Abstract
Objective
Therapy-induced tumour microenvironment (TME) remodelling poses a major hurdle for cancer cure. As the majority of patients with hepatocellular carcinoma (HCC) exhibits primary or acquired resistance to antiprogrammed cell death (ligand)-1 (anti-PD-[L]1) therapies, we aimed to investigate the mechanisms underlying tumour adaptation to immune-checkpoint targeting.
Design
Two immunotherapy-resistant HCC models were generated by serial orthotopic implantation of HCC cells through anti-PD-L1-treated syngeneic, immunocompetent mice and interrogated by single-cell RNA sequencing (scRNA-seq), genomic and immune profiling. Key signalling pathway was investigated by lentiviral-mediated knockdown and pharmacological inhibition, and further verified by scRNA-seq analysis of HCC tumour biopsies from a phase II trial of pembrolizumab (NCT03419481).
Results
Anti-PD-L1-resistant tumours grew >10-fold larger than parental tumours in immunocompetent but not immunocompromised mice without overt genetic changes, which were accompanied by intratumoral accumulation of myeloid-derived suppressor cells (MDSC), cytotoxic to exhausted CD8+ T cell conversion and exclusion. Mechanistically, tumour cell-intrinsic upregulation of peroxisome proliferator-activated receptor-gamma (PPARγ) transcriptionally activated vascular endothelial growth factor-A (VEGF-A) production to drive MDSC expansion and CD8+ T cell dysfunction. A selective PPARγ antagonist triggered an immune suppressive-to-stimulatory TME conversion and resensitised tumours to anti-PD-L1 therapy in orthotopic and spontaneous HCC models. Importantly, 40% (6/15) of patients with HCC resistant to pembrolizumab exhibited tumorous PPARγ induction. Moreover, higher baseline PPARγ expression was associated with poorer survival of anti-PD-(L)1-treated patients in multiple cancer types.
Conclusion
We uncover an adaptive transcriptional programme by which tumour cells evade immune-checkpoint targeting via PPARγ/VEGF-A-mediated TME immunosuppression, thus providing a strategy for counteracting immunotherapeutic resistance in HCC.
Keywords: CANCER IMMUNOBIOLOGY, HEPATOCELLULAR CARCINOMA, IMMUNOTHERAPY, PPAR GAMMA
WHAT IS ALREADY KNOWN ON THIS TOPIC
Adaptive immune resistance has emerged as a basis for therapeutic evasion in cancer.
Despite breakthroughs in immune-checkpoint blockade (ICB) therapies for advanced hepatocellular carcinoma (HCC), most patients still succumb to the primary or acquired resistance.
ICB-resistant HCC has been primarily associated with an immunosuppressive tumour microenvironment (TME), characterised by myeloid-derived suppressor cell (MDSC) expansion and CD8+ T cell dysfunction.
A better understanding of how cancer cells adapt to ICB-induced immune attack may present new opportunities to avert ICB resistance.
WHAT THIS STUDY ADDS
On ICB-induced immune attack, HCC cells harness transcriptional adaptation—rather than genetic selection—to acquire immune evasion capacities.
Tumour cell-intrinsic peroxisome proliferator-activated receptor-gamma (PPARγ) upregulation orchestrates an MDSC-enriched and T cell-dysfunctional TME through vascular endothelial growth factor-A (VEGF-A) trans-activation.
Genetic or pharmacological inhibition of PPARγ augments antitumour immunity and overcomes ICB resistance in multiple immune-cold HCC models.
PPARγ upregulation correlates with poor response in cancer patients undergoing ICB monotherapies.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
We established two mouse models of adaptive ICB resistance that recapitulate the immune landscape of human ‘cold’ HCC and enable identification of actionable targets to improve ICB response.
Our studies provide mechanistic insights into how targeting an upstream driver of VEGF augments antitumour immunity and ICB efficacy.
Our findings may be of great value to accelerate clinical development of PPARγ inhibitors for combined immunotherapy in HCC and other PPARγ-expressing malignancies.
Introduction
Immune-checkpoint blockade (ICB) therapies have transformed the treatment landscapes of solid malignancies including hepatocellular carcinoma (HCC), which is currently the sixth most common cancer and the third leading cause of cancer death worldwide.1 However, the strong immunosuppressive tumour microenvironment (TME) prohibits sufficient cytotoxic T lymphocyte infiltration, thus restricting the ICB responsiveness to a minority of patients with HCC.2–4 Notably, a recent front-line phase III IMbrave150 trial of combination ICB therapy by atezolizumab, a programmed death ligand 1 (PD-L1) inhibitor, and bevacizumab, a monoclonal antibody targeting the vascular endothelial growth factor (VEGF), has improved the objective response rate (ORR) to 27%,5 which could be attributable to antiangiogenic immune modulation of the TME.6 Despite this breakthrough, a high proportion of patients with HCC still do not benefit from this new standard of care due to primary or acquired resistance.7
Adaptive resistance mediated by the upregulation of PD-L1 in cancer cells, and the resulting T cell dysfunction in the TME, is a classical tumour immune evasion mechanism.8 During cancer progression, diverse cancer cell plasticity programmes result from microenvironmental cues, stochastic genetic and epigenetic alterations, thereby contributing to tumour heterogeneity and therapeutic resistance.9 10 Given the evolving nature of the tumour-immune interactions, the tumour adaptive immune responses could clinically manifest as primary or acquired resistance to PD-L1/programmed cell death protein 1 (PD-1) blockade.11 However, our understanding of how cancer cells adapt to ICB-induced immune attack remains incomplete, particularly in HCC TME.
Single-cell RNA sequencing (scRNA-seq) is one approach for dissecting tumour heterogeneity, cellular and molecular wiring of TME, and cell state transitions that may affect therapeutic responses, particularly in the context of cancer immunotherapy.12 There is currently a paucity of single-cell analyses in preclinical models and clinical studies involving HCC. Prior scRNA-seq studies of human HCC largely focused on the blood samples or treatment-naïve/pretreatment biopsies,13–15 due in part to challenges associated with obtaining high-quality HCC biopsies at multiple treatment time points. As the unique milieu of the TME can have a significant impact on response to therapy, the commonly used subcutaneous syngeneic graft models do not reflect the complex organ-specific aspects of tumour development that also influence immunotherapeutic response.16 While genetically engineered mouse models may better approximate the TME, they tend to have low mutation burden that may limit their translational potential. Advances in modelling the dynamic tumour-immune interactions will be critical to understanding mechanisms of ICB resistance.16
Here, we address the mechanisms of ICB therapy-induced tumour adaptation by serial orthotopic implantation of mouse HCC cells through anti-PD-L1-treated syngeneic, immunocompetent hosts. Leveraging the baseline and on-treatment biopsy samples collected from patients with HCC in an ongoing phase II trial of pembrolizumab (NCT03419481), our integrative single-cell analysis has revealed a tumorous adaptive transcriptional programme that evades therapeutic pressure of PD-L1/PD-1 blockade by immunosuppressive TME remodelling. We further demonstrate that cotargeting the emerging peroxisome proliferator-activated receptor gamma (PPARγ) signalling can avert ICB resistance in multiple orthotopic and spontaneous HCC models. This study will provide a new strategy of precision cancer immunotherapy tailored to block a specific immune escape pathway of HCC cells.
Methods
Mice
Six- to 8-week-old male C57BL/6 and BALB/c nude mice were obtained and maintained at Laboratory Animal Services Centre of CUHK under specific pathogen-free condition. All animal experiments were performed in accordance with the protocols approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong (CUHK-AEEC).
Human studies
Patients with HCC with a confirmed diagnosis of hepatitis B virus (HBV) were recruited for the single-arm phase II trial of pembrolizumab (NCT03419481) in the Prince of Wales Hospital in Hong Kong, China. Pretreatment and on-treatment tumour biopsies after two cycles of pembrolizumab were collected. Tumour biopsies from 26 patients were analysed by scRNA-seq, standard histopathology assessment or coimmunofluorescence staining. A radiologist conducted blinded evaluation of clinical imaging to define ORRs and clinical benefit according to Response Evaluation Criteria in Solid Tumours (RECIST) V.1.1 criteria. Written informed consent was provided by all patients prior to biopsy acquisition.
A detailed description of all methods used in this study can be found in online supplemental file 1.
gutjnl-2022-328364supp001.pdf (21.3MB, pdf)
Results
HCC cells acquire immune evasion capability on serial implantation in ICB-treated mice
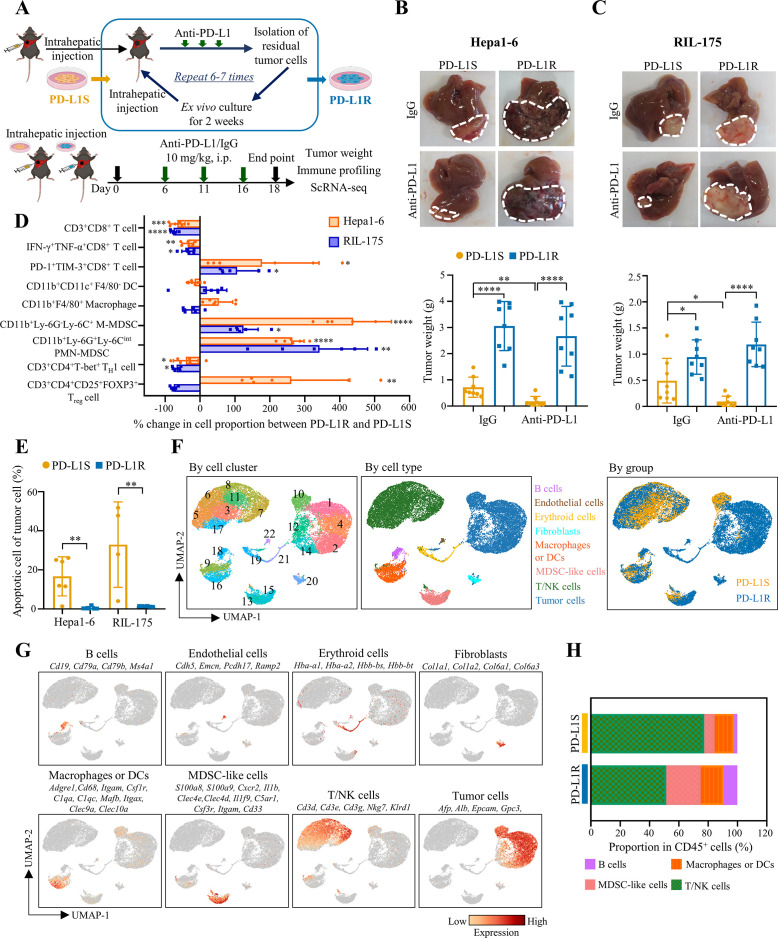
We established two mouse models of ICB resistance in HCC through serial orthotopic implantation into C57BL/6 immunocompetent hosts for anti-PD-L1 therapy (figure 1A). Tumours formed by syngeneic Hepa1-62 or RIL-17517 HCC cells initially shrank in response to the anti-PD-L1 treatment, and the residual tumours were dissociated into single-cell suspensions for ex vivo expansion before reinoculation into the livers of new recipient mice. In contrast to the parental cell lines, both Hepa1-6 and RIL-175 tumours became non-responsive to anti-PD-L1 therapy after six and seven treatment cycles, respectively (figure 1B,C). Moreover, the PD-L1-resistant (PD-L1R) tumours grew significantly larger than the parental PD-L1-sensitive (PD-L1S) tumours in both anti-PD-L1-treated (>10-fold) and IgG isotype control (~2-fold) C57BL/6 mice. However, the PD-L1R and PD-L1S HCC cells exhibited similar tumour weights in immunocompromised nude mice (online supplemental figure 1). These data suggest that PD-L1R HCC cells acquire a capacity to evade immune attack elicited by ICB therapy, which is not due to enhanced growth ability per se.
