Abstract
Consistent with earlier analyses of human cytomegalovirus UL36 mRNA, we find that the UL36 protein is present throughout infection. In fact, it is delivered to the infected cell as a constituent of the virion. Curiously, much less UL36 protein accumulated in cells infected with the AD169 strain of human cytomegalovirus than in cells infected with the Towne or Toledo strain, and localization of the protein in cells infected with AD169 is strikingly different from that in cell infected with the Towne or Toledo strain. The variation in steady-state level of the proteins results from different stabilities of the proteins. The UL36 proteins from the three viral strains differ by several amino acid substitutions. However, this variability is not responsible for the different half-lives because the AD169 and Towne proteins, which exhibit very different half-lives within infected cells, exhibit the same half-life when introduced into uninfected cells by transfection with expression plasmids. We demonstrate that the UL36 protein is nonessential for growth in cultured cells, and we propose that the ability of the virus to replicate in the absence of UL36 function likely explains the striking strain-specific variation in the half-life and intracellular localization of the protein.
Human cytomegalovirus (HCMV), a betaherpesvirus, typically causes asymptomatic infections in healthy people but can produce life-threatening disease in immunocompromised individuals (reviewed in reference 3). The first set of HCMV genes to be expressed after infection are termed immediate-early genes, and many of their products have been shown to modulate transcription in transient transfection assays (reviewed in reference 10). This activity is consistent with the observation that the early and late classes of HCMV genes are not transcribed after infection in the presence of drugs that prevent expression of immediate-early proteins (reviewed in reference 10).
The HCMV UL36-38 locus encodes four known mRNAs (9, 17). Three of these mRNAs are expressed during the immediate-early phase of the viral growth cycle. One of the immediate-early mRNAs encodes the UL36 protein, one encodes the UL37 protein, and another is bicistronic, producing both the UL38 protein and a polypeptide comprising the first exon of the UL37 coding region when translated in vitro (17). The fourth mRNA from this locus is expressed at early and late times; it encodes the UL38 protein (17). UL37 is a type 1 membrane glycoprotein (1). The intracellular localizations of the other proteins encoded by this locus have not been reported.
Relatively little is known of the functions of the UL36, UL37, and UL38 proteins. The UL36-38 locus is required for efficient transient complementation of DNA replication that is dependent on the HCMV origin of DNA replication (7, 11), but the specific gene product or products encoded by the locus that are responsible for the effect have not been identified. The UL36-38 locus might contribute to DNA replication indirectly, activating the expression of genes directly involved in the process. Transient transfection assays have revealed that the UL36-38 block of coding regions cooperates with other immediate-early viral gene products to activate viral and cellular promoters (5, 7).
We have produced a monoclonal antibody and examined the UL36 protein (pUL36). Consistent with earlier analyses of UL36 mRNA (9, 17), we find that the protein is present throughout infection. However, the half-life and localization of pUL36 differ between cells infected with the AD169 strain of HCMV and cells infected with one of two other HCMV strains, Towne and Toledo. These differences are not intrinsic properties of the proteins since the AD169 and Towne pUL36 exhibit the same half-life when introduced into uninfected cells by transfection. We find that pUL36 is not required for HCMV replication in cultured cells, and we suspect that the variation that we have observed has resulted from genetic drift during passage in the laboratory.
MATERIALS AND METHODS
Biological reagents.
Primary human foreskin fibroblasts (HFFs) between passages 6 and 12 were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum.
Wild-type HCMV strains AD169 (14), Towne (12), and Toledo (13) were used in this study. To construct the AD169 UL36-deficient derivative, ADsubUL36, the HindIII fragments of cosmid pCM1015 (6) were subcloned into pGEM-7Zf(−) (Promega), and then the HindIII-J clone was further subcloned by ligation of the 5-kb fragment generated by BamHI cleavage into pGEM. The resulting clone was digested with SalI; the ends were blunted with the Klenow fragment of DNA polymerase I and then treated with phosphatase. This DNA was joined using ligase to a blunt-ended DNA fragment containing the coding regions for the green fluorescent protein (GFP) and a puromycin resistance marker that were separated by an internal ribosomal entry site. The resulting plasmid contains the HCMV DNA sequences flanking the UL36 coding region surrounding the marker cassette that is situated in a reverse orientation compared to the direction of transcription of the UL36-38 locus. The marker cassette surrounded by UL36 flanking sequences was removed from the plasmid by digestion with FspI plus XbaI and then transfected into HFFs together with AD169 virion DNA plus a pp71 expression vector (2). The transfected cells were plated, puromycin was added 24 h later, and after an additional 24 h during which puromycin-sensitive cells were killed, the drug was removed and fresh fibroblasts were added to regenerate confluent cultures. GFP-positive plaques were isolated, and virus was prepared and used to infect fresh HFF cultures. After three cycles of puromycin selection followed by isolation of GFP-positive plaques, pure clones of mutant virus were obtained, and the structure of the mutant DNA was confirmed by Southern blot assay. To produce virus stocks, HFFs were infected at a multiplicity of 0.01 PFU/cell with mutant or wild-type viruses. After 5 to 7 days, when the cells showed full cytopathic effect, the medium was harvested and cellular debris was removed by centrifugation at 5,000 × g for 20 min at 4°C. The supernatants were stored at −80°C and served as virus stocks which were titered by plaque assay on HFFs. Virus particles were purified by centrifugation through a sorbitol cushion as previously described (15).
