Abstract
The herpes simplex virus virion host shutoff (vhs) protein (UL41 gene product) is a component of the HSV virion tegument that triggers shutoff of host protein synthesis and accelerated mRNA degradation during the early stages of HSV infection. Previous studies have demonstrated that extracts from HSV-infected cells and partially purified HSV virions display vhs-dependent RNase activity and that vhs is sufficient to trigger accelerated RNA degradation when expressed as the only HSV protein in an in vitro translation system derived from rabbit reticulocytes. We have used the rabbit reticulocyte translation system to characterize the mode of vhs-induced RNA decay in more detail. We report here that vhs-dependent RNA decay proceeds through endoribonucleolytic cleavage, is not affected by the presence of a 5′ cap or a 3′ poly(A) tail in the RNA substrate, requires Mg2+, and occurs in the absence of ribosomes. Intriguingly, sites of preferential initial cleavage were clustered over the 5′ quadrant of one RNA substrate that was characterized in detail. The vhs homologue of pseudorabies virus also induced accelerated RNA decay in this in vitro system.
Herpes simplex virus (HSV) is a large enveloped DNA virus that replicates in the nuclei of infected mammalian cells. HSV executes a complex genetic regulatory program during lytic infection of permissive host cells (16); reviewed in references 37 and 50). Five immediate-early (IE) genes are expressed first, and four of these encode nuclear regulatory proteins that act at transcriptional and posttranscriptional levels to stimulate the expression of the viral early (E) and late (L) genes. Expression of the second temporal class of HSV genes, the E genes, leads to synthesis of the seven proteins that comprise the viral DNA replicative machinery. Viral DNA replication then augments expression of the L genes that encode most of the structural components of the virus particle.
The HSV virion contains a variety of regulatory proteins that prime the newly infected cell to support efficient virus replication. These virion-associated regulators are located in the tegument—the space between the viral envelope and the nucleocapsid—and are therefore presumably delivered into the cytoplasm after fusion of the viral envelope with the host cell plasma membrane. The best characterized of these virion regulators is VP16, an abundant tegument protein that stimulates transcription of the five IE genes (reviewed in references 37 and 50). The tegument also contains the virion host shutoff protein (vhs), which triggers rapid shutoff of host cell protein synthesis, disruption of preexisting polysomes, and degradation of host mRNAs in the absence of de novo viral gene expression (10, 13–15, 18–21, 28–31, 34, 36, 42, 46–49).
Three lines of evidence demonstrate that the vhs protein encoded by the HSV UL41 gene is both necessary and sufficient for virion-induced host shutoff. First, Read and Frenkel (34) isolated viable HSV mutants deficient in virion-induced shutoff, and one of these mutations (vhs1) was subsequently mapped to the UL41 open reading frame (ORF) (21, 26). Confirming this assignment, targeted disruptions of the UL41 gene produce a vhs-deficient phenotype (11, 35, 42). Second, viral recombinants in which the UL41 gene of HSV-1 has been replaced by the corresponding gene from HSV-2 display the more robust shutoff phenotype characteristic of HSV-2 (12). Third, vhs suffices to block reporter gene expression when it is expressed as the only HSV protein in transiently transfected mammalian cells (18, 32). The UL41 gene product has been identified as a 58-kDa phosphoprotein that is packaged into the virion tegument (35, 40).
vhs destabilizes most if not all of the viral and cellular mRNAs during infection (14, 20, 21, 30, 31, 34, 46). However, rRNAs and tRNAs are spared (19, 20, 30, 52), raising the possibility that one or more features common to most mRNAs [such as the 5′ cap or 3′ poly(A) tail] play a role in selectively targeting mRNA for degradation. The rapid decline in host mRNA levels triggered by vhs presumably helps viral mRNAs gain access to the cellular translational apparatus. In addition, the relatively short half-life of viral mRNAs contributes to the sharp transitions between the successive phases of viral protein synthesis, by more tightly coupling changes in the rate of synthesis of viral mRNAs to altered mRNA levels (20, 30, 31, 46). These effects probably enhance virus replication and may account for the finding that vhs mutants display a ca. 10-fold reduction in virus yield in tissue culture (34, 42) and severe defects in the nervous system of mice (45). Although viral mRNAs belonging to all three temporal classes are significantly destabilized by vhs, Fenwick and Owen have provided strong evidence that the vhs activity of the infecting virion is partially downregulated by a newly synthesized viral protein, allowing viral mRNAs to accumulate after host transcripts have been degraded (14). vhs directly binds to VP16 (41), raising the possibility that VP16 modulates vhs activity. Consistent with this hypothesis, VP16 null mutants undergo vhs-induced termination of viral protein synthesis at intermediate times postinfection (23).
Although the mechanism of vhs action has yet to be completely defined, the available data strongly suggest that vhs is an RNase that triggers shutoff by degrading mRNA. First, vhs displays weak but significant amino acid sequence homology to the fen-1 family of nucleases (8), which are involved in DNA replication and repair (reviewed in reference 24). Second, vhs-dependent mRNA degradation can be reproduced in cytoplasmic extracts prepared from HSV-infected cells (19, 43) and in extracts of partially purified HSV virions (52). Moreover, Zelus et al. (52) have shown that vhs induces accelerated RNA turnover when it is expressed as the only HSV protein in a rabbit reticulocyte lysate (RRL) in vitro translation system. The vhs-dependent RNase activity detected in extracts of partially purified virions is inhibited by anti-vhs antibodies (52), suggesting that vhs is an integral and required component of this nuclease.
We used the rabbit reticulocyte-based vhs activity assay system (52) to further characterize the mode of vhs-induced RNA degradation. We confirmed that vhs induces accelerated RNA decay in this system and further showed that it severely inhibits the translation of reporter RNAs. RNA decay proceeded through endoribonucleolytic cleavage events, was not affected by the presence of a 5′ cap or 3′ poly(A) tail, and occurred in the absence of ribosomes. Detailed characterization of the mode of decay of one RNA substrate revealed that the preferred sites of initial cleavage were clustered over the 5′ quadrant of the transcript. Finally, we show that the vhs homologue of pseudorabies virus (PrV) also induced accelerated RNA decay in this system.
MATERIALS AND METHODS
Plasmids.
A vhs in vitro translation vector was constructed by inserting a 1.8-kb NcoI-EcoRI fragment containing the vhs ORF from pCMVvhs (18) between the NcoI and EcoRI sites of pSPUTK (9). The resulting plasmid (pSP6vhs) bears the vhs ORF fused to a modified derivative of the 5′ untranslated region (UTR) of Xenopus laevis β-globin (termed UTK) which has been engineered to contain a consensus Kozak translational initiation signal. This construct bears an SP6 RNA polymerase promoter immediately upstream of the modified UTR. A control plasmid (pSP6vhs1) bearing the inactivating vhs1 point mutation (21, 34) was constructed in the same manner, using pCMVvhs1 (18) as the source of vhs1 sequences. In vitro translation vectors encoding active, doubly tagged (HA and His8) versions of vhs were constructed by modifying the previously described plasmids pN138, pN138-HA, pS344, and pS344-HA (18). pN138 and pS344 encode active modified versions of vhs that bear in-frame insertions of an XhoI linker following codons 138 and 344 respectively, driven from the human cytomegalovirus IE promoter. pN138-HA and pS344-HA were derived from pN138 and pS344 by inserting sequences encoding an influenza virus hemagglutinin (HA) epitope at these newly introduced XhoI sites. Analogous plasmids bearing sequences encoding eight tandem histidine residues (pN138-HIS and pS344-HIS) were generated by inserting annealed complementary oligonucleotides 5′-TCGACATCATCATCATCATCATCATCA and 5′-TCGATGATGATGATGATGATGATGATG into the XhoI sites of pN138 and pS344. Doubly tagged (HA-His8) derivatives were then generated by exchanging appropriate restriction fragments between these plasmids. pN138HA-S344His was constructed by replacing a 646-bp BamHI-EcoRI fragment of pN138-HA with a 673-bp BamHI-EcoRI fragment of pS344-His containing the His8 tag. pN138His-S344HA was constructed by replacing a 646-bp BamHI-EcoRI fragment of pN138-His with a 679-bp BamHI-EcoRI fragment of pS344-HA containing the HA tag. The doubly tagged vhs ORFs were then transferred from these cytomegalovirus vectors to pSPUTK as described above for pSP6vhs to yield plasmids 1.1vhs (pSPN138HA-S344His) and 2.1vhs (pSPN138His-S344HA). Control derivatives bearing the vhs1 point mutation (1.1vhs1 and 2.1vhs1) were generated by replacing a 583-bp SmaI-BamHI fragment of 1.1vhs and 2.1vhs with the same fragment of pCMVvhs1 (18).
