SUMMARY
Many toxic man-made compounds have been introduced into the environment, and bacterial strains that are able to grow on them are ideal model systems for studying the evolution of metabolic pathways and regulatory systems. Acidovorax sp. strain JS42 is unique in its ability to use 2-nitrotoluene as a sole carbon, nitrogen, and energy source for growth. The LysR-type transcriptional regulator NtdR activates expression of the 2-nitrotoluene degradation genes not only when nitroaromatic compounds are present, but also in the presence of a wide range of aromatic acids and analogs. The molecular determinants of inducer specificity were identified through comparative analysis with NagR, the activator of the naphthalene degradation pathway genes in Ralstonia sp. strain U2. Although NagR is 98% identical to NtdR, it does not respond to nitrotoluenes. Exchange of residues that differ between NagR and NtdR revealed that residues at positions 227 and 232 were key for the recognition of nitroaromatic compounds, while the amino acid at position 169 determined the range of aromatic acids recognized. Structural modeling of NtdR suggests that these residues are near the predicted inducer binding pocket. Based on these results, an evolutionary model is presented that depicts the stepwise evolution of NtdR.
INTRODUCTION
Nitroaromatic compounds are a diverse group of chemicals used in a wide variety of industrial and consumer products such as hair dyes, drugs, pesticides, and explosives. Their prevalent use has resulted in contamination of soil and groundwater, where they are resistant to biodegradation due to their toxicity and chemical stability (Spain et al., 2000). Environmental contamination is further exacerbated by the tendency of the nitro-groups to undergo reduction to form aromatic amines, which not only are more toxic, but in some cases are known carcinogens (Cerniglia and Somerville, 1995). Although many of these compounds are synthetic and have been present in the environment for relatively short periods of time, bacterial strains capable of using specific nitroaromatic compounds as sole carbon, nitrogen, and energy sources for growth have been isolated from contaminated environments (Nishino and Spain, 2004).
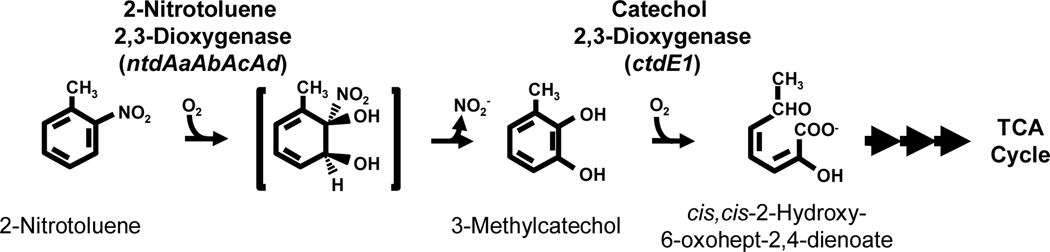
In Acidovorax sp. strain JS42, degradation of nitrobenzene and 2-nitrotoluene is initiated by enzymatic dioxygenation to produce (3-methyl)catechol and nitrite. Metabolism of the catechols proceeds by a standard meta ring-cleavage pathway that funnels intermediates into the citric acid cycle (Fig. 1) (Haigler et al., 1994). The enzyme responsible for the initial oxidation is 2-nitrotoluene 2,3-dioxygenase (2NTDO), which belongs to the naphthalene family of Rieske non-heme iron oxygenases (An et al., 1994; Parales et al., 2005). 2NTDO is a multicomponent enzyme consisting of an iron-sulfur flavoprotein reductase and a Rieske ferredoxin that transfer electrons from NAD(P)H to the catalytic oxygenase, an α3β3 heterohexamer (An et al., 1994; Parales et al., 2005). The four genes that encode 2NTDO (ntdAaAbAcAd) are divergently transcribed from ntdR, which encodes a LysR-type regulatory protein that is required for expression of the ntd operon and for growth of JS42 on 2-nitrotoluene (Lessner et al., 2003).
Fig. 1.
2-Nitrotoluene degradation pathway in Acidovorax sp. strain JS42.
Based on sequence comparisons, it is apparent that the genes and enzymes of this and related pathways for the degradation of nitroarene substrates have ancestral origins common to the naphthalene degradation pathway of Ralstonia sp. strain U2 (Fuenmayor et al., 1998; Jones et al., 2003; Ju and Parales, 2006; Lessner et al., 2002; Lessner et al., 2003; Parales, 2000; Zhou et al., 2001). This evolutionary link is further supported by investigations focusing on the regulation of these pathways. In both Acidovorax sp. strain JS42 and Ralstonia sp. strain U2, expression of each dioxygenase operon is activated by the divergently transcribed LysR-regulator in the presence of recognized inducer compounds (Jones et al., 2003; Lessner et al., 2003). Sequence comparisons revealed that the regulator in strain JS42 (NtdR) differs from the regulator in strain U2 (NagR) by only five amino acids. The LysR-binding sites and promoters are identical in both strains, and both regulators activate gene expression in the presence of salicylate, which is an intermediate of the naphthalene degradation pathway and the natural inducer of the naphthalene degradation genes in strain U2 (Jones et al., 2003; Lessner et al., 2003). NtdR, however, has acquired the ability to activate transcription of the ntd operon in response to 2-nitrotoluene and other nitroaromatic compounds (Lessner et al., 2003).
In this study, we examined the role of NtdR in the physiological response of strain JS42 towards nitroaromatic compounds by evaluating the dynamics of nitroarene dioxygenase gene expression in mutants of JS42 blocked at different steps of 2-nitrotoluene metabolism. Rational mutagenesis was then used to examine the effects of single and multiple mutations in NtdR and NagR on gene induction in the JS42 strain background. Variants of JS42 expressing these mutant regulators were tested with a suite potential inducer compounds to determine the chemical properties required for inducer recognition, as well as the critical amino acids that mediate this specificity. Based on these results, we propose a likely evolutionary history for NtdR, i.e., the order in which the mutations accumulated in the development of the ability to detect nitroarene compounds as inducers.
RESULTS
Time course of induction by NtdR in response to salicylate, 2-nitrotoluene, and 2-nitrobenzyl alcohol.
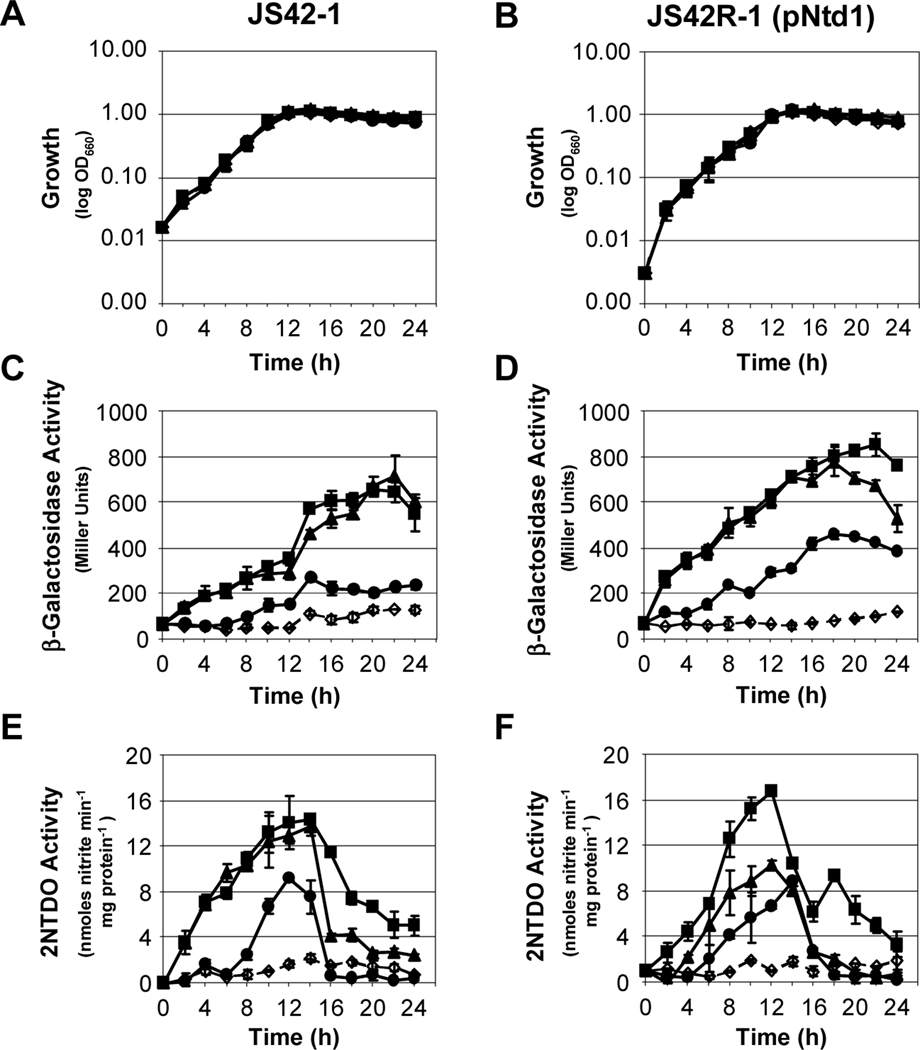
Temporal expression of the ntd operon encoding 2NTDO was monitored in growing cultures to determine the timing of induction. 2NTDO and β-galactosidase activities were measured in liquid cultures of wild-type JS42 carrying a chromosomal ntd-lacZ transcriptional fusion (JS42–1), an ntdR mutant (JS42R-1[pBBR1MCS]), and a complemented ntdR mutant (JS42R-1[pNtd1]) grown in the presence of salicylate (2-hydroxybenzoate), 2-nitrotoluene, or 2-nitrobenzyl alcohol. Previous studies had shown that 3-methylcatechol and nitrite, the primary products of 2-nitrotoluene oxidation, did not induce dioxygenase gene expression (Lessner et al., 2003). However, a small amount of the dead-end metabolite 2-nitrobenzyl alcohol is also formed during 2-nitrotoluene oxidation (Haigler et al., 1994), so this compound was tested as a potential inducer. All cultures had similar growth profiles, reaching maximal cell densities approximately 14 hours after inoculation (Fig. 2A, B). Both JS42–1 and JS42R-1(pNtd1), each of which carries a wild-type ntdR gene, either in single copy on the chromosome or on a multi-copy plasmid, showed similar induction profiles (Fig. 2C-F)). In contrast, JS42R-1(pBBR1MCS), which has no functional ntdR gene, had no detectable β-galactosidase or 2NTDO activity (data not shown). All three compounds caused induction of transcription from the reporter, although 2-nitrobenzyl alcohol was not as strong an inducer as salicylate or 2-nitrotoluene (Fig. 2C-F). 2NTDO activity peaked between 12 and 14 hours before sharply declining as cells entered stationary phase (Fig. 2E, F). In comparison, β-galactosidase activity did not decrease until later stages of growth. The persistent β-galactosidase activity may reflect the inherent stability of the enzyme in JS42. Regardless, these results demonstrate that β-galactosidase measurements provide an accurate representation of the induction of the ntd operon up to 14 hours into growth. Therefore, for all subsequent experiments, cultures were harvested after growth for between 12 and 14 h.
Fig. 2.
