Abstract
NS5A of the hepatitis C virus (HCV) is a highly phosphorylated protein involved in resistance against interferon and required most likely for replication of the viral genome. Phosphorylation of this protein is mediated by a cellular kinase(s) generating multiple proteins with different electrophoretic mobilities. In the case of the genotype 1b isolate HCV-J, in addition to the basal phosphorylated NS5A (designated pp56), a hyperphosphorylated form (pp58) was found on coexpression of NS4A (T. Kaneko, Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno, Biochem. Biophys. Res. Commun. 205:320–326, 1994). Using a comparative analysis of two full-length genomes of genotype 1b, competent or defective for NS5A hyperphosphorylation, we investigated the requirements for this NS5A modification. We found that hyperphosphorylation occurs when NS5A is expressed as part of a continuous NS3-5A polyprotein but not when it is expressed on its own or trans complemented with one or several other viral proteins. Results obtained with chimeras of both genomes show that single amino acid substitutions within NS3 that do not affect polyprotein cleavage can enhance or reduce NS5A hyperphosphorylation. Furthermore, mutations in the central or carboxy-terminal NS4A domain as well as small deletions in NS4B can also reduce or block hyperphosphorylation without affecting polyprotein processing. These requirements most likely reflect the formation of a highly ordered NS3-5A multisubunit complex responsible for the differential phosphorylation of NS5A and probably also for modulation of its biological activities.
Hepatitis C virus (HCV) is the major causative agent of sporadic and transfusion-associated non-A, non-B hepatitis (10, 24). Although most infections are inapparent or initially associated with only mild symptoms, due to the high persistence the long-term effects are dramatic. About 50% of all infections lead to chronic liver disease, which can range from an apparently healthy carrier state to chronic active hepatitis, liver cirrhosis, or hepatocellular carcinoma. It is estimated that 100 million to 200 million people worldwide are infected with this insidious agent.
HCV was classified as the distinct genus Hepacivirus together with the genera Flavivirus and Pestivirus in the family Flaviviridae (41). These viruses are characterized by an enveloped virion harboring a plus-strand RNA genome. In case of HCV, this genome has a length of ca. 9.6 kb and carries a single long open reading frame (ORF) flanked at the 5′ and 3′ ends by nontranslated regions required for RNA translation and replication (for reviews, see references 4, 9, and 45). The viral genes are expressed as a polyprotein, just 3,000 amino acids in length, which is cleaved co- and posttranslationally by host cell signal peptidases and two viral proteinases. At least 10 different cleavage products have been identified, which are ordered within the polyprotein (from the amino to the carboxy terminus): NH2-core-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. The structural proteins core, envelope protein 1 (E1), and E2 are the major constituents of the virion. The function of p7, a small highly hydrophobic peptide is not known. NS2 and the amino-terminal domain of NS3 constitute the NS2-3 proteinase responsible for cleavage at the NS2/3 junction (18, 20). NS3 is a bifunctional molecule. The amino-terminal domain carries a serine-type proteinase required for cleavage at the NS3/4A, NS4A/B, NS4B/5A, and NS5A/B junctions (5, 12, 19, 20, 39, 55). The carboxy-terminal two-thirds constitute a nucleoside triphosphatase-RNA helicase (23, 30, 50, 51) belonging to the superfamily class II helicases (27). NS4A is a proteinase cofactor forming a stable complex with NS3 and enhancing proteolytic activity (6, 7, 13, 31, 34, 38, 53, 57). Mutation analyses have shown that sequences close to the amino terminus of NS3 (residues 1027 to 1049) and a 14-residue-long sequence in the center of NS4A (residues 1678 to 1691) are sufficient for interaction and full proteinase activation (7, 14, 32, 33, 47–49, 56). Except for its hydrophobic nature, virtually nothing is known about NS4B. The highly phosphorylated NS5A can interfere with the function of the interferon (IFN)-induced protein kinase PKR, accounting for the IFN resistance observed at least with some genotypes (16, 17). Whether NS5A has an additional, more direct role in formation or activity of the replicase complex is not known. NS5B is the RNA-dependent RNA polymerase (8, 37).
Labeling experiments with 32Pi and phosphoamino acid analyses have shown that NS5A is phosphorylated on serine and, to a much lesser extent, on threonine residues (25, 28, 44). For the genotype 1b HCV-J isolate, on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at least two NS5A phosphorylation variants can be found, migrating with apparent molecular masses of 56 and 58 kDa; they were designated the basal and hyperphosphorylated forms, respectively (28, 54). Two distinct NS5A domains located around the center and close to the carboxy terminus (residues 2200 to 2250 and 2350 to 2419, respectively [1, 54]) were shown to be required for basal phosphorylation. Although phosphate acceptor sites have not directly been mapped, mutation analyses indicate that three serine residues in the central region (at positions 2197, 2201, and 2204) are important for hyperphosphorylation (54). At least for the HCV-J isolate, hyperphosphorylation of NS5A requires the presence of fully processed NS4A (28, 52, 54). Coprecipitation studies have shown that NS4A and NS5A can form a complex (1). Mutational ablation of this interaction, in which amino-terminal NS5A sequences, in particular residues 2135 to 2139, play an important role, also affects hyperphosphorylation (1). However, this observation does not account for all HCV isolates. For the HCV-H isolate, belonging to genotype 1a, multiple phosphorylated NS5A species were found even when the protein was expressed in the absence of other nonstructural proteins (44). The enzyme responsible for NS5A phosphorylation is not known. Results from an in vitro kinase assay clearly show that one or several cellular enzymes are responsible (25, 44), and their inhibitor profiles identify them as members of the CMGC group of serine-threonine kinases (44).
Using a comparative analysis of two full-length HCV genomes of genotype 1b, differing with respect to NS5A hyperphosphorylation, we found that a continuous NS3-5A region is required for this NS5A modification. Various amino acid substitutions or small deletions in the NS3-4B region not affecting proteolytic processing severely reduced formation of pp58. Our results suggest that hyperphosphorylation of NS5A requires a higher-order protein complex forming only in cis and imply that different mechanisms operate in NS5A hyperphosphorylation of different HCV isolates.
MATERIALS AND METHODS
Cells and viruses.
