Abstract
Microglia reactivity entails a large-scale remodeling of cellular geometry, but the behavior of the microtubule cytoskeleton during these changes remains unexplored. Here we show that activated microglia provide an example of microtubule reorganization from a non-centrosomal array of parallel and stable microtubules to a radial array of more dynamic microtubules. While in the homeostatic state, microglia nucleate microtubules at Golgi outposts, and activating signaling induces recruitment of nucleating material nearby the centrosome, a process inhibited by microtubule stabilization. Our results demonstrate that a hallmark of microglia reactivity is a striking remodeling of the microtubule cytoskeleton and suggest that while pericentrosomal microtubule nucleation may serve as a distinct marker of microglia activation, inhibition of microtubule dynamics may provide a different strategy to reduce microglia reactivity in inflammatory disease.
In brief
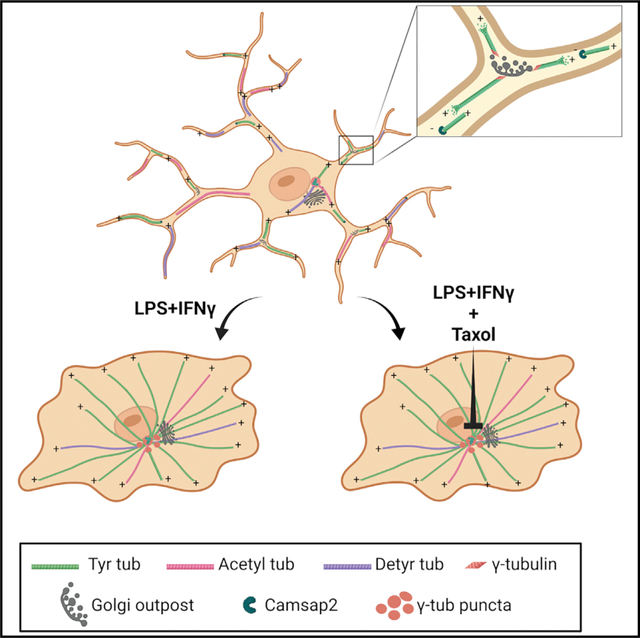
Rosito et al. find that in microglia cells, the transition between homeostatic and reactive states is characterized by a dramatic reorganization of the microtubule cytoskeleton from a Golgi-outpost-nucleated acentrosomal array to a pericentrosomal radial array to enable proper cytokine release.
Graphical Abstract
INTRODUCTION
Microglia are the brain’s primary innate immune cells. In their homeostatic state in the healthy brain, they exhibit a ramified morphology and continuously patrol the local environment by extension and retraction of highly motile processes1 that act to clear cellular debris, reshape synapses, and provide neurotrophic factors.2–8 However, when activated by neuronal inflammation and injury, and in neurodegenerative disorders, microglia exhibit dramatically altered gene expression and morphology, displaying an ameboid shape.9–11 In this activated state, reactive microglia exhibit phagocytic activities that can promote tissue remodeling, and if overactivated are widely thought to contribute to brain damage and neurodegeneration.2,12
In eukaryotes, changes in cellular symmetry are associated with massive reorganization of both the actin and microtubule (MT) cytoskeletons. While actin and actin-based motor proteins are required for breaking the symmetry in most cells, specification of neuronal polarity depends on MTs and MT-associated proteins.13 Early studies suggested changes in MT spatial organization and stability also with microglia activation.14 Despite these observations, however, only remodeling of the actin cytoskeleton has been extensively studied in microglia, and the role of the MT cytoskeleton in breaking cellular polarity during the transition from homeostatic to a reactive state has not been explored.
MTs are intrinsically polarized polymers composed of α/β-tubulin heterodimers arranged in a head-to-tail fashion.15 They are characterized by a fast-growing plus end and a slow-growing minus end that in non-neuronal cells is often attached to the centrosome, which acts as the MT organizing center (MTOC). Directional transport is enabled by the structural polarity of MTs, which is recognized by motor proteins that drive transport to either the minus end (dynein) or plus end (most kinesins) (for reviews, see Klinman and Holzbaur16 and Reck-Petersen et al.17). MTs are generally highly dynamic structures constantly undergoing stochastic transitions from polymerization to depolymerization (catastrophe events) and vice versa (rescue events), with the two dynamic states exhibiting characteristic rates of growth or shrinkage.18 When stabilized, MTs resist disassembly and become substrates of tubulin-modifying enzymes that add molecular moieties on either the α- or β-tubulin subunit. The combinatorial nature of these modifications provides a “tubulin code” which controls a variety of functions, including organelle transport and the mechanical properties of the MT lattice (for reviews, see Gadadhar et al.,19 Janke and Magiera,20 and Janke and Bulinski21).
MT orientation, density, and post-translational modifications all respond and contribute to breaking cellular symmetry.22,23 Establishment of cell polarity can be achieved through centrosome repositioning or by the formation of non-radial MT arrays, in which MTs are not preferentially nucleated at the centrosome.24–26 During this transition, the centrosome typically loses its maximal MT nucleating activity while intracellular membranes or self-organizing assemblies of MT nucleating material distant from the centrosome serve as non-centrosomal MTOCs, with members of the calmodulin-regulated spectrin-associated protein (CAMSAP) family often capping and stabilizing released free MT minus ends.27,28 The local stabilization of non-centrosomal MTs and relocation of the MTOCs away from the centrosome and the cell center establish asymmetric MT arrays that are critical to cell differentiation.
We hypothesized that rearrangement of the MT cytoskeleton might be required for the morphological changes that guide microglia transition from surveilling/homeostatic to reactive states. Here we show that activated microglia engage a unique example of MT transition from a non-centrosomal array of parallel and stable MTs in the homeostatic state to a radial array of more dynamic MTs in which all MT minus ends are anchored to a pericentrosomal region. We further find that in the homeostatic state, Golgi outposts are sites of non-centrosomal MT nucleation and that an activating challenge leads to the recruitment of pericentriolar material (PCM) to the centrosome, a process inhibited by MT stabilization. Our results unveil the unique rearrangement of the MT cytoskeleton in activated microglia and indicate that while pericentriolar material relocalization and assembly offer a means to observe the dynamics of microglial reactivity in tissue, inhibition of MT dynamics may provide a potential target of microglia treatment in inflammatory disease.
RESULTS
Homeostatic, activated, and alternatively activated primary microglia differ in MT distribution, stability, and dynamic behavior
To investigate the organization of the MT cytoskeleton in homeostatic and reactive microglia, we used primary mouse microglia cultures in which the presence of ramified cells was maintained by growth factors secreted by astrocytes.29 With this approach we prepared a nearly pure population of primary microglia comprising 99% of Iba1-positive cells. To steer microglia toward different reactivity states such as an activated or an alternatively polarized microglia state (defined as alternatively activated), cells were challenged with either lipopolysaccharide-interferon-γ (LPS-IFNγ) (100 ng/mL LPS, 20 ng/mL IFNγ, 48 h for activated) or interleukin-4 (IL-4) (20 ng/mL, 48 h for alternatively activated) and measured for the expression of their signature activation genes (Figure 1A). As revealed by Iba1 staining (Figure 1B), polarized microglia underwent dramatic morphological changes. We classified cell morphology as ramified (≥3 ramifications), ameboid, or bipolar based on number of cellular processes, cell area, and solidity, a measure of cell shape complexity (Figures 1B–1D). Analysis of morphology distribution under homeostatic, activated, and alternatively activated conditions revealed that ramified cells were enriched in untreated microglia (35% ± 3%) (Figures 1B and 1D), while ameboid cells represented a large majority after activating stimulation (82% ± 3%) (Figures 1B and 1D). Conversely, when cells were challenged with an alternatively activating stimulus, microglia mostly acquired a unipolar or bipolar rod-shape morphology (54% ± 3%) characterized by the presence of a lamellipodium and a trailing edge, or uropod (Figures 1B and 1D). To further detail the structural changes associated with reactive microglia states via single-cell analyses, we chose to select for comparison only the most representative morphology of each in vitro phenotype (ramified for homeostatic, ameboid for activated, and bipolar for alternatively activated microglia).
Figure 1. Molecular and morphological characterization of homeostatic, activated, and alternatively activated primary microglia.
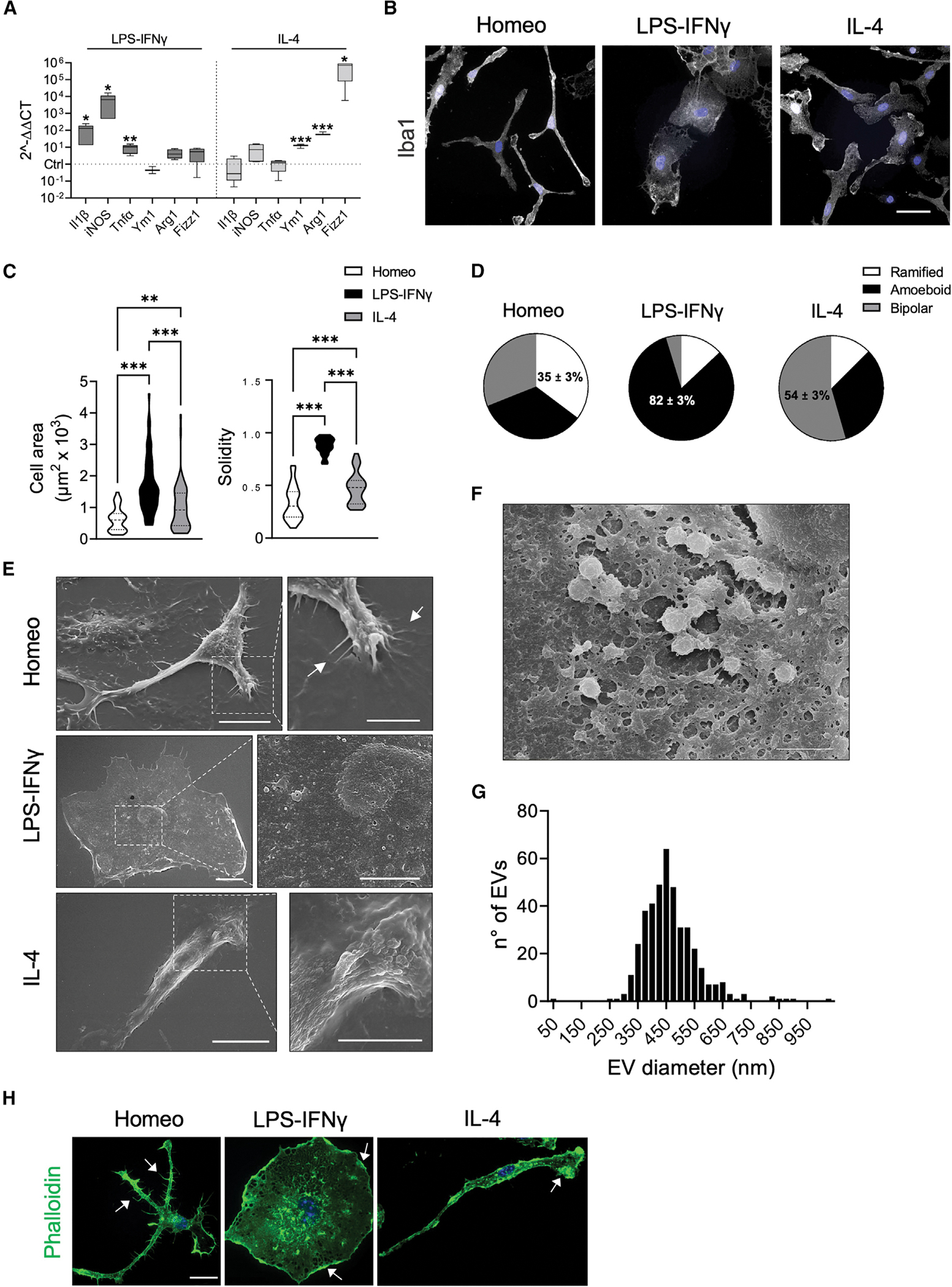
(A) Real-time qPCR reveals increased expression of pro-inflammatory (Il1b, iNOS, Tnfα) and anti-inflammatory genes (Ym1, Arg1, Fizz1) upon LPS-IFNγ or IL-4 challenge, respectively. Gene expression is normalized to the housekeeping gene Gapdh, n = 4 independent cultures. Box plots indicate the median, 25th, and 75th percentiles; whiskers show the minimum and maximum. ***p < 0.001, **p < 0.01, *p < 0.05, Student’s t test.
(B) Representative images showing Iba1 immunostaining (gray) of microglia in homeostatic (Homeo) condition and following LPS-IFNγ or IL-4 treatment. Hoechst for nuclei visualization, blue. Scale bar, 20 μm.
(C) Violin plots showing cell surface area (left) and solidity coefficient (right) related to Homeo (white), LPS-IFNγ (black), or IL-4 (gray) challenged cells (n = 70 cells for each condition, from 3 independent experiments). ***p < 0.001, **p < 0.01; left: Kruskall-Wallis with Dunn’s multiple comparisons test for cell area; right: one-way ANOVA with Tukey’s multiple comparisons test for solidity.
(D) Pie charts illustrating the distribution of cell morphology in Homeo, LPS-IFNγ, and IL-4 conditions. Ramified cells are enriched in Homeo (35% ± 3%); ameboid cells in LPS-IFNγ-treated microglia (82% ± 3%); and bipolar cells in IL-4-treated microglia (54% ± 3%). n = 3 independent experiments.
(E) Representative scanning electron micrographs of microglia in Homeo (top), LPS-IFNγ (middle), and IL-4 (bottom) conditions. Ramified cells (Homeo) show filopodia extensions as indicated by arrows, ameboid cells (LPS-IFNγ) exhibit numerous extracellular vesicles (EVs) on the cell surface, while bipolar cells (IL-4) are characterized by extensive membrane ruffling on the cell surface and leading edge. Scale bars, 10 μm (5 μm in zoom images).
(F) Scanning electron micrographs showing EVs on the LPS-IFNγ cell surface at higher magnification. Scale bar, 2 μm.
(G) Distribution of EV size measured on the ameboid cell surface. n = 22 cells.
