Abstract
Background:
Toxoplasmosis is a parasitic disease caused by compilation protozoan agent Toxoplasma gondii, leading to significant financial and quality-adjusted life-year losses. Overcooked or raw meat consumption has been a considerable transmission route. The present study was conducted to determine the seropositivity rate of T. gondii in sheep and goats by serological and molecular tests and genotyping of obtained isolates in northeast Iran.
Methods:
Blood and tissue samples (diaphragm, heart) of 296 animals (including 168 sheep and 128 goats) were collected from the slaughterhouse in Quchan Country from august 2016 to April 2017. Modified agglutination test (MAT) and the PCR method performed to detect parasite DNA on tissues.PCR-RFLP method of GRA6 gene was used to determine the genotype of T. gondii. In addition, sequencing analysis was performed to evaluate the Toxoplasma type strains.
Results:
Serum positive for MAT results were found in 27.4% of sheep and 23.4% of goats. Positive PCR of B1 gene results in diaphragm and heart tissues of sheep and goats was 47.8% and 26.1%, 40% and 23.3%, respectively. PCR of GRA6 gene results were positive in 10 samples that RFLP technique results using MseI enzyme revealed genotype I. Sequencing and phylogenetic analysis revealed DNA of all samples was closely related to Toxoplasma type I.
Conclusion:
Concerning the high seropositivity rate of toxoplasmosis, undertaking an appropriate preventive program for reducing the prevalence of T. gondii infection by raw or undercooked meat consumption of livestock is recommended. Our study supports the notion that these animals' consumption of raw and undercooked meat can be a probable source of human toxoplasmosis.
Keywords: Toxoplasma gondii, Sheep, Goat, MAT, Genotyping, Iran
Introduction
Toxoplasma gondii is an obligate intracellular protozoan widely prevalent in humans and other animals (1,2). Felines, mainly cats, are definitive hosts in the life cycle of T. gondii and excrete millions of resistant oocysts after primary infection into the environment. Almost all warm-blooded plays a role in transmission cycle as intermediate hosts such as sheep, goat, cattle, pigs and camels or aberrant hosts as humans (3).
Even though most cases of human infection are asymptomatic or mild clinical symptoms, the parasite can cause severe complications such as encephalitis in congenitally infected children and immunocompromised individuals (4,5). Reactivations of latent infection in immunocompromised individuals can cause fatal toxoplasmic encephalitis, pneumonitis and myocarditis. Acquired infections during pregnancy are associated with severe damage to the fetus including stillbirths or abortions. Humans are usually infected by consuming undercooked meat containing tissue cysts or cyst contaminated water (6,7).
T .gondii is broadly spread among farm animals and humans. Overall assessed frequency is reported with variable seroprevalence rates of 75% in dogs, 11–36% in pigs,11–61% in goats, less than 10% in cows, 35–73% in cats, and 35–73% in humans (8,9). In comparison, this value in Iran and humans has been reported 29% to 55% (10). In Khorasan Razavi Province, the seroprevalence rate of toxoplasmosis in sheep was found 15.5% (11). Also, another recent study in Sabzavar City in Khorasan Razavi Province revealed that 60% of sheep, 52.5% of goats and 65% of camels were infected by T. gondii.(12).
According to the published data related to the census of animal husbandry in Iran, (https://www.amar.org.ir/), Khorasan Razavi Province is the most important provinces in livestock breeding, and Quchan City is the animal husbandry hub of this province. Therefore, it is essential to evaluate the prevalence of T. gondii in livestock as humans’ food. Lack of accurate and comprehensive data concerning to livestock prevalence of toxoplasmosis in this area is the main obstacle to control and design preventive plan. Therefore, the present study was conducted to survey the frequency of T. gondii in livestock meat (sheep and goats) by using Modified Agglutination Test (MAT) test and compare with PCR methods as well as to determine genetically diversity infecting strains of T. gondii.