Cell monolayers of the BHK-21 cell line were grown in Dulbecco’s modified minimal essential medium (DMEM) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin, 100 μg of streptomycin, and 10% fetal calf serum. The recombinant vaccinia virus vTF7-3 expressing the RNA polymerase of bacteriophage T7 (15) was obtained from B. Moss (National Institutes of Health, Bethesda, Md.). Stocks of recombinant vaccinia viruses were grown in human TK 143 cell monolayers, and titers of infectious progeny were determined by plaque assay using the same cell line.
Construction of full-length NC and C genomes.
Two full-length HCV genomes of genotype 1b were cloned by reverse transcription-PCR (RT-PCR) methodologies from serum and total liver RNA of a chronically infected German patient who had undergone liver transplantation. The serum-derived genome was obtained by conventional RT-PCR performed with RNA isolated from several blood samples. Total nucleic acids were isolated from 350 μl of serum by digestion with proteinase K (40 μg/ml) in a buffer containing 10 mM Tris-HCl (pH 8.3), 0.5% SDS, and 100 mM NaCl in a total volume of 900 μl. After 2 h at 56°C, samples were extracted with phenol and phenol-chloroform and nucleic acids were precipitated with ethanol. Samples were resuspended in 20 μl of RNase-free water, and one half was denatured for 10 min at 80°C in the presence of an HCV-specific primer. cDNA synthesis was performed with Superscript reverse transcriptase (Life Technologies, Karlsruhe, Germany) at 42°C for 1 h. The cDNA mixture was treated with RNase H (Boehringer Mannheim), and the enzymes were inactivated by 10 min of incubation at 95°C. Nested PCRs were performed with Taq DNA polymerase (Perkin-Elmer, Weiterstadt, Germany) under standard conditions using two rounds of 35 cycles and HCV-specific primers derived from published genome sequences. Cloned PCR fragments were sequenced, and the complete ORF was reconstituted from overlapping fragments, which were combined via convenient restriction sites or by PCR according to the method of Ho et al. (22). The complete ORF was inserted into the vector pTM1-2 (7) via engineered NcoI and SpeI sites at the 5′ and 3′ ends, respectively. Due to this cloning strategy, the core protein starts with MG instead of MS whereas translation stops at the authentic UGA codon of NS5B at position 9372 of our isolate. The resulting nonconsensus (NC) sequence plasmid was designated pTM-NC1. Corresponding polypeptides are termed NS5ANC, etc.
The second genome was cloned by long-distance RT-PCR using total RNA prepared from a piece of the explanted liver of the same patient. This genome represents an isolate-specific consensus (C) sequence derived from comparisons of sequences of several cloned PCR fragments. Details of the cloning procedure and reconstitution of this full-length genome will be described elsewhere. The complete ORF of this genome was cloned via engineered NcoI and SpeI sites into pTM1-2 and designated pTM-C1. Corresponding polypeptides are termed NS5AC, etc.
Plasmid constructions.
Standard recombinant DNA techniques were used for generation of the expression plasmids (46). Construction of the basic expression vector pTM1-2 directing the transcription of HCV sequences under control of the bacteriophage T7 RNA polymerase and allowing the translation under control of the internal ribosome entry site of the encephalomyocarditis virus has been described elsewhere (7). All HCV sequences were inserted into pTM1-2 via NcoI and SpeI sites introduced by oligonucleotides used for PCR or, in case of pTM2-5BC, by using an authentic NcoI site present at the exact 5′ end of NS2. The following conserved restriction sites within the NS3-5B region were used to generate chimeras between the NC and the C sequences (all numbers refer to the nucleotide positions of our HCV isolates): SfiI (3622), SalI (4725), StuI (5032), NsiI (5286), StuI (5869), EcoRI (6699), and NsiI (8726). Site-directed mutagenesis was done by generating overlapping PCR fragments carrying the desired nucleotide exchange within the overlap. Fragments were combined by second PCR and subcloned into the appropriate plasmids. In-frame deletions of NS4B sequences were generated in the same way, using oligonucleotides carrying the desired deletion. All HCV sequences amplified by PCR were completely sequenced after subcloning.
Protein expression with the vaccinia virus-T7 hybrid system, immunoprecipitation, and protein analyses.
Transient expression of HCV proteins in cell cultures with the vaccinia virus-T7 hybrid system and metabolic labeling have been described elsewhere (7). In brief, 0.75 × 105 cells seeded in 15-mm-diameter dishes were infected with vTF7-3 with a multiplicity of infection of 5 to 10 for 1 h at room temperature. After removal of the inoculum and a 30-min incubation in DMEM supplemented with 10% fetal calf serum and 2 mM glutamine, cells were transfected with 350 ng of purified plasmid DNA and 2.5 μl of Lipofectamine (Life Technologies, Eggenstein, Germany) as instructed by the manufacturer. After 3 h at 37°C, cells were washed with prewarmed DMEM lacking methionine and incubated for 4 h in DMEM supplemented with 2 mM glutamine and 100 μCi of 35S protein labeling mixture (Express; NEN Life Science, Köln, Germany) per ml. For radiolabeling of proteins with [32P]orthophosphate (Amersham Pharmacia Biotech, Freiburg, Germany), cells were transfected as described above, washed three times with phosphate-free medium (Sigma, Deisenhofen, Germany), and labeled for 4 h in the same medium supplemented with 100 μCi of [32P]orthophosphate per ml. Cells were washed several times, and HCV-specific proteins were isolated by immunoprecipitation from cell lysates under nondenaturing or denaturing conditions as described recently (6, 7). Proteins were separated by SDS-PAGE or Tricine-SDS-PAGE. For optimal separation of the two NS5A phosphoprotein variants, SDS-PAGE using gels with an acrylamide:bisacrylamide ratio of 12.5%:0.08% was performed. Dephosphorylation of NS5A was performed with calf intestinal phosphatase (CIP) or lambda protein phosphatase (both purchased from New England Biolabs GmbH, Schwalbach/Taunus, Germany), using immunoprecipitated proteins as instructed by the manufacturers. Immunocomplexes were washed three times with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) and once with the corresponding phosphatase assay buffer. Samples were treated with 400 U of phage λ phosphatase or 10 U of CIP. After 1 h at 30°C, samples were centrifuged, supernatants were removed, and dephosphorylated immunocomplexes were analyzed by SDS-PAGE and autoradiography.
