eosinophilia. In β2-microglobulin-deficient mice (which lack functional CD8+ T cells), priming with the RSV F protein was shown to result in marked eosinophilia upon RSV challenge (27). In addition, when mice are primed with an immunogen that induces primarily CD4+ T cells, the response to RSV infection is predominantly Th2 type and is associated with pulmonary eosinophilia. We suspect that the FI-RSV-immunized mice developed a vigorous CD4+ T-cell response to the G and/or SH protein that either overwhelms or inhibits CD8+ memory T cells induced by other viral proteins. Consequently, when the G and/or SH protein was not present in the challenge virus, fewer CD4+ T cells were induced, allowing the CD8+ T-cell response to progress and abate the Th2 and eosinophilic responses. The G protein has been shown to be ineffective in inducing CD8+ T cells (23). Similarly, we suspect that pcDNA-F and pcDNA-G favor induction of CD4+ over CD8+ T cells, and preliminary data from our laboratory suggest that this is the case.
Cytokine expression is an important determinant in the type of immune and inflammatory response to infection and can affect disease outcome for a number of infections, including RSV. For example, evidence suggests that a Th2-type cytokine response is important in the development of enhanced disease following FI-RSV vaccination (6, 11, 34). The cytokine pattern seen with the absence of the G and/or SH protein is consistent with the earlier observations that the G protein can prime for a Th2-type cytokine response (1, 2). When G and SH proteins were present in the challenge virus, we saw a higher percentage of cells expressing IL-4 and IL-5 (Th2 cytokines) and lower percentage expressing IL-2 (Th1) cytokine.
A scarcity of information exists with respect to the frequency of RSV-specific CTLp generated following primary infection (30), and no studies have examined secondary infection. The CTLp results suggest that the response to CP52 is delayed. This delay could be the result of a number of factors, including the less efficient replication of this virus, the lack of G and/or SH protein epitopes available to induce CTLp, or other factors such as a difference in cytokine milieu associated with CP52 compared to B1.
In summary, our data demonstrate that the G and/or SH protein has a pronounced impact on the immune response to RSV infection, both as immunogen and as protein in the challenge virus. Our data do not allow us to differentiate between the effects of the G and SH proteins; however, previous studies lead us to hypothesize that the effect is probably attributable to the G protein. As noted above, the G protein has been shown to prime for Th2 cytokines and eosinophilia (3, 12, 13, 16, 25). Our speculation is that the G and/or SH protein modifies the kinetics, pattern, or magnitude of chemokines produced in the inflammatory response to RSV infection. The consequence is altered recruitment and trafficking of innate immune cells to the site of infection, thus providing a temporary advantage for the virus. This hypothesis is currently under investigation. Independent of which protein (G and/or SH) is responsible for the findings presented here, these data advance our understanding of the pathogenesis of RSV disease and suggest an intriguing direction for future studies of RSV disease.
ACKNOWLEDGMENTS
We thank Stephen Whitehead and Brian Murphy at LID, National Institute of Allergy and Infectious Diseases, Bethesda, Md., for providing CP52, Robert L. Coffman at DNAX for providing the RB6-8C5 B cell hybridoma, and Terry Tumpey for helpful comments and suggestions regarding the RB6-8C5 antibody.
Wayne Sullender was supported by PHS grants AI37197 and AI33425.