Timing of ntd gene expression during growth of JS42 derivatives. Growth as measured by culture turbidity at 660 nm (A, B), β-galactosidase activity measured from the chromosomal ntdA-lacZ fusion (C, D), and 2NTDO activity (E, F) were followed over a 24-hour period for JS42–1 and JS42R-1(pNtd1). Cultures were grown in MSB with 10 mM succinate (open diamonds, dashed lines), or 10 mM succinate supplemented with 500 μM of salicylate (squares), 2-nitrotoluene (triangles), or 2-nitrobenzyl alcohol (circles) as inducers. n = 6. Error bars indicate standard deviations.
NtdR controls expression of the ntd operon by direct detection of 2-nitrotoluene in wild-type JS42 and catabolic mutants.
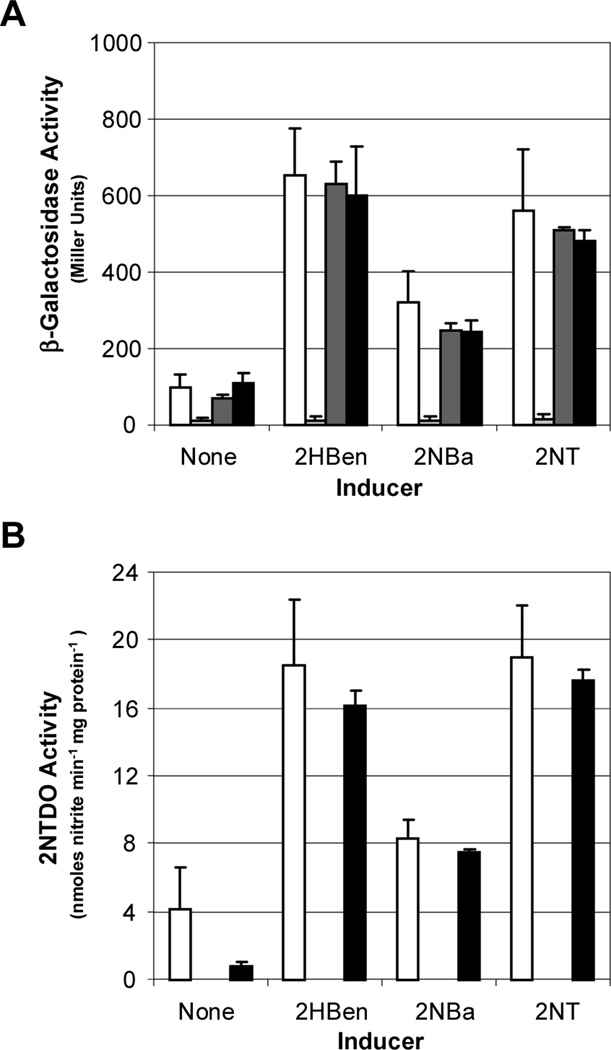
Results presented in Fig. 2 show that 2-nitrobenzyl alcohol, a product of 2-nitrotoluene oxidation by 2NTDO, functions as an inducer. To determine whether NtdR also senses 2-nitrotoluene, we compared induction in strains blocked at different steps in 2-nitrotoluene catabolism. We constructed a mutant strain in which the ntdAc gene, which encodes the α subunit of 2NTDO, was inactivated (JS42Ac). A second mutant (JS42E1) with a streptomycin resistance cassette inserted in the ctdE1 gene, which encodes catechol 2,3-dioxygenase (Fig. 1), was also constructed. The same transposable ntdA-lacZ fusion present in JS42–1 and JS42R-1 (Lessner et al., 2003) was introduced into the two mutant strains to generate JS42Ac-1 and JS42E1–1. As reported previously (Lessner et al., 2003), JS42R-1, which lacks a functional NtdR protein, was unresponsive to all compounds, and β-galactosidase activities were below those of JS42–1 grown in the absence of inducer (Fig. 3A). As expected, there was no detectable 2NTDO activity in JS42R-1, which cannot induce the genes encoding 2NTDO, or in JS42Ac-1 (Fig. 3B). Both JS42Ac-1 and JS42E1–1 showed similar levels of β-galactosidase as JS42–1, and 2NTDO activities were comparable in JS42E1–1 and JS42–1 (Fig. 3). These results indicate that 2-nitrotoluene metabolism is not required for ntd operon induction, and therefore 2-nitrotoluene is directly detected as an inducer.
Fig. 3.
Expression of the ntd genes in wild type and catabolic and regulatory mutants of Acidovorax sp. JS42. β-Galactosidase activity measured from the chromosomal ntdA-lacZ fusion (A) and 2NTDO activity (B) in cultures of JS42–1 (wild type; white bars), JS42R-1 (ntdR::Km; light grey bars), JS42Ac-1 (ntdAc::Km; dark grey bars), and JS42E1–1 (ctdE1::Sm; black bars) grown on succinate alone (none) or with 500 μM of salicylate (2HBen), 2-nitrobenzyl alcohol (2NBa) or 2-nitrotoluene (2NT) provided as inducers. n = 6. Error bars indicate standard deviations.
NtdR and NagR inducer specificity.
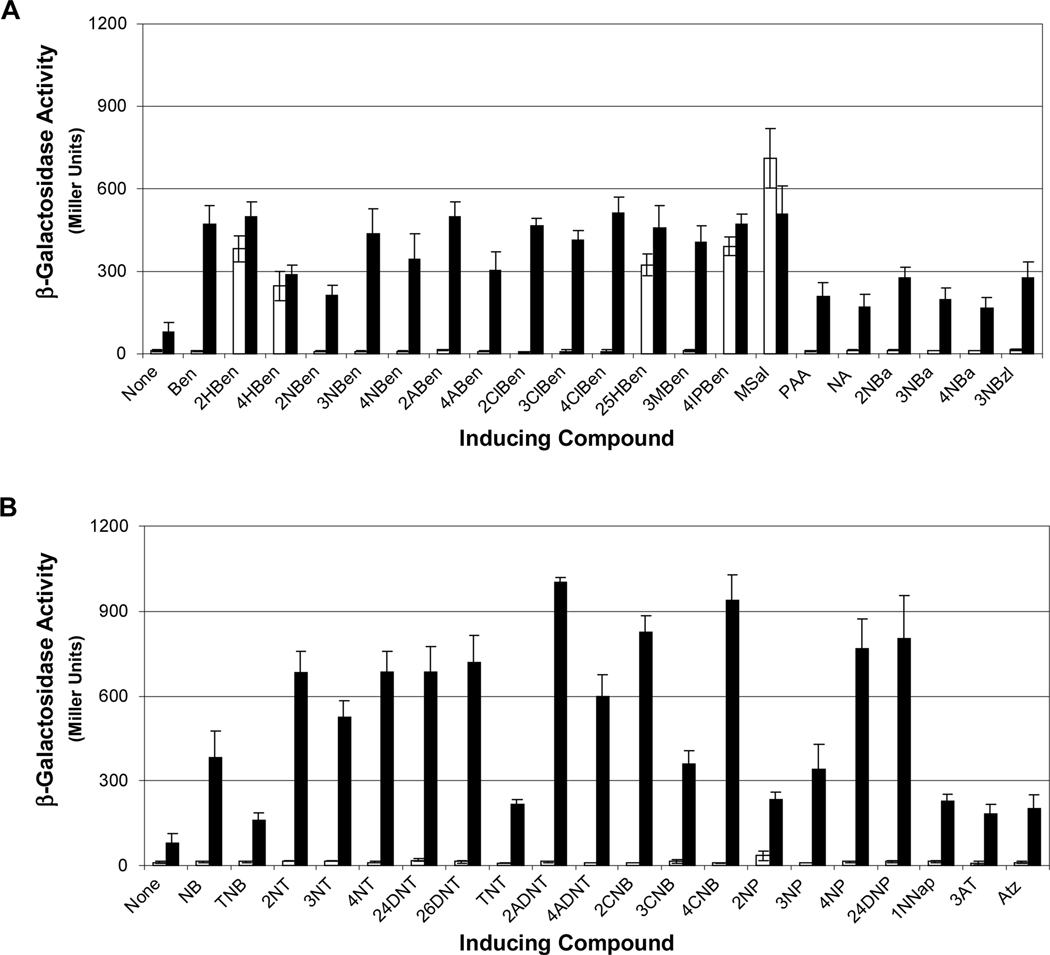
In the JS42 strain background, NtdR was previously shown to recognize nitrobenzene, mono- and dinitrotoluenes, aminodinitrotoluenes, salicylate, and anthranilate (2-aminobenzoate), while NagR recognized only salicylate (Lessner et al., 2003). To further evaluate the range of inducers recognized by NtdR and NagR, JS42R-1(pNtd1) and JS42R-1(pNag1) were tested with a panel of 63 compounds (Supplementary Tables 1-8). Both β-galactosidase and 2NTDO assays were carried out and induction results were similar in all cases. For simplicity, only the β-galactosidase activities are reported here. Of the 63 tested compounds, only five (salicylate, gentisate, 4-hydroxybenzoate, 4-isopropylbenzoate, and methyl salicylate) were recognized by NagR, while 41 of the 63 compounds elicited at least 2-fold induction with NtdR (Fig. 4). The negative control strain JS42R-1(pBBR1MCS) had no β-galactosidase activity with any of the tested compounds (data not shown).
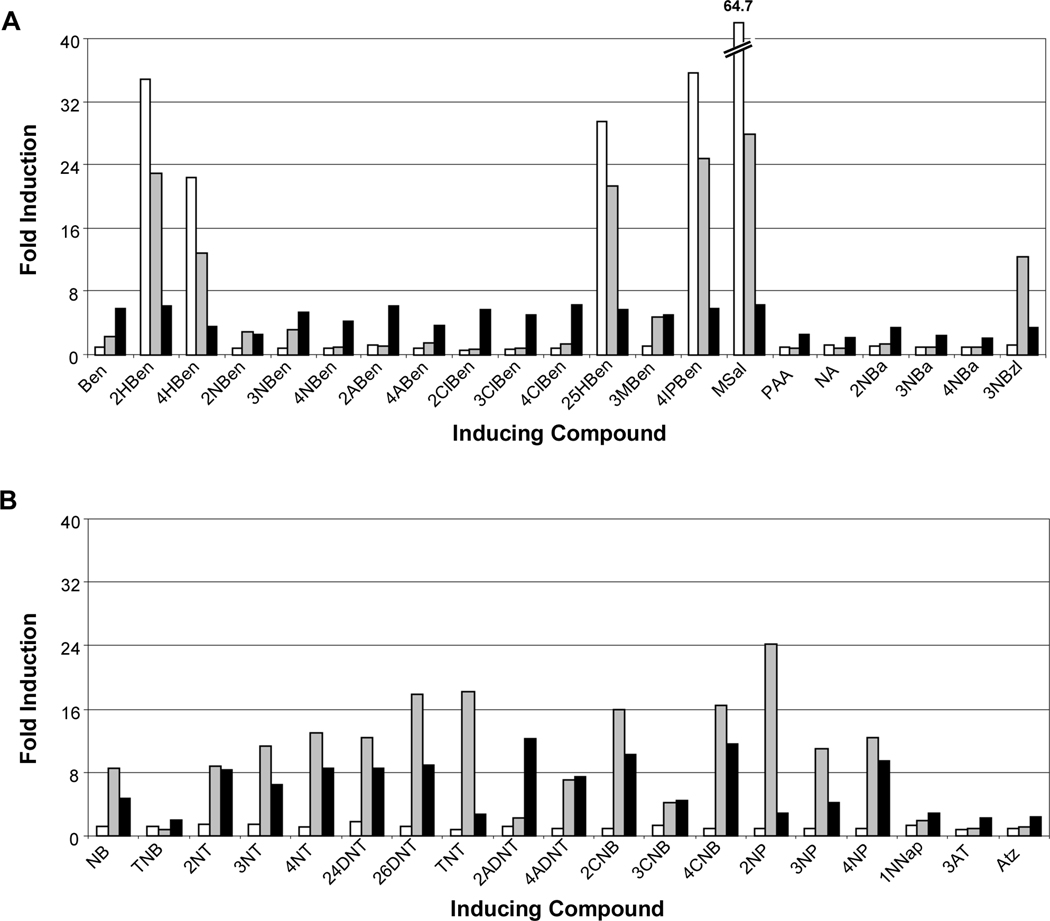
Fig. 4.