Derivatives of pCGN, which fuses a nine-amino-acid epitope derived from the influenza virus hemagglutinin protein to the amino terminus of proteins expressed under control of the HCMV major immediate-early promoter (16), were constructed by inserting PCR-amplified UL36-specific cDNA sequences. The PCR primers incorporated KpnI or BamHI restriction endonuclease cleavage sites into the 5′ and 3′ ends, respectively, of the amplified UL36-specific DNA fragment, facilitating its incorporation into pCGN.
To produce a pUL36-specific monoclonal antibody, the carboxyl-terminal 196-amino-acid coding region of UL36 was cloned into the pGTK bacterial expression vector, and it produced a 45-kDa glutathionine S-transferase fusion protein. After purification, the soluble fusion protein was used to immunize mice. Antibody 5G11, which specifically recognizes the 52-kDa pUL36, was generated. The IE1-specific 1B12 monoclonal antibody was used as previously described (18), and anti-hemagglutinin tag mouse monoclonal 12CA5 (8) was used to visualize epitope-tagged proteins.
Transfections.
HFFs were grown to approximately 90% confluence, released from culture plates by trypsinization, and washed twice with DMEM containing 10% fetal calf serum. The washed cells were resuspended at a concentration of 107 cells/ml in DMEM containing 10% fetal calf serum, and 0.4 ml was added to a electroporation cuvette (4.0-mm electrode gap). Viral (2 μg) and/or plasmid (1 to 15 μg) DNAs were added to the cell suspension and mixed thoroughly. DNA was electroporated into cells (at 260 V and 960 μF) in a Gene Pulser (Bio-Rad).
Assays for gene structure and expression.
For Western blots, cell extracts were prepared by adding buffer containing 100 mM Tris (pH 6.8), 10 mM EDTA, 4% sodium dodecyl sulfate (SDS), 40% glycerol, 5% β-mercaptoethanol, and 0.015% bromophenol blue directly to cells in tissue culture dishes. After electrophoresis in an SDS-containing 8% polyacrylamide gel, proteins were transferred to an Immobilon-P membrane (Millipore) and blocked with 10% dry milk in PBST (phosphate-buffered saline [PBS] containing 0.1% Triton X-100 and 0.05% Tween 20). The immobilized proteins were reacted sequentially with primary mouse monoclonal antibody and secondary horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham). The Amersham ECL (enhanced chemiluminescence) detection system was then used for visualization of protein bands.
For Northern analysis, total cellular RNA was isolated by using the TRIzol Reagent (Gibco-BRL); then 5-μg aliquots of RNA were resolved by electrophoresis in a 1% formaldehyde-agarose gel, transferred to a Nytran membrane (Schleicher & Schuell), and cross-linked to the membrane with UV light. RNA on membranes was probed with DNA labeled by random priming in the presence of [32P]dCTP (Pharmacia).
For Southern blot analysis of virion DNA, purified DNA (8 μg) was digested with restriction enzymes, subjected to electrophoresis in a 0.8% agarose gel, transferred to a membrane, and probed as described for Northern blots.
