Highlights
-
•
Colorectal cancer (CRC) is one of the most common cancers and the second leading cause of cancer-related deaths globally.
-
•
Only a limited number of CRC patients have a positive response due to radiotherapy.
-
•
BBOX1-AS1 promotes radiosensitivity by negatively regulating glycolysis and affecting cell cycle in CRC cells.
-
•
BBOX1-AS1 can be used as a potential prognostic biomarker and radiotherapy sensitization target for CRC.
Keywords: Colorectal cancer, Phosphofructokinase-1, BBOX1-AS1, Radiotherapy, Colo205 cells, HCT116 cells
Abstract
Purpose
Our study explored the effect of long noncoding RNA BBOX1-AS1 on colorectal cancer (CRC) radiosensitivity in vivo and in vitro.
Methods
Differentially expressed lncRNAs in CRC were screened using a bioinformatics database and an online prediction website. The expression of BBOX1-AS1 in tissue samples was analyzed via real-time quantitative PCR (RT-qPCR). Subcellular localization of BBOX1-AS1 in CRC cells was analyzed using fluorescence in situ hybridization (FISH). The correlation between BBOX1-AS1 and PFK1 expression levels in CRC tissues was analyzed via Pearson's correlation coefficient. The effect of BBOX1-AS1 on PFK1 stability was investigated using RNA and protein stability testing. RNA Binding Protein Immunoprecipitation (RIP) and RNA pull-down assays were used to confirm the binding of BBOX1-AS1 to PFK1.
Results
BBOX1-AS1 was highly expressed in CRC and associated with poor prognosis. Similarly, it was highly expressed in CRC tissues and CRC cell lines. In addition, BBOX1-AS1 promoted the proliferation, invasion, migration, and glycolysis of CRC cells and inhibited apoptosis. RIP and RNA pull-down experiments confirmed that BBOX1-AS1 bound to PFK1. RNA stability and protein stability experiments showed that BBOX1-AS1 affected the stability of PFK1 mRNA and protein. Furthermore, we confirmed that BBOX1-AS1 increased radiation resistance through the regulation of PFK1 expression.
Conclusions
BBOX1-AS1 promoted the proliferation, invasion, migration, and glycolysis of CRC cells through stabilization of the expression of PFK1. BBOX1-AS1 also inhibited CRC cell apoptosis and increased radiotherapy resistance in CRC cells.
Introduction
Colorectal cancer (CRC) is one of the most common cancers and the second leading cause of cancer-related deaths globally [1]. CRC incidence is steadily increasing and shifting toward younger individuals [2]. Different factors including genetic background, age, and unhealthy lifestyle factors have been implicated in colorectal tumorigenesis [3].
Radiotherapy is a common locoregional therapy for cancer and is a main treatment modality in CRCs [4], [5], [6]. The guidelines formulated by the National Comprehensive Cancer Network (NCCN) in the United States recommend radiotherapy for all patients with advanced rectal cancer of T3 or higher stages and for those with lymph node metastasis without systemic metastasis; palliative radiotherapy is also feasible for some patients with advanced colon cancer. Ionizing radiation kills cancer cells via direct and indirect DNA damage, causing decreased proliferation. However, due to tumor heterogeneity, only a limited number of CRC patients have a positive response (reduced tumor volume, hypoxia, dysregulation of various genes) due to radiotherapy [7]. Thus, it is urgent to further reveal the underlying mechanisms of radiation resistance in CRC and identify new targets to increase radiotherapy sensitivity.
The development of RNA sequencing technology and integration of genomics studies have shown that long noncoding RNAs (lncRNAs) are RNA transcripts longer than 200 nucleotides that have limited or no protein-coding potential [8]. However, a growing number of lncRNAs have been shown to play crucial roles in various biological processes such as cell proliferation, apoptosis, invasion, and metastasis [9,10]. In addition, some studies have found that lncRNAs can regulate radiotherapy sensitivity [11,12]. Regarding CRC, studies have found that differential lncRNA expression affects a wide range of associated biological activities [13], [14], [15]. However, only a small number of lncRNAs are well annotated in CRC, and lncRNAs implicated in CRC remain to be characterized. In addition, there are few studies on the mechanism by which lncRNA affects radiotherapy sensitivity by influencing glycolysis.
In the present study, we demonstrated that the BBOX1-AS1/PFK1 axis may play an important role in CRC radiotherapy sensitivity. The application of RNAi targeting BBOX1-AS1 is a potential treatment strategy in cases with resistance to ionizing radiation.
Methods
Analysis of differentially expressed lncRNAs between cancer and normal tissues
The GEPIA2 database (http://gepia2.cancer-pku.cn/#index) is an open access dataset used to analyze RNA sequencing expression data. In this study, a total of 786 differentially expressed genes (DEG) in COAD (colon adenocarcinoma) and 1759 DEGs in READ (rectal cancer) were identified using the following dataset setting: |log2FC| ≥ 2 and q-value < 0.01. LncRNAs < 2000 nt in length and rarely reported were selected for further study.
The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) is a powerful cancer genomics program which we used to download the latest expression data and clinical follow-up information on patients with colon adenocarcinoma (COAD).
Candidate lncRNA-PFK1 interactions
IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA), an open resource, enables enhanced and customizable RNA–RNA interaction prediction [16]. catRAPID (http://service.tartaglialab.com/page/catrapid_group), is a useful website which can predict protein-RNA interactions by detecting binding regions through physico-chemical features [17].
Patients and tissue specimens
CRC and adjacent normal tissue samples were collected from 38 patients at the Third Xiangya Hospital, Central South University (Changsha, China) from October 2019 to January 2020. None of these patients had been treated with preoperative therapy. The clinicopathological features, including the patient's age, gender, tumor size, and TNM stage, were summarized from the medical records of the patients (Table S1). The collected tissues were immediately frozen at −80 °C after surgery. This study was approved by the Third Xiangya Hospital Institutional Ethical Review Board, and informed consent was obtained from each patient.
Immunofluorescence
All CRC samples were cut into 3-μm sections, fixed in formalin, and embedded in paraffin. The sections were deparaffinated in xylene followed by microwave treatment for 10 min in phosphate-buffered saline (PBS, pH 6.0). After blocking with 5% BSA, the sections were incubated overnight at 4 °C with anti-Ki-67 antibodies (1:1000; 27,309-1-AP, Santa Cruz, CA, USA), anti-Cyclin D1 antibodies (1:100; ab16663, Santa Cruz, CA, USA), anti-Caspase-3 antibodies (1:50; ab32351, Santa Cruz, CA, USA), and anti-PFKM antibodies (1:100; sc-377,346, Santa Cruz, CA, USA). Detection of immunostaining was performed using the UltraSensitive™ S-P kit (KALANG, Shanghai, China) and 3, 3-diaminobenzidine (Santa Cruz, CA, USA) as the chromogen, according to the manufacturer's specifications. Finally, cells were observed and analyzed under a confocal microscope (Olympus, Tokyo, Japan).
Cell lines
Seven human CRC cell lines (SW480, SW620, HCT116, Colo205, HT29, caco-2, and Lovo), the human normal colon mucosal cell line (NCM460) and human normal colorectal epithelial cells (FHC) were purchased from the American Type Culture Collection, Manassas, VA, USA. The cells were cultured in Dulbecco's modified eagle medium (DMEM) containing 10% fetal bovine serum and incubated at 37 °C with 5% CO2.
Cell transfection
Three siRNAs targeting BBOX1-AS1 (siRNA-BBOX1-AS1-318, siRNA-BBOX1-AS1-705, and siRNA-BBOX1-AS1-832) were designed to silence endogenous BBOX1-AS1. All siRNAs and si-NC were designed and chemically synthesized by Guangzhou RiboBio Co., Ltd. (RiboBio, Guangzhou, China). The BBOX1-AS1 overexpression plasmid pCDNA3.1-BBOX1-AS1 (pc-BBOX1-AS1) was obtained from GenePharma Co., Ltd. (Shanghai, China). The HCT116 and Colo205 cells were seeded into 6-well plates for 24 h, transfected with the siRNAs and overexpression plasmids using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and incubated in a CO2 incubator at 37 °C for 3–5 h.