Materials and Methods
Ethics approval
The study was approved by the Ethics Committee of the Kerman University of Medical Sciences in Iran (Ethical number: 94/389).
Study area
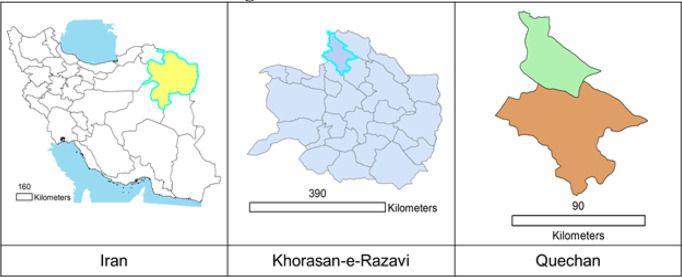
This Cross-Sectional study that was conducted in Quchan City in KhorasanRazavi Province in northeastern Iran. It has an area of about 523400 hectares and a population of over 180,000 peoples. The city is located in a mountainous area, elevation 1149 meters above sea level and has cold winters and mild summers. Its rainfall is 200–150mm/year and lies between 37.11° latitude and 58.51 ° E longitude.(Available at https://www.worldatlas.com/as/ir/30/where-is-quchan.html, https://en.wikipedia.org/wiki/Quchan#Geography). Fig.1 shows ArcGIS geographic location of Iran and Quechan.
Fig. 1:
The situation of Khorasan Razavi Province in Iran and location of study area in Quchan
Sample collection
Based on statistical advice and previous studies, 296 animals including 168 sheep and 128 goats were sampled from the slaughterhouse in Quchan City for this study. This descriptive cross-sectional study was implemented from August 2016 to April 2017 and samples were collected in four stages in the spring, summer, autumn and winter seasons.
According to the seasonal pattern, 65, 68, 104 and samples were collected in spring; summer, autumn in winter respectively. Livestock was numbered and randomly selected, whereas their blood samples (jugular vein) were obtained from numbered livestock. At the same time, age and gender were also recorded. We had from each livestock a blood sample, a heart sample and a diaphragm sample. Blood samples were centrifuged without anticoagulant tubes at 8000 rpm for 5–10 minutes and sera were transferred to 1.5 ml micro-tubes. Until to performing serological and molecular tests, all sera and tissue samples were stored at −20 degree.
Serological examination
Sera of sheep and goats were examined for anti- Toxoplasma gondii antibodies by the modified agglutination test (MAT) (Toxo screen DA, bi-omerieux®, France) as described by Dubey and Desmonts(13).
DNA extraction
All positive samples of MAT test were investigated by PCR assay on the heart and diaphragm the same animals. The DNA extraction was performed using (Gene All, Exgene, Cell SV mini, Korea) kit and according to the manufacturer`s instruction.
Nested-PCR for B1 gene
The Nested-PCR assays were accomplished using two repeated genomic targets, B1 to detect T. gondii DNA in contaminated tissues (13). Two PCR primer pairs of the B1 gene, S1 (5′-CGACAGAAAGGGAGCAAGAG-3′) and AS1 (5′-ACGCTGTGTCTCCTCTAGGC-3′), S2 (5′-TCTTCCCAGACGTGGATTTC-3′) and AS2 (5′-CTCGACAATACGCTGCTTGA-3′), eventually amplifying a 531 bp fragment were used. The first amplification was carried out in 20 μl of reaction mixture containing 1 μl of each primer (S1 and AS1), 10 μl Master mix (Ampliqon Company, Denmark), 2 μl extracted DNA from heart or diaphragm samples and 6μl Distilled water sterilized. The first PCR was performed in a thermocycler (Flex Cycler) for initial denaturation at 94 C for 3 min, this step was followed by 35 cycles of denaturation at 94 degree for 30 s, annealing at 60 degree for 30 s, extension at 72 degree for 2 min and a final extension step at 30 degree for 1 min. The second amplification was performed in 20 μl reaction mixture. The first PCR product was diluted with a ratio of 1:40 to distilled water and then used as a template. Twenty-μl reaction mixture was containing 1 μl of each primer (S2 and AS2), 8μl Master mix (Ampliqon Company, Denmark), 1 μl of our new template and 9μl distilled water sterilized. The second PCR was performed in 30 cycles.