Figure 1.
HCC cells acquire immune evasion capability in ICB-resistant orthotopic mouse models. (A) Schematic diagram of establishment of ICB-resistant models. (B) Representative liver tumour photos and tumour weights of Hepa1-6-derived and (C) RIL-175-derived PD-L1S and PD-L1R tumours at the endpoint are shown (n=8). (D) Proportions of CD3+CD8+ T cells, CD11b+CD11c+F4/80- DCs, CD11b+F4/80+ macrophages, CD11b+Ly-6G-Ly-6C+ M-MDSCs, CD11b+Ly-6G+Ly-6Cint PMN-MDSCs, CD3+CD4+T-bet+ TH1 cells and CD3+CD4+CD25+FOXP3+ Treg cells in tumour-infiltrating CD45+ leucocytes, and IFN-γ+TNF-α+ cells and PD-1+TIM-3+ cells among tumour-infiltrating CD8+ T cells were determined by flow cytometry. Percentage changes of indicated immune cell proportions between anti-PD-L1-treated PD-L1R versus PD-L1S tumours from Hepa1-6 and RIL-175 models (n=4 to 6). (E) Percentages of apoptotic cells indicated by Annexin V+/PI+ in CD45- cells isolated from anti-PD-L1 treated tumours are shown (n=4 to 6). (F) ScRNA-seq analysis of anti-PD-L1-treated PD-L1S and PD-L1R tumours (n=2 per group) from Hepa1-6 orthotopic model. UMAP projection of 25 396 single cells isolated from tumour tissues, coloured by graph-based cell clusters, inferred cell types or experimental groups, respectively. (G) UMAP plots showing the mRNA expression and distribution of canonical markers for major cell types. Each cell was coloured based on normalised mRNA level of indicated genes. (H) Percentage of tumour-infiltrating immune populations identified in (F) is shown as mean in each group. Data represent as mean±SD. Statistical significance was determined by unpaired two-tailed Student’s t-test. *P<0.05; **p<0.01; ***p<0.001; ****p<0.0001. DC, dendritic cell; HCC, hepatocellular carcinoma; ICB, immune-checkpoint blockade; MDSC, myeloid-derived suppressor cell; PD-L1R, programmed death-1-ligand-1 resistant; PD-L1S, programmed death-1-ligand-1 sensitive; PMN, polymorphonuclear; UMAP, Uniform Manifold Approximation and Projection.
To investigate the tumour immune evasion in our ICB-resistant models, we compared the immune profiling of PD-L1R and PD-L1S in anti-PD-L1-treated C57BL/6 mice using multi-colour flow cytometry (online supplemental figure 2A). In contrast to the TME of the PD-L1S tumours, the PD-L1R tumours exhibited reduced CD4+ T helper 1 (TH1) cells and CD8+ T cells but increased monocytic (M)-and polymorphonuclear (PMN)-myeloid-derived suppressor cells (MDSCs) in both Hepa1-6 and RIL-175 models (figure 1D and online supplemental figure 2B). Moreover, the CD8+ T cells expressed higher levels of immune-checkpoint molecules such as PD-1 and T cell immunoglobulin and mucin-domain containing-3 (TIM-3), but lower levels of cytotoxic cytokines interferon gamma (IFN-γ) and tumour necrosis factor alpha (TNF-α) (online supplemental figure 2C), consistent with a phenotypic shift from cytotoxic to exhausted CD8+ T cells in the PD-L1R tumours (figure 1D). However, the proportions of regulatory T cells (Treg cells), dendritic cells (DCs) and macrophages were not consistently changed. In general, the DC and macrophage phenotypes were not altered in the PD-L1R tumours, except a concordant increase in CD80 costimulatory molecule expression (online supplemental figure 3). Although no increase in PD-L1 expression was observed in both tumour and immune cells such as PMN-MDSCs in PD-L1R compared with PD-L1S tumours (online supplemental figure 4), tumour cell apoptosis was dramatically reduced (figure 1E and online supplemental figure 5). These data suggest that PD-L1R HCC cells survive under ICB therapeutic pressure by remodelling a T cell-excluded and immunosuppressive TME. Moreover, we observed consistent CD8+ T cell reduction and PMN-MDSC induction in blood and spleen of PD-L1R tumour-bearing mice (online supplemental figure 6), suggesting that ICB-resistant tumour can cause a systemic immunosuppression. Considering genetic mutation as a key driver of ICB resistance,18 we performed whole-exome sequencing using the PD-L1R and PD-L1S tumour cells. We observed highly overlapping mutational profiles in both Hepa1-6 and RIL-175 models (online supplemental figure 7), indicating non-genetic adaptive pathways for ICB resistance.10
To probe the transcriptional drivers underlying therapeutic evasion, we leveraged scRNA-seq to enable cell type-specific profiling of Hepa1-6-PD-L1R and Hepa1-6-PD-L1S tumours from C57BL/6 mice (n=2 per group) on anti-PD-L1 therapy. After initial quality control, 25 396 cells with a median of 924 genes detected per cell were used for Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction and unsupervised graph-based clustering. Twenty-two distinct clusters were identified (figure 1F), which were further grouped into eight major cell populations, namely B cells, endothelial cells, erythroid cells, fibroblasts, macrophages or DCs, MDSC-like cells, T/natural killer (NK) cells and tumour cells based on their canonical marker gene expressions (figure 1F,G). Similar to the findings by flow cytometry, the immune microenvironment of PD-L1R tumours harboured relatively less T/NK lymphocytes but more MDSC-like cells than the PD-L1S tumours (figure 1H), further supporting that TME remodelling is a key immune evasion mechanism in our ICB-resistant models.
Adaptive upregulation of PPARγ orchestrates TME remodeling to resist ICB therapy
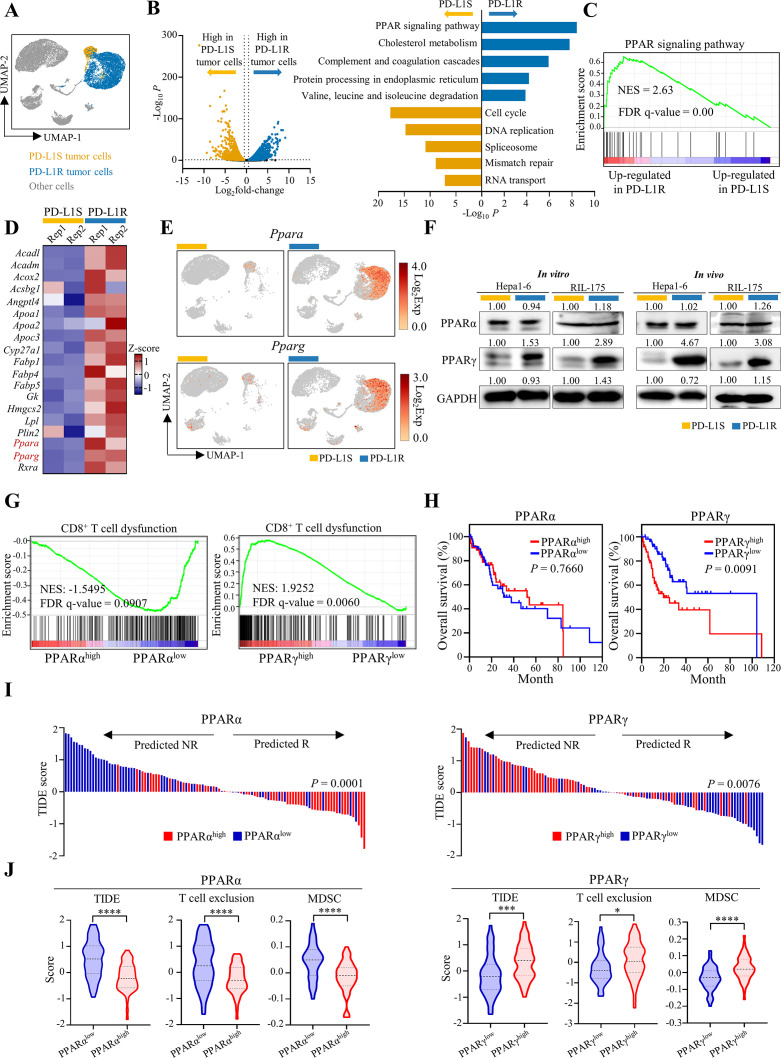
Our scRNA-seq analysis showed that the ICB-sensitive and ICB-resistant tumour cells exhibited distinctive transcriptomic profiles (figure 2A), supporting dynamic transcriptional adaptation but not selection of pre-existing programme in therapeutic evasion.10 The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the 1024 significantly upregulated genes in the PD-L1R tumour cells revealed PPAR signalling as the most significantly enriched pathway (adjusted p<1 × 10−5; figure 2B and online supplemental table 1). Gene Set Enrichment Analysis further confirmed the significant enrichment of PPAR signalling pathway (figure 2C), which was consistent with single-cell analysis from the isolated PD-L1R and PD-L1S HCC cell lines (online supplemental figure 8). PPARs (α, β, γ and δ isoforms) are a family of nuclear receptor transcription factors controlling diverse metabolic, inflammatory and oncogenic programmes.19 Among the PPARs, PPARα and PPARγ were present in the 19-gene list of the enriched PPAR signalling pathway (figure 2B,D) and primarily expressed in the tumour cell clusters (figure 2E). Critically, PPARγ but not PPARα protein level was prominently up-regulated in both Hepa1-6 and RIL-175 models in vitro and in vivo (figure 2F).
Figure 2.