RT-qPCR
Total RNA was isolated from tissues and cells via TRIzol®, quantified using a NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Waltham, USA), and reverse-transcribed into cDNA using the Reverse Transcription Kit (Takara Biotechnology, Co., Ltd., Dalian, China). Next, real-time quantitative polymerase chain reaction (RT-qPCR) was carried out using SYBR Green PCR Master Mix (Takara Biotechnology, Co., Ltd., Dalian, China). β-actin /18S was used as the internal control, and relative gene expression was calculated using the 2−∆∆Ct method [18].
The primers were designed as follows: BBOX1-AS1, 5′- GCCTTCTGCCATGATTGTGT-3′ (forward) and 5′- GCGTTAGGTTTGGAGTTGCA-3′ (reverse); PFK1, 5′- CCGTTCTGAGTGGAGTGACT-3′ (forward) and 5′- AGAGTCAGTGCCAATGGTCA-3′ (reverse); β-actin, 5′- ACCCTGAAGTACCCCATCGAG-3′ (forward) and 5′- AGCACAGCCTGGATAGCAAC-3′ (reverse); and 18S, 5′- CAGCCACCCGAGATTGAGCA-3′ (forward) and 5′- TAGTAGCGACGGGCGGTGTG-3′ (reverse).
Fluorescence in situ hybridization
The BBOX1-AS1 FISH probe was synthesized by Guangzhou RiboBio Co., Ltd. (RiboBio, Guangzhou, China). Cells were seeded in culture plates, fixed with 4% paraformaldehyde (Sigma-Aldrich, CA, USA), and then blocked with a pre-hybridization buffer for 30 min at 37 °C. After discarding the pre-hybridization buffer, the FISH probe was added into the hybridization mixture and incubated overnight. Then, cells were washed in a washing buffer containing saline sodium citrate and PBS in the dark. Cells were stained using 4′,6-diamidino-2-phenylindole (DAPI) for 10 min and observed via fluorescence microscope (Olympus, Tokyo, Japan) in the dark.
MTT assay
HCT116 and Colo205 cells were plated in 96-well plates containing 100 µL DMEM/well at a density of 1 × 104 cells/well. Cell proliferation was evaluated via MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazo-lium bromide] assay after incubation for 0 h, 12 h, 24 h, 36 h, 48 h, 36 h and 72 h. Fifty microliters of diluted MTT reagent was added to each well and the cells were incubated at 37 °C for another 4 h. After removing the MTT solution, 150 µL dimethyl sulfoxide was added to each well to dissolve the purple formazan crystals. The optical density (OD) of each well was measured at a wavelength of 570 nm (OD 570 nm) via a Microplate Reader (Bio-Rad, CA, USA). The cell survival rate was then calculated using the following formula: Cell viability (%) = (OD value of test wells/OD value of control cells) × 100%.
Western blotting
Tissue or cell protein was extracted using the Total Protein Extraction Kit (ProMab Biotechnologies, Richmond, USA, Cat. No: SJ-200,501) or a RIPA lysis buffer, and then centrifuged at 9000 rpm at 4 °C for 10 min. Nucleoprotein was extracted on ice via the NucBuster TM Protein Exaction Kit (Merck Millipore, USA, Cat. No: 71,183-3) and then centrifuged at 16,000 g at 4 °C for 5 min. Protein concentration was determined by BCA protein assay kit (Auragene, Changsha, China). The primary antibodies used were as follows: anti-PFK1 (santa, Cat: sc-377,346); anti-MCM10 (ptgcn, Cat:12,251-1-AP); anti-MCM6 (ptgcn, Cat:13,347-2-AP); anti-LDHA (ptgcn, Cat:21,799-1-AP); anti-GLUT1 (ptgcn, Cat:21,829-1-AP); anti-γ-H2AX (Abcam, Cat: ab26350); and anti-β-actin (ptgcn, Cat:66,009-1-Ig).
Cell migration assay
Colo205 and HCT116 cells were inoculated into 6-well plates at a density of 1 × 105 cells/well and cultured until more than 90% confluent or covering the bottom of the well. Then, a zigzag scratch was made in the middle of the cell using a 20-μL sterile pipette tip, and each well was washed with serum-free medium three times to remove the scratched cells. Finally, plates were photographed after culturing with various treatments. The percent of wound closure was calculated using the following formula: Scratch healing rate = (width between edges of migrated scratches/width between edges of initial scratches) × 100%.
Transwell assay
Before conducting the assay, the upper chamber of the Transwell was coated with 50 µL Matrigel diluent (354,480; BD Biosciences) and incubated at 37 °C for 30 min. Then, 5 × 104 Colo205 or HCT116 cells were resuspended in 200 µL FBS-free culture medium and added into the upper compartment; the bottom chamber was filled with 1 ml culture medium containing 20% FBS. After incubation, cells in the upper chamber and Matrigel were eliminated with a cotton swab. Following fixation in 4% paraformaldehyde and staining with 0.5% crystal violet, the invasive cells were counted under an inverted microscope.
X-ray irradiation
The radiotherapy equipment was equipped with an American Varian TrueBeam linear accelerator (Varian, CA, USA). The cells or nude mice were irradiated with 2 Gy.
Glycolysis analysis
Changes in glycolysis were measured with a glucose assay kit or lactic acid assay kit (Nanjing, China) according to the manufacturer's instructions.
Flow cytometry
Apoptosis was analyzed with the Annexin V-FITC Apoptosis Detection Kit (KeyGEN BioTECH Corp., Ltd, CHINA). Briefly, the cells were detached using EDTA‐free Trypsin and washed with PBS twice via centrifugation at 300 × g for 10 min at 4 °C. After treatment, the cells were adjusted to 1 × 106 cells/ml using 250-μl 1 × Binding Buffer. Later, 5-μl Annexin V/PE and 5-μl 7-AAD were added to the cells, followed by cell incubation at room temperature in the dark for 10 min. Within 1 h, the cells were transferred onto a flow cytometer (Beckman Coulter, Inc., USA).
Cell cycle analysis assay
Cells were adjusted to 1 × 106 cells/ml, digested using Trypsin, and washed with PBS at 1000 × g for 5 min. Next, the cells were fixed using ethanol (75%) for 4 h at 4 °C. The ethanol was then discarded, and the cells were washed with PBS again. After resuspending the cells, 100 μl PI (R40432, Sigma, USA) and 100 μl RNase A (R4875, Sigma, USA) were added to the cells. Later, the cells were incubated at 4 °C for 30 min and transferred onto a flow cytometer (Beckman Coulter, Inc., USA), and the cell cycle was analyzed using Modfit software (Version LT 4.1, Verity Software House).
RIP assay
RIP was carried out using the EZ-Magna RIP kit (Merck Millipore, German) according to the recommended instructions. Briefly, the prepared cell lysate was mixed with magnetic beads conjugated with anti- SNRNP70 antibody or anti-immunoglobulin G (IgG) antibody on a shaker overnight. Then, the RNA was purified using proteinase K. Subsequently, the target RNA was detected by conventional cDNAs reverse transcription and q-PCR.
RNA pull-down
The interaction between BBOX1-AS1 and the PFK1 protein was detected using a Pierce™ Magnetic RNA-Protein Pull-Down Kit (20164Y, Thermo Fisher Scientific, MA, USA). In brief, cell lysis fluid was prepared, and no less than 2 mg/mL protein was used for pull-down assays. The labeled RNA was pre-washed and bound to Streptavidin Magnetic Beads or RNA-binding proteins. Beads were then washed to remove RNA-binding protein complexes. Finally, the pulled down proteins were identified via western blotting.
Animal assays
All animal experiments and handling were approved by the animal center of the Central South University (Changsha, China) according to international guidelines and procedures. To produce tumors in vivo, 1 × 106 treated HCT116 cells were injected subcutaneously into male BALB/ C nude mice (4–6 weeks of age). Tumor volume (longest diameter A and shortest diameter B) was measured once every 3 days for 6 consecutive days. When the tumor volume reached 100 mm3, the experimental group was irradiated with 2 Gy. After 25 days, the mice were sacrificed and tumor volume was calculated according to the formula: tumor volume (mm3) = (the longest diameter × the shortest diameter 2) / 2.