For pulse-chase analysis of proteins, cells were starved for methionine and cysteine and then labeled for 1.5 h in DMEM lacking methionine and cysteine and supplemented with 2% dialyzed fetal bovine serum plus a mixture of 35S-labeled methionine and cysteine (0.5 mCi/10-cm-diameter plate). A chase was performed after cells were washed and fed with DMEM containing 10% fetal calf serum. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 8]) with Complete protease inhibitors (Boehringer Mannheim). Equal amounts of radioactive protein in 500 μl of RIPA buffer were subjected to immunoprecipitation with a monoclonal antibody followed by capture with protein A-Sepharose (Pharmacia). After three washes with RIPA buffer, proteins were separated by electrophoresis in an SDS-containing 8% polyacrylamide gel, and radioactivity in bands was quantified with a PhosphoImager (Molecular Dynamics).
For immunofluorescent staining, cells growing on cover slips were fixed in PBS containing 4% paraformaldehyde for 10 min at room temperature, permeabilized with PBS containing 0.1% Triton X-100 plus 0.05% Tween 20, and blocked with 1% bovine serum albumin in PBS for 30 min at room temperature. All antibody dilutions were done in PBS containing 0.1% bovine serum albumin. pUL36 was detected with mouse monoclonal antibody 5G11 followed by fluorescein isothiocyanate (FITC)-coupled goat anti-mouse secondary antibody.
Analysis of virion proteins.
Virions were purified and treated with trypsin as described earlier (2). Virion proteins were separated by electrophoresis in an SDS-containing 8% polyacrylamide gel, and pUL36 and pp28 were visualized by Western blotting using monoclonal antibodies.
Viral DNA replication assays.
Cells were infected with wild-type or mutant virus in 35-mm-diameter dishes and were harvested by scraping into cold PBS at various times after infection. These were pelleted, lysed in buffer containing 50 mM Tris (pH 8), 10 mM EDTA, 1% SDS, 100 μg of proteinase K per ml, and 100 μg of RNase A per ml, and incubated for 1 h at 37°C and then overnight at 55°C. A set of samples was also processed with the original amount of virus added for each infection, in order to monitor the amount of input viral DNA, and glycogen was added as carrier. DNA was extracted with phenol-chloroform, ethanol precipitated, and resuspended overnight in 100 μl of buffer containing 10 mM Tris (pH 8) and 1 mM EDTA; 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was added to samples to bring the final concentration to 10× SSC, and DNA was denatured by boiling. A slot blot apparatus (Minifold II; Schleicher & Schuell) was used to apply samples onto a nylon membrane. The membrane was UV cross-linked and then processed with radioactive probe as described above.
RESULTS
Marked differences in UL36 expression and localization among HCMV strains.
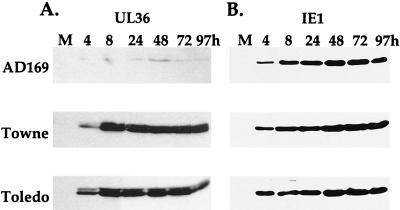
A pUL36-specific monoclonal antibody was produced by using the C-terminal 196-amino-acid segment of the protein as an immunogen, and it was used to characterize the expression of pUL36 in cells infected with three strains of HCMV. AD169 (14) and Towne (12) are laboratory-adapted strains, and Toledo (13) is a clinical isolate that has been passaged in cultured cells to a minimal extent. These viruses were used to infect HFFs at a multiplicity of 3 PFU/cell, and extracts were prepared for Western analysis at various times after infection. Although the three strains of virus were used to infect cells at the same input multiplicity, pUL36 accumulated to markedly lower levels in AD169-infected cells than in cells receiving the Towne or Toledo strain (Fig. 1A). This was not the case for all virus-encoded proteins; AD169 directed the accumulation of the same quantity of IE1 protein as did the other two HCMV strains (Fig. 1B). The pUL36-specific antibody recognized a doublet of proteins migrating at the predicted molecular mass for the viral protein, 52 kDa. The doublet was evident in cells infected with the Towne and Toledo strains (Fig. 1A), and it was also seen in the AD169-infected cell sample when longer exposures of the autoradiogram were examined (data not shown). Given their size and the fact that both bands are lost when a mutant virus lacking the UL36 gene is tested (see below), we conclude that both bands contain pUL36. The doublet could result from posttranslational modification of the protein. For all three HCMV strains, pUL36 was evident at 4 h after infection, the earliest time assayed (Fig. 1A), consistent with the earlier demonstration that its mRNA can be detected during the immediate-early phase of AD169 infection (9, 17).
FIG. 1.
Lower levels of accumulation of pUL36 in AD169-infected HFFs than in Towne- or Toledo-infected HFFs. Extracts were prepared at the indicated times after infection, and pUL36 and IE1 proteins were visualized by Western blotting. M, mock-infected cells.