A PrV vhs in vitro translation vector was generated as follows. pPRV41 (3) was digested with DraI and EcoRI, and a 1,580-nucleotide (nt) fragment extending from 20 nt downstream of the vhs initiation codon (the DraI site) into 3′-flanking sequences (EcoRI) was purified by gel electrophoresis. This fragment was then ligated to the complementary oligonucleotides 5′-CATGGGGCTCTTTGGCCTTTT and 5′-AAAAGGCCAAAGAGCCCG to regenerate the 5′-most 20 bp of the PrV vhs ORF and place an engineered NcoI site at the initiation codon. The modified PrV vhs ORF was then inserted between the NcoI and EcoRI sites of pSPUTK, yielding pSPPRVvhs.
pSPSR19N contains a complete cDNA encoding the canine signal recognition particle α subunit (SRPα), initiating at an engineered NcoI site, inserted into pSPUTK (51). pMAC39 contains the bovine preprolactin (PPL) ORF inserted in the same vector (9). pPRL3′UTRpA contains the PPL ORF and 3′ UTR, followed by an engineered 35-nt poly(A) tail, inserted into pSPUTK. The poly(A) tail is flanked by a 5′ SspI and 3′ EagI site. pBlueK(coreD) contains a 4.5-kbp EcoRI fragment of human SH-2 containing inositol 5′ phosphatase (hSHIP) cDNA cloned at the EcoRI site downstream of the T7 promoter of the vector pBluescript (courtesy of Peter Whyte, McMaster University).
In vitro transcription and RNA labeling.
Transcription reactions were carried out with the Riboprobe in vitro transcription system (Promega) as specified by the vendor. mRNAs destined for in vitro translation (vhs, vhs1, PPL, and PrV vhs) were generated by transcription of 3 to 5 μg of supercoiled plasmid DNA (pSP6vhs, pSP6vhs1, pMAC39, and pSPPRVvhs) in a 50-μl reaction mixture for 30 mins at 30°C with 40 U of SP6 RNA polymerase in the presence of 0.5 mM cap primer 7mG(5′)ppp(5′)G (Pharmacia), 12.5 μM GTP, and 0.25 mM each CTP, ATP, and UTP. Following digestion of plasmid DNA with 5 U of RQ1 DNase (Promega), the reaction mixture was extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform. The resulting solution was made 2.5 M ammonium acetate, and the mRNA was precipitated with 95% ethanol. The mRNA pellet was then washed with 70% ethanol, dried, and resuspended in RNase-free water.
Capped, internally labeled reporter RNAs were generated as above, except that the template was linearized at an appropriate site prior to transcription and 1 μCi of [α-32P]-GTP was added to the transcription reaction mixture. Uncapped, internally labeled reporter RNAs were produced in a similar fashion, except that the cap primer was omitted and the GTP concentration was raised to 0.25 mM. SRPα reporter mRNA was generated with SP6 polymerase and EcoRV-linearized pSPSR19N plasmid DNA as a template to yield a 2.4-kb runoff transcript. The SRP antisense transcript was generated by using T7 RNA polymerase and SnaBI-linearized pSPSR19N to yield a 2.2-kb runoff transcript. HindIII-linearized pBlueK(coreD) plasmid DNA was transcribed by using T7 RNA polymerase to yield a 4.5-kb runoff hSHIP transcript. PPL RNA containing a 35-nt poly(A) tail was generated by using SP6 polymerase and EagI-linearized pPRL3′UTRpA plasmid DNA. A poly(A)− derivative of the same RNA was generated by linearizing the template with SspI. A vhs runoff transcript was generated by using SP6 RNA polymerase and EcoRI-linearized pSP6vhs.
Cap-labeled reporter RNAs were generated from uncapped unlabeled runoff transcripts by using vaccinia virus guanylyltransferase in the presence of [α-32P]GTP. Approximately 500 ng of RNA in 50 mM Tris-HCl(pH 7.9)–1.25 mM MgCl2–6 mM KCl–2.5 mM dithiothreitol (DTT)–0.1 mg of I RNase-free bovine serum albumin per ml–1 U of RNasin per μl–0.1 mM S-adenosyl-l-methionine was combined with 1 to 3 U of guanylyltransferase (Gibco-BRL) and 50 μCi of [α-32P]GTP in a total reaction volume of 30 μl. Following a 45-min reaction at 37°C, the reaction mixture was extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform and the RNA was recovered by ethanol precipitation.
HeLa cell translation extracts.
HeLa cell translation extracts were prepared by the method described by Carroll and Lucas-Lernard (6) with the following modifications. (i) HeLa S3 cells were grown in suspension culture in Joklik minimal essential medium supplemented with 5% fetal bovine serum, 1% vitamin mix (Gibco-BRL), and 1% nonessential amino acids (Gibco-BRL). (ii) A 300-μl volume of the postmitochondrial supernatant was mixed with 3 μl of 100 mM CaCl2 and 60 U of micrococcal nuclease. Following a 15-min incubation at room temperature, 10 μl of 100 mM EGTA was added to the nuclease-treated lysate, and samples were snap frozen in liquid N2 and stored at −70°C.
In vitro translation.
Approximately 5 to 10 μg of vhs mRNA was translated in a 50-μl RRL (Promega or Novagen) reaction mixture containing 40 μCi of [35S]methionine, as specified by the vendor. Translation reactions were carried out for 20 min (experiment in Fig. 1) or 1 h (all the remaining experiments) at 30°C. Blank RRL controls were generated as above, except that mRNA was omitted from the translation reactions. vhs was synthesized in vitro translation extracts derived from HeLa cells by combining 75 μl of nuclease-treated extract (above) with 10 μl of 10× HeLa cell E-Mix (20 mM HEPES-KOH [pH7.4], 100 mM potassium acetate, 2.2 mM magnesium acetate, 2.0 mM DTT, 12.5 mM ATP, 2.5 mM GTP, 375 mM creatine phosphate, 2 mM spermidine, 0.2 mg of calf liver tRNA per ml, 0.1 mM amino acids minus methionine, 62.5 U of creatine kinase), 80 μCi of [35S]methionine, 80 U of RNasin, and 5 to 10 μg of capped vhs RNA (total volume, of 100 μl). HeLa translation reactions were carried out at 30°C for 1 h. Samples of the translation reaction products were assessed for [35S]methionine incorporation by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis analysis (22).
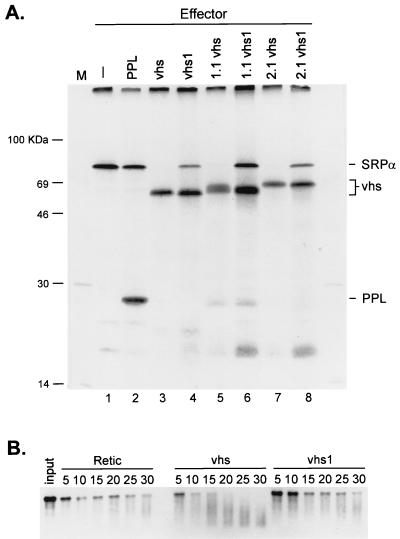
FIG. 1.
HSV-1 vhs induces translational arrest and mRNA degradation in vitro. (A) RRL were programmed with the indicator effector mRNAs, and translation was allowed to proceed for 20 mins in the presence of [35S]methionine. Lysates were then challenged with an equal amount of capped SRPα reporter mRNA, and the reactions were allowed to continue for an additional 60 min. The translation products were resolved on an SDS–12% polyacrylamide gel, and the 35S signal was detected by autoradiography with Kodak X-Omat AR film. 1.1 and 2.1, doubly tagged active vhs variants; vhs1, 1.1 vhs1, and 2.1 vhs1, inactive vhs point mutant derivatives of vhs, 1.1, and 2.1. (B) RRL were programmed with vhs RNA (lanes vhs), vhs1 RNA (lanes vhs1), or no RNA (control, lanes Retic), and translation was allowed to proceed for 20 min. The lysates were then challenged with capped, internally labeled SRPα mRNA. Samples were recovered at the indicated times (numbers above lanes, in minutes), and the RNA reaction products were resolved on a 1% agarose–6% formaldehyde gel, transferred to a Nytran Plus membrane, and detected by autoradiography with Kodak X-Omat AR film.
vhs activity assay.