(H) Representative confocal images of microglia labeled with Alexa Fluor 488-conjugated phalloidin (green). Ramified cells (Homeo) exhibit filopodia-like structures (as indicated by arrows) while ameboid (LPS-IFNγ) and bipolar cells (IL-4) are characterized by membrane ruffles along cell borders and at lamellipodia (as indicated by arrows). Hoechst for nuclei visualization, blue. Scale bar, 25 μm.
We employed scanning electron microscopy and confocal microscopy to identify defined ultrastructural elements typical of each functional state (Figure 1E). Homeostatic microglia exhibited many branched processes extending outward from the cell body and multiple filopodia-like structures (Figure 1E) that were also positive for phalloidin staining (Figure 1H). Upon activating challenge, microglia retracted most of their processes and acquired a flattened and round morphology (Figures 1E and 1H). Scanning electron microscopy imaging further revealed that activated microglia displayed numerous tethered extracellular vesicles (EVs) blebbing from the cell surface (Figure 1F). Analysis of EV diameter showed a distribution of size ranging from 250 to 650 nm (Figure 1G), consistent with microglia-shedded microvesicles.30 Alternatively activated microglia were characterized by extensive membrane ruffling at both uropod and leading edge, which appeared as sheet-like structures on the dorsal cell surface (Figures 1E and 1H).
We began to analyze the MT cytoskeleton in each functional state by immunofluorescence (IF) staining of tyrosinated α-tubulin (Tyr tub), a bulk tubulin marker labeling the entire MT network. MTs appeared to be packed in a parallel fashion in all the cellular branches extending from the cell body in both homeostatic and alternatively activated microglia (Figure 2A). However, MTs distributed radially from a perinuclear region in activated microglia (Figure 2A). Radial profiling of Tyr tub intensity (Figure 2A), a measure of the distribution of tubulin signal that is independent of cell shape, confirmed that MT staining was uniformly distributed along the cell profile in homeostatic and alternatively activated microglia (Figure 2B), while in activated cells Tyr tub signal rapidly decayed at increasing distances from a perinuclear region (Figure 2B; exponential decay constant kLPS-IFNγ = 0.046 ± 0.001; khomeo = 0.013 ± 0.002; kIL-4 = 0.015 ± 0.002).
Figure 2. Homeostatic, activated, and alternatively activated primary microglia differ in MT distribution, stability, and dynamic behavior.
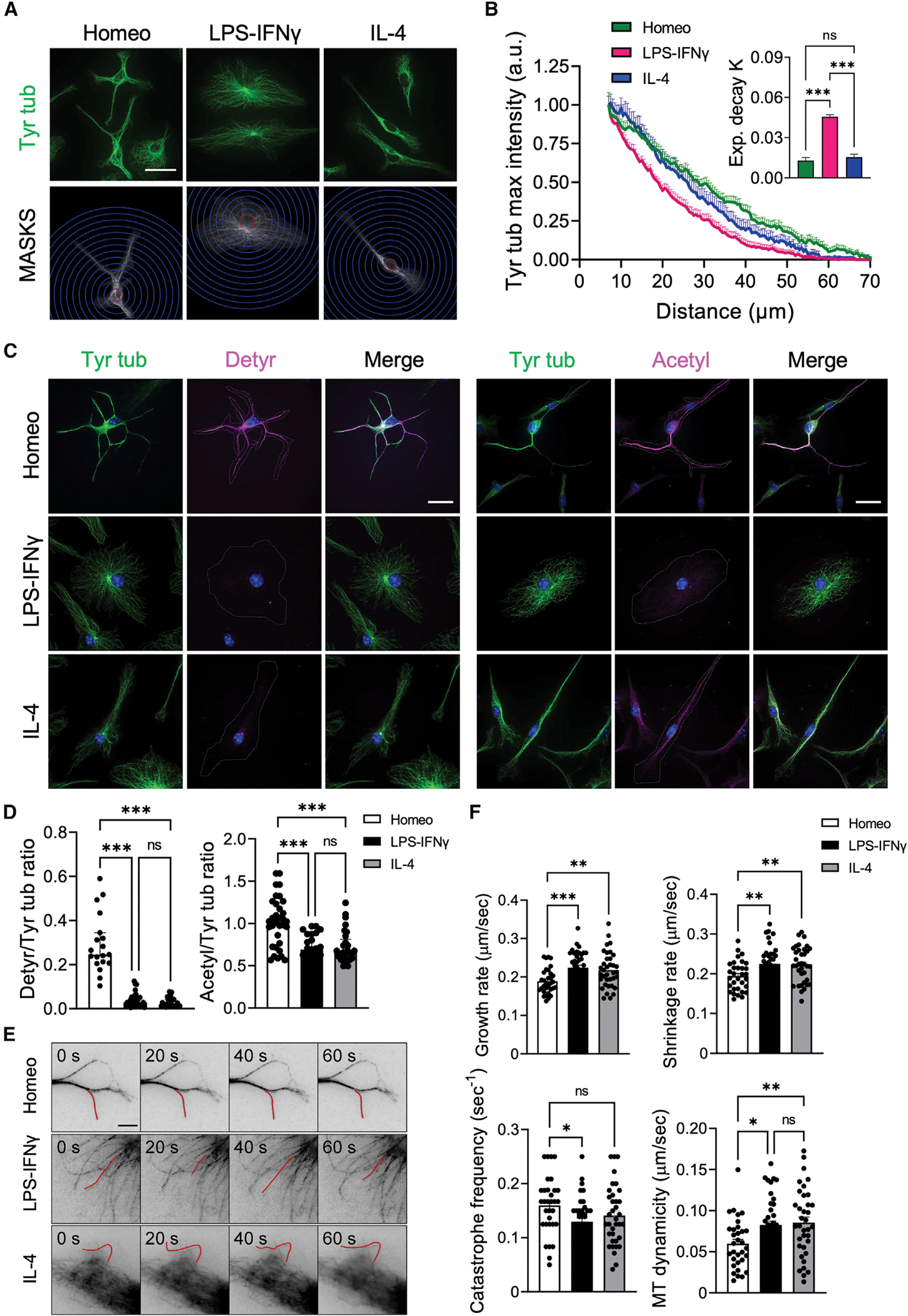
(A) Representative IF images of tyrosinated α-tubulin (Tyr tub, green) staining in homeostatic (Homeo), activated (LPS-IFNγ), and alternatively activated (IL-4) microglia (top; scale bar, 20 μm) and corresponding masks (MASKS) used for the analysis (bottom), with radial scale (unit = 5 μm; blue circles) centered at the centroid of the cell nucleus. Red circle indicates the radius corresponding to the largest intensity value.
(B) Plot showing maximum fluorescence intensity values of Tyr tub versus the radial distance from cell nucleus, obtained with radial profiling, in Homeo (n = 38, green), LPS-IFNγ (n = 31, magenta), or IL-4 (n = 38, blue) treated microglia. Curve fit was performed using a single exponential decay function. Inset: bar chart reporting the exponential decay constant values (K) for each condition (values are expressed as mean ± SEM from 4 independent experiments; ns, not significant; ***p < 0.001, one-way ANOVA with Tukey’s multiple comparison test). Note faster decay of Tyr tub signal in activated microglia.
(C) Representative IF images of Homeo-, LPS-IFNγ-, or IL-4-treated microglia. Left: co-staining of Tyr tub (green) and detyrosinated tubulin (Detyr, magenta) (Hoechst for nuclei visualization, blue; scale bar: 20 μm). Right: co-staining of Tyr tub (green) and acetylated tubulin (Acetyl, magenta) (Hoechst for nuclei visualization, blue; scale bar, 20 μm).
(D) Scatter dot plots showing IF signal quantification of detyrosinated/tyrosinated (Detyr/Tyr) tub ratio (left; Homeo n = 19, LPS-IFNγ n = 32, IL-4 n = 28 cells from 3 independent experiments) and acetylated/tyrosinated (Acetyl/Tyr) tubulin ratio (right; Homeo n = 33, LPS-IFNγ n = 25, IL-4 n = 29 cells from 3 independent experiments). Values are expressed as median ± interquartile range. ns, not significant; ***p < 0.001, Kruskall-Wallis with Dunn’s multiple comparisons test. Note that microglia challenged with LPS-IFNγ or IL-4 have reduced tubulin PTM levels compared with Homeo cells.
(E) Representative inverted contrast wide-field frames from time-lapse acquisitions of SiR-Tubulin in Homeo-, LPS-IFNγ-, or IL-4-treated microglia at four different time points (0 s, 20 s, 40 s, and 60 s). Red lines highlight MT length changes ≥0.5 μm between frames. Scale bar, 5 μm.
(F) Scatter dot plots representing growth rate (top, left), shrinkage rate (top, right), catastrophe frequency (bottom, left) and MT dynamicity (bottom, right) in Homeo, LPS-IFNγ, and IL-4 microglia. Values are expressed as mean ± SEM (Homeo n = 30, LPS-IFNγ n = 27, IL-4 n = 29 cells from 4 independent experiments). ns, not significant; ***p < 0.001, **p < 0.01, *p < 0.05, one-way ANOVA with Dunnett’s multiple comparisons test.
To evaluate whether MTs differed in stability according to the reactive state, we analyzed levels and distribution of detyrosinated and acetylated tubulins, two independent tubulin post-translational modifications (PTMs) associated with MT longevity.20,31 Semiquantitative IF analyses revealed that homeostatic cells had the highest level of both detyrosinated (Figure 2C, left) and acetylated (Figure 2C, right) tubulin compared with activated or alternatively activated microglia (detyrosinated/Tyr tub 0.30 ± 0.03, 0.04 ± 0.01, 0.03 ± 0.01; acetylated/Tyr tub 1.02 ± 0.05, 0.69 ± 0.04, 0.72 ± 0.04 in homeostatic, activated, and alternatively activated microglia, respectively; Figure 2D), suggesting that homeostatic microglia display more stable MTs.
We observed the behavior of SiR-Tubulin-labeled MTs32 in shallow peripheral sections of the cell to measure MT plus-end dynamics using time-lapse wide-field fluorescence microcopy (Figure 2E). No change was observed among the three different microglia states in rescue frequency (frequency of transitions from shrinkage to growth) or the fraction of time spent in pausing or shrinkage. However, while in activated and alternatively activated microglia, MTs exhibited a moderate yet significant drop in catastrophe frequency (frequency of transitions from growth to shrinkage); they also significantly enhanced their growth rates and acquired a nearly 2.5-fold increase in rates of shrinkage, resulting in an overall net rise in MT dynamicity compared with MTs of homeostatic cells (0.06 ± 0.01, 0.08 ± 0.01, 0.09 ± 0.01 in homeostatic, activated, and alternatively activated microglia, respectively; Figure 2F, Table 1, and Videos S1, S2, and S3). In summary, and consistent with our analysis of tubulin PTMs, these data suggest that microglia acquisition of activated and alternatively activated phenotypes is characterized by loss of MT stability and a marked increase in MT dynamics.
Table 1.
Parameters of MT dynamics in homeostatic and activated microglia
| Homeo | LPS-IFNγ | IL-4 | |
|---|---|---|---|
|
| |||
| Growth rate (μm/s) | 0.19 ± 0.01 | 0.22 ± 0.01*** | 0.22 ± 0.01** |
| Shrinkage rate (μm/s) | 0.9 ± 0.01 | 0.23 ± 0.01** | 0.22 ± 0.01** |
| % Growth | 15 ± 1 | 20 ± 1* | 18 ± 1 |
| % Shrinkage | 15 ± 1 | 16 ± 1 | 19 ± 1 |
| % Pausing | 70 ± 2 | 64 ± 2 | 63 ± 2 |
| Catastrophe frequency (s−1) | 0.16 ± 0.01 | 0.13 ± 0.01* | 0.14 ± 0.01 |
| Rescue frequency (s−1) | 0.14 ± 0.06 | 0.15 ± 0.01 | 0.13 ± 0.01 |
| MT dynamicity (μm/s) | 0.06 ± 0.01 | 0.08 ± 0.01* | 0.09 ± 0.01** |
| MTs (n) | 32 | 40 | 36 |
| Cells (n) | 30 | 27 | 29 |
Parameters of MT dynamics obtained from wide-field fluorescence time-lapse analysis of SiR-Tubulin in homeostatic (Homeo), activated (LPS-INFγ), and alternatively activated (IL-4) microglia. Values are expressed as mean ± SEM from Homeo (n = 32/30 MTs/cells), LPS-INFγ (n = 40/27 MTs/cells), and IL-4 (n = 36/29 MTs/cells) arising from 4 independent experiments.
p < 0.001
p < 0.01
p < 0.05, one-way ANOVA with Dunnett’s multiple comparisons test.
Homeostatic, activated, and alternatively activated microglia differ in MT orientation
In most dividing and motile cells the centrosome is responsible for MT nucleation and anchoring, leading to the formation of radial MT arrays in which all MT minus ends are attached to the centrosome while MT plus ends extend toward the cell periphery. In contrast, in most differentiated, stationary, and axially polarized cells, MTs are more stable and organized in non-centrosomal arrays that are non-radially anchored at the centrosome.27
We hypothesized that in microglia, the transition from the homeostatic phenotype to a migrating reactive state would be paralleled by prominent changes in cell polarity driven by the remodeling of MT anchoring and orientation. To test this, we analyzed the localization and expression of MT plus-end end binding (EB) and minus-end CAMSAP markers in homeostatic, activated, and alternatively activated microglia cells that had been selected according to their most representative morphology (Figures 3 and S1). While EB proteins are widely adopted markers of actively growing MT plus ends (comets), members of the CAMSAP family regulate the formation and stability of non-centrosomal MT arrays by capping free MT minus ends.24,33
Figure 3. Homeostatic, activated, and alternatively activated primary microglia differ in MT orientation.