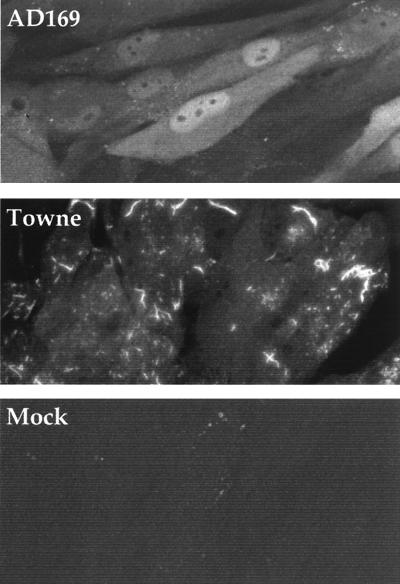
We also examined the intracellular localization of pUL36 by using the monoclonal antibody (Fig. 2). HFFs were assayed at 24 h after infection or mock infection. Little fluorescent signal was evident in mock-infected cells, and AD169-infected cells displayed uniform nuclear as well as cytoplasmic staining with occasional intense fluorescent spots in the cytoplasm. In marked contrast, Towne-infected cells exhibited significantly less nuclear fluorescence. Rather, the cytoplasm of Towne-infected cells contained intensely fluorescent spots and worm-shaped structures. The fluorescent analysis was repeated at various times after infection (data not shown). Towne does not exhibit a localization similar to that observed for AD169 during an earlier phase of infection, and AD169-infected cells do not eventually accumulate the worm-like structures observed for Towne. The Toledo strain was also tested, and it exhibited a fluorescent pattern similar to that seen for Towne (data not shown). We have attempted to colocalize this structure with several organelle markers. The pUL36 Towne structure does not colocalize with the Golgi apparatus, lysosomes, mitochondria, recycling and early endosomes, endoplasmic reticulum, or death effector filaments (data not shown). Consistent with these findings, treatment of Towne-infected cells with brefeldin A does not change the pUL36 structure (data not shown).
FIG. 2.
Difference in localization of pUL36 in AD169- compared to Towne-infected HFFs. Immunofluorescent staining was done with a pUL36 monoclonal antibody plus an anti-mouse, FITC-conjugated secondary antibody.
Different half-lives for pUL36 in AD169-infected compared to Towne-infected cells.
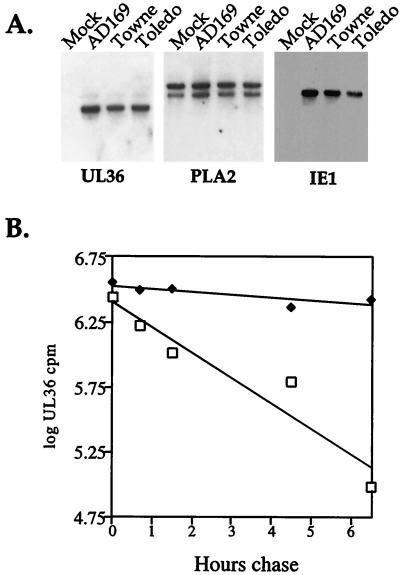
To ascertain whether the HCMV strain-specific differences in the level of pUL36 might result from differences in mRNA accumulation, we performed a Northern blot analysis. The three virus strains produced similar amounts of UL36 mRNA at 24 h after infection (Fig. 3A), a time at which the protein encoded by the mRNA has accumulated to much lower levels in AD169-infected cells than in Towne- or Toledo-infected cells (Fig. 1A). Duplicate RNA samples were also probed for a cellular mRNA (phospholipase A2 [19]) as a loading control and for IE1 mRNA to monitor the virus infection (Fig. 3A).
FIG. 3.