Enrichment analysis
The relevant CRC data were downloaded from the TCGA database (https://portal.gdc.cancer.gov/), and analyzed using Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis with the R package cluster profile [19].
Statistical analysis
The experimental data were analyzed using GraphPad Prism 9.0. Student's t-test was used to analyze two sets of measurement data, and ANOVA was used to analyze more than two sets of measurement data. Spearman's correlation analysis was used to analyze the correlation between BBOX1-AS1 and PFK1. All experiments were repeated three times and all experimental results are expressed as mean ± standard deviation. P < 0.05 was considered statistically significant.
Results
BBOX1-AS1 is upregulated in CRC and negatively correlated with clinical prognosis
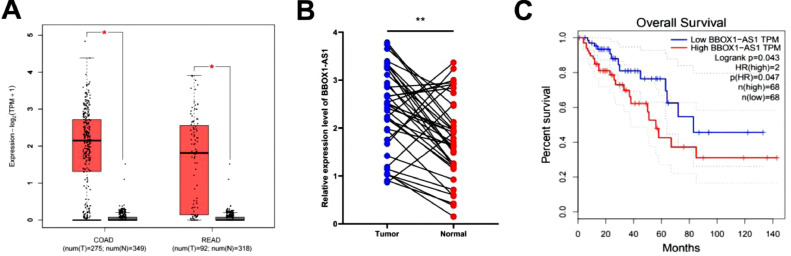
There were 786 differentially expressed genes (DEGs) in colon cancer and 1759 common DEGs in rectal cancer when the P value was set to <0.01 and the absolute difference value was (Log2FC) ≥2 in the GEPIA2 database (http://gepia2.cancer-pku.cn/). After searching the National Center for Biotechnology Information (NCBI), we identified 24 non-coding RNAs. Among these genes, we excluded ncRNAs larger than 2000 nt and obtained the following genes: CASC9, CRNDE, LINC01123, UCA1, LINC00511, TRPM2-AS, HNF1A-AS1, VPS9D1-AS1, and BBOX1-AS1. By reading the literature extensively and excluding genes that have been studied extensively, BBOX1-AS1 was selected as a candidate lncRNA. Analysis in the GEPIA2 database showed that BBOX1-AS1 was highly expressed in CRC tissues compared with para-cancer tissues (Fig. 1A). Then, we collected 38 pairs of fresh colon cancer tissues and analyzed the expression of BBOX1-AS1 in cancer tissues and adjacent normal tissues by qRT-PCR. As shown in Fig. 1B, BBOX1-AS1 was differentially expressed in CRC and para-cancer tissues (P < 0.01). Additionally, BBOX1-AS1 expression was negatively correlated with the OS of CRC patients in GEPIA2 (Fig. 1C). These results suggest that BBOX1-AS1 is highly expressed in CRC and negatively correlated with prognosis (Table S1).
Fig. 1.
BBOX1-AS1 was highly expressed in colorectal cancer and negatively correlated with prognosis. (A) Expression level of BBOX1-AS1 in tumor tissues and adjacent tissues in COAD and READ in the GEPIA2 database. (B) The expression level of BBOX1-AS1 in 38 pairs of colorectal cancer tissues and adjacent tissues. (C) Prediction of the relationship between BBOX1-AS1 and prognosis of colorectal cancer patients in the GEPIA2 database. *P < 0.05, **P < 0.01.
BBOX1-AS1 promoted CRC cell proliferation, invasion, migration, and G0/G1 cell cycle arrest but inhibited apoptosis in vitro
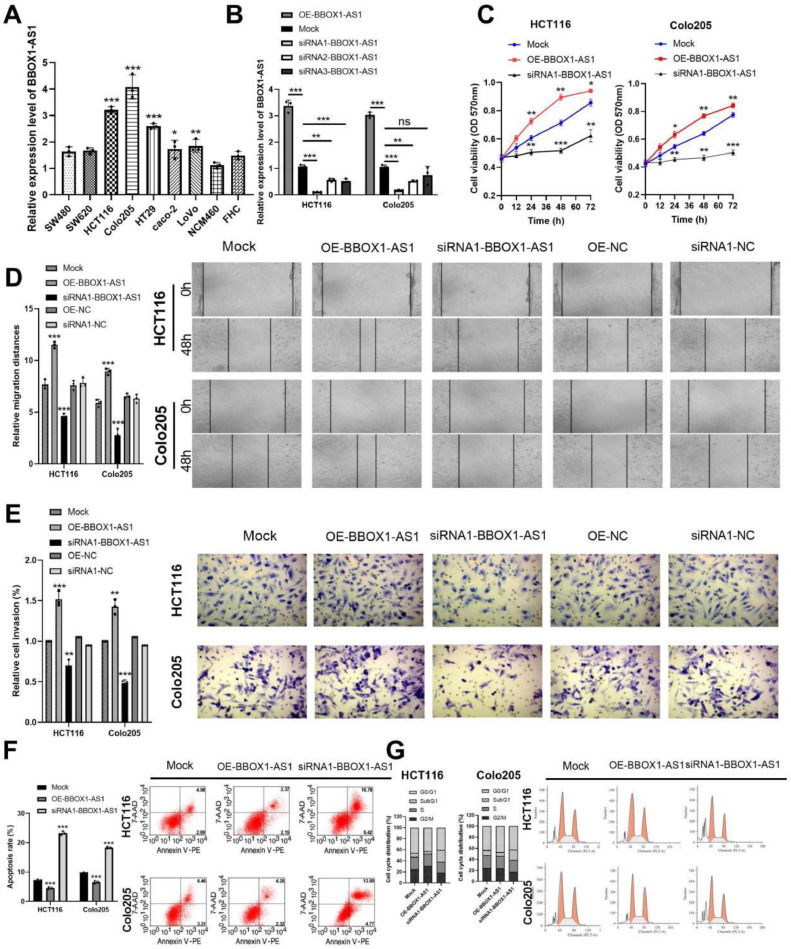
BBOX1-AS1 expression was detected in SW480, SW620, HCT116, Colo205, HT29, caco-2, and LoVo CRC cell lines, as well as in the NCM460 normal human colon and FHC rectal cell lines using RT-qPCR. Results showed that BBOX1-AS1 mRNA was significantly upregulated in the CRC cell lines compared with normal human colon and rectal cell lines (Fig. 2A). Among the seven CRC cell lines, HCT116 and Colo205 showed the highest BBOX-AS1 expression and were selected for further studies.
Fig. 2.
BBOX1-AS1 promoted CRC cell proliferation, invasion, migration, and G0/G1 cell cycle arrest but inhibited apoptosis in vitro. (A) Expression level of BBOX1-AS1 in CRC cell lines. (B) The expression level of BBOX1-AS1 by RT-qPCR. (C) The effect of BBOX1-AS1 on CRC cell proliferation. (D) The invasion and migration capacity of CRC cells. (E) BBOX1-AS1 inhibited apoptosis in CRC cells. (F) BBOX1-AS1 inhibited apoptosis in CRC cells. (G) The expression of BBOX1-AS1 affected the cell cycle in CRC cells. Mean ± SD (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the function of BBOX1-AS1 in CRC, three small interfering RNAs (siRNAs) against BBOX1-AS1 expression (siRNA1-BBOX1-AS1, siRNA2-BBOX1-AS1 and siRNA3-BBOX1-AS1) were applied to silence BBOX1-AS1, and one plasmid (OE-BBOX1-AS1) was applied to overexpress BBOX1-AS1. RT-qPCR revealed that siRNA1-BBOX1-AS1 significantly suppressed BBOX1-AS1 expression in HCT116 and Colo205 cells (Fig. 2B). Thus, we used siRNA1-BBOX1-AS1 in further studies and renamed it si-BBOX1-AS1.
