Abstract
Many cancers originate from stem or progenitor cells hijacked by somatic mutations that drive replication, exemplified by rapid adenomatous transformation of pulmonary alveolar type II (AT2) cells. Here, we demonstrate a different scenario: expression of KrasG12D in fully differentiated AT1 cells reprograms them slowly and asynchronously back into AT2 cells that go on to generate indolent tumors. Like human lepidic adenocarcinoma, the tumor cells slowly spread along intact alveolar walls in a non-destructive manner and have low ERK activity. We find that AT1 and AT2 cells act as distinct cells of origin and manifest divergent responses to concomitant Wnt activation and KrasG12D induction, which dramatically increases AT2-derived but inhibits AT1-derived adenoma proliferation. Pharmacological augmentation of ERK activity in KrasG12D-induced AT1 cells dramatically increases transformation efficiency, proliferation, and likelihood of progression from pure lepidic to mixed tumor histology. Overall, we have identified a novel cell of origin for lung adenocarcinoma, the AT1 cell, which recapitulates features of human lepidic cancer. In so doing, we also uncover a capacity for oncogenic Kras to reprogram a differentiated and quiescent cell back into its parent stem cell en route to adenomatous transformation. Our work further reveals that irrespective of a given cancer’s current molecular profile and driver oncogene, the cell of origin exerts a pervasive and perduring influence on its subsequent behavior.
One-Sentence Summary:
Oncogenic Kras reprograms differentiated AT1 cells into AT2 cells that generate indolent and functionally constrained tumors that share features with human lepidic lung adenocarcinoma.
Introduction
Lung adenocarcinoma (LuAd) is the leading cancer killer worldwide1. It arises in the distal gas exchange region of lung and is comprised of tumor cells with cytological and molecular features of alveolar epithelial type II (AT2) cells2. Multiple groups have demonstrated that AT2 cells are a key cell of origin for LuAd, rapidly generating adenomatous tumor nodules upon induction of KrasG12D in mouse 3-5. Genetic mouse models recapitulate key aspects of aggressive human LuAd, including nodular growth, metastasis, and lethality6. Yet, there is a remarkably benign subtype within the human LuAd spectrum whose origin remains enigmatic. This outlier was previously considered as a separate class altogether, called bronchioalveolar carcinoma (BAC)7. However, because this term was not applied consistently, BAC was subsumed under the LuAd spectrum as the lepidic subtype in 20118. Lepidic lung adenomas are marked by indolent progression, infrequent metastasis, and non-destructive spread of tumor cells as a monolayer along intact alveolar septae9. Like in other histologic subtypes of LuAd, the tumor cells in lepidic LuAds cytologically and molecularly resemble AT2 cells. Yet, because they harbor driver mutations common to other LuAd subtypes, lepidic LuAds cannot be convincingly ascribed to an oncogene-specific effect on AT2 cells. Indeed, no mouse models of LuAd targeting AT2 (or any other lung cell types) have recapitulated benign alveolar tumors that slowly expand in a lepidic manner. Deep mutational and transcriptional profiling of human LuAd tumors has also not generated any fundamental molecular insights. Hence, both the cellular origin and molecular basis for the benign phenotype of lepidic tumors remains unknown.
We hypothesized that given its overlapping mutational spectrum with other LuAd subtypes yet highly distinctive biology, lepidic cancer might arise from a different (non-AT2) cell of origin. Because it is the only other alveolar epithelial cell type, the AT1 cell was a prime candidate. AT1 cells are ultra-thin, expansive cells that comprise the gas-exchange surface of the distal lung and are interspersed with cuboidal, surfactant-producing AT2 cells. AT1 cells are believed to be terminally differentiated and regenerated throughout life by AT2 stem cells10. We thus asked if AT1 cells were responsive to conditional expression of KrasG12D, a common LuAd oncogene found in multiple histological subtypes, including lepidic.
Results
KrasG12D reprograms terminally differentiated AT1 cells into AT2 stem cells
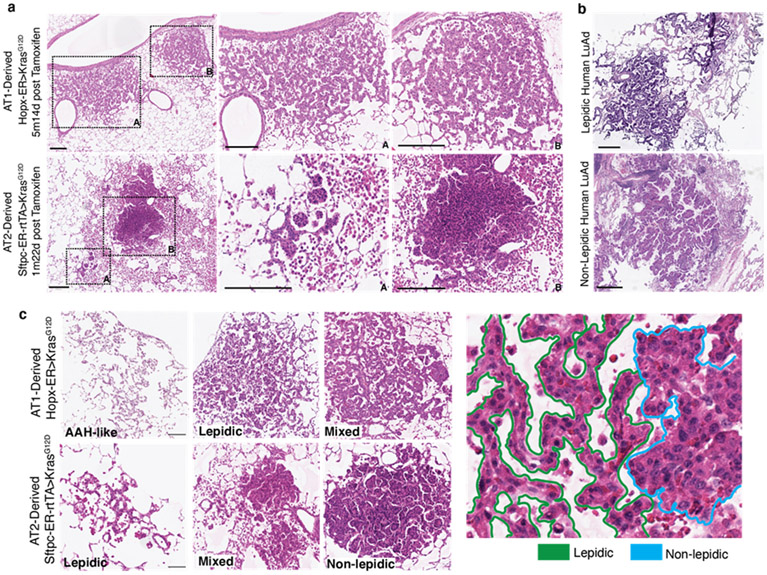
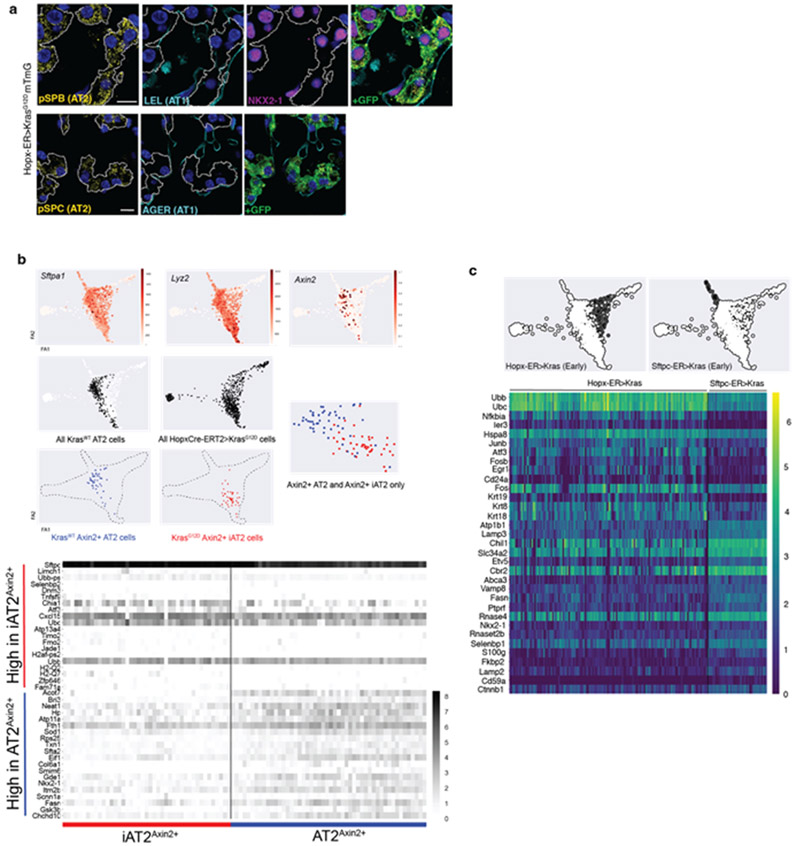
Hopx encodes a Homeodomain-only protein that is highly expressed by AT1 cells. We targeted AT1 cells for expression of oncogenic Kras along with a membrane GFP lineage tag by administering tamoxifen to adult mice harboring Hopx-CreER11, KrasLSL-G12D12, and R26mTmG13 alleles. Immunohistological analysis revealed normal-appearing flat mGFP+ AT1 cells in control lungs but discrete foci ranging from small clusters of cuboidal mGFP+ cells to expansive hyperplasia spanning up to 10 contiguous alveoli at 6 months after KrasG12D induction (Figure 1a). mGFP+ cells with morphologies intermediate between squamous and cuboidal first emerged several weeks after tamoxifen administration. A notable feature of this process was the asynchronous response of AT1 cells following ‘pulsed’ induction of KrasG12D by a one-time dose of tamoxifen. This temporally heterogeneous response facilitated reconstruction of the process of AT1 ‘unflattening’ and adoption of a cuboidal morphology (Figure 1b).
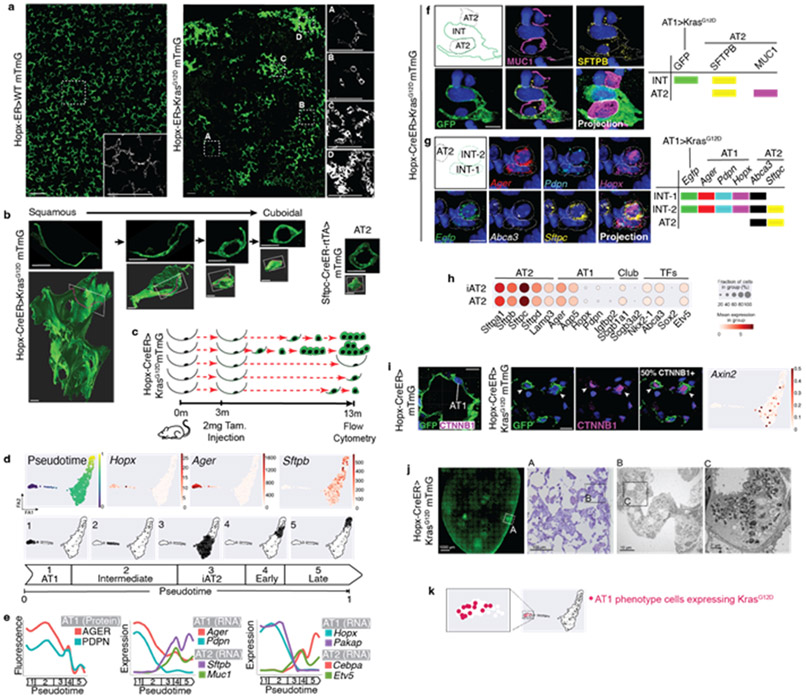
Figure 1: KrasG12D asynchronously reprograms AT1 cells into AT2Axin2 stem cells.
(a) AT1 membrane marking (GFP, green) shows uniformly flat AT1 cells in control lung (left) and cells with variable morphologies 6m after Kras induction (right). (b) 2D sections (top) and 3D reconstructions (below) of Kras-activated AT1 cells at progressive stages of morphological transition and an AT2 cell (far right). (c) Schematic of scRNA sequencing experiment for Hopx-CreER>KrasG12D mTmG mouse showing asynchronous AT1-to-iAT2 transition and tumor initiation. (d) PAGA of 696 AT1-lineage cells 10m after Kras induction with pseudotemporal ordering and feature plots of representative genes. (e) Levels of AT1 protein markers (left), AT1 and AT2 cell type markers (middle) and transcription factors (right) over pseudotime. Multimodal appearance to AGER and PDPN expression as well as transient drops in expression of Sftpb, Muc1, Cebpa, and Etv5 between pseudotime groups 4 and 5 are an artifact of splining. (f, g) AT1-to-AT2 intermediates (INT) in intact lung by immunostaining and (g) multiplex in situ hybridization. (h) Dot plot comparing lung cell type marker expression in AT2 and iAT2 cells. (i) Representative images show absence of nuclear ß-catenin in AT1 cells (left) and presence in cuboidal Kras-induced AT1 cells (middle, arrowheads). Axin2 feature plot shows absence in AT1 and AT1-to-iAT2 intermediate cells with subsequent emergence at iAT2 stage. (j) Pre-fixation endogenous fluorescence, toluidine blue stained semi-thin section, and transmission electron microscopy images demonstrating lamellar bodies in AT1-derived tumor cells. (k) PAGA of Hopx-ER>KrasG12D mTmG mouse highlighting 13 AT1-phenotype cells with KrasG12D mutant transcripts 10 months after recombination.