Adaptive upregulation of PPARγ correlates with immune dysregulation in mouse models and patients with HCC. (A) ScRNA-seq analysis of tumour cell clusters from Hepa1-6-PD-L1S and PD-L1R tumours with anti-PD-L1 treatment (n=2 per group). UMAP plot of the identified tumour cells. (B) Volcano plot of differentially expressed genes (DEGs; left; Log2fold-change >0.5 and p<0.05) and KEGG pathway enrichment analysis for top five enriched pathways of DEGs (right; Log2fold-change>0.5, p<0.05 and normalised expression >0.1) in anti-PD-L1-treated PD-L1S (orange bar) or PD-L1R tumour cells (blue bar), respectively. (C) GSEA plot of PPAR signalling pathway (KEGG) in PD-L1R tumour cells compared with PD-L1S tumour cells. (D) Heatmap showing z-score transformed expression of PPAR-related genes identified in (B) in PD-L1S or PD-L1R tumour cells from each sample. (E) UMAP plots of expression patterns of Ppara (top) and Pparg (bottom) in PD-L1S and PD-L1R single cells. (F) Representative western blot analysis of PPARα and PPARγ in Hepa1-6 or RIL-175-PD-L1S and -PD-L1R cell lines and anti-PD-L1-treated tumour tissues. GAPDH served as loading control. (G) TCGA HCC samples with high (n=55) and low (n=55) mRNA levels of PPARA or PPARG that were stratified by top and bottom 15% in 369 patients were selected for subsequent analysis. GSEA plots of CD8+ T cell dysfunction signature in patients with TCGA HCC with high and low expressions of PPARA or PPARG. (H) Kaplan-Meier curves of overall survival in patients with HCC according to the expression of PPARA or PPARG. (I) Prediction of potential clinical ICB response in patients with PPARαhigh versus PPARαlow or PPARγhigh versus PPARγlow HCC using the TIDE signature. (J) Analysis of TIDE, T cell exclusion and MDSC scores by TIDE algorithm in patients with PPARαhigh and PPARαlow or PPARγhigh and PPARγlow HCC. Statistical significance was assessed by two-sided log-rank (Mantel-Cox) test for (H), by two-sided χ² test for (I) and by unpaired two-tailed Student’s t-test for (J). *P<0.05; ***p<0.001; ****p<0.0001. DEGs, differentially expressed genes; GSEA, Gene Set Enrichment Analysis; HCC, hepatocellular carcinoma; ICB, immune-checkpoint blockade; KEGG, Kyoto Encyclopedia of Genes and Genomes; MDSC, myeloid-derived suppressor cell; PD-L1R, programmed death-1-ligand-1 resistant; PD-L1S, programmed death-1-ligand-1 sensitive; PPARγ, peroxisome proliferator-activated receptor-gamma; TIDE, Tumor Immune Dysfunction and Exclusion; UMAP, Uniform Manifold Approximation and Projection.
Using TCGA dataset, we found that patients with HCC with high expression of PPARγ, but not PPARα, was associated with a dysfunctional CD8+ T cell signature20 (FDR q-value <0.01; figure 2G) and poorer survival (p<0.01; figure 2H). Moreover, the Tumor Immune Dysfunction and Exclusion (TIDE) analysis, which integrates the expression signatures of T cell dysfunction and T cell exclusion to predict ICB response,21 showed that the patients with PPARγhigh HCC were significantly more susceptible to ICB resistance (p<0.01; figure 2I) based on their higher TIDE (p<0.001), T cell exclusion (p<0.05) and MDSC signature scores (p<0.0001) when compared with the patients with PPARγlow HCC (figure 2J). Consistent with the previous findings that PPARα exerts multiple hepatoprotective effects in metabolic homeostasis,22 high PPARα expression in HCC was significantly associated with better ICB response with lower TIDE, T cell exclusion and MDSC signature scores (p<0.0001; figure 2I,J).
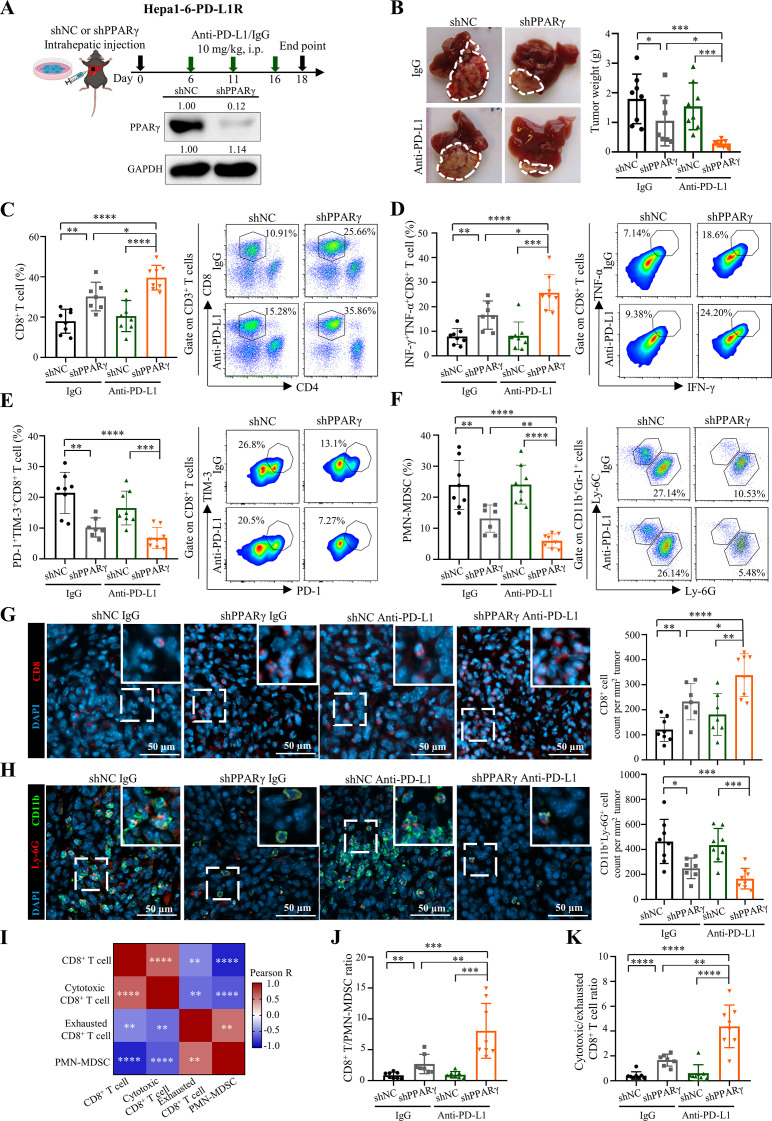
Our mouse and human data highlight the potential role of tumorous PPARγ upregulation in immune evasion, an essential process for ICB resistance. We thus reasoned that inhibition of PPARγ might improve antitumour immune responses and resensitise tumour to ICB therapy. To test this, we established PPARγ-knockdown (KD) and control sublines of the Hepa1-6-PD-L1R tumour cells using short-hairpin RNA (shRNA), followed by orthotopic implantation, anti-PD-L1 treatment, tumour size measurement and immune cell profiling (figure 3A). Compared with the control tumour-bearing mice that did not respond to anti-PD-L1 therapy, tumour cell-intrinsic PPARγ inhibition resulted in a significant reduction in tumorigenicity on ICB treatment (p<0.001; figure 3B). Dual inhibition of PPARγ and PD-L1 significantly increased the intratumoral levels of total and cytotoxic CD8+ T cells (p<0.0001; figure 3C,D and online supplemental figure 9A,B) and decreased the exhausted CD8+ T cells (p<0.01; figure 3E and online supplemental figure 9C) and PMN-MDSCs (p<0.0001; figure 3F and online supplemental figure 9D), which were further validated by immunofluorescence staining of tumour tissues, showing concordant changes in CD8-positive T cells (figure 3G) and CD11b/Ly-6G double-positive PMN-MDSCs (figure 3H).
Figure 3.
Tumour-intrinsic PPARγ orchestrates TME remodelling to resist ICB therapy. (A) Treatment schedule for anti-PD-L1 antibody (10F.9G2) or isotype control (LTF-2) in Hepa1-6-PD-L1R-short hairpin RNA against negative control sequence (shNC)-tumour or shPPARγ-tumour bearing mice. In brief, 5×106 tumour cells were intrahepatically injected into C57BL/6 mice, which were then treated with anti-PD-L1 or isotype control antibodies via i.p. injection at day 6, 11 and 16. Tumour samples were collected at day 18-post tumour implantation for further analysis. Representative western blot images of PPARγ in Hepa1-6-PD-L1R-shNC or -shPPARγ stable cell lines are shown. GAPDH served as loading control. (B) Representative liver tumour photos and tumour weights of PD-L1R-shNC-tumour or shPPARγ-tumour bearing mice with treatment of anti-PD-L1 or isotype control at the endpoint are shown (n=7 to 8). (C) Representative flow cytometry dot plots and proportions of CD8+ T cells in CD45+ cells, (D) IFN-γ+TNF-α+ cells and (E) PD-1+TIM-3+ cells in CD8+ T cells, as well as (F) PMN-MDSCs in CD45+ cells in tumours from indicated groups are shown (n=7 to 8). (G) Representative immunofluorescence images of CD8 (red), (H) CD11b (green)/Ly-6G (red) and quantification dot plot bar graphs in liver tumours from indicated groups (n=7 to 8). DAPI (blue) indicates the nuclei staining. Scale bars, 50 µm. (I) Correlation heatmap of immune cell proportions calculated by data from C to F. (J) The ratios of CD8+ T/PMN-MDSCs and (K) cytotoxic/exhausted CD8+ T cell in tumours (n=7 to 8). Data represent as mean±SD. Statistical significance was determined by unpaired two-tailed Student’s t-test. Two-tailed Pearson’s correlation was used to describe the correlation between variables. *P<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ICB, immune-checkpoint blockade; IFN, interferon; MDSC, myeloid-derived suppressor cell; PD-L1, programmed death-1-ligand-1; PMN, polymorphonuclear; PPARγ, peroxisome proliferator-activated receptor-gamma; TIM, T cell immunoglobulin; TME, tumour microenvironment; TNF, tumour necrosis factor.
As an evidence of MDSC-driven CD8+ T cell suppression, the proportion of tumour-infiltrating PMN-MDSCs exhibited significantly negative correlation with the total and cytotoxic CD8+ T cells (R=−0.864, p<0.0001 and R=−0.657, p<0.0001; figure 3I). Critically, PPARγ KD alone resulted in significantly increased CD8+ T cell to PMN-MDSC (p<0.01; figure 3J) and cytotoxic to exhausted CD8+ T cell ratios (p<0.0001; figure 3K), which were further alleviated by combined anti-PD-L1 treatment (p<0.01; figure 3J,K). Indeed, PMN-MDSCs from PPARγ-KD tumours exhibited reduced proliferation (indicated by Ki-67 level; p<0.01) and expressions of immunosuppressive mediators arginase-1 (Arg-1; p<0.05) and reactive oxygen species (p<0.01; online supplemental figure 10A), which could impair their T cell suppressive activities in coculture experiments when compared with PMN-MDSCs from control tumours (online supplemental figure 10B).
