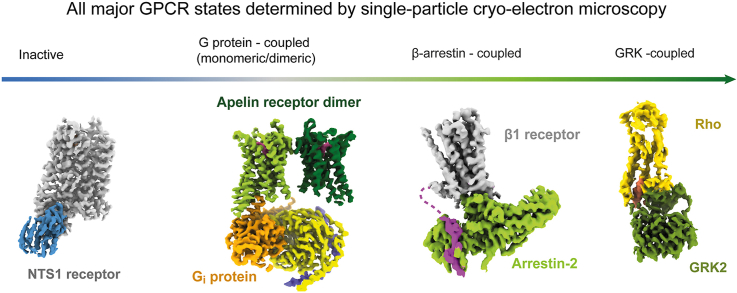
Abstract
Over the past three years (2020–2022) more structures of GPCRs have been determined than in the previous twenty years (2000–2019), primarily of GPCR complexes that are large enough for structure determination by single-particle cryo-EM. This review will present some structural highlights that have advanced our molecular understanding of promiscuous G protein coupling, how a G protein receptor kinase and β-arrestins couple to GPCRs, and GPCR dimerisation. We will also discuss advances in the use of gene fusions, nanobodies, and Fab fragments to facilitate the structure determination of GPCRs in the inactive state that, on their own, are too small for structure determination by single-particle cryo-EM.
Graphical abstract
Highlights
-
•
Promiscuous GPCR coupling shows mechanism of secondary coupling.
-
•
Cryo-EM structures of GPCR dimers have been determined for Class A, C and D receptors.
-
•
The first GPCR-GRK structure has been determined.
-
•
Arrestin-coupled GPCR structures provide new insights for the development of biased agonists.
-
•
Methodologies have been developed for determining cryo-EM structures of inactive state GPCRs.
Introduction
GPCRs play a pivotal role in intercellular signalling throughout the human body and are the targets of 34% of FDA approved drugs [1]. Only a proportion of all GPCRs have been drugged and there is intense scrutiny of other GPCRs to develop novel therapeutics for the treatment of diseases such as diabetes, cancer and neurodegeneration [2]. Structural biology plays a key role in drug development through either providing a structure suitable for screening in silico ultra-large drug libraries [3••] or through providing a mechanistic understanding of fundamental molecular processes such as receptor and G protein activation [4,5]. Here we highlight a few of the fundamental molecular insights that underpin complexities in GPCR pharmacology that have been uncovered by the wealth of structures determined by cryo-EM over the past few years.
Structural mechanisms in promiscuous GPCR-G protein coupling
GPCRs signal through heterotrimeric G proteins and the type of α-subunit determines the downstream signalling cascade affected. There are four major families of G proteins in humans, Gs, Gi/o, Gq/11 and G12/13 that signal through different pathways. Although some GPCRs are specific and activate a single type of G protein, at least 50% of GPCRs activate two or more G proteins [6, 7, 8]. Promiscuous coupling activates different G proteins with varying efficacies and kinetics, generating a fingerprint-like signalling profile within the cell [9], thus enhancing the complexity of GPCR signalling and providing new therapeutic opportunities.
Cryo-EM structures of eleven GPCRs have been determined with each GPCR coupled to two or more distinct G proteins: GCGR, β1AR, ADGRF1 and 5HT4R coupled to Gs and Gi/o [10, 11, 12, 13], NK1R coupled to Gs and Gq/11 [14], CCKAR coupled to Gq, Gi1 and Gs [15,16], ADGRL3 coupled to Gs, Gi, Gq and G12 [17••] and four receptors coupled to Gi/o and Gq/11 (GSHR [18,19], CCKBR [20], GPR139 [21] and MRGPRX2 [22]). Several trends arise from analysing this set of structures [23].