Reporter RNA substrates generated by in vitro transcription were added to rabbit reticulocyte or HeLa cell lysates containing pretranslated vhs, and the reaction mixture was incubated at 30°C. Aliquots (5 μl) of the reaction mixture were removed at various times and immediately added to 200 μl of Trizol (Gibco-BRL) containing 20 μg of carrier Escherichia coli tRNA (Sigma). The samples were extracted after the addition of 40 μl of chloroform, and the resulting aqueous phase was extracted with chloroform. RNA was recovered by isopropanol precipitation, resuspended in 100 μl of RNase-free water, and reprecipitated with 95% ethanol. Following a 70% ethanol wash, the RNA pellet was dried and resuspended in RNase-free water. The RNA samples were then analyzed by electrophoresis through agarose-formaldehyde or polyacrylamide sequencing gels or by primer extension.
Agarose gel electrophoresis and Northern blot analysis.
RNA samples were resuspended in 4.5 μl of RNase-free water and then combined with 2 μl of 10× MOPS buffer (200 mM 3-n-morpholinopropanesulfonic acid [pH 7.0], 50 mM sodium acetate, 5 mM EDTA), 10 μl of deionized formamide, and 3.5 μl of 37% formaldehyde solution. Following a 10-min incubation at 75 to 80°C, the solution was combined with 6 μl of RNA loading buffer (50% glycerol, 1 mM EDTA, 10 mg of xylene cyanol per ml, 10 mg of bromophenol blue per ml) and subjected to electrophoresis through a 1% agarose gel containing 6% formaldehyde. Electrophoresis was carried out at approximately 5 V/cm for 3 to 4 h in 1× MOPS buffer containing 6% formaldehyde. The gel was then washed in water for 10 min, treated with 50 mM NaOH–10 mM NaCl for 20 min, and neutralized with 100 mM Tris-HCl for 20 min. RNA was then transferred to a Nytran Plus membrane in 20× SSC (3 M sodium chloride, 0.3 M sodium citrate). Following UV cross-linking (Stratalinker 2400; Stratagene), 32P-labeled RNA fragments were detected by exposure to Kodak X-Omat AR film at −70°C.
Unlabeled SRPα RNA fragments were detected by Northern blot analysis (7). Briefly, the Nytran Plus membrane was prehybridized in Church buffer (250 mM sodiumphosphate buffer [pH7.2], 7% SDS, 1% bovine serum albumin, 1 mM EDTA) at 62°C for 1 h. The membrane was then hybridized to a 400-nt EcoRV-EcoRI fragment of pSPSR19N corresponding to the 3′-most portion of the SRPα transcript. The probe was 32P labeled by random priming. Hybridization was carried out in Church buffer at 62°C for 13 to 17 h. The membrane was then washed twice for 10 min in 2× SSC–0.1% SDS and twice for 10 min in 0.1× SSC–0.1 SDS and subjected to autoradiography.
Primer extension.
RNA samples were suspended in 10 μl of 10 mM Tris-HCl (pH7.9)–1 mM EDTA–250 mM KCl containing 50,000 Cerenkov cpm of a 5′-32P-labeled oligonucleotide (5′-GGTGAAGAAGTCGACCATGGTAGAT-3′) complementary to nt 60 to 84 of the SRPα RNA. After annealing for 1 h at 65°C, the samples were combined with 25 μl of PE buffer (20 mM Tris-HCl [pH 8.7], 10 mM MgCl2, 5 mM DTT, 330 μM each deoxynucleoside triphosphate, 10 μg of actinomycin D per ml, 10 U of SuperScript II [Gibco-BRL] reverse transcriptase) and the extension reaction was carried out for 1 h at 42°C. Nucleic acids were precipitated with 95% ethanol, washed with 70% ethanol, dried, and resuspended in water. The samples were then combined with an equal volume of sequencing-gel-loading buffer, heated to 80°C for 2 to 3 min, and resolved on 8% polyacrylamide sequencing gels. The radioactive signal was detected by autoradiography.
Markers.
RNA size markers were generated by in vitro transcription with SP6 RNA polymerase and pSPSR19N DNA linearized with EcoRV, PvuII, SmaI, NruI, or SnaBI to yield runoff transcripts of 2,422, 1,628, 800, 429, and 298 nt, respectively. DNA size markers were generated by Klenow filling of HpaII-digested pBR322 plasmid DNA in the presence of [α-32P]dCTP.
RESULTS
HSV-1 vhs induces translational arrest and mRNA degradation in vitro.
As reviewed in the introduction, vhs suffices to trigger accelerated RNA degradation of reporter RNAs when it is expressed as the only HSV protein in an RRL in vitro translation system (52). Here we report the results of experiments that use this RRL-based in vitro system to examine the mechanism of vhs-induced RNA degradation in more detail.
We first established that vhs displays shutoff activity in vitro under our experimental conditions. To this end, RRL were programmed with in vitro transcripts encoding control bovine PPL, three active forms of vhs (wild type and two doubly tagged variants, 1.1vhs and 2.1vhs), and three inactive derivatives bearing the inactivating vhs1 point mutation (vhs1, 1.1vhs1, and 2.1vhs1). Translation was allowed to proceed for 20 min in the presence of [35S]methionine, and the lysates were then challenged with a 2.4-kb capped reporter RNA encoding SRPα. Translation was allowed to continue for an additional 60 min, and the translation products were analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 1A). As expected, pretranslation of control PPL mRNA had little effect on subsequent translation of the SRPα reporter RNA (compare lane 1 with lane 2). In striking contrast, translation of the reporter transcript was severely inhibited in lysates that contained active vhs (lanes 3, 5, and 7). This inhibitory effect was reduced by the vhs1 mutation (lanes 4, 6, and 8), arguing for the biological relevance of the results. To determine if translational arrest was accompanied by accelerated decay of the reporter RNA, the experiment was repeated with capped internally labeled reporter RNA (Fig. 1B). The results indicated that the reporter RNA decayed at an accelerated rate in the presence of wild-type vhs (see also below). These data confirm that vhs induces accelerated RNA degradation in the RRL in vitro translation system (52) and that this activity severely inhibits translation of a reporter RNA.
vhs-induced degradation of SRPα RNA involves early endonucleolytic cleavage events clustered in the 5′ quadrant of the transcript.
Previous studies have established that vhs induces RNA degradation in both HSV-infected cells and in vitro systems. However, little information is available about the mode of vhs-induced RNA decay. Zelus et al. (52) suggested that vhs extracted from partially purified virions preferentially degrades the 3′ end of globin RNA, possibly by targeting sequences within or close to the poly(A) tail. These authors also suggested that RNA degradation probably proceeds through endoribonucleolytic cleavage, although (as acknowledged by the authors) the data advanced to support this conclusion were not definitive. We therefore examined the mode of vhs-induced RNA decay in more detail (Fig. 2).
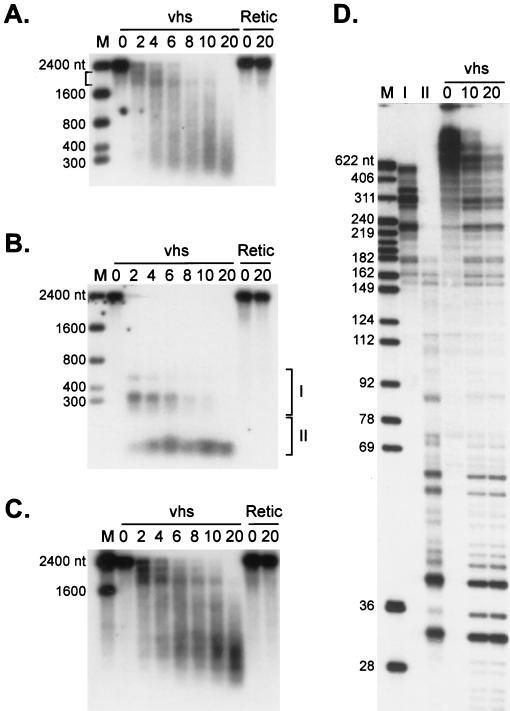
FIG. 2.