Scale bars, 100μm (a), 10μm (b, f, g, i), or as indicated (j).
iAT2, induced AT2; m, months; INT-1, intermediate cell 1; INT-2, intermediate cell 2; m, months; PAGA, Partition-Based Graph Abstraction; Tam, tamoxifen; TFs, transcription factors.
We next examined the molecular status of KrasG12D-induced AT1 cells. Considering the notably asynchronous AT1 cell response, we reasoned that single cell RNA sequencing (scRNA-seq) would illuminate a broad diversity of molecular stages. We therefore performed scRNA-seq of mGFP+ cells using SmartSeq2 at 10 months after tamoxifen, then ordered individual cells in pseudotime using PAGA14 (Figure 1c). This analysis supported a direct molecular reversion from AT1 to AT2 cell phenotype driven by KrasG12D (Figure 1d). The transcriptomes of cells upon cessation of AT1 marker expression closely matched that of normal AT2 cells, so we refer to KrasG12D-expressing AT1 cells at this pseudotime stage as in vivo induced AT2 (iAT2) cells (Figure 1d). Charting dynamic changes in AT1 marker proteins, AT1 and AT2 marker genes, and AT1 and AT2 cell transcription factors all indicated progressive loss of AT1 with acquisition of AT2 molecular identity (Figure 1e). Interestingly, several recently identified genes transiently expressed during physiologic AT2 to AT1 transdifferentiation were not expressed in KrasG12D-activated AT1 cells during reversion back into an AT2 cell phenotype. We next used antibody staining and multiplex in situ hybridization15 to correlate cytological, molecular and lineage features of single cells in intact lung, which revealed co-expression of selected AT1 and AT2 markers in AT1 lineage (mGFP+) cells morphologically intermediate between squamous and cuboidal (Figures 1f-g). The iAT2 group of cells were remarkably transcriptionally similar to normal AT2 cells (Figure 1h). We also assessed the Wnt status of iAT2 stage cells and found that a significant proportion were Wnt-active, indicated by nuclear ß-catenin protein and Axin2 expression (Figure 1i) This observation is significant because the Axin2-expressing subset of AT2 cells is enriched for stem cell activity in normal lung and Wnt activity is required for their proliferation10,16,17. When we specifically compared Axin2+ iAT2 cells with Axin2+ AT2 stem cells, we observed that cells from each population were intermixed in the PAGA (Figure ED2b). We also performed transmission electron microscopy which revealed the presence of lamellar bodies, the hallmark feature of mature AT2 cells, in Kras-induced AT1-lineage cells (Figure 1j). Together, these findings suggest that at least a subset of AT1 cells was fully reprogrammed into an AT2 stem cell phenotype at the molecular and ultrastructural level.
We noted that some AT1 lineage cells from the Hopx-CreER>LSL-KrasG12D, mTmG lungs had maintained an AT1 cell molecular phenotype at the transcriptomic level. To determine if these cells had merely recombined the lineage tag without inducing KrasG12D, we manually inspected the Exon2 reads from the Kras gene in this group. We were surprised to discover that a portion of these cells were actively expressing KrasG12D to which they were apparently unresponsive over the 10 months after tamoxifen induction (Figure 1k).
KrasG12D-induced AT1 cells generate indolent adenomas that grow in a lepidic manner and have differential MAPK pathway activation from KrasG12D-induced AT2 adenomas
To directly compare the oncogenic capacity of AT1 and AT2 cells, we employed an Sftpc-CreER18 knock-in allele to express the same conditional KrasLSL-G12D and R26mTmG alleles in AT2 cells. As expected, Kras-induced AT2 cells rapidly expanded as nodular lesions compressing surrounding alveoli. By contrast, Kras-induced AT1 cells gradually adopted a cuboidal morphology then slowly spread in a monolayer along alveolar walls. Adenomas arising from AT2 cells contained a much higher proportion of proliferating cells, and the vast majority were nodules at 2 months after tamoxifen. By contrast, at 6 months after KrasG12D induction adenomas arising from AT1 cells were pure lepidic or in some cases had acquired a minor nodular component (Figure 2a-b, Extended Data 1a-c). Notably, Hopx-CreER>KrasLSL-G12D mice never displayed signs of respiratory distress even at 10 months post-tamoxifen. By contrast, Sftpc-CreER>KrasLSL-G12D mice uniformly met criteria for euthanasia due to tumor burden within several months after receiving tamoxifen.
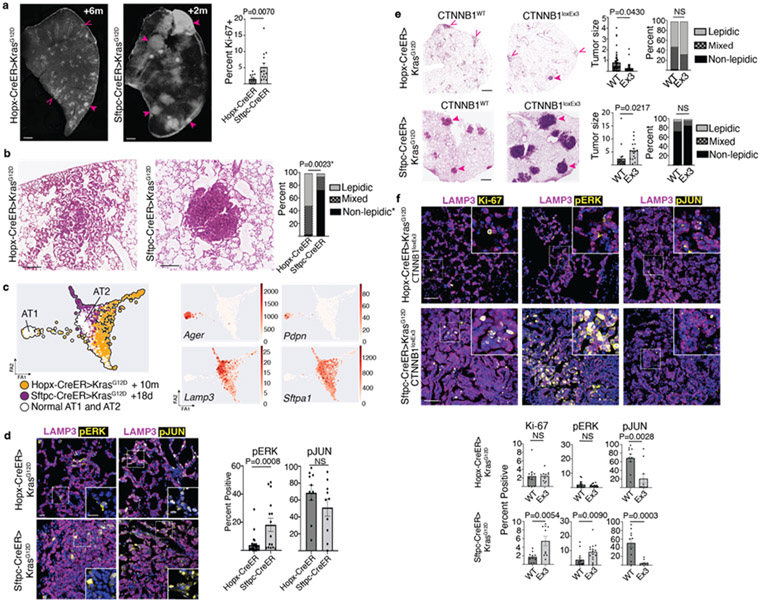
Figure 2: AT1 derived lung adenomas are histologically, molecularly, and functionally distinct from AT2 derived adenomas.
(a) Lung lobe sections 6m after Kras induction in AT1 cells (left) or 2m after induction in AT2 cells (middle) with non-solid (open arrowheads) and solid (closed arrowheads) tumors, and quantitation of tumor cell proliferation (right) (n=3 mice per group, five tumors per mouse) (b) Representative close-up H&E stain of an AT1-derived mixed histology adenoma and an AT2-derived solid adenoma with quantification of histology (n=3 mice per group, 9-18 tumors per mouse). (c) PAGA of scRNA-seq profiles of indicated populations with feature plots of AT1 and AT2 markers - 722 Hopx-CreER>KrasG12D, 305 Sftpc-CreER>KrasG12D, and 428 KrasWT cells. (d) Co-staining for LAMP3 (purple) and pERK (left, yellow) or pc-JUN (right, yellow) in AT1 (top) and AT2 (bottom) derived adenomas with quantitation. (n=4 mice for Sftpc-CreER, n=5 mice for Hopx-CreER; 2-5 tumors per mouse; 1-3m after tamoxifen for Sftpc-CreER and 4-6 months after tamoxifen for Hopx-CreER). (e) H&E stain of lungs with AT1 (top) and AT2 (bottom) derived tumors in controls (WT) or with concomitant activation of Wnt signaling (loxEx3) at 4m after Kras induction with quantitation of tumor size and histology (n=2 mice per genotype for Sftpc-CreER, n=11 mice per genotype for Hopx-CreER; 2-10 tumors per mouse; 2m or 4-6m after Kras induction for Sftpc-ER and Hopx-ER mice, respectively). Open arrowheads denote lepidic tumors, closed arrowheads denoted non-lepidic tumors. (f) Representative images and quantitation of proliferation (Ki67), pERK and pJUN in AT1 (top) and AT2 (bottom) derived tumors with (Ex3) and without (WT) activation of Wnt signaling (n= 2 littermate-paired mice per genotype, 5 tumors per mouse, total of 2,041-12,069 tumor cells per group, 2m and 4m after Kras induction for Sftpc-ER and Hopx-ER mice, respectively).
Tumor size is represented as 1x105 μm2.
Scale bars, 500 μm (a), 200 μm (b), 10 μm (d), 1mm (e), 20 μm (f)
d, days; m, months; ns, not significant; PAGA, Partition-Based Graph Abstraction; pERK, phosphorylated ERK; pJUN, phosphorylated c-JUN/JUND; scRNA-seq, single cell RNA sequencing.
Data are mean±s.e.m. P values calculated by Student’s unpaired t test.
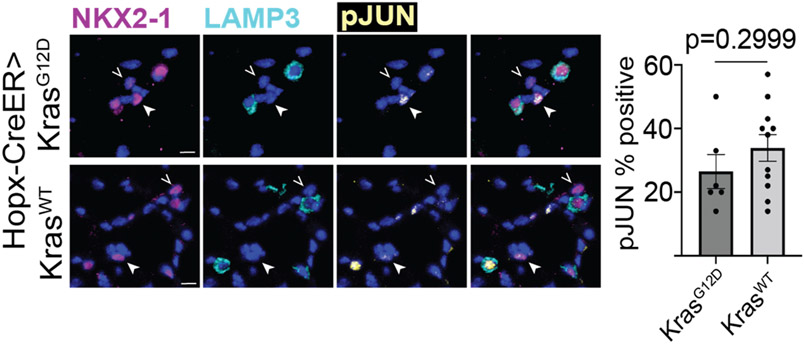
Given these very different outcomes, we next focused on Kras-induced AT1 cells at the pseudotime stage immediately after the iAT2 stage, which we refer to as ‘early’ indicating early tumor stage. We directly compared the transcriptomes of these ‘early’ stage Kras-induced AT1 cells with ‘early’ stage Kras-induced AT2 cells isolated at 18 days after tamoxifen. The results showed that ‘early’ stage Kras-induced AT1 and AT2 cells were entirely non-overlapping and embarked on separate pseudotime trajectories (Figure 2c and ED2). To explore the basis for the transcriptionally divergent responses of AT1 and AT2 cells to the identical oncogenic stimulus, we examined activity of the mitogen-activated protein kinase (MAPK) pathway downstream of Kras in AT1- and AT2-derived tumor cells by immunostaining. The results showed no difference in phosphorylated JUN (pJUN), a readout of JNK activity, which was detected in many AT1- and AT2-derived adenoma cells, whereas phosphorylated ERK (pERK) was present in significantly fewer AT1-derived adenoma cells (Figure 2d).