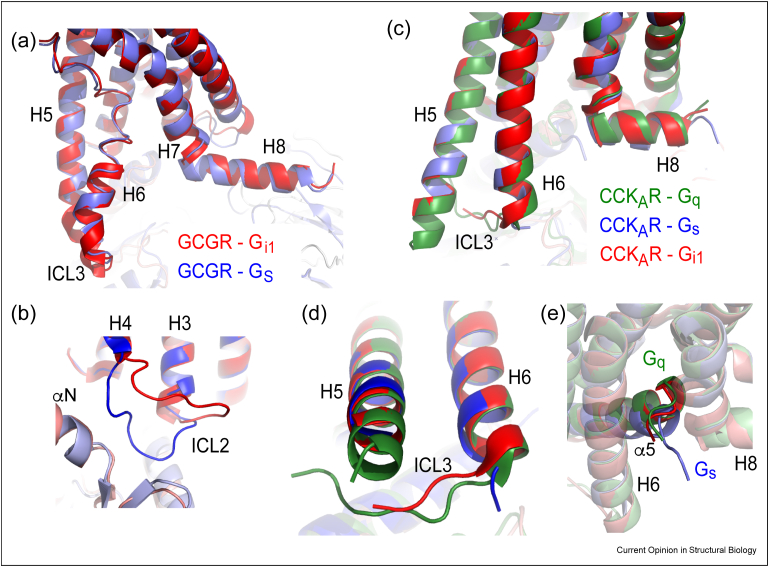
The outward movement of the cytoplasmic end of transmembrane helix TM6 is a hallmark of GPCR activation and is thought to determine the size and shape of the intracellular cleft where the cytoplasmic end of helix α5 of the G protein α-subunit couples [24]. Structures of many different GPCRs coupled to G proteins suggested initially that the magnitude of TM6 displacement correlated with the type of G protein. A large outward movement of TM6 forms a wide intracellular cleft that is required typically for Gs coupling, whilst smaller movements of TM6 form a narrower cleft characteristic of Gi/o-Gq/11 coupling [25,26]. However, recent new structures show that this is not always the case when they are the secondary couplers, with Gs sometimes coupling to a narrow cleft and Gi or Gq coupling to a wide cleft. Structures of the same GPCR coupled to either Gs or another G protein suggest that the movement of TM6 is usually the same regardless of the secondary G protein coupled i.e. the secondary G protein has to use a similar intracellular cleft for coupling as the primary G protein (Figure 1a–e). For example, the primary coupler to GCGR is Gs and the GCGR-Gs cryo-EM structure shows a wide intracellular cleft; the receptor structure coupled to its secondary coupler Gi/o shows an equally wide cleft to when Gs is coupled, and not a narrow cleft as might be expected [10]. Conversely, CCKAR and NK1R couple primarily to Gq and adopt a narrow intracellular cleft upon activation, and the secondary G protein Gs also couples to this narrow cleft. In some instances, such as for CCKAR, this forces the G protein to adopt ‘non-standard’ conformations where the α-subunit shows an unwinding of the ‘wavy hook’ in the α5 helix C-terminus, which protrudes outwards from the receptor intracellular cavity (Figure 1e). Primary coupling of Gi/o and Gq/11 results in a similar narrow intracellular cleft, which may explain the high abundance of Gi/o-Gq/11 promiscuous couplings [7].
Figure 1.
Structural snapshots of promiscuous GPCR-G protein coupling. Structural superposition of the GCGR coupled to Gs (blue) and Gi1 (red) showing similarities in TM6 (a) and differences in ICL2 (b) [10]. Structural superposition of the CCKAR coupled to Gq (green), Gi1 (red) and Gs (blue) showing similarities in TM6 position (c), differences in the ordering of ICL3 depending on the coupled G protein (d) and differences in the engagement mode of the α-subunit C-terminal ‘wavy hook’ for Gs vs Gq(e) [15,16].
The intracellular loops (ICLs) of GPCRs are the elements that differ most when coupling to different G proteins. However, there appears to be no correlation with the type of ICL rearrangement and the type of G protein or primary/secondary couplings. ICL3 takes a prominent role in promiscuous G protein coupling in MRGPRX2, 5-HT4R, ADGRF1, GSHR, GPR139, and CCKAR where it makes different interactions to different G proteins (Figure 1d.) ICL2 also changes conformation or interactions in most GPCR-G protein complexes (e.g. GCGR and GSHR, Figure 1b), whereas ICL1 differential interactions have only been observed in GCGR. The loop between TM7 and H8 also varies in β1AR coupled to either Gs or Gi. Such differences in ICLs contribution to promiscuous G protein coupling were supported by mutagenesis and functional assays, where alterations in the CCKAR ICL3 had a major impact on Gq but not GS or Gi signalling [15]. Similarly, alterations in the GCGR ICL3 and ICL1 showed a greater impact on Gi compared to GS signalling [10].
GPCR structures coupled to GRK or arrestin
One mechanism in the cell to terminate GPCR-G protein signalling at the plasma membrane is through receptor phosphorylation by GRKs, recruitment of arrestin via the phosphorylated C-terminus/ICL3 and then clathrin-mediated endocytosis mediated by arrestin-clathrin/AP2 interactions [27]. Arrestin interacts with GPCRs in two distinct ways. Arrestin binds first to the phosphorylated C-terminus/ICL3 of the receptor, causing a conformation change in arrestin that subsequently facilitates coupling of arrestin to the receptor [28, 29, 30]. Arrestin couples to GPCRs using the same intracellular cleft that binds the C-terminal α5 helix of the G protein [31] and results in activation of the intracellular ERK1/2 signalling cascade. It is crucial to understand the molecular differences between coupling of G proteins, GRKs and arrestins, because the therapeutic effect and side effects of drugs may arise through different signalling pathways [32]. There is thus intense interest in developing biased ligands that specifically activate/inhibit only one specific pathway.