Inducing compounds detected by NagR and NtdR in Acidovorax sp. JS42R-1. (A) β-Galactosidase activity measured from the chromosomal ntdA-lacZ fusion in response to aromatic acids and related compounds detected by NagR (white bars) and NtdR (black bars). None: no inducer added; Ben: benzoate; HBen: 2-, 4-hydroxybenzoate; NBen: 2-, 3-, 4-nitrobenzoate; ABen: 2-, 4-aminobenzoate; ClBen: 2-, 3-, 4-chlorobenzoate; 25HBen: 2,5-dihydroxybenzoate (gentisate); 3MBen: 3-methylbenzoate; 4IPBen: 4-isopropylbenzoate; MSal: methyl salicylate; PAA: phenylacetic acid; NA: nicotinic acid; NBa: 2-, 3-, 4-nitrobenzyl alcohol; 3NBzl: 3-nitrobenzaldehyde. (B) β-Galactosidase activity in response to nitrobenzenes, nitrotoluenes, nitrophenols, and related compounds by NagR (white bars) and NtdR (black bars). None: no inducer added; NB: nitrobenzene; TNB: 1,3,5-trinitrobenzene; NT: 2-, 3-, 4-nitrotoluene; DNT: 2,4-, 2,6-dinitrotoluene; TNT: 2,4,6-trinitrotoluene; 2ADNT: 2-amino-4,6-dinitrotoluene; 4ADNT: 4-amino-2,6-dinitrotoluene; CNB: 2-, 3-, 4-chloronitrobenzene; NP: 2-, 3-, 4-nitrophenol; 24DNP: 2,4-dinitrophenol; 1NNap: 1-nitronaphthalene; AT: 3-aminotoluene; Atz: atrazine. Compounds not detected by either protein were 3-hydroxybenzoate, 3-aminobenzoate, benzyl alcohol, 2-hydroxybenzylalcohol, 2-aminobenzyl alcohol, benzaldehyde, 3-hydroxybenzaldehyde, benzene, aminobenzene, chlorobenzene, toluene, 2- and 4-chloroaminobenzene, phenol, 2-aminophenol, catechol, 3- and 4-methylcatechol, cyanuric acid, and naphthalene. N = 6. Error bars indicate standard deviations.
The carboxylic acid functional group appeared to be key for binding aromatic acids and in most cases, the presence of additional functional groups did not affect recognition. Methoxylation of the terminal alcohol group of salicylate (methyl salicylate) did not change induction activity, but derivatives of benzoate, salicylate, and anthranilate lacking the carbonyl oxygen (benzyl alcohol, 2-hydroxybenzyl alcohol, and 2-aminobenzyl alcohol) did not function as inducers (Fig. 4). However, the presence of a nitro group on the aromatic ring was able to supersede the need for a carboxylic acid group, as all three nitrobenzyl alcohol isomers were recognized as inducers. Although phenol, chlorobenzene, benzaldehyde and 3-hydroxybenzaldehyde were not inducers, nitrophenols, chloronitrobenzenes, and 3-nitrobenzaldehyde were recognized by NtdR (Fig. 4). Similarly, although naphthalene was not recognized as an inducer, 1-nitronaphthalene did elicit a modest induction (Fig. 4B). Increasing the distance of the carboxylic acid group from the benzene ring (benzoate versus phenylacetic acid) reduced induction activity by half. Consistent with previous results (Lessner et al., 2003), NtdR recognized nitrobenzene and various nitrotoluenes (Fig. 4B). Other environmental contaminants including the explosives 1,3,5-trinitrobenzene (TNB) and 2,4,6-trinitrotoluene (TNT), and the s-triazine herbicide atrazine were also recognized as inducers by NtdR (Fig. 4B).
Residues at positions 169 and 227 affect gene expression in the absence of inducer.
The previous study reported a 30-fold difference in uninduced expression levels in JS42R-1(pNtdR) and JS42R-1(pNagR) (Lessner et al., 2003). We found that one of the components of Balch’s vitamins, 4-aminobenzoate, is actually recognized by NtdR (Fig. 4A), so for all experiments reported in this study, we grew Acidovorax strains in a modified medium in which 4-aminobenzoic acid was omitted. However, even in the absence of 4-aminobenzoate, uninduced expression in strains carrying NtdR was 7-fold higher than in those with NagR (Fig. 4, Fig. 5).
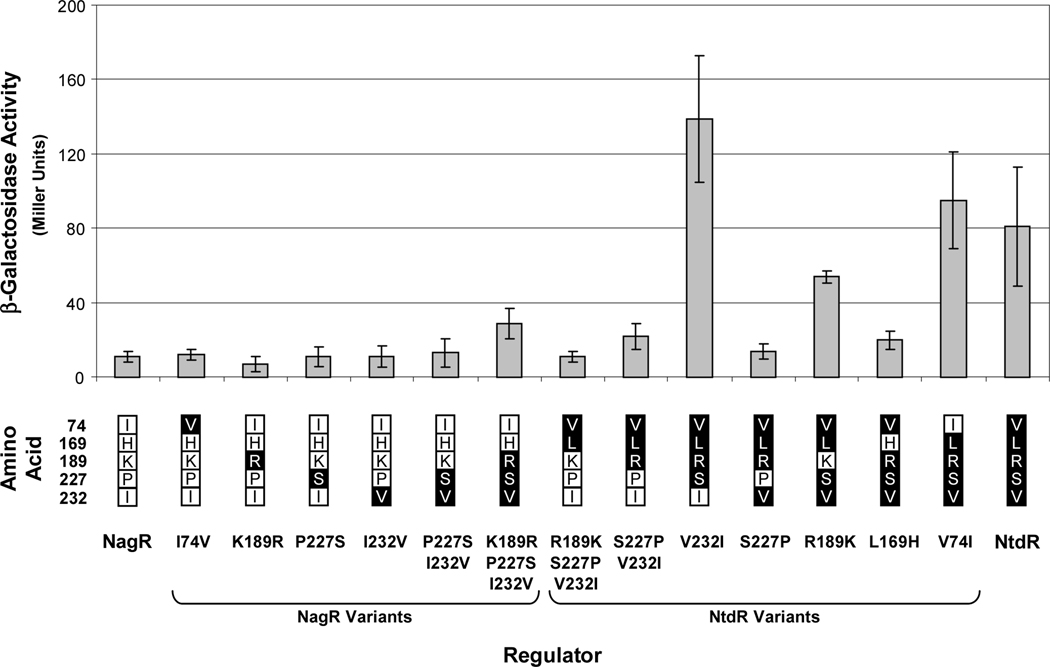
Fig. 5.
Uninduced levels of expression from the ntdA promoter mediated by wild-type NagR and NtdR and the 13 active derivatives. β-Galactosidase activities from the chromosomal ntdA-lacZ fusion were measured in cultures of JS42R-1 expressing NtdR-NagR variants from plasmid pBBR1-MCS after growth in minimal medium containing succinate and chloramphenicol. N = 6. Error bars indicate standard deviations. No activity was detected with NagR H169L (data not shown). The amino acid substitutions in each NagR and NtdR variant are indicated below the graph. Residues native to NagR and NtdR are indicated in white and black boxes, respectively.
To clarify the contributions of the five differing amino acids to inducer specificity and induction activity, site-directed mutagenesis was used to replace each of the amino acids present in one protein with the corresponding amino acid of the other protein in single and multiple combinations. The 14 mutant ntdR and nagR constructs were then expressed in JS42R-1, and the resulting strains were assayed for β-galactosidase and 2NTDO activities after growth under various conditions. When uninduced activities were compared, we found that the NtdR L169H variant had 4-fold lower activity than NtdR, and the S227P substitution in NtdR resulted in expression comparable to that of NagR (Fig. 5). It also appeared that the residues at positions 189 and 232 slightly affected uninduced expression levels in certain protein contexts (Fig. 5).
The residues at positions 169, 227, and 232 control inducer recognition.
The results presented in Fig. 4 demonstrate that NtdR is a generalist, recognizing 41 chemical inducers, while NagR is much more specific and lacks the ability to detect any nitroaromatic compounds. Acidovorax sp. JS42R-1 expressing NagR, NtdR, and the 14 variant NagR-NtdR proteins were assayed for β-galactosidase and 2NTDO activities after growth in the presence of the same set of 63 test compounds to evaluate the roles of the differing amino acids in inducer recognition. Complete β-galactosidase assay results with all tested inducers are presented in Tables S1-S8. Results for all NagR-NtdR derivatives with a representative subset of inducers (2-nitrotoluene, TNT, salicylate, and benzoate) are shown in Fig. 6. Single amino acid substitutions at positions 74 and 189 in either NagR or NtdR did not cause major changes in the range of inducers recognized, but in some cases caused slight (typically less than 2-fold) changes in the strength of induction (Fig. 6; Tables S1-S8). The NtdR R189K and NtdR V74I proteins demonstrated the same specificity as NtdR with the following exceptions. Both lost the ability to recognize the weak inducers 1,3,5-trinitrobenzene and 3-aminotoluene (Table S5), and the NtdR R189K protein gained the ability to recognize three compounds (benzaldehyde, 9-fold induction; naphthalene, 5-fold induction; and 3-aminobenzoate, 3.5-fold induction) that did not function as inducers with either NtdR or NagR (Tables S1, S3, S7).
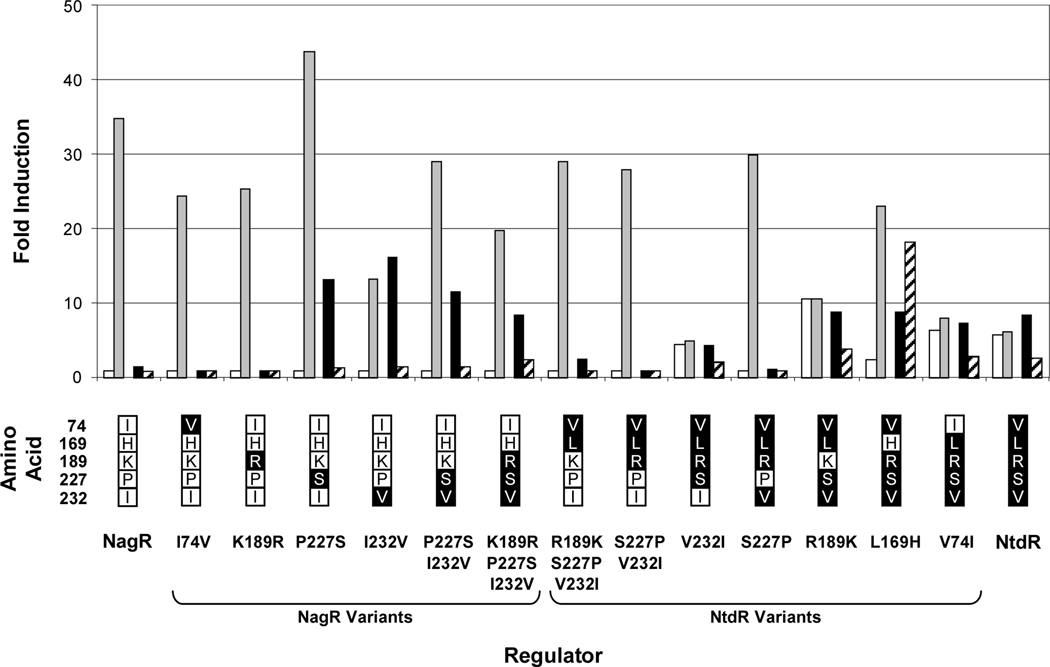
Fig. 6.