Next, MTT assays were performed to examine the effects of BBOX1-AS1 on CRC cell proliferation. Proliferation was significantly increased in the OE-BBOX1-AS1 group and suppressed in the si-BBOX1-AS1 group (Fig. 2C). Additionally, transwell and wound healing assays were performed to determine invasion and migration capacity in CRC cells. Results showed that invasion and migration of CRC cells were decreased in the si-BBOX1-AS1 group and increased in the OE-BBOX1-AS1 group compared with the Mock group, and the difference was not statistically significant among the Mock group, siRNA1-NC group and OE-NC group. Figure 2(D, E). In addition, apoptosis assays showed that BBOX1-AS1 inhibited apoptosis in CRC cells (Fig. 2F). Furthermore, flow cytometry revealed that BBOX1-AS1 knockdown resulted in a substantial accumulation of HCT116 and Colo205 cells in the SubG1 phase (representing apoptotic cells), which suggested that BBOX1-AS1 can inhibit apoptosis in CRC cells (Fig. 2G). These results suggested that BBOX1-AS1 is a cancer promoting gene in CRC.
BBOX1-AS1 promoted CRC cell glycolysis
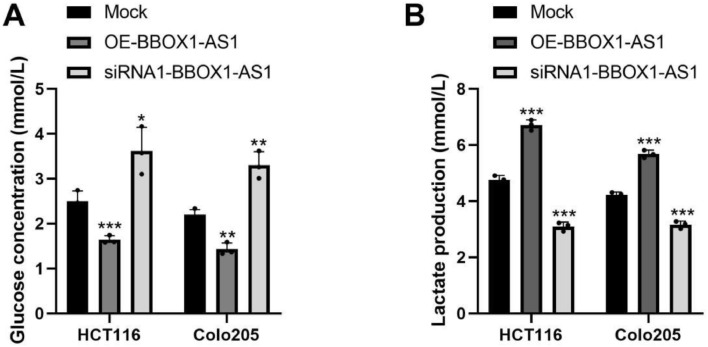
To investigate the function of BBOX1-AS1 in CRC cell glycolysis, we used glucose and lactic acid assay kits to detect changes in glucose and lactic acid content, respectively. Results showed that glucose consumption and lactic acid content in the OE- BBOX1-AS1 group were significantly increased compared with the blank control group. However, glucose consumption and lactic acid content were depressed in the si-BBOX1-AS1 group. The differences were statistically significant (Fig. 3). These results suggested that BBOX1-AS1 promotes CRC cell glycolysis.
Fig. 3.
BBOX1-AS1 promoted CRC cell glycolysis. (A) Glucose concentration in CRC cells. (B) Lactate production in CRC cells. *P < 0.05, **P < 0.01, ***P < 0.001.
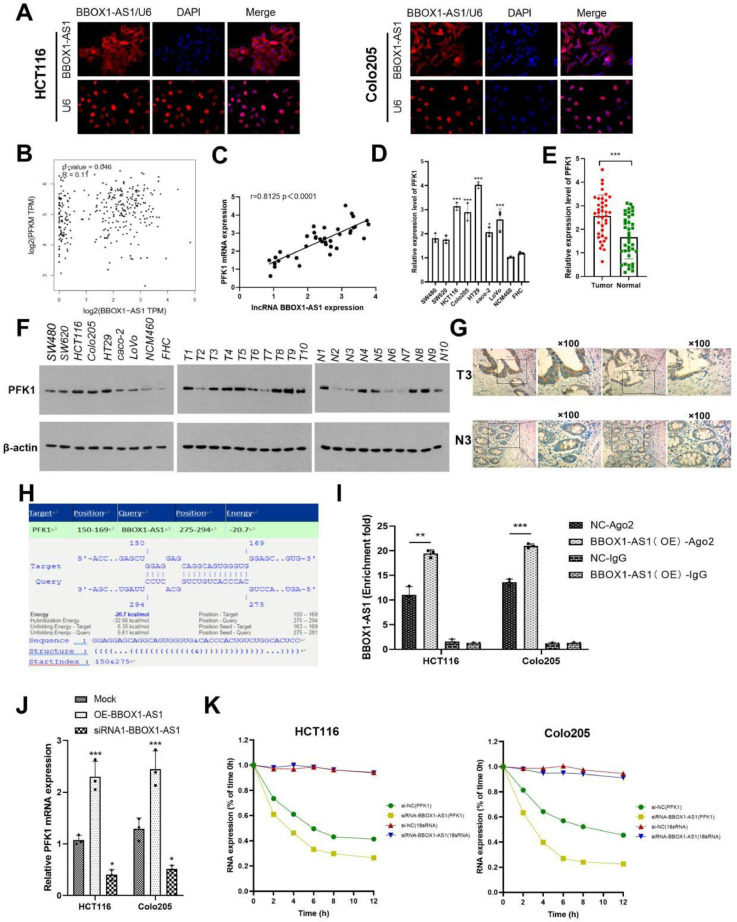
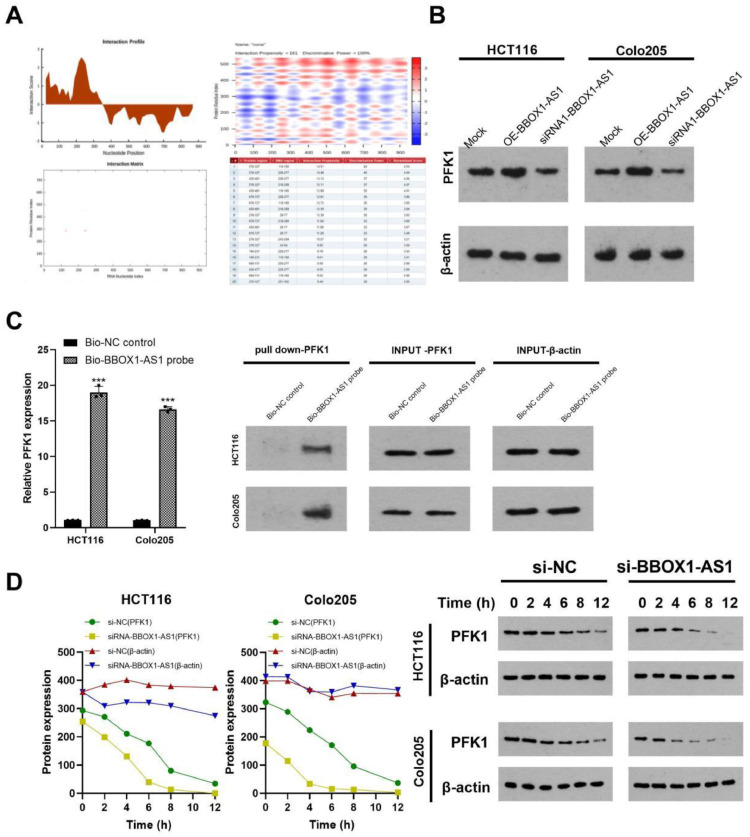
BBOX1-AS1 specifically interacted with PFK1
Binding to a specific protein is important to lncRNA implementing its functions [20]. To further study the function of BBOX1-AS1, we detected the localization of BBOX1-AS1 in CRC cells by FISH assay. Results showed that BBOX1-AS1 was mainly expressed in the cytoplasm (Fig. 4A). The GEPIA2 website showed that BBOX1-AS1 was significantly positively correlated with PFKM expression in colon cancer, as shown in Fig. 4B. In addition, correlation analysis confirmed that BBOX1-AS1 was positively correlated with PFKM expression (Fig. 4C). We analyzed PFK1 expression in cells via RT-qPCR and western blotting and found that it is significantly highly expressed in CRC (Fig. 4D, E). The above conclusion was also confirmed by analysis of collected samples (Fig. 4F) and further verified by immunohistochemistry (Fig. 4G). After investigating the interaction between BBOX1-AS1 and PFKM via IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA), we found that BBOX1-AS1 may interact with the 5 '-UTR of PFKM (Fig. 4H). RIP assays verified that the PFK1 protein bound with BBOX1-AS1 (Fig. 4I). Knockdown of BBOX1-AS1 significantly downregulated PFK1 mRNA, while overexpression of BBOX1-AS1 had the opposite effect (Fig. 4J). To explore the effect of BBOX1-AS1 on the stability of PFK1 mRNA, RNA and protein stability tests were carried out. Compared with the negative control group, the expression of PFK1 mRNA in the siRNA1-BBOX1-AS1 group was significantly decreased and the half-life of PFK1 mRNA was shortened, indicating that BBOX1-AS1 affected the stability of PFK1 mRNA (Fig. 4K).
Fig. 4.