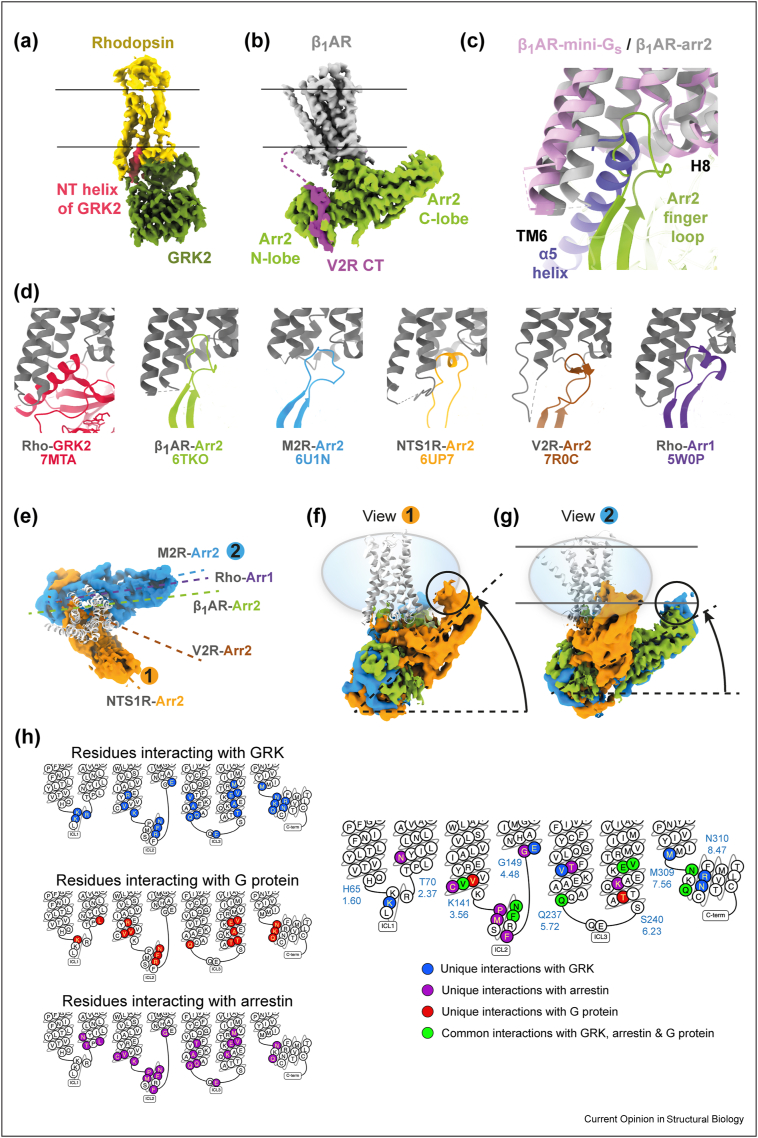
Structure determination of a GPCR-GRK complex has been difficult, however, stabilisation of the rhodopsin-GRK1 complex by a combination of crosslinking, binding of two Fabs and lipids resulted in the first low resolution structures [33••]. The receptor was in its active state, with the N-terminus of GRK1 forming an α-helix that binds to the intracellular cleft like G proteins and arrestin (Figure 2a,d). Comparison between the conformation of rhodopsin when coupled to either GRK, arrestin or the G protein transducin shows that they are virtually identical (RMSDs of 0.9–1.0 Å) and that the binding sites on rhodopsin overlap significantly (Figure 2h). There are eight residues that interact with all three coupled proteins (Val1393.54, Asn14534.53, Phe14634.54, Gln2375.72, Glu2496.32, Val2506.33, Asn3108.47, Gln3128.49) and a further subset of residues (Figure 2h) that interact only with GRK1 (6 residues), visual arrestin (8 residues) or transducin (2 residues).
Figure 2.
Variations in coupling of arrestins and GRK2 to GPCRs. (a) Cryo-EM density (EMDB-23979) of rhodopsin coupled to GRK1 [33]. Density for the Fab required for structure determination has been removed for clarity. (b) Cryo-EM density of β1AR in a lipid nanodisc coupled to β-arrestin1 (EMDB-10515) [37]. Density for Fab30 required for structure determination has been removed for clarity. (c) Superposition of β1AR coupled to mini-Gs (purple; PDB code 7JJO [72]) and β1AR (grey) coupled to β-arrestin1 (green; PDB code 6TKO [37]). (d) Different conformations of the GRK coupling helix and arrestin finger loop when coupled to different receptors. (e–g) Variation in the angle of arrestin coupled to different receptors (see main text for references): (e) a view perpendicular to the membrane plane; panels (f–g) are views parallel to the membrane plane in positions 1 and 2, respectively, as defined in panel (e). (h) Snake plots of bovine rhodopsin with amino acid residues within 3.9 Å (inclusive) of either GRK, G protein or arrestin coloured appropriately. PDB codes for the complexes are as follows: rhodopsin-GRK, 7MTB [33••]; rhodopsin-G protein, 6OYA [73]; rhodopsin-arrestin, 5W0P [74]. The panels were made using GPCRdb [75].