Given the apparent robust response of JNK to Kras activation in AT1 cells, we reasoned that evaluating the phosphorylation of JUN in ‘unresponsive’ AT1 cells would shed light on the mechanism of their resistance. The results showed no difference in the pJUN staining of AT1 phenotype (NKX2-1+/LAMP3−) cells in KrasG12D and KrasWT mice, suggesting that the mechanism of resistance to AT2 reprogramming was upstream of MAPK activation (Figure ED3).
AT1-derived adenomas are selectively resistant to growth enhancement by Wnt activation
Wnt activity has been shown to be essential for proliferation of AT2 stem cells and in KrasG12D and BrafV600E driven lung tumors, with substantially increased growth upon experimental activation19-21. We therefore asked whether activating Wnt would have the same pro-tumor effects on KrasG12D-induced AT1 cells and if it impacted MAPK activity in AT1- or AT2-derived tumors. To test this, we combined the ß-catenin-flox-Exon322 and KrasLSL-G12D alleles with the Hopx-CreER (AT1) or Sftpc-CreER (AT2) drivers. Exon 3 encodes the phosphorylation sites for GSK-3ß that activates degradation of ß-catenin protein, so its conditional deletion results in ß-catenin accumulation and constitutive Wnt signaling in the targeted cells. Using this genetic strategy, we found that concomitant KrasG12D induction and Wnt activation in AT1 cells resulted in significantly smaller tumors compared with Kras-only controls. On the other hand, AT2-derived adenomas more than doubled in size compared to Kras-only controls without Wnt activation. There was no significant effect on tumor histology for either AT1 or AT2 cells (Figure 2e). Tumor staining showed no significant difference in proliferation or pERK staining of AT1-derived adenomas with concomitant Wnt activation. By contrast, AT2 derived adenomas showed almost triple the rate of proliferation and a significant increase in pERK+ tumor cells. Both AT1 and AT2 tumors showed a major reduction in the proportion of pJUN+ cells (Figure 2f). Thus, Wnt activation drove AT2 adenoma proliferation and enhanced growth with a concomitant increase in ERK and reduction in JNK activity. In KrasG12D-induced AT1 cells, by contrast, Wnt failed to induce ERK but did reduce JNK activity, resulting in low overall MAPK activity which was associated with significantly reduced adenoma growth. Thus, the AT1 versus AT2 cell of origin dictates a divergent anti- versus pro-tumor response, respectively, to Wnt activation on KrasG12D-driven lung adenomas.
ERK augmentation increases AT1 cell transformation efficiency, adenoma size and histological progression
To map the molecular progression of AT1 lung adenomas, we analyzed the scRNA-seq expression of selected genes in tumor cells from Hopx-CreER, KrasLSL-G12D, R26mTmG mice over ‘early’ and ‘late’ tumor pseudotime stages. In addition to MAPK pathway target genes, we considered AT1, AT2, and lung embryonic progenitor markers; gastric epithelial markers; and AT2-to-AT1 ‘transitional’ markers that are transiently upregulated in intermediates during AT2 to AT1 transdifferentiation after acute lung injury. This analysis revealed the emergence and progressive increase of early (Jun, Fos) and late (Clu) ERK target genes23 that correlated with increased proliferation in ‘late’ stage AT1 tumor cells (Figure 3a-b). We therefore hypothesized that ERK is a key driver of AT1 adenoma progression and that its relative lack of activity in KrasG12D-induced AT1 cells underlies their indolent growth. To test this idea, we provided chow containing either PLX4720, an agonist of ERK signaling; PD0325901, an inhibitor of MEK1/2 (directly upstream of ERK); or no additives to Hopx-CreER, KrasLSL-G12D mice for 11 weeks starting one week after tamoxifen administration24. Staining for pERK confirmed successful augmentation and inhibition in ERK in AT1 tumors, and quantitation revealed a strong correlation of adenoma number and size with ERK activity level. Histological analysis showed that ERK augmentation more than doubled the proportion of lepidic adenomas containing a nodular component (Figure 3c). This experiment thus revealed that ERK pathway activation both promotes adenomatous transformation of AT1 cells and drives tumor progression.
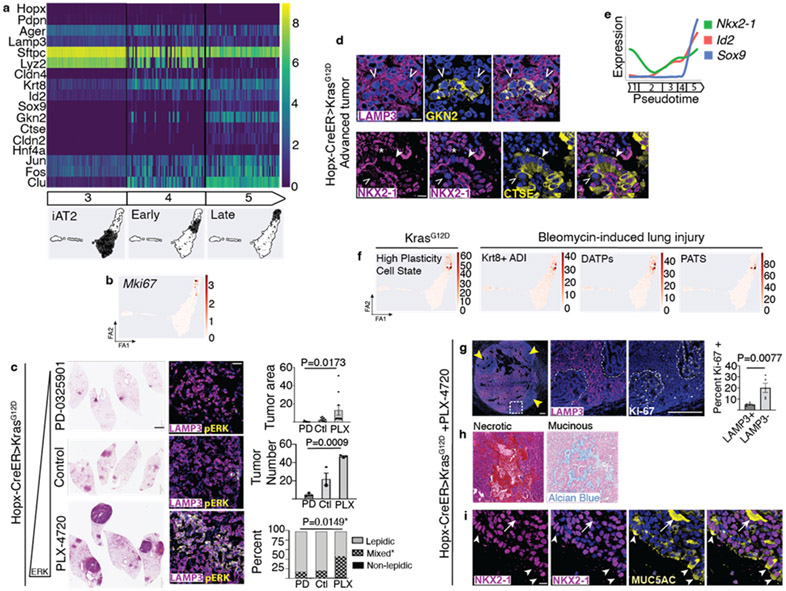
Figure 3: ERK drives AT1 adenoma growth, molecular and histological progression.
(a) Single cell profiles over pseudotime reveal emergence of cell-to-cell molecular heterogeneity at Early tumor stage with progressive increase in ERK target genes, lung progenitor and gastric markers. (b) Feature plot shows strong enrichment of Mki67 at Late tumor stage. (c) ERK pathway modulation in Hopx-CreER>KrasG12D mice with representative H&E stains, pERK staining, and quantitation of tumor area, number and histology (n=3 mice per group, 3m post tamoxifen with continual chow provision, ten largest tumors per group for tumor area). (d) Co-staining shows a cluster of GKN2 expressing cells within a LAMP3+ AT1-derived adenoma (above) and AT1-derived tumor cells with normal (arrowhead), reduced (open arrowhead) and absent (asterisk) NKX2-1 expression all positive for CTSE. (e) Maintenance of Nkx2-1 with induction of distal lung embryonic progenitor markers Sox9 and Id2 at Late tumor stage. (f) Gene scores derived from marker genes for high plasticity state in advanced AT2-derived Kras LuAd and for AT2-to-AT1 transitional intermediates in lung injury show enrichment in the same cluster of cells bridging Early and Late AT1 tumor stages. (g) Large ERK-activated AT1 LuAd with loss of Lamp3 in clusters correlating with increased proliferation, with quantitation. (n=5 areas LAMP3−/+). (h) Representative ERK-activated AT1 LuAds with central necrosis (left) or mucinous transformation (right). (i) Co-staining shows MUC5AC in the cytoplasm of NKX2-1+ tumor cells (arrowheads) and filling a lumen (arrow).
Scale bars, 10 μm (d, i), 2mm and 20 μm (f), 200 μm (h).
PD, PD-0325901 compounded chow; PLX, PLX-4720 compounded chow; Ctl, control chow.
Data are mean±s.e.m. P values calculated by ordinary one-way ANOVA (f) and Student’s t test (g).
Examination of other classes of transcripts at the ‘late’ tumor pseudotime stage revealed that AT1 tumors became molecularly dysregulated with induction of gastric epithelial markers mirroring late stages of AT2-derived lung adenomas25,26. However, one notable finding here is that Nkx2-1 continued to be expressed when gastric markers emerged, including Gkn2, Ctse, and Hnf4a, which we corroborated at the protein level by antibody staining (Figure 3a, d). This result differs from what has been reported previously, where emergence of gastric markers corresponds with lung-to-gastric reprogramming from the loss of Nkx2-125,27. Our ‘late’ tumor stage analysis also revealed emergence of transcription factors expressed by embryonic lung epithelial tip progenitors, Id228 and Sox929 (Figure 3e) and of genes normally expressed by cellular intermediates transitioning from AT2 to AT1 phenotype after bleomycin lung injury, including Cldn4 and Krt830-32. These AT2-to-AT1 ‘transitional’ markers overlap with a panel of LuAd ‘plasticity’ markers recently identified in advanced stage AT2-derived tumors26. We used the defining markers from each of four primary scRNA-seq studies as a reference gene set of the respective transitional and plasticity states. We scored these gene sets in our scRNA seq data and found strong and exclusive enrichment at the ‘late’ tumor pseudotime stage (Figure 3f, Supplemental Data). Interestingly, as mentioned above, the AT2-to-AT1 ‘transitional’ markers were not induced in intermediates during the earlier AT1-to-AT2 reversion pseudotime stage. Overall, rather than switching from lung to an alternate endodermal lineage identity, advanced stage AT1 adenoma cells failed to maintain repression of embryonic lung progenitor and gastric epithelial genes. Another notable finding was that significant transcriptional heterogeneity between individual AT1 adenoma cells rapidly emerged after the iAT2 pseudotime stage. This molecular diversity may correlate with a recently described transient plasticity state that emerged upon loss of a stable AT2 cell state in a KrasG12D induction and Trp53 deletion model of LuAd33.
We next correlated molecular with histological features by staining AT1-derived adenomas in which ERK activity had been augmented. The results showed highly proliferative tumor cells that lacked the AT2 cell marker LAMP3 in the nodular component of mixed adenomas (Figure 3g). Necrosis and mucinous transformation of large adenocarcinoma tumors was also evident (Figure 3h). Loss of the AT2 marker LAMP3 also correlated with induction of mucin gene expression, but as with gastric marker expression this was not associated with loss of Nkx2-1 (Figure 3i).
Human lepidic and non-lepidic LuAd share molecular features with AT1- and AT2-derived adenomas, respectively
The idea that human lepidic LuAd may be similarly regulated (and perhaps also originate from AT1 cells) is appealing because they share a unique histology and remarkable indolence with AT1-derived mouse lepidic LuAd. We collected lepidic and non-lepidic human LuAd surgical specimens and characterized MAPK signaling by immunostaining as we did for AT1- and AT2- derived mouse tumors. In patients, lepidic LuAds are only rarely resected before they have developed an additional low grade histological component, which is typically acinar34. We therefore considered lepidic LuAd as the lepidic portion of any resected cancer that was interpreted by a pathologist as having a significant lepidic component. We considered non-lepidic LuAd as a resected cancer that entirely lacked a lepidic component on comprehensive pathologic examination. This analysis revealed that human lepidic LuAd was significantly less proliferative with minimal ERK activity in tumor cells compared with non-lepidic LuAd. By contrast, JNK was equally active in lepidic and non-lepidic cancers (Figure 4a). Notably, our human LuAd specimens included diverse genotypes with mutations in KRAS, EGFR, TP53, MET, MAPK21, NF1, NF2, CDH1, STK11, ARID1A, and KEAP1.