REFERENCES
- 1.Alwan W H, Kozlowska W J, Openshaw P J. Distinct types of lung disease caused by functional subsets of antiviral T cells. J Exp Med. 1994;179:81–89. doi: 10.1084/jem.179.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alwan W H, Record F M, Openshaw P J. Phenotypic and functional characterization of T cell lines specific for individual respiratory syncytial virus proteins. J Immunol. 1993;150:5211–5218. [PubMed] [Google Scholar]
- 3.Bembridge G P, Garcia-Beato R, Lopez J A, Melero J A, Taylor G. Subcellular site of expression and route of vaccination influence pulmonary eosinophilia following respiratory syncytial virus challenge in BALB/c mice sensitized to the attachment G protein. J Immunol. 1998;161:2473–2480. [PubMed] [Google Scholar]
- 4.Chanock R M, Parrott R H, Connors M, Collins P L, Murphy B R. Serious respiratory tract disease caused by respiratory syncytial virus: prospects for improved therapy and effective immunization. Pediatrics. 1992;90:137–143. [PubMed] [Google Scholar]
- 5.Chin J, Magoffin R L, Shearer L A, Schieble J H, Lennette E H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 6.Connors M, Giese N A, Kulkarni A B, Firestone C Y, Morse III H C, Murphy B R. Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J Virol. 1994;68:5321–5325. doi: 10.1128/jvi.68.8.5321-5325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowe J E, Jr, Bui P T, Firestone C-Y, Connors M, Elkins W R, Chanock R M, Murphy B R. Live subgroup B respiratory syncytial virus vaccines that are attenuated, genetically stable, and immunogenic in rodents and nonhuman primates. J Infect Dis. 1996;173:829–839. doi: 10.1093/infdis/173.4.829. [DOI] [PubMed] [Google Scholar]
- 8.Crowe J E, Jr, Bui P T, London W T, Davis A R, Hung P P, Chanock R M, Murphy B R. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine. 1994;12:691–699. doi: 10.1016/0264-410x(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 9.Dudas R A, Karron R A. Respiratory syncytial virus vaccines. Clin Microbiol Rev. 1998;11:430–439. doi: 10.1128/cmr.11.3.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fixler D E. Respiratory syncytial virus infection in children with congenital heart disease: a review. Pediatri Cardiol. 1996;17:163–168. doi: 10.1007/BF02505206. [DOI] [PubMed] [Google Scholar]
- 11.Graham B S, Henderson G S, Tang Y W, Lu X, Neuzil K M, Colley D G. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol. 1993;151:2032–2040. [PubMed] [Google Scholar]
- 12.Hancock G E, Speelman D J, Heers K, Bortell E, Smith J, Cosco C. Generation of atypical pulmonary inflammatory responses in BALB/c mice after immunization with the native attachment (G) glycoprotein of respiratory syncytial virus. J Virol. 1996;70:7783–7791. doi: 10.1128/jvi.70.11.7783-7791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussell T, Baldwin C J, O’Garra A, Openshaw P J. CD8+ T cells control Th2-driven pathology during pulmonary respiratory syncytial virus infection. Eur J Immunol. 1997;27:3341–3349. doi: 10.1002/eji.1830271233. [DOI] [PubMed] [Google Scholar]
- 14.Hussell T, Khan U, Openshaw P. IL-12 treatment attenuates T helper cell type 2 and B cell responses but does not improve vaccine-enhanced lung illness. J Immunol. 1997;159:328–334. [PubMed] [Google Scholar]
- 15.Hussell T, Openshaw P J. Intracellular IFN-g expression in natural killer cells precedes lung CD8+ T cell recruitment during respiratory syncytial virus infection. J Gen Virol. 1998;79:2593–2601. doi: 10.1099/0022-1317-79-11-2593. [DOI] [PubMed] [Google Scholar]
- 16.Johnson T R, Johnson J E, Roberts S R, Wertz G W, Parker R A, Graham B S. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J Virol. 1998;72:2871–2880. doi: 10.1128/jvi.72.4.2871-2880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhasz K, Whitehead S S, Bui P T, Biggs J M, Crowe J E, Boulanger C A, Collins P L, Murphy B R. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J Virol. 1997;71:5814–5819. doi: 10.1128/jvi.71.8.5814-5819.1997. . (Erratum, 71:8953.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian R H, Freeman J D, Mager D L, Takei F. Role of conserved glycosylation site unique to murine class I MHC in recognition by Ly-49 NK cell receptor. J Immunol. 1998;161:2301–2306. [PubMed] [Google Scholar]
- 20.McIntosh K, Fishaut J M. Immunopathologic mechanisms in lower respiratory tract disease of infants due to respiratory syncytial virus. Progr Med Virol. 1980;26:94–118. [PubMed] [Google Scholar]