To demonstrate the function of PMN-MDSCs in PPARγ-induced ICB resistance, we treated the RIL-175-PD-L1R tumour-bearing mice with anti-Ly-6G antibody (online supplemental figure 11A). Blockade of PMN-MDSCs by anti-Ly-6G antibody significantly reduced tumorigenicity (p<0.05) and led to a remarkable tumour regression when combined with anti-PD-L1 antibody (p<0.0001; online supplemental figure 11B,C), which was accompanied by significant increases in total and cytotoxic tumour-infiltrating CD8+ T cells with IFN-γ, TNF-α, granzyme B (GzmB) and CD107a expressions (online supplemental figure 11D–F) and decrease in the exhaustion phenotype (online supplemental figure 11G). We further determined the function of PMN-MDSCs by adoptive transfer, which restored their intratumoral level and abolished tumour regression by PPARγ KD (online supplemental figure 12A–C) through cytotoxic-to-exhausted CD8+ T cell conversion (online supplemental figure 12D–G). Consistent with the importance of cold-to-hot conversion of TME in ICB efficacy,23 our findings suggest that adaptive upregulation of PPARγ in tumour cells orchestrates TME immunosuppression and CD8+ T cell dysfunction to resist ICB therapy.
Tumour-intrinsic PPARγ transcriptionally activates VEGF-A production to remodel TME
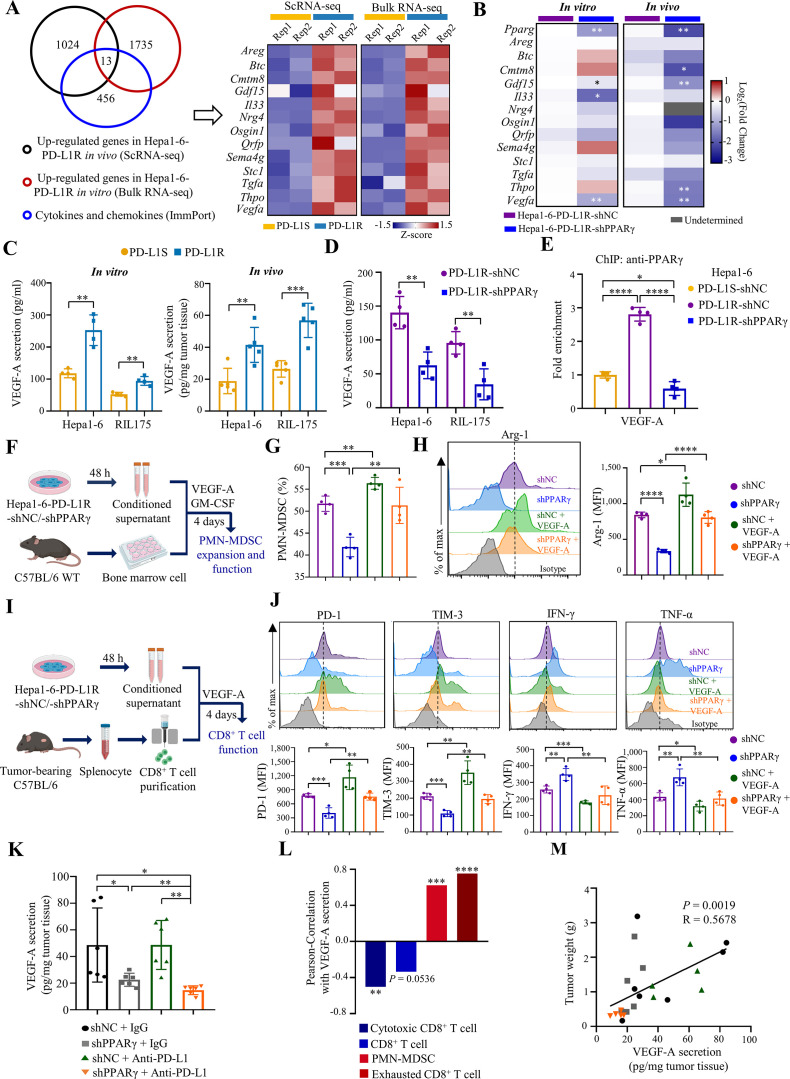
The positive correlation between PMN-MDSCs and exhausted CD8+ T cells in vivo (R=0.561, p<0.01; figure 3I) prompted us to speculate a secretable factor that mediates TME remodelling by PPARγ. We thus compared the upregulated genes in Hepa1-6 PD-L1R tumour cells from in vivo scRNA-seq and in vitro bulk RNA-seq. From a registry of cytokines and chemokines from ImmPort, an open repository of immunology data,24 we identified 13 candidate secretable factors (figure 4A). Further RT-qPCR analysis from PPARγ-KD/control tumour cells and tissues revealed concordant downregulation of growth differentiation factor 15 (Gdf15) and Vegfa when PPARγ was blocked in vitro and in vivo (figure 4B). As we have previously reported GDF15 as a direct target of PPARγ in HCC cells,25 in this study we focused on the role of VEGF-A. Consistent with its roles in MDSC immunosuppression and T cell exhaustion,26 27 high VEGFA expression in HCC of the TCGA dataset was associated with poorer patient survival (p<0.01; online supplemental figure 13A) and poorer predicted ICB response (p<0.05; online supplemental figure 13B) based on higher TIDE, T cell exclusion and MDSC signature scores (p<0.05; online supplemental figure 13C). We next confirmed the significant secretion of VEGF-A by PD-L1R relative to PD-L1S tumour cells in vitro and in vivo (figure 4C). To further evaluate the regulation of VEGF-A by PPARγ, we performed shRNA-mediated KD of PPARγ in both Hepa1-6 and RIL-175 models (figure 3A and online supplemental figure 14A) and observed a significant reduction in Vegfa mRNA expression and protein secretion (figure 4D and online supplemental figure 14B). KD of PPARγ in PD-L1R tumour cells also significantly reduced its own occupancy at the Vegfa promoter similar to the level of PD-L1S tumour cells (figure 4E), indicating that VEGF-A is a direct PPARγ target in anti-PD-L1-resistant tumour cells.
Figure 4.
Tumour-intrinsic PPARγ transcriptionally activates VEGF-A production to remodel TME. (A) Venn diagram of upregulated genes in Hepa1-6-PD-L1R tumour cell clusters (scRNA-seq), Hepa1-6-PD-L1R cell lines from bulk RNA-seq data and cytokines/chemokines from ImmPort. Heatmap showing normalised and z-scored expression of the 13 overlapped genes in scRNA-seq or bulk RNA-seq data, respectively (n=2). (B) Heatmap of relative mRNA levels of Pparg and the 13 indicated candidate genes in Hepa1-6-shNC-tumour and shPPARγ tumour cells (n=4) or tissues (n=5 to 6) are shown. (C) ELISA analysis of VEGF-A secretion levels in cell lines (n=4) and anti-PD-L1-treated tumour tissues (n=5) from Hepa1-6 and RIL-175 models. (D) VEGF-A protein concentration in cell lysates of Hepa1-6 or RIL-175-shNC and shPPARγ cell lines determined by ELISA (n=4). (E) ChIP-qPCR analysis of PPARγ occupancy on the Vegfa promoter region in the indicated cell lines (n=4). Data are normalised to PD-L1S control level. (F) Schematic illustration of PMN-MDSC expansion and functional analysis ex vivo. (G) Proportions of PMN-MDSC in CD45+ cells in indicated groups were assessed by flow cytometry (n=4). (H) Representative overlay histogram and expression levels determined by mean fluorescence intensity (MFI) of Arg-1 in PMN-MDSCs (n=4). (I) Schematic illustration of CD8+ T cell functional assay ex vivo. (J) Representative overlay histogram and MFI of PD-1, TIM-3, IFN-γ and TNF-α in CD8+ T cells (n=4). Dashed line indicates the peak of the Hepa1-6-shNC sample. (K) C57BL/6 mice were intrahepatically injected with Hepa1-6-PD-L1R-shNC or shPPARγ cells (5×106), followed by three doses of treatment with anti-PD-L1 antibody 10F.9G2 or isotype control LTF-2 (10 mg/kg, i.p., every 5 days). Tumours were harvested at the experimental endpoint. VEGF-A secretion levels in tumour tissues were measured from indicated groups. (L) Pearson correlation of immune cell proportions with VEGF-A secretion in tumour tissues. (M) Correlation between VEGF-A secretion in tumour tissues and tumour weight. Data represent as mean±SD. Statistical significance was determined by unpaired two-tailed Student’s t-test. Single-tailed Pearson’s correlation was used to describe the correlation between variables. *P<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ICB, immune-checkpoint blockade; IFN, interferon; MDSC, myeloid-derived suppressor cell; PD-1, programmed cell death protein 1; PMN, polymorphonuclear; PPARγ, peroxisome proliferator-activated receptor-gamma; TME, tumour microenvironment; TNF, tumour necrosis factor.
To determine the importance of VEGF-A in tumorous PPARγ-induced TME remodelling, we performed ex vivo experiments with the conditional media (CM) of PPARγ-KD or control PD-L1R tumour cells (figure 4F). We found that the expansion of CD11b+Gr-1+Ly-6G+Ly-6CInt PMN-MDSCs from mouse bone marrow cells were significantly reduced when cultured with the CM of PPARγ-KD relative to control tumour cells (figure 4G). Of note, VEGF-A fully restored the PMN-MDSC expansion and the expression of Arg-1 (figure 4G,H). Similar results were obtained when we cultured peripheral blood mononuclear cells with the CM (online supplemental figure 15). In concord, we observed significant reduction in PMN-MDSCs in blood and spleen of PPARγ-KD tumour-bearing mice relative to control tumour counterparts (online supplemental figure 16A), while the circulating proportion of Ki-67+PMN-MDSCs was also significantly reduced (online supplemental figure 16B). However, we observed no significant change in the proportions and phenotypes of DCs and macrophages in the ex vivo model (online supplemental figure 17), which was consistent with data from the mouse models (figure 1D and online supplemental figure 3). Overall, these results suggest that tumour-intrinsic PPARγ signalling may increase tumour-infiltrating PMN-MDSCs by promoting MDSC expansion and proliferation.