Effect of selected inducers on activity from the ntdA promoter in JS42R-1 expressing NtdR-NagR variants. β-Galactosidase activities from the chromosomal ntdA-lacZ fusion were measured in cultures grown as described in the legend to Fig. 5 in response to benzoate (white bars); salicylate (grey bars); 2NT, 2-nitrotoluene (black bars); TNT, 2,4,6-trinitrotoluene (striped bars). Activities are reported as fold induction over background for each strain. N = 6. Standard deviations are reported in Tables S1-S8. No activity was detected with NagR H169L (data not shown). The amino acid substitutions in each NagR and NtdR variant are indicated below the graph. Residues native to NagR and NtdR are indicated in white and black boxes, respectively.
Substitutions at positions 169, 227, and 232 in NtdR and NagR resulted in the greatest changes in specificity, suggesting that the residues at these positions may be directly involved in binding inducers. None of the NagR variants gained the ability to detect benzoate, but all NtdR mutants with a S227P substitution lost the ability, and the NtdR L169H variant had a reduced response to benzoate. All NagR variants with P227S and I232V substitutions acquired the ability to detect 2-nitrotoluene, while the S227P substitution in NtdR changed the specificity with these four test inducers to mirror that of wild-type NagR (Fig. 6). One particularly interesting mutant (NtdR L169H) showed very strong induction in response to TNT (Fig. 6). At the same time, the L169H substitution eliminated half of the compounds recognized by wild-type NtdR, including almost all of the aromatic acids not recognized by NagR (Fig. 7A). These results suggest that the residue at position 169 is particularly important for recognition of aromatic acids. Recognition of many nitroaromatic inducers was retained, however, with the exception of 4-nitrobenzoate, 2-, 3- and 4-nitrobenzyl alcohol, 1,3,5-trinitrobenzene, and 1-nitronaphthalene (Fig. 7). Therefore, NtdR L169H had increased specificity for nitroaromatic inducers: of the 41 compounds recognized by NtdR, 25 were nitroaromatic compounds; NtdR L169H recognized 19 nitroaromatic compounds out of a total of 26 recognized inducers. NtdR L169H had the largest responses to 2,6-DNT, TNT, and 2NP (18-, 18-, and 24-fold induction, respectively, compared to 9-, 2.7-, and 2.8-fold induction by NtdR; Fig. 7B, Tables S5 and S7). Interestingly, while the response to TNT was improved in NtdR L169H, the response to 2-aminodinitrotoluene was substantially reduced (Fig. 7B). Fold induction with most compounds was significantly higher for NtdR L169H than NtdR (Fig. 7) due to the lower uninduced activity with NtdR L169H (Fig. 5). In contrast to the L169H mutation in NtdR, the opposite mutation in NagR (H169L) resulted in a complete loss of induction with all tested compounds (data not shown).
Fig. 7.
Inducing compounds detected by NagR, NtdR, and NtdR L169H in Acidovorax sp. JS42R-1. (A) β-Galactosidase activity measured from the chromosomal ntdA-lacZ fusion in response to aromatic acids and related compounds. (B) β-Galactosidase activity in response to nitrobenzenes, nitrotoluenes, nitrophenols, and related compounds. Abbreviations are as in the legend to Fig. 4. NagR (white bars) NtdR L169H (grey bars) and NtdR (black bars). N = 6. Standard deviations are reported in Tables S1-S8.
The amino acids at positions 227 and 232 were particularly important for the recognition of nitroaromatic compounds. The single P227S substitution in NagR resulted in the recognition of 21 additional compounds, including 16 nitroaromatic compounds (Tables S2, S4, S6, S8). The NagR I232V variant gained the ability to detect six nitroaromatic compounds. Although both proteins allowed for induction by nitrobenzene, 2-nitrotoluene, aminodinitrotoluenes, and 2-nitrophenol, NagR P227S recognized additional nitroaromatic compounds such as 2- and 3-nitrobenzoates, 3- and 4-nitrotoluenes, 2,4- and 2,6-dinitrotoluenes, all of the tested nitrophenols, and 1-nitronaphthalene. Interestingly, both NagR P227S and NagR I232V also responded to naphthalene, which was not recognized by either NagR or NtdR (Tables S2, S4, S6, S8).
NagR P227S I232V had a similar inducer recognition profile to that of NagR P227S, but the double mutant gained the ability to respond to 4-chloronitrobenzene and 2- and 3-nitrobenzyl alcohol. In the context of the NagR P227S I232V variant, the residue at position 189 played a role in determining inducer recognition, as the NagR K189R P227S I232V triple mutant gained the ability to detect TNT, and it had stronger overall responses to several nitroaromatic inducers, including dinitrotoluenes, aminodinitrotoluenes, and 1-nitronaphthalene (Tables S2, S4, S6, S8).
In the NtdR background, the S227P mutation dramatically narrowed the specificity to a profile similar to that of NagR, with only salicylate, 2-chlorobenzoate, gentisate, and 4-isopropylbenzoate recognized as inducers (Tables S1, S3, S5, S7). Despite losing the ability to induce the ntd genes in response to 1,3,5-trinitrobenzene and 1-nitronaphthalene, the overall inducer profile of the NtdR V232I mutant was similar to that of NtdR. In addition, this mutant protein gained the ability to recognize naphthalene, which is not detected by either wild-type protein. In combination, the S227P and V232I substitutions in NtdR reduced the number of recognized inducers to only nine compounds. NtdR R189K S227P V232I recognized a similar subset of inducers as NtdR S227P V232I (Tables S1-S8).
These results show that serine at position 227 is the major amino acid controlling recognition of nitroaromatic compounds and aromatic acids. Valine 232 appeared to act synergistically with serine 227 to extend the range of detected nitroaromatic compounds.
Thresholds of detection and apparent regulatory protein-effector affinities.
To determine the minimum detection concentrations (MDC) and apparent affinities (K’s) of the variant regulators for inducers, JS42R-1 strains expressing the LysR protein variants were assayed after growth on succinate in the presence of varying concentrations of selected inducers. The inducers salicylate, 2-nitrotoluene, and TNT were chosen to represent a “universal” inducer, a catabolically relevant inducer, and an important environmental contaminant, respectively. The maximum induction activities (Vmax values; Table 1) were consistent with the activity values in Tables S1-S8, indicating that an absence of induction was not due to an insufficient amount of compound present, but to true differences in inducer specificity.
Table 1.
Coefficients of induction and the thresholds of detection for salicylate, 2-nitrotoluene, and TNT
| Regulator | Activity Coefficientsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Salicylate |
2-Nitrotoluene |
TNT |
|||||||
| MDCb (μM) | K’s (μM) | Vmax (Miller units) | MDC (μM) | K’s (μM) | Vmax (Miller units) | MDC (μM) | K’s (μM) | Vmax (Miller units) | |
| NagR (wild type) | 1 | 44.9 ± 5.35 | 386 ± 46 | --- c | --- | --- | --- | --- | --- |
| NagR I74V | 1 | 4.21 ± 0.31 | 389 ± 29 | --- | --- | --- | --- | --- | --- |
| NagR K189R | 1 | 9.79 ± 1.13 | 147 ± 17 | --- | --- | --- | --- | --- | --- |
| NagR P227S | 0.1 | 7.23 ± 0.64 | 432 ± 38 | 10 | 29.9 ± 5.45 | 170 ± 31 | --- | --- | --- |
| NagR I232V | 0.1 | 0.17 ± 0.03 | 133 ± 21 | 0.5 | 1.78 ± 0.30 | 196 ± 33 | --- | --- | --- |
| NagR P227S I232V | 0.1 | 0.71 ± 0.08 | 443 ± 51 | 1 | 7.02 ± 1.18 | 160 ± 27 | --- | --- | --- |
| NagR K189R P227S I232V |
1 | 152 ± 10.1 | 735 ± 49 | 1 | 2.88 ± 0.44 | 247 ± 38 | 10 | 9.3 ± 1.3 | 99 ± 19 |
| NtdR R189K S227P V232I |
1 | 10.8 ± 1.33 | 358 ± 44 | --- | --- | --- | --- | --- | --- |
| NtdR S227P V232I | 1 | 7.35 ± 0.39 | 580 ± 31 | --- | --- | --- | --- | --- | --- |
| NtdR V232I | 0.1 | 0.24 ± 0.01 | 734 ± 45 | 0.01 | 0.23 ± 0.02 | 588 ± 53 | 20 | 4.8 ± 0.8 | 301 ± 42 |
| NtdR S227P | 10 | 21.6 ± 2.10 | 396 ± 39 | --- | --- | --- | --- | --- | --- |
| NtdR R189K | 0.1 | 0.13 ± 0.01 | 623 ± 35 | 0.5 | 1.79 ± 0.17 | 509 ± 47 | 20 | 17 ± 2.9 | 312 ± 38 |
| NtdR L169H | 0.1 | 0.26 ± 0.05 | 482 ± 62 | 10 | 53 ± 8.74 | 194 ± 32 | 10 | 9.9 ± 1.2 | 371 ± 29 |
| NtdR V74I | 0.1 | 0.24 ± 0.02 | 716 ± 63 | 0.5 | 2.6 ± 0.12 | 768 ± 34 | 10 | 7.6 ± 2.1 | 299 ± 38 |
| NtdR (wild type) | 0.1 | 0.12 ± 0.01 | 516 ± 41 | 10 | 66.5 ± 4.85 | 741 ± 54 | 20 | 30 ± 7.2 | 525 ± 48 |
R2 ≥ 0.93
MDC, minimum detection concentration
---, not a recognized inducer
For most of the LysR-variants, the lower threshold of detection ranged from 0.05 to 1 μM (Table 1). NtdR mutants containing the S227P substitution had the greatest changes in sensitivity and relative affinity with salicylate. The NtdR S227P single mutant showed a 100-fold decrease in sensitivity and a 180-fold decrease in affinity compared to wild-type NtdR. Addition of the V232I or V232I /R189K mutations to NtdR S227P decreased the threshold of detection to 1 μM, but the half-saturation concentration was still between 7 and 11 μM.
All substitutions that made NagR more like NtdR improved the affinity for salicylate except for the K189R P227S I232V triple mutant, which had a three-fold decrease in affinity. Changes at positions 227 and 232 had the greatest influence on induction levels. The P227S mutation increased the sensitivity of salicylate detection 10-fold and the affinity 6-fold, and the I232V mutation improved NagR affinity for salicylate to a level similar to that of NtdR. Combining both mutations resulted in intermediate threshold and affinity values compared to the single mutants, but an elevated Vmax that approached that of NtdR.