Seven structures of GPCRs coupled to arrestins have now been determined. The first high-resolution structure of a GPCR-arrestin complex was a crystal structure of constitutively active mutant of human rhodopsin fused to a preactivated form of mouse arrestin 1 (visual arrestin) [34]. A variety of different strategies were required for cryo-EM structure determination of non-visual arrestins coupled to activated receptors, including combinations of the following: fusion with the C-terminus of phosphorylated V2 receptor, arrestin mutants, cross-linking, binding of Fab30 to stabilise the active state of arrestin and the use of lipid-mimicking environments. Structures of complexes with arrestin 2 (Arr2; also called β-arrestin1; Figure 2b,c) were determined coupled to NTS1R [35•,36], β1AR [37••], M2R [38•], V2R [39•] and 5HT2BR [40••].
G proteins couple to different receptors in a relatively conserved way [25], but in contrast arrestins have shown a wide variation in their binding poses. Significant variations occur in the structure of the finger loop of arrestin inserted into the intracellular cleft of the receptor (Figure 2d) and the angle of interaction between arrestin and the GPCR when viewed both perpendicular to the membrane plane and parallel to the membrane plane (Figure 2e–g).
Two GPCR-arrestin structures (β1AR [37] and 5HT2BR [40]) have been determined at 3.3 Å resolution where there is good density for the ligand in the orthosteric binding pocket. Comparison with the receptor bound to the same ligand but coupled either to a G protein (5HT2BR) or a G protein-mimetic nanobody (β1AR) showed similar weakening of interactions between the ligand and H5, explaining the weaker ligand affinity in the arrestin-coupled state compared to the G protein coupled state. There are also other differences between a G protein-coupled receptor and arrestin-coupled receptor, the most obvious one being the difference in outward movement of H6, although in β1AR this is less than in the G protein coupled state whereas for 5HT2BR it is greater than in the G protein coupled state. The differences observed between structures could be used in the development of biased agonists.
GPCR dimers
The existence and functional role of obligate class C and class D GPCR dimers are well-established, both structurally and functionally [41,42]. However, for Class A receptors there is no consensus on whether dimerisation is a ubiquitous mechanism in regulating Class A GPCR function. Some class A GPCRs are accepted to form transient dimers and higher order oligomers, although their physiological role is often uncertain [43,44]. Any structural dimer composed of parallel protomers observed in either X-ray crystal structures [45] or cryo-EM has the potential to be physiologically relevant, but careful validation is required by biochemistry and pharmacology to support this.
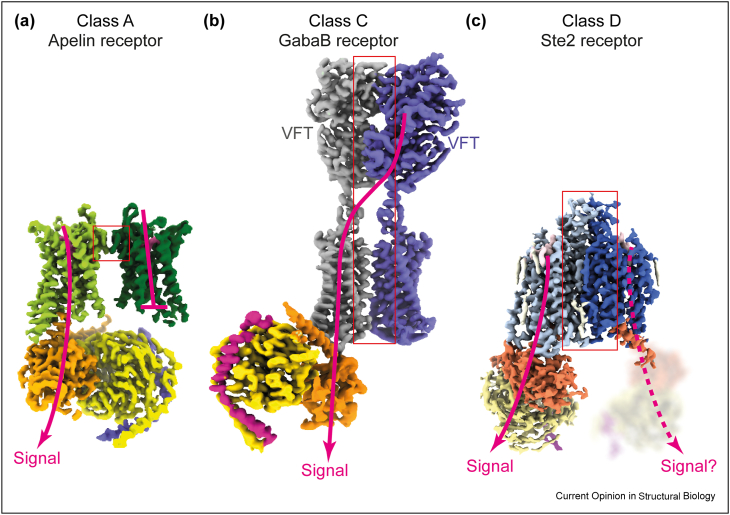
Humans possess 22 Class C GPCRs and there are now 76 cryo-EM structures, determined between 2019 and 2022, bound to either antagonist, agonist, positive allosteric modulator (PAM), negative allosteric modulator (NAM), regulator of G protein signalling (RGS) protein and/or G protein. Due to space constraints, we will discuss only those receptors where a fully active G protein-coupled state has been determined (Table 1), namely the GABAB receptor [46••,47•] and metabotropic glutamate receptors (mGluRs) [48•]. The common feature of Class C dimers is that they are maintained dimeric predominantly through interactions in the extracellular Venus fly trap domain (VFT; Figure 3b) that binds agonists. The agonist-induced conformational change in the VFT is transmitted via a linker region to the transmembrane regions, ultimately resulting in a rotation of one helical bundle with respect to the other. In the GABAB receptor, this changes the dimer interface from being formed by predominantly H5-H5 to H6-H6 [46••,47•] and in the mGluRs from mainly H4-H4 to H6-H6 [48•]. A number of variations between these states have also been described, highlighting the plasticity of these receptors and a number of different solutions for how PAMs can promote the formation of active-like states [46••,48•,49,50•]. Extensive pharmacological and biochemical studies have determined that only one protomer in the dimer couples to a G protein and that signalling is transmitted from the VFT of one receptor in the dimer to the G protein coupling site on the adjacent dimer [41]. This is recapitulated in the asymmetric active-state dimer structures where only a single G protein is coupled per dimer, via a coupling site formed through interactions primarily to ICL2, which is distinct to that found in other GPCR families [47•,48•,51•].