Nested PCR for GRA6 gene
The positive samples of Nested-PCR of B1 gene included in analyzing by Nested-PCR of GRA6 gene. GRA6, ahighly polymorphicgene is repeated in the genome of the T. gondii. This gene is suited to distinguish between three typesI, II and III from each other, primarily type III that is close to type I. Two PCR primer pairs of the GRA6 gene, GRA6FO (5′GGCAAACAAAACGAAGTG-3′) and GRA6RO (5′-CGACTACAAGACATAGAGTG-3′) used in first amplification, and GRA6R (5′- GTAGCGTGCTTGTTGGCGAC-3′) and GRA6 (5′TACAAGACATAGAGTGCCCC-3′) used in second amplification. The first amplification was carried out in 25 μl of reaction mixture containing 1 μl of each primer (GRA6FO and GRA6RO), 8μl Master mix (Ampliqon Company, Denmark), 5 μl extracted DNA of heart or diaphragm samples and 10μl Distilled water sterilized. The first PCR was performed in a thermocycler (Flex Cycler) for initial denaturation at 94 degree for 5 min, this step was followed by 35 cycles of denaturation at 94 degree for 30 s, annealing at 54 degree for 60 s, extension at 72 degree for 90 s and a final extension step at 72 degree for 7 min[14].The second amplification was performed in 25 μl reaction mixture. The first PCR product used as a template while diluted with a ratio of 1:10 to distilled water. Twenty-five microlitres reaction mixture contained 1 μl of each primer (GRA6R and GRA6), 8 μl Master mix, 1μl of our new template and 14 μl Distilled water sterilized.
The second PCR was performed at the annealing temperature of 60 degree for the 60s[15]. The PCR products were electrophoresed in a 1.5% agarose gel in tris-borate-EDTA 0.5X (TBE 0.5X) buffer and stained with ethidium bromide. To differentiate the three types (I, II, III) of T. gondii, all positive samples of Nested PCR for GRA6 gene were used to performing PCR-RFLP technique.
PCR-RFLP Assay
The GRA6 gene amplified product was digested with MseI with MseI restriction endonuclease (10 U/μl, 300 units), (Fermentas, Thermo Scientific, USA).
Sequencing
The GRA6 gene amplified product (with suitable quality in PCR-RFLP) was sent to Macrogen Company (South Korea) for sequence analysis and to obtain more accurate results from the genotype of the T. gondii (I, II, III). Outcomes were aligned with BioEdit and sequence Scanner program and compared to the following sequence data available from GenBank: AJ635332, AF239283, AF239292 and AF239284. The maximum-likelihood analysis was employed to estimate phylogenetic relationships among genotypes. Additionally, Mega6 and BioEdit software were used to construct the phylogeny tree to compare our collected isolates against types submitted in GenBank as well as to demonstrate homology of obtained sequences respectively.
Statistical Analysis
Differences in T. gondii prevalence with variables such as season, sex and age were analyzed using Pearson Chi-square test and crosstab. Statistical analysis was performed using SPSS version 23 (IBM Corp., Armonk, NY, USA). The P-values less than 0.05 were considered statistically significant.
Results
Serological, molecular and risk factor
T. gondii antibodies (MAT titers≥1:20) were found in 46 (27.4%) of 168 sheep and 30 (23.4%) of 128 goats (Table 1). The samples were assayed at dilution from 1:20 to 1:640 (Table 2). In addition, positive results were categorized at dilution of ≥1:20 based on the seasons, sex and age (Tables 3 and 4).