Nucleotide sequence accession number.
The nucleotide and amino acid sequences of the HCV NC and C genomes used in this study are deposited in the EMBL nucleotide sequence database and can be retrieved under accession no. AJ238800 (NC1) and AJ238799 (Con1), respectively.
RESULTS
Phosphorylation of NS5A expressed from two different full-length HCV genomes.
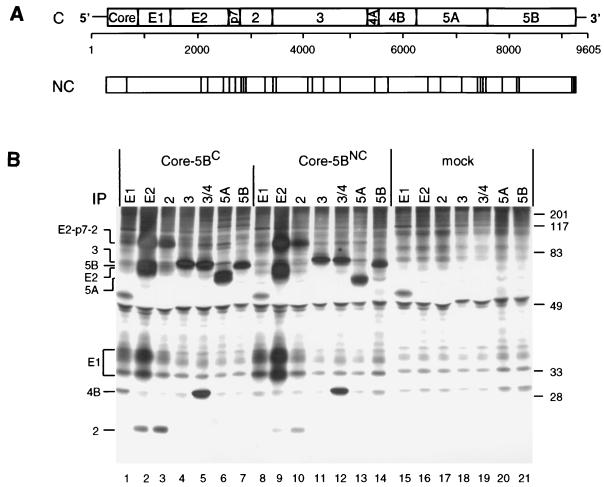
In an attempt to construct a functional HCV genome, we cloned two full-length sequences of genotype 1b from a chronically infected German patient. The first genome was cloned by conventional RT-PCR from multiple serum samples and reconstituted from 11 different fragments. The second genome, cloned by long-distance RT-PCR from total liver RNA, represents a consensus sequence established by sequence analysis of several PCR fragments each spanning half of the genome. Therefore, the sequence of the first genome is referred to as the NC sequence; it differs from the C genome at 30 positions scattered throughout the polyprotein (Fig. 1A).
FIG. 1.
(A) Structure of the HCV genome and comparison of the C and NC polyproteins. (A) Schematic representation of the HCV genome organization, with the structural proteins core to E2, p7, and the nonstructural proteins NS2 to NS5B shown at the top. Numbers below refer to the nucleotide positions of our HCV isolate. Amino acid deviations of the NC polyprotein from the C sequence are indicated by vertical lines. (B) Expression of the complete ORFs of the NC and C isolates in BHK-21 cells and detection of cleavage products. Cells infected with the vTF7-3 vaccinia virus recombinant were transfected with plasmids directing expression of the complete polyprotein of the NC or C genome. After metabolic radiolabeling with [35S]methionine-cysteine, HCV-specific proteins were isolated from the cell lysate under nondenaturing conditions by immunoprecipitation (IP) using antisera with specificities given above the lanes. Identification of HCV-specific proteins and the positions of protein molecular mass standards (in kilodaltons) are given on the left and right, respectively.
To analyze functionality with respect to polyprotein processing, the full-length ORFs of both genomes were transiently expressed with the vaccinia virus-T7 hybrid system in BHK-21 cells. Proteins were radiolabeled metabolically with [35S]methionine-cysteine, and production of HCV proteins was analyzed by nondenaturing immunoprecipitation, SDS-PAGE, and autoradiography. As shown in Fig. 1B, cleavage products of correct sizes, immunoreactivities, and coprecipitation properties were found, with two major differences between the genomes. First, the apparent molecular weight of E2C was higher than that of E2NC (compare lane 2 with lane 9), most likely due to the presence of an additional glycosylation site at amino acid position 576, conserved between most HCV genomes but absent in the NC sequence. Second, while NS5ANC appeared as a single protein species, two anti-NS5A-reactive proteins with apparent molecular masses of approximately 56 and 58 kDa were found with the C ORF (compare lane 13 with lane 6). Since NS5A is a phosphoprotein, described for HCV genotype 1b as a 56-kDa hypophosphorylated species and a 58-kDa hyperphosphorylated form (28, 54), this result suggested a selective defect of NS5ANC hyperphosphorylation.
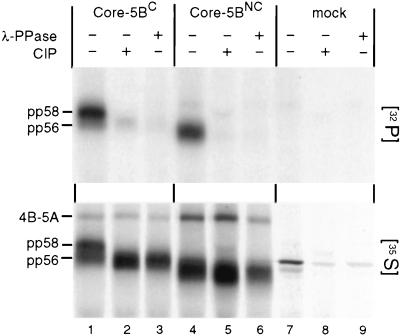
To further substantiate this assumption, we expressed the complete polyproteins of both genomes with the vaccinia virus T7-hybrid system in BHK-21 cells and radiolabeled proteins with either [32P]orthophosphate or [35S]methionine-cysteine. NS5A was isolated by immunoprecipitation and analyzed either directly by SDS-PAGE or after treatment with phage λ phosphatase or CIP. As shown in the top panel of Fig. 2, NS5A expressed from both genomes underwent basal phosphorylation. However, the heavily labeled hyperphosphorylated pp58 form was found only with NS5AC and not with NS5ANC (compare lane 1 with lane 4). All 32P-labeled NS5A proteins were sensitive to treatment with phosphatases. The loss of the signals after dephosphorylation could not be ascribed to contaminating proteinases present in the phosphatase preparations, because no degradation of the 35S-labeled proteins was found (bottom panel). In summary, these results show that the slower-migrating form of NS5AC (pp58) is a phosphatase-sensitive hyperphosphorylated species and that only the basal phosphorylated form is generated with NS5A of the nonconsensus genome. It should be noted that the overall electrophoretic mobility of NS5ANC was slightly faster than that of NS5AC. Since this difference was not affected by dephosphorylation, it most likely can be ascribed to differences in the amino acid compositions rather than to different levels of phosphorylation (see below).
FIG. 2.
Hyperphosphorylation of NS5A encoded by the C genome. Cells previously infected with vTF7-3 were transfected with plasmids encoding the core-5B sequence of the C or NC genome or with the plasmid vector (mock). Four hours after transfection, proteins were metabolically labeled with [32P]orthophosphate (top) or [35S]methionine-cysteine (bottom). HCV-specific proteins were isolated by immunoprecipitation, and one third of the immunocomplexes was loaded directly onto the gel. One half of the remainder was treated with CIP, and the other half was treated with phosphatase from phage λ (λ-PPase) prior to SDS-PAGE.