Induction of β-galactosidase activity required at least 10 μM 2-nitrotoluene in cultures expressing wild-type NtdR (Table 1), and the affinity of NtdR for 2-nitrotoluene was 550-fold weaker than for salicylate. The L169H mutation in NtdR decreased the maximum induction activity with 2-nitrotoluene, but substitutions at 74, 189, or 232 lowered the threshold of detection and increased the apparent affinity. NtdR V232I was the most sensitive protein, activating gene expression in the presence of 0.01 μM 2-nitrotoluene, with an induction activity near that of wild-type NtdR. Substitutions at positions 227 and 232 of NagR resulted in the ability to recognize 2-nitrotoluene. As with salicylate, the I232V mutation increased sensitivity and affinity to a greater extent than the P227S mutation, and the combination of both changes resulted in intermediate properties.
Only five of the 14 NagR-NtdR-variants were able to induce gene expression with TNT, with the lower threshold of detection ranging between 10 and 20 μM (Table 1). Mutations at positions 74, 169, 189, and 232 increased the apparent affinity of NtdR towards TNT, but decreased the maximal induction activity. The increase in affinity and lower threshold for TNT detection for the NtdR L169H protein are consistent with the significant increase in fold induction (Fig. 6; Fig 7B). Although the NagR K189R P227S I232V mutant gained the ability to recognize TNT, its maximal activity was only about 20% of the activity of wild-type NtdR.
Acidovorax sp. JS42 growth studies.
JS42 was tested for growth on the 63 potential inducers as sole carbon sources. The only compounds that supported growth were nitrobenzene, 2-nitrotoluene, catechol, 3-methylcatechol, and 4-methylcatechol. JS42R-1 expressing NtdR and NagR variants were examined for differences in the ability to grow on 2-nitrotoluene. JS42R-1 strains containing variant regulators that recognized 2-nitrotoluene (Fig. 6; Tables S5 and S6) showed growth within three to four days (similar to wild type), with no apparent differences in colony morphology. When salicylate was included in the medium, all of the strains except the one expressing the NagR H169L variant were able to grow on 2-nitrotoluene. Although salicylate is not a growth substrate for JS42, its inclusion as a universal inducer of the ntd operon bypassed the need for induction by 2-nitrotoluene. The inability of the strain carrying the NagR H169L variant to grow was consistent with its lack of induction activity under all tested conditions.
DISCUSSION
Transcriptional control systems are thought to evolve because of the non-specificity of pre-existing promoters and regulatory proteins. The new pathway is commonly expressed at a constitutive level prior to recruitment of a regulatory system; eventually the suboptimal control system is fine-tuned to respond more specifically to the new signal (Cases and de Lorenzo, 2001; de Lorenzo and Perez-Martin, 1996). The 2NTDO regulation system in JS42 appears to be at an intermediate stage of evolution, with relatively high background gene expression in the absence of inducers, as well as gratuitous induction in response to both relevant and non-metabolizable compounds due to the extensive broadening of inducer specificity. Because evolution of specificity is unlikely to occur in a single step, it is rare to identify mutant proteins with actual changes in specificity; it is much more typical for specificity to become relaxed, (Galvao and de Lorenzo, 2006). This concept of protein promiscuity in which specificity to old substrates is retained even as new specificity is acquired has been addressed in both enzymes and regulatory proteins (Aharoni et al., 2005; Galvao et al., 2007; Tokuriki and Tawfik, 2009). NtdR appears to have regressed to a protein with broad specificity, as seen with experimentally-evolved forms of the XylR protein. The evolved XylR proteins retained the ability to recognize the native inducer, but acquired responsiveness to several nitro- and chloroaromatic compounds after selection for the ability to respond to 2,4-dinitrotoluene (Galvao et al., 2007).
NtdR differs from other LysR-type transcriptional regulators in its ability to activate gene expression in response to synthetic nitrotoluenes, as well as in the unusually wide range of inducer compounds it recognizes. There are, however, several examples of promiscuous transcriptional regulators that control expression of various aromatic compound degradation pathway genes in other regulatory protein families. In particular, regulatory systems that respond to primary substrates rather than metabolic intermediates tend to recognize a wider range of inducers (Shingler, 2003). For example, the two-component TodST protein pair is responsible for activation of the toluene degradation genes in Pseudomonas putida F1 and DOT-E1 in response to more than 20 single-ring aromatic hydrocarbons (including the growth substrates toluene, benzene and ethylbenzene), as well as trichloroethylene (Busch et al., 2007; Cho et al., 2000; Lacal et al., 2006; Shingleton et al., 1998). Similarly, IpbR, a member of the AraC family that functions as a regulator of the isopropylbenzene degradation genes in P. putida RE204, recognizes isopropylbenzene and more than 35 other aromatic and alkane inducers (Selifonova and Eaton, 1996).
A limited number of studies have examined the inducer specificity of LysR-type regulators that control expression of degradation pathways for aromatic compounds. Substitution of Asn 169 to Asp in NahR, the activator of the naphthalene degradation pathway in Pseudomonas putida G7, expanded the inducer specificity to include benzoate in addition to the native inducer salicylate, and also amplified the response to 2- and 3-chlorobenzoates (Cebolla et al., 1997). Mutations at positions 132 and 248 of NahR also expanded inducer specificity; the corresponding residues are predicted to be near the binding pocket for benzoate in BenM, the transcriptional activator of the benzoate degradation genes in Acinetobacter baylyi ADP1 (Ezezika et al., 2007), and the predicted inducer binding pocket in DntR, a transcriptional regulator that responds to salicylate from the 2,4-dinitrotoluene degrading strain Burkholderia sp. strain DNT (Lonneborg et al., 2007; Smirnova et al., 2004; Spanggord et al., 1991). In a separate study, converting His169 to Val in DntR resulted in an increased response to benzoate. When combined with a Phe111 to Leu substitution, the resulting double mutant demonstrated modest increases in activity with benzoate and 4-nitrobenzoate compared to wild-type DntR. Similarly, a F111L H169V double mutant of DntR gained the ability to respond to 2,4-dinitrotoluene; however, induction was only 1.6-fold higher than background (Lonneborg et al., 2007). Substitutions at positions 129, 199, 226, 246, and 267 in TfdT, the transcriptional regulator of the chlorocatechol operon in Burkholderia sp. strain NK8 increased the range of inducers recognized (Lang and Ogawa, 2009). Based on sequence alignments, residues 129 and 246 in TfdT are quite close to residues 132 and 248 in NahR.
In this study, the key amino acids responsible for the wide inducer specificity of NtdR were identified through comparison to NagR, the LysR-type regulator from the naphthalene degrading bacterium Ralstonia sp. strain U2 (Jones et al., 2003). The inducer specificity of NagR was expanded by changing the differing amino acids to those present in NtdR, whereas specificity of NtdR became restricted when the differing residues were changed to those present in NagR. Substitutions at positions 74 and 189 mainly changed levels induction, whereas substitutions of residues 169, 227, and 232 changed regulator specificity. The residue at position 169 was found to be critical for recognition of aromatic acids, and positions 227 and 232 were important for recognition of nitroaromatic compounds. Induction by 1,3,5-trinitrobenzene, TNT, atrazine, 4-chloronitrobenzene, and 1-nitronaphthalene required multiple amino acid substitutions, suggesting that some of the positions may modulate effector recognition in a synergistic fashion.
Sequence comparisons showed that conserved amino acids are present at some of these positions in related LysR-type regulators (Table 2). Amino acids at positions 74 and 169 appear to be quite variable, while charged or polar residues are consistently present at position 189. Pro227 is conserved in NagR, NahR (the activator of naphthalene and salicylate catabolic operons in P. putida G7), BphR2 (the activator of salicylate catabolic genes in Pseudomonas pseudoalcaligenes KF707 (Fujihara et al., 2006; Schell, 1985)), and DntR. Given the functional and sequence similarities between these regulators, it is possible that changes at positions 227 and 232 of NahR, BphR2, and DntR may also allow recognition of nitroaromatic compounds.
Table 2.
Amino acids at selected positions in different LysR-type transcriptional activators based on sequence alignments
| Protein | Accession No. | Strain | Sequence Identity to NtdR (%) | Amino acid residue at positiona | ||||
|---|---|---|---|---|---|---|---|---|
| 74 | 169 | 189 | 227 | 232 | ||||
| NtdR | AAP70492 | Acidovorax sp. JS42 | 100 | Val | Leu | Arg | Ser | Val |
| NbzR | AAP70491 | Comamonas sp. JS765 | 100 | Val | Leu | Arg | Ser | Val |
| NagR | AAG13636 |
Ralstonia sp. U2 |
98b | Ile | His | Lys | Pro | Ile |
| DntR | AAP70493 | Burkholderia sp. DNT | 97 | Ile | His | Lys | Pro | Ile |
| NahR | P10183 | Pseudomonas putida G7 | 61 | Thr | Asn | Glu | Pro | Val |
| BphR2 | AAZ08063 | Pseudomonas pseudoalcaligenes KF707 | 59 | Ala | Asn | Glu | Pro | Val |
| BenM | AAC46441 | Acinetobacter baylyi ADP1 | 15 | Arg | Glu | Asnc | Arg | Ala |
Numbering is based on the NtdR sequence.
NagR is identical to NtdR / NbzR except for the amino acids at the five listed positions.
According to sequence alignments, the amino acid at position 189 of DntR, NagR, and NtdR aligns to Val182 of BenM. However, based on structure comparisons between DntR and BenM, Asn185 is the correct corresponding amino acid, not Val182 (Ezezika et al., 2007).
The common domain structure of all LysR-type regulators includes the presence of a helix-turn-helix or winged helix-turn-helix DNA binding motif at the N-terminus, and an effector binding domain (EBD) in the C-terminus. To gain insight into how the amino acids that differ between NtdR and NagR alter the inducer specificity of these two regulators, a homology model of NtdR was generated based on the available protein structure of DntR (Fig. 8). DntR was selected as the template for modeling due to its high sequence identity with NtdR (97%) and NagR (99%). Although inducer specificity studies have shown that DntR does not activate gene expression in the presence of nitroaromatic compounds (Lonneborg et al., 2007), and neither salicylate nor 2,4-dinitrotoluene could be co-crystallized with the EBD (Smirnova et al., 2004), it is still a useful platform for comparative analysis of our LysR-variants since DntR is identical in sequence to NagR except at positions 46 and 48, which are located in the N-terminal DNA binding domain. Although the full-length DntR protein was crystallized, only the structure of the EBD was determined (Smirnova et al., 2004), and therefore the DntR structure is expected to equivalent to that of NagR. The homology model of NtdR was compared to the structure of BenM (Ezezika et al., 2007), which is the only LysR-type regulator of an aromatic compound degradation pathway that has been successfully co-crystallized with bound inducers (Fig. 8). While the amino acid sequence of BenM is only 15% identical to NtdR, benzoate is recognized as an inducer by both proteins. The structure of the BenM EBD and the predicted structure of the NtdR EBD are quite similar (Fig. 8). Position 74 in NtdR is located in the linker helix that connects the helix-turn-helix DNA binding motif to the EBD (not shown in the structure), while residue 189 is in the region of the EBD that has been postulated to function in tetramerization (Ezezika et al., 2007). Their distal locations from the identified inducer binding pockets in BenM (see below) are consistent with results showing that amino acid changes at 74 and 189 have relatively minor effects on inducer specificity.