Table 1.
Details of GPCR structures discussed in the main text.
| Receptor | Dimer type | Class | PDB | Agonist (Ag), antagonist (Ant), PAM, NAM | G protein family | Stabilising antibodies and fusions | Reference | |
|---|---|---|---|---|---|---|---|---|
| Apelin | Homo | A | 7W0N | Ag | Gi | scFv16 + BRIL | [52••] | |
| 7W0L | Ag | Gs | scFv16 +BRIL | |||||
| Ste2 | Homo | D | 7AD3 | Ag | Gpa1 | – | [53••] | |
| 7QB9 | – | – | – | [54••] | ||||
| 7QA8 | Ant | – | – | |||||
| 7QBC | Ag | – | – | |||||
| 7QBI | Ag | – | – | |||||
| GABAB | Hetero | C | 7EB2 | Ag | Gi | scFv16 | [47•] | |
| C | 7CA3 | PAM | – | – | [49] | |||
| C | 7CA5 | – | – | – | ||||
| C | 7CUM | Ant + NAM | – | – | ||||
| C | 6UO8 | Ag + PAM | – | – | [50•] | |||
| C | 6UO9 | Ag | – | – | ||||
| C | 6UOA | Ag | – | – | ||||
| C | 6VJM | APO | – | – | ||||
| C | 7C7S | Ant | – | – | [46••] | |||
| C | 7C7Q | Ag + PAM | Gi1 | – | ||||
| C | 6WIV | – | – | – | [55] | |||
| C | 6W2X | Ant + NAM | – | – | [56] | |||
| Homo | C | 6W2Y | Ant + NAM | – | – | |||
| Metabotropic glutamate receptors | mGlu1 | Homo | C | 7DGD | – | – | – | [57] |
| 7DGE | Ag | Nb43 | ||||||
| mGlu2 | C | 7E9G | Ag + PAM | Gi | scFv16 + Nb | [48•] | ||
| mGlu2 | Homo | C | 7MTQ | Ant | – | – | [51•] | |
| C | 7MTR | Ago-PAM + Ag | – | – | ||||
| C | 7MTS | Ago-PAM | Gi | |||||
| mGlu2 | Homo | C | 7EPA | – | – | – | [58•] | |
| 7EPB | Ag | Nb-RON | ||||||
| mGlu7 | Homo | C | 7EPC | – | – | – | ||
| mGlu2mGlu7 | Hetero | C | 7EPD | – | – | – | ||
| mGlu5 | Homo | C | 6N52 | – | [59] | |||
| Homo | 6N51 | Ag | Nb43 | |||||
| mGlu5-5M | Homo | C | 7FD8 | Ag | – | – | [60] | |
| Homo | C | 7FD9 | Ant | |||||
| mGlu3 | Homo | C | 7WI8 | Ant | – | – | [61] | |
| 7WI6 | Ag + NAM | – | – | |||||
| 7WIH | Ag | – | – | |||||
| mGlu4 | Homo | C | 7E9H | Ag | Gi3 | scFv16 | [48•] | |
Figure 3.
Signalling routes in GPCR dimers. (a) Cryo-EM density of the apelin receptor (EMDB-32243) shows that there is sufficient room for only one G protein to couple per dimer, and the C-terminus of the adjacent protomer binds in the G protein-coupling cleft in an auto-inhibitory mechanism [52]. The dimer interface is shown by in the GABAB receptor dimer (EMDB-21534) is from the VFT domain of one protomer through the transmembrane helices of the adjacent protomer that can couple to G protein. The structures of two transmembrane helical bundles are not identical and the G protein coupling site forms only in one protomer [46,47]. The dimer interface is shown by the red box. (c) The Ste2 dimer (EMDB-11720) contains two protomers of identical conformation that are both capable of coupling to G proteins simultaneously, although one G protein is highly mobile, with the exception of the α5 helix that is ordered where it contacts the receptor [53]. The tilt of the G protein with respect to the receptor is over 50° different from that observed in G protein-coupling to Class A receptors, thus allowing two G proteins to couple simultaneously. The signalling pathways through the receptor are assumed to follow the paradigm of a monomeric receptor, however it is unclear whether both G proteins can signal to the same extent and there could be crosstalk between protomers across the dimer interface [54]. The dimer interface is shown by the red box.
In contrast to Class C receptors, the cryo-EM structure of the class D receptor homodimeric GPCR Ste2 (Figure 3c) showed that it couples to two G proteins simultaneously [53••]. The density for one G protein was well-resolved, but the density for the adjacent G protein was diffuse and molecular dynamics simulations showed that each G protein underwent phases of mobility, with only one G protein being ordered at any one time. The interface between the two protomers is also dynamic [54••], even though it has a very large surface area in the active state (2500 Å2) and is composed of interactions between the N-terminus, ECL1 and H1. Cryo-EM structures of five different receptor conformations showed that Ste2 activation upon binding the native agonist α-factor involved an increase in the strength of the interface and a 20 Å movement of the cytoplasmic end of H7 [54••]. The movement of H7 unblocked the G protein coupling site and then formed additional contacts at the dimer interface in a mechanism currently unique to Ste2.