A continuous NS3-5A sequence is required for hyperphosphorylation of NS5A.
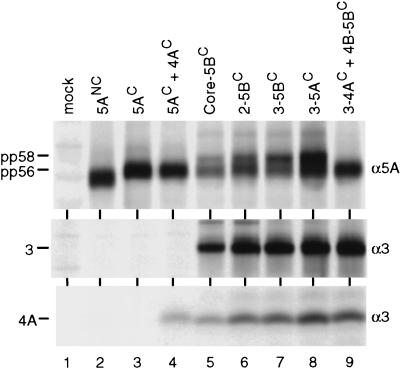
Having confirmed the different phosphorylation states of the two NS5A proteins, we assumed that the lack of NS5ANC hyperphosphorylation was caused by particular deviations from the C sequence within the NS5A region. To simplify the analysis, we determined the phosphorylation patterns of both NS5A proteins after transient expression in the absence of other nonstructural proteins, because it was shown that expression of NS5A alone is sufficient for generation of multiple phosphorylation forms with some genotypes (44). However, under these conditions only the faster-migrating form of NS5AC (pp56) was generated, suggesting that additional nonstructural proteins are required for hyperphosphorylation (Fig. 3, lane 3). The most likely candidate was NS4A, because for the HCV-J isolate, belonging to genotype 1b like our isolate, it is required for hyperphosphorylation of NS5A (1, 28, 54). However, no hyperphosphorylation was found even when NS4AC was coexpressed with NS5AC (lane 4), suggesting that other or additional HCV proteins are required.
FIG. 3.
A continuous NS3-5A sequence is required for NS5A hyperphosphorylation. vTF7-3-infected BHK-21 cells were transfected with plasmids directing the expression of proteins given above the lanes, and HCV-specific proteins were analyzed after radiolabeling by immunoprecipitation under nondenaturing conditions, allowing the coprecipitation of NS4A with NS3. Specificities of antisera are given at the left; identities of the HCV proteins are shown at the right.
To identify such additional cofactors, we analyzed various amino- and carboxy-terminal truncations, introduced into the parental core-5B polyprotein for NS5AC hyperphosphorylation. pp58 was found with the NS2-5B, NS3-5B, and NS3-5A polyproteins, demonstrating that the structural proteins, p7, NS2, and NS5B are dispensable for NS5A hyperphosphorylation (Fig. 3, lane 6 to 8). In fact, a gradual increase of the pp58/pp56 ratio was found with the amino-terminally truncated polyproteins, suggesting that sequences amino terminal of NS3 negatively affect hyperphosphorylation.
To analyze whether hyperphosphorylation of NS5AC can be mediated in trans, a series of cotransfection experiments was performed with constructs directing the expression of individual HCV proteins or polyprotein fragments. An example of this type of analysis is given in Fig. 3. When the NS3/4A proteinase was coexpressed with an NS4B-5B polyprotein, no hyperphosphorylation of NS5A was found (lane 9). Similarly, coexpression of NS3 with an NS4A-5B polyprotein did not allow formation of pp58 (not shown). In an extensive series of cotransfection experiments, every possible combination of individual nonstructural proteins and polyprotein fragments was tested, but we never observed hyperphosphorylation of NS5A (not shown), demonstrating that this modification occurs only when NS5A is expressed in the context of a continuous NS3-5A polyprotein.
Influence of the NS3-4B region on NS5A hyperphosphorylation.
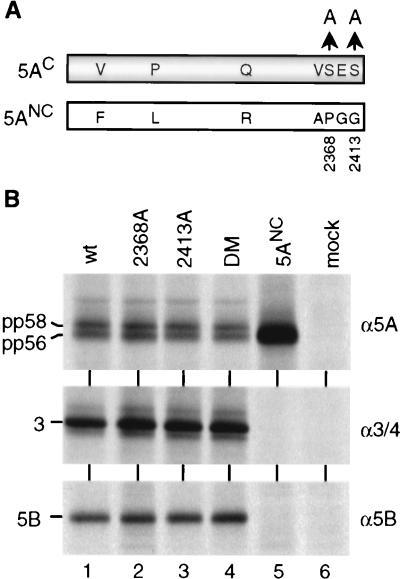
Using an amino acid sequence comparison between NS5AC and NS5ANC, we found seven differences, among them two serine residues close at the carboxy terminus (Fig. 4A; Table 1). Since NS5A is preferentially phosphorylated at serine residues with one cluster most likely located close to the carboxy terminus (residues 2350 to 2419 [54]), we assumed that the absence of these two serine residues with NS5ANC might be responsible for the lack of hyperphosphorylation. In this case, mutations affecting serine residues 2368 and 2413 of NS5AC should block hyperphosphorylation. Therefore, single or double alanine substitutions for the serine residues were introduced into NS5AC, and the mutants were expressed in BHK-21 cells in the context of the NS3-5B polyprotein. As shown in Fig. 4B, both the single substitutions and the double mutation did not affect production of pp58, suggesting that other differences between the two genomes were responsible for the selective hyperphosphorylation of NS5AC.
FIG. 4.
Removal of two serine residues at the carboxy terminus of NS5AC does not affect hyperphosphorylation. (A) Amino acid sequence differences between NS5A of the C (shaded bar) and NC (light bar) genomes (see also Table 1). Arrows point to alanine residues substituting for serine residues of 5AC at positions 2368 and 2413. (B) Cells were transfected with constructs directing the expression of NS3-5BC polyproteins carrying the indicated amino acid substitutions within NS5AC. HCV-specific proteins were radiolabeled and analyzed by immunoprecipitation using antisera given to the right and SDS-PAGE. wt, wild type; DM, double mutant.
TABLE 1.