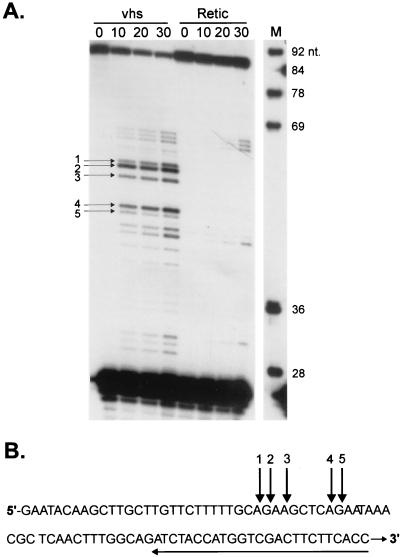
Analysis of vhs-induced degradation intermediates of SRPα RNA. (A and B) Internally labeled (A) and cap-labeled (B) SRPα mRNAs were added to RRL containing pretranslated vhs (lanes vhs) or RRL control (lanes Retic). RNA degradation products were recovered at the indicated times (minutes) and analyzed by agarose-formaldehyde gel electrophoresis. (C) The membrane in panel B was hybridized to a 32P-labeled DNA probe corresponding to the 3′-most 400 nt of SRPα RNA (after the radioactive signal from the cap label had been allowed to decay for six half-lives). The bound probe was then detected by autoradiography with Kodak X-Omat AR film. Numbers to the left of panels A, B, and C represent the sizes of RNA markers (lanes M) in nucleotides. (D) Cap-labeled SRPα RNA was added to RRL-vhs, and the RNA degradation intermediates recovered at 10 min were resolved on a 1% agarose–6% formaldehyde gel. RNA fragments contained in the gel slices indicated by brackets I and II in panel C were eluted and resolved on an 8% polyacrylamide sequencing gel (lanes I and II) along with the unfractionated products of a vhs reaction on cap-labeled SRPα RNA (sampled at 0, 10, and 20 min). Numbers to the left of panel D represent the sizes of DNA markers (lane M) in nucleotides.
Internally labeled SRPα RNA gave rise to several discrete high-molecular-weight intermediates, ranging in length from ca. 1,800 to ca. 2,200 nt, early during the reaction (Fig. 2A, bracket; also see Fig. 4A and 6B). These intermediates subsequently decayed to lower-molecular-weight products as the reaction proceeded. In principle, the 1,800 to 2,200-nt intermediates could arise either from exonucleolytic decay initiated at one or both ends of the transcript or from endonucleolytic cleavage at several sites located close to one or both ends of the 2,400-nt substrate RNA. We distinguished between these possibilities by monitoring the fate of the 5′ and 3′ ends of the transcript during the course of the reaction. To this end, 5′-cap-labeled SRPα RNA was added to RRL containing pretranslated vhs and the resulting degradation products were analyzed by agarose-formaldehyde gel electrophoresis and autoradiography (Fig. 2B). The 32P signal from the cap label was then allowed to decay for six half-lives, and the membrane was hybridized to a probe corresponding to the 3′-most 400 nt of SRPα RNA (Fig. 2C). The 3′ probe revealed a pattern of degradation intermediates similar to that observed with internally labeled RNA (compare Fig. 2C with Fig. 2A): discrete fragments of 1,800 to 2,200 nt were observed at early times, and these were reduced in size as the reaction proceeded (Fig. 2C). In contrast, the 5′ cap label was recovered in considerably smaller products at the early time points (Fig. 2B). To obtain a more accurate estimate of the sizes of these 5′ products, RNA was recovered from the gel slices indicated by brackets in Fig. 2B and resolved on an 8% polyacrylamide sequencing gel (Fig. 2D). The 5′ fragments recovered from gel slices I and II ranged in size from ca. 200 to 700 nt and ca. 30 to 40 nt, respectively. No prominent larger cap-labeled products were observed in repeated trials. The 30- to 40-nt fragments accumulated throughout the course of the reaction, while the 200- to 700 nt fragments detected at earlier times decayed as the reaction proceeded (Fig. 2B). Taken in combination, these data exclude the possibility that vhs-induced RNA degradation proceeds exclusively through a 5′-3′ or 3′-5′ exonucleolytic pathway and are completely consistent with an endonucleolytic mode of RNA decay. In particular, the early generation of sets of 5′ and 3′ intermediates whose sum roughly yields the length of the intact substrate (Fig. 2B and C, 2-min time point) suggests that a substantial fraction of the RNA molecules are initially cleaved at a limited number of sites clustered in the 5′ quadrant of the RNA. The data do not, however, demonstrate that all molecules are initially cleaved at these positions and do not exclude the possibility that strong cleavage sites are also located very close to the 3′ end of the RNA.
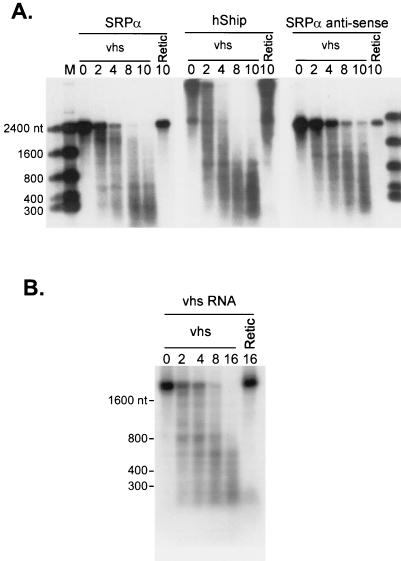
FIG. 4.
vhs induces degradation of a variety of RNA substrates. (A) Internally labeled SRPα (2.4 kb), hSHIP (4.5 kb), and SRPα antisense (2.2 kb) RNAs were added to RRL containing vhs (lanes vhs) or RRL control (lanes Retic), and samples recovered at the indicated times (minutes) were analyzed by agarose-formaldehyde gel electrophoresis as in Fig. 1B. (B) Internally labeled vhs RNA (1.8 kb) was reacted with RRL containing vhs or control RRL, and samples recovered at the indicated times (minutes) were analyzed as in panel A.
FIG. 6.
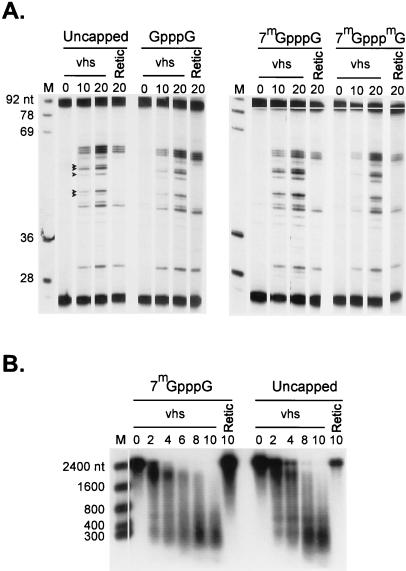
vhs-induced RNA degradation is cap independent. (A) Unlabeled SRPα RNAs bearing the indicated 5′ cap structures were added to RRL-vhs (lanes vhs) and control RRL (lanes Retic), and samples were recovered at the indicated times (minutes). The RNA reaction products were then analyzed by primer extension with 5′ 32P-labeled oligonucleotide complementary to residues 60 to 84 of the SRPα RNA, as in Fig. 3. Solid arrowheads indicate the mobilities of the vhs-induced products. Numbers to the left of panel A indicate the sizes of DNA markers (lanes M) in nucleotides. (B) Internally labeled capped (7mGpppG) and uncapped SRPα RNAs were added to RRL containing vhs (lanes vhs) and RRL control (lanes Retic). Samples recovered at the indicated times (minutes) were then analyzed by agarose-formaldehyde gel electrophoresis. Numbers to the left of panel B indicate the sizes of RNA markers (lane M) in nucleotides.
If, as argued above, vhs-induced decay proceeds through endoribonucleolytic cleavage, novel 5′ and 3′ RNA termini should be generated at each of the putative sites of endonucleolytic cleavage. We tested this prediction by high-resolution analysis of the events at the extreme 5′ end of SRPα RNA. As shown in Fig. 2D, the four 5′-most vhs-induced digestion products generated from cap-labeled SRPα RNA migrate on a sequencing gel with apparent lengths of 30, 32, 38, and 39 nt when measured against DNA size markers. The 30- and 38-nt products were the most prominent of these. Taking the 5′ cap into account, these data localize putative sites of endonucleolytic cleavage to positions +29, +31, +37, and +38. To test this interpretation, we asked if we could detect, by primer extension, the predicted novel 5′ termini of the 3′ products produced by cleavage at these sites. Unlabeled capped SRPα RNA was added to RRL containing pretranslated vhs, and samples extracted at various times were analyzed by primer extension with a 32P-labeled oligonucleotide complementary to nt 60 to 84 of the SRPα mRNA (Fig. 3A). We detected major primer extension products of 55, 53, and 48 nt and minor products of 56 and 47 nt (Fig. 3A). The precise lengths of these products were determined by comparison to a DNA sequencing ladder produced with the same primer (data not shown). These novel 5′ ends would correspond to endoribonucleolytic cleavage events at positions +29, +31, and +38 (major) and +28 and +39 (minor) on SRPα mRNA. The excellent agreement between the two data sets provides a very strong indication that vhs-induced RNA degradation activity occurs through an endonucleolytic process. The minor discrepancies between the relative abundance of the products detected by these two assays might stem from exonucleolytic fraying of some of the 3′ and 5′ ends generated by these endonucleolytic cleavages. The positions of the novel 5′ ends detected by primer extension are displayed on the sequence of the SRPα transcript in Fig. 3B. Interestingly, all five of these 5′-most cleavages occur within GA or AG dinucleotides. Further experiments are required to determine if the vhs-induced activity displays marked sequence specificity.