Table 1:
Toxoplasma infection in sheep and goats by MAT method at ≥1:20 dilution
| Animal | MAT | Total | Percent (%) | |
|---|---|---|---|---|
|
| ||||
| Positive | Negative | |||
| Sheep | 46 | 122 | 168 | 27.4 |
| Goats | 30 | 98 | 128 | 23.4 |
Table 2:
Toxoplasma infection in sheep and goats by MAT at 1:20 to 1:640 dilution
| Variable | Sheep | Goats | |
|---|---|---|---|
| MAT | 1:20 | 18 | 8 |
| 1:40 | 13 | 10 | |
| 1:80 | 9 | 8 | |
| 1:160 | 4 | 3 | |
| 1:320 | 1 | - | |
| 1:640 | 1 | 1 | |
| Total | 46 | 30 | |
Table 3:
Toxoplasma infection in sheep and goats by MAT method at ≥1:20 dilution based on seasons and sex
| Variable | Season | Sex | |||||
|---|---|---|---|---|---|---|---|
| Spring | Summer | Autumn | Winter | Male | Female | ||
| Sheep | Positive | 5 | 8 | 17 | 16 | 20 | 26 |
| Negative | 40 | 32 | 38 | 12 | 88 | 34 | |
| Total | 45 | 40 | 55 | 28 | 108 | 60 | |
| Seroprevalence (%) | 11.1 | 20 | 30.9 | 57.1 | 18.5 | 43.3 | |
| Goats | Positive | 2 | 5 | 13 | 10 | 15 | 15 |
| Negative | 18 | 23 | 36 | 21 | 65 | 33 | |
| Total | 20 | 28 | 49 | 31 | 80 | 48 | |
| Seroprevalence (%) | 10 | 17.9 | 26.5 | 32.3 | 18.8 | 31.3 | |
Table 4:
Toxoplasma infection in sheep and goats by MAT method at ≥1:20 dilution based on ages
| Age(yr) | Sheep | Goat | ||||
|---|---|---|---|---|---|---|
| MAT | Percent (%) | MAT | Percent (%) | |||
| Positive | Negative | Positive | Negative | |||
| <1 | 1 | 24 | 4 | 1 | 32 | 3 |
| 1–3 | 15 | 59 | 20.3 | 13 | 44 | 22.8 |
| 3–5 | 21 | 32 | 39.6 | 8 | 13 | 38.1 |
| >5 | 9 | 7 | 56.3 | 8 | 9 | 47.1 |
| Total | 46 | 122 | 27.4 | 30 | 98 | 23.4 |
The comparison of collected seropositivity data and different seasonal patterns in sheep indicate significant differences (P = 0.002), while there was no significant difference in goats. Our analyzed data showed a statistically significant difference between age associations and seropositivity in sheep and goats (P-value = 0.001). In addition, the results of statistical analysis showed a significant difference between sex associations with seropositive in sheep (P-value = 0.0001); whereas there was no significant difference in goat (P-value = 0.01).
Table 5 and Fig. 2 depict the results of Nested-PCR for B1 gene on tissue samples of the same animals with previously positive MAT reports. The analyzed data indicated a significant difference between serum dilutions and positive results of Nested-PCR for B1 gene in sheep and goats (P-value < 0.05). Table 6 and Fig.3 demonstrate the related results about Nested-PCR for GRA6 gene that was performed on positive samples of Nested-PCR of B1 gene. Fig. 4 exhibits the results PCR-RFLP technique (to determine the genotypes of T. gondi) that carry out on positive samples of Nested-PCR for GRA6 gene.