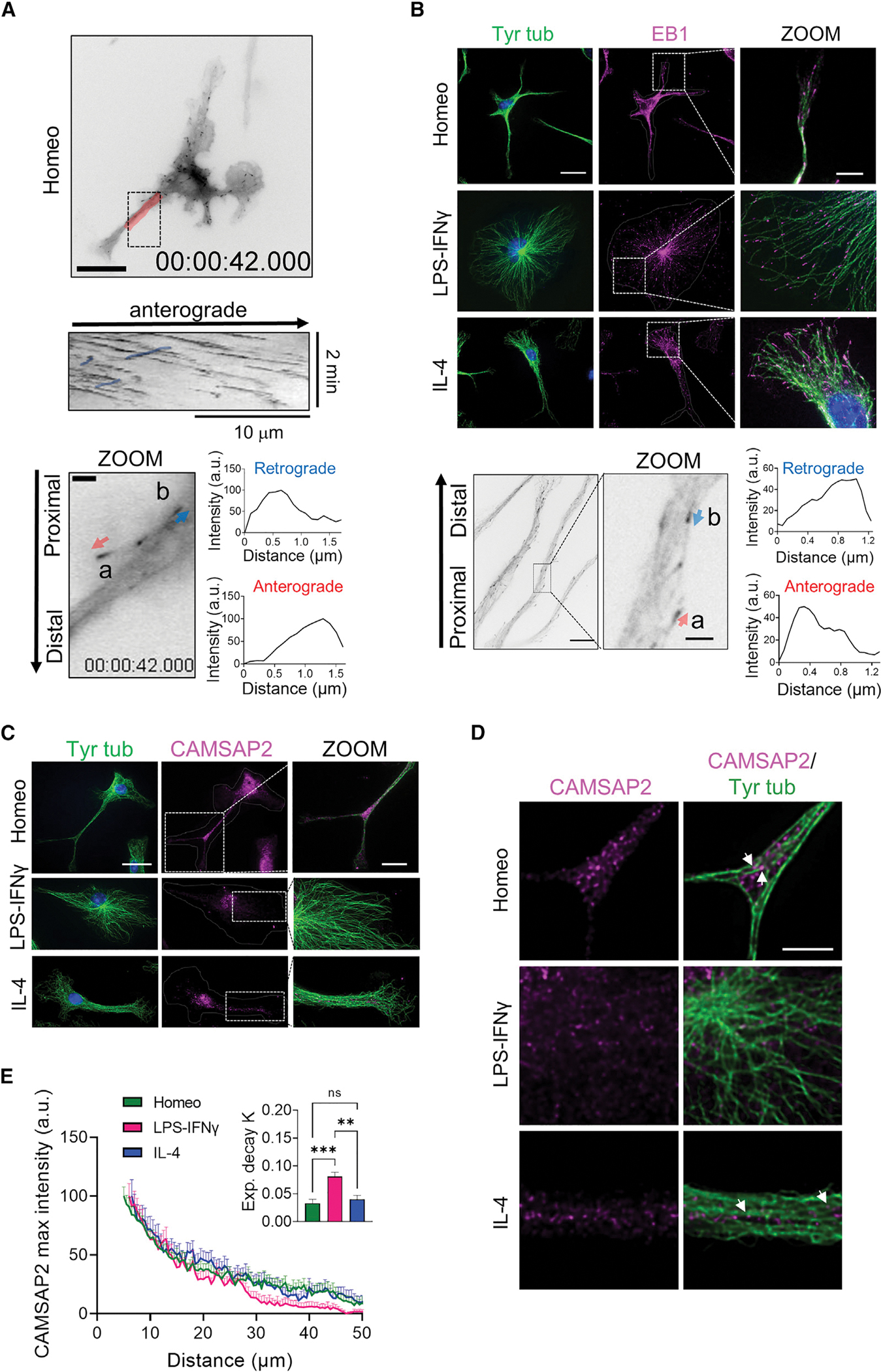
(A) Top: representative inverted contrast wide-field frames from time-lapse acquisitions of EB3-EGFP-infected homeostatic (Homeo) microglia (scale bar, 20 μm) and kymograph of the selected region in red (middle). Note that retrograde comets in the kymograph are highlighted in blue. Bottom: inverted contrast single frame image of EB3-EGFP at higher magnification (scale bar, 2 μm). Relative orientation of EB3 comet peaks with respect to the cell nucleus was used to distinguish between EB3 anterograde (a, red arrow) and retrograde (b, blue arrow) comets.
(B) Top: representative IF images of EB1 (magenta) and tyrosinated α-tubulin (Tyr tub) (green) in homeostatic (Homeo), activated (LPS-IFNγ), and alternatively activated (IL-4) microglia. Hoechst for nuclei visualization, blue. Scale bar, 20 μm (5 μm in zoom images). Bottom left: representative inverted contrast single plane image of EB1 IF (scale bar, 10 μm). Bottom middle: direction of EB1 signal gradient relative to the cell nucleus was used to identify EB1 anterograde (a, red arrow) and retrograde (b, blue arrow) comets (scale bar, 2 μm). Bottom right: intensity profiles of anterograde (lower) and retrograde (upper) comets.
(C) Representative z-projection confocal images showing CAMSAP2 (magenta) and Tyr tub (green) signal in Homeo-, LPS-IFNγ-, or IL-4-treated microglia. Hoechst for nuclei visualization, blue. Scale bar, 20 μm (5 μm in zoom images).
(D) Single confocal planes at higher magnification of CAMSAP2 (magenta) and Tyr tub (green) signal in Homeo-, LPS-IFNγ-, or IL-4-treated microglia. Note that CAMSAP2 signal is present in microglia processes in Homeo- and IL-4-treated cells. Scale bar, 5 μm.
(E) Plot showing maximum fluorescence intensity values of CAMSAP2 versus the radial distance from cell nucleus, obtained with radial profiling, in Homeo (n = 18 cells, green), LPS-IFNγ (n = 14 cells, magenta), or IL-4 (n = 19 cells, blue) treated microglia. Curve fit was performed using single exponential decay function. Inset: bar chart reporting the exponential decay constant values (K) for each condition (values are expressed as mean ± SEM from 4 independent experiments; ns, not significant; ***p < 0.001, **p < 0.01, one-way ANOVA with Tukey’s multiple comparisons test).
Low-efficiency lentiviral expression of EB3-EGFP showed 13% (n = 13 cells) of retrograde EB3 comets in homeostatic ramified microglia and 12% (n = 6 cells) in alternatively activated bipolar microglia, suggesting the existence of bioriented arrays (Figures 3A and S1A; Videos S4 and S5). We had no success in infecting microglia that had been activated with LPS and IFNγ. In addition, in infected homeostatic microglia, values of cell area (1,123 ± 111 cm2) and solidity (0.39 ± 0.03) appeared slightly increased compared with non-infected cells (see Figure 1C), indicating that microglia reactivity might be affected by viral infection. To identify MT polarity in non-infected cells, we thus exploited the comet-shaped accumulation of EB proteins at MT ends, with comet-tail intensity decaying exponentially after the maximum at the MT end that is toward the MT lattice.34 First, we measured fluorescence intensity gradients of EB comets from single frames of our movies (Figure 3A, arrows) and confirmed that this approach was reliable to identify anterograde and retrograde comet orientation. Next, we applied this analysis to non-infected microglia cells that had been fixed and immunostained using EB1 and tyrosinated tubulin antibodies to identify endogenous EB comets and MTs, respectively. Confocal immunofluorescence analysis showed that in activated and alternatively activated microglia, EB1 decorated most free MT ends (Tyr tub stained) (activated, 88%; alternatively activated, 82%) that extended toward the cell periphery (Figure 3B), confirming the existence of a prominent pool of dynamic MTs arranged radially with their minus ends attached to a perinuclear region of the cell. EB1 comets were also clearly visible at MT ends in homeostatic microglia although to a lesser extent (homeostatic 63% of MTs, p < 0.001, see Figure S1B for contingency analysis). Western blot analysis of whole-cell lysates showed that total EB1 protein levels did not change in reactive microglia compared with homeostatic cells (Figure S1C). Analysis of fluorescence intensity gradients of EB-positive comets (Figure 3B, arrows) confirmed the presence of a population of retrograde comets in homeostatic microglia and alternatively activated microglia as opposed to activated microglia in which all the comets were oriented away from the cell nucleus and toward the cell periphery (23.4%, 12.5%, and 0.5%, respectively, p < 0.001; Figure S1D).
Detection of a pool of retrograde comets in homeostatic microglia suggested the presence of non-centrosomal MT arrays, which we investigated by analyzing the expression and subcellular distribution of endogenous CAMSAP2. Western blot analysis of whole-cell lysates revealed endogenous expression of CAMSAP2 in all three microglia phenotypes, with higher protein content in homeostatic microglia (Figure S1E). However, while in homeostatic and alternatively activated cells CAMSAP2 were often distributed to isolated and clustered puncta along cell ramifications (Figures 3C and 3D, arrows), cytosolic CAMSAP2 signal was detectable only around the perinuclear region in activated microglia (Figures 3C and 3D). Radial profiling of CAMSAP2 fluorescence intensity (Figure 3E), used as a measure of CAMSAP2 distribution in the cytosol, confirmed that CAMSAP2 signal decayed more rapidly at increasing distances from the perinuclear region in activated microglia than in homeostatic and alternatively activated cells (Figure 3E and inset).
Altogether, these data indicate that homeostatic and alternatively activated microglia display a mixed MT polarity pattern resembling neuronal MTs in dendrites and that the acquisition of an activated phenotype represents a unique example of remodeling of the MT cytoskeleton from an array of parallel non-centrosomal MTs to a radial array of MTs all anchored to pericentrosomal MTOCs through their minus ends.
Homeostatic microglia nucleate non-centrosomal MTs from Golgi outposts
CAMSAP2 is necessary for the tethering of newly nucleated non-centrosomal MT minus ends during the establishment of polarity in many cell types.33,35,36 We investigated the distribution of γ-tubulin, the major MT nucleator in eukaryotic cells, in homeostatic and reactive microglia by confocal microscopy and found that while in homeostatic and alternatively activated cells γ-tubulin displayed a punctate and often slightly diffuse distribution around the perinuclear region and along cellular processes, in activated microglia γ-tubulin signal was most prominent in the pericentrosomal area as distinct puncta (Figures 4A, S2A, 5, and S3). Normalized radial profiling (Figure 4B) of γ-tubulin fluorescence intensity, used as a measure of γ-tubulin signal distribution in the cell, confirmed that γ-tubulin signal decayed more rapidly at increasing distances from the perinuclear region in activated microglia compared with homeostatic and alternatively activated cells (Figure 4B and inset). This observation was confirmed by the analysis of γ-tubulin signal over the cell area (Figure S2C) and detection of higher γ-tubulin levels in homeostatic and alternatively activated microglia compared with activated cells (Figure S2D).
Figure 4. Homeostatic microglia nucleate non-centrosomal MTs from Golgi outposts.
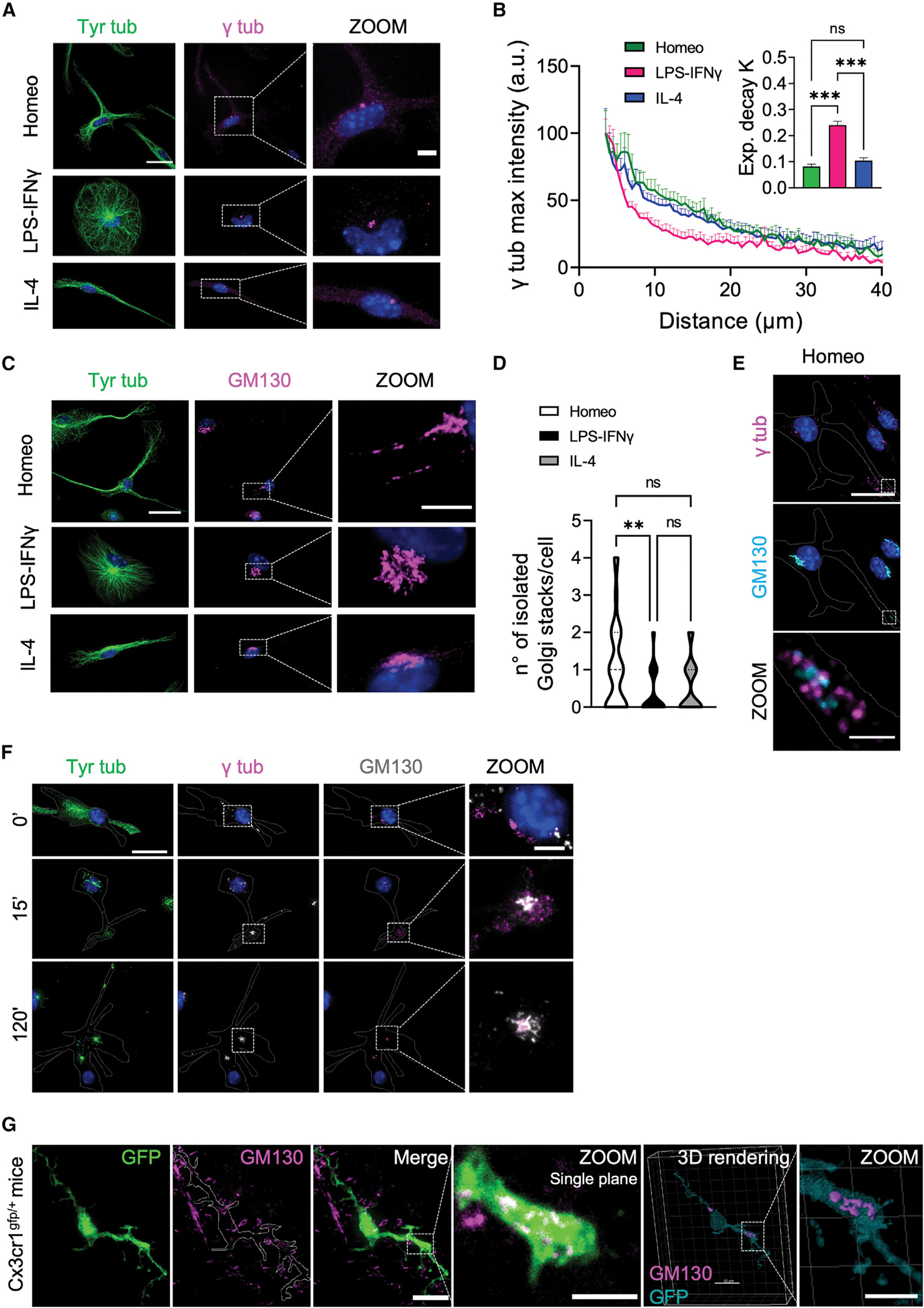
(A) Representative IF images of homeostatic (Homeo), activated (LPS-IFNγ), and alternatively activated (IL-4) microglia stained for tyrosinated α-tubulin (Tyr tub, green) and γ-tubulin (γ tub, magenta). Hoechst for nuclei visualization, blue. Scale bar, 20 μm (5 μm in zoom images).