BBOX1-AS1 bound to PFK1 mRNA and increased its stability. (A) BBOX1-AS1 was mainly expressed in the cytoplasm as determined by FISH assay. (B) BBOX1-AS1 was significantly positively correlated with PFKM expression in colon cancer as determined by GEPIA2 analysis. (C) BBOX1-AS1 was positively correlated with PFKM expression in the collected specimens. (D) PFK1 was significantly highly expressed in CRC as determined by RT-qPCR. (E) PFK1 was significantly highly expressed in CRC as determined by western blotting. (F) The expression of PFK1 in collected CRC samples as determined by RT-qPCR. (G) Immunohistochemistry confirmed the expression of PFK1 in colorectal cancer. (H) IntaRNA 2.0 predicted binding sites between BBOX1-AS1 and PFKM mRNA. (I) RIP assay verified that BBOX1-AS1 bound to PFK1 mRNA. (J) RT-qPCR showed that knocking down BBOX1-AS1 significantly downregulated PFK1 mRNA. (K) RNA stability experiments showed that BBOX1-AS1 affected PFK1 mRNA stability. *P < 0.05, **P < 0.01, ***P < 0.001.
After searching catRAPID (http://service.tartaglialab.com/page/ catrapid_ group), we predicted that BBOX1-AS1 may directly bind with the PFKM protein (Fig. 5A). Western blot assays showed that knockdown of BBOX1-AS1 downregulated the protein expression of PFK1, while overexpression of BBOX1-AS1 resulted in the opposite result. Figure 5(B). RNA pull-down assays showed that compared with the negative control probe, the BBOX1-AS1/PFKM 5 '-UTR probe enriched more PFKM 5′-UTR (P < 0.01) (Fig. 5C). Moreover, western blotting showed that the stability of PFK1 was decreased in the BBOX1-AS1 knockdown group compared to the control group (Fig. 5D). In summary, BBOX1-AS1 bound to PFK1 and increased its expression.
Fig. 5.
BBOX1-AS1 bound to PFK1 to increase its stability. (A) The catRAPID website predicted that BBOX1-AS1 may directly bind PFKM protein. (B) Western blotting showed that knocking down BBOX1-AS1 significantly downregulated PFK1 protein level. (C) RNA pull down test showed that BBOX1-AS1 interacted with the PFK1 protein. (D) Protein stability test showed that BBOX1-AS1 affected the stability of PFK1. Mean ± SD (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
BBOX1-AS1 increased radiotherapy resistance by regulating PFK1 in colorectal cells in vitro
In previous studies, our team confirmed that PFK1 is related to radiotherapy resistance in colorectal cancer, and 2 Gy was selected for further research [21]. However, how PFK1 affects the sensitivity of CRC to radiotherapy remains to be determined.
Gene Ontology (GO) analysis was performed on genes positively correlated with PFKM expression in CRC using the TCGA database. We found that the genes positively correlated with PFKM were mainly involved in glycolysis, cell proliferation, regulation of DNA replication initiation, and the G1/S transition of the mitotic cell cycle. We analyzed MCM10, MCM6, LDHA and GluT1 in 10 CRC and para-cancer tissues by western blotting. The result showed that MCM10, MCM6, LDHA and GluT1 were higher in CRC tissues and relatively lower in para-cancer tissues. This indicated that BBOX1-AS1 may influence the radiotherapy sensitivity of CRC by affecting PFKM expression, subsequently affecting CRC proliferation, DNA replication, and mitotic cell cycle regulation. These data are presented in the supplemental results (Figure S1).
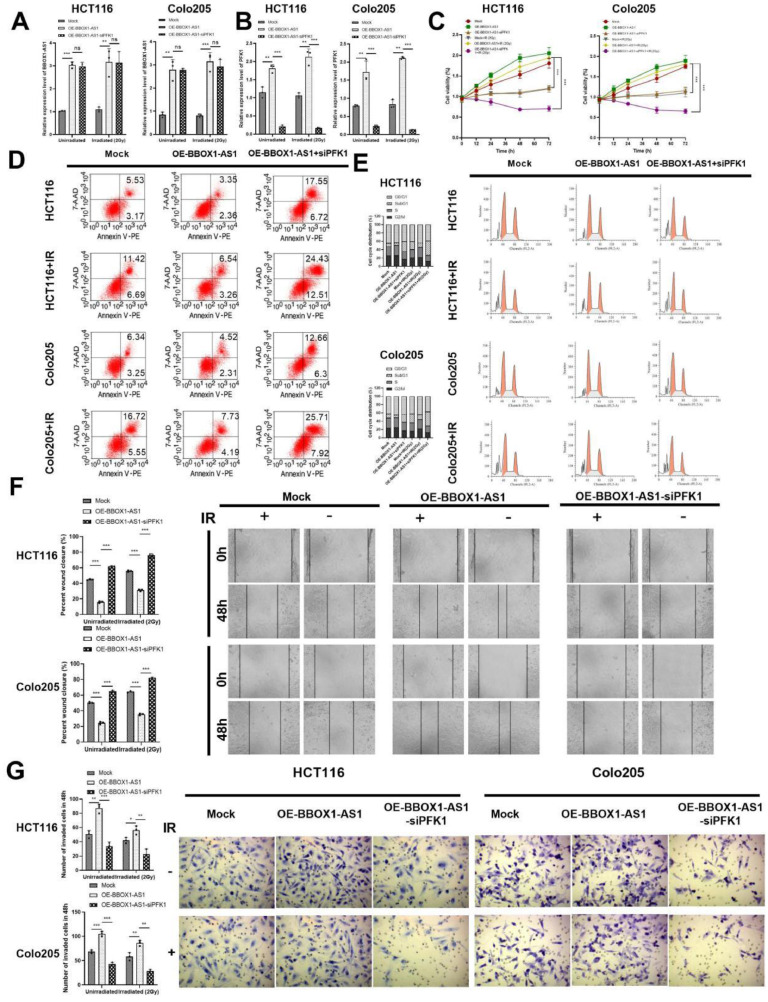
To verify the mechanism by which BBOX1-AS1 may increase the radiotherapy resistance of CRC cells by regulating PFK1, we carried out the following experiments. We found that compared with the blank control group, the expression of BBOX1-AS1 was significantly increased in the OE- BBOX1-AS1 group, where there was no significant difference in the expression of BBOX1-AS1 in the OE-BBOX1-AS1-siPFK1 group (Fig. 6A). The expression of PFK1 in the OE-BBOX1-AS1 group was significantly higher than in the blank control group, where the OE-BBOX1-AS1-siPFK1 group offset the promoting effect of BBOX1-AS1 overexpression on PFK1 expression in the same way (Fig. 6B). The same results were obtained in each group after 2 Gy radiation. Additionally, after radiation, the proliferation of cells in the OE-BBOX1-AS1 group was significantly increased compared with the blank control group, and the proliferation of cells in the OE-BBOX1-AS1-siPFK1 group was significantly lower than in the OE-BBOX1-AS1 group (P < 0.001). Furthermore, knockdown of PFK1 expression alone reduced the promoting effect of BBOX1-AS1 overexpression on cell proliferation (Fig. 6C).
Fig. 6.
BBOX1-AS1 increased radiotherapy resistance by regulating PFK1 in colorectal cells in vitro. (A) RT-qPCR detected the expression of BBOX1-AS1. (B) RT-qPCR detected the expression of PFK1. (C) The proliferation of CRC cells. (D) Apoptosis assays in CRC cells. (E) The effect of BBoX1-AS1 on the cell cycle. (F) CRC invasion was analyzed via scratch assay. (G) The migration of CRC cells was detected by transwell assays. Mean ± SD (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
Regardless of exposure to radiation, overexpression of BBOX1-AS1 and knockdown of PFK1 reversed the inhibitory effect of overexpression of BBOX1-AS1 on CRC apoptosis (Fig. 6D). In addition, the proportion of cells in the SubG1 phase in the OE-BBOX1-AS1 group was significantly lower than in the blank control group, and the effect of inhibition of BBOX1-AS1 on apoptosis was reversed by knockout of PFK1 (Fig. 6E). Additionally, cell invasion and migration were detected by scratch assay and transwell assay. Compared to the blank control group, invasion and migration significantly increased in the OE-BBOX1-AS1 group, regardless of radiation exposure. However, overexpression of BBOX1-AS1 and simultaneous deletion of PFK1 expression reversed the promoting effect of BBOX1-AS1 on invasion and metastasis in CRC cells (Fig. 6F, G).