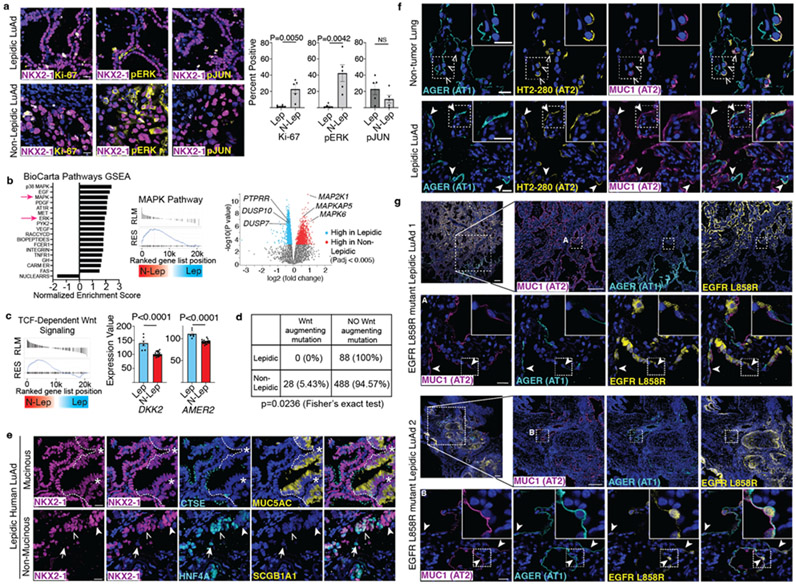
Figure 4: Human lepidic LuAds share histological and molecular features with mouse AT1 derived LuAds.
(a) Representative images of human lepidic (top) and non-lepidic (bottom) LuAd stained for proliferation (Ki-67), pERK and pc-JUN (n=5 individuals per group). (b) GSEA evaluating BioCarta pathways of cDNA microarray data from Zabeck et al. of lepidic and non-lepidic LuAds shows enrichment of MAPK and ERK activity in non-lepidic tumors (n=6 lepidic and 21 non-lepidic LuAds) (left). GSEA plot of the BioCarta MAPK pathway (center). Volcano plot of differentially expressed genes in the lepidic and non-lepidic groups highlighting members of the peptidyl-tyrosine dephosphorylation GO, PTPRR, DUSP10, and DUSP7, as well as constituents of MAPK signaling MAP2K1, MAPKAP5, MAPK6. (c) GSEA evaluating for canonical Wnt signaling revealed enrichment in the non-lepidic group (left) while several Wnt inhibitors, including DKK2 and AMER2, were more highly differentially expressed in the lepidic group (right). (d) A comprehensive mutation analysis by Caso et al. into lepidic and non-lepidic tumors reveals a lack of Wnt augmenting mutations in lepidic tumors with a significant number of Wnt augmenting mutations in non-lepidic human LuAd. (e) Human lepidic mucinous (top) and non-mucinous (bottom) LuAd both contain NKX2-1+ cells co-expressing intestinal markers (CTSE, HNF4A, left side of image at top and open arrowhead on bottom panel). The non-mucinous tumor also contains NKX2-1+HNF4A− (solid arrowhead) and NKX2-1−HNF4A+ (arrow) cells. Mucinous (MUC5AC+) region is NKX2-1Lo (asterisk). (e) Co-staining for AT1 (AGER) and AT2 (MUC1, HT2-280) membrane antibodies shows co-exclusive marking of AT1 and AT2 cells (open arrowheads) in non-tumor human lung (top) but co-expression of AT1 and AT2 markers by squamous cells in human lepidic LuAd (bottom, closed arrowheads). (f) Co-staining of AT1 (AGER), AT2 (MUC1) and driver mutation (EGFR L858R) antibodies in human lepidic LuAd showing ubiquitous oncogene and AT2 marker staining of the tumor but also flat cells in the periphery positive for the driver mutation that co-express both AT1 and AT2 markers (arrowheads) (n=2 individuals).
Scale bars, 20 μm (a, e, f, g rows 2 and 4) and 200 μm (g rows 1 and 3)
GO, gene ontology; GSEA, Gene Set Enrichment Analysis; Lep, lepidic; N-Lep, non-lepidic; NS, not significant; pERK, phosphorylated ERK; pJUN, phosphorylated c-JUN/JUND
Data in bar graphs of a and c are mean±s.e.m. P values calculated by Student’s t test except where indicated otherwise.
We next searched The Cancer Genome Atlas (TCGA) and published literature for human LuAd gene expression datasets with profiles of only lepidic tumor cells. We identified a single report that had employed laser capture microdissection with accompanying histological annotation35. We identified and segregated LuAds from this dataset into two groups: those that were > 80% lepidic (lepidic) or no evidence of lepidic histology (non-lepidic). We evaluated the transcriptional profile of these two groups with Gene Set Enrichment Analysis (GSEA) and conducted unbiased pathway enrichment analysis of differentially expressed genes between the two groups with g:Profiler. GSEA with BioCarta pathway gene sets demonstrated that MAPK and ERK signaling are enriched in non-lepidic tumors. Indeed, several of the most highly differentially expressed genes in non-lepidic human LuAd cells were components of the MAPK pathway, namely MAPK6, MAPKAP5, and MAP2K1. This biology closely matches that of the mouse, in which non-lepidic AT2-derived tumors have much higher levels of MAPK signaling. Interestingly, unbiased pathway enrichment analysis of differentially expressed genes in the lepidic tumor group revealed that peptidyl-tyrosine dephosphorylation was enriched in lepidic tumors, suggesting a potential role of phosphatase activity in suppressing receptor tyrosine signaling (Figure 4b).
We reasoned that if human lepidic and non-lepidic tumors respectively originate from AT1 and AT2 cells, they might replicate the biology of mouse tumors, namely that Wnt signaling inhibits the former and promotes the latter (see Figure 2e). We thus performed GSEA of human LuAd which revealed TCF-dependent Wnt signaling was highly enriched in non-lepidic tumors. We then examined the list of most highly differentially expressed genes between these groups for Wnt pathway genes. This analysis revealed two inhibitors of Wnt signaling, DKK2 and AMER2, whose expression was enriched in lepidic human tumors (Figure 4c). We also identified a human lung cancer dataset that included detailed histological annotation of tumors with comprehensive mutational profiling36. We regrouped these samples into those with lepidic and non-lepidic histology and extracted all the Wnt pathway mutations in each group. The results showed that none of the 88 lepidic tumors but 28 out of 516 non-lepidic tumors harbored OncoKB-annotated driver mutations in Wnt pathway genes. Remarkably, each of the 28 mutations was annotated by OncoKB as augmenting Wnt signaling (Figure 4d, Supplemental Data). Hence, the human tumor analysis strongly corroborated multiple aspects of the mouse biology including elevated MAPK activity and Wnt signaling, and enrichment for Wnt augmenting mutations in non-lepidic tumors.
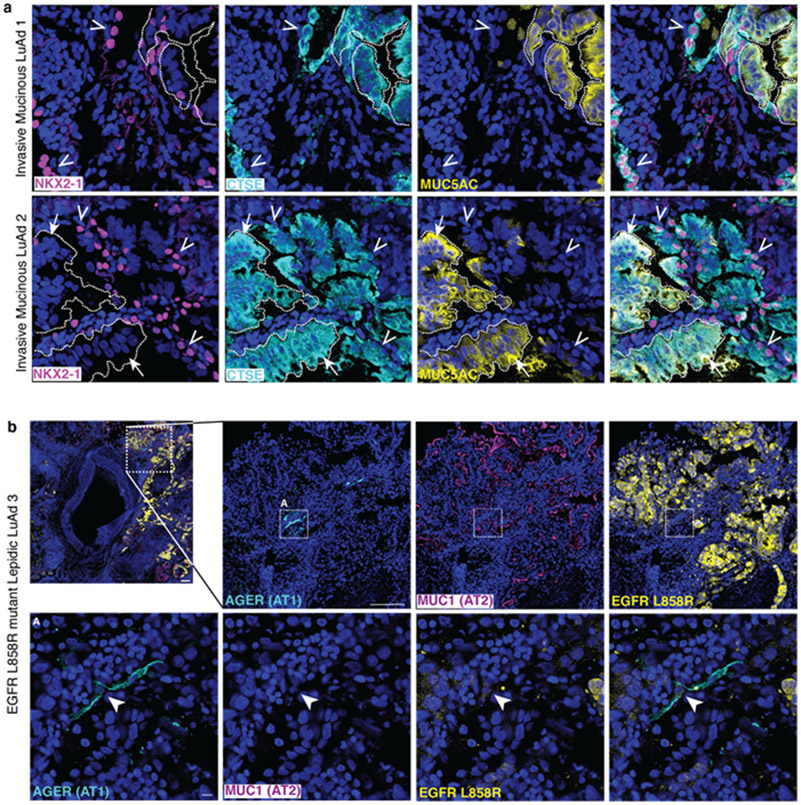
Our molecular analysis of mouse LuAd identified aberrant co-expression of lung and gastric markers by individual AT1-derived tumor cells. We therefore asked if tumor cells with combined lung and gastric gene expression were also present in human lepidic LuAd. We did so by co-staining for NKX2-1, MUC5AC and gastric epithelial markers Cathepsin E (CTSE) and HNF4A in human lepidic tumors, both with and without mucinous transformation. This analysis revealed that the mucinous portion of human lepidic tumors expressed lower levels of NKX2-1, matching the observation in mouse that Nkx2-1 haploinsufficiency results in mucinous transformation (Figure 4e)37. Invasive mucinous human LuAd foci by contrast were marked by loss of NKX2-1 and co-expression of MUC5AC and the gastric marker CTSE, mirroring the gastric transdifferentiation found in mouse to result from loss of Nkx2-125,27 (Figure ED 4a). However, we also repeatedly observed in human LuAd the aberrant population we discovered in mouse, namely tumor cells expressing gastric markers without having lost lung identity. These “lung/gastric hybrid” cells were present in mucinous and non-mucinous human lepidic LuAd (Figure 4e) and in invasive mucinous non-lepidic LuAd (Figure ED 4a) and therefore appear to be a previously unrecognized constituent of human LuAds.
Human lepidic LuAd tumors harbor squamous cells co-expressing AT1 and AT2 cell markers that also carry the driver oncogene
Lastly, we focused on examining the possibility of an AT1 cell of origin for human lepidic LuAd within the confines of what is experimentally tractable by retrospective analysis. We began by analyzing the expression of AT1 and AT2 cell type markers in tumor-remote (histologically normal) human lung and at the tumor margins of lepidic and non-lepidic LuAds. We used only antibodies against membrane proteins to illuminate cell morphologies and mitigate the risk of ‘false negatives’ from trying to visualize the miniscule cytoplasmic compartment of flat AT1 cells. We confirmed appropriate co-exclusive staining of flat AT1 and cuboidal AT2 cells in tumor-remote alveolar regions (Figure 4d, top row). By contrast, we found squamous cells co-expressing AT1 and AT2 markers at the periphery of human lepidic LuAd tumors (Figure 4d, bottom row). Intriguingly, these cells could represent residual tumor progenitors analogous to the AT1-derived morphological and molecular ‘intermediates’ that co-express AT1 and AT2 markers with KrasG12D induction in mouse (see Figure 1f-h). However, they could also be non-tumor cells whose aberrant gene expression profile results from the altered microenvironment of the tumor.