Differences in amino acid sequence of the NS3-5B region between the NC and C genomes
| Position of amino acid differencea
| ||||
|---|---|---|---|---|
| NS3 (1027–1657b) | NS4A (1658–1711) | NS4B (1712–1972) | NS5A (1973–2419) | NS5B (2420–3010) |
| 1028 (H→P) | 1687 (V→I) | 1792 (V→A) | 2047 (F→V) | 2536 (S→N) |
| 1061 (M→V) | 2119 (L→P) | 2626 (T→A) | ||
| 1263 (S→G) | 2260 (R→Q) | 2629 (S→A) | ||
| 1285 (T→I) | 2360 (A→V) | 2979 (T→I) | ||
| 1373 (V→I) | 2368 (P→S) | 2981 (R→H) | ||
| 1496 (I→M) | 2383 (G→E) | |||
| 2413 (G→S) | ||||
Amino acids are given in single-letter code, with the arrow pointing to the residue found in the consensus sequence.
Amino- and carboxy-terminal boundaries.
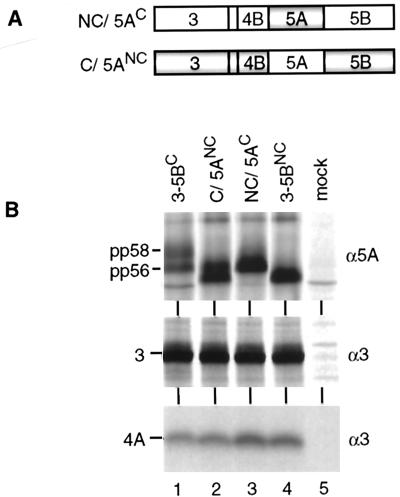
To analyze whether any of the other amino acid sequence differences might be responsible, the complete NS5A sequences of both isolates were exchanged in the context of the NS3-5B polyproteins, and the resulting chimeras (Fig. 5A) were examined as described above. Surprisingly, NS5AC expressed in the context of NS3-5BNC sequence was no longer hyperphosphorylated (Fig. 5B, lane 3), whereas NS5ANC expressed in the context of the NS3-5BC sequence became hyperphosphorylated (lane 2). Thus, NS5ANC was competent for hyperphosphorylation and the failure to generate pp58, observed with the NC genome, was due to mutations in the NS3-4B region.
FIG. 5.
Effect of flanking sequences on NS5A hyperphosphorylation. (A) Schematic representation of the NC and C polyproteins carrying exact substitutions for 5AC and 5ANC, respectively. Shaded and light areas indicate the C and NC sequences, respectively. (B) Expression of chimeric polyproteins in BHK-21 cells and detection of radiolabeled HCV proteins by immunoprecipitation under nondenaturing conditions allowing the coprecipitation of NS4A with NS3. Specificities of antisera are given at the right; identities of HCV proteins are shown at the left.
A single amino acid substitution in the amino terminus of NS3 affects NS5A hyperphosphorylation.
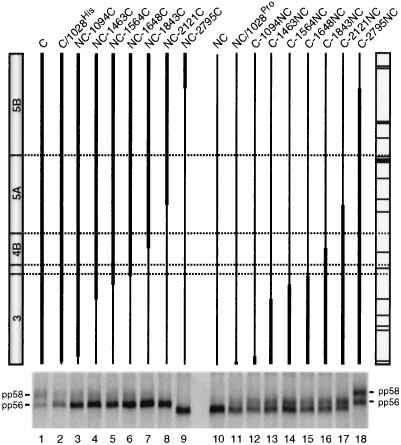
To further narrow down the sequence(s) or protein(s) responsible for the defect of NS5ANC hyperphosphorylation, a panel of NS3-5B chimeras was constructed by exchanging sequences of variable lengths between the C and NC genomes (Fig. 6). These constructs were transiently expressed in BHK-21 cells and analyzed for the production of pp58. Surprisingly, the smallest change, replacement of only a proline residue at the amino terminus of NS3C (position 1028 of the polyprotein [Table 1]) with the NC histidine residue, significantly reduced production of pp58 (compare lane 1 with lane 2). Additional, more extensive replacements of C sequences with NC sequences, including a C valine residue at position 1061, further reduced hyperphosphorylation of NS5AC (lane 3). No pp58 was found when most of the NS3 sequence of the C genome was replaced by the NC sequence (lane 4 to 6). Conversely to this loss of NS5A hyperphosphorylation, a gain of hyperphosphorylation was found when NS3 sequences of the NC genome were replaced by corresponding sequences of the C genome. Introduction of the C proline residue into the amino terminus of NS3NC led to production of pp58 (compare lane 10 with lane 11). Increasing the size of the transferred NS3C sequence led to an increase of NS5A hyperphosphorylation, which was at maximum when almost the complete NS3NC was replaced by the corresponding C sequence (lane 12 to 15). In summary, these data demonstrate that NS3 strongly affects hyperphosphorylation of NS5A.
FIG. 6.
Hyperphosphorylation of NS5A is influenced by NS3. A schematic representation of the chimeric NS3-5B polyproteins expressed with the vaccinia virus-T7 hybrid system in BHK-21 cells is given above each lane. Thick lines refer to the C sequence; thin lines indicate the NC sequence. The individual constructs are identified above the lines. Schematic drawings of the corresponding C and NC polyproteins are given at the left and right, respectively, with dotted horizontal lines indicating the cleavage sites. Amino acid deviations of the NC sequence from the C sequence are marked with horizontal lines in the NC ORF. The phosphorylation patterns of NS5A proteins expressed from given constructs are shown below. The two phosphoprotein variants pp56 and pp58 are marked.
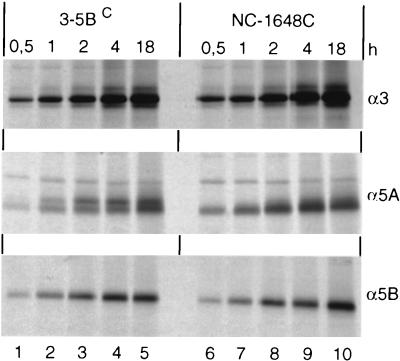
Phosphorylation of NS5A most likely takes place after release from the polyprotein precursor (53, 54). Therefore, it was possible that the effect of NS3 on NS5A hyperphosphorylation was due to different efficiencies with which these chimeric polyproteins were cleaved. For example, a delay of NS5A processing might lead to a low hyperphosphorylation rate. Although the amounts of cleavage products accumulating during the 4-h labeling period did not differ significantly between the various chimeric polyproteins (not shown), it was possible that we missed more subtle effects on processing kinetics. To rule out this possibility, a pulse-labeling experiment was performed with the parental NS3-5BC and a chimera producing very low amounts of pp58 (NC-1648C). The comparison shown in Fig. 7 demonstrates that the two polyproteins were processed with similar kinetics, ruling out the possibility that the different degrees of hyperphosphorylation were due to different cleavage efficiencies. Since enzymatic activities of the two proteinases were comparable, yet hyperphosphorylation of NS5A was found only with the C polyprotein, these results suggest that NS3 acts as a structural component in pp58 formation.