(B) Plot showing maximum fluorescence intensity values of γ tub versus the radial distance from cell nucleus, obtained with radial profiling, in Homeo (n = 13 cells, green), LPS-IFNγ (n = 14 cells, magenta), or IL-4 (n = 14 cells, blue) treated microglia. Curve fit was performed using single exponential decay function. Inset: bar chart reporting the exponential decay constant values (K) for each condition (values are expressed as mean ± SEM from 3 independent experiments; ns, not significant; ***p < 0.001, one-way ANOVA with Tukey’s multiple comparisons test). Note faster decay of γ tub signal in activated microglia.
(C) Representative confocal images showing co-staining of tyrosinated tubulin (Tyr tub) (green) and GM130 (magenta) in Homeo-, LPS-IFNγ-, or IL-4-treated microglia. Hoechst for nuclei visualization, blue. Scale bar, 20 μm (5 μm in zoom images).
(D) Violin plot showing number of isolated Golgi stacks per cell in the three phenotypes. Values are expressed as mean ± SEM of Homeo (n = 57), LPS-IFNγ (n = 34) and IL-4 (n = 36) cells from 3 independent experiments. ns, not significant; **p < 0.01, Kruskall-Wallis with Dunn’s multiple comparisons test.
(E) Representative IF images showing GM130 (cyan) and γ tub (magenta) staining in Homeo microglia. Note the presence of Golgi outposts in microglia processes. Hoechst for nuclei visualization, blue. Scale bar, 20 μm (2 μm in zoom images).
(F) Representative confocal images of the time course of the MT renucleation assay after nocodazole washout in Homeo microglia stained for Tyr tub (green), GM130 (gray), and γ tub (magenta). Time 0′ represents the MT depolymerizing effect of nocodazole in homeostatic cells without free tubulin extraction. Note that MTs nucleate from distal Golgi outposts that are positive for gtub. Hoechst for nuclei visualization, blue. Scale bar, 20 μm (5 μm in zoom images).
(G) Left: Representative z-projection confocal images of retinal slices (50 μm thickness) from cx3cr1gfp/+ mice, expressing GFP in microglia cells, stained with GM130 (magenta) to visualize Golgi outposts. Scale bar, 5 μm. Zoom is of a single confocal plane of a microglia ramification stained for GM130 (scale bar, 5 μm). Right: 3D rendering of sample retinal GFP+ microglia (cyan) stained for GM130 (magenta). Scale bar, 10 μm (5 μm in zoom image).
Figure 5. Redistribution of pericentriolar material is a hallmark of activated microglia in vitro and in vivo.
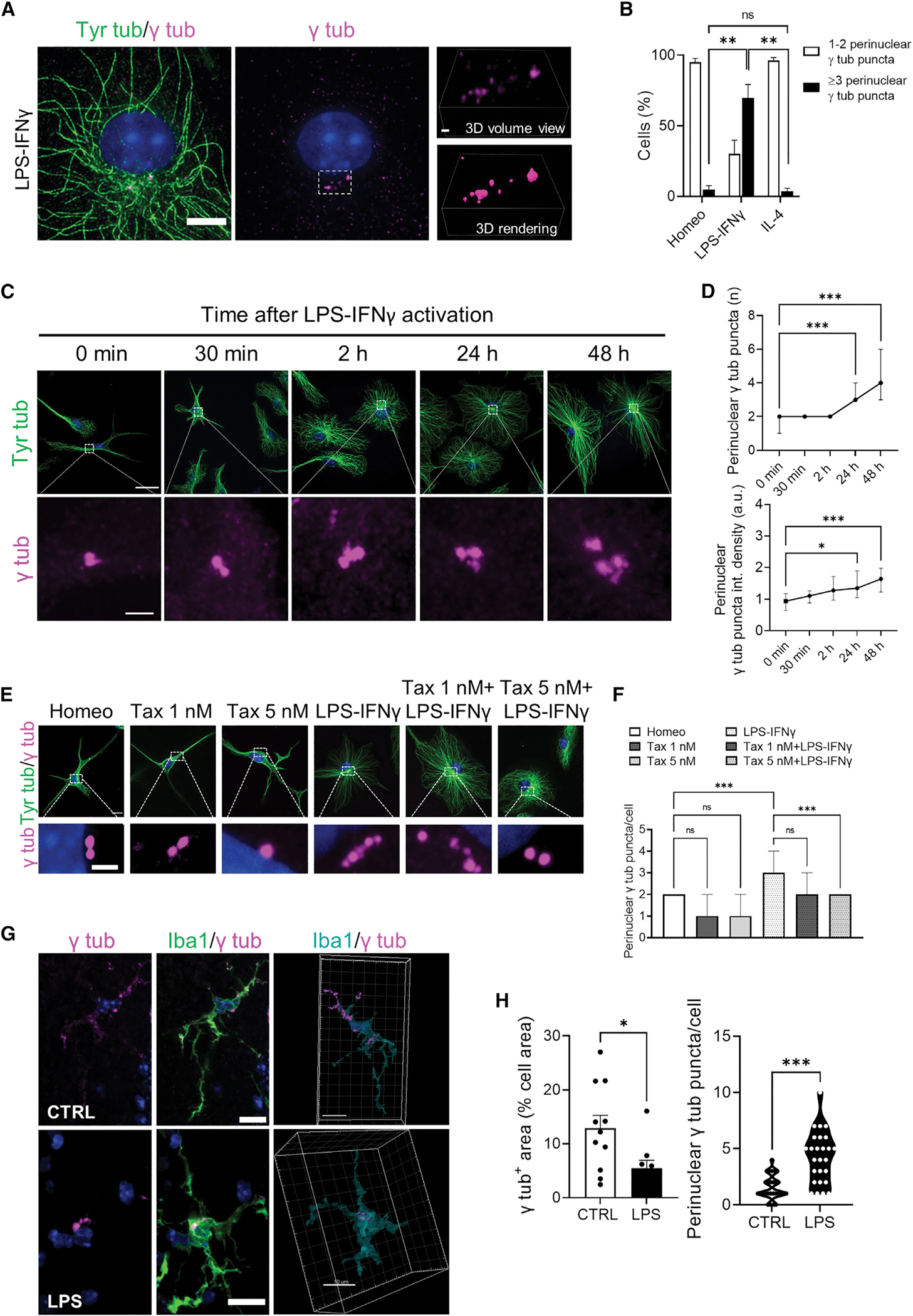
(A) Representative confocal images showing γ-tubulin (γ tub) puncta (magenta) and tyrosinated α-tubulin (left; Tyr Tub, green) immunolabeling in activated (LPS-IFNγ) microglia (middle). Hoechst for nuclei visualization, blue. Scale bar, 5 μm. Right: relative volume view (top) and 3D rendering (bottom) of γ tub puncta (magenta) acquired via structured illumination microscopy. Scale bar 1 μm.
(B) Bar chart reporting the percentage of cells displaying 1–2 γ tub puncta (white bars) or >3 γ tub puncta (black bars) in homeostatic (Homeo), activated (LPS-IFNγ), or alternatively activated (IL-4) microglia. Values are expressed as mean ± SEM from 3 independent experiments. ns, not significant; **p < 0.01, one-way ANOVA with Dunnett’s multiple comparisons test.
(C) Time course of γ tub redistribution during microglia activation: representative IF images showing γ tub (magenta) and tyrosinated α-tubulin (Tyr tub, green) staining at different time points (0 min, 30 min, 2 h, 24 h, and 48 h). Hoechst for nuclei visualization, blue. Scale bar, 20 μm (2 μm in zoom images).
(D) Time course of number of γ tub puncta per cell (top) and quantification of γ tub puncta fluorescence intensity (bottom) during microglia activation. Values are expressed as median ± interquartile range (T0 n = 32; T30 min n = 40; T2h n = 44; T24 h n = 41; T48 h n = 33; from 3 independent experiments). ***p < 0.001, **p < 0.01, *p < 0.05, Kruskal-Wallis with Dunn’s multiple comparisons test with respect to T0.
(E) Representative IF images of tyrosinated Tyr tub (green) and γ tub (magenta) in homeostatic (Homeo), taxol 1 nM treated (Tax 1 nM), Taxol 5 nM treated (Tax 5 nM), activated (LPS-IFNγ), Taxol 1 nM + LPS-IFNγ treated (Tax 1 nM + LPS-IFNγ), and Taxol 5 nM + LPS-IFNγ treated (Tax 5 nM + LPS-IFNγ) microglia. Hoechst for nuclei visualization, blue. Scale bar, 10 μm (2 μm in zoom images).
(F) Bar chart reporting the number of γ tub puncta per cell in Homeo, Tax 1 nM, Tax 5nM, LPS-IFNγ, Tax 1 nM + LPS-IFNγ, and Tax 5 nM + LPS-IFNγ microglia. Values are expressed as median ± interquartile range from 3 independent experiments. ns, not significant; ***p < 0.001, Kruskall-Wallis with Dunn’s multiple comparisons test.
(G) Left: representative maximum intensity projections of images in retinal slices (50 μm thickness) from control (CTRL) and LPS-treated mice stained for γ tub (magenta) and Iba-1 (green) antibodies. Hoechst for nuclei visualization, blue. Scale bar, 10 μm. Right: 3D rendering highlighting the intracellular distribution of γ tub in CTRL (sham) and LPS-treated mice. Scale bars, 10 μm.
(H) Left: scatter dot plot showing the γ tub signal over the cell area of retinal microglia from CTRL (sham) and LPS-treated mice. Values are expressed as mean ± SEM of n = 11/3 cells/mice (CTRL) and n = 9/3 cells/mice (LPS). *p < 0.05, Student’s t test. Right: violin plot showing the number of γ tub puncta per microglia in retinal slices from CTRL and LPS-treated mice of n = 28/3 cells/mice (CTRL) and n = 26/3 cells/mice (LPS). ***p < 0.001, Student’s t test.
Altogether, these data demonstrate that the acquisition of an activated phenotype is characterized by increased localization of γ-tubulin to a pericentrosomal area, which is necessary to establish radial MT arrays. The presence of relatively higher γ-tubulin signal in cell ramifications of homeostatic microglia further suggests that these microglia are alternatively enriched in non-centrosomal MT nucleation sites, which are necessary to establish non-centrosomal bioriented MT arrays.
Golgi outposts can serve as acentrosomal MTOCs in other highly polarized brain cells, such as neurons and oligodendrocytes.37–39 We thus analyzed the distribution of the Golgi marker GM130, a scaffolding protein peripherally associated with Golgi membranes and a marker of Golgi outposts.40 Co-staining of GM130 with Tyr tub demonstrated that the presence of Golgi outposts is a feature of homeostatic microglia and that their presence was dramatically reduced in activated cells (Figures 4C and 4D). Importantly, most of the isolated GM130-positive mini-stacks (73.6%) were decorated by γ-tubulin (Figure 4E). To determine whether Golgi outposts could function as MTOCs in homeostatic microglia, MT nucleation was evaluated in situ by analyzing MT renucleation after nocodazole washout (Figure S2E). We found that both at 15 and 120 min after washout to allow MT regrowth and Golgi reassembly after nocodazole-induced MT depolymerization, MTs (stained with Tyr tub) emerged from Golgi membranes (GM130 positive) both at pericentrosomal sites and at Golgi outposts located far from the centrosome (Figure 4F). Importantly, γ-tubulin was localized to nucleation-competent Golgi outposts, indicating that non-centrosomal MT renucleation did not occur spontaneously at these sites but was dependent on the presence of a γ-tubulin nucleation complex (Figure 4F).
These data demonstrate that in homeostatic microglia, Golgi outposts can function as sites of acentrosomal MT nucleation and suggest that γ-tubulin-dependent non-centrosomal nucleation is necessary to establish an asymmetric MT array in these cells.
To assess whether these in vitro observations were representative of MT nucleation in microglia residing in tissue, we analyzed the subcellular distribution of GM130 in retinal microglia. Retina and brain share a common embryological origin and similar cell types.41–46 Moreover, retinal neurons are arranged in distinct layers, and microglia are usually restricted to the retinal ganglion cell layer, thus offering an accessible structure for the imaging of their MT cytoskeleton. To identify the microglial cytoskeleton in retina we used cx3cr1gfp/+ mice, which constitutively express GFP in microglia. As expected, retinal microglia from control cx3cr1gfp/+ mice displayed a highly ramified morphology (Figure S2F), typical of homeostatic surveillant cells.6,47,48 More importantly, confocal immunofluorescence analysis of GM130 signal in retinal GFP-positive microglia confirmed the presence of isolated Golgi outposts also in the processes of homeostatic microglia residing in tissue (5 ± 1 per cell, n = 11; Figures 4G and S2G; Video S6).
Altogether, these results support the notion that MT organization in homeostatic microglia resembles the MT architecture typical of highly polarized, terminally differentiated cells and strongly suggest that γ-tubulin-dependent non-centrosomal MT nucleation at Golgi outposts is a bona fide feature of homeostatic, surveilling microglia in vitro and in vivo.
Pericentrosomal redistribution of microtubule-nucleating material is a hallmark of activated microglia
The recruitment of PCM to the centrosome has been described as a functional step for macrophage activation upon activating stimuli.49 We thus investigated whether the recruitment of γ-tubulin to a pericentrosomal area was also a hallmark of activated microglia.