To determine whether these ‘intermediate’ cells were within the tumor lineage or merely bystanders, we took advantage of a commercially available antibody against EGFR L858R, a relatively common LuAd driver oncogene. EGFR L858R carries a point mutation in the sequence encoding the activation loop that results in markedly increased kinase activity38. We identified three lepidic LuAd patient samples with this driver mutation which we stained for EGFR L858R along with AT1 and AT2 cell membrane markers. EGFR L858R strongly stained the cuboidal tumor epithelial cells that expressed AT2 but not AT1 cell type markers. However, at the margins of all three specimens we detected rare but distinct squamous cells carrying the driver oncogene that also co-expressed the AT1 and AT2 markers (Figure 4e, Extended Data 4b). These results represent only a cytologic and molecular ‘snapshot’ and do not provide any information about whether these ‘intermediates’ evolved into versus derived from the tumor. Nonetheless, they do confirm a lineage relationship between phenotypically distinct cells in human lepidic LuAd that matches the prediction of an AT1 cell of origin model.
Discussion
Mouse models employing cell type specific oncogene induction have been invaluable for uncovering the strong propensity of stem or progenitor cells in multiple tissues to generate cancers39. Here, we identify a surprising capacity of KrasG12D, a prototypical pro-proliferative oncogene, to drive adenomatous transformation of terminally differentiated AT1 cells. The mechanism by which oncogenic Kras accomplishes this feat is by first directly reprogramming AT1 cells into AT2 stem cells, which are canonical LuAd progenitors. Our work raises the question of whether acquisition of a stem cell state may be a generalized mechanism by which non-stem cells can generate cancer. In any case, cell type specific vulnerabilities are increasingly being implicated in the development of LuAd40. Regarding KrasG12D, it has been shown to induce expression of the AT2 cell marker, Sftpc, in airway Club cells, a process accelerated by MAPK signaling24,41. This positive association of MAPK with AT2 cell state recapitulates lung development, where FGF signaling drives AT2 cell differentiation of a bipotent embryonic progenitor3,42,43. For AT1 cells, which unlike Club cells share a direct lineage relationship with AT2 cells, the AT2 reprogramming by KrasG12D is remarkably comprehensive (see Figure 1e). In another MAPK signaling context it was shown that co-induction of BRAFV600E and Phosphoinositide 3-Kinase (PI3K) signaling in AT2 cells rapidly drove dedifferentiation44.
While AT1-derived LuAd cells molecularly resemble their AT2-derived counterparts, they nonetheless behave much differently. AT1 tumors are indolent, spread in a lepidic manner, and are paradoxically inhibited by Wnt activation. Their slow growth and the opposite response to Wnt of KrasG12D induced AT1 versus AT2 cells is presumably due to differential molecular circuitry (‘hardwiring’) and epigenetic landscape, respectively. The idea that AT2-like cells generated from AT1 cells produce less aggressive cancers than primary AT2 cells is somewhat intuitive since terminally differentiated cells presumably have a smaller repertoire of gene programs available due to silencing by methylation during terminal differentiation.
Are AT1 cells the source of human lepidic LuAd? While this cannot be definitively answered, several lines of evidence make this idea appealing. The first is the close biological similarities we uncover between AT1-derived mouse and human lepidic LuAds. Both advance indolently in a non-destructive manner and are ERK low compared with AT2-derived and non-lepidic LuAds. Wnt inhibits progression of lepidic LuAd in mouse, and human lepidic LuAds are strongly enriched for expression of Wnt inhibitors, which could result from positive selection for a Wnt-low state. AT2-derived mouse LuAds, by contrast, are proliferatively responsive to Wnt, which is a feature of normal AT2 cells from which they likely arise16. We also identified transcriptomic and mutational signatures in human lepidic and non-lepidic LuAds that recapitulate the biology and predictions of our mouse model. Another intriguing observation is that the periphery of human lepidic LuAds harbor cells carrying the driver oncogene that resemble mouse Kras-induced AT1 cells in the process of AT2 reprogramming. Both populations are squamous and co-express AT1 and AT2 markers. It is also worth noting that although they are less prevalent than AT2 cells, flat AT1 cells cover > 95% of the alveolar surface area and are thus continually exposed to inhaled toxins45. In this regard, it is interesting that a subset of AT1 cells appeared to be entirely unresponsive to expression of oncogenic KrasG12D over ten months. It may be that initiation of AT2 cell reprogramming and subsequent adenoma formation by AT1 cells requires a specialized intrinsic capacity and/or a second signal. Such a “two-step” requirement would serve to protect exposed and long-lived AT1 cells against adenomatous transformation.
Perhaps our most provocative result is that despite achieving a transcriptional state closely resembling AT2 cells, Kras-induced AT1 cells demonstrate an opposite and inhibitory response to Wnt activation from Kras-induced AT2 cells. This observation reveals that mouse lepidic LuAd tumor cells remain constrained in their emergent malignant potential. Could human LuAds initiating as lepidic tumors harbor a similar perduring constraint that transcriptionally comparable tumor cells in non-lepidic cancers do not have? Current society guidelines recommend to only consider the invasive portion of a lung adenoma when measuring the tumor size for staging, leaving out the lepidic portion46,47. However, multiple groups including ours have reported that the presence of even a small ground-glass tumor component visualized on CT scan, which corresponds to lepidic histology, predicts markedly improved survival compared to a pure non-lepidic tumor at the same stage48-51. While there is consideration of incorporating the predominant and high grade histological patterns into LuAd staging52, integrating the ground glass or lepidic features of LuAds remains controversial53. If the human biology matches the mouse, there may be a molecular explanation for why non-lepidic LuAds with a small lepidic component might portend a more favorable outcome for some patients, namely, that it betrays an AT1 cell of origin.
Materials and Methods
Transgenic mice
Expression of estrogen-inducible Cre recombinase (Cre-ERT2) for conditional, cell-type specific targeting in vivo in mixed strain backgrounds used the following gene-targeted Cre alleles: Hopx-CreER1, expressed by AT1 cells, and Sftpc-CreER2 or Sftpc-CreER-rtTA3, expressed by AT2 cells. Conditional KrasG12D induction in AT1 and AT2 cells was achieved through Cre-dependent removal of a stop codon preceding a G12D mutant Kras allele in the endogenous locus4. Conditional activation of Wnt signaling was achieved through Cre-dependent deletion of Exon3 of Ctnnb1 using a “floxed” allele1. Cells were fluorescently tagged using the R26mTmG Cre-dependent reporter allele, in which recombination switches expression from membrane-targeted Tomato to mGFP5. Induction of Cre-ERT2 alleles was accomplished by intraperitoneal injection of 1-4 mg tamoxifen (Sigma) dissolved in corn oil by sonication and stored in aliquots at −80C. Genotyping was performed by PCR of DNA extracted from ear or toe clips using published primer sets. All experiments were performed using adult animals, between two and 14 months of age. Mice were housed in filtered cages and all experiments were approved by the Institutional Animal Care and Use Committee at Stanford University.
Human tissue
Human tissue was obtained from excess tissue from lung resections carried out at Stanford Hospital for suspected or confirmed lung adenocarcinoma. Informed consent was obtained from adult participants under a protocol approved by the Stanford Institutional Review Board.
Lung isolation, fixation, staining, and imaging
Mice were euthanized by carbon dioxide inhalation, the abdominal aorta severed, and sternotomy performed. Phosphate buffered saline (PBS; Ca2+- and Mg2+-free, pH 7.4) was perfused at a flow rate of 7.5 ml/min into the right ventricle using a 25-gauge needle attached to a digital pump (Ismatec) to clear the pulmonary vasculature. Following cannulation of the trachea, the lungs were gently inflated with 2% ultra-low gelling temperature agarose (Sigma) until visualization of complete distention of the accessory lobe, then removed en bloc and submerged in ice-cold PBS. Individual lobes were dissected then processed for fixation. For whole mount imaging, the left and right upper lobes were separated and trimmed to create a flat base, glued to a tissue disk with cyanoacrylate, then submerged in ice-cold PBS and sliced at a thickness of 600μm using a vibrating microtome (Leica). For direct imaging without immunostaining, slices were fixed in paraformaldehyde (PFA, 4% in PBS) overnight at 4C then stored in 80% glycerol at 4C. For a subset of specimens, the right middle lobe was fixed in paraformaldehyde (PFA, 4% in PBS) overnight at 4C then stored in Scale clearing agents. For whole mount immunofluorescence staining, slices were fixed in methanol:dimethylsulphoxide (DMSO) (4:1) with rocking overnight at 4C, then dehydrated in methanol and stored at −20C until staining. For immunohistochemistry and RNA in situ hybridization, the right middle or accessory lobes were immediately fixed in 4% PFA in PBS with 5 mM EDTA, 10 mM Citrate, and PhosSTOP (Roche) overnight at 4C, transferred to 30% sucrose in PBS at 4C overnight, then submerged in optimal cutting temperature compound (OCT, Fisher) in an embedding mold, frozen on dry ice, and stored at −80C. 14μm sections were then obtained using a cryostat (Leica CM3050) and collected on glass slides. For H&E, the right lower lobe was fixed overnight in 4% PFA in PBS overnight then transferred to 70% ethanol prior to embedding in paraffin.
For transmission electron microscopy, the agarose-inflated right middle lobe and left lobe were sectioned on a vibratome, then sections examined on a widefield microscope for areas of mGFP+ cells were identified. These sections of interest were then fixed for one hour with Karnovsky’s fixative: 2% Glutaraldehyde (EMS Cat# 16000) and 4% Formaldehyde (EMS Cat# 15700) in 0.1M Sodium Cacodylate (EMS Cat# 12300) at pH 7.4. The sections were then placed in cold/aqueous 1% Osmium tetroxide (EMS Cat# 19100) and allowed to warm to room temperature (RT) over two hours rotating in a hood. They were then washed three times with ultrafiltered water, then stained en bloc in 1% Uranyl Acetate at RT for two hours while rotating. Samples were dehydrated in 50% EtOH at room temperature, then 70% EtOH at room temperature, and then moved to 4°C overnight. They were next submerged in ice cold 95% EtOH and allowed to warm to RT, changed to 100% two times, then placed in Propylene Oxide (PO) for 15 min. Samples were infiltrated with EMbed-812 resin (EMS Cat#14120) mixed 1:2, 1:1, and 2:1 with PO for two hours each while leaving samples in 2:1 resin:PO overnight rotating at RT in the hood. The samples were then placed into EMbed-812 for two to four hours then transferred into molds with fresh resin, oriented and placed into 65°C oven overnight. Sections were made to around 80nm using a UC7 microtome (Leica, Wetzlar, Germany), picked up on formvar/Carbon coated 100 mesh Cu grids, stained for 40 seconds in 3.5% Uranyl Acetate in 50% Acetone followed by staining in Sato’s Lead Citrate for two minutes. Tissue was examined using the JEOL JEM-1400 120kV and images obtained using a Gatan Orius 832 4k X 2.6k digital camera with 9um pixel.
Human tissue specimens were fixed in 10% neutral buffered formalin with 5 mM EDTA, 10 mM Citrate, and PhosSTOP (Roche) overnight at 4C. They were placed in 30% sucrose in PBS overnight then submerged in OCT in an embedding mold, frozen on dry ice, and stored at −80C. 8μm (for IHC) and 5μm (for H&E) sections were obtained using a cryostat (Leica CM3050) and collected on glass slides.