FIG. 7.
Comparison of processing kinetics of NS3-5BC and the chimeric NC-1648C polyprotein. BHK-21 cells infected with vTF7-3 were transfected with the indicated constructs, and proteins were pulse-labeled with [35S]methionine-cysteine for various times. HCV proteins were analyzed after immunoprecipitation by SDS-PAGE and autoradiography.
Influence of NS4A on NS5A hyperphosphorylation.
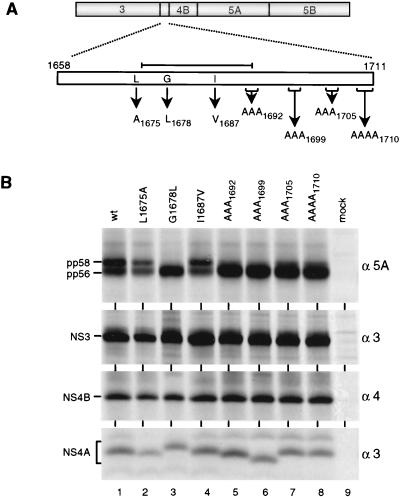
Although the data described so far show that NS3 is a determinant of NS5A hyperphosphorylation, they do not rule out the contribution of the other nonstructural proteins, in particular NS4A. First, it was shown for the HCV-J isolate that NS4A is required for production of pp58, although in this case it can be mediated in trans by NS4A alone (1, 28, 54). Second, the serine-type proteinase located in the amino-terminal NS3 domain forms a stable complex with NS4A, which most likely leads to conformational changes within NS3 (31, 38, 57) that might also affect the function of NS3 with respect to NS5A hyperphosphorylation. Therefore, a panel of amino acid substitutions within NS4AC was generated and expressed in BHK-21 cells in the context of the NS3-5B polyprotein of the consensus isolate (Fig. 8A). The mutations were selected to preserve functional domains of NS4A: an amino-terminal transmembrane helix, ca. 20 residues in length (57), and the central proteinase activation domain (residues 1678 to 1691) [7, 31, 34, 48, 49, 53, 56]). We also included a valine substitution for the isoleucine at position 1687, corresponding to the only amino acid sequence difference within NS4A between the two isolates (Table 1). As shown in Fig. 8B, the alanine substitution for the leucine residue amino terminal of the activation domain as well as the substitution converting the NS4AC sequence to the NC sequence did not affect NS5A hyperphosphorylation (lanes 2 and 4, respectively). In case of the substitution in the central proteinase activation domain (G1678L), previously shown not to affect polyprotein processing (7), production of pp58 was completely abolished (lane 3). Interestingly, the multiple alanine substitutions carboxy terminal of the activation domain drastically reduced NS5A hyperphosphorylation (lane 5 to 8). These results suggest that a proper structure of NS4A is required for hyperphosphorylation of NS5A.
FIG. 8.
Modulation of NS5A hyperphosphorylation by NS4A. (A) A schematic representation of the NS3-5BC sequence is given at the top; a magnification of NS4A is drawn below. Amino acid residues in NS4A selected for site-directed mutagenesis are given in single-letter code, with arrows pointing to the substituting residue. Subscript numbers refer to the positions of the amino acids (in case of multiple substitutions of the last residue) within the complete polyprotein. The central proteinase activation domain of NS4A is indicated by the line above the magnified NS4A ORF. (B) Cells infected with the vTF7-3 recombinant vaccinia virus were transfected with NS3-5BC constructs noted above the lanes; after metabolic radiolabeling, HCV proteins were isolated from cell lysates by immunoprecipitation under nondenaturing conditions. Therefore, NS4A is coprecipitated with NS3. Specificities of antisera and the identities of HCV proteins are given at the right and left, respectively. wt, wild type.
Essential role of NS4B for NS5A hyperphosphorylation.
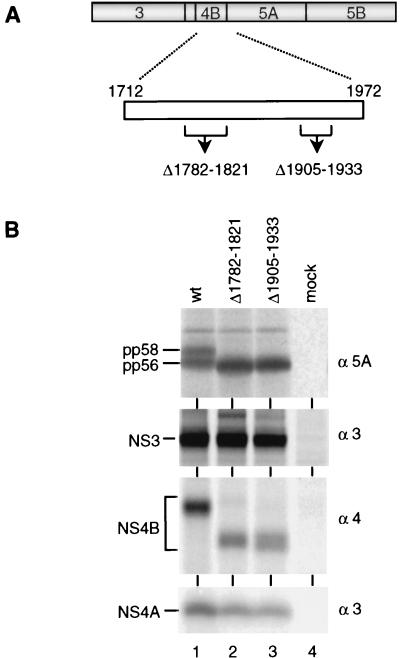
Having shown that both NS3 and NS4A are required for hyperphosphorylation of NS5A, we anticipated that NS4B might play a role as well, because formation of pp58 was found only with a continuous NS3-5A polyprotein. To validate this assumption, two NS4B in-frame deletions were constructed (Fig. 9A). By using secondary structure predictions and the hydrophobicity profile, the mutations were designed not to affect the overall structure of the protein. When expressed in the context of the NS3-5BC polyprotein, all cleavage products were found, indicating proper processing. However, formation of pp58 was completely blocked, demonstrating that an intact NS4B is also required for hyperphosphorylation of NS5A.
FIG. 9.
Hyperphosphorylation of NS5A is influenced by NS4B. (A) Schematic drawing of the NS3-5BC sequence, with a magnification of the NS4B given below. The positions of in-frame deletions (▵) are drawn below. Numbers refer to the first and the last residue of the deleted sequence. (B) Analysis of immunoprecipitated HCV proteins after transient expression of given NS3-5BC constructs in BHK-21 cells. Identification of HCV proteins is given at the left; specificities of antisera are shown at the right.