As revealed by super-resolution microscopy, activated microglia exhibited multiple γ-tubulin+ puncta that localized to a perinuclear region (Figure 5A). Quantification of the number of γ-tubulin+ puncta indicated that most activated cells had more than three puncta (70% ± 10%; Figure 5B). Conversely, almost all homeostatic and alternatively activated microglia displayed only one or two γ-tubulin+ puncta (95% ± 3% and 96% ± 2%, respectively; Figure 5B). γ-tubulin localization to pericentrosomal puncta showed a time-dependent increase of both number and fluorescence integrated density upon LPS-INFg challenge (Figures 5C and 5D). Notably, in most activated cells with >2 γ-tubulin+ puncta (65% ± 10%, Figure S3A), the centrosomal marker centrin-3 localized only to <2 γ-tubulin+ puncta. In addition, while a quarter of activated cells was proliferating (23% ± 4%; Figures S3C and S3D), most activated microglia displayed >2 γ-tubulin+ puncta (70% ± 10%; Figure 5B) and <2 centrin+ puncta (Figure S3B). Conversely, PCM localization to γ-tubulin+ puncta was confirmed by co-immunostaining with pericentrin (86% ± 5% of co-localizing puncta), a conserved PCM scaffold protein necessary for MTOC assembly and maturation50 (Figure S3E). Indeed, both centrin+ and centrin− γ-tubulin+ puncta localized to the center of MT asters (Figure S3F), and MT regrowth after nocodazole washout (Figures S3G and S3H) revealed that de novo MT nucleation occurred at γ-tubulin+ puncta (Figure S3H).
We found that recruitment of γ-tubulin+ puncta was dependent on a dynamic MT cytoskeleton and was rate limiting for cytokine release: while a low dose of taxol was sufficient to inhibit it (Figures 5E and 5F), hampering PCM maturation with the selective PLK4 inhibitor centrinone51 (Figure S4A) increased the number (4.7-fold; Figure S4C, left) and reduced the diameter (by 30%; Figure S4C, right) of EVs blebbing from the cell surface, with significant inhibition of IL-1b expression (Figures S4B and S4C). Importantly, perinuclear γ-tubulin redistribution was also observed in activated microglia in a mouse model of retinal inflammation. For this, we took advantage of a well-established protocol of acute inflammatory uveitis52–57 induced by intravitreal injection of LPS (Figure S5A) to activate retinal microglia toward the activated phenotype. Microglia residing in retinal slices from LPS-treated mice acquired an ameboid morphology with reduced branching complexity, as revealed by skeleton analysis of Iba1-positive cells (Figures S5B and S5C). Moreover, co-immunolabeling with Iba1 and γ-tubulin demonstrated that while in control (sham) mice retinal microglia displayed punctate diffuse γ-tubulin staining along cellular ramifications (Figure 5G and Video S7), in LPS-treated mice microglia clearly exhibited a condensed γ-tubulin pattern clustered around a perinuclear region (Figure 5G and Video S8). This was confirmed by quantitative analysis of γ-tubulin signal over the cell area and of the number of γ-tubulin+ puncta per cell (Figure 5H).
Altogether, these results demonstrated that: (1) pericentrosomal MTOC maturation is a bona fide feature of activated microglia in vitro and in vivo; (2) γ-tubulin reorganization is dependent on MT dynamics, ascribed to PCM maturation but not centrosome duplication, and regulatory of cytokine release; and (3) de novo generated PCM puncta can act as MTOCs.
DISCUSSION
Here we describe the reorganization of the microglial MT cytoskeleton that characterizes the transitions between homeostatic, activated, and alternatively activated states, and demonstrate the functional interplay between microglial PCM maturation and reactivity. Our findings demonstrate that activated microglia orchestrate a so far unique rearrangement of the MT cytoskeleton, from a non-centrosomal array of parallel and stable MTs nucleated at Golgi outposts characteristic of the homeostatic state to a radial array in which MTs are anchored to de novo formed pericentrosomal MTOCs through their minus ends. Through in vitro phenotyping and in vivo validation, we report four main findings summarized in our graphical abstract. (1) Homeostatic microglia possess stable MT arrays, while microglia reactivity increases MT dynamic behavior. (2) Non-centrosomal MT organization in arrays with mixed polarity is a feature of homeostatic microglia, like the architecture typical of highly specialized cells such as neurons and oligodendrocytes. (3) Microglia activation results in enhanced γ-tubulin localization to puncta around the centrosome and de novo PCM and MTOC maturation, providing a distinct marker of microglia reactivity in live-imaging studies. (4) PCM maturation in activated microglia is inhibited by MT stabilization.
To date, only circumscribed evidence has suggested that ramified microglia possess more acetylated and detyrosinated MTs than ameboid activated cells.14,58 Here, we show that homeostatic ramified microglia display higher levels of tubulin acetylation and detyrosination, two indirect indicators of “older,” i.e., more stable and less dynamic, MT subpopulations.31,59 Moreover, we report that during classical and alternative activation obtained with either LPS-IFNγ or IL-4 stimulation, respectively,60–62 microglia MTs become less stable and more dynamic, suggesting that the acquisition of new cellular functions induces changes in MT stability via modulation of MT dynamics.63
We describe that homeostatic/ramified microglial MTs exhibit an asymmetric dendrite-like organization characterized by EB1 comets arranged in mixed polarity with the minus-end capping protein CAMSAP2 localized at branching points and cell ramifications. In alternatively activated microglia, we find lower levels of CAMSAP2 that localize in a similar fashion in bipolar processes. The presence of CAMSAP2 in microglia processes might suggest that, as in neurons,33,64–68 stabilization of non-centrosomal MTs at their minus ends is important in achieving elongated bipolar morphology, typical of alternatively activated microglia, and for the formation of long-branched cellular extensions patrolling brain parenchyma in homeostatic microglia.
We find that the acquisition of an activated phenotype disrupts the cellular asymmetry of homeostatic microglia and reduces the pool of non-centrosomal, parallel, and mixed oriented MTs, leading to their rearrangement into a radial array of uniformly oriented MTs characteristic of the ameboid shape. In addition, while in activated microglia all the MTs are anchored to a centrosomal region, homeostatic microglia nucleate acentrosomal MTs from Golgi outposts located far from the cell body at the branching points of microglia ramifications, resembling the structure of the dendritic tree of mature neurons33,69,70 or the organization of oligodendrocytic myelin sheaths.38,39 This Golgi outpost-dependent non-centrosomal nucleation contrasts with activated and alternatively activated microglia in which the Golgi apparatus displays a compact perinuclear location,71 suggesting that in reactive microglia cellular arborization is reduced by restricting the Golgi to a region adjacent to the centrosome, which acts as the major MT nucleator in these cells. Indeed, we observed that the acquisition of an activated phenotype is characterized by γ-tubulin redistribution to puncta located in a pericentrosomal region, a feature we confirmed in retinal microglia residing in tissue. Co-localization of γ-tubulin with pericentrin and de novo nucleation of radial MTs from γ-tubulin+ puncta upon nocodazole washout strongly suggests that these protein assemblies are composed of PCM and act as pericentrosomal MTOCs. Importantly, γ-tubulin redistribution during the transition to an activated phenotype did not derive from centriolar duplication during cell division,72,73 as no more than two γ-tubulin+ puncta co-localized with centrin-3 in the same cell, an abundant protein associated with the centrosome.74
We found that PCM maturation occurring during microglia activation was dependent on a dynamic MT cytoskeleton and that its inhibition impacted on the IL-1b secretory pathway. These data suggest that pericentrosomal redistribution of MT nucleating material in microglia may provide a druggable target for modulating cytokine release while providing a valuable live-imaging marker to detect progression of neuroinflammatory disease and efficacy of therapeutics over time.
In summary, we identify a heretofore unique example of MT reorganization from a non-centrosomal array of MTs with mixed polarity to a radial array in which all the MTs are uniformly oriented and anchored either at the centrosome or pericentrosomal MTOCs. Our structural, functional, and tissue analyses further demonstrate that acentrosomal MT nucleation at Golgi outposts may play an important role in supporting the patrolling phenotype of microglia cells, that tubulin remodeling enables microglia reactivity in vitro and in tissue, and that targeting PCM maturation in reactive microglia may represent a valid approach to limit tissue damage during neurodegenerative disease in which microgliosis contributes to neuronal injury and cognitive decline.2,12 In addition, given the recently identified role for a population of spinal CD11c+ microglia in the remission and recurrence of neuropathic pain,75 it will become critical to determine the contribution of spinal microglial MT dysfunction in the peripheral neuropathy caused by chemotherapeutic drugs, most of which target the MT cytoskeleton.
Limitations of the study
We acknowledge that our in vitro system poses several limitations, including having no success in infecting microglia that had been activated with LPS and IFNγ. In addition, given the short duration of the centrinone treatment imposed by our delicate microglia culturing conditions, we could not evaluate whether prolonged loss of PLK4 activity also resulted in inhibition of centrosome duplication and/or additional microglia-specific effects. Clearly, there is potential for additional signaling complexity and regulation during reorganization of the MT array in reactive microglia and in the molecular axis that leads from PCM maturation to cytokine release.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Silvia Di Angelantonio. silvia.diangelantonio@uniroma1.it.
Materials availability
This study did not generate new unique reagents.
Data and code availability
Original western blots, flow cytometry and microscopy raw data reported in this paper will be shared by the lead contact upon request.
The original code has been deposited at Zenodo and is publicly available as of the date of publication at https://doi.org/10.5281/zenodo.7516751.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were housed in standard cages in a group of a maximum of 5 animals, with light–dark cycles of 12 h at 22 ± 2°C. Wild type C57BL-6 male and pregnant mice were purchased from Charles River and pups (P0-P2) were used to obtain primary glial cultures. Cx3cr1gfp/gfp male mice were purchased from The Jackson Laboratory company (B6.129P2(Cg)-Cx3cr1tm1Litt/J); the colony was established in our animal facility, and progenitors were bred to C57BL6J to obtain cx3cr1gfp/+ mice as we previously reported.81
Primary murine microglia culture and treatment
Primary cortical glial cells were prepared from 0- to 2-d-old mice as previously described.82 Briefly, cerebral cortices were chopped and digested in 30 U/mL papain for 40 min at 37°C followed by gentle trituration. The dissociated cells were washed, suspended in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma-Aldrich by Merck KGaA, Darmstadt, Germany) with 10% FBS (Gibco by Life Technologies, Carlsbad, CA, USA) and 2 mM L-glutamine, plated at a density of 9–10 × 105 in 175 cm2 cell culture flasks and cultured for 10–12 days.
Study approval
All procedures performed using laboratory animals were in accordance with the Italian and European guidelines and were approved by the Italian Ministry of Health in accordance with the guidelines on the ethical use of animals from the European Communities Council Directive of September 20, 2010 (2010/63/UE). All efforts were made to minimize suffering and number of animals used.
METHOD DETAILS
Primary murine microglia culture treatment
At confluence (10–12 DIV), glial cells were shaken for 2 h at 37°C to detach and collect microglial cells. These procedures gave an almost pure (<1% astrocyte contamination) microglial cell population. Microglia cells were plated at a density of 7 × 103/cm2 (to prevent cell contact activation) in astrocytes conditioned medium/DMEM 2.5% FBS (1:1). The day after plating microglia cells were treated for 48 h with IFNγ (20 ng/mL) and LPS (100 ng/mL) or with IL-4 (20 ng/mL) to obtain the activated or alternatively activated phenotype, respectively. To disassemble MTs, homeostatic and activated microglia were treated with 20 μm nocodazole (Sigma-Aldrich) added to the culture medium for 1 h at 37°C. Samples were then kept on ice and washed 5x times with ice-cold medium. MTs were allowed to regrow in conditioned medium without nocodazole for 5 min or 15 min and 120 min at 37°C. Right before fixation, free tubulin was rapidly extracted using an MT-preserving extraction buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 0.1% saponin, pH 6.983 for 20 s at 37°C. Cells were subsequently fixed with methanol at −20°C for 4 min and processed for immunofluorescence staining. To stabilize MTs, microglia cells were treated for 24 h with 1 nM and 5 nM Taxol (Sigma-Aldrich) alone or together with IFNγ (20 ng/mL) and LPS (100 ng/mL).
IF staining on fixed cells
Methanol fixation at −20°C was elected for preserving an intact MT cytoskeleton: culture medium was removed and cells were fixed with pre-cooled 100% methanol at −20°C for 4 min prior to re-hydration with Phosphate-buffered saline (PBS, 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride, pH 7.4, at 25°C, Sigma-Aldrich) for at least 30 min at RT To preserve membrane associated components, cells were fixed with 4% paraformaldehyde (PFA)/PBS for 15 min at RT and then washed with PBS. When PFA fixed, cells were permeabilized with 0.1% Triton X-100/PBS for 1 to 3 min. After 2 washes in PBS, cells were blocked with 3% BSA (BSA, Sigma-Aldrich) in PBS for 1 h at RT Primary antibodies (Rabbit Camsap1L1, Novus Biologicals, Englewood, CO, USA, 1:200; rabbit γ-tubulin, Invitrogen, Waltham, MA, USA, 1: 5000; mouse γ-tubulin, Sigma-Aldrich, 1:1000; rat Tyrosinated tubulin YL 1/2, Merck-Millipore, 1:1000; mouse Acetylated tubulin clone 6–11B-1, Sigma-Aldrich, 1:1000; mouse α-tubulin clone DM1A, Sigma-Aldrich, 1:500; rabbit Detyrosinated α-tubulin, Merck-Millipore, 1:1000; mouse EB1, BD Biosciences, San Jose, CA, USA 1:100; rabbit Pericentrin, Abcam, Cambridge, UK, 1:1500; mouse Centrin3, Abnova, Taipei City, Taiwan, 1:100; mouse GM130, BD Biosciences, 1:600; rabbit IBA-1, FujiFilm Wako, Richmond, VA, 1:300; Atto 488 Phalloidin, Sigma-Aldrich, 1:50) were incubated in 1.5% BSA in PBS for 2 h (RT) or overnight (+4°C). Cells were then extensively washed and stained with fluorophore-conjugated secondary antibodies in PBS (Alexa Fluor 488 goat anti-mouse, 488 goat anti-rat, 488 goat anti-rabbit, 594 goat anti-mouse, 647 goat anti-rabbit, Invitrogen; CF 594 goat anti-rat, Sigma-Aldrich; 1:500) and Hoechst (Sigma-Aldrich) for nuclei visualization for 1 h at RT prior to wash and mounting using Ibidi Mounting Medium. Cell outlines shown for display only were obtained by increasing the tubulin signal to identify the cell perimeter and are indicated by white dashed lines.