- 21.Oien N L, Brideau R J, Thomsen D R, Homa F L, Wathen M W. Vaccination with a heterologous respiratory syncytial virus chimeric FG glycoprotein demonstrates significant subgroup cross-reactivity. Vaccine. 1993;11:1040–1048. doi: 10.1016/0264-410x(93)90131-g. [DOI] [PubMed] [Google Scholar]
- 22.Openshaw P J. Immunity and immunopathology to respiratory syncytial virus. The mouse model. Am J Respir Crit Care Med. 1995;152:S59–S62. doi: 10.1164/ajrccm/152.4_Pt_2.S59. [DOI] [PubMed] [Google Scholar]
- 23.Openshaw P J, Clarke S L, Record F M. Pulmonary eosinophilic response to respiratory syncytial virus infection in mice sensitized to the major surface glycoprotein G. Int Immunol. 1992;4:493–500. doi: 10.1093/intimm/4.4.493. [DOI] [PubMed] [Google Scholar]
- 24.Parrott R H, Kim H W, Brandt C D, Chanock R M. Potential of attenuated respiratory syncytial virus vaccine for infants and children. Dev Biol Stand. 1975;28:389–399. [PubMed] [Google Scholar]
- 25.Sparer T E, Matthews S, Hussell T, Rae A J, Garcia-Barreno B, Melero J A, Openshaw P J. Eliminating a region of respiratory syncytial virus attachment protein allows induction of protective immunity without vaccine-enhanced lung eosinophilia. J Exp Med. 1998;187:1921–1926. doi: 10.1084/jem.187.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spender L C, Hussell T, Openshaw P J. Abundant IFN-gamma production by local T cells in respiratory syncytial virus-induced eosinophilic lung disease. J Gen Virol. 1998;79:1751–1758. doi: 10.1099/0022-1317-79-7-1751. [DOI] [PubMed] [Google Scholar]
- 27.Srikiatkhachorn A, Braciale T J. Virus-specific CD8+ T lymphocytes downregulate T helper cell type 2 cytokine secretion and pulmonary eosinophilia during experimental murine respiratory syncytial virus infection. J Exp Med. 1997;186:421–432. doi: 10.1084/jem.186.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srikiatkhachorn A, Braciale T J. Virus-specific memory and effector T lymphocytes exhibit different cytokine responses to antigens during experimental murine respiratory syncytial virus infection. J Virol. 1997;71:678–685. doi: 10.1128/jvi.71.1.678-685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullender W. Antigenic analysis of chimeric and truncated G proteins of respiratory syncytial virus. Virology. 1995;209:70–79. doi: 10.1006/viro.1995.1231. [DOI] [PubMed] [Google Scholar]
- 30.Tripp R A, Anderson L J. Cytotoxic T-lymphocyte precursor frequencies in BALB/c mice after acute respiratory syncytial virus (RSV) infection or immunization with a formalin-inactivated RSV vaccine. J Virol. 1998;72:8971–8975. doi: 10.1128/jvi.72.11.8971-8975.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripp R A, Hou S, McMickle A, Houston J, Doherty P C. Recruitment and proliferation of CD8+ T cells in respiratory virus infections. J Immunol. 1995;154:6013–6021. [PubMed] [Google Scholar]
- 32.Tripp R A, Sarawar S R, Doherty P C. Characteristics of the influenza virus-specific CD8+ T cell response in mice homozygous for disruption of the H-21Ab gene. J Immunol. 1995;155:2955–2959. [PubMed] [Google Scholar]
- 33.Tristram D A, Welliver R C, Mohar C K, Hogerman D A, Hildreth S W, Paradiso P. Immunogenicity and safety of respiratory syncytial virus subunit vaccine in seropositive children 18–36 months old. J Infect Dis. 1993;167:191–195. doi: 10.1093/infdis/167.1.191. [DOI] [PubMed] [Google Scholar]
- 34.Waris M E, Tsou C, Erdman D D, Day D B, Anderson L J. Priming with live respiratory syncytial virus (RSV) prevents the enhanced pulmonary inflammatory response seen after RSV challenge in BALB/c mice immunized with formalin-inactivated RSV. J Virol. 1997;71:6935–6939. doi: 10.1128/jvi.71.9.6935-6939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weibel R E, Stokes J, Jr, Leagus M B, Mascoli C C, Hilleman M R. Respiratory virus vaccines. V. Field evaluation for efficacy of heptavalent vaccine. Am Rev Respir Dis. 1966;94:362–379. doi: 10.1164/arrd.1966.94.3.362. [DOI] [PubMed] [Google Scholar]
- 36.Wertz G W, Stott E J, Young K K, Anderson K, Ball L A. Expression of the fusion protein of human respiratory syncytial virus from recombinant vaccinia virus vectors and protection of vaccinated mice. J Virol. 1987;61:293–301. doi: 10.1128/jvi.61.2.293-301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehead S S, Juhasz K, Firestone C Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Delgado R, Xu L, Todd R F, Nabel E G, Sanchez A, Nabel G J. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]