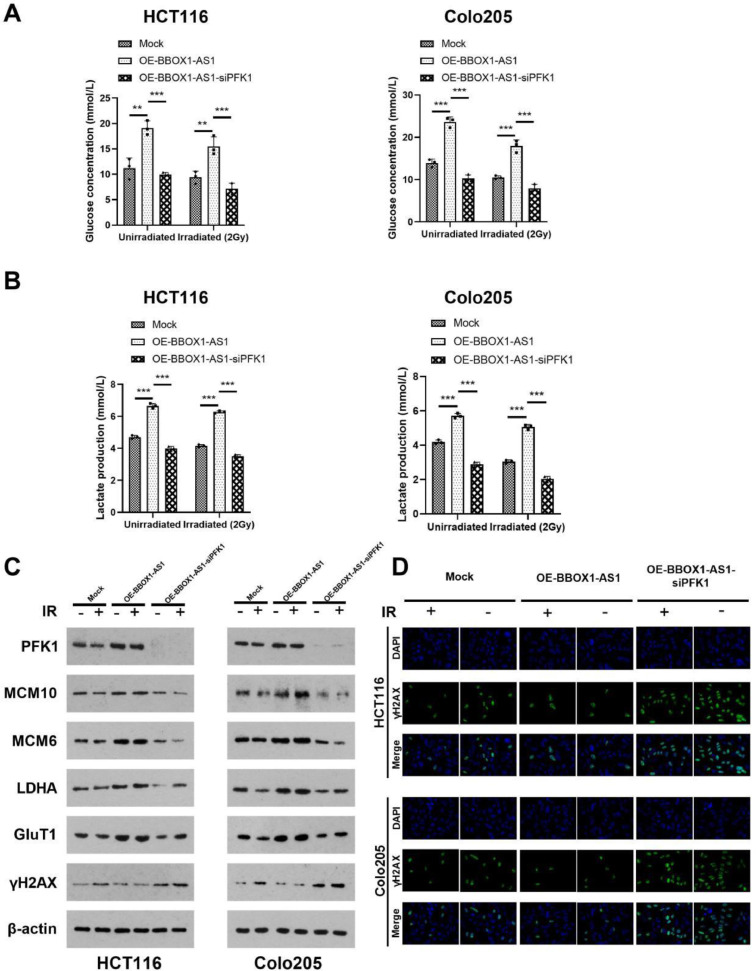
In previous studies, we found that PFK1 can regulate sensitivity to radiotherapy in colorectal cancer by affecting glycolysis [20]. In this experiment, we found that overexpression of BBOX1-AS1 enhanced glycolysis in CRC cells, while overexpression of BBOX1-AS1 and simultaneous knockdown of PFK1 expression inhibited the promoting effect of BBOX1-AS1 overexpression on glycolysis levels in CRC cells (Fig. 7A, B). Expression of the glycolytic-related genes PFK1, LADH, GluT1 and cell replication-related genes MCM10 and MCM6 were significantly increased, where γH2AX, which reflects radiotherapy sensitivity, was significantly decreased in the OE-BBOX1-AS1 group. In addition, γH2AX increased significantly in the OE-BBOX1-AS1-siPFK1 group, especially after radiotherapy (Fig. 7C, D).
Fig. 7.
BBOX1-AS1 affected CRC glycolysis and the cell cycle. (A) Glucose concentration in cells. (B) Lactate production in cells. (C) The expression of PFK1, LADH, GluT1, MCM10, MCM6, and γH2AX were analyzed by western blotting. (D) The expression of γH2AX in cells. Mean ± SD (n = 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001.
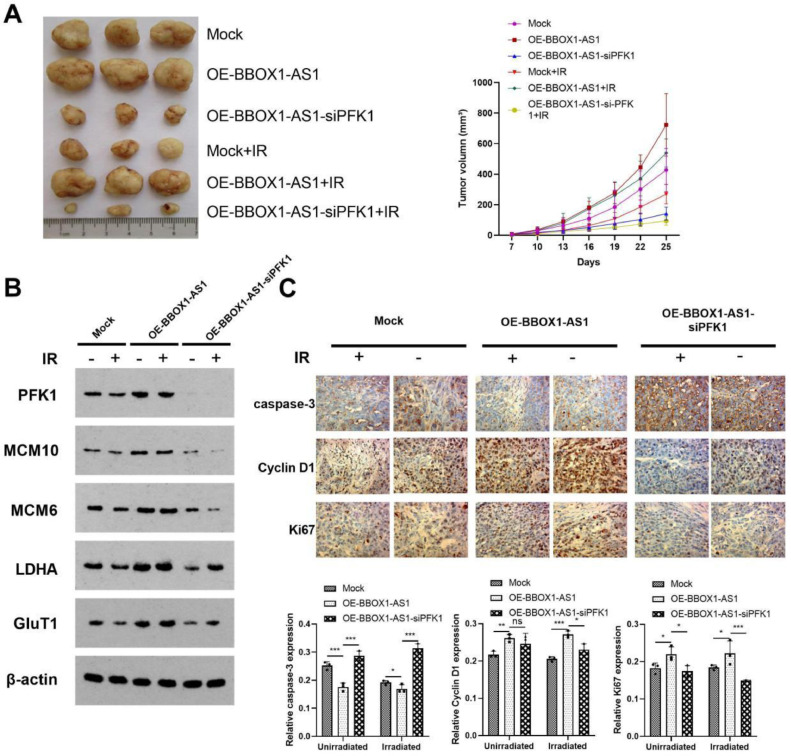
BBOX1-AS1 increased radiotherapy resistance in vivo
In this experiment, we found that tumor volume in xenograft mice increased significantly in the OE-BBOX1-AS1 group, regardless of radiation exposure. Alternatively, tumor volume in the OE-BBOX1-AS1-siPFK1 group decreased compared with the OE-BBOX1-AS1 group (Fig. 8A). In addition, MCM10, MCM6, PFK1, LDHA, and GluT1 were highly expressed in the OE-BBOX1-AS1 group. However, knockdown of PFK1 expression decreased the expression of these genes (Fig. 8B). Therefore, overexpression of BBOX1-AS1 may increase tumor cell radiation resistance by inhibiting radiation-induced apoptosis (Fig. 8C).
Fig. 8.
BBOX1-AS1 increased radiotherapy resistance in vivo. (A) Photographs of tumors and tumor growth curves from different nude mouse treatment groups. (B) The expression of PFK1, MCM10, MCM6, LDHA, and GluT1. (C) The expression of caspase-3, Cyclin D1, and Ki-67 following various treatments. Mean ± SD (n = 5/group). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Several studies have shown that lncRNAs are involved in various physiological processes such as proliferation, apoptosis, migration, and aging, and associated with the occurrence and development of cancer [22,23]. Using the GEPIA database, we found that BBOX1-AS1 was highly expressed in CRC, and we verified that BBOX1-AS1 was overexpressed in CRC tissues and colorectal cancer cell lines. Studies have found that BBOX1-AS1 promotes the development of cervical cancer, gastric cancer, ovarian cancer, and CRC using competing endogenous RNA (ceRNA) [24], [25], [26]. However, there have been few studies on colorectal cancer. Our results confirmed that BBOX1-AS1 promoted the proliferation, invasion, migration, and glycolysis of CRC cells and inhibited apoptosis in vivo and in vitro. However, the effect of BBOX1-AS1 on radiotherapy sensitivity in CRC cells is unknown. It is well known that radiotherapy resistance often leads to a poor response to therapy [27,28]. Therefore, our findings provide novel ideas for the clinical treatment of radiotherapy resistance.
Radiotherapy is an important CRC treatment, however, some patients are insensitive to radiotherapy and develop radiation resistance. Many researchers are investigating radiosensitive tumor markers to improve the efficacy of radiotherapy. Many studies have shown that ncRNAs can regulate radiotherapy sensitivity by affecting aerobic glycolysis [29], [30], [31]. Our study proved that BBOX1-AS1 stabilized the expression of PFK1 mRNA and protein levels. We further confirmed that BBOX1-AS1 promoted the proliferation, invasion, migration, and glycolysis of CRC cells by regulating PFK1, and inhibited CRC apoptosis.