We next examined the effect of the PPARγ/VEGF-A axis on T cell dysfunction by culturing splenocytes from tumour-bearing mice with the CM (figure 4I) and found that PPARγ-KD tumour cells significantly reduced the expressions of inhibitory receptors PD-1 and TIM-3 but increased the cytokine expressions of IFN-γ and TNF-α in CD8+ T cells (figure 4J). Importantly, the conversion of exhausted to cytotoxic CD8+ T cells was completely abolished by VEGF-A (figure 4J), suggesting that PPARγ impairs antitumour function of CD8+ T cells in a VEGF-A-dependent manner. Using the tumour lysates from the ICB-resistant model, we found that PPARγ-KD tumours exhibited significantly lower VEGF-A levels than the control tumours in both anti-PD-L1-treated (p<0.01) and IgG control mice (p<0.05; figure 4K). Moreover, the levels of VEGF-A were significantly correlated with the proportions of VEGF receptor 2 (VEGFR2)-expressing PMN-MDSCs (R=0.6248, p<0.001) and exhausted CD8+ T cells (R=0.7531, p<0.0001; figure 4L and online supplemental figure 18) and tumour burden in vivo (R=0.5678, p<0.01; figure 4M).
We further determined the functional significance of the PPARγ/VEGF-A axis in vivo by establishing a VEGF-A-KD subline of the RIL-175-PD-L1R tumour cells for orthotopic implantation and anti-PD-L1 therapy (online supplemental figure 19A). We found that tumour cell-intrinsic downregulation of VEGF-A resensitised tumours to anti-PD-L1 treatment, resulting in tumour regression to a similar extent with the PPARγ-KD tumours (online supplemental figure 19B). Flow cytometry analysis revealed that VEGF-A-KD tumours displayed an increase in the total and cytotoxic CD8+ T cells but a decrease in exhausted CD8+ T cells and PMN-MDSCs (online supplemental figure 19C–F). Moreover, coblockade of PD-L1 could further enhance the above antitumour immune responses in VEGF-A-KD tumours, which were in parallel with the PPARγ-KD tumours (online supplemental figure 19C–F). Collectively, our data highlight VEGF-A as a major regulator responsible for PPARγ-driven MDSC-mediated immunosuppression and CD8+ T cell dysfunction.
A PPARγ antagonist averts ICB resistance in multiple HCC models
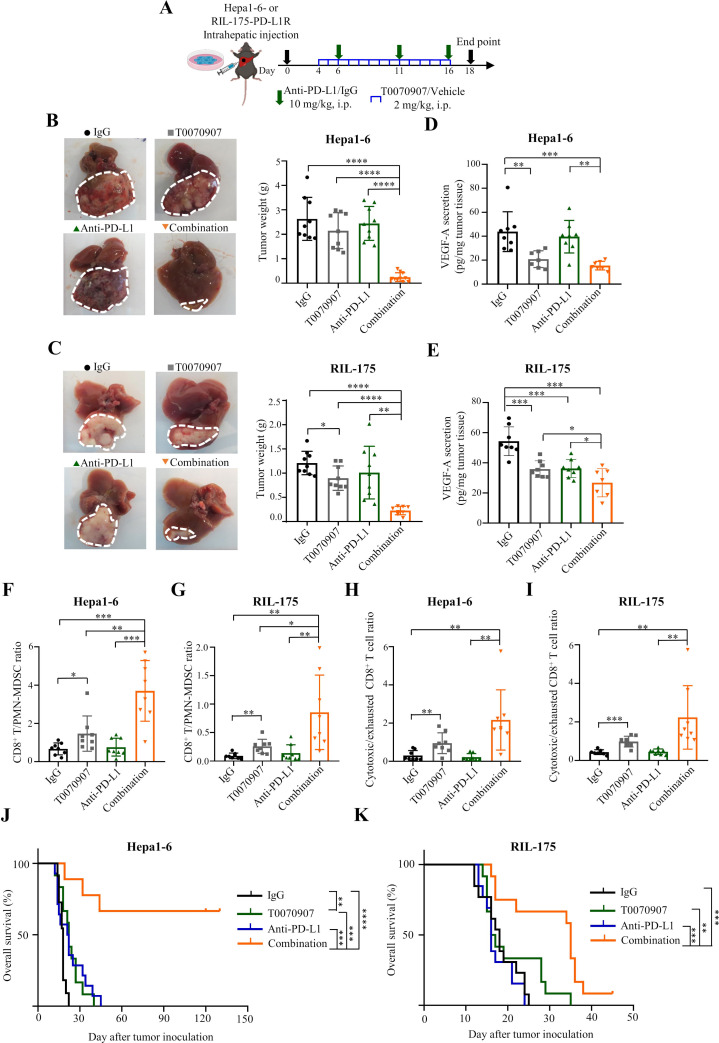
Given the functional significance of PPARγ signalling in TME remodelling and ICB resistance, we next assessed the therapeutic efficacy of a selective PPARγ antagonist T007090728 to overcome the therapeutic resistance in both Hepa1-6 and RIL-175 models (figure 5A). While treatment with the PPARγ antagonist alone showed limited effect on tumour burden in the resistant models (figure 5B,C), tumour growth was substantially abrogated when T0070907 treatment was combined with ICB (p<0.0001; figure 5B,C). Similar to the effects of tumour cell-intrinsic PPARγ-KD (figures 3C–F and 4K), pharmacological PPARγ inhibition reduced tumorous VEGF-A levels (figure 5D,E) and converted an immunosuppressive TME into a T cell-inflamed TME (online supplemental figure 20). Importantly, the CD8+ T/PMN-MDSC (p<0.05 and p<0.01; figure 5F,G) and cytotoxic/exhausted CD8+ T cell ratios (p<0.01 and p<0.001; figure 5H,I) were significantly increased by T0070907, and further elevated by combined anti-PD-L1 treatment (figure 5F–I). In the Hepa1-6-resistant model, while T0070907 alone slightly prolonged survival compared with the vehicle control, the combination treatment led to remarkable durable response and survival benefit, in which 67% of mice (6/9) survived over 4 months (figure 5J). Similar pattern of durable survival benefit of the combination therapy could also be observed in the RIL-175 resistant model (figure 5K). These data suggest that PPARγ inhibition primes a favourable TME for effective and durable ICB therapy via diminishing MDSC immunosuppression and T cell exhaustion. Furthermore, the single or combined treatment with T0070907 and anti-PD-L1 antibody was well tolerated, as we did not observe any body weight loss (online supplemental figure 21A), hepatic dysfunction (online supplemental figure 21B) or abnormalities of internal organs such as liver, spleen, kidney and heart (online supplemental figure 21C–F).
Figure 5.
PPARγ antagonist T0070907 averts ICB resistance in orthotopic HCC models. (A) Combinatory treatment schedule of T0070907 and anti-PD-L1 antibody 10F.9G2 in mice bearing Hepa1-6 or RIL-175-PD-L1R tumours. (B) Representative liver tumour photos and tumour weights of indicated groups in Hepa1-6 and (C) RIL-175-derived ICB-resistant models (n=8 to 10). (D) ELISA analysis of VEGF-A secretion levels of indicated groups in Hepa1-6 and (E) RIL-175-derived ICB-resistant models (n=7 to 8). (F) The ratios of CD8+ T/PMN-MDSC in Hepa1-6 and (G) RIL-175 resistant tumours from indicated groups (n=8). (H) The ratios of cytotoxic/exhausted CD8+ T cell in Hepa1-6 and (I) RIL-175 resistant tumours from indicated groups (n=8). (J) Kaplan-Meier survival analysis of mice from indicated groups in Hepa1-6 and (K) RIL-175-derived ICB-resistant models (n=9 to 14). Data represent as mean±SD. Statistical significance was assessed by unpaired two-tailed Student’s t-test for (B)–(I), and by two-sided log-rank (Mantel-Cox) test for (J) and (K). *P<0.05; **p<0.01; ***p<0.001; ****p<0.0001. IFN, interferon; MDSC, myeloid-derived suppressor cell; MFI, mean fluorescence intensity; PD-L1R, programmed death-1-ligand-1 resistant; PMN, polymorphonuclear; PPARγ, peroxisome proliferator-activated receptor-gamma; TIM, T cell immunoglobulin; TME, tumour microenvironment; TNF, tumour necrosis factor; VEGF, vascular endothelial growth factor.
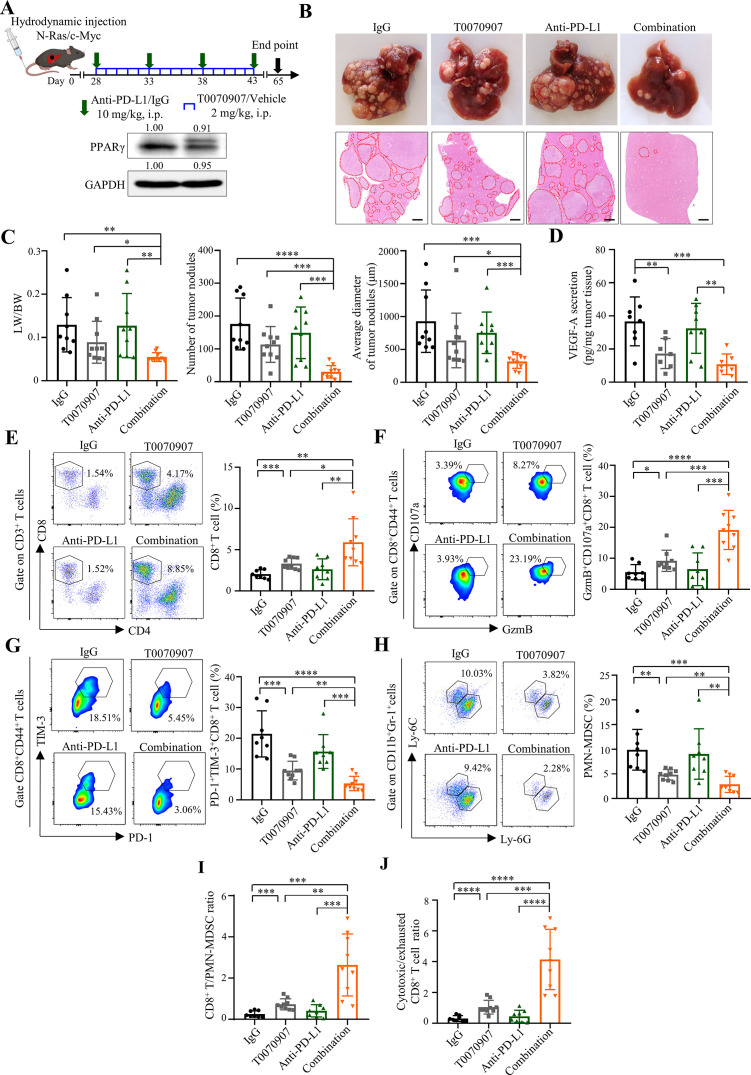
Given the intricate relationship between tumour heterogeneity and immunotherapy responses,29 orthotopic models using cell lines may not fully resemble immune surveillance during treatment. We thus employed our combination immunotherapy in a spontaneous HCC model induced by hydrodynamic tail-vein injection (HDTVi) of N-Ras and c-Myc-luciferase encoding plasmids together with a sleeping beauty transposase construct,30 which may recapitulate the aberrant activation of Ras/mitogen-activated protein kinase pathway documented in more than 50% of human HCCs.31 In this aggressive tumour model which exhibited PPARγ overexpression, multiple nodules were evident at 28 day-post HDTVi and further developed into anti-PD-L1-resistant tumours (figure 6A,B and online supplemental figure 22). As depicted by the liver weight/body weight, tumour nodule number and size (in diameter), T0070907 could resensitise tumour to anti-PD-L1 therapy (p<0.01), resulting in further tumour regression when compared with T0070907 alone (p<0.05; figure 6C). Consistent with the reduction in tumorous VEGF-A levels (p<0.01; figure 6D), immune profiling revealed that PPARγ inhibition significantly alleviated the T cell-excluded and MDSC-enriched TME (p<0.05), which was further enhanced by PD-L1 coblockade (p<0.05; figure 6E–H), resulting in superior elevation of the ratios of CD8+ T/PMN-MDSC and cytotoxic/exhausted CD8+ T cell (p<0.01; figure 6I,J). These findings underline the importance of TME normalisation in overcoming ICB resistance by this combinatorial approach.