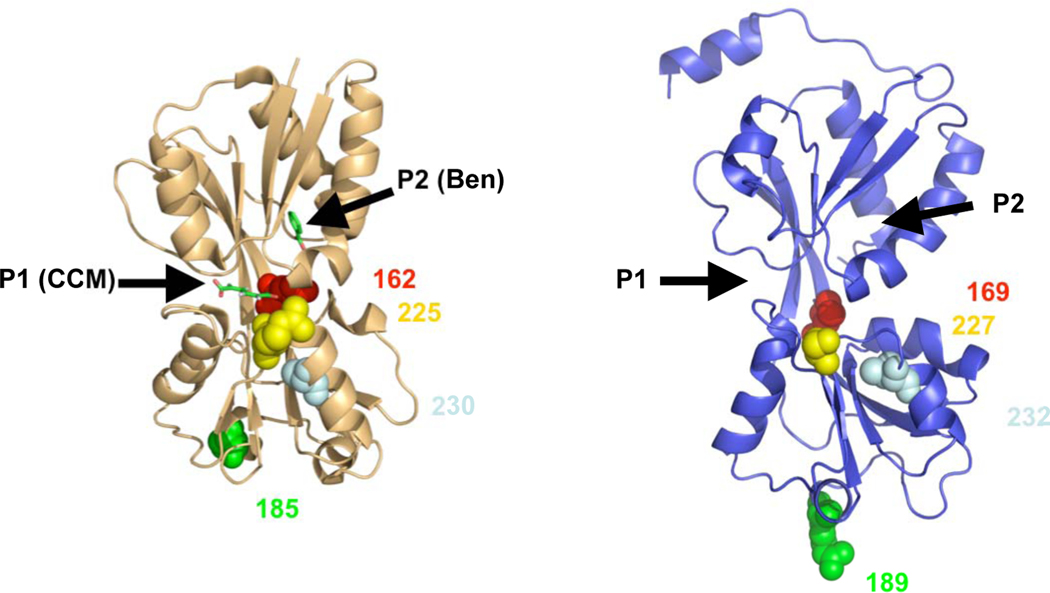
Fig. 8.
Crystal structure of the effector binding domain of BenM (tan) and model of NtdR (blue) based on the crystal structure of DntR. The natural inducers recognized by BenM, cis,cis-muconate (CCM) in pocket 1 (P1) and benzoate (Ben) in pocket 2 (P2), are shown in green. The amino acid residues 169 (red), 189 (green), 227 (yellow), and 232 (light blue) that were changed in this study (corresponding to 162, 185, 225, 230 in BenM), are indicated in space fill format. The root mean-square deviation of the modeled NtdR was 0.06 Å using 227 C α atoms from DntR and NtdR (residues 75–301).
In BenM, two distinct regions allow for the differential binding of cis,cis-muconate (pocket 1) and benzoate (pocket 2) (Ezezika et al., 2007). Based on the model of NtdR, cavities that appear to be similar to the inducer binding sites within BenM are present at analogous positions in NtdR. Residues 227 and 232 of NtdR are positioned to influence the conformation of binding pocket 1 (Fig. 8), and it is known that the residue analogous to Val227 in BenM coordinates the hydroxyl groups of cis,cis-muconate (Ezezika et al., 2007). The benzoate binding site in BenM (pocket 2) appears to be fairly well conserved in NtdR (Fig. 8). Residue 169 of NtdR, which mediates specificity towards aromatic acids, is positioned towards the binding pocket, while the analogous residue (Glu162) in BenM faces outward. The smaller side-group of Leu169 in NtdR compared to the histidine in DntR and NagR may increase the size of pocket, which could explain the wide range of aromatic acids recognized by NtdR, while DntR and NagR are restricted to a limited number of inducers. Analysis of BenM suggested that Glu 162 (corresponding to position 169 in all of the other regulators; Table 2) may influence both binding pockets due to its physical location within the core of the protein (Ezezika et al., 2007).
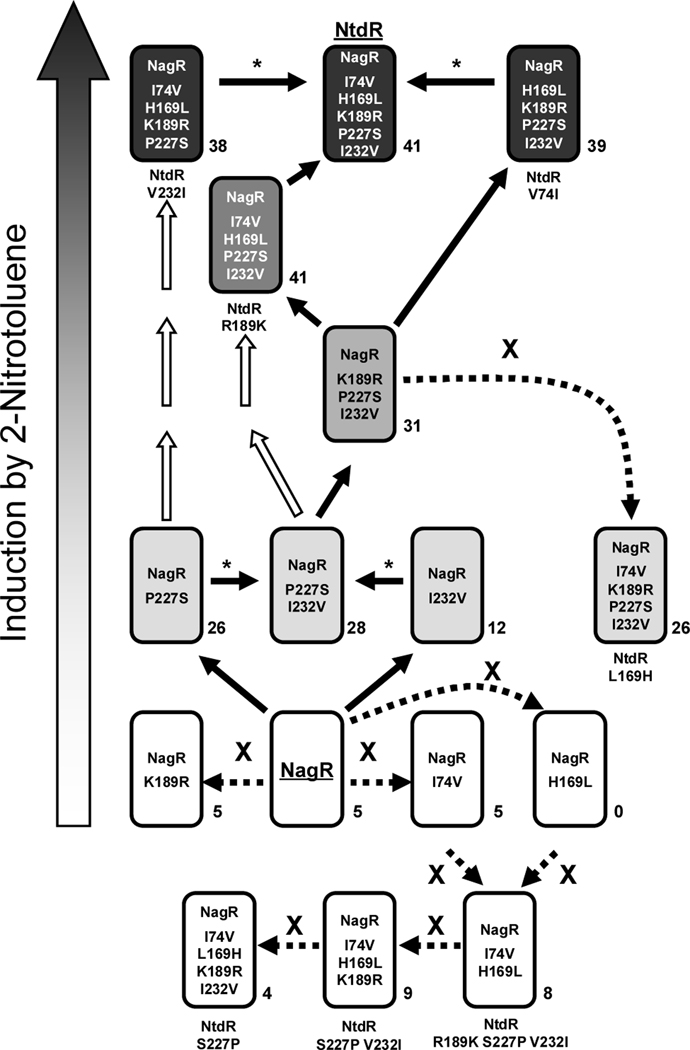
Although our analysis has only explored 16 out of the 32 total possible combinations of the five different amino acids between NagR and NtdR, it is possible to predict the order in which the mutations may have occurred. Based on the premise that increased expression in response to 2-nitrotoluene is advantageous and assuming that the mutations accumulated individually, a model of the evolutionary pathway from NagR to NtdR can be generated based on the measured induction activities of our mutant regulators in response to 2-nitrotoluene (Fig. 9). Evolution of NagR to NtdR would not have been initiated by single mutations resulting in K189R, I74V, or H169L substitutions, as these changes do not allow for induction by 2-nitrotoluene. In an evolving population of cells, only beneficial mutations would be retained, and combinations that provide no benefit or cause deleterious effects would be rapidly swept out of the population. The P227S and I232V single mutations each allowed the regulator to induce gene expression to similar levels when 2-nitrotoluene is present. After combining these changes in the P227S I232V double mutant, the most straightforward path leading to NtdR would be the sequential addition of the K189R substitution, followed by H169L, and finally I74V (Fig. 9). With each of these changes, the level of induction with 2-nitrotoluene is improved. In contrast, addition of the I74V change to the K189R P227S I232V triple mutant before H169L lowered induction activity and consequently the fitness of the host.
Fig. 9.
Model depicting the possible pathway by which mutations could have accumulated during the evolution of NtdR from an ancestral NagR based on measured levels of β-galactosidase activity during induction with 2-nitrotoluene. For clarity, all of the NtdR-variants are indicated in the boxes as mutants of NagR (i.e. NtdR = NagR I74V H169L K189R P227S I232V; for reference, the mutant names that are used in the text are indicated below each boxed protein). The increased shading of the boxed NagR proteins (bottom to top of the figure) represents increasing levels of gene expression in response to 2-nitrotoluene; boxed NtdR variants shown in white do not respond to 2-nitrotoluene. Solid arrows indicate steps that lead to increased induction, and white arrows indicate other possible routes to improved induction that were not explored in this study. Asterisks (*) indicate changes that do not increase activity in response to 2-nitrotoluene, but increase the total number of inducers recognized. Dashed arrows labeled with an “X” indicate routes to regulators with decreased or no activity that would result in a loss in fitness for the host strain during evolution for growth on 2-nitrotoluene. The total number of inducers that elicited significant induction (minimum 2-fold induction with standard deviations less than 20–25%) for each NtdR-NagR-variant is indicated next to the box.
Interestingly, NtdR V232I (NagR I74V H169L K189R P227S in Fig. 9) and NtdR V74I (NagR H169L K189R P227S I232V in Fig. 9) demonstrated similar induction activities with 2-nitrotoluene as NtdR, but each had lower thresholds for detection and higher affinities for 2-nitrotoluene (Table 1). It is possible that Acidovorax sp. JS42 evolved in an environment where 2-nitrotoluene was not limiting and selection was primarily for fast turnover of 2-nitrotoluene rather than for sensitive detection (i.e. in a highly contaminated environment and/or in enrichment culture). Also, evolution of NtdR may have occurred before 2NTDO was optimized for 2-nitrotoluene oxidation, and increased production of an inefficient ancestral 2-nitrotoluene-oxidizing enzyme might have been favored. Acidovorax sp. JS42 was isolated from a contaminated environment (Haigler et al., 1994) that may have contained mixtures of chemicals including those recognized by NtdR (mononitrotoluenes, dinitrotoluenes, chloroaromatic compounds, etc). The high level of expression mediated by NtdR in the absence of inducers may have also played a role in mitigating the toxicity of these or other compounds by increasing the level of 2NTDO, which has a broad substrate range (Parales et al., 1998a). Hydroxylation of aromatic compounds not only decreases their toxicity, but also allows them to be removed by physical sorption processes; alternatively, these oxidized products may be growth substrates for other bacteria.
We have described routes by which mutations could occur in NagR that would lead to the generation of NtdR, but are they representative of what actually occurs within a strain evolving the ability to grow on 2-nitrotoluene, where both regulator and dioxygenase may evolve in parallel? Also unknown is whether the lower-pathway for 2-nitrotoluene degradation (3-methylcatechol to TCA cycle intermediates) is coordinately regulated with the upper pathway and if expression of the genes has been integrated into other regulatory networks such as a catabolite repression system. Finally, the biochemical interactions of NtdR with target promoters and with RNA polymerase remain unexplored. This study has provided insight into the molecular origins of nitrotoluene sensing and gene expression, and lays the groundwork for revealing the answers to these and other questions about the evolution of nitroarene catabolic pathways.
EXPERIMENTAL PROCEDURES
Chemicals.