FIG. 3.
Primer extension analysis of the 5′-most degradation products of SRPα RNA. (A) Unlabeled, capped SRPα RNA was added to RRL-vhs (lanes vhs) or RRL control (lanes Retic), and RNA reaction products were recovered at the indicated time points (minutes). The RNA reaction products were then analyzed by primer extension with 5′-32P-labeled oligonucleotide complementary to nt 60 to 84 of the SRPα RNA. Primer extension products were resolved on an 8% polyacrylamide sequencing gel and detected by autoradiography. Numbered arrows indicate the positions of primer extension products representing vhs-induced novel 5′ ends. Numbers to the right of lane M (marker) indicate the sizes of DNA markers in nucleotides. (B) Sequence of the extreme 5′ 84 nt of SRPα RNA. Numbered arrows above the sequence correspond to those in panel A and indicate the positions of vhs-induced cleavage at the 5′ end of SRPα RNA. The arrow under the sequence indicates the position of the oligonucleotide primer used.
vhs induces degradation of a variety of RNA substrates and displays activity in an in vitro translation system derived from HeLa cells.
vhs induces global inhibition of cellular protein synthesis during HSV infection and destabilizes both viral and cellular mRNAs in vivo (10, 13–15, 18–21, 28–31, 34, 36, 42, 46–49). To determine if vhs displays a similar lack of selectivity in our in vitro system, we compared the overall degradation profiles of internally labeled uncapped in vitro transcripts encoding SRPα (2.4 kb), hSHIP (4.5 kb), and vhs itself (Fig. 4). In addition, we tested an antisense transcript of the SRPα ORF (Fig. 4A, SRPα antisense; 2.2 kb). These RNA substrates are entirely unrelated in sequence, with the exception that the vhs and the SRPα RNAs share 69 nt at their extreme 5′ ends, upstream of the respective ORFs. All of these transcripts were markedly destabilized in RRL containing pretranslated vhs (Fig. 4). We have not yet characterized the mode of degradation of these additional RNAs in detail and therefore do not know if they contain preferred sites for initial cleavage such as those detected above in the SRPα transcript.
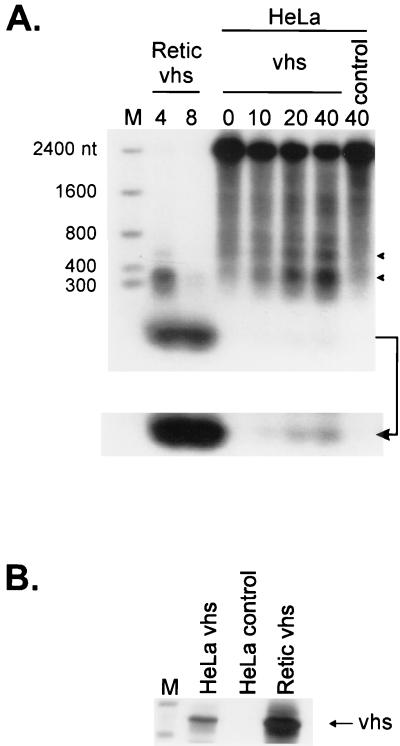
Krikorian and Read have shown that extracts of infected HeLa cells display vhs-dependent RNA-destabilizing activity in vitro (19). It was therefore of interest to determine if vhs induces accelerated RNA turnover when it is expressed as the only HSV protein in an in vitro translation system derived from HeLa cells and, if so, whether the mode of RNA degradation resembles that observed in the RRL system. As shown in Fig. 5, cap-labeled SRPα RNA was less stable in HeLa cell translation extracts containing pretranslated vhs than in control extracts lacking vhs. Moreover, some of the vhs-dependent degradation intermediates observed in the HeLa extract displayed electrophoretic mobilities similar to those obtained in the RRL system. However, the overall rate of vhs-induced RNA decay was substantially lower in the HeLa extract than in the RRL system. This difference is most probably due to the large difference in the amount of vhs protein produced in the two in vitro translation systems (Fig. 5B). These data demonstrate that vhs suffices to induce accelerated RNA turnover when it is expressed as the only HSV protein in extracts derived from human cells and offer a preliminary suggestion that the mode of vhs-induced RNA decay may be similar in the two systems.
FIG. 5.
vhs synthesized in a HeLa cell translation extract induces degradation of SRPα RNA. (A) HeLa cell translation extracts and RRL were programmed with capped unlabeled vhs RNA, and translation was allowed to proceed for 60 min in the presence of [35S]methionine. Extracts were then challenged with cap-labeled SRPα RNA, and samples were recovered at the indicated times (minutes). Reaction products were then analyzed by agarose-formaldehyde gel electrophoresis as in Fig. 1B. The bottom of panel A shows an overexposure of the lower portion of the membrane in panel A (indicated by an arrow). Lane control, HeLa cell extracts lacking vhs RNA. Numbers to the left of panel A indicate the sizes of RNA markers (lane M) in nucleotides. Solid arrowheads indicate the positions of vhs-induced RNA degradation products. (B) Samples of the translation reaction products used in panel A were resolved on an SDS–12% polyacrylamide gel, and the 35S signal was detected by autoradiography.
vhs-induced RNA degradation does not require a 5′ cap or a 3′ poly(A) tail in the RNA substrate.
Although vhs appears to destabilize most if not all mRNAs in infected cells, rRNA is not degraded (19, 20, 30, 52). This observation raises the possibility that some feature(s) specific to mRNA molecules renders them susceptible to vhs-induced cleavage. Two obvious candidates are the 5′ cap structure and the 3′ poly(A) tract. To investigate whether the presence of a 5′ cap influences vhs-induced RNA degradation, we compared the rate and mode of degradation of capped and uncapped SRPα RNA (Fig. 6). In the first experiment, equal amounts of unlabeled SRPα RNA bearing a 5′ triphosphate terminus (uncapped) or one of three different cap structures (GpppG, 7mGpppG, and 7mGpppmG) were added to RRL containing vhs and RNA samples recovered at the indicated times were analyzed by primer extension as in Fig. 3 (Fig. 6A). The results indicated that the presence of a cap structure did not greatly influence the rate of cleavage at the extreme 5′ end of the substrate. The level of background vhs-independent cleavages was greater in this experiment than in that in Fig. 3. We also compared the overall degradation profile of internally labeled uncapped and 7mGpppG-capped SRPα RNA (Fig. 6B). Again, the presence of a 5′ cap structure did not greatly alter the rate of RNA decay or the nature of the degradation intermediates. Consistent with these findings, we found that vhs activity was not altered when the cap binding protein eukaryotic initiation factor 4E (eIF4E) was depleted from the extracts by using 7mGTP-Sepharose resin (data not shown).
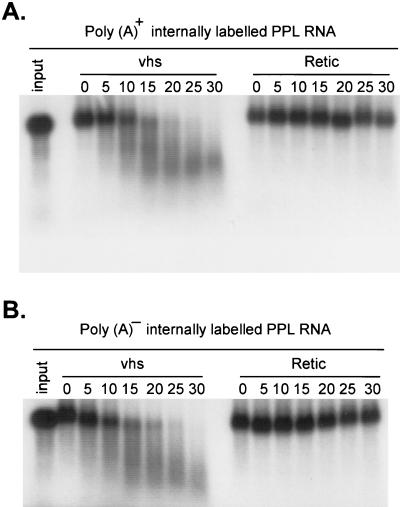
None of the RNA substrates examined above contained a 3′ poly(A) tail, indicating that this feature is not required for substrate recognition. To directly assess whether the presence of a 3′ poly(A) tract alters the rate of the reaction, we compared the degradation profile of internally labeled uncapped bovine PPL RNA containing a 35-nt poly(A) tract (Fig. 7A) to that of a derivative lacking the poly(A) tract (Fig. 7B). The presence of the poly(A) tail had little or no effect on the rate or course of reaction.