Figure 6.
PPARγ antagonist T0070907 overcomes ICB resistance in the spontaneous HCC model. (A) Combinatory treatment schedule of T0070907 and anti-PD-L1 antibody 10F.9G2 in N-Ras/c-Myc-induced spontaneous HCC model (top). Representative western blot images of PPARγ in N-Ras/c-Myc-induced tumours (bottom). GAPDH served as loading controls. (B) Representative photos (top) and H&E staining images (bottom) of liver tumours of indicated groups (n=9 to 10). Scale bars, 500 µm. Tumour area is circled by red dotted line. (C) Tumour burden in indicated groups was evaluated by liver versus body weight ratios (LW/BW; left), numbers (middle) and average diameters (right) of tumour nodules per mouse from H&E images. (D) VEGF-A secretion levels in tumour tissues from indicated groups (n=7 to 8). (E) The proportions of CD8+ T cells, (F) GzmB+CD107a+ and (G) PD-1+TIM-3+ cells in tumorous CD8+ T cells as well as (H) PMN-MDSCs in tumorous CD45+ cells from indicated groups (n=8 to 9). (I) The ratios of CD8+ T/PMN-MDSC and (J) cytotoxic/exhausted CD8+ T cell in indicated groups. Data represent as mean±SD. Statistical significance was determined by unpaired two-tailed Student’s t-test. *P<0.05; **p<0.01; ***p<0.001; ****p<0.0001. BW, body weight; HCC, hepatocellular carcinoma; ICB, immune-checkpoint blockade; LW, liver weight; MDSC, myeloid-derived suppressor cell; PD-L1, programmed death-1-ligand-1; PMN, polymorphonuclear; PPARγ, peroxisome proliferator-activated receptor-gamma; TIM, T cell immunoglobulin; VEGF, vascular endothelial growth factor.
ICB-resistant patients with HCC exhibit concurrent tumour cell PPARγ induction and immunosuppressive TME
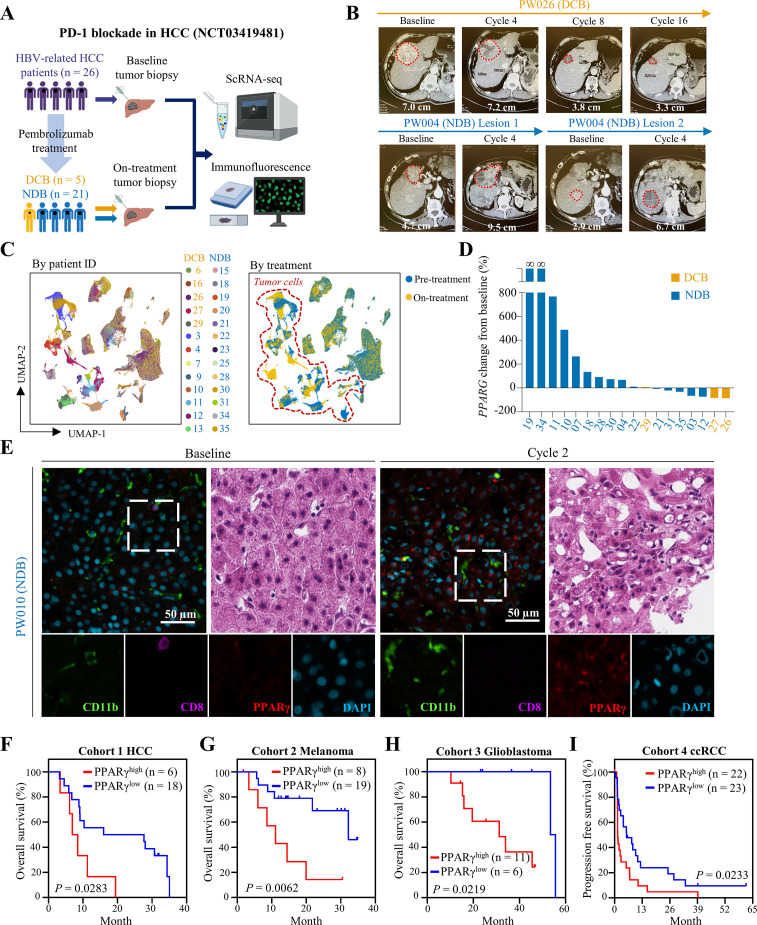
To validate the clinical relevance of our findings, we performed scRNA-seq using tumor biopsies from our single-arm phase II study of an anti-PD-1 antibody pembrolizumab (200 mg every 3 weeks infusion) in 26 patients with HBV-related HCC (NCT03419481) (figure 7A). Five out of the 26 patients exhibited durable clinical benefits (DCB) to pembrolizumab treatment, that is, partial response or stable disease for at least 6 months according to RECIST V.1.1, whereas the remaining 21 patients showed no durable benefits (NDB) based on CT scan (figure 7A,B). Leveraging the 26 baseline and 20 on-treatment (after 2 cycles of pembrolizumab) biopsies, we interrogated the ICB-induced dynamic alterations of tumour ecosystem. Analysis of >210 000 single-cell transcriptomes (~1000 genes per cell) by UMAP and marker gene annotation identified tumour cells for downstream investigation (figure 7C). To probe the involvement of PPARγ in ICB therapeutic evasion, we compared the tumour cell-intrinsic PPARG expressions between the paired baseline and on-treatment biopsies of 18 patients with HCC, since 2 patients with DCB did not have enough on-treatment tumour cells (< 20) for analysis (figure 7D). Notably, while all 3 remaining patients with DCB showed no change or decrease in PPARγ expression, 40% (6/15) of NDB patients displayed PPARG upregulation (>2-fold) on ICB therapy (figure 7D and online supplemental figure 23A). We further validated the protein expression of PPARγ and its relationship with immune cells using coimmunofluorescence. Compared with the baseline biopsies of a NDB patient, we found higher immunoreactivity of PPARγ and CD11b/CD15, markers of PMN-MDSCs,17 as well as lower CD8 positivity in the on-treatment samples (figure 7E and online supplemental figure 23B). In line with the in vitro and in vivo findings, we observed a significant correlation between PPARG and VEGFA expressions in the tumour cells of our in-house and a public14 dataset of human liver cancer scRNA-seq (R>0.6, p<2e−10; online supplemental figure 24). Collectively, these clinical data support the roles of ICB-induced PPARγ on TME immunosuppression and therapeutic resistance in a proportion of patients with HCC.
Figure 7.
High PPARγ expression correlates with immunosuppressive TME and poor ICB response in human HCC and other cancer types. (A) Schematic overview of a phase II clinical trial of pembrolizumab in patients with HBV-related HCC (NCT03419481) and tumour biopsy collection for analyses. (B) Longitudinal CT scans of a patient with DCB (PW026) and another patient with NDB (PW004) at indicated time points. The target lesion and its longest diameter are shown. Treatment for PW004 was stopped at cycle 4 due to disease progression. (C) UMAP plot of 210 153 single cells coloured by patient ID and treatment status. (D) Waterfall plot of individual patient-level percentage change in tumour cell-intrinsic PPARG mRNA levels from baseline to two-cycle of pembrolizumab (on-treatment). Baseline PPARG expression of PW019 and PW034 is numerically equal to 0. (E) Representative coimmunofluorescence images of CD11b, CD8 and PPARγ, as well as H&E images of paired baseline and on-treatment biopsies from a patient with NDB (PW010) are shown. Scale bars, 50 µm. (F) Kaplan-Meier survival analyses of patients with HCC, (G) melanoma, (H) glioblastoma and (I) ccRCC (CheckMate 010) undergone ICB treatment according to their baseline PPARG expression levels. Statistical significance was assessed by two-sided log-rank (Mantel-Cox) test. ccRCC, clear cell renal cell carcinoma; DCB, durable clinical benefits; HCC, hepatocellular carcinoma; ICB, immune-checkpoint blockade; NDB, no durable benefits; PPARγ, peroxisome proliferator-activated receptor-gamma; TME, tumour microenvironment; UMAP, Uniform Manifold Approximation and Projection.
High PPARγ expression correlates with poor ICB response in multiple human cancers
Finally, we evaluated the significance of PPARγ in patients’ response to ICB therapy. To compare the survival outcomes in ICB cohorts, we used the optimal cut-off determined by the maximal z-score in Cox proportional hazard models. In a small HCC cohort which includes baseline tumor transcriptomic data,32 we found that patients with higher expression of PPARG were significantly associated with poorer survival on anti-PD-1/PD-L1 treatment when compared with those with lower PPARG expression (p<0.05; figure 7F). Using independent ICB-treated cancer patient cohorts of which transcriptomic and clinical data were available, we further found that higher baseline PPARG expression was significantly associated with worse overall survival of patients with melanoma33 (p<0.01; figure 7G) and glioblastoma34 (p<0.05; figure 7H), as well as progression-free survival in clear cell renal cell carcinoma35 (p<0.05; figure 7I). These results implicate an important role of PPARγ in primary resistance of ICB and prognosis of patients in multiple cancer types, supporting the translational potential of PPARγ cotargeted ICB therapy.