There is currently only one high-resolution structure of a Class A GPCR dimer, the active state of the apelin receptor [52••]. This is different from dimers of Class C and Class D receptors as the interface is extremely small (140 Å2), comprising residues at the extracellular end of H3 (Figure 3a). Only one of the protomers is coupled to a G protein, and there are no contacts between the G protein and the adjacent protomer. Mutation of a key residue at the dimer interface (F101 A3.24) significantly reduced dimer formation and had a profound effect on the pharmacology of the apelin receptor, increasing basal activity and Emax significantly.
Inactive GPCR structures by cryo-EM
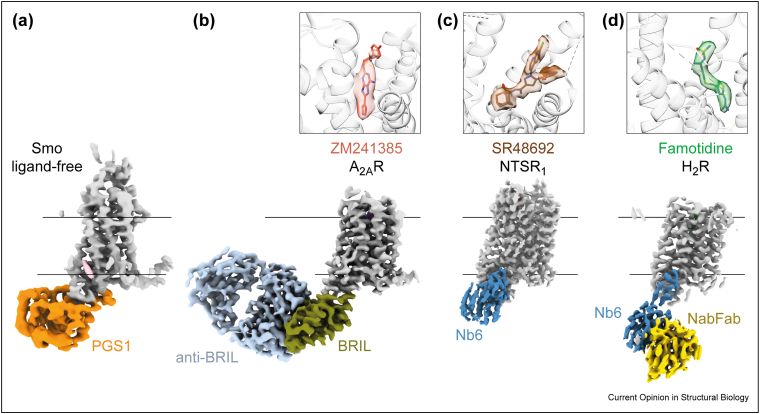
The inactive state of GPCRs may only consist of 35–40 kDa of ordered protein, which is embedded in a detergent micelle typically ∼100 kDa in size and makes processing of cryo-EM images of these small membrane proteins highly challenging. To circumvent this problem, extra mass needs to be added to the receptor that can extend beyond the detergent micelle and facilitate particle alignment during image processing. An obvious solution is to repurpose successful strategies in engineering GPCRs for X-ray structure determination through either binding an antibody Fab fragment [62], nanobody [63] or insert a small soluble protein such as BRIL in ICL3 [64].
One recent approach was to graft a section of H5-ICL3-H6 from the mu opioid receptor (MOR) into a target GPCR and then bind nanobody Nb6 that specifically recognises this region [65,66]. This resulted in sub-3 Å resolution structures of the inactive states of NTS1R, H2R (Figure 4c,d) and somatostatin receptor 2 [67•]. Another approach was to insert BRIL in place of ICL3 in Frizzled5 and then use an anti-BRIL Fab/Nb complex to increase the mass further; the structure was determined by single-particle cryo-EM to 3.7 Å resolution, with the low resolution being explained by the flexibility of the GPCR-BRIL fusion points [68•]. This methodology was explored further [69•] to determine the structure of thermostabilised A2AR-BRIL bound to an anti-BRIL Fab to 3.4 Å resolution (Figure 4b) and a Smoothened ICL3 chimera fused to Pyrococcus glycogen synthase (PGS) at 3.7 Å resolution (Figure 4a). A recent innovative strategy to create a three-point linkage between the heterodimer calcineurin and the β2AR facilitated the structure determination of the receptor either in the ligand-free state or bound to antagonist/agonist with overall resolutions between 3.5 and 3.9 Å [70•].
Figure 4.
Examples of strategies to determine structures of GPCR inactive states. (a) Cryo-EM density of ligand-free Smoothened (EMDB-27062) [69]. (b) Cryo-EM density (EMDB-25648) of the adenosine A2A receptor with a BRIL insertion in ICL3 and bound to an anti-BRIL Fab fragment [69]. (c) Cryo-EM density (EMDB-26589) of the neurotensin receptor NTSR1 engineered to contain the H5-ICL3-H6 region of MOR and bound to the anti-MOR nanobody Nb6 [67]. (d) Cryo-EM density (EMDB-26590) of the histamine H2 receptor engineered to contain the H5-ICL3-H6 region of MOR, bound to the anti-MOR nanobody Nb6 and the anti-nanobody Fab (NabFab) [67]. Ligand density in the orthosteric binding pocket is shown above each receptor.
Conclusions
The incredible advances in all the technology involved in single particle cryo-EM have made the structure determination of GPCR complexes in all conformational states considerably easier than using X-ray crystallography [71]. There are more advances in the cryo-EM pipeline and so the future holds rich promise for improving the throughput of GPCR structure determination, making it the premier tool for structure-based drug design and the determination of novel GPCR structures. A concerted effort over the coming years will undoubtedly determine structures of all human non-olfactory GPCRs.
Declaration of competing interest
CGT is a shareholder and SAB member of Sosei Heptares. None of the other authors have any conflicts to declare.