FIG. 7.
vhs-induced RNA degradation is not influenced by a 3′ poly(A) tail. Internally labeled PPL RNA containing (A) or lacking (B) a ∼35-residue poly(A) tail followed by GU was incubated with RRL vhs (lanes vhs) and RRL control (lanes Retic) for the indicated times (minutes). RNA reaction products were then analyzed by agarose-formaldehyde gel electrophoresis as in Fig. 1. Lanes 1. input, untreated RNAs.
Taken in combination, these data demonstrate that neither the 5′ cap nor the 3′ poly(A) tail detectably influence vhs-induced decay of substrate RNAs in this RRL-based system.
vhs-induced RNA degradation occurs in the absence of ribosomes, and requires magnesium.
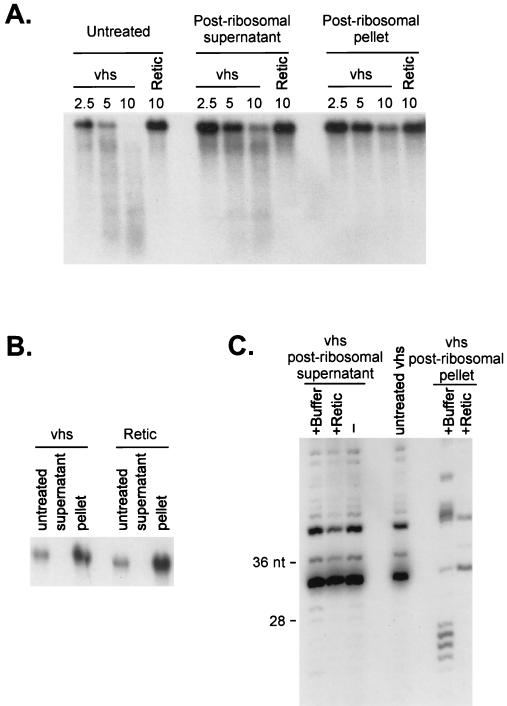
vhs-induced mRNA decay in HSV-1-infected cells occurs in the presence of drugs that block translational initiation and elongation, suggesting that substrate mRNAs need not be engaged in ongoing translation in order to be degraded (13, 39). Furthermore, Sorenson et al. (43) reported that vhs-dependent RNA degradation activity partitions with the postribosomal fraction of extracts of HSV-1-infected cells. To determine if ribosomes are required to recruit vhs activity to substrate RNAs in our in vitro system, we removed the ribosomes from the RRL (after translating vhs) by centrifugation at 160,000 × g for 50 min. Northern blot analysis revealed that all of the 18S rRNA was removed from the postribosomal supernatant (Fig. 8B), confirming that the procedure effectively depleted the extract of ribosomes. The postribosomal supernatant and ribosomal pellet were then assayed for vhs activity, using uncapped internally labeled SRPα RNA as the substrate (Fig. 8A). This experiment indicated that the vhs activity was associated predominantly with the postribosomal fraction. This conclusion was confirmed in a experiment where the postribosomal supernatant and ribosomal pellet were assayed for activity on 5′-cap-labeled SRPα RNA and the reaction products were displayed on an 8% polyacrylamide sequencing gel (Fig. 8C).
FIG. 8.
vhs-induced RNA degradation does not require ribosomes. RRL containing (vhs) or lacking (Retic) vhs were centrifuged at 160,000 × g for 50 min at 4°C to pellet the ribosomes. The ribosomal pellet was resuspended in Retic buffer (1.6 mM Tris acetate [pH 7.8], 80 mM potassium acetate, 2 mM magnesium acetate, 0.25 mM ATP, 0.1 mM DTT). (A) Untreated lysates, postribosomal supernatants, and pellets were mixed with internally labeled SRPα RNA, and samples recovered at various times (minutes) were analyzed by agarose-formaldehyde gel electrophoresis as in Fig. 1B. (B) A Northern blot analysis of the postribosomal supernatant and pellet fractions was performed with a rabbit 18S rRNA-specific 5′-32P-labeled oligonucleotide probe. (C) Cap-labeled SRPα RNA was added to untreated RRL vhs and to postribosomal supernatants and pellets that had been mixed with an equal volume of Retic buffer (+buffer) or naive RRL (+Retic). RNA samples were recovered after 10 min and then resolved on an 8% polyacrylamide sequencing gel.
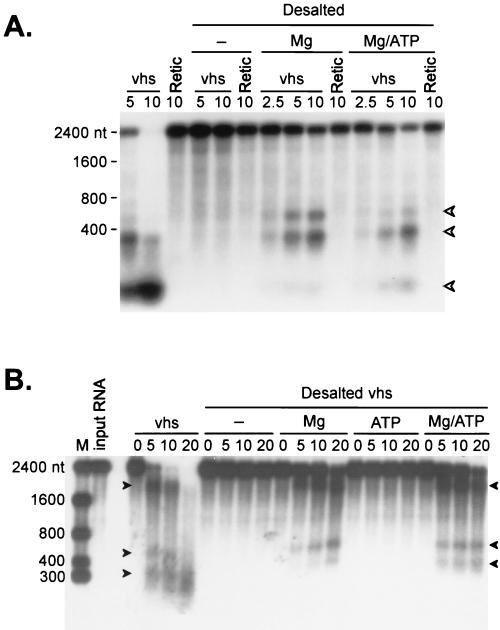
Kirkorian and Read (19) reported that the vhs-dependent RNA degradation activity observed in extracts of HSV-1-infected cells requires Mg2+ but not ATP. We examined the ATP and Mg2+ requirements of the RRL system. Small molecules were removed from lysates (after translating vhs) by passage over a Sephadex G-25 spin column. The resulting desalted extracts were then assayed for vhs activity before and after reconstitution with 0.25 mM ATP and/or 2 mM magnesium acetate. In the experiment in Fig. 9, cap-labeled SRPα (Fig. 9A) and uncapped internally labeled SRPα (Fig. 9B) RNAs were used as substrates for vhs activity in intact, desalted, and desalted and reconstituted extracts. The desalted extracts were devoid of activity, which was partially restored by adding Mg2+ ions. ATP had no effect by itself (Fig. 9B) but marginally increased the rate of the reaction when it was added in combination with Mg2+ ions (Fig. 9). The overall reduction in activity after desalting may be due to dilution effects and loss of some of the vhs protein. Alternatively, it is possible that additional cofactors are required for optimal activity. These data demonstrate that the vhs-induced activity requires Mg2+ ions (as previously reported [19]) and suggest that ATP may accelerate the rate of RNA degradation in the presence of Mg2+ ions.
FIG. 9.
vhs-induced RNA degradation requires magnesium. RRL containing (vhs) or lacking (Retic) vhs were desalted on Sephadex G-25 spin columns at 4 ml of packed resin per 100 μl of lysate. The resin was swollen in Retic buffer lacking magnesium acetate and ATP, loaded in a glass wool-plugged 5-ml syringe, and precentrifuged for 5 min at 4°C and 1,750 × g in a clinical centrifuge equipped with a swinging-bucket rotor. Samples of the desalted lysates were then combined with and equal volume of Retic buffer containing 4 mM magnesium acetate (lanes Mg), 0.5 mM ATP (lanes ATP) or 4 mM magnesium acetate and 0.5 mM ATP (lanes Mg/ATP). Substrate SRPα RNA was then added, and samples withdrawn at the indicated times (minutes) were analyzed by formaldehyde-agarose gel electrophoresis. (A) Analysis of cap-labeled SRPα RNA. (B) Analysis of internally labeled SRPα RNAs. Arrowheads indicate the mobilities of some of the vhs-dependent RNA degradation intermediates. Numbers to the left of the panels indicate the sizes of RNA markers (lanes M) in nucleotides.
The PrV vhs homologue induces RNA degradation in vitro.