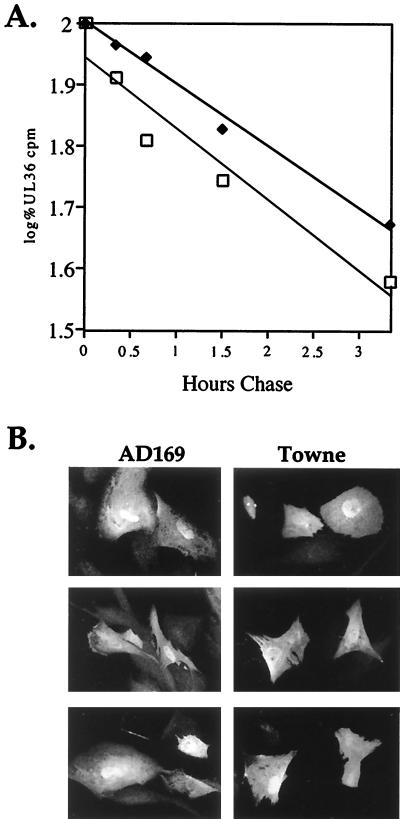
Stability of pUL36 varies among HCMV strains. (A) Northern blot demonstrating the UL36-specific RNA accumulates to similar levels in AD169-, Towne-, and Toledo-infected HFFs. Total cellular RNA was prepared at 24 h after infection, and identical blots were probed with 32P-labeled plasmid DNA containing viral UL36 or IE1 sequences or, as a loading control, cellular cytoplasmic phospholipase A2 (PLA2) sequence. (B) Pulse-chase analysis of pUL36 expressed from AD169 (□)- and Towne (⧫)-infected HFFs. At 24 h after infection, cells were labeled with [35S]methionine-cysteine for 1.5 h and then chased in unlabeled medium for various time periods. pUL36 was immunoprecipitated from cell lysates and subjected to electrophoresis, and radioactivity in the pUL36-specific bands was quantified with a PhosphoImager.
Finding no difference in mRNA accumulation, we next examined the stability of pUL36 in cells infected with the AD169 and Towne strains of HCMV. A pulse-chase experiment was performed at 24 h after infection of HFFs (Fig. 3B). 35S-labeled protein was immunoprecipitated and subjected to electrophoresis, and pUL36-specific radioactivity was quantified. The two pUL36-specific bands comprising the 52-kDa doublet exhibited the same stability. The half life for pUL36 in AD169-infected cells was 1.5 h. In contrast, the pUL36 half life was 13.5 h in Towne-infected cells. In a second independent experiment (data not shown), the half-lives of pUL36 were determined to be 1.5 and 10.5 h in cells infected with AD169 and Towne, respectively. Thus, the different steady-state levels of pUL36 result from differences in its stability.
Different half-lives are not intrinsic properties of pUL36 encoded by different strains of HCMV.
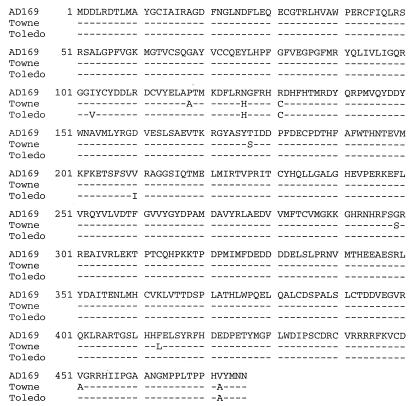
UL36-specific cDNAs were prepared by reverse transcription and PCR amplification, and two independently produced clones for each strain were sequenced. The protein encoded by Towne has eight single amino acid substitutions compared to the AD169 protein, and the protein encoded by Toledo has five substitutions (Fig. 4). The Towne and Toledo proteins share three of the amino acid differences in comparison to AD169 pUL36, and it seemed possible that one or more of these substitutions is responsible for the altered stability and localization of pUL36 observed for Towne and Toledo versus AD169. To test this supposition, HFFs were transfected with plasmids expressing the pUL36 variants. Pulse-chase analysis revealed that pUL36 from AD169 and Towne decayed at similar rates, exhibiting half-lives of 2.6 and 3.2 h, respectively (Fig. 5A). We also examined the localization of pUL36 variants in transfected cells by immunofluorescence, and both displayed uniform nuclear and cytoplasmic staining (Fig. 5B).
FIG. 4.
Sequence comparison of HCMV UL36 open reading frame translated from sequenced cDNAs. Reverse transcription-PCR was used to amplify UL36 from AD169-, Towne-, and Toledo-infected cellular RNAs. Two independently isolated clones were sequenced, and the translated open reading frames are shown. Identical matches of Towne and Toledo to AD169 are shown by dashes, and amino acid changes are represented in the single-letter code.
FIG. 5.
The half-life and localization of AD169 and Towne UL36 proteins are similar when expressed in uninfected HFFs. (A) Half-lives of AD169 (□) and Towne (⧫) proteins, determined 48 h after transfection. Cells were labeled with [35S]methionine-cysteine for 1.5 h and then chased in unlabeled medium for various time periods. pUL36 was immunoprecipitated from cell extracts and subjected to electrophoresis, and the radioactivity in the pUL36-specific bands was quantified. (B) Intracellular localization of AD169 and Towne proteins expressed in uninfected HFFs, determined 48 h after transfection. Immunofluorescent staining was done with a pUL36 monoclonal antibody plus an anti-mouse, FITC-conjugated secondary antibody.