Table 5:
Results of Nested-PCR for B1 gene on the heart and diaphragm tissues that their MAT results at ≥1:20 dilution were positive
| Animal | No. | Tissue | No. tissue | No. positive samples at Nested-PCR with using B1 gene | Percent (%) infection in each tissue | No. infection in each animal at Nested-PCR with using B1 gene | Percent(%) in each animal |
|---|---|---|---|---|---|---|---|
| Sheep | 46 | Diaphragm | 46 | 22 | 47.8 | 27 | 58.7 |
| Heart | 46 | 12 | 26.1 | ||||
| Goats | 30 | Diaphragm | 30 | 12 | 40 | 16 | 53.3 |
| Heart | 30 | 7 | 23.3 |
Fig. 2:
Electrophoretic pattern of the PCR products of B1 gene (531bp) from tissue samples. Lines 1–6: diaphragm tissue of sheep, lanes 7–11: heart tissues of sheep, lanes 12–16: diaphragm tissues of goats, lanes 17&18: heart tissues of goats, lane 18: positive control, lane 19: negative control, lane M: DNA marker
Table 6:
Nested-PCR results for GRA6 gene on the positive samples of Nested-PCR of B1 gene
| Animal | No. | Tissue | No. tissue | No. positive samples at Nested- PCR with using GRA6 gene |
|---|---|---|---|---|
| Sheep | 27 | Diaphragm | 22 | 6 |
| Heart | 12 | 1 | ||
| Goats | 16 | Diaphragm | 12 | 3 |
| Heart | 7 | - |
Fig. 3:
Electrophoretic pattern of the PCR products of GRA6 gene from tissue samples. Lanes 1–6: diaphragm tissue of sheep, lane 7: heart tissues of sheep, lanes 8–11: diaphragm tissues of goats, lane 12: negative control, lane 13: positive control, lane M: DNA marker
Fig. 4:
PCR-RFLP analysis of GRA6 gene coding region with MseI endonuclease. Lane 9 is DNA marker, Lanes 1–8 are Toxoplasma gondii, type I (RH)
Additionally, our analyzed results represent a significant difference between dilutions of serums and positive results of Nested-PCR for GRA6 gene in sheep and goats (P-value =0.02).
Sequencing and phylogenetic analysis
As shown in Fig.5 phylogenetic analysis of 10 sequenced products confirmed that all isolates belonged to type1with high similarity in sister clade and their sequences are available in GenBank with accession numbers: MG976038 to MG976047. Homology of identified sequence compared with gene-bank sequences (Fig. 6).
Fig. 5:
The comparison of sequencing of GRA6 gene isolated from sheep and goats with gene bank sequences
Fig. 6:
The phylogenic tree was constructed by maximum likelihood method using the nucleotide sequence of reference strains and our isolates (indicated with colorful shapes behind them). The scale bar indicates a 2% nucleotide difference
Discussion
This study was conducted to determine the seropositivity and molecular detection of T. gondii in sheep and goats in northeastern Iran. Goats and sheep are the most important livestock in societies such as Iran and their products consider as main food sources for humans. Despite some progress in diagnosing and treating toxoplasmosis, the disease remains a significant public health and economic loss.
Based on a previous comprehensive study, maximum and minimum worldwide seropositivity of sheep was reported 4.4% (in China), 99.2% (in France)and the same result for goats was founded in ranged from 3.7 to 81% (16). In contrast, these values in Iran for sheep and goats were recorded between 13–35% and 13–30%, respectively [17].
Our study revealed that the seropositivity of Toxoplasma in sheep and goats was 27.4% and 23.4%, respectively. These values are in agreement with finding in various area of Iran including 18.8% and 29.5% in Fars province, 24.7% and 15.8% in Kerman province for sheep and goats respectively; as well as 21.1 % of sheep in Urmia(18–20). Comparing our findings with previous studies from other parts revealed more or fewer similarities/differences. For instance, in two studies of China serologic evidence of infection was found in 12.71% of sheep and 20.3% of goats (24,25). Additionally, in an effort by Dubey et al. to estimate seropositivity of 234 goats in USA, it was shown 53.4% of goats were positive (13).
It is well known that discrepancies in overall seropositivity results for toxoplasmosis in animals may be attributed to kind of used serological test, sample size, ecological status, management and hygienic standards and other factors. Generally, MAT was selected as a first choice method to evaluate seropositivity of Toxoplasma in animals, whereas in several studies molecular methods were used to determine the rate of toxoplasmosis in animals (18).