In vivo pharmacological agonism and inhibition of ERK signaling
Hopx-CreER > KrasLSL-G12D, R26mTmg mice between the ages of 2.5 and 9 months were administered a single intraperitoneal injection of tamoxifen at 0.1 mg/gm. Eight days after injection, mice were initiated on a diet of either control chow (n=3, AIN-76A) or chow compounded with PLX4720 (n=6, 417mg/kg, Plexxikon) or PD0359501 (n=5, 7mg/kg, ApexBio). Chow was compounded by Research Diets in New Brunswick, NJ. The mice were sacrificed and processed as above 90 days after tamoxifen administration.
Immunohistochemistry
Immunofluorescence was performed as previously described6. Secondary antibodies conjugated to an Alexa Fluor dye (A488, A555, or A633; Invitrogen) or streptavidin conjugated to Atto490LS were used. DAPI (5 ng/ml, Sigma) was included during incubation with secondary antibodies. For detection of phosphorylated c-JUN/JUND and ERK, a horseradish peroxidase conjugated secondary antibody was used (Invitrogen) against a phospho-specific antibody and detected by tyramide signal amplification (TSA, Biotium). Lycopersicon esculentum lectin (1:500, Vector DL11781) was pre-conjugated to DyLight-649. Primary antibodies were against the following antigens: AGER (rat, 1:500, R&D MAB1179), AGER (goat, 1:250, R&D AF1145), p-cJUN (rabbit, 1:100, Cell Signaling 9164), CTNNB1-FITC (mouse, 1:250, BD Transduction Laboratories 14), CTSE (goat, 1:250, R&D AF1130), EGFR L858R (rabbit, 1:100, Cell Signaling 43B2), p-ERK1/2 (rabbit, 1:100, Cell Signaling 9101), FITC-Alexa488 (goat, 1:250, Invitrogen A11096), GFP (chicken, 1:500, Abcam 13970), GKN2 (rabbit, 1:250, Abcam EPR15377B), HT2-280 (mouse, 1:500, Terrace Biotech), Ki67 (rabbit, 1:100, Abcam 16667), LAMP3 (guinea pig, 1:500, Synaptic Systems #391005), MUC1 (rabbit, 1:500, Novus NBP160046), NKX2-1 (rabbit, 1:100, Abcam EPR5955), NKX2-1 (mouse, 1:100, Leica Novocastra SPT24), PDPN (hamster, 2 μg/ml, DSHB 8.1.1), PDPN-biotin (hamster, 1:250, eBioScience 8.1.1), pro-SPB-biotin (mouse, 1:500, Labvision SPB01), pro-SPC (rabbit, 1:500, Millipore AB3786).
In situ RNA detection
Multiplex in situ RNA hybridization was performed by Proximity Ligation In Situ Hybridization (PLISH) as previously described7. The following sets of primer pairs were used to detect transcripts of the indicated genes:
Abca3
5’AGGTCAGGAATATTCATATTGGTACGG tcgttcaggcttagcttcag3’
5’gcacaaccgttctaccatac TCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGG accagcgtcatgtggtagcc3’
5’gattgcagtgcggttctttt TCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtgggggggcgcgtcggagcg3’
5’gctccggctcctggagggggTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtcagcctggtataagctgtg3’
5’cgacaggatgttgaatatggTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtaaacagtggcattgggcac3’
5’cctggatggactggtccgggTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtcatacaccttggagagctc3’
5’caaggagcggtgctcgctggTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtgaagtcgcctggcgtcagc3’
5’tgtcactgcaaggctacgggTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGttgatcttgattccagctac3’
5’ggaacaccttggagagatgcTCCAGCATGGAAGATTATAGCCAGGTT3’
5’AGGTCAGGAATATTCATATTGGTACGGtcccgccactggatgccagt3’
5’cgttgacagggttgaggaggTCCAGCATGGAAGATTATAGCCAGGTT3’
Ager
5’AGGTCAGGAATACTTAGCTATTGATGGtcatcgacaattccagtggc3’
5’cccgacaccggaaagtccccTTCTGTGTAGACGACTATAGCCAGGTT3’
5’AGGTCAGGAATACTTAGCTATTGATGGtttgccatcgggaatcagaa3’
5’tcttccttcacgagtgtttcTTCTGTGTAGACGACTATAGCCAGGTT3’
Egfp
5’AGGTCAGGAATATGACTTTTCCGCGGGtgctggtagtggtcggcgag3’
5’cgtcgccgatgggggtgttcTCTTTTTGTAACAATTATAGCCAGGTT3’
5’AGGTCAGGAATATGACTTTTCCGCGGGatagacgttgtggctgttgt3’
5’ttctgcttgtcggccatgatTCTTTTTGTAACAATTATAGCCAGGTT3’
5’AGGTCAGGAATATGACTTTTCCGCGGGttgtacagctcgtccatgcc3’
5’tagagtcgcggccgctttacTCTTTTTGTAACAATTATAGCCAGGTT3’
5’AGGTCAGGAATATGACTTTTCCGCGGGacgtcgccgtccagctcgac3’
5’cgctgaacttgtggccgtttTCTTTTTGTAACAATTATAGCCAGGTT3’
5’AGGTCAGGAATATGACTTTTCCGCGGGaggtcagggtggtcacgagg3’
5’gctgaagcactgcacgccgtTCTTTTTGTAACAATTATAGCCAGGTT3’
Hopx
5’AGGTCAGGAATAAGCGACAAGTTTGGG actaggttgctttccaaggt3’
5’ttggaatccatagctgcta TAGGAGCGACGACGATATAGCCAGGTT3’
5’AGGTCAGGAATAAGCGACAAGTTTGGGatgaattactactggaaggt3’
5’ctctgaaggggatggtccatTAGGAGCGACGACGATATAGCCAGGTT3’
5’AGGTCAGGAATAAGCGACAAGTTTGGGtgaccgccgccactctgcca3’
5’cattccgaaggcaagccttcTAGGAGCGACGACGATATAGCCAGGTT3’
5’AGGTCAGGAATAAGCGACAAGTTTGGGtgtctgcttaaatagcctgc3’
5’aagcagcggatgctggtgccTAGGAGCGACGACGATATAGCCAGGTT3’
5’AGGTCAGGAATAAGCGACAAGTTTGGGtgggagtgcacgctcaagca3’
5’tcgcacgttttatttacgccTAGGAGCGACGACGATATAGCCAGGTT3’
Pdpn
5’AGGTCAGGAATAGGACAACAAGGTCGGatgttaaaaatttagttttc3’
5’aatcatcgatcataacatttTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtaccagaggtgccttgccag3’
5’ccacgctctctctgcgttggTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtgaacccatggttacagttg3’
5’gtaatagggactggccagggTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtcggtctgacttcctatact3’
5’aggcaggtgttagaaactggTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtaccaacacagacgaccaag3’
5’gatgtctacactgtattgggTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtcgggaggaggttggaaggc3’
5’caggcccagccgggaagagcTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtcgttggagcctcagcagct3’
5’gcacggtccacatcttgatcTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGatgattccaaccagggtgac3’
5’tggctaacaagacgccaactTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGattttcagatgtgttgggag3’
5’ggcaagttggaagctctcttTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGtgagggcttgagtcctaggg3’
5’ccagaaagatgcagctgatcTCAGCGCTAATCACATATAGCCAGGTT3’
Sftpc
5’AGGTCAGGAATAGGACAACAAGGTCGG tggcccgtaggagagacacc3’
5’cttttgtgataggatccccc TCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGaatccggtggactctccatc3’
5’gctcctgggacctgccgagtTCAGCGCTAATCACATATAGCCAGGTT3’
5’AGGTCAGGAATAGGACAACAAGGTCGGatgtttttgactcatgtgga3’
5’atctcaaggaccatctcagtTCAGCGCTAATCACATATAGCCAGGTT3’
After staining, vibratome slices were transferred onto Chambered Coverglass (#1.5, Lab-Tek) and submerged in Vectashield (Vector). Cryotome sections were sealed by #1.5 Coverglass (Fisher) with nail hardener (Sally Hansen). Images were acquired using a Leica Sp8 inverted laser scanning confocal microscope with LAS X Software. Tiled images in Figure 2A were acquired using the THUNDER imaging system (Leica) on an inverted microscope. Images were processed (pseudocolored, contrast adjusted, overlaid) using ImageJ (NIH) and Imaris 9.3-9.9 (Oxford). Quantification of H&E tumor characteristics as well as immunohistochemistry was performed in QuPath (University of Edinburgh).8
Fluorescence Activated Cell Sorting (FACS)
Digestion buffer was prepared with elastase 4 U/mL (Alfa Aesar aqueous porcine elastase), Collagenase I 200 U/mL, Dispase 5 U/mL (Corning), and DNAse 1 4 U/mL (Worthington) in HBSS with Calcium and Magnesium. Mice were sacrificed and dissected as above. After the trachea was cannulated, approximately 2.5 mL of digestion buffer was used to inflate the lungs, after which the trachea was tied off and the lungs dissected out en bloc and placed in a petri dish at RT for 40 minutes. The trachea and large airways were then excised, and the lobes separated followed by mincing with a razor. The cells were passed through 100 μm and 40 μm filters pre-wetted with FACS buffer (2% fetal bovine serum in PBS). The cells were treated with ACK lysis buffer for three minutes prior to being stained with the following fluorochrome-conjugated antibodies: CD45 (BD, clone 30-F11), TER119 (BD, clone 17), CD31 (BD, clone 13.3), EPCAM (BD, clone G8.8), PDPN (Biolegend, clone 8.1.1), AGER (Novus, MAB1179). Sytox (Invitrogen) was used for staining dead cells. Live, lineage-tagged epithelial cells were sorted into 96 well plates holding lysis buffer using a BD FACS Aria II instrument.