We conclude that the differences in half-life and localization for UL36 observed in cells infected with different strains of HCMV are not intrinsic properties of the proteins. The differences are observed only in infected cells; variations in stability and intracellular location require the activity of additional viral gene products.
UL36 is not essential for HCMV growth in cultured cells.
One explanation for the marked variability in the half-life and localization of pUL36 in cells infected by different HCMV strains could be that the protein is not required and does not function during replication of the virus in cultured cells. To test this possibility, we constructed a mutant derivative of AD169. We replaced the UL36 coding region with a DNA segment expressing GFP and a puromycin resistance marker under control of the simian virus 40 early promoter to produce a mutant termed ADsubUL36 (Fig. 6A).
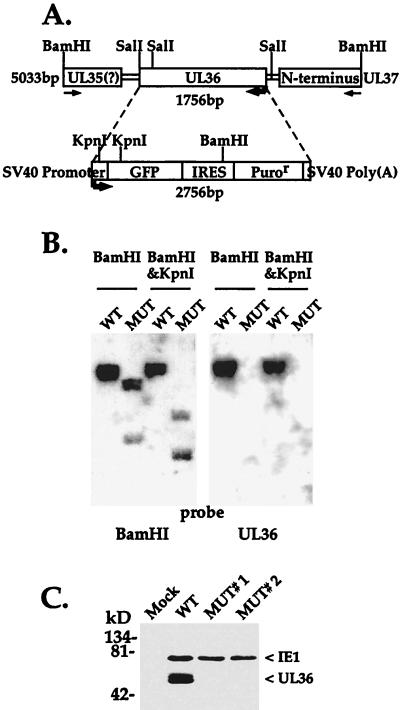
FIG. 6.
ADsubUL36 lacks the UL36 coding region. (A) Diagrammatic representation of the UL36 locus on the HCMV chromosome and the puromycin resistance (Puror) marker cassette used to generate a substitution mutation. The mutant lacks 1,755 bp located between HCMV positions 48082 and 49837. SV40, simian virus 40; IRES, internal ribosomal entry site. (B) Southern blot analysis of BamHI-plus-KpnI-cleaved wild-type AD169 (WT) and ADsubUL36 (MUT) DNAs. The blot shown on the left was probed with a DNA segment encompassing 5 kb (including the UL36 coding region) corresponding to HCMV positions 46434 to 51468; the blot on the right was probed with a UL36 cDNA as a probe. (C) Western blot analysis of pUL36 expression at 24 h after infection of HFFs with wild-type or mutant virus. As a control, IE1 expression was also monitored. The position at which marker proteins migrated are indicated to the left. Two independent isolates of ADsubUL36, MUT#1 and MUT#2, were tested.
Southern analysis of virion DNA cut with BamHI and KpnI confirmed that the UL36 coding region was replaced with the GFP-puromycin resistance cassette in the mutant virus. Wild-type and ADsubUL36 DNAs were distinguished by two different probes. One probe, derived from the region adjacent to and containing the UL36 locus, identified the altered restriction enzyme-generated fragments in the mutant predicted for the substitution (Fig. 6B, left); the other probe, corresponding to the UL36 cDNA, detected a UL36-specific band in the digest for wild-type but not mutant DNA (Fig. 6B, right). The mutant virus was also tested for expression of pUL36 to be certain that a functional copy of the gene had not been inserted by an aberrant recombination event at a new location within the mutant genome (Fig. 6C). Whereas an extract prepared at 24 h after infection at a multiplicity of 3 PFU/cell with wild-type virus contained the protein, no pUL36 was detected in extracts of cells infected with two independent isolates of the mutant. The extracts from mutant-infected cells contained normal amounts of IE1 proteins, confirming that the cells were infected.