Discussion
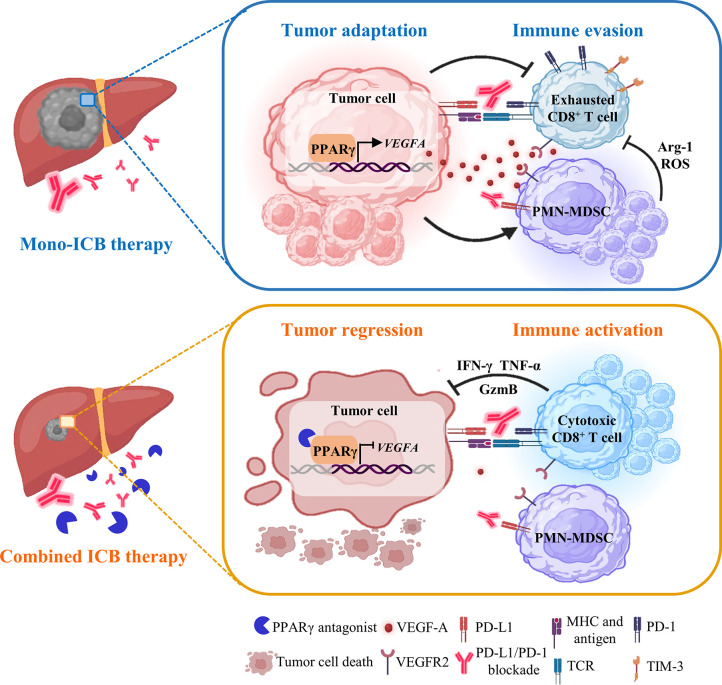
Understanding tumor cell adaptation on ICB therapy may present new opportunities to obviate the emergence of refractory cell states and enable specific targeted agents to improve clinical outcomes for patients.8 Compared with the genetic changes leading to loss of antigen expression and presentation and insensibility to T cells,11 the importance of non-genetic mechanisms in tumour adaptation remains underappreciated.10 Here, our integrative single-cell and functional analysis has shown a tumorous adaptive transcriptional programme that evades ICB therapeutic pressure by immunosuppressive TME remodelling (figure 8). Using refined ICB-resistant orthotopic HCC preclinical models, we demonstrate that tumour cell-intrinsic upregulation of PPARγ directly binds to the VEGFA promoter and activates its production to create an MDSC-enriched and T cell-dysfunctional microenvironment. Our results suggest a form of transcriptional adaptation—rather than subclonal genetic diversification and selection—can support a striking gain in immune evasion capabilities. Notably, concordant reconfigured transcription factor expression and microenvironmental changes are observed in a subset of patients with HCC developing resistance to ICB therapy. This non-mutational adaptive process is analogous to the occurrence of drug-tolerant transcriptional states on targeted therapeutic challenge.36 More importantly, the transcriptional plasticity can be pharmacologically exploited for prevention of therapeutic resistance.10 36
Figure 8.
A working model of a tumorous adaptive transcriptional programme to evade immune-checkpoint targeting. During mono-ICB therapy, HCC cells adapt by PPARγ upregulation to orchestrate an MDSC-enriched and T cell-dysfunctional TME via VEGF-A secretion. Selective targeting of PPARγ signalling abrogates the adaptive immune-evasive programme in TME to avert ICB resistance, leading to tumour regression. HCC, hepatocellular carcinoma; ICB, immune-checkpoint blockade; MDSC, myeloid-derived suppressor cell; PPARγ, peroxisome proliferator-activated receptor-gamma; TME, tumour microenvironment; VEGF, vascular endothelial growth factor.
To date, there is a lack of preclinical mouse models to explore mechanisms of immunotherapy resistance in HCC. The two pairs of murine HCC cells we report here enables rapid production of tumours that recapitulate the immune landscape of human ‘cold’ HCC in syngeneic, immunocompetent mouse models. A range of immune evasion pathways that we identified in the PD-L1R/S models are highly relevant to ICB resistance in patients with HCC, primarily the expansion of PMN-MDSCs and exclusion of CD8+ T cells, which express higher levels of T cell exhaustion markers such as PD-1 and TIM-3 but less cytotoxic cytokines IFN-γ and TNF-α.2 4 26 Using these models and ex vivo studies, we have been able to identify the transcriptional cascade that derives from immune attack and dissect the critical role of PPARγ/VEGF-A signalling in the PMN-MDSC and CD8+ T cell remodelling. Genetic KD of tumour cell-intrinsic PPARγ abrogates VEGF-A secretion to convert the immunosuppressive TME into an immunostimulatory milieu, thereby overcoming the anti-PD-L1 therapeutic resistance. Our results are consistent with and providing mechanistic rationale on the recent clinical findings on synergism of combining atezolizumab and bevacizumab in HCC.5 It is noteworthy that other cellular components such as cancer-associated fibroblasts (CAFs) have been reported to play a role in immunotherapy resistance.37 Although our scRNA-seq data showed preferential enrichment (>10-fold) of CAFs in the PD-L1R tumours, the relatively small number of CAFs (<400) precluded comprehensive subtype and gene ontology analyses. More in-depth characterisations are required to unveil the potential CAF-mediated immunosuppression in our mouse models.
Data from preclinical mouse models have shown that the superior therapeutic activity of combining immune-checkpoint inhibitors with drugs targeting VEGF-dependent signalling stems, in part, from restoration of antitumour functions of T cells, reduction in immunosuppressive myeloid cells and Treg cells in the TME.6 These clinical and preclinical studies lend support to the notion that prevention of the emergence of TME remodelling by inhibiting the tumour cell adaptive mechanisms might augment the effectiveness of ICB therapy. Although substantially increasing the ORR, the IMbrave150 study has limited applicability to patients at high risk of gastrointestinal and variceal bleeding and also reported increased toxicities, particularly grade 3/4 immune-related adverse events in more than half of the anti-PD-L1/anti-VEGF-A treated patients.38 Integration of findings from our preclinical and human studies has enabled us to identify an alternative actionable target of PPARγ for combined immunotherapy, especially for patients with HCC who would fail VEGF-based therapy due to intolerable adverse events or lack of VEGF-A expression. Our results may open up exciting possibility for a new mechanism-based combination ICB therapy by cotargeting an upstream transcriptional driver to circumvent limitations associated with the conventional approach.
As a lipid-activated nuclear transcription factor and metabolic sensor, PPARγ has been reported to modulate tumour immunity by regulating lipid metabolism.39 In glucose-limited and hypoxic TME, fatty acids (FAs) are essential for survival of both tumour cells and CD8+ T cells.40 41 Recent reports further demonstrated aberrant lipid metabolism and lipid droplet accumulation in the development of immunotherapy resistance.42 43 In concord, we also observed evident lipid accumulation in the PD-L1R tumours, which may contribute to adaptive PPARγ upregulation (data not shown). Reflecting the lipid-sensing nature of PPARγ, our scRNA-seq analysis of ICB-resistant tumour cells showed transcriptional upregulation of PPARγ downstream genes involved in FA transport (FABP4 and LPL) and oxidation (ACADL and ACADM).44–46 This transcriptional adaptation may give tumour cells an edge over the effector T cells for the metabolic competition to further restrict the effectiveness of cancer immunotherapy.41
In addition, PPARγ is a critical transcriptional regulator for various immunosuppressive and protumoral activities. In contrast to PPARα which promotes FA catabolism in T lymphocytes for effector functions,40 PPARγ drives M2 macrophage polarisation and DC tolerisation by triggering FA oxidation.47 48 Of note, both PPARγ agonist and antagonist have been shown to enhance anti-PD-(L)1 efficacy in cancer models,49–51 which may depend on the CD274 trans-activation effect of PPARγ in different cell types.49 51 In addition to anti-PD-L1 therapy, we found that cotargeting PPARγ with anti-PD-1 antibody can also counteract the adaptive resistance phenotype (data not shown). As we did not observe changes in tumorous and myeloid PD-L1 expression in our models, it is plausible that the notable ICB-enhancing effects of the selective PPARγ antagonist T0070907 in HCC may extend beyond the tumour cell-intrinsic pathways to the multi-faceted cell-specific roles of PPARγ in the immunosuppressive TME.
There are limitations to our study. Although we have applied single-cell sequencing and immune profiling to characterise the tumour adaptation mechanisms, we cannot exclude the possibility of the existence of an initially resistant clone. Recent technological advancements such as integrated barcoding/scRNA-seq52 53 may enable simultaneous analysis of single-cell transcriptomes and their clonal trajectory during the acquisition of ICB resistance. As tumour heterogeneity may influence drug-resistance mechanisms,29 orthotopic mouse models using cell lines may not fully reflect the antitumour responses during treatment. Using an HDTVi spontaneous HCC model, we have validated the therapeutic benefits of PPARγ/PD-L1 coblockade. On the basis of TME complexity, it needs to be emphasised that VEGF-A overexpression in HCC can be multifactorial14 and is not solely driven by PPARγ upregulation. The role of other PPARγ downstream targets in immunosuppressive and metabolic remodelling warrants further investigation. Furthermore, additional pre-ICB and on-ICB treatment biopsies from patients are required to consolidate the transcriptional adaptive process.
In summary, our study identified PPARγ upregulation as a tumour adaptation mechanism against ICB-induced immune attack. Targeted PPARγ inhibition yielded an immunostimulatory TME with a high cytotoxic CD8+ T/PMN-MDSC to avert the therapeutic resistance with no evidence of toxicity. In accordance, the clinical data showed tumour cell PPARγ induction in ~40% of patients with HCC who had lack of response to anti-PD-1 therapy. As higher baseline PPARγ expression in multiple cancer types was associated with poorer ICB response in terms of patient survival, future studies are warranted to elucidate the effects of PPARγ/PD-1 coblockade in these tumours. On the basis of these findings, we demonstrate a druggable pathway that improves responses to immunotherapy in preclinical models of HCC. Our findings may be of great value to accelerate clinical development of PPARγ inhibitors for combination immunotherapy in HCC and other PPARγ-expressing malignancies.
Acknowledgments
We acknowledge Professor Lars Zender and Professor Tim F Greten for their kind gift of the RIL-175 HCC cell line. We would also thank Professor Xin Chen and Professor Stephanie K Y Ma for providing us with HDTVi plasmids (pT3-EF1a-c-Myc, pT3-EF1a-NRasV12, pCMV-SB13). The scRNA-seq was performed at the Single Cell Omics Core, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong. The immunofluorescence staining was performed at the State Key Laboratory of Translational Oncology, The Chinese University of Hong Kong.
Footnotes
ZX, SLC and JZ contributed equally.