Single Cell RNA Sequencing
Single cells were sorted into 2uL of lysis buffer containing RNAse inhibitor (1 U/μL, Clontech, cat. #2313B), 0.1% Triton X-100 (Servo, cat # 39795.01), 2.5 mM dNTP (Thermo Fisher Scientific, cat. #10297018), 2.5 μM oligo dT30VN (Integrated DNA Technologies, 5′-AAGCAGTGGTATCAACGCAGAGTACT30VN-3’), and 1:600,000 ERCC (external RNA controls consortium) RNA spike-in mix at 1:600,000 (Thermo Fisher Scientific, cat. #4456739) in UltraPure water (Thermo Fisher Scientific, cat. #10977015)1. Immediately after storing, plates were centrifuged at 4200 RPM for 30 seconds, snap frozen on dry ice and then stored at −80°C. Reverse transcription (RT) and cDNA pre-amplification was performed using the SmartSeq2 protocol. Briefly, lysis plates were thawed on ice and incubated at 72°C for 3 minutes then immediately snap chilled on ice to anneal the oligo dT30VN primer. 3 μL of RT mix (SMARTScribe™ Reverse Transcriptase (Takara # 639538), 0.5 U/μL recombinant RNase inhibitor (Takara, cat. #2313B), First Strand Buffer, 8 mM dithiothreitol (DTT) (Promega, cat. #P1171), 1M Betaine (), 2.5 mM MgCl2 (Thermo Fisher Scientific, cat. #AM9530G), and 2 μM TSO (Integrated DNA Technologies, 5′-AAGCAGTGGTATCAACGCAGAGTGAATrGrGrG-3′) in UltraPure water) was added to each well. The plates were incubated in a C1000 Touch Thermal Cycler (BioRad) at 42°C for 90 min, and then 70°C for 15 min to terminate the reaction. 7.5 uL of PCR mix(KAPA HiFi HotStart ReadyMix (Kapa Biosystems, cat. #KK2602) and 0.1 μM IS PCR primer (Integrated DNA Technologies, 5′-AAGCAGTGGTATCAACGCAGAGT-3′) in UltraPure water) was added to each well and cycled using the following program: (1) 98°C for 3 min, (2) denaturing at 98°C for 20 sec, (3) annealing at 67°C for 15 sec, (4) elongation at 72°C for 6 minutes, (5) repeat from step 2 28 times, and (6) 72°C for 5 minutes. Preamplified cDNA was then purified using 0.8X volume of calibrated AMPure XP beads (Beckman Coulter, cat. #A63882) to remove residual reaction components and oligos smaller than 400 base pairs, and eluted in 12.5 μL UltraPure water. From the purified cDNA, 1 μL was taken for quality control for each well. cDNA concentration and size distribution for each well was determined on a capillary electrophoresis-based Fragment Analyzer (Advanced Analytical). Wells with a concentration less than 1.7 ng/μL were excluded; this cutoff was determined by measuring the concentration blank wells with ERCC but no sorted cell. The wells within the 96 well plates with cDNA concentration above the cutoff value were then consolidated and reformatted to a new 384 well plate using the Mosquito X1 liquid handler (SPT Labtech), such that in the destination 384 well plate each well’s concentration was also normalized to a desirable concentration range of 1.7-4.0 ng/μL by diluting with UltraPure water. Normalized cDNA was used to prepare Illumina sequencing libraries. Tagmentation was performed by combining 0.4 μL cDNA with 1.2 μL Illumina Nextera XT (cat. #FC-131-1096) ATM+TD buffers. After incubation at 55 °C for 10 minutes, 0.4 μL NT buffer was added to each well to stop the reaction. Indexing PCR reactions were performed by adding 0.4 μL of 5 μM i5 indexing primer, 0.4 μL of 5 μM i7 indexing primer (Integrated DNA Technologies, custom made 7680-plex unique dual index-primer set), and 1.2 μL NPM buffer. PCR amplification was performed on a C1000 Touch Thermal Cycler (BioRad) using the following program: (1) 72°C for 3 min, (2) 95°C for 30 sec, (3) denaturing at 98°C for 10 sec, (4) annealing at 67°C for 30 sec, (5) elongation at 72°C for 60 sec, (6) repeat from step 3 10 times. For each 384 well plate, 1 μL was taken from each well for pooling, followed by purification using 0.8X volume of AMPure XP beads (Beckman Coulter, cat. #A63882). The 384-cell library pool from each plate was analyzed for concentration and size distribution using Fragment Analyzer (Advanced Analytical). Six 384-cell library pools were then normalized for concentration, further pooled to get a 2100-plex library pool, which was purified once more and concentrated using 0.8X AMPure beads. The final 2100-plex library pool was then sequenced on a NovaSeq 6000 S4 flow cell (Illumina) to obtain ~1-2 million 2×150 base-pair paired-end reads per cell.
Sequences were demultiplexed using bcl2fastq version 2.19.0.316. 3’ adapter sequences were removed from reads using skewer v0.2.2,3 and aligned to the M27 genome (Gencode version GRCm39) with STAR aligner version 2.6.1d using 2-pass mapping. Briefly, as a first pass, reads for every cell were aligned using STAR genome index generated using the Gencode transcript annotation for the murine genome. Mapped splice junctions for each cell from the first-pass mapping were extracted, aggregated together and a new STAR index was created where any newly discovered splice junctions were included in addition to the existing Gencode annotation during genome index generation. The new STAR index with all known and newly identified splice-junctions was then used for second pass read mapping. Parameters used for STAR mapping were adapted from the ENCODE long-mRNA-pipeline (https://github.com/ENCODE-DCC/long-read-rna-pipeline) recommendations, also detailed in the STAR manual. In addition to the ENCODE recommended options we also used the “--quantMode TranscriptomeSAM” option during second-pass mapping to generate a bam file containing a catalog of all reads mapped to the transcriptome. This bam file was used as input to calculate expression levels of either genes (sum total of expression levels of all known transcript variants) or individual transcripts using RSEM version 1.3.3 with settings “--single-cell-prior.” Gene count tables were combined with metadata using the Scanpy python package v.1.8.2.6 We filtered out genes expressed in fewer than 3 cells, as well as cells with fewer than 250 detected genes. Cells with greater than 30% mitochondrial gene reads or greater than 60% ERCC reads or greater than 10% ribosomal gene reads were removed from the dataset. The data were normalized using size factor normalization so that every cell has 10,000 read counts, log transformed, and scaled to a maximum value of 10. Highly variable genes were computed using default parameters. We then performed principal component analysis, computed the neighborhood graph, and clustered the data using the Leiden method. PAGA was used to visualize data, as well as reconstruct gene expression changes along pseudotemporal trajectories. Step-by-step instructions to reproduce preprocessing and analysis of data are available on GitHub.
For the feature plots in Figure 3f, score was created using the “score_genes” function in Scanpy based upon markers stated in the indicated papers (see Supplemental Data).
Human LuAd Microarray Re-analysis
Microarray gene expression data from micro-dissected lung cancer tissues with different growth patterns (GSE57772) were reclassified according to the percentage of lepidic histology.9 Tumors with 80% or greater lepidic histology were considered “Lepidic” (n=6), and tumors with no lepidic component were considered “Non-lepidic” (n=21). A list of top differentially expressed genes, UMAP, and volcano plot were retrieved with GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Benjamini & Hochberg method was used for P-value adjustment for multiple testing and the significance level cut-off for the volcano plot was set at 0.005. Unbiased pathway enrichment analysis was conducted using g:Profiler by entering the top 200 differentially expressed genes in each group with a term size upper limit of 500.10 For Gene Set Enrichment Analysis (GSEA) of the same data, a ranked gene list was generated, with non-lepidic having a higher rank. GSEA was performed by using the R package clusterProfiler (version 4.4.4).11 Gene sets from BioCarta ranging in size from 25 to 500 were used to probe for pathway activity. The P-value cutoff was 0.05, and Benjamin-Hochberg correction was done to correct for multiple comparisons. Mutation data from 604 histologically annotated tumors were accessed by cBioPortal and selecting ‘Lung Adenocarcinoma (MSK, J Thorac Oncol 2020)’ (https://www.cbioportal.org/).12 The somatic mutations with OncoKB driver annotation and predominant histologic subtypes of samples were downloaded and grouped into lepidic (n=88) and non-lepidic (n=516) tumors. Number of patients with Wnt augmenting mutations in these two groups were used to form a 2 x 2 contingency table. Significance was determined with Fisher’s exact test.
Kras Mutation Analysis
Integrative Genomics Viewer v. 2.15.4 was used to manually interrogate Kras transcripts in the SmartSeq2 single cell RNA sequencing data.13 The .bam files from individual cells were loaded into the software and Exon 2 was examined for the presence or absence of transcripts. A cell was considered positive for the mutation if three transcripts included the G12D-instigating mutation or if a single transcript included the G12D-instigating mutation in addition to the two other synonymous base pair differences between the knocked-in human Kras gene and the murine wildtype Exon 2.
Extended Data
Extended Data Figure 1: Histological features of AT1- and AT2-derived lung adenomas.
(a) H&E stains of Hopx-ER>KRASG12D and Sftpc-ER-rtTA>KRASG12D mouse lungs at 5m14d and 1m22d after Kras induction, respectively, showing examples of the lepidic-predominant histology of AT1 derived adenomas (top) and the non-lepidic histology of AT2 derived adenomas (bottom). Note massive macrophage infiltration of the alveolar lumens in the lepidic AT1 tumors that extend multiple alveolar spaces beyond the solid AT2 tumors. (b) H&E stains showing examples of human lepidic and non-lepidic KRAS-driven LuAds. (c) Exemplary H&E stains of histological classifications of AT1- and AT2-derived tumors as quantified in the body of the manuscript.
Scale bars, 200 μm (a, c) and 400 μm (b)
Extended Data Figure 2: Comparison of AT2 and iAT2 cells and their resulting tumors.
(a) Co-staining of early-stage AT1 adenomas shows absence of AT1 (LEL, AGER) and presence of AT2 (SFTPB, SFTPC) markers as well as cuboidal morphologies. (b) Feature plots localizing Axin2+ AT2 and iAT2 in single cell RNA sequencing data demonstrating co-clustering (above) with a heatmap of differentially expressed genes between the two groups (below). (c) Heatmap of the most differentially expressed genes between AT2>Kras cells 18d after Kras induction and early-stage AT1>Kras cells. Note on PAGA that the early-stage AT1 and AT2 tumor populations are distinct from normal AT2 and iAT2 (AT1>Kras) cells and that their trajectories diverge from each other.
Scale bars, 10 μm (a)
LEL, Lycopersicon esculentum lectin
Extended Data Figure 3: Phospho-JUN expression in AT1 phenotype cells in Hopx-CreER>KrasG12D and Hopx-CreER>KrasWT mice.
Immunostaining of AT1 phenotype cells (NKX2-1+/LAMP3−) in Hopx-CreER>KrasG12D and Hopx-CreER>KrasWT mice for phosphorylated JUN with quantification of pJUN+ (arrowhead) and pJUN− (open arrowhead) cells (n=2 WT mice and 4 KrasG12D mice, 3 25x fields per mouse).
Scale bars, 10 μm
Extended Data Figure 4: Heterogeneous molecular states in invasive mucinous LuAd and driver mutation-carrying squamous cell in human lepidic LuAd.
(a) Co-staining of human invasive mucinous LuAd showing co-expression of MUC5AC and CTSE by NKX2-1− tumor cells (arrows). Non-mucinous tumor regions are molecularly diverse, containing NKX2-1+CTSE+ (open arrowheads), NKX2-1−CTSE+, and NKX2-1+CTSE− cells (n=2 individuals). (b) Co-staining of an EGFR L858R mutant human lepidic LuAd reveals an isolated squamous AGER+Muc1− cell in the tumor periphery that is faintly positive for the mutation-specific antibody (arrowhead).
Scale bars, 20 μm (a, b lower row) 200 μm (b, upper row)
Supplementary Material
Acknowledgments
The authors wish to thank Roel Nusse, Monte Winslow, Sylvia Plevritis, Laura Attardi, and members of the Desai and Nusse labs for valuable discussions; the CZI Biohub; Pauline Chen of the Human Histology Core; Doreen Wu of the Animal Histology Service; Michael Eckart at the Stanford Protein and Nucleic Acid Facility; Catherine Carswell Crumpton and Cheng Pan at the Stanford University Stem Cell Institute FACS Core facility; and John Perrino of Stanford’s Cell Science Imaging Facility.
This research was supported in part by the Virginia and D.K. Ludwig Fund for Cancer Research.