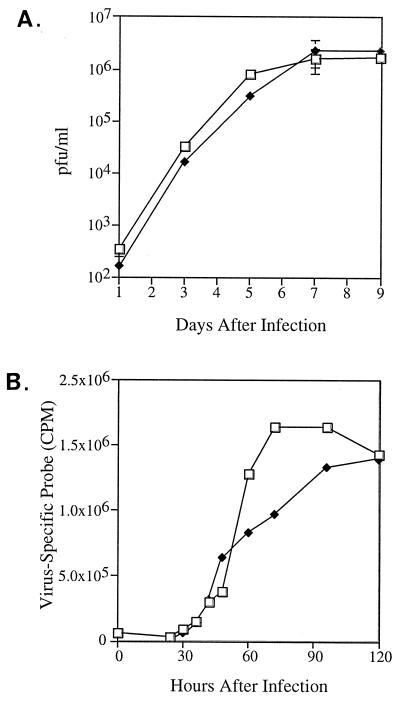
The mutant virus was successfully propagated without supplying UL36 function to generate virus stocks with infectious titers similar to wild-type virus. To more carefully evaluate the growth characteristics of ADsubUL36, HFFs were infected at an input multiplicity of 1, and the production of progeny virus was monitored at intervals for the next 9 days. The growth kinetics observed for mutant and wild-type viruses were indistinguishable (Fig. 7A), demonstrating that pUL36 is dispensable for growth of HCMV in HFFs. The accumulation of viral DNA was also monitored in cells infected with wild-type versus ADsubUL36 (Fig. 7B), and no differences greater than twofold were observed. These results also were obtained for cells infected at a multiplicity of 0.1 or 0.01 (data not shown).
FIG. 7.
Similar growth and viral DNA replication kinetics of AD169 (□) and ADsubUL36 (⧫). (A) Growth kinetics of wild-type and mutant virus on HFFs infected at a multiplicity of 1 PFU/ml. Cultures were harvested at various times after infection, and infectious virus was quantitated by plaque assay on HFFs. (B) Accumulation of wild-type and mutant DNA in HFFs infected at a multiplicity of 1 PFU/ml. Cells were harvested at various times after infection, and viral DNA was quantified by slot blot analysis using an HCMV-specific DNA probe.
pUL36 is packaged with AD169 and Towne virions.
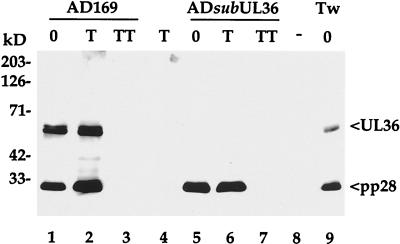
Western blot assay revealed that pUL36 is associated with purified HCMV virions, and a similar amount of the protein was found in preparations of AD169 compared to Towne particles in spite of the substantial differences in pUL36 accumulation and localization within cells infected with these two strains (Fig. 8). To determine whether pUL36 is likely located inside of the virion envelope, intact and detergent disrupted virions were treated with trypsin. Whereas pUL36 was degraded in the preparation of disrupted virions (lane 3), it was resistant to proteinase treatment of intact virions (lane 2), consistent with the view that pUL36 is packaged within virus particles. The mutant virus, ADsubUL36, lacks pUL36 but contains similar amounts of another virion protein, pp28, as the wild-type AD169.
FIG. 8.
pUL36 is present in wild-type particles. Purified AD169, ADsubUL36, and Towne (Tw) virions were incubated alone (0), with trypsin (T), or with trypsin and Triton X-100 (TT). Virions were then solubilized in SDS-containing buffer, and proteins were analyzed by Western blotting using a monoclonal antibody to pUL36, and, as a control, a monoclonal antibody to pp28. The positions at which marker proteins migrated are indicated to the left.
DISCUSSION
As predicted by earlier studies demonstrating that UL36 encodes an immediate-early mRNA (9, 17), pUL36 can be detected at 4 h after HCMV infection and then continues to be present in infected cells through the early and late phases of infection (Fig. 1A). The protein consistently migrates as a doublet in SDS-containing polyacrylamide gels (Fig. 1A and 6C). Since there is no evidence for alternatively spliced versions of UL36 mRNA and a cloned cDNA directs the synthesis of both species, it seems likely that the protein is posttranslationally modified, but we have not identified the nature of the presumptive modification. HCMV strain AD169-infected cells contain much less pUL36 than cells infected with the Towne or Toledo strain (Fig. 1A). The steady-state levels of the protein are different because the half-life of the protein is shorter in cells infected with AD169 compared to Towne (Fig. 3B). Even though the primary sequence of pUL36 differs among HCMV strains (Fig. 4), the difference in stability is not an intrinsic property of pUL36. When pUL36 was introduced into cells by transfection of expression plasmids, the half-lives for the AD169 and Towne proteins were very similar (Fig. 5). The difference in half-life is evident only in infected cells. Possibly, the differences in the pUL36 sequence from various HCMV strains cause the protein to interact differently with another viral protein or with a cell protein that is induced or modified by viral infection. The differences in such an interaction could, in turn, modulate the stability of the protein. Alternatively, there might be a mutation at a second locus in the viral genome that causes another viral gene product to interact with pUL36 differently, irrespectively of the alterations that we have observed.