In this regard, our molecular results via B1 gene shows 58.7% and 53.3% of studied sheep and goats were infected with T. gondii and this is in agreement with the finding of 56.66% and 44.16 % for sheep and goats in Kerman province respectively (23). In line with our finding in Iran, 38% of studied sheep in Chaharmahal va Bakhtiary province were infected with Toxoplasma (18). As well as similar results were taken in the Fars Province in the investigation of 56 sheep and 22 goats tissue samples with total molecular prevalence of Toxoplasma was 33.3% (24).
Our molecular prevalence data are comparable with other worldwide reports; for example in Tunisia, 33.3% and 32.5% of B1 gene Nested PCR tests were positive for ewes and goats, respectively (25). Routinely, genotyping as a crucial determinant for pathogenesis and virulence of Toxoplasma was performed in recent studies. There was a correlation between of T. gondii genotype and clinical symptoms and pathogenesis profile of the parasite (26,27). Besides, genotype I known as acutely virulent, whereas genotype II and III are significantly less virulent that can establish latent toxoplasmosis (28).
In our study, the Nested-PCR of GRA6 gene for 10 tissue samples (7 sheep and 3 goats) was positive and RFLP technique approved all of them cluster into genotype I. These results are in agreement with two previous surveys of different parts of Iran (29,30). Predominant genotype in animal especially ruminants in Iran is genotype II; Whereas prior European investigation demonstrated the most predominant genotype belongs to type III (31). In our study, variables analysis in sheep and goats showed that infection rate of toxoplasmosis increased with age. This age-related variation can be because older animals have been exposed to risk factors for a longer time. In addition, infection rate in sheep is higher in dry than wet seasons. Likewise, female sheep has more chance of acquiring toxoplasmosis than male. Infection occurs more often in the wet season, and since IgG antibodies can persist a long time, so the high infection in dry season might be due to the carry-over effect from preceding wet season infections. Analysis of age and sex variables showed that seropositivity was higher in adults than younger animals and also higher in females than males (8).
The finding related to influence of age and seasons on seropositivity was verified by parallel researches (32,33). Collectively, discovering series factors such as seropositivity of toxoplasmosis, the biological properties of parasite and other risk factors in sheep and goats as main human foods are tied to establish efficient prevention and control health programs against toxoplasmosis.
Conclusion
This study can serve as a road map and valuable information source to clarify the quantitative risk assessment of toxoplasmosis in humans as a foodborne disease. The presence of T. gondii DNA in the tissues of sheep and goats from northeastern Iran implicates that meat consumption might pose the risk of human infection. This investigation depicts new perspectives about the genotyping map of T. gondii, which is an indispensable factor for evaluation of meaningful control and prevention strategies.
Acknowledgments
This study has been extracted from the thesis in the School of Medicine Kerman University of Medical Sciences (Grant No: 94/389). Authors would like to acknowledge all staff in parasitology and mycology department of Kerman University of Medical Sciences.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interests.