The project described was supported, in part, by NIH S10 Award Number 1S10OD028536-01, titled "OneView 4kX4k sCMOS camera for transmission electron microscopy applications" from the Office of Research Infrastructure Programs (ORIP)). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the National Institutes of Health.”
Funding:
NHLBI 5R01HL14254902, NIH 5UG3HL14562302 (TJD)
Ludwig Cancer Institute (TJD, NHJ, RS)
NIH 1T32HL129970-01A1, 5T32HL129970-03 (NHJ)
NIH Loan Repayment Program (NHJ)
NHLBI 1 F32 HL147417-01(NHJ)
Stanford Medical Scholars Fellowship Program (MCM)
Woods Family Endowed Faculty Scholar in Pediatric Translational Medicine of the Stanford Child Health Research Institute (TJD)
Robert A. and Gertrude T. Hudson Endowed Professorship (TJD)
Footnotes
Competing interests:
The authors have no competing interests to declare.
Research Animals:
Authors have complied with all relevant ethic regulations regarding the care and use of laboratory animals. The included experimental protocols were approved by the Stanford Institutional Animal Care and Use Committee (protocol 22988).
Human Research Participants:
Human tissues assayed for this manuscript were obtained with informed consent through a protocol approved by the Stanford University Institutional Review Board (protocols 15166 and 18891).
Supplementary Materials
Supplemental data description:
Characteristics of tumors included in the manuscript; gene sets used for Figure 3f; and patient/mutation information from cBioPortal.
Data Availability Statement
Tissue microarray data from Zabeck et al 2018 reanalyzed and presented in Figure 4 were obtained from the NCBI's Gene Expression Omnibus under accession number GSE58772. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58772). Histologically annotated mutation data from Caso et al 2020 reanalyzed and presented in Figure 4 was obtained from cBioPortal (www.cbioportal.org; ‘Lung Adenocarcinoma (MSK, J Thorac Oncol 2020’). scRNA sequencing data presented in this manuscript will be deposited in the Gene Expression Omnibus prior to publication.
References
- 1.Siegel RL, Miller KD, Fuchs HE & Jemal A Cancer statistics, 2022. CA Cancer J Clin 72, 7–33 (2022). 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Desai TJ Developmental Insights into Lung Cancer. Annual review of cancer biology 5, 351–369 (2021). 10.1146/annurev-cancerbio-070820-032858 [DOI] [Google Scholar]
- 3.Desai TJ, Brownfield DG & Krasnow MA Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014). 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C et al. Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One 7, e53817 (2012). 10.1371/journal.pone.0053817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci U S A 109, 4910–4915 (2012). 10.1073/pnas.1112499109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFadden DG et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci U S A 113, E6409–e6417 (2016). 10.1073/pnas.1613601113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okubo K et al. Bronchoalveolar carcinoma: clinical, radiologic, and pathologic factors and survival. J Thorac Cardiovasc Surg 118, 702–709 (1999). 10.1016/S0022-5223(99)70016-4 [DOI] [PubMed] [Google Scholar]
- 8.Travis WD et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6, 244–285 (2011). 10.1097/JTO.0b013e318206a221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones KD Whence lepidic?: the history of a Canadian neologism. Arch Pathol Lab Med 137, 1822–1824 (2013). 10.5858/arpa.2013-0144-HP [DOI] [PubMed] [Google Scholar]
- 10.Juul NH, Stockman CA & Desai TJ Niche Cells and Signals that Regulate Lung Alveolar Stem Cells In Vivo. Cold Spring Harb Perspect Biol (2020). 10.1101/cshperspect.a035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda N et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011). 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson EL et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muzumdar MD, Tasic B, Miyamichi K, Li L & Luo L A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 14.Wolf FA et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20, 59 (2019). 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagendran M, Andruska AM, Harbury PB & Desai TJ Advances in Proximity Ligation in situ Hybridization (PLISH). Bio-protocol 10, e3808 (2020). 10.21769/BioProtoc.3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabhan A, Brownfield DG, Harbury PB, Krasnow MA & Desai TJ Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science (2018). 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zacharias WJ et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018). 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock JR et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 108, E1475–1483 (2011). 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tammela T et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017). 10.1038/nature22334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacheco-Pinedo EC et al. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest 121, 1935–1945 (2011). 10.1172/jci44871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juan J, Muraguchi T, Iezza G, Sears RC & McMahon M Diminished WNT -> beta-catenin -> c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev 28, 561–575 (2014). 10.1101/gad.233627.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada N et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 18, 5931–5942 (1999). 10.1093/emboj/18.21.5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlitz F et al. An immediate-late gene expression module decodes ERK signal duration. Mol Syst Biol 13, 944 (2017). 10.15252/msb.20177986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cicchini M et al. Context-Dependent Effects of Amplified MAPK Signaling during Lung Adenocarcinoma Initiation and Progression. Cell Rep 18, 1958–1969 (2017). 10.1016/j.celrep.2017.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder EL et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell 50, 185–199 (2013). 10.1016/j.molcel.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marjanovic ND et al. Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell 38, 229–246.e213 (2020). 10.1016/j.ccell.2020.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tata PR et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Dev Cell 44, 679–693 e675 (2018). 10.1016/j.devcel.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlins EL, Clark CP, Xue Y & Hogan BL The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 (2009). 10.1242/dev.037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichane M et al. Isolation and 3D expansion of multipotent Sox9(+) mouse lung progenitors. Nat Methods 14, 1205–1212 (2017). 10.1038/nmeth.4498 [DOI] [PubMed] [Google Scholar]
- 30.Strunz M et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun 11, 3559 (2020). 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi J et al. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 27, 366–382.e367 (2020). 10.1016/j.stem.2020.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi Y et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol 22, 934–946 (2020). 10.1038/s41556-020-0542-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 185, 1905–1923 e1925 (2022). 10.1016/j.cell.2022.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi Y et al. Rethinking a Non-Predominant Pattern in Invasive Lung Adenocarcinoma: Prognostic Dissection Focusing on a High-Grade Pattern. Cancers 13 (2021). 10.3390/cancers13112785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zabeck H et al. Molecular signatures in IASLC/ATS/ERS classified growth patterns of lung adenocarcinoma. PLoS One 13, e0206132 (2018). 10.1371/journal.pone.0206132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caso R et al. The Underlying Tumor Genomics of Predominant Histologic Subtypes in Lung Adenocarcinoma. J Thorac Oncol 15, 1844–1856 (2020). 10.1016/j.jtho.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda Y et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest 122, 4388–4400 (2012). 10.1172/jci64048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun CH et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11, 217–227 (2007). 10.1016/j.ccr.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayakawa Y, Nakagawa H, Rustgi AK, Que J & Wang TC Stem cells and origins of cancer in the upper gastrointestinal tract. Cell Stem Cell 28, 1343–1361 (2021). 10.1016/j.stem.2021.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Concepcion CP et al. Smarca4 Inactivation Promotes Lineage-Specific Transformation and Early Metastatic Features in the Lung. Cancer Discov 12, 562–585 (2022). 10.1158/2159-8290.CD-21-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutherland KD et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 111, 4952–4957 (2014). 10.1073/pnas.1319963111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z et al. MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Rep 16, 1810–1819 (2016). 10.1016/j.celrep.2016.07.020 [DOI] [PubMed] [Google Scholar]
- 43.Treutlein B et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Veen JE et al. Mutationally-activated PI3'-kinase-alpha promotes dedifferentiation of lung tumors initiated by the BRAF(V600E) oncoprotein kinase. Elife 8 (2019). 10.7554/eLife.43668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weibel ER On the tricks alveolar epithelial cells play to make a good lung. Am J Respir Crit Care Med 191, 504–513 (2015). 10.1164/rccm.201409-1663OE [DOI] [PubMed] [Google Scholar]
- 46.Nicholson AG et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J Thorac Oncol 17, 362–387 (2022). 10.1016/j.jtho.2021.11.003 [DOI] [PubMed] [Google Scholar]
- 47.Travis WD et al. The IASLC Lung Cancer Staging Project: Proposals for Coding T Categories for Subsolid Nodules and Assessment of Tumor Size in Part-Solid Tumors in the Forthcoming Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 11, 1204–1223 (2016). 10.1016/j.jtho.2016.03.025 [DOI] [PubMed] [Google Scholar]
- 48.Berry MF et al. Presence of Even a Small Ground-Glass Component in Lung Adenocarcinoma Predicts Better Survival. Clin Lung Cancer 19, e47–e51 (2018). 10.1016/j.cllc.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 49.Ye T et al. Lung Adenocarcinomas Manifesting as Radiological Part-Solid Nodules Define a Special Clinical Subtype. J Thorac Oncol 14, 617–627 (2019). 10.1016/j.jtho.2018.12.030 [DOI] [PubMed] [Google Scholar]
- 50.Hattori A et al. Distinct Clinicopathologic Characteristics and Prognosis Based on the Presence of Ground Glass Opacity Component in Clinical Stage IA Lung Adenocarcinoma. J Thorac Oncol 14, 265–275 (2019). 10.1016/j.jtho.2018.09.026 [DOI] [PubMed] [Google Scholar]
- 51.Hou Y et al. The presence of lepidic and micropapillary/solid pathological patterns as minor components has prognostic value in patients with intermediate-grade invasive lung adenocarcinoma. Translational lung cancer research 11, 64–74 (2022). 10.21037/tlcr-21-934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreira AL et al. A Grading System for Invasive Pulmonary Adenocarcinoma: A Proposal From the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 15, 1599–1610 (2020). 10.1016/j.jtho.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joubert P & Travis WD Prognostic Impact of Ground-Glass Opacity/Lepidic Component in Pulmonary Adenocarcinoma: A Hazy Staging Dilemma. J Thorac Oncol 17, 19–21 (2022). 10.1016/j.jtho.2021.10.012 [DOI] [PubMed] [Google Scholar]
References
- 1.Takeda N et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011). 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rock JR et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 108, E1475–1483 (2011). 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman HA et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–2862 (2011). 10.1172/jci57673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson EL et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muzumdar MD, Tasic B, Miyamichi K, Li L & Luo L A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 6.Desai TJ, Brownfield DG & Krasnow MA Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014). 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagendran M, Riordan DP, Harbury PB & Desai TJ Automated cell type classification in intact tissues by single-cell molecular profiling. Elife 7 (2018). 10.7554/eLife.30510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bankhead P et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zabeck H et al. Molecular signatures in IASLC/ATS/ERS classified growth patterns of lung adenocarcinoma. PLoS One 13, e0206132 (2018). 10.1371/journal.pone.0206132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raudvere U et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47, W191–W198 (2019). 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caso R et al. The Underlying Tumor Genomics of Predominant Histologic Subtypes in Lung Adenocarcinoma. J Thorac Oncol 15, 1844–1856 (2020). 10.1016/j.jtho.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26 (2011). 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tissue microarray data from Zabeck et al 2018 reanalyzed and presented in Figure 4 were obtained from the NCBI's Gene Expression Omnibus under accession number GSE58772. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58772). Histologically annotated mutation data from Caso et al 2020 reanalyzed and presented in Figure 4 was obtained from cBioPortal (www.cbioportal.org; ‘Lung Adenocarcinoma (MSK, J Thorac Oncol 2020’). scRNA sequencing data presented in this manuscript will be deposited in the Gene Expression Omnibus prior to publication.