In addition to changes in half-life, there are significant differences in the localization of pUL36 within AD169 compared to Towne- or Toledo-infected cells (Fig. 2). We have not been able to colocalize the striking worm-like structures evident in Towne-infected cells with known markers of the endoplasmic reticulum, Golgi apparatus, lysosomes, endosomes, mitochondria, or death effector filaments. We would guess that the localization is influenced by the same event that leads to altered half-lives, but we cannot be certain until the modulatory factor has been identified.
An HCMV mutant (Fig. 6) unable to express pUL36 proved to be viable, growing with the same kinetics as its wild-type parent (Fig. 7). This observation is consistent with an earlier report that the murine cytomegalovirus ie2 gene is dispensable for viral replication in cultured mouse cells (4), since the first exon of the HCMV UL36 coding region exhibits homology to the ie2 gene encoded by the murine virus. UL36 is a member of the UL22 gene family, a group of 13 HCMV coding regions predicted to encode proteins with several shared motifs (4). Conceivably, another member of the UL22 family performs the same or a similar function and substitutes for pUL36. Alternatively, pUL36 might execute a function that is needed for efficient viral replication within its infected host, the human, but not for replication in cultured cells. Whatever its role, pUL36 might begin to act immediately after infection before the viral genome becomes transcriptionally active since the protein is packaged in virions (Fig. 8).
The lack of an essential role for pUL36 in cultured cells might explain why the AD169-coded protein has a reduced half-life and altered localization compared to the protein in cells infected with the Towne or Toledo strain. Although we cannot be certain, we suspect that the longer half-life and predominantly cytoplasmic localization in worm-like structures seen for the Towne and Toledo strains represent the wild-type phenotype for UL36 since the Toledo strain has not been extensively passaged in the laboratory. We propose that one or more mutations have arisen in the AD169 strain as a consequence of genetic drift and are responsible for the altered pUL36 accumulation and localization.
ACKNOWLEDGMENTS
We thank P. Schaffer for advice on the construction of mutant herpesviruses, H. Zhu for antibody to HCMV IE1 protein, and G. Tullis for plasmid constructs. We also thank M. Marlow and J. Goodhouse for assistance with the production of monoclonal antibodies and confocal microscopy.
C.E.P. was supported by a fellowship from the American Heart Association, and T.S. is an American Cancer Society Professor and an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Al-Barazi H O, Colberg-Poley A M. The human cytomegalovirus UL37 immediate-early regulatory protein is an integral membrane glycoprotein which traffics through the endoplasmic reticulum and Golgi apparatus. J Virol. 1996;70:7198–7208. doi: 10.1128/jvi.70.10.7198-7208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 4.Cardin R D, Abenes G B, Stoddart C A, Mocarski E S. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology. 1995;209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 5.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 7.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodziej P A, Young R A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 10.Mocarski E S J. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 11.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plotkin S A, Furukawa T, Zygraich N, Huygelen C. Candidate cytomegalovirus strain for human vaccination. Infect Immun. 1975;12:521–527. doi: 10.1128/iai.12.3.521-527.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinnan G V, Jr, Delery M, Rook A H, Frederick W R, Epstein J S, Manischewitz J F, Jackson L, Ramsey K M, Mittal K, Plotkin S A, et al. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann Intern Med. 1984;101:478–483. doi: 10.7326/0003-4819-101-4-478. [DOI] [PubMed] [Google Scholar]
- 14.Rowe W P, Hartley J W, Waterman S, Turner H C, Huebner R J. Cytopathic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc Soc Exp Biol Med. 1956;92:418–424. [PubMed] [Google Scholar]
- 15.Stinski M F. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J Virol. 1976;19:594–609. doi: 10.1128/jvi.19.2.594-609.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 17.Tenney D J, Colberg P A. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zupan L A, Steffens D L, Berry C A, Landt M, Gross R W. Cloning and expression of a human 14-3-3 protein mediating phospholipolysis; identification of an arachidonoyl-enzyme intermediate during catalysis. J Biol Chem. 1992;267:8707–8710. [PubMed] [Google Scholar]