Acknowledgements
The work in JGN's laboratory is funded by the Ministerio de Ciencia, Innovación y Universidades (PID2020-113359GA-I00), the Spanish Ramón y Cajal program and the Fondo Europeo de Desarrollo Regional (FEDER). The work in CGT's laboratory is supported by the Medical Research Council, as part of United Kingdom Research and Innovation (also known as UK Research and Innovation) [MC_U105197215]. For the purpose of open access, the MRC Laboratory of Molecular Biology has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising.
This review comes from a themed issue on Membranes (2023)
Edited by Simon Newstead and Robert Tampé
Data availability
No data was used for the research described in the article.
References
- 1.Hauser A.S., Attwood M.M., Rask-Andersen M., Schiöth H.B., Gloriam D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Congreve M., de Graaf C., Swain N.A., Tate C.G. Impact of GPCR structures on drug discovery. Cell. 2020;181:81–91. doi: 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 3••.Sadybekov A.A., Sadybekov A.V., Liu Y., Iliopoulos-Tsoutsouvas C., Huang X.P., Pickett J., Houser B., Patel N., Tran N.K., Tong F., et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature. 2022;601:452–459. doi: 10.1038/s41586-021-04220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; In silico screening of GPCRs using an ultra-large virtual library was shown to be an exceedingly effective tool for finding rapidly novel small molecules that bind to the orthosteric binding site. Very often, these initial hits already bound with nanomolar affinity and required little further diversification to obtain potent ligands.
- 4.Flock T., Ravarani C.N.J., Sun D., Venkatakrishnan A.J., Kayikci M., Tate C.G., Veprintsev D.B., Babu M.M. Universal allosteric mechanism for Galpha activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatakrishnan A.J., Deupi X., Lebon G., Heydenreich F.M., Flock T., Miljus T., Balaji S., Bouvier M., Veprintsev D.B., Tate C.G., et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature. 2016;536:484–487. doi: 10.1038/nature19107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avet C., Mancini A., Breton B., Le Gouill C., Hauser A.S., Normand C., Kobayashi H., Gross F., Hogue M., Lukasheva V., et al. Effector membrane translocation biosensors reveal G protein and betaarrestin coupling profiles of 100 therapeutically relevant GPCRs. Elife. 2022;11 doi: 10.7554/eLife.74101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser A.S., Avet C., Normand C., Mancini A., Inoue A., Bouvier M., Gloriam D.E. Common coupling map advances GPCR-G protein selectivity. Elife. 2022;11 doi: 10.7554/eLife.74107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue A., Raimondi F., Kadji F.M.N., Singh G., Kishi T., Uwamizu A., Ono Y., Shinjo Y., Ishida S., Arang N., et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell. 2019;177:1933–1947. doi: 10.1016/j.cell.2019.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuho I., Ostrovskaya O., Kramer G.M., Jones C.D., Xie K., Martemyanov K.A. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci Signal. 2015;8:ra123. doi: 10.1126/scisignal.aab4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao A., Han S., Li X., Li Z., Zhao P., Dai A., Chang R., Tai L., Tan Q., Chu X., et al. Structural basis of Gs and Gi recognition by the human glucagon receptor. Science. 2020;367:1346–1352. doi: 10.1126/science.aaz5346. [DOI] [PubMed] [Google Scholar]
- 11.Alegre K.O., Paknejad N., Su M., Lou J.S., Huang J., Jordan K.D., Eng E.T., Meyerson J.R., Hite R.K., Huang X.Y. Structural basis and mechanism of activation of two different families of G proteins by the same GPCR. Nat Struct Mol Biol. 2021;28:936–944. doi: 10.1038/s41594-021-00679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S., Xu P., Shen D.D., Simon I.A., Mao C., Tan Y., Zhang H., Harpsoe K., Li H., Zhang Y., et al. GPCRs steer Gi and Gs selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors. Mol Cell. 2022;82:2681–2695. doi: 10.1016/j.molcel.2022.05.031. [DOI] [PubMed] [Google Scholar]
- 13.Qu X., Qiu N., Wang M., Zhang B., Du J., Zhong Z., Xu W., Chu X., Ma L., Yi C., et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature. 2022;604:779–785. doi: 10.1038/s41586-022-04580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thom C., Ehrenmann J., Vacca S., Waltenspuhl Y., Schoppe J., Medalia O., Pluckthun A. Structures of neurokinin 1 receptor in complex with Gq and Gs proteins reveal substance P binding mode and unique activation features. Sci Adv. 2021;7 doi: 10.1126/sciadv.abk2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q., Yang D., Zhuang Y., Croll T.I., Cai X., Dai A., He X., Duan J., Yin W., Ye C., et al. Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor. Nat Chem Biol. 2021;17:1238–1244. doi: 10.1038/s41589-021-00841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobbs J.I., Belousoff M.J., Harikumar K.G., Piper S.J., Xu X., Furness S.G.B., Venugopal H., Christopoulos A., Danev R., Wootten D., et al. Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Qian Y., Ma Z., Liu C., Li X., Zhu X., Wang N., Xu Z., Xia R., Liang J., Duan Y., et al. Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling. Mol Cell. 2022;82:4340–4352. doi: 10.1016/j.molcel.2022.10.009. [DOI] [PubMed] [Google Scholar]; This is a detailed comparison between the structures of a single receptor coupled to a member of each of the four major G protein families. The structures highlight the key residues at the C-terminus of the G protein that determine coupling specificity and mutations have been made in the receptor that enhance signalling down one pathway over another.