Contributors: Study concept and design: ZX, SLC, JZ, JJ-YS. and AS-LC. Data acquisition and analysis: ZX, SLC, JZ, TTK, XZ, YT, YF, WY, PP-CW, WW-YS-T, XL, JW, WT, ZL, JL, KML, J-TL, MW-YC, HHWL, JJ-YS. and AS-LC. Clinical resources: SLC, AWHC and K-FT. ScRNA-seq platform: YMDL and JSLV. Bioinformatics analysis: HW, JC and KY-LY. Writing of manuscript: ZX, SLC, JZ and AS-LC. Critical review of manuscript: SLC, JZ, KY-LY, YMDL, JJ-YS and AS-LC. Supervision: JJ-YS and AS-LC. Funding acquisition: SLC, JZ, YMDL, JJ-YS and AS-LC. Guarantor: AS-LC.
Funding: This project is supported by the Collaborative Research Fund (C4045-18W to AS-LC), the General Research Fund (14115820 and 14120621 to AS-LC), the Li Ka Shing Foundation (grant number not applicable to YMDL, AS-LC), the Health and Medical Research Fund (07180556 to JZ), the Terry Fox Foundation—Terry Fox Run, Hong Kong (I1008 to SLC, AS-LC), the CUHK Strategic Seed Funding for Collaborative Research Scheme (grant number not applicable to AS-LC) and the Charlie Lee Charitable Foundation (grant number not applicable). We also acknowledge support (funding and study medications) by Merck Sharp and Dohme (MSD-IIS 55253 to SLC) for the clinical trial (NCT03419481).
Competing interests: The authors declare no conflicts of interest that pertain to this work except the following declarations. YMDL is a scientific cofounder of Grail, receives royalties from Illumina, Grail, Sequenom, Xcelom, DRA and Take2, serves as a consultant to Decheng Capital and holds equity in Illumina/Grail, DRA and Take2. SLC serves as a consultant to and receives honoraria from Astra-Zeneca, Eisai and MSD.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Written informed consent was provided by all patients prior to biopsy acquisition. All clinical records in this study were obtained with the approval of the joint CUHK-NTEC Clinical Research Ethics Committee (CREC 2017.465-T). Participants gave informed consent to participate in the study before taking part.
References
- 1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301–14. 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 2. Zhou J, Liu M, Sun H, et al. Hepatoma-intrinsic CCRK inhibition diminishes myeloid-derived suppressor cell immunosuppression and enhances immune-checkpoint blockade efficacy. Gut 2018;67:931–44. 10.1136/gutjnl-2017-314032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu M, Zhou J, Liu X, et al. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut 2020;69:365–79. 10.1136/gutjnl-2018-317257 [DOI] [PubMed] [Google Scholar]
- 4. Yang W, Feng Y, Zhou J, et al. A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci Transl Med 2021;13:eaaz6804. 10.1126/scitranslmed.aaz6804 [DOI] [PubMed] [Google Scholar]
- 5. Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 6. Zhu AX, Abbas AR, de Galarreta MR, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med 2022;28:1599–611. 10.1038/s41591-022-01868-2 [DOI] [PubMed] [Google Scholar]
- 7. Pinter M, Scheiner B, Peck-Radosavljevic M. Immunotherapy for advanced hepatocellular carcinoma: a focus on special subgroups. Gut 2021;70:204–14. 10.1136/gutjnl-2020-321702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim TK, Vandsemb EN, Herbst RS, et al. Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities. Nat Rev Drug Discov 2022;21:529–40. 10.1038/s41573-022-00493-5 [DOI] [PubMed] [Google Scholar]
- 9. Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov 2020;19:39–56. 10.1038/s41573-019-0044-1 [DOI] [PubMed] [Google Scholar]
- 10. Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20:743–56. 10.1038/s41568-020-00302-4 [DOI] [PubMed] [Google Scholar]
- 11. Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis-Marcisak EF, Deshpande A, Stein-O’Brien GL, et al. From bench to bedside: single-cell analysis for cancer immunotherapy. Cancer Cell 2021;39:1062–80. 10.1016/j.ccell.2021.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chuah S, Lee J, Song Y, et al. Uncoupling immune trajectories of response and adverse events from anti-PD-1 immunotherapy in hepatocellular carcinoma. J Hepatol 2022;77:683–94. 10.1016/j.jhep.2022.03.039 [DOI] [PubMed] [Google Scholar]
- 14. Ma L, Hernandez MO, Zhao Y, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 2019;36:418–30. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Q, He Y, Luo N, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 2019;179:829–45. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 16. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity 2020;52:17–35. 10.1016/j.immuni.2019.12.011 [DOI] [PubMed] [Google Scholar]
- 17. Kwong TT, Wong CH, Zhou JY, et al. Chemotherapy-induced recruitment of myeloid-derived suppressor cells abrogates efficacy of immune checkpoint blockade. JHEP Rep 2021;3:100224. 10.1016/j.jhepr.2020.100224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andrews MC, Wargo JA. Cancer evolution during immunotherapy. Cell 2017;171:740–2. 10.1016/j.cell.2017.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer 2012;12:181–95. 10.1038/nrc3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, van der Leun AM, Yofe I, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 2019;176:775–89. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550–8. 10.1038/s41591-018-0136-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francque S, Verrijken A, Caron S, et al. PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol 2015;63:164–73. 10.1016/j.jhep.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 23. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18:197–218. 10.1038/s41573-018-0007-y [DOI] [PubMed] [Google Scholar]
- 24. Bhattacharya S, Dunn P, Thomas CG, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data 2018;5:180015. 10.1038/sdata.2018.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu J, Shen B, Chu ESH, et al. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology 2010;51:2008–19. 10.1002/hep.23550 [DOI] [PubMed] [Google Scholar]
- 26. Kim CG, Jang M, Kim Y, et al. VEGF-A drives TOX-dependent T cell exhaustion in anti-PD-1-resistant microsatellite stable colorectal cancers. Sci Immunol 2019;4:eaay0555. 10.1126/sciimmunol.aay0555 [DOI] [PubMed] [Google Scholar]
- 27. Fukumura D, Kloepper J, Amoozgar Z, et al. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol 2018;15:325–40. 10.1038/nrclinonc.2018.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shang J, Mosure SA, Zheng J, et al. A molecular switch regulating transcriptional repression and activation of PPARγ. Nat Commun 2020;11:956. 10.1038/s41467-020-14750-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res 2015;21:1258–66. 10.1158/1078-0432.CCR-14-1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wen L, Xin B, Wu P, et al. An efficient combination immunotherapy for primary liver cancer by harmonized activation of innate and adaptive immunity in mice. Hepatology 2019;69:2518–32. 10.1002/hep.30528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest 2015;45:609–23. 10.1111/eci.12441 [DOI] [PubMed] [Google Scholar]
- 32. Hsu C-L, Ou D-L, Bai L-Y, et al. Exploring markers of exhausted CD8 T cells to predict response to immune checkpoint inhibitor therapy for hepatocellular carcinoma. Liver Cancer 2021;10:346–59. 10.1159/000515305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2017;168. 10.1016/j.cell.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 34. Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med 2019;25:462–9. 10.1038/s41591-019-0349-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Braun DA, Hou Y, Bakouny Z, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 2020;26:909–18. 10.1038/s41591-020-0839-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rambow F, Rogiers A, Marin-Bejar O, et al. Toward minimal residual disease-directed therapy in melanoma. Cell 2018;174:843–55. 10.1016/j.cell.2018.06.025 [DOI] [PubMed] [Google Scholar]
- 37. Kieffer Y, Hocine HR, Gentric G, et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov 2020;10:1330–51. 10.1158/2159-8290.CD-19-1384 [DOI] [PubMed] [Google Scholar]
- 38. Sangro B, Sarobe P, Hervás-Stubbs S, et al. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:525–43. 10.1038/s41575-021-00438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christofides A, Konstantinidou E, Jani C, et al. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism 2021;114:154338. 10.1016/j.metabol.2020.154338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Kurupati R, Liu L, et al. Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 2017;32:377–91. 10.1016/j.ccell.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kishton RJ, Sukumar M, Restifo NP. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab 2017;26:94–109. 10.1016/j.cmet.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Denis M, Grasselly C, Choffour P-A, et al. In vivo syngeneic tumor models with acquired resistance to anti-PD-1/PD-L1 therapies. Cancer Immunol Res 2022;10:1013–27. 10.1158/2326-6066.CIR-21-0802 [DOI] [PubMed] [Google Scholar]
- 43. Ramapriyan R, Caetano MS, Barsoumian HB, et al. Altered cancer metabolism in mechanisms of immunotherapy resistance. Pharmacol Ther 2019;195:162–71. 10.1016/j.pharmthera.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 44. Hotamisligil GS, Bernlohr DA. Metabolic functions of FABPs -- mechanisms and therapeutic implications. Nat Rev Endocrinol 2015;11:592–605. 10.1038/nrendo.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagashima K, Lopez C, Donovan D, et al. Effects of the PPARgamma agonist pioglitazone on lipoprotein metabolism in patients with type 2 diabetes mellitus. J Clin Invest 2005;115:1323–32. 10.1172/JCI23219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang WH, Lai AG. The pan-cancer mutational landscape of the PPAR pathway reveals universal patterns of dysregulated metabolism and interactions with tumor immunity and hypoxia. Ann N Y Acad Sci 2019;1448:65–82. 10.1111/nyas.14170 [DOI] [PubMed] [Google Scholar]
- 47. Liu S, Zhang H, Li Y, et al. S100A4 enhances protumor macrophage polarization by control of PPAR-γ-dependent induction of fatty acid oxidation. J Immunother Cancer 2021;9:e002548. 10.1136/jitc-2021-002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao F, Xiao C, Evans KS, et al. Paracrine Wnt5a-β-catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity 2018;48:147–60. 10.1016/j.immuni.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gutting T, Hauber V, Pahl J, et al. PPARγ induces PD-L1 expression in MSS+ colorectal cancer cells. Oncoimmunology 2021;10:1906500. 10.1080/2162402X.2021.1906500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu B, Sun X, Yuan B, et al. PPARγ inhibition boosts efficacy of PD-L1 checkpoint blockade immunotherapy against murine melanoma in a sexually dimorphic manner. Int J Biol Sci 2020;16:1526–35. 10.7150/ijbs.42966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu B, Sun X, Gupta HB, et al. Adipose PD-L1 modulates PD-1/PD-L1 checkpoint blockade immunotherapy efficacy in breast cancer. Oncoimmunology 2018;7:e1500107. 10.1080/2162402X.2018.1500107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang CA, Jen J, Jiang S, et al. Ontogeny and vulnerabilities of drug-tolerant persisters in HER2+ breast cancer. Cancer Discov 2022;12:1022–45. 10.1158/2159-8290.CD-20-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baron CS, van Oudenaarden A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat Rev Mol Cell Biol 2019;20:753–65. 10.1038/s41580-019-0186-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328364supp001.pdf (21.3MB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.