Catechol (>99.5%), 3-methylcatechol (98%), 4-methylcatechol (>95%), 2-nitrobenzyl alcohol (97%), 3-nitrobenzyl alcohol (98%), 4-nitrobenzyl alcohol (99%), 4-nitrotoluene (4NT; 99%), and 2,6-dinitrotoluene (98%) were purchased from Aldrich (Milwaukee, Wisc.). Naphthalene (99%) and nitrobenzene (99%) were obtained from Acros Organics (Morris Plains, N.J.), and 2-nitrotoluene (99%), 3-nitrotoluene (99%), and 2,4-dinitrotoluene (97%) were from Avocado (Heysham, Lancashire, UK). Atrazine was generously provided by Dr. Larry Wackett (University of Minnesota). All other chemicals were of the highest purity commercially available. Oligonucleotides were synthesized by MWG-Biotech (Greensboro, N.C.).
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 3. Strain and plasmid constructions are described in detail in the Supplementary Materials. Escherichia coli strains were grown in Luria-Bertani broth (Bertani, 1951) at 37° C unless otherwise indicated. Tryptone-yeast extract medium (TY; 1% tryptone, 0.5% yeast extract), and minimal-salts broth (MSB) (Stanier et al., 1966) containing 1% vol/vol Balch’s vitamins (Gerhardt et al., 1994) solution (lacking thiamine and 4-aminobenzoate) were used for growth of Acidovorax strains as described below. For plates, MSB was solidified with 1.8% (wt/vol) Noble agar, while LB and TY were solidified with 1.6% (wt/vol) Bacto agar. Antibiotics were added at the following concentrations for plasmid selection and maintenance: ampicillin, 200 μg ml−1; streptomycin, 200 μg ml−1; kanamycin, 50 μg ml−1; chloramphenicol, 30 μg ml−1; tetracycline, 20 μg ml−1; gentamicin, 7.5 μg ml-1.
TABLE 3.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Cloning host; thi | Invitrogen |
| S17–1 | Host for plasmid mobilization, thi | (Simon et al., 1983) |
| S17–1 λ-pir | Host for plasmid mobilization, contains λ-pir; thi | (de Lorenzo et al., 1993) |
| Acidovorax sp. | ||
| JS42 | Wild-type 2-nitrotoluene degrader; naturally ApR | (Haigler et al., 1994) |
| JS42–1 | Derivative of JS42 containing ntdA-lacZ; GmR | (Lessner et al., 2003) |
| JS42Ac | ntdAc mutant of JS42; KmR | This study |
| JS42Ac-1 | JS42Ac containing ntdA-lacZ; KmR; GmR | This study |
| JS42R-1 | ntdR mutant of JS42–1; KmR; GmR | (Lessner et al., 2003) |
| JS42E1 | ctdE1 mutant of JS42; SmR | This study |
| JS42E1–1 | JS42E1 containing ntdA-lacZ; SmR; GmR | This study |
| Plasmids | ||
| pBBR1MCS | Broad-host-range plasmid; CmR | (Kovach et al., 1994) |
| pBBR1MCS2 | Broad-host-range plasmid; KmR | (Kovach et al., 1995) |
| pBBR1MCS5 | Broad-host-range plasmid; GmR | (Kovach et al., 1995) |
| pHP45Ω | Carries SmR cassette for insertional mutagenesis | (Prentki and Krisch, 1984) |
| pK18 | Cloning vector; KmR | (Pridmore, 1987) |
| pRK415 | Broad-host-range plasmid; TcR | (Keen et al., 1988) |
| pNtd1 | pBBR1MCS containing ntdR; CmR | (Lessner et al., 2003) |
| pNag1 | pBBR1MCS containing nagR; CmR | (Lessner et al., 2003) |
| pDTG800 | pUC18 containing ntdAaAbAcAd;ApR | (Parales et al., 1996) |
| pDTG850 | pUC13 containing ntdAaAbAcAd; ApR | (Parales et al., 1998b) |
| pDTG850-Km | Derivative of pDTG850; ntdAaAbAc::KmAd; AmpR, KmR | This study |
| pRK415–850-Km | Derivative of pRK415; ntdAaAbAc::KmAd; KmR, TcR | This study |
| pDTG903 | pUC19 containing 4.5 kb-fragment carrying cdoE from Comamonas sp. strain JS765; ApR | (Parales et al., 1997) |
| pDTG928 | pK18 containing 4.5 kb-fragment from pDTG903; KmR | This study |
| pDTG929 | Derivative of pDTG928 containing cdoE::Sm; KmR, SmR | This study |
| pDTG930 | pRK415 containing cdoE::Sm from pDTG929; SmR, TcR | This study |
| pDTG931 | pUTminiTn5-Gm carrying ntdA-lacZ; ApR GmR | (Lessner et al., 2003) |
| pJVP1 | pBBR1MCS containing nagR I74V; CmR | This study |
| pJVP2 | pBBR1MCS containing nagR H169L; CmR | This study |
| pJVP3 | pBBR1MCS containing nagR K189R; CmR | This study |
| pJVP4 | pBBR1MCS containing nagR P227S; CmR | This study |
| pJVP5 | pBBR1MCS containing nagR I232V; CmR | This study |
| pJVP6 | pBBR1MCS containing nagR P227S I232V; CmR | This study |
| pJVP7 | pBBR1MCS containing ntdR V74I; CmR | This study |
| pJVP8 | pBBR1MCS containing ntdR L169H; CmR | This study |
| pJVP9 | pBBR1MCS containing ntdR R189K; CmR | This study |
| pJVP10 | pBBR1MCS containing ntdR S227P; CmR | This study |
| pJVP11 | pBBR1MCS containing ntdR V232I; CmR | This study |
| pJVP12 | pBBR1MCS containing ntdR S227P V232I; CmR | This study |
| pKSJ33 | pBBR1MCS containing nagR K189R P227S I232V; CmR | This study |
| pKSJ34 | pBBR1MCS containing ntdR R189K S227P V232I; CmR | This study |
| pKSJ44 | pBBR1MCS5 containing ntdAaAbAcAd from pDTG800 | This study |
| pKSJ113 | pUC18 containing ctdE1 from JS42; ApR | This study |
| pKSJ126 | pBBR1MCS2 containing ctdE1 from pKSJ113; KmR | This study |
ApR, ampicillin resistance; KmR, kanamycin resistance; GmR, gentamicin resistance; TcR, tetracycline resistance; CmR, chloramphenicol resistance; SmR, streptomycin resistance
DNA manipulations.
Standard methods were used to manipulate plasmids and DNA fragments (Maniatis et al., 1982). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs (Beverly, Mass). Plasmids were purified using commercial kits or as previously described (Lee and Rasheed, 1990). DNA fragments were purified with a QIAquick Gel Extraction kit (Qiagen), and a Puregene DNA purification kit was used to isolate genomic DNA (Gentra Systems, Minneapolis, Minn.). Fluorescent automated DNA sequencing was carried out at the University of California, Davis, sequencing facility with an Applied Biosystems 3730 automated sequencer.
E. coli strains were transformed with plasmid DNA by standard procedures (Maniatis et al., 1982). E. coli strains S17–1 and S17–1 λ-pir were used to mobilize plasmids and reporter fusions into JS42 by conjugation. Donor strains were cross-struck with recipient JS42 strains on TY plates and incubated at 30° C. After 48 hours, cells were scraped from plates, resuspended in 10 ml of M9 medium (Miller, 1975), homogenized by vortexing, and plated on MSB plates containing 10 mM succinate, vitamins and the appropriate antibiotics. Exconjugants were purified by repeated single colony isolation on selective medium containing ampicillin (to which JS42 is naturally resistant) to eliminate donor E. coli strains. The presence of plasmids was confirmed by purification and diagnostic restriction-digestions.
Induction assays.
Starter cultures of JS42–1, JS42Ac-1, JS42E1–1, JS42R-1, and JS42R-1 carrying plasmids expressing LysR-regulators were grown overnight in 250-ml Fernbach flasks containing 50-ml of MSB, 10 mM succinate, 0.05% wt/vol yeast extract, and the appropriate antibiotics, in a shaking incubator at 28–30°C and 200 RPM. Assay cultures were similarly grown in 125-ml Erlenmeyer flasks containing 25-ml of the above medium, inducer compound, and 0.5-ml of the starter cultures as the inoculum (initial OD660 ~ 0.025). Induction by the LysR regulator variants was determined by measuring 2NTDO and β-galactosidase activities (see below) after growth in the presence of the following suite of compounds. Unless specified, inducers were added to a final concentration of 500 μM from 1,000-x aqueous (for aromatic acids) or methanolic stock solutions. Tested compounds included: benzoate (Ben); 2-, 3-, 4-hydroxybenzoate (HBen); 2-, 3-, 4-nitrobenzoate (NBen); 2-, 3-, 4-chlorobenzoate (ClBen); 2-, 3-, 4-aminobenzoate (ABen); 2,5-dihydroxybenzoate (25HBen); 3-methylbenzoate (3MBen); 4-isopropylbenzoate (4IPBen); methyl salicylate (MSal); phenylacetic acid (PAA); nicotinic acid (NA); benzyl alcohol (BA); 2-hydroxybenzyl alcohol (2HBa); 2-aminobenzyl alcohol (2ABa); 2-, 3-, 4-nitrobenzyl alcohol (NBa); benzaldehyde (Bzl); 3-hydroxybenzaldehylde (3HBzl, 100 μM); 3-nitrobenzaldehyde (3NBzl, 100 μM); benzene (Bz); nitrobenzene (NB); 1,3,5-trinitrobenzene (TNB, 20 μM); aminobenzene (AB); chlorobenzene (ClB); toluene (Tol); 2-, 3-, 4-aminotoluene (AT); 2-, 3-, 4-nitrotoluene (NT); 2,4-dinitrotoluene (24DNT, 100 μM); 2,6-dinitrotoluene (26DNT, 100 μM); 2,4,6-trinitrotoluene (TNT, 20 μM); 2-amino-4,6-dinitrotoluene (2ADNT; 100 μM); 4-amino-2,6-dinitrotoluene (4ADNT; 100 μM); 2-, 3-, 4-chloronitrobenzene (CNB); 3-, 4-chloroaminobenzene (CAB); phenol (Phe); 2-, 3-, 4-nitrophenol (NP, 100 μM); 2-aminophenol (AP, 100 μM); 2,4-dinitrophenol (24DNP, 100 μM); atrazine (Atz, 100 μM); cyanuric acid (CA, 100 μM); catechol (Cat, 10 μM); 3-, 4-methylcatechol (MC, 10 μM); naphthalene (Nap, 100 μM); 1-nitronaphthalene (1NNap, 100 μM). Stock solutions for catechols, benzyl alcohols, and benzaldehydes were freshly prepared just prior to inoculation of cultures.
In time course experiments, multiple assay cultures were initiated at the same time and activity was measured every two hours over a 24-hour period. For inducer specificity experiments, assay cultures were analyzed after 12–14-hours of growth (OD660 ~ 0.90). Limits of detection were determined with inducer concentrations ranging from 1 nM to 1 mM. All cultures were harvested by centrifugation at 10,000 RPM for 10 minutes at 4° C, resuspended in Z-buffer to an OD660 between 0.40–0.50 and β-galactosidase activity was measured as described previously (Miller, 1975). β-Galactosidase activities reported in this study represent the averages of three independent biological replicates, each containing two technical replicates (n = 6). The β-galactosidase activities were analyzed using the Michaelis-Menten model (Michaelis and Menten, 1913) to obtain estimates of the relative affinities (K’s) of variant regulators for the three inducers and the maximum activity with each inducer (Vmax) by non-linear regression as previously described (Ramos et al., 1990).