MT dynamics
For live fluorescence imaging to measure MT dynamics, cells were incubated with 100 nM SiR-Tubulin (SpiroChrome, Stein-amRhein, Switzerland) at 37°C for 30 min and washed with conditioned medium prior to visualization to improve signal to noise ratio. 10 μm Verapamil was added to inhibit efflux pumps and improve labeling. Live wide-field fluorescence imaging of SiRTub-labeled MTs was performed on an Olympus IX73 microscope, LDI laser source and CoolSNAP Myo camera, 4.54 μm pixels (Photometrics, Tucson, AZ, USA) with a built-in incubator, maintaining the temperature at 37°C during recordings. Acquisitions were performed for 4 min (1 frame/4 s) with a UPLSXAPO100x/1.45 oil objective and then analyzed with ImageJ software (see image preparation and analysis).
Lentiviral infection of microglia cells
Production of lentiviral particles was conducted using the second generation packaging system as described in.84 Briefly, HEK293T were transfected with lentiviral DNA plasmid and the packaging vectors pLP1, pLP2, and pLP-VSV-G (Thermo Fisher) using the calcium phosphate transfection method. At 24, 36, and 48 h after transfection, the virus was collected, filtered through 0.45 mm filter, and further concentrated using lentiviral precipitation solution as recommended by the manufacturer (ALSTEM). Concentrated virus was aliquoted and stored at −80°C.
EB3 comets analysis
Homeostatic microglia cells were infected with EB3-EGFP lentivirus the day after plating for 24 h and live-cell imaging of EB3 comets was performed (2 s/frame for 3 min) in astrocytes conditioned medium/DMEM 2.5% FBS (1:1) using an epifluorescence microscope (Nikon Eclipse Ti) equipped with an Orca II ER charge-coupled device (CCD) camera (Hamamatsu, Hamamatsu, Japan) and a temperature-controlled (37°C) CO2 incubator using a 603/1.40NA objective. Kymographs were generated with ImageJ software.
Intravitreal injection and EIU
Adult C57BL6/J mice were intravitreally injected with sterile PBS (vehicle) or 5 ng/mL LPS from E. Coli (O55:B5, Sigma Aldrich). Intravitreal injection of LPS has been previously reported as a model of endotoxin induced uveitis (EIU) activating microglia in the retina.52–54,56,57 Animals were anesthetized with 100 μg/kg methadomidine and 0.25 μg/kg ketamine. Pupils were dilated using 1% tropicamide and 2.5% phenylephrine (Chauvin, Essex, UK) and a small guide hole was made under the limbus with a 30G needle. The eye was gently massaged with a cotton swab to remove a portion of the vitreous to avoid a post-injection reflux of vitreous and/or drug solution. Then, 1 μL of vehicle or LPS solution was intravitreally injected through the initial hole using a 34G Hamilton syringe.
IF staining on retinal tissue
Cx3cr1gfp/+ control mice were sacrificed at P70. CTRL (sham) and LPS intravitreally injected adult C57BL6/J mice were sacrificed 20 h after the injection procedure. Eyes were removed and kept in 4% PFA solution overnight. Eyes were then cryoprotected in 30% sucrose and, after precipitation, frozen in isopentane prior to storage at 80°C. Frozen eyes were cut in 50-μm-thick sections with a Leica cryostat and processed for IF as published.55 Briefly, slices were immersed for 30 min in a boiling 1 mm EDTA solution (pH = 8.0) for antigen retrieval, then incubated with blocking solution (0.1% Triton X-100, 3% BSA and 0.05% Tween 20 in PBS) for 1 h at RT. Sections were incubated with primary antibodies (Iba1, FujiFilm Wako, 1:500; γ-tubulin, clone GTU-88, Sigma-Aldrich, 1:500; GM130, BD bioscience, 1:500) in diluted blocking solution overnight at 4°C and 1 h at RT with fluorophore-conjugated secondary antibodies (Alexa Fluor 488 goat anti-rabbit, 594 goat anti-mouse) and Hoechst for nuclei visualization. The sections were mounted with anti-fade mounting medium (Invitrogen).
Confocal spinning disk and structured illumination (SIM) microscopy
For fluorescence imaging of fixed samples, images were collected with spinning disk confocal microscopy on a Nikon Eclipse Ti equipped with X-Light V2 spinning disk (CrestOptics, Rome, Italy), combined with a VCS (Video Confocal Super resolution) module (CrestOptics) based on structured illumination, and an LDI laser source (89 North, Williston, VT, USA) and Prime BSI Scientific CMOS (sCMOS) camera, 6.5 μm pixels (Photometrics) or a CoolSNAP Myo camera, 4.54 μm pixels (Photometrics), with a 10x/0.25 NA Plan E air objective, 40x/0.75 PlanApo l air objective, a 60x/1.4 PlanApo l oil objective and a 100x/1.45 Plan E oil objective. The used Z step size was 0.2 μm for spinning disk and 0.1 μm for VCS. In order to achieve super-resolution, raw data obtained by the VCS module have been processed with a modified version of the joint Richardson-Lucy (jRL) algorithm,85–87 where the out of focus contribution of the signal has been explicitly added in the image formation model used in the jRL algorithm, and evaluated as a pixel-wise linear “scaled subtraction”88 of the raw signal. Retinal sections images were acquired on an Olympus IX73 microscope equipped with X-Light V3 spinning disk (CrestOptics), LDI laser source and a Prime BSI Scientific CMOS (sCMOS), 6.5 μm pixels (Photometrics) with a UPLSXAPO100x/1.45 oil objective. All the images were acquired by using Metamorph software version 7.10.2. (Molecular Devices, Wokingham, UK) and then analyzed with ImageJ software (see image preparation and analysis).
Image preparation and analysis
For image preparation, we used the open-source software ImageJ77 for adjustments of levels and contrast, maximum intensity projections, and thresholding signals for fluorescence intensity analysis.
Radial profile analysis
For tyrosinated α-tubulin, CAMSAP2 and γ-tubulin distribution analysis, microglia cells were fixed in methanol at −20°C for 4 min or PFA 4% for 15 min and then stained with an anti-tyrosinated tubulin, CAMSAP2 or γ-tubulin antibody according to the immunofluorescence protocol, and Hoechst for nuclei visualization. Images obtained by confocal microscopy were analyzed with ImageJ to identify the coordinates of the center of the nucleus in each cell and to generate single-cell masks based on the morphology of each cell. A Python script (see Supplementals) was written to apply an Otsu threshold to the images89 and to perform a radial scanning of fluorescence values, starting from the center of the nucleus of each cell, with a resolution of 0.065 μm. Maximum value radial profile was defined as the maximum fluorescence intensity (a.u.) for each concentric circle with an increasing distance from the nucleus center. For each analyzed cell the radial profile of the maximum value of fluorescence intensity (a.u.) was computed and plotted. Plots were smoothed with a resolution of 0.5 μm. All data points were exported into a Microsoft Excel 2010 compatible format. In CAMSAP2 analysis, only cytoplasmic staining was analyzed. Curve fit was performed using a single exponential decay function on GraphPad Prism 9.0 (Y=(Y0 − Plateau)*exp(−K*X) + Plateau).
MT dynamics analysis
Analysis of MT dynamics was performed bacing the lengths of the MTs via the “freehand line” tracing tool in ImageJ. Changes in length between successive frames were exported into an Excel sheet to determine the growth, shortening and pause events for each MT. Only changes >0.5 μm were considered growth or shortening events.90,91 MT dynamics parameters were defined as follows: growth/shrinkage rate: distance (mm) covered in growth or shrinkage per second; % pause/growth/shrinkage: number of frames in pause/growth/shrinkage divided total number of frames X 100; catastrophe/rescue frequency (sec-1): number of catastrophe or rescue events divided by the product of the time of analysis and the percentage of growth or shrinkage; MT dynamicity: the sum of total length in growth and shortening divided by the time of analysis.
In vitro cell morphology analysis
Cell morphology analysis was performed using a quantitative measurement of cell area; cell solidity is expressed as the ratio between cell area and convex area. Measurements were obtained with the Particle Analysis tool and images were processed with ImageJ.
Extracellular vesicle analysis
For the statistical analysis of EV blebbing from the surface of microglia, 20 microglia cells per sample, collected in four different areas of the support, were randomly selected and scanned to count and measure the visualized vesicles. The ImageJ software was used to count and measure the vesicle major axis.
IF signal quantification
Cells were selected based on their representative morphology: ramified for homeostatic, ameboid for activated and bipolar for alternatively activated states. Detyr/Tyr tubulin ratio and Acetyl/Tyr tubulin ratio were calculated from the mean gray values of the respective IF signals, obtained from sum slices z-projections of 15 confocal planes after background subtraction (calculated as mean gray value of three circle background areas). EB1 anterograde or retrograde comets were defined from the EB1 fluorescence signal gradient from single plane images, measured with the “plot profile” tool of ImageJ. For Golgi stacks analysis, GM130 maximum intensity z-projection IF images were uniformly thresholded on ImageJ by setting the same minimum values (‘Default’ threshold) to identify single Golgi stacks; a single Golgi stack was defined as a non-round object (roundness <0.9) with a major axis length >0.5 μm. For GM130- γ-tubulin co-staining analysis, GM130 and tubulin signals from max intensity z-projections were uniformly processed among different images increasing the ‘brightness’ and ‘contrast’ parameters by the same percentage. γ-tubulin signal over cell area was calculated as percentage of cell area covered by γ-tubulin signal; γ-tubulin signal threshold was uniformly applied on MetaMorph analysis software by setting the same minimum values to all images. Two or more distinct γ-tubulin+ puncta were identified by counting the peaks of fluorescence intensity on a linescan drawn through the centroid of each puncta using the free-hand tool on ImageJ; the Find Peaks ImageJ plugin was used to identify the peaks by setting the minimum peak amplitude value at 100 gray values. Number of puncta was measured as the number of distinct peaks of fluorescence intensity in the perinuclear region after uniformly thresholding max intensity z-projections (‘Default’ threshold); integrated density was calculated in ImageJ as mean gray value*thresholded area. Pericentrin and Centrin-3− γ-tubulin co-localization analysis was performed by defining Pericentrin and Centrin-3+ puncta as described above for γ-tubulin+ puncta.
Retinal microglia cell skeleton analysis
Morphology of microglia cells in retinal sections was analyzed on max intensity z-projections; only entirely visible cells inside the acquisition field were analyzed; cells were isolated and then skeletonized on binary images, using the dedicated ImageJ plug-in; branches, endpoints and junction number was calculated from the skeletonized image.
Real-time PCR
RNA was extracted from microglia cells with the Quick RNA MiniPrep (Zymo Research, Freiburg, DE) and retrotranscribed with iScript Reverse Transcription Supermix for Real-time PCR (RT-PCR) (Bio-Rad, Hercules, CA, USA). RT-PCR was carried out using Sybr Green (Bio-Rad) according to the manufacturer’s instructions. The PCR protocol consisted of 40 cycles of denaturation at 95°C for 30 s and annealing/extension at 60°C for 30 s. For quantification, the comparative Threshold Cycle (Ct) method was used. The Ct values from each gene were normalized to the Ct value of GAPDH in the same RNA samples. Relative quantification was performed using the 2−ΔΔCt method92 and expressed as fold change in arbitrary values.
Scanning electron microscopy (SEM) analysis
Samples were fixed in a solution of 1.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 h at RT and post-fixed in 1% osmium tetroxide in Milli q (MQ) H2O for 2 h. After several washes in MQ H2O, the samples were subsequently dehydrated in rising concentrations of ethanol in H2O solutions (from 30% to 100%), 1:1 ethanol:hexamethyldisilazane (HMDS, Sigma-Aldrich) and 100% HMDS and dried overnight in air. Finally, the samples were sputtered with a 10 nm gold layer and analyzed using a JEOL JSM-6490LA SEM operating at 10 kV of accelerating voltage.
Western blot
Cells were lysed in Laemmli sample buffer and boiled at 95°C for 5 min. Proteins were separated by 4–12% Bis-Tris gel (Invitrogen) and transferred onto nitrocellulose membrane. After blocking in 5% milk/TBS (Tris 20 mM, NaCl 150 mM), membranes were incubated with primary antibodies at 4°C overnight prior to 1 h incubation with secondary antibodies and signal detected using a commercial chemiluminescent assay (Immun-Star WesternC Kit; Bio-Rad). Image acquisition was performed with ChemiDoc MP imaging system (Bio-Rad) and densitometric analysis was performed with Quantity One software (Bio-Rad).
Cell cycle analysis
Microglia were collected following trypsin treatment. Cells were rinsed twice with phosphate buffered saline (PBS pH 7.4) and collected by centrifugation. Pellets were resuspended in ice-cold 70% ethanol and stored at 4°C for 1 h. Cells were collected by centrifugation, rinsed twice in PBS and resuspended in 20 mg/mL propidium iodide (PI) in PBS with 50 mg/mL RNase A for a minimum of 30 min. After PI incubation, Flow cytometry analysis of DNA content was performed and analyzed using a BD LSRFortessa (BD Biosciences). The percentage of cells in different phases of the cell cycle was determined using the FlowJo V10.7.1 computer software (TreeStar, Ashland, OR, USA). At least 10.000 events for each sample were acquired.