Furthermore, GO analysis showed that the genes positively correlated with PFKM mainly play a role in glycolysis, cell proliferation, regulation of DNA replication initiation, and the G1/S transition of the mitotic cell cycle. Studies have also reported that ionizing radiation can damage DNA and induce DNA single or double strand breaks, and the repair of DNA double strand breaks is related to the radiosensitivity of tumor cells [32,33]. H2AX at position 139 of Serine can be phosphorylated by the ataxic telangiectasia mutant gene (ATM) to form γ-H2AX after DNA damage. Since γ-H2AX is considered a marker of DNA double-strand breaks, it can be used to evaluate repair efficiency following DNA damage [34,35]. In this study, we found that after exposure to radiation, the expression rate of γ-H2AX in the OE-BBOX1-AS1 group was lower than in the blank control group, while the expression rate of γ-H2AX was significantly increased in the OE-BBOX1-AS1 and si-PFK1 groups. We also observed that the MCM10 and MCM6 genes were highly expressed in CRC. Both MCM10 and MCM6 belong to the microchromosome maintenance protein family, which plays a key role in the regulation of DNA replication and repair [36], [37], [38]. The aberrant expression of MCM family protein genes can promote abnormal cell proliferation and lead to the occurrence of cancer. In addition, overexpression of MCM proteins is considered a proliferation marker in a variety of tumor types [36]. Therefore, we speculate that BBOX1-AS1 can increase CRC cell radiotherapy resistance by promoting DNA damage repair, and that MCM family proteins may be involved. To the best of our knowledge, our study is the first to elucidate that BBOX1-AS1 can regulate radiosensitivity in CRC.
With the widespread application of radiotherapy in cancer, an increasing number of studies have investigated how to improve the efficacy of radiotherapy. Our study demonstrated that BBOX1-AS1 may increase radiotherapy resistance of CRC cells by promoting glycolysis, repairing DNA damage, and inhibiting radiation-induced apoptosis. Thus, targeting BBOX1-AS1 is a novel and promising treatment strategy to enhance the radiosensitivity of CRC.
BBOX1-AS1 is crucial for CRC radiosensitivity. However, the effect of downregulating BBOX1-AS1 expression by transfecting siRNA is not well established, which may be due to the transfection efficiency of siRNA. In future studies, we plan to explore this avenue and use shRNA transfection instead of siRNA transfection to silence BBOX1-AS1 for better and more stable sensitization.
Conclusions
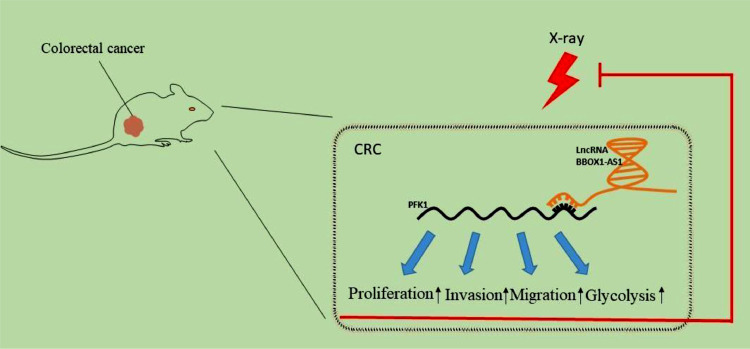
We demonstrated that BBOX1-AS1 promoted the occurrence and development of CRC and increased radiotherapy resistance of CRC by stabilizing PFK1 expression in vivo and in vitro (Fig. 9). BBOX1-AS1 can be used as a potential prognostic biomarker and radiotherapy sensitization target for CRC, providing new therapeutic clues and ideas for treatment of patients with radioresistant CRC. However, only a single dose of X-ray was adopted in the present study. Identifying the optimal dose and investigating further mechanisms will be the focus in the future.
Fig. 9.
Molecular mechanisms of BBOX1-AS1 involved in regulating PFK1 expression, which promotes the proliferation, invasion, migration, and glycolysis of CRC cells and thereby increases radiation resistance.
Ethics approval and consent to participate
The clinical samples were collected from patients after informed consent was obtained. Experiments were performed with the approval of the IRB of Third Xiangya Hospital, Central South University.
Consent for publication
All authors are aware of and agree to the content of the paper, as well as to being listed as co-authors of the paper.
Availability of data and material
All the data and material in this paper are available upon request.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Qi Wang: Conceptualization, Investigation, Funding acquisition. Xiao-Fei Li: Data curation, Writing – original draft. Ying-Hui Zhou: Data curation. Xiang-Hong Qin: Formal analysis. Li-Hui Wang: Methodology. Meng-Qing Xiao: Software, Visualization. Ke Cao: Resources, Supervision. John K. Ma: Validation. Cheng-Hui Huang: Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding
This work was financially supported by the Natural Science Foundation of Hunan Province (2022JJ70151).
Acknowledgments
We thank patients for providing clinical samples and LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101751.
Appendix. Supplementary materials
Reference
- 1.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 2.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. Epub 2019 Aug 27. [DOI] [PubMed] [Google Scholar]
- 3.Puglisi C., Giuffrida R., Borzì G., Di Mattia P., Costa A., Colarossi C., Deiana E., Picardo M.C., Colarossi L., Mare M., Marino L., Di Grazia A., Forte S. Radiosensitivity of cancer stem cells has potential predictive value for individual responses to radiotherapy in locally advanced rectal cancer. Cancers (Basel) 2020;12(12):3672. doi: 10.3390/cancers12123672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gollins S., Sebag-Montefiore D. Neoadjuvant treatment strategies for locally advanced rectal cancer. Clin. Oncol. (R. Coll. Radiol.) 2016;28(2):146–151. doi: 10.1016/j.clon.2015.11.003. Epub 2015 Nov 29. [DOI] [PubMed] [Google Scholar]
- 5.Martella A., Willett C., Palta M., Czito B. The selective use of radiation therapy in rectal cancer patients. Curr. Oncol. Rep. 2018;20(6):43. doi: 10.1007/s11912-018-0689-7. [DOI] [PubMed] [Google Scholar]
- 6.Benson A.B., Venook A.P., Al-Hawary M.M., Arain M.A., Chen Y.J., Ciombor K.K., Cohen S., Cooper H.S., Deming D., Garrido-Laguna I., Grem J.L., Gunn A., Hoffe S., Hubbard J., Hunt S., Kirilcuk N., Krishnamurthi S., Messersmith W.A., Meyerhardt J., Miller E.D., Mulcahy M.F., Nurkin S., Overman M.J., Parikh A., Patel H., Pedersen K., Saltz L., Schneider C., Shibata D., Skibber J.M., Sofocleous C.T., Stoffel E.M., Stotsky-Himelfarb E., Willett C.G., Johnson-Chilla A., Gurski L.A. NCCN guidelines insights: rectal cancer, version 6.2020. J. Natl. Compr. Canc. Netw. 2020;18(7):806–815. doi: 10.6004/jnccn.2020.0032. [DOI] [PubMed] [Google Scholar]
- 7.Couwenberg A.M., Burbach J.P.M., van Grevenstein W.M.U., Smits A.B., Consten E.C.J., Schiphorst A.H.W., Wijffels N.A.T., Heikens J.T., Intven M.P.W., Verkooijen H.M. Effect of neoadjuvant therapy and rectal surgery on health-related quality of life in patients with rectal cancer during the first 2 years after diagnosis. Clin. Colorectal Cancer. 2018;17(3):e499–e512. doi: 10.1016/j.clcc.2018.03.009. Epub 2018 Mar 21. [DOI] [PubMed] [Google Scholar]
- 8.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., Poliakov A., Cao X., Dhanasekaran S.M., Wu Y.M., Robinson D.R., Beer D.G., Feng F.Y., Iyer H.K., Chinnaiyan A.M. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. Epub 2015 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Li C.H., Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013;45(8):1895–1910. doi: 10.1016/j.biocel.2013.05.030. Epub 2013 Jun 4. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z., Wang Y.H., Xu H., Yuan C.S., Zhou H.H., Huang W.H., Wang H., Zhang W. LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death Dis. 2021;12(1):69. doi: 10.1038/s41419-020-03302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Wang L., He X., Zhang J., Zhu Z., Zhang M., Li X. lncRNA CCAT2 promotes radiotherapy resistance for human esophageal carcinoma cells via the miR‑145/p70S6K1 and p53 pathway. Int. J. Oncol. 2020;56(1):327–336. doi: 10.3892/ijo.2019.4929. Epub 2019 Dec 2. [DOI] [PubMed] [Google Scholar]