2NTDO activity was determined by dispensing 1-ml of cell suspensions into glass culture tubes, adding one drop of nitrobenzene, and incubating at 30°C with shaking at 200 RPM for 30 minutes. The nitrite released from nitrobenzene oxidation was quantified as previously described (Ju and Parales, 2006). Total protein was determined by the method of Bradford (Bradford, 1976). Benzyl alcohols and benzaldehydes are known to undergo abiotic and enzymatic oxidations under aerobic conditions (James et al., 2000; Streitwieser et al., 1998). To verify that benzyl alcohols and benzaldehydes were stable during the induction experiments, supernatants from cultures containing these inducers were extracted with ethyl acetate and analyzed by GC-MS as previously described (Ju and Parales, 2006).
Homology modeling.
A homology model of the inducer binding domain of NtdR was generated with the SWISS-MODEL server (Arnold et al., 2006) using the crystal structure of the inducer binding domain of DntR (PDB ID: 1UTB, chain B) as the template (Smirnova et al., 2004). SuperPose (Maiti et al., 2004) and PyMol (DeLano Scientific) were used for analysis and creation of molecular representations (DeLano Scientific).
Growth assays.
The ability of JS42R-1 complemented with variant LysR regulators to grow on 2-nitrotoluene was tested by streaking the strains onto MSB plates with or without 100 μM salicylate, and incubating at 28–30° C in an atmosphere of 2-nitrotoluene.
The ability of JS42 to grow on the above inducer compounds as sole carbon sources was assayed in MSB containing 2 mM of each inducer. Cultures were incubated at 28–30° C with shaking (200 RPM) and monitored for increases in turbidity compared to controls lacking added substrates over a period of 14 days. Growth was also tested on MSB plates supplemented with select inducers. Aromatic acids were added to molten agar (2 mM final concentration), while nitrophenols, naphthalene, 1-nitronaphthalene, aminotoluenes, nitrotoluenes, chlorobenzene, nitrobenzene, benzene, and toluene were supplied in vapor form. Dinitrotoluene- and aminodinitrotoluene-containing media were prepared as previously described (Nishino et al., 2000).
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported in part by funds from the Strategic Environmental Research and Development Program (project CU1212), and the National Science Foundation (MCB 02627248). K.-S.J. was supported by an NIH Traineeship in Molecular and Cellular Biology (NIH TM32 GM070377) and a University of California Toxic Substances Research and Teaching Program graduate fellowship (http://tsrtp.ucdavis.edu/). We thank Dan Lessner for constructing JS42E1.
REFERENCES
- Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, and Tawfik DS (2005) The ‘evolvability’ of promiscuous protein functions. Nat. Genet 37: 73–76. [DOI] [PubMed] [Google Scholar]
- An D, Gibson DT, and Spain JC (1994) Oxidative release of nitrite from 2-nitrotoluene by a three-component enzyme system from Pseudomonas sp. strain JS42. J. Bacteriol 176: 7462–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Busch A, Lacal J, Martos A, Ramos JL, and Krell T. (2007) Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc. Natl. Acad. Sci. U.S.A 104: 13774–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I, and de Lorenzo V. ( 2001) The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Sousa C, and de Lorenzo V. (1997) Effector specificity mutants of the transcriptional activator NahR of the naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem 272: 3986–3992. [DOI] [PubMed] [Google Scholar]
- Cerniglia CE, and Somerville CC (1995) Reductive metabolism of nitroaromatic and nitropolycyclic aromatic hydrocarbons. In Biodegradation of nitroaromatic compounds. Vol. 49. Spain JC (ed). New York: Plenum Press, pp. 99–115. [Google Scholar]
- Cho MC, Kang D-O, Yoon BD, and Lee K. (2000) Toluene degradation pathway from Pseudomonas putida F1: substrate specificity and gene induction by 1-substituted benzenes. J. Ind. Microbiol. Biotechnol 25: 163–170. [Google Scholar]
- de Lorenzo V, Cases I, Herrero M, and Timmis KN (1993) Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol 175: 6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, and Perez-Martin J. (1996) Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol 19: 1177–1184. [DOI] [PubMed] [Google Scholar]
- Ezezika OC, Haddad S, Clark TJ, Neidle EL, and Momany C. (2007) Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J. Mol. Biol 367: 616–629. [DOI] [PubMed] [Google Scholar]
- Fuenmayor SL, Wild M, Boyles AL, and Williams PA (1998) A gene cluster encoding steps in the conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol 180: 2522–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara H, Yoshida H, Matsunaga T, Goto M, and Furukawa K. (2006) Cross-regulation of biphenyl- and salicylate-catabolic genes by two regulatory systems in Pseudomonas pseudoalcaligenes KF707. J. Bacteriol 188: 4690–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao TC, and de Lorenzo V. (2006) Transcriptional regulators a la carte: engineering new effector specificities in bacterial regulatory proteins. Curr. Opin. Biotechnol 17: 34–42. [DOI] [PubMed] [Google Scholar]
- Galvao TC, Mencía M, de Lorenzo V,., and (2007) Emergence of novel functions in transcriptional regulators by regression to stem protein types. Mol. Microbiol 65: 907–919. [DOI] [PubMed] [Google Scholar]
- Gerhardt P, Murray RGE, Wood WA, and Krieg NR, (eds) (1994) Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology. [Google Scholar]
- Haigler BE, Wallace WH, and Spain JC (1994) Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol 60: 3466–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KD, Hughes MA, and Williams PA (2000) Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol 182: 3136–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Britt-Compton B, and Williams PA (2003) The naphthalene catabolic (nag) genes of Ralstonia sp. strain U2 are an operon that is regulated by NagR, a LysR-type transcriptional regulator. J. Bacteriol 185: 5847–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju K-S, and Parales RE (2006) Control of substrate specificity by active site residues in nitrobenzene 1,2-dioxygenase. Appl. Environ. Microbiol 72: 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, and Trollinger D. (1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70: 191–197. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop II RM, and Peterson KM (1994) pBBR1MCS: a broad host range cloning vector. BioTechniques 16: 800–802. [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, and Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- Lacal J, Busch A, Guazzaroni ME, Krell T, and Ramos JL (2006) The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins. Proc. Natl. Acad. Sci. USA 103: 8191–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GH, and Ogawa N. (2009) Mutational analysis of the inducer recognition sites of the LysR-type transcriptional regulator TfdT of Burkholderia sp. NK8. Appl. Microbiol. Biotechnol 83: 1085–1094. [DOI] [PubMed] [Google Scholar]
- Lee S-Y, and Rasheed S. (1990) A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques 9: 676–679. [PubMed] [Google Scholar]
- Lessner DJ, Johnson GR, Parales RE, Spain JC, and Gibson DT (2002) Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol 68: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner DJ, Parales RE, Narayan S, and Gibson DT (2003) Expression of nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol 185: 3895–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonneborg R, Smirnova I, Dian C, Leonard GA, and Brzezinski P. (2007) In vivo and in vitro investigation of transcriptional regulation by DntR. J. Mol. Biol 372: 571–582. [DOI] [PubMed] [Google Scholar]
- Maiti R, Van Domselaar GH, Zhang H, and Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic. Acids Res 32: W590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, and Sambrook J. (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Michaelis L, and Menten ML (1913) Die kinetik der invertinwirkung. Biochem. Z 49: 333–369. [Google Scholar]
- Miller JH (1975) Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory. [Google Scholar]
- Nishino SF, Paoli GC, and Spain JC (2000) Aerobic degradation of dinitrotoluenes and the pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol 66: 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino SF, and Spain JC (2004) Catabolism of nitroaromatic compounds. In Pseudomonas. Vol. 3. Ramos J-L (ed). New York: Kluwer Academic / Plenum Publishers, pp. 575–608. [Google Scholar]
- Parales JV, Kumar A, Parales RE, and Gibson DT (1996) Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181: 57–61. [DOI] [PubMed] [Google Scholar]
- Parales JV, Parales RE, Resnick SM, and Gibson DT (1998a) Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α subunit of the oxygenase component. J. Bacteriol 180: 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE, Ontl TA, and Gibson DT (1997) Cloning and sequence analysis of a catechol 2,3-dioxygenase gene from the nitrobenzene-degrading strain Comamonas sp. JS765. J. Ind. Microbiol. Biotechnol 19: 385–391. [DOI] [PubMed] [Google Scholar]
- Parales RE, Emig MD, Lynch NA, and Gibson DT (1998b) Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol 180: 2337–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE (2000) Molecular biology of nitroarene degradation. In Biodegradation of nitroaromatic compounds and explosives. Spain JC, Hughes JB and Knackmuss H-J (eds). Boca Raton, FL.: CRC Press, pp. 63–89. [Google Scholar]
- Parales RE, Huang R, Yu C-L, Parales JV, Lee FKN, Ivkovic-Jensen MM, Liu W, Lessner DJ, Friemann R, Ramaswamy S, and Gibson DT (2005) Purification, characterization, and crystallization of the components of the nitrobenzene and 2-nitrotoluene dioxygenase enzyme systems. Appl. Environ. Microbiol 71: 3806–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P, and Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313. [DOI] [PubMed] [Google Scholar]
- Pridmore RD (1987) New and versatile cloning vectors with kanamycin-resistance marker. Gene 56: 309–312. [DOI] [PubMed] [Google Scholar]
- Ramos JL, Michan C, Rojo F, Dwyer D, and Timmis K. (1990) Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J. Mol. Biol 211: 373–382. [DOI] [PubMed] [Google Scholar]
- Schell MA (1985) Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene 36: 301–309. [DOI] [PubMed] [Google Scholar]
- Selifonova OV, and Eaton RE (1996) Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl. Environ. Microbiol 62: 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingler V. (2003) Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ. Microbiol 5: 1226–1241. [DOI] [PubMed] [Google Scholar]
- Shingleton JT, Applegate BM, Nagel AC, Bienkowski PR, and Sayler GS (1998) Induction of the tod operon by trichloroethylene in Pseudomonas putida TVA8. Appl. Environ. Microbiol 64: 5049–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, and Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1: 784–789. [Google Scholar]
- Smirnova IA, Dian C, Leonard GA, McSweeney S, Birse D, and Brzezinski P. (2004) Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J. Mol. Biol 340: 405–418. [DOI] [PubMed] [Google Scholar]
- Spain JC, Hughes JB, and Knackmuss H-J (2000) Perspectives of bioelimination of polynitroaromatic compounds. In Biodegradation of nitroaromatic compounds and explosives. Lenke H, Achnich C. and Knackmuss H-J (eds). Boca Raton, FL.: CRC Press, pp. 91–126. [Google Scholar]
- Spanggord RJ, Spain JC, Nishino SF, and Mortelmans KE (1991) Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol 57: 3200–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Palleroni NJ, and Doudoroff M. (1966) The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol 43: 159–271. [DOI] [PubMed] [Google Scholar]
- Streitwieser A, Heathcock C, and Kosower E. (1998) Introduction into Organic Chemistry, 4th edition: Prentice Hall. [Google Scholar]
- Tokuriki N, and Tawfik DS (2009) Protein dynamism and evolvability. Science 324: 203–207. [DOI] [PubMed] [Google Scholar]
- Zhou N-Y, Fuenmayor SL, and Williams PA (2001) nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol 183: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.