QUANTIFICATION AND STATISTICAL ANALYSIS
The n number for each experiment and details of statistical analyses are described in the figure legends or main text. Data are reported as mean ± SEM; when not normally distributed, data are reported as median ± interquartile range. Origin 6 and GraphPad Prism 9 software were used for statistical analysis. Normality tests were performed with Prism 9 and nonparametric tests were used when appropriate. Significant differences are indicated in the figures by *p < 0.05, **p < 0.01, ***p < 0.001. Notable non-significant differences are indicated in the figures by ns.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Camsap2 (rabbit polyclonal) | Novus Biologicals | Cat# NBP1-21402; RRID: AB_2068823 |
| γ Tubulin (rabbit polyclonal) | Invitrogen | Cat# PA1-28042; RRID: AB_2256789 |
| γ Tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T5326; RRID: AB_532292 |
| Tyr Tubulin (rat monoclonal) | Millipore | Cat# MAB1864; RRID: AB_2128189 |
| Acetylated Tub (Mouse monoclonal) | Sigma-Aldrich | Cat# T7451; RRID: AB_609894 |
| α-Tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T6199; RRID: AB_477583 |
| De-tyr Tubulin (Rabbit polyclonal) | Millipore | Cat# AB3201; RRID: AB_177350 |
| EB1 (Mouse monoclonal) | BD Biosciences | Cat# 610535; RRID: AB_397892 |
| Pericentrin (Rabbit polyclonal) | Abcam | Cat# Ab4448; RRID: AB_304461 |
| Centrin3 (Mouse monoclonal) | Abnova | Cat# H00001070-M01; RRID: AB_304461 |
| GM130 (Mouse monoclonal) | BD Biosciences | Cat# 610822; RRID: AB_398141 |
| IBA1 (Rabbit polyclonal) | FujiFilm | Cat# 019-19741; RRID: AB_839504 |
| Atto488 Phalloidin | Sigma-Aldrich | Cat# 49409 |
| Alexa Fluor 488 goat anti-mouse | Invitrogen | Cat# A11001; RRID: AB_2534069 |
| 488 goat anti-rat | Invitrogen | Cat# A11006; RRID: AB_141373 |
| 488 goat anti-rabbit | Invitrogen | Cat# A11008; RRID: AB_143165 |
| 594 goat anti-mouse | Invitrogen | Cat# A11032; RRID: AB_2534091 |
| 647 goat anti-rabbit | Invitrogen | Cat# A32733; RRID: AB_2633282 |
| CF 594 goat anti-rat | Sigma-Aldrich | Cat# SAB4600111 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| LPS from E.Coli O55:B5 | Sigma-Aldrich | Cat# L5418 |
| LPS from E.Coli O111:B4 | Sigma-Aldrich | Cat# L4391 |
| Murine recombinant IL-4 | PEPROTECH | Cat# 214-14 |
| Murine recombinant IFNγ | PEPROTECH | Cat# 315-05 |
| Taxol | Sigma-Aldrich | Cat# PHL89806 |
| Centrinone (PLK4 inhibitor) | Tocris | Cat# 5687 |
| Nocodazole | Sigma-Aldrich | Cat# 31430-18-9 |
| Hoechst | Sigma-Aldrich | Cat# 23491-45-4 |
| Propidium Iodide | Sigma-Aldrich | Cat# 25535-16-4 |
| Poly-L-lysine hydrobromide | Sigma-Aldrich | Cat# P6282 |
| Hexamethyldisilazane | Sigma-Aldrich | Cat# 999-97-3 |
| Dulbecco’s Modified Eagle’s Medium (DMEM) | Sigma-Aldrich | Cat# D6546 |
| Fetal bovine serum (FBS) | Gibco | Cat# 11573397 |
| Phosphate-Buffered Saline (PBS) tablets | Sigma-Aldrich | Cat# P4417 |
| Saponin | Sigma-Aldrich | Cat# 47036-50G-F |
| Lentiviral precipitation solution | ALSTEM | Cat# VC100 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Quick RNA MiniPrep | Zymo Research, Freiburg, DE | Cat# R1055 |
| iScript Reverse Transcription Supermix | Bio-Rad, Hercules, CA, USA | Cat# 1708840 |
| Sybr Green | Bio-Rad, Hercules, CA, USA | Cat# 172-5124 |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.0 mm, Mini Protein | Invitrogen | Cat# NP0321PK2 |
| SiR-Tubulin Kit | Cytoskeleton | Cat# CY-SC002 |
| Immun-Star Western Chemiluminescence Kit | Bio-Rad | Cat#170-5070 |
|
| ||
| Recombinant DNA | ||
|
| ||
| EB3-EGFP | Pero et al.76 | N/A |
|
| ||
| Deposited data | ||
|
| ||
| Max Intensity Radial Profile | This paper | https://github.com/ggosti/maxIntensityRadialProfile |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | Charles River Laboratories | 027C57BL/6 |
| Mouse: Cx3cr1gfp/gfp | Jackson Laboratories | B6.129P2(Cg)-Cx3cr1tm1Litt/J bred with C57BL6/j |
|
| ||
| Oligonucleotides | ||
|
| ||
| GAPDH forward: TCGTCCCGTAGACAAAAT GG | Sigma-Aldrich | Custom |
| GAPDH reverse: TTGAGGTCAATGAAGGGGTC | Sigma-Aldrich | Custom |
| Ym1 forward: CAGGTCTGGCAATTCTTCTGAA | Sigma-Aldrich | Custom |
| Ym1 reverse: GTCTTGCTCATGTGTGTAAGTGA | Sigma-Aldrich | Custom |
| Fizz1 forward: CCAATCCAGCTAACTATCCCTCC | Sigma-Aldrich | Custom |
| Fizz1 reverse: ACCCAGTAGCAGTCATCCCA | Sigma-Aldrich | Custom |
| Tnfα forward: GTGGAACTGGCAGAAGAG | Sigma-Aldrich | Custom |
| Tnfα reverse: CCATAGAACTGATGAGAGG | Sigma-Aldrich | Custom |
| IL1β forward: GCAACTGTTCCTGAACTCAACT | Sigma-Aldrich | Custom |
| IL1β reverse: ATCTTTTGGGGTCCGTCAACT | Sigma-Aldrich | Custom |
| iNOS forward: ACATCGACCCGTCCACAGTAT | Sigma-Aldrich | Custom |
| iNOS reverse: CAGAGGGGTAGGCTTGTCTC | Sigma-Aldrich | Custom |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism v9.0 | GraphPad Software Inc. | https://www.graphpad.com |
| FIJI/ImageJ 1.53c | Schindelin et al.77 | http://www.imagej.nih.gov/ij |
| ImageJ 1.50i | Wayne Radband, National institute of Health, USA | http://imagej.nih.gov/ij |
| Metamorph 7.10.2 | Molecular Devices | https://www.moleculardevices.com/ |
| Imaris 8.1.2 | Oxford Instruments | https://imaris.oxinst.com/ |
| FlowJo v10.7.1 | TreeStar, Ashland, OR, USA | https://www.flowjo.com/ |
| Quantity One software | Biorad | https://www.bio-rad.com |
| NIS-Elements AR 4.40.00 | Nikon | https://www.microscope.healthcare.nikon.com/ |
| Anaconda3 | Anaconda Inc. | https://www.anaconda.com |
| Python 3.8.5 | Python Software Foundation. | http://www.python.org |
| Numpy 1.19.2 | Harris et al.78 | https://numpy.org |
| Matplotlib 3.3.2 | Hunter,79 | https://matplotlib.org |
| Scikit-image 0.17.2 | Van Der Walt et al.80 | https://scikit-image.org |
|
| ||
| Other | ||
|
| ||
| Killik O.C.T. Compound | Bio-Optica | Cat# 05-9801 |
| μ-Slide 8 Well | Ibidi | Cat# 80826 |
| Ibidi Mounting Medium | Ibidi | Cat# 50001 |
| ProLong Glass Antifade Mountant | Invitrogen | Cat# P36982 |
| Superfrost ultra plus slides | Thermo Scientific | Cat# 10149870 |
Highlights.
Reactive microglia dramatically remodel their microtubule cytoskeleton
Golgi outposts nucleate acentrosomal microtubules in homeostatic microglia
Control of microtubule dynamics enables proper cytokine release in reactive microglia
ACKNOWLEDGMENTS
The authors wish to thank the Animal Facility of Physiology and Pharmacology Department of Sapienza University, the Center for Life Nano- and Neuro-Science Imaging and Cell Sorting Facilities, Istituto Italiano di Tecnologia, and the Taub Institute for Research on Alzheimer’s Disease and the Aging Brain at Columbia University. The graphical abstract was created using BioRender.com. We are grateful to David Sulzer for comments on the manuscript and useful discussions. We thank Gregg G. Gundersen for access to his microscopes and to Alessandro Comincini for sharing the EGFP-EB3-expressing lentivirus for microglia infection. This study was supported by NIH/NIA RF1AG050658 and NIH/NINDS R21 NS120076–01 grants to F.B. and a Fulbright Award to F.B. and S.D.A (FSP-P005556). This research was also funded by the CrestOptics-IIT JointLab for Advanced Microscopy and the D-Tails-IIT JointLab (to S.D.A. and G.C.R.), the Regione Lazio FSE 2014–2020 (19036AP000000019 and A0112E0073) grants (to S.D.A.), by Sapienza University grants (RM118163E0297F84, PH12017270934C3C, and MA32117A7B698029 to S.D.A.), and Fondazione Istituto Italiano di Tecnologia (to M.R. and C.S.). C.S. was also supported by the PhD program in Life Science at Sapienza University in Rome and by H2CU, Honors Center of Italian Universities for housing in New York City. The research leading to these results has been also supported by European Research Council Synergy Grant ASTRA (855923 to G.C.R.). G.G. was supported by Regione Lazio PORFESR “Gruppi di Ricerca 2020” (A0375–2020- 36549).
Footnotes
DECLARATION OF INTERESTS
M.G. is an employee of CrestOptics S.p.A. S.D.A. is member of the scientific advisory board of D-Tails s.r.l.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112104.
REFERENCES
- 1.Bolasco G, Weinhard L, Boissonnet T, Neujahr R, and Gross CT (2018). Three-dimensional nanostructure of an intact microglia cell. Front. Neuroanat. 12, 105. 10.3389/fnana.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colonna M, and Butovsky O (2017). Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468. 10.1146/annurev-immunol-051116-052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nimmerjahn A, Kirchhoff F, and Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 4.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 5.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR, and Gan WB (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609. 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basilico B, Pagani F, Grimaldi A, Cortese B, Di Angelantonio S, Weinhard L, Gross C, Limatola C, Maggi L, and Ragozzino D (2019). Microglia shape presynaptic properties at developing glutamatergic synapses. Glia 67, 53–67. 10.1002/glia.23508. [DOI] [PubMed] [Google Scholar]
- 7.Bernier LP, Bohlen CJ, York EM, Choi HB, Kamyabi A, Dissing-Olesen L, Hefendehl JK, Collins HY, Stevens B, Barres BA, et al. (2019). Nanoscale surveillance of the brain by microglia via cAMP-regulated filopodia. Cell Rep. 27, 2895–2908.e4. 10.1016/j.celrep.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Basilico B, Ferrucci L, Ratano P, Golia MT, Grimaldi A, Rosito M, Ferretti V, Reverte I, Sanchini C, Marrone MC, et al. (2022). Microglia control glutamatergic synapses in the adult mouse hippocampus. Glia 70, 173–195. 10.1002/glia.24101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, and Gan WB (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758. 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 10.Madry C, Kyrargyri V, Arancibia-Cárcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial ramification, surveillance, and interleukin-1β release are regulated by the two-pore domain K+ channel THIK-1. Neuron 97, 299–312.e6. 10.1016/j.neuron.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kettenmann H, Hanisch U-K, Noda M, and Verkhratsky A (2011). Physiology of microglia. Physiol. Rev. 91, 461–553. 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 12.Heneka MT, Kummer MP, and Latz E (2014). Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 14, 463–477. 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 13.Li R, and Gundersen GG (2008). Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9, 860–873. 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 14.Ilschner S, and Brandt R (1996). The transition of microglia to a ramified phenotype is associated with the formation of stable acetylated and detyrosinated microtubules. Glia 18, 129–140. 10.1002/(SICI)1098-1136. [DOI] [PubMed] [Google Scholar]
- 15.Nogales E, Whittaker M, Milligan RA, and Downing KH (1999). High-resolution model of the microtubule. Cell 96, 79–88. 10.1016/S0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 16.Klinman E, and Holzbaur ELF (2018). Walking forward with kinesin. Trends Neurosci. 41, 555–556. 10.1016/j.tins.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Reck-Peterson SL, Redwine WB, Vale RD, and Carter AP (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19, 382–398. 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 19.Gadadhar S, Bodakuntla S, Natarajan K, and Janke C (2017). The tubulin code at a glance. J. Cell Sci. 130, 1347–1353. 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- 20.Janke C, and Magiera MM (2020). The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 21, 307–326. 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- 21.Janke C, and Bulinski JC (2011). Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12, 773–786. 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 22.Etienne-Manneville S (2013). Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 29, 471–499. 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 23.Meiring JCM, and Akhmanova A (2020). Microtubules keep large cells in shape. J. Cell Biol. 219, e202004031. 10.1083/JCB.202004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhmanova A, and Hoogenraad CC (2015). Microtubule minus-end-targeting proteins. Curr. Biol. 25, R162–R171. 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Alieva IB, Berezinskaya T, Borisy GG, and Vorobjev IA (2015). Centrosome nucleates numerous ephemeral microtubules and only few of them participate in the radial array. Cell Biol. Int. 39, 1203–1216. 10.1002/cbin.10492. [DOI] [PubMed] [Google Scholar]
- 26.Vinogradova T, Miller PM, and Kaverina I (2009). Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle 8, 2168–2174. 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartolini F, and Gundersen GG (2006). Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119, 4155–4163. 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 28.Akhmanova A, and Steinmetz MO (2019). Microtubule minus-end regulation at a glance. J. Cell Sci. 132, jcs227850. 10.1242/jcs.227850. [DOI] [PubMed] [Google Scholar]
- 29.Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, and Barres BA (2017). Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94, 759–773.e8. 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herr DR, Yam TYA, Tan WSD, Koh SS, Wong WSF, Ong WY, and Chayaburakul K (2020). Ultrastructural characteristics of DHA-induced pyroptosis. Neuromolecular Med. 22, 293–303. 10.1007/s12017-019-08586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster DR, and Borisy GG (1989). Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 92, 57–65. 10.1242/jcs.92.1.57. [DOI] [PubMed] [Google Scholar]