DISCUSSION
In this study, we investigated the requirements for NS5A phosphorylation. Taking advantage of the different phosphorylation patterns of NS5A observed with two cloned full-length genomes, a genetic analysis was performed. Our results show that a continuous NS3-5A sequence is required for NS5A hyperphosphorylation. Mutations at various positions in the NS3-4B region, not affecting polyprotein processing, can reduce or enhance this NS5A modification. Thus, structural integrity of each of these proteins, forming most likely a multisubunit protein complex, is essential for differential phosphorylation of NS5A.
Although phosphorylation of NS5A is a biochemical trait conserved among all HCV isolates analyzed so far, the conditions required for this modification appear to differ between various genotypes and even between different isolates of the same genotype. In case of the genotype 1a HCV-H isolate, the phosphorylation patterns of NS5A expressed on its own or in the context of an NS2-5B polyprotein were almost identical, suggesting that no viral proteins other than NS5A itself are involved (44). For the HCV-J isolate, a member of genotype 1b, two distinct phosphorylation variants were described (28): a basal phosphorylated form with an apparent molecular mass of 56 kDa and a 58-kDa hyperphosphorylated form. Production of the latter occurred only in the presence of a fully processed NS4A which could be provided in trans to restore hyperphosphorylation (28). Results from coprecipitation analyses showed that NS4A and NS5A form a complex (1). Since mutational ablation of this interaction also affected production of pp58, complex formation appears to be required for hyperphosphorylation of NS5A. In case of the HCV-BK isolate, also belonging to genotype 1b, NS2 appears to be important for NS5A hyperphosphorylation (36). For the HCV isolate described in the present report, much more complex requirements were found. First, NS5A expressed on its own is not hyperphosphorylated; second, hyperphosphorylation cannot be restored by coexpression of NS4A or other nonstructural proteins; third, NS3 and NS4B are required in addition to NS4A. The explanation for these various observations is not clear, but isolate-specific differences appear to play an important role. The HCV C genome analyzed in this study belongs to genotype 1b and therefore is more closely related to the HCV-J than the HCV-H isolate. This similarity is partly reflected by the dependence of NS5A hyperphosphorylation on additional viral factors, namely, NS4A for HCV-J or a continuous NS3-5A polyprotein for the genome described here. It remains to be determined whether analogous complex requirements will be found for NS5A hyperphosphorylation with other HCV isolates and whether there is a correlation with the genotype.
The finding that several mutations affecting NS3, NS4A, or NS4B can reduce or enhance NS5A hyperphosphorylation suggests the formation of a highly ordered multisubunit protein complex, containing most likely also cellular proteins, in particular the protein kinase(s) associated with NS5A (25, 44). Although NS5B is not required for hyperphosphorylation, this result does not exclude the possibility that NS5B also participates in this complex. In fact, it was shown that NS5A can also interact with NS5B (21), suggesting that the cleavage products of the NS3-5B precursor form a multiprotein complex. The formation of such a complex has been proposed by others. Using coprecipitation studies, Lin and coworkers (35) have shown that NS4A of the HCV-H isolate forms a nonionic detergent-stable complex with an uncleaved NS4B-5A substrate as well as with NS3 and NS4B. In a similar approach, a coprecipitation of NS5B with NS3 and NS4A has been shown after transient expression of individual proteins in cell culture (26). Thus, formation of a multisubunit protein complex, most likely constituted of NS3-5B plus a cellular protein(s), is a feature found with several HCV isolates.
The role of NS5A phosphorylation in the viral life cycle is not known. The conservation of this biochemical trait among divergent isolates may indicate an important role. Interestingly, for the NS5 protein of dengue virus type 2, two phosphorylated forms were predominantly found in infected cells (29). While the hyperphosphorylated form was located primarily in the nucleus, the hypophosphorylated form was found in the cytoplasm. Coprecipitation studies showed that NS3 interacts with NS5 but only with the hypophosphorylated form (29). Based on these results and immunofluorescence studies, it was suggested that NS3 and NS5, the key players in RNA replication, form a complex tightly associated with endoplasmic reticulum membranes. Due to hyperphosphorylation of NS5 at a late stage of replication, the NS3/5 interaction would be disrupted and NS5 be transported to the nucleus (29).
As discussed recently (44), regulation of virus replication by phosphorylation has also been shown for other viruses. In case of the well-studied vesicular stomatitis virus, a sequential phosphorylation of the P protein is required for transcription. Using an in vitro reconstitution system composed of a vesicular stomatitis virus mRNA, purified L protein (the RNA polymerase), and recombinant P protein, it was shown that transcription occurred only after in vitro phosphorylation of P protein with casein kinase II followed by a second phosphorylation mediated by a kinase(s) associated with the purified L protein (2, 3). Similar requirements have been described for other nonsegmented minus-strand RNA viruses such as parainfluenza virus and human respiratory syncytial virus (11, 40). Due to the lack of an efficient cell culture system, the importance of NS5A hyperphosphorylation for the HCV life cycle is unclear. However, the aforementioned conservation of this modification among HCV isolates as well as the flaviviruses and pestiviruses (43) together with numerous examples from other virus systems strongly suggests an important role.
Apart from its putative function for virus replication, NS5A has been implicated in modulation of the host IFN-stimulated antiviral response. It was shown that NS5A isolated from certain genotypes can interact with the IFN-induced double-stranded RNA-dependent protein kinase PKR (17). Interaction requires a 66-residue sequence around the center of NS5A (16) and includes two of the three serine residues shown by genetic studies to be important for hyperphosphorylation (54). It is not known whether phosphorylation of NS5A contributes to the interaction with PKR or to some other function(s) interfering with host defense mechanisms. If this is the case, then mutations outside the IFN sensitivity-determining region (ISDR) or even outside NS5A itself may influence the cellular antiviral response. As exemplified by the single amino acid substitution at the amino terminus of NS3 or in the central NS4A domain, mutations in nonstructural proteins other than NS5A can affect the phosphorylation state of NS5A and in this way might alter its biological activity. This observation may in part explain the contradictory results on the correlation between mutations in the ISDR and IFN resistance. It seems likely that resistance can be achieved in many ways. Apart from mutations within the ISDR or somewhere else in NS5A, directly affecting its ability to interfere with PKR, the same effect might be caused by alterations within other nonstructural proteins affecting in turn NS5A functions. In agreement with this assumption recent epidemiological data suggest that HCV-1b resistance to IFN might be conferred by mutations located at different positions throughout the viral genome (42).