References
- 1.Dubey JP. Toxoplasmosis in pigs—the last 20 years. Vet Parasitol. 2009;164(2–4):89–103. [DOI] [PubMed] [Google Scholar]
- 2.Shojaee S, Firouzeh N, Keshavarz H, AZAMI SJ-P, Salimi M, Mohebali M. Nanosilver Colloid Inhibits Toxoplasma gondii Tachyzoites and Bradyzoites in Vitro. Iran J Parasitol. 2019;14(3):362–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Verhelst D, De Craeye S, Vanrobaeys M, Czaplicki G, Dorny P, Cox E. Seroprevalence of Toxoplasma gondii in domestic sheep in Belgium. Vet Parasitol. 2014;205(1–2):57–61. [DOI] [PubMed] [Google Scholar]
- 4.Opsteegh M, Teunis P, Züchner L, Koets A, Langelaar M, van der Giessen J. Low predictive value of seroprevalence of Toxoplasma gondii in cattle for detection of parasite DNA. Int J Parasitol. 2011;41(3–4):343–54. [DOI] [PubMed] [Google Scholar]
- 5.Teimouri A, Azami SJ, Keshavarz H, et al. Anti-toxoplasma activity of various molecular weights and concentrations of chitosan nanoparticles on tachyzoites of RH strain. Int J Nanomedicine. 2018;13:1341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarean M, Shafiei R, Gholami M, et al. Seroprevalence of anti–Toxoplasma gondii antibodies in healthy voluntary blood Donors from Mashhad City, Iran. Arch Iran Med. 2017;20(7):441–5. [PubMed] [Google Scholar]
- 7.Tavakoli kareshk A, Keyhani A, Asadi A, Zia-Ali N, Mahmoudvand H, Mohammadi AR. Seroprevalence of Toxoplasma gondii infection among childbearing age women in Kerman city, southeastern Iran. J Parasit Dis. 2016;40(4):1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegegne D, Abdurahaman M, Yohannes M. Seroepidemiology and associated risk factors of Toxoplasma gondii in sheep and goats in Southwestern Ethiopia. BMC Vet Res. 2016;12(1):280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonouhewa ABN, Akpo Y, Sessou P, et al. Toxoplasma gondii infection in meat animals from Africa: Systematic review and meta-analysis of sero-epidemiological studies. Vet World. 2017;10(2):194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkari B, Asgari Q, Bagherian N, et al. Molecular and serological evaluation of Toxoplasma gondii infection in reared turkeys in Fars Province, Iran. Jundishapur J Microbiol. 2014;7(7):e11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rassouli M, Razmi GR, Bassami MR, Movassaghi AR, Azizzadeh M. Study on ovine abortion associated with Toxoplasma gondii in affected herds of Khorasan Razavi Province, Iran based on PCR detection of fetal brains and maternal serology. Parasitology. 2011;138(6):691–7. [DOI] [PubMed] [Google Scholar]
- 12.Aliabadi J, Ziaali PN. Survey of Toxoplasma gondii in Livestocks’ Meat (Sheep, Goat, Camel), Using Nested PCR Method in Sabzavar District. Eur Online J Nat Soc Sci. 2016;5(2):368. [Google Scholar]
- 13.Dubey JP, Rajendran C, Ferreira LR, et al. High prevalence and genotypes of Toxoplasma gondii isolated from goats, from a retail meat store, destined for human consumption in the USA. Int J Parasitol. 2011;41(8):827–33. [DOI] [PubMed] [Google Scholar]
- 14.Habibi GR, Imani AR, Gholami MR, et al. Detection and identification of Toxoplasma gondii type one infection in sheep aborted fetuses in Qazvin Province of Iran. Iran J Parasitol. 2012;7(3):64–72. [PMC free article] [PubMed] [Google Scholar]
- 15.Fazaeli A, Carter PE, Darde ML, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasitol. 2000;30(5):637–42. [DOI] [PubMed] [Google Scholar]
- 16.Guo M, Dubey JP, Hill D, et al. Prevalence and risk factors for Toxoplasma gondii infection in meat animals and meat products destined for human consumption. J Food Prot. 2015;78(2):457–76. [DOI] [PubMed] [Google Scholar]
- 17.Tavakoli Kareshk A, Mahmoudvand H, Keyhani A, et al. Molecular detection and genetic diversity of Toxoplasma gondii in different tissues of sheep and goat in Eastern Iran. Trop Biomed. 2017;34(3):681–690. [PubMed] [Google Scholar]