- 32.Lukinavičius G, Reymond L, D’Este E, Masharina A, Göttfert F, Ta H, Güther A, Fournier M, Rizzo S, Waldmann H, et al. (2014). Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 11, 731–733. 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 33.Yau KW, vanBeuningen SFB, Cunha-Ferreira I, Cloin BMC, van-Battum EY, Will L, Schätzle P, Tas RP, vanKrugten J, Katrukha EA, et al. (2014). Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron 82, 1058–1073. 10.1016/j.neuron.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Bieling P, Kandels-Lewis S, Telley IA, Van Dijk J, Janke C, and Surrey T (2008). CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulinbinding sites. J. Cell Biol. 183, 1223–1233. 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka N, Meng W, Nagae S, and Takeichi M (2012). Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of non-centrosomal microtubules. Proc. Natl. Acad. Sci. USA 109, 20029–20034. 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin M, Veloso A, Wu J, Katrukha EA, and Akhmanova A (2018). Control of endothelial cell polarity and sprouting angiogenesis by noncentrosomal microtubules. Elife 7, e33864. 10.7554/eLife.33864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ori-McKenney KM, Jan LY, and Jan YN (2012). Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76, 921–930. 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu MM, McAlear TS, Nguyen H, Oses-Prieto JA, Valenzuela A, Shi RD, Perrino JJ, Huang TT, Burlingame AL, Bechstedt S, et al. (2019). The Golgi outpost protein TPPP nucleates microtubules and is critical for myelination. Cell 179, 132–146.e14. 10.1016/j.cell.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valenzuela A, Meservey L, Nguyen H, and Fu MM (2020). Golgi outposts nucleate microtubules in cells with specialized shapes. Trends Cell Biol. 30, 792–804. 10.1016/j.tcb.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Wu J, and Akhmanova A (2017). Microtubule-organizing centers. Annu. Rev. Cell Dev. Biol. 33, 51–75. 10.1146/annurev-cellbio-100616-060615. [DOI] [PubMed] [Google Scholar]
- 41.Sernagor E, Eglen SJ, and Wong RO (2001). Development of retinal ganglion cell structure and function. Prog. Retin. Eye Res. 20, 139–174. 10.1016/S1350-9462(00)00024-0. [DOI] [PubMed] [Google Scholar]
- 42.Kavcic V, Vaughn W, and Duffy CJ (2011). Distinct visual motion processing impairments in aging and Alzheimer’s disease. Vis. Res. 51, 386–395. 10.1016/j.visres.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta VB, Chitranshi N, den Haan J, Mirzaei M, You Y, Lim JK, Basavarajappa D, Godinez A, Di Angelantonio S, Sachdev P, et al. (2021). Retinal changes in Alzheimer’s disease- integrated prospects of imaging, functional and molecular advances. Prog. Retin. Eye Res. 82, 100899. 10.1016/j.preteyeres.2020.100899. [DOI] [PubMed] [Google Scholar]
- 44.Javaid FZ, Brenton J, Guo L, and Cordeiro MF (2016). Visual and ocular manifestations of Alzheimer’s disease and their use as biomarkers for diagnosis and progression. Front. Neurol. 7, 55. 10.3389/fneur.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang LYL, Lowe J, Ardiles A, Lim J, Grey AC, Robertson K, Danesh-Meyer H, Palacios AG, and Acosta ML (2014). Alzheimer’s disease in the human eye. Clinical tests that identify ocular and visual information processing deficit as biomarkers. Alzheimers Dement. 10, 251–261. 10.1016/j.jalz.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Bambo MP, Garcia-Martin E, Otin S, Pinilla J, Larrosa JM, Polo V, and Pablo LE (2015). Visual function and retinal nerve fibre layer degeneration in patients with Alzheimer disease: correlations with severity of dementia. Acta Ophthalmol. 93, e507–e508. 10.1111/aos.12635. [DOI] [PubMed] [Google Scholar]
- 47.Cordella F, Sanchini C, Rosito M, Ferrucci L, Pediconi N, Cortese B, Guerrieri F, Pascucci GR, Antonangeli F, Peruzzi G, et al. (2021). Antibiotics treatment modulates microglia–synapses interaction. Cells 10, 2648. 10.3390/cells10102648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimaldi A, Brighi C, Peruzzi G, Ragozzino D, Bonanni V, Limatola C, Ruocco G, and Di Angelantonio S (2018). Inflammation, neurodegeneration and protein aggregation in the retina as ocular biomarkers for Alzheimer’s disease in the 3xTg-AD mouse model. Cell Death Dis. 9, 685. 10.1038/s41419-018-0740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vertii A, Ivshina M, Zimmerman W, Hehnly H, Kant S, and Doxsey S (2016). The centrosome undergoes plk1-independent interphase maturation during inflammation and mediates cytokine release. Dev. Cell 37, 377–386. 10.1016/j.devcel.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman WC, Sillibourne J, Rosa J, and Doxsey SJ (2004). Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657. 10.1091/mbc.e03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang XD, Li W, Zhang S, Wu D, Jiang X, Tan R, Niu X, Wang Q, Wu X, Liu Z, et al. (2020). PLK 4 deubiquitination by Spata2-CYLD suppresses NEK7-mediated NLRP3 inflammasome activation at the centrosome. EMBO J. 39, e102201. 10.15252/embj.2019102201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenbaum JT, Woods A, Kezic J, Planck SR, and Rosenzweig HL (2011). Contrasting ocular effects of local versus systemic endotoxin. Invest. Ophthalmol. Vis. Sci. 52, 6472–6477. 10.1167/iovs.11-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becker MD, Garman K, Whitcup SM, Planck SR, and Rosenbaum JT (2001). Inhibition of leukocyte sticking and infiltration, but not rolling, by antibodies to ICAM-1 and LFA-1 in murine endotoxin-induced uveitis. Invest. Ophthalmol. Vis. Sci. 42, 2563–2566. [PubMed] [Google Scholar]
- 54.Wang M, Ma W, Zhao L, Fariss RN, and Wong WT (2011). Adaptive Müller cell responses to microglial activation mediate neuroprotection and coordinate inflammation in the retina. J. Neuroinflammation 8, 173. 10.1186/1742-2094-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y, Zheng X, Ding Y, Zhou M, Wei Z, Liu T, and Liao K (2020). The molecular chaperone Hsp90a deficiency causes retinal degeneration by disrupting Golgi organization and vesicle transportation in photoreceptors. J. Mol. Cell Biol. 12, 216–229. 10.1093/jmcb/mjz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu CJ, Gardner PJ, Copland DA, Liyanage SE, Gonzalez-Cordero A, Kleine Holthaus SM, Luhmann UFO, Smith AJ, Ali RR, and Dick AD (2016). Multimodal analysis of ocular inflammation using the endotoxin-induced uveitis mouse model. Dis. Model. Mech. 9, 473–481. 10.1242/dmm.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bell OH, Copland DA, Ward A, Nicholson LB, Lange CAK, Chu CJ, and Dick AD (2019). Single eye mRNA-seq reveals normalisation of the retinal microglial transcriptome following acute inflammation. Front. Immunol. 10, 3033. 10.3389/fimmu.2019.03033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Z, Yang L, Yang Y, Chen S, Sun D, Xu H, and Fan X (2018). Epothilone B benefits nigral dopaminergic neurons by attenuating microglia activation in the 6-hydroxydopamine lesion mouse model of Parkinson’s disease. Front. Cell. Neurosci. 12, 324. 10.3389/fncel.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baas PW, Rao AN, Matamoros AJ, and Leo L (2016). Stability properties of neuronal microtubules. Cytoskeleton 73, 442–460. 10.1002/cm.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durafourt BA, Moore CS, Zammit DA, Johnson TA, Zaguia F, Guiot MC, Bar-Or A, and Antel JP (2012). Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia 60, 717–727. 10.1002/glia.22298. [DOI] [PubMed] [Google Scholar]
- 61.Gordon S, and Taylor PR (2005). Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964. 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 62.Perry VH, Nicoll JAR, and Holmes C (2010). Microglia in neurodegenerative disease. Nat. Rev. Neurol. 6, 193–201. 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- 63.Li L, and Yang XJ (2015). Tubulin acetylation: responsible enzymes, biological functions and human diseases. Cell. Mol. Life Sci. 72, 4237–4255. 10.1007/s00018-015-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AFM, Heck AJR, Hoogenraad CC, et al. (2014). Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell 28, 295–309. 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 65.Baas PW, Deitch JS, Black MM, and Banker GA (1988). Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc. Natl. Acad. Sci. USA 85, 8335–8339. 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baas PW, Slaughter T, Brown A, and Black MM (1991). Microtubule dynamics in axons and dendrites. J. Neurosci. Res. 30, 134–153. 10.1002/jnr.490300115. [DOI] [PubMed] [Google Scholar]
- 67.Burton PR, and Paige JL (1981). Polarity of axoplasmic microtubules in the olfactory nerve of the frog. Proc. Natl. Acad. Sci. USA 78, 3269–3273. 10.1073/pnas.78.5.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heidemann SR, Landers JM, and Hamborg MA (1981). Polarity orientation of axonal microtubules. J. Cell Biol. 91, 661–665. 10.1083/jcb.91.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horton AC, Rácz B, Monson EE, Lin AL, Weinberg RJ, and Ehlers MD (2005). Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron 48, 757–771. 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Nguyen MM, McCracken CJ, Milner ES, Goetschius DJ, Weiner AT, Long MK, Michael NL, Munro S, and Rolls MM (2014). γ-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell 25, 2039–2050. 10.1091/mbc.E13-09-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Ahat E, and Wang Y (2019). Golgi structure and function in health, stress, and diseases. Results Probl. Cell Differ. 67, 441–485. 10.1007/978-3-030-23173-6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuller SD, Gowen BE, Reinsch S, Sawyer A, Buendia B, Wepf R, and Karsenti E (1995). The core of the mammalian centriole contains γ-tubulin. Curr. Biol. 5, 1384–1393. 10.1016/S0960-9822(95)00276-4. [DOI] [PubMed] [Google Scholar]
- 73.Alvarado-Kristensson M (2018). Γ-tubulin as a signal-transducing molecule and meshwork with therapeutic potential. Signal Transduct. Target. Ther. 3, 24. 10.1038/s41392-018-0021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Middendorp S, Küntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, and Bornens M (2000). A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148, 405–416. 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kohno K, Shirasaka R, Yoshihara K, Mikuriya S, Tanaka K, Takanami K, Inoue K, Sakamoto H, Ohkawa Y, Masuda T, et al. (2022). A spinal microglia population involved in remitting and relapsing neuropathic pain. Science 376, 86–90. 10.1126/science.abf6805. [DOI] [PubMed] [Google Scholar]
- 76.Pero ME, Meregalli C, Qu X, Shin GJE, Kumar A, Shorey M, Rolls MM, Tanji K, Brannagan TH, Alberti P, et al. (2021). Pathogenic role of delta 2 tubulin in bortezomib-induced peripheral neuropathy. Proc. Natl. Acad. Sci. USA 118, e2012685118. 10.1073/pnas.2012685118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, et al. (2020). Array programming with NumPy. Nature 585, 357–362. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hunter JD (2007). Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 80.Van Der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, and Yu T; scikit-image contributors (2014). Scikit-image: image processing in python. PeerJ 2, e453. 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pagani F, Paolicelli RC, Murana E, Cortese B, Di Angelantonio S, Zurolo E, Guiducci E, Ferreira TA, Garofalo S, Catalano M, et al. (2015). Defective microglial development in the hippocampus of Cx3cr1 deficient mice. Front. Cell. Neurosci. 9, 111. 10.3389/fncel.2015.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosito M, Deflorio C, Limatola C, and Trettel F (2012). CXCL16 orchestrates adenosine A 3 receptor and MCP-1/CCL2 activity to protect neurons from excitotoxic cell death in the CNS. J. Neurosci. 32, 3154–3163. 10.1523/JNEUROSCI.4046-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu X, and Kaverina I (2011). Quantification of asymmetric microtubule nucleation at subcellular structures. Methods Mol. Biol. 777, 235–244. 10.1007/978-1-61779-252-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Pasini S, Shelanski ML, and Greene LA (2014). Activating transcription factor 4 (ATF4) modulates post-synaptic development and dendritic spine morphology. Front. Cell. Neurosci. 8, 177. 10.3389/fncel.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ingaramo M, York AG, Hoogendoorn E, Postma M, Shroff H, and Patterson GH (2014). Richardson-Lucy deconvolution as a general tool for combining images with complementary strengths. Chemphyschem 15, 794–800. 10.1002/cphc.201300831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ströhl F, and Kaminski CF (2015). A joint Richardson-Lucy deconvolution algorithm for the reconstruction of multifocal structured illumination microscopy data. Methods Appl. Fluoresc. 3, 014002. 10.1088/2050-6120/3/1/014002. [DOI] [PubMed] [Google Scholar]
- 87.Chakrova N, Rieger B, and Stallinga S (2016). Deconvolution methods for structured illumination microscopy. J. Opt. Soc. Am. Opt Image Sci. Vis. 33, B12–B20. 10.1364/josaa.33.000b12. [DOI] [PubMed] [Google Scholar]
- 88.Heintzmann R, and Benedetti PA (2006). High-resolution image reconstruction in fluorescence microscopy with patterned excitation. Appl. Opt. 45, 5037–5045. 10.1364/AO.45.005037. [DOI] [PubMed] [Google Scholar]
- 89.Otsu N (1979). A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66. 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 90.Dhamodharan R, and Wadsworth P (1995). Modulation of microtubule dynamic instability in vivo by brain microtubule associated proteins. J. Cell Sci. 108, 1679–1689. 10.1242/jcs.108.4.1679. [DOI] [PubMed] [Google Scholar]
- 91.Rusan NM, Fagerstrom CJ, Yvon AM, and Wadsworth P (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 12, 971–980. 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original western blots, flow cytometry and microscopy raw data reported in this paper will be shared by the lead contact upon request.
The original code has been deposited at Zenodo and is publicly available as of the date of publication at https://doi.org/10.5281/zenodo.7516751.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