Finally, as suggested in recent reports (43, 44), the possibility that hyperphosphorylation of NS5A is an epiphenomenon not relevant for virus replication or interference with the host defense cannot be ruled out; e.g., NS5A might be required to trap a cellular kinase(s), which itself participates in RNA replication. In this case, the distinct requirements for hyperphosphorylation described here might reflect an alteration of NS5A structure upon interaction with NS3-4B proteins. Such interactions could induce conformational changes of NS5A, thereby exposing target sequences for cellular kinases or phosphatases. Further studies requiring the development of a cell-based HCV replication system are necessary to address these questions.
ACKNOWLEDGMENTS
We are grateful to U. Herian for excellent technical assistance and to L. Theilmann for gifts of patient serum and liver sample. We thank H. Müller and A. Heller for the gift of the NS2-specific antiserum, B. Moss for providing the recombinant vaccinia virus vTF7-3, and L. Tomei for the gift of the NS4-specific antiserum.
This work was supported by grants from the German Research Community (BA1505/1-2) and the Federal Government for Research and Technology (01 KI 96539).
REFERENCES
- 1.Asabe S I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–796. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barik S, Banerjee A K. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik S, Banerjee A K. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J Virol. 1992;66:1109–1118. doi: 10.1128/jvi.66.2.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R. Candidate targets for hepatitis C virus-specific antiviral therapy. Intervirology. 1997;40:378–393. doi: 10.1159/000150570. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartenschlager R, Lohmann V, Wilkinson T, Koch J O. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens S E, Tomei L, DeFrancesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. [DOI] [PubMed] [Google Scholar]
- 10.Cuthbert J A. Hepatitis C: progress and problems. Clin Microbiol Rev. 1994;7:505–532. doi: 10.1128/cmr.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De B P, Gupta S, Banerjee A K. Cellular protein kinase C isoform zeta regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckart M R, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo Q L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 13.Failla C, Tomei L, DeFrancesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Failla C, Tomei L, DeFrancesco R. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J Virol. 1995;69:1769–1777. doi: 10.1128/jvi.69.3.1769-1777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale M J, Blakely S M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanism of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 18.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Hong Z, Ferrari E, Wright-Minogue J, Chase R, Risano C, Seelig G, Lee C G, Kwong A D. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells by using the herpes simplex virus amplicon system. J Virol. 1996;70:4261–4268. doi: 10.1128/jvi.70.7.4261-4268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe P M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 25.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-α catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 26.Ishido S, Fujita T, Hotta H. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem Biophys Res Commun. 1998;244:35–40. doi: 10.1006/bbrc.1998.8202. [DOI] [PubMed] [Google Scholar]
- 27.Kadare G, Haenni A L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 30.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 31.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O’Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 32.Koch J O, Lohmann V, Herian U, Bartenschlager R. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology. 1996;221:54–66. doi: 10.1006/viro.1996.0352. [DOI] [PubMed] [Google Scholar]
- 33.Landro J A, Raybuck S A, Luong Y C, O’Malley E T, Harbeson S L, Morgenstern K A, Rao G, Livingston D J. Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping. Biochemistry. 1997;36:9340–9348. doi: 10.1021/bi963054n. [DOI] [PubMed] [Google Scholar]
- 34.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C, Wu J W, Hsiao K, Su M S. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J Virol. 1997;71:6465–6471. doi: 10.1128/jvi.71.9.6465-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Bhat A B, Prince A M, Zhang P. The hepatitis C virus NS2 protein generated by NS2-3 autocleavage is required for NS5A phosphorylation. Biochem Biophys Res Commun. 1999;254:572–577. doi: 10.1006/bbrc.1998.9986. [DOI] [PubMed] [Google Scholar]
- 37.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Margosiak S, Dagostino E, Hostomska Z. The conformation of hepatitis C virus NS3 proteinase with and without NS4A: a structural basis for the activation of the enzyme by its cofactor. Clin Diagn Virol. 1998;10:151–156. doi: 10.1016/s0928-0197(98)00036-1. [DOI] [PubMed] [Google Scholar]
- 39.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yosida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 40.Mazumder B, Barik S. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 41.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Classification and nomenclature of viruses: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 424–426. [Google Scholar]
- 42.Pawlotsky J M, Germanidis G, Neumann A U, Pellerin M, Frainais P O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed K E, Gorbalenya A E, Rice C M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72:6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe P M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Satoh S, Tanji Y, Hijikata M, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus nonstructural protein 3 (NS3) is essential for stable complex formation with NS4A. J Virol. 1995;69:4255–4260. doi: 10.1128/jvi.69.7.4255-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu Y, Yamaji K, Masuho Y, Yokota T, Inoue H, Sudo K, Satoh S, Shimotohno K. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J Virol. 1996;70:127–132. doi: 10.1128/jvi.70.1.127-132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinkühler C, Tomei L, DeFrancesco R. In vitro activity of hepatitis C virus protease NS3 purified from recombinant baculovirus-infected Sf9 cells. J Biol Chem. 1996;271:6367–6373. doi: 10.1074/jbc.271.11.6367. [DOI] [PubMed] [Google Scholar]
- 50.Suzich J A, Tamura J K, Palmer H F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tai C L, Chi W K, Chen D S, Hwang L H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. Hepatitis C virus polyprotein processing: kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J Virol. 1994;68:8418–8422. doi: 10.1128/jvi.68.12.8418-8422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomei L, Failla C, Santolini E, DeFrancesco R, LaMonica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomei L, Failla C, Vitale R L, Bianchi E, DeFrancesco R. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J Gen Virol. 1996;77:1065–1070. doi: 10.1099/0022-1317-77-5-1065. [DOI] [PubMed] [Google Scholar]
- 57.Yan Y W, Li Y, Munshi S, Sardana V, Cole J L, Sardana M, Steinkühler C, Tomei L, DeFrancesco R, Kuo L C, Chen Z G. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 angstrom resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]