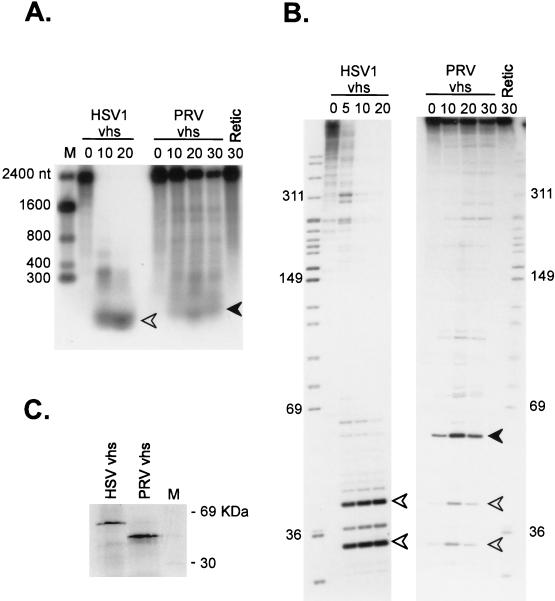
Homologues of vhs have been found in all of the alphaherpesvirus genomes characterized to date (for example, see reference 3). However, with the exception of the proteins encoded by HSV-1 and HSV-2, it is not yet clear if these vhs homologues trigger accelerated RNA turnover. To address this question, we asked if the vhs homologue of PrV (3) displays activity in the reticulocyte lysate system. Cap-labeled SRPα RNA was added to lysates containing pretranslated HSV-1 and PrV vhs, and the reaction products were analyzed on an agarose-formaldehyde gel (Fig. 10A), and an 8% polyacrylamide sequencing gel (Fig. 10B). We found that the RNA substrate was destabilized in lysates containing PrV vhs relative to the blank RRL control (Fig. 10A). However, the rate of induced decay was substantially lower than that provoked by HSV-1 vhs. Inasmuch as the PrV lysate contained at least as much vhs protein as the HSV-1 sample did (Fig. 10C), these data may indicate that the PrV vhs homologue displays reduced activity relative to its HSV-1 counterpart. Degradation induced by the PrV protein appeared to proceed through intermediates that were, for the most part, different from those induced by the HSV-1 vhs (Fig. 10A and B). The only common intermediates detected were the 30- and 40-nt 5′ fragments, which accumulated to a lesser extent in reaction mixtures containing Prv vhs (Fig. 10B).
FIG. 10.
PrV vhs displays RNA degradation activity in vitro. RRL were programmed with RNAs encoding HSV1 or PrV vhs, and translation was allowed to proceed for 60 min. Lysates were then challenged with cap-labeled SRPα RNA, and samples recovered at the indicated times (minutes) were analyzed by electrophoresis through an agarose-formaldehyde gel (A) and an 8% polyacrylamide sequencing gel (B). Numbers to the sides of the panels indicate the sizes of marker fragments in nucleotides (lanes M; RNA and DNA in panels A and B, respectively). Solid and open arrowheads indicate the positions of corresponding RNA fragments in the two different gel systems. (C) SDS-polyacrylamide gel electrophoresis of the HSV and PrV vhs proteins produced in the translation reactions in panels A and B.
DISCUSSION
Previous reports demonstrated that the HSV-1 vhs protein is necessary and sufficient to trigger accelerated RNA turnover in vivo and in extracts prepared from mammalian cells and partially purified HSV virions (18, 19, 32, 43, 52). Although highly informative, these studies left many basic questions about the mechanism of vhs action unanswered. In particular, the overall mode of vhs-induced RNA decay had not been defined. We therefore conducted a detailed examination of the mechanism of vhs-dependent mRNA degradation in an RRL-based in vitro system (52).
Our data establish that vhs-induced RNA decay proceeds (at least in part) through endoribonucleolytic cleavage events (Fig. 2 and 3). The strongest evidence supporting this conclusion emerged from a high-resolution analysis of the vhs-dependent events at the extreme 5′ end of SRPα RNA, where we were able to detect matching novel 5′ and 3′ termini at several of the putative sites of endonucleolytic cleavage (Fig. 2 and 3). Our conclusion that vhs induces endoribonucleolytic cleavage is in accord with a previous suggestion that the vhs-dependent RNase present in virion extracts acts as an endonuclease (52). However, as acknowledged by the authors, the data presented in that earlier report did not definitively establish this point.
vhs destabilizes mRNAs but not rRNAs in vivo (19, 30, 31, 52), an observation that raises the possibility that one or more features common to most mRNAs [such as the 5′ cap structure or 3′ poly(A) tail] specifically target mRNAs for selective degradation. However, we found that the in vitro reaction was not detectably influenced by the presence of a 5′ cap or 3′ poly(A) tail in the RNA substrate. These observations indicate that neither of these two characteristic features of mRNAs plays a major role in substrate recognition in our in vitro system. Moreover, the majority of the endoribonuclease activity partitioned with the postribosomal fraction, arguing against the possibility that ribosomes serve to selectively deliver the nuclease to mRNAs. The cap and ribosome independence of our in vitro system mirrors earlier data indicating that the vhs-dependent RNase present in virion extracts is cap independent (52) and that the RNA-destabilizing activity detected in extracts of HSV-infected HeLa cells partitions with the postribosomal fraction (43). In addition, our observation that poly(A)+ and poly(A)− RNA substrates are degraded at comparable rates in vitro is consistent with an earlier report that histone H3 and H4 mRNAs [which lack poly(A) tails (25)] are destabilized in HSV-infected cells (39). Thus, the basis for the apparently selective destruction of mRNAs in vivo remains obscure. Perhaps, as suggested by Zelus et al. (52), mRNAs are selectively targeted in vivo because they are relatively free of secondary structure and are packaged into ribonucleoprotein structures that are more accessible to nuclease attack than ribosomes.
As reviewed in the introduction, the currently available evidence strongly suggests that vhs either is an RNase or serves as a required subunit of an RNase that also contains one or more cellular subunits. Our finding that the HSV-1 and PrV vhs homologues induce the formation of partially overlapping yet distinct sets of 5′-terminal degradation products of SRPα RNA is consistent with this suggestion, since it demonstrates that the choice of cleavage sites depends on the nature of the vhs protein. A definitive determination of whether vhs has RNase activity in the absence of cellular proteins will require the purification of biologically active vhs to homogeneity.
Our detailed characterization of the degradation intermediates of SRPα RNA indicated that many of the most prominent sites of initial cleavage induced by HSV-1 vhs are clustered over the 5′ quadrant of this RNA, in an interval extending from approximately nt 200 to 700 from the 5′ end. The basis for this apparent clustering remains unknown, and it is not yet clear whether many or all RNA substrates display a similar profile. Apparently arguing against this possibility, Zelus et al. (52) suggested that the vhs-dependent RNase activity present in virion extracts preferentially cleaves globin mRNA close to the 3′ end. High-resolution mapping of the cleavage sites at the extreme 5′ end of SRPα RNA provided little evidence of sequence specificity, aside from a tendency to cleave between purine residues (Fig. 3). Moreover, the data of Zelus et al. (52) suggest that cleavage can also occur within AC and CU dinucleotides. We have recently mapped a number of additional cleavage sites at high resolution (8a). Of 30 sites analyzed (including the 4 indicated in Fig. 3), 15 correspond to AG or GA dinucleotides (the others are 6 GC, 5 UG, and 1 each of GU, UU, UC, CG, and AC). Thus, the available data argue that the vhs-induced endoribonuclease displays a relatively relaxed sequence specificity. This suggests that other features, such as RNA secondary structure, may play a major role in defining the sites of preferential cleavage.
vhs displays limited but significant amino acid homology to the fen-1 family of nucleases (8) that are involved in DNA replication and repair (reviewed in reference 24). Although fen-1 was initially identified as a DNase, Stevens (44) recently reported that it also cleaves RNA substrates in a cap-independent reaction that requires magnesium. The preferred sites of cleavage in several RNA substrates are confined to the 5′-most 200 nt of the transcript and are located at the 5′ base of predicted stem-loop structures. Based on these results, Stevens (44) proposed that fen-1 loads onto the 5′ end of the RNA and then tracks in a 3′ direction until it encounters secondary-structure elements that trigger cleavage. Similarly, it is possible that the vhs-dependent endoribonuclease loads at one or both ends of the RNA and migrates until it encounters preferred cleavage determinants. Although the vhs-dependent cleavage sites that we have mapped at the extreme 5′ end of SRPα RNA do not obviously correlate with predicted features of RNA secondary structure (data not shown), we have not yet precisely mapped the more prominent sites of initial cleavage that are located ca. 200 to 700 nt from the 5′ end of the RNA. In this context, we have recently found that inserting a picornavirus internal ribosome entry site (IRES) at a variety of sites throughout the SRPα transcript provokes novel vhs-dependent endoribonucleolytic cleavage events in the sequences located immediately downstream of the inserted IRES (8a). Inasmuch as IRES elements exhibit extensive secondary structure (27, 33, reviewed in reference 38), this observation is consistent with (but does not prove) the hypothesis that secondary structure may play a major role in dictating the sites of vhs-induced cleavage.