- 13.Xu W., Zhou G., Wang H., Liu Y., Chen B., Chen W., Lin C., Wu S., Gong A., Xu M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int. J. Cancer. 2020;146(10):2901–2912. doi: 10.1002/ijc.32747. Epub 2019 Nov 23. [DOI] [PubMed] [Google Scholar]
- 14.Dai M., Chen X., Mo S., Li J., Huang Z., Huang S., Xu J., He B., Zou Y., Chen J., Dai S. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci. Rep. 2017;7:46572. doi: 10.1038/srep46572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shademan M., Naseri Salanghuch A., Zare K., Zahedi M., Foroughi M.A., Akhavan Rezayat K., Mosannen Mozaffari H., Ghaffarzadegan K., Goshayeshi L., Dehghani H. Expression profile analysis of two antisense lncRNAs to improve prognosis prediction of colorectal adenocarcinoma. Cancer Cell Int. 2019;19:278. doi: 10.1186/s12935-019-1000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann M., Wright P.R., Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45(W1):W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livi C.M., Klus P., Delli Ponti R., Tartaglia G.G. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 2016;32(5):773–775. doi: 10.1093/bioinformatics/btv629. Epub 2015 Oct 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Shi W., Li C., Wartmann T., Kahlert C., Du R., Perrakis A., Brunner T., Croner R.S., Kahlert U.D. Sensory ion channel candidates inform on the clinical course of pancreatic cancer and present potential targets for repurposing of FDA-approved agents. J. Pers. Med. 2022;12(3):478. doi: 10.3390/jpm12030478. Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin G., Tu X., Li H., Cao P., Chen X., Song J., Han H., Li Y., Guo B., Yang L., Yan P., Li P., Gao C., Zhang J., Yang Y., Zheng J., Ju H.Q., Lu L., Wang X., Yu C., Sun Y., Xing B., Ji H., Lin D., He F., Zhou G. Long noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology. 2020;71(1):112–129. doi: 10.1002/hep.30793. Epub 2019 Aug 12. [DOI] [PubMed] [Google Scholar]
- 21.Tian R.F., Li X.F., Xu C., Wu H., Liu L., Wang L.H., He D., Cao K., Cao P.G., Ma J.K., Huang C.H. SiRNA targeting PFK1 inhibits proliferation and migration and enhances radiosensitivity by suppressing glycolysis in colorectal cancer. Am. J. Transl. Res. 2020;12(9):4923–4940. [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y., Li J., Yu P., Sun J., Hu Y., Meng X., Xiang L. Targeting lncRNAs in programmed cell death as a therapeutic strategy for non-small cell lung cancer. Cell Death Discov. 2022;8(1):159. doi: 10.1038/s41420-022-00982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishore C., Karunagaran D. Non-coding RNAs as emerging regulators and biomarkers in colorectal cancer. Mol. Cell Biochem. 2022;477(6):1817–1828. doi: 10.1007/s11010-022-04412-5. Epub 2022 Mar 24. [DOI] [PubMed] [Google Scholar]
- 24.Xu J., Yang B., Wang L., Zhu Y., Zhu X., Xia Z., Zhao Z., Xu L. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361-3p and HuR to drive cervical cancer progression. Cell Prolif. 2020;53(7):e12823. doi: 10.1111/cpr.12823. Epub 2020 Jun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao H., Chen R., Yang Y., Jiang J. LncRNA BBOX1-AS1 aggravates the development of ovarian cancer by sequestering miR-361-3p to augment PODXL expression. Reprod. Sci. 2021;28(3):736–744. doi: 10.1007/s43032-020-00366-5. Epub 2020 Nov 6. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Yu Q., Li B., Guan R., Huang C., Yang X. BBOX1-AS1 accelerates gastric cancer proliferation by sponging miR-3940-3p to upregulate BIRC5 expression. Dig. Dis. Sci. 2021;66(4):1054–1062. doi: 10.1007/s10620-020-06308-0. Epub 2020 May 11. [DOI] [PubMed] [Google Scholar]
- 27.Meng W., Palmer J.D., Siedow M., Haque S.J., Chakravarti A. Overcoming radiation resistance in gliomas by targeting metabolism and DNA repair pathways. Int. J. Mol. Sci. 2022;23(4):2246. doi: 10.3390/ijms23042246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cáceres-Gutiérrez R.E., Alfaro-Mora Y., Andonegui M.A., Díaz-Chávez J., Herrera L.A. The influence of oncogenic RAS on chemotherapy and radiotherapy resistance through DNA repair pathways. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.751367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan F., Qin Q., Yu H., Yue X. Effect of glycolysis inhibition by miR-448 on glioma radiosensitivity. J. Neurosurg. 2019;132(5):1456–1464. doi: 10.3171/2018.12.JNS181798. [DOI] [PubMed] [Google Scholar]
- 30.Cao K., Li J., Chen J., Qian L., Wang A., Chen X., Xiong W., Tang J., Tang S., Chen Y., Chen Y., Cheng Y., Zhou J. microRNA-33a-5p increases radiosensitivity by inhibiting glycolysis in melanoma. Oncotarget. 2017;8(48):83660–83672. doi: 10.18632/oncotarget.19014. Jul 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan L., Huang C., Li J., Gao T., Lin Z., Yao T. Long non‑coding RNA urothelial cancer associated 1 regulates radioresistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int. J. Mol. Med. 2018;42(4):2247–2259. doi: 10.3892/ijmm.2018.3778. Epub 2018 Jul 13. [DOI] [PubMed] [Google Scholar]
- 32.Nakano T., Akamatsu K., Tsuda M., Tujimoto A., Hirayama R., Hiromoto T., Tamada T., Ide H., Shikazono N. Formation of clustered DNA damage in vivo upon irradiation with ionizing radiation: visualization and analysis with atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 2022;119(13) doi: 10.1073/pnas.2119132119. Epub 2022 Mar 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo Y., Tamari K., Takahashi Y., Minami K., Tatekawa S., Isohashi F., Suzuki O., Akino Y., Poly O.K. ADP-ribose) polymerase inhibitors sensitize cancer cells to hypofractionated radiotherapy through altered selection of DNA double-strand break repair pathways. Int. J. Radiat. Biol. 2022;98(7):1222–1234. doi: 10.1080/09553002.2022.2020357. Epub 2022 Jan 4. [DOI] [PubMed] [Google Scholar]
- 34.Mladenov E., Magin S., Soni A., Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front. Oncol. 2013;3:113. doi: 10.3389/fonc.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subedi P., Gomolka M., Moertl S., Dietz A. Ionizing radiation protein biomarkers in normal tissue and their correlation to radiosensitivity: a systematic review. J. Pers. Med. 2021;11(2):140. doi: 10.3390/jpm11020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Chen H., Zhang J., Cheng A.S.L., Yu J., To K.F., Kang W. MCM family in gastrointestinal cancer and other malignancies: from functional characterization to clinical implication. Biochim. Biophys. Acta Rev. Cancer. 2020;1874(2) doi: 10.1016/j.bbcan.2020.188415. Epub 2020 Aug 19. [DOI] [PubMed] [Google Scholar]
- 37.Zeng T., Guan Y., Li Y.K., Wu Q., Tang X.J., Zeng X., Ling H., Zou J. The DNA replication regulator MCM6: an emerging cancer biomarker and target. Clin. Chim. Acta. 2021;517:92–98. doi: 10.1016/j.cca.2021.02.005. Epub 2021 Feb 18. [DOI] [PubMed] [Google Scholar]
- 38.Baxley R.M., Bielinsky A.K. Mcm10: a dynamic scaffold at eukaryotic replication forks. Genes (Basel) 2017;8(2):73. doi: 10.3390/genes8020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data and material in this paper are available upon request.
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.