- 18.Asgari Q, Sarkari B, Amerinia M, Panahi S, Mohammad-pour I, Sarvestani AS. Toxoplasma Infection in farm animals: a seroepidemiological survey in Fars province, south of Iran. Jundishapur J Microbiol. 2013;6(3):269. [Google Scholar]
- 19.Kamyabi H, Bahrieni M, Beigzadeh M, Harandi MF, Zia-Ali N. Risk Factors Analysis Associated with Seropositivity to Toxoplasma gondii in Sheep and Goats in Southeastern Iran Using Modified Agglutination Test (MAT). Iran J Parasitol. 2008;3:1–2. [Google Scholar]
- 20.Raeghi S, Akaberi A, Sedeghi S. Seroprevalence of Toxoplasma gondii in sheep, cattle and horses in Urmia North-West of Iran. Iran J Parasitol. 2011;6(4):90–4. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Wang S, Wang D, et al. Seroprevalence of Toxoplasma gondii infection and risk factors in domestic sheep in Henan province, central China. Parasite. 2016;23:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin MY, Wang JL, Huang SY, et al. Seroprevalence and risk factors of Toxoplasma gondii in Tibetan Sheep in Gansu province, Northwestern China. BMC Vet Res. 2015;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavakoli kareshk A, Tavakoli oliaee R, mahmoudvand H, et al. The First Survey of Isolation and Molecular Typing of Toxoplasma gondii by Bioassay and PCR Method in BALB/c Mice in Camels (Camelus dromedarius) from Eastern Iran. Iran J Parasitol.2018; 13(3):382–91. [PMC free article] [PubMed] [Google Scholar]
- 24.Azizi H, Shiran B, Boroujeni AB, Jafari M. Molecular survey of Toxoplasma gondii in sheep, cattle and meat products in Chaharmahal va Bakhtiari Province, Southwest of Iran. Iran J Parasitol. 2014;9(3):429–34. [PMC free article] [PubMed] [Google Scholar]
- 25.Amdouni Y, Rjeibi MR, Rouatbi M, Amairia S, Awadi S, Gharbi M. Molecular detection of Toxoplasma gondii infection in slaughtered ruminants (sheep, goats and cattle) in Northwest Tunisia. Meat Sci. 2017;133:180–4. [DOI] [PubMed] [Google Scholar]
- 26.Taylor S, Barragan A, Su C, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314(5806):1776–80. [DOI] [PubMed] [Google Scholar]
- 27.da Silva IB, de Andrade Batista TP, Martines RB, et al. Genotyping of Toxoplasma gondii: DNA extraction from formalin-fixed paraffin-embedded autopsy tissues from AIDS patients who died by severe disseminated toxoplasmosis. Exp Parasitol. 2016;165:16–21. [DOI] [PubMed] [Google Scholar]
- 28.Genot S, Franck J, Forel J-M, et al. Severe Toxoplasma gondii I/III recombinant-genotype encephalitis in a human immunodeficiency virus patient. J Clin Microbiol. 2007;45(9):3138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danehchin L, Razmi G, Naghibi A. Isolation and genotyping of Toxoplasma gondii strains in ovine aborted fetuses in Khorasan Razavi Province, Iran. Korean J Parasitol. 2016;54(1):15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armand B, Solhjoo K, Kordshooli MS, Davami MH, Pourahmad M, Orfaee V. Toxoplasma gondii Type I, predominant genotype isolated from sheep in South of Iran. Vet World. 2017;10 (4): 386–392. In Abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zia-Ali N, Fazaeli A, Khoramizadeh M, Ajzenberg D, Dardé M, Keshavarz-Valian H. Isolation and molecular characterization of Toxoplasma gondii strains from different hosts in Iran. Parasitol Res. 2007;101(1):111–5. [DOI] [PubMed] [Google Scholar]
- 32.Xu P, Li X, Tang F, et al. Research Note Seroprevalence and risk factors for Toxoplasma gondii in sheep and goats in Jinzhou, Northeastern China. Trop Biomed. 2015;32(3):563–7. [PubMed] [Google Scholar]
- 33.Gebremedhin EZ, Abdurahaman M, Hadush T, Tessema TS. Seroprevalence and risk factors of Toxoplasma gondii infection in sheep and goats slaughtered for human consumption in Central Ethiopia. BMC Res Notes. 2014;7(1):696. [DOI] [PMC free article] [PubMed] [Google Scholar]