Our data strongly suggest that vhs-dependent RNA decay is initiated by endoribonucleolytic cleavage and establish that 5′ fragments produced early during the reaction can be recleaved by the vhs-dependent endonuclease as the reaction proceeds (Fig. 2). However, they do not exclude the possibility that other cellular endo- and exoribonucleases also contribute to the subsequent decay of the primary cleavage products, particularly in vivo. Thus, as previously proposed (32, 43, 52), it is possible that vhs achieves translational arrest by endonucleolytic cleavage of mRNAs at limited number of sites and that the products of these initial cleavages are then further processed by existing cellular mRNA surveillance pathways (reviewed in reference 1. An analogous pathway regulates the stability of human transferrin receptor mRNA and Xenopus Xlhbox2B and Xoo1 mRNAs (4, 5). These mRNAs undergo endonucleolytic cleavage in their 3′ UTRs, generating unstable 5′ and 3′ products that are probably further processed by cellular exonucleases. These initiating endonucleolytic cleavages resemble the vhs activity described in this report, in that they do not require a 5′ cap structure, 3′ poly(A) tail, or ongoing translation. This model might help explain how the limited amount of vhs delivered by the infecting HSV virion is able to trigger global shutoff of host protein synthesis.
The PrV vhs homologue shares most of the amino acid sequences that are conserved among the vhs proteins encoded by alphaherpesviruses (3, 18). Despite this conservation, host shutoff induced by infection with PrV requires de novo viral protein synthesis (2, 17), suggesting that PrV vhs is either less active than its HSV-1 counterpart or not packaged into virions. Our finding that PrV vhs induces RNA decay in vitro substantially less efficiently than its HSV-1 counterpart strongly argues for the former possibility and illustrates the sensitivity of our in vitro assay.
Our findings provide a glimpse into the mechanism of vhs activity and begin to define some of the requirements of the vhs-induced RNA degradation reaction. Further studies of the vhs-induced RNA decay pathway may ultimately lead to a better understanding of analogous cellular pathways.
ACKNOWLEDGMENTS
We thank Joanne Duncan and Carol Lavery for superb technical assistance, David Andrews for many gifts of plasmids and advice on in vitro translation systems, Peter Whyte for pBlueK(coreD), Alberto Epstein for pPRV41, and Evan Llewelyn for help constructing the PrV vhs expression vector.
This work was supported by a grant from the National Cancer Institute of Canada [NCI(C)]. J.R.S. was a Terry Fox Senior Scientist of the NCI(C).
REFERENCES
- 1.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Porat T, Rakusanova T, Kaplan A S. Early functions of the genome of herpesvirus. II. Inhibition of formation of cell-specific polysomes. Virology. 1971;46:890–899. doi: 10.1016/0042-6822(71)90089-4. [DOI] [PubMed] [Google Scholar]
- 3.Berthomme H, Jacquemont B, Epstein A. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology. 1993;193:1028–1032. doi: 10.1006/viro.1993.1221. [DOI] [PubMed] [Google Scholar]
- 4.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown B D, Zipkin I D, Harland R M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 6.Carroll R, Lucas-Lenard J. Preparation of a cell-free translation system with minimal loss of initiation factor eIF-2/eIF-2B activity. Anal Biochem. 1993;212:17–23. doi: 10.1006/abio.1993.1284. [DOI] [PubMed] [Google Scholar]
- 7.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Elgadi, M. M., and J. R. Smiley. Picornavirus IRES elements target RNA cleavage events induced by the herpes simplex virus vhs protein. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 9.Falcone D, Andrews D W. Both the 5′ untranslated region and the sequence surrounding the start site contribute to efficient initiation of translation in vitro. Mol Cell Biol. 1991;11:2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenwick M L, Clark J. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J Gen Virol. 1982;61:121–125. doi: 10.1099/0022-1317-61-1-121. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 12.Fenwick M L, Everett R D. Transfer of UL41, the gene controlling virion-associated host cell shutoff, between different strains of herpes simplex virus. J Gen Virol. 1990;71:411–418. doi: 10.1099/0022-1317-71-2-411. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 14.Fenwick M L, Owen S A. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J Gen Virol. 1988;69:2869–2877. doi: 10.1099/0022-1317-69-11-2869. [DOI] [PubMed] [Google Scholar]
- 15.Fenwick M L, Walker M J. Suppression of synthesis of cellular macromolecules by herpes simplex virus. J Gen Virol. 1978;41:37–51. doi: 10.1099/0022-1317-41-1-37. [DOI] [PubMed] [Google Scholar]
- 16.Honess R, Roizman B. Regulation of herpes macromolecular synthesis I. Cascade regulation of synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihara S, Feldman L, Watanabe S, Ben-Porat T. Characterization of the immediate-early functions of pseudorabies virus. Virology. 1993;131:437–454. doi: 10.1016/0042-6822(83)90510-x. [DOI] [PubMed] [Google Scholar]
- 18.Jones F E, Smibert C A, Smiley J R. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J Virol. 1995;69:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krikorian C R, Read G S. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 24.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 25.Marzluff W F, Pandey N B. Multiple regulatory steps control histone mRNA concentration. Trends Biochem Sci. 1988;13:49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- 26.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson R, Pelletier J, Le S Y, Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol. 1991;65:5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishioka Y, Silverstein S. Degradation of cellular mRNAs during infection by herpes simplex virus. Proc Natl Acad Sci USA. 1977;74:2370–2374. doi: 10.1073/pnas.74.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishioka Y, Silverstein S. Requirement of protein synthesis for the degradation of host mRNA in friend erythroleukemia cells infected with herpes simplex virus type 1. J Virol. 1978;27:619–627. doi: 10.1128/jvi.27.3.619-627.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 34.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate-early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–60. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roizman B, Borman G S, Rousta M-K. Macromolecular synthesis in cells infected with herpes simplex virus. Nature. 1965;206:1374–1375. doi: 10.1038/2061374a0. [DOI] [PubMed] [Google Scholar]
- 37.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 38.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 39.Schek N, Bachenheimer S L. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of vero cells. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 41.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smibert C A, Smiley J R. Differential regulation of endogenous and transduced beta-globin genes during infection of erythroid cells with a herpes simplex virus type 1 recombinant. J Virol. 1990;64:3882–3894. doi: 10.1128/jvi.64.8.3882-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens A. Endonucleolytic cleavage of RNA at 5′ endogenous stem structures by human flap endonuclease 1. Biochem Biophys Res Commun. 1998;251:501–508. doi: 10.1006/bbrc.1998.9499. [DOI] [PubMed] [Google Scholar]
- 45.Strelow L I, Leib D A. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J Virol. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strom T, Frenkel N. Effect of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sydiskis R J, Roizman B. The disaggregation of host polyribosomes in productive and abortive infection with herpes simplex virus. Virology. 1967;32:678–686. doi: 10.1016/0042-6822(67)90043-8. [DOI] [PubMed] [Google Scholar]
- 48.Sydiskis R J, Roizman B. Polysomes and protein synthesis in cells infected with a DNA virus. Science. 1966;153:76–78. doi: 10.1126/science.153.3731.76. [DOI] [PubMed] [Google Scholar]
- 49.Sydiskis R J, Roizman B. The sedimentation profiles of cytoplasmic polyribosomes in mammalian cells productively and abortively infected with herpes simplex virus. Virology. 1968;34:562–565. doi: 10.1016/0042-6822(68)90075-5. [DOI] [PubMed] [Google Scholar]
- 50.Wagner E K, Guzowski J F, Singh J. Transcription of the herpes simplex virus genome during productive and latent infection. Prog Nucleic Acid Res Mol Biol. 1995;51:123–165. doi: 10.1016/s0079-6603(08)60878-8. [DOI] [PubMed] [Google Scholar]
- 51.Young J C, Ursini J, Legate K R, Miller J D, Walter P, Andrews D W. An amino terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor alpha subunit to the beta subunit on the endoplasmic reticulum membrane. J Biol Chem. 1995;270:15650–15657. doi: 10.1074/jbc.270.26.15650. [DOI] [PubMed] [Google Scholar]
- 52.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]