- 18.Qin J., Cai Y., Xu Z., Ming Q., Ji S.Y., Wu C., Zhang H., Mao C., Shen D.D., Hirata K., et al. Molecular mechanism of agonism and inverse agonism in ghrelin receptor. Nat Commun. 2022;13:300. doi: 10.1038/s41467-022-27975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Guo S., Zhuang Y., Yun Y., Xu P., He X., Guo J., Yin W., Xu H.E., Xie X., et al. Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor. Nat Commun. 2021;12:5064. doi: 10.1038/s41467-021-25364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., He C., Wang M., Zhou Q., Yang D., Zhu Y., Feng W., Zhang H., Dai A., Chu X., et al. Structures of the human cholecystokinin receptors bound to agonists and antagonists. Nat Chem Biol. 2021;17:1230–1237. doi: 10.1038/s41589-021-00866-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Daver H., Trapkov B., Wu L., Wu M., Harpsoe K., Gentry P.R., Liu K., Larionova M., Liu J., et al. Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139. Cell Res. 2022;32:210–213. doi: 10.1038/s41422-021-00591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao C., Kang H.J., Singh I., Chen H., Zhang C., Ye W., Hayes B.W., Liu J., Gumpper R.H., Bender B.J., et al. Structure, function and pharmacology of human itch GPCRs. Nature. 2021;600:170–175. doi: 10.1038/s41586-021-04126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrión-Antolí Á., Mallor-Franco J., Arroyo-Urea S., García-Nafría J. Structural insights into promiscuous GPCR-G protein coupling. Prog Mol Biol Transl Sci. 2023;195:137–152. doi: 10.1016/bs.pmbts.2022.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Rose A.S., Elgeti M., Zachariae U., Grubmüller H., Hofmann K.P., Scheerer P., Hildebrand P.W. Position of transmembrane helix 6 determines receptor G protein coupling specificity. J Am Chem Soc. 2014;136:11244–11247. doi: 10.1021/ja5055109. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Nafria J., Tate C.G. Cryo-EM structures of GPCRs coupled to Gs, Gi and Go. Mol Cell Endocrinol. 2019;488:1–13. doi: 10.1016/j.mce.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Glukhova A., Draper-Joyce C.J., Sunahara R.K., Christopoulos A., Wootten D., Sexton P.M. Rules of engagement: GPCRs and G proteins. ACS Pharmacol Transl Sci. 2018;1:73–83. doi: 10.1021/acsptsci.8b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbaum D.M., Rasmussen S.G., Kobilka B.K. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurevich V.V., Benovic J.L. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 29.Gurevich V.V., Gurevich E.V. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Ranjan R., Dwivedi H., Baidya M., Kumar M., Shukla A.K. Novel structural insights into GPCR-β-arrestin interaction and signaling. Trends Cell Biol. 2017;27:851–862. doi: 10.1016/j.tcb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Seyedabadi M., Gharghabi M., Gurevich E.V., Gurevich V.V. Receptor-arrestin interactions: the GPCR Perspective. Biomolecules. 2021;11:218. doi: 10.3390/biom11020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whalen E.J., Rajagopal S., Lefkowitz R.J. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Chen Q., Plasencia M., Li Z., Mukherjee S., Patra D., Chen C.-L., Klose T., Yao X.-Q., Kossiakoff A.A., Chang L., et al. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature. 2021;595:600–605. doi: 10.1038/s41586-021-03721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structure of a GRK coupled to a GPCR. The cryo-EM structure was difficult to determine, presumably due to the low affinity interactions between the GRK and GPCR, and the flexibility of the complex. The authors had to develop antibodies and cross-linking strategies to be able to get sufficiently good density to build a model.
- 34.Kang Y., Zhou X.E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T.A., Yefanov O., Han G.W., et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Huang W., Masureel M., Qu Q., Janetzko J., Inoue A., Kato H.E., Robertson M.J., Nguyen K.C., Glenn J.S., Skiniotis G., et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature. 2020;579:303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; A higher resolution structure of the NTSR1-arrestin-2 complex was determined than the first structure [36•]. The structure gives details of the arrestin residues that form the interface with the receptor and also the role of the specific lipid phosphatidylinositol-4,5-bisphosphate.
- 36•.Yin W., Li Z., Jin M., Yin Y.-L., de Waal P.W., Pal K., Yin Y., Gao X., He Y., Gao J., et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019;29:971–983. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Lee Y., Warne T., Nehmé R., Pandey S., Dwivedi-Agnihotri H., Chaturvedi M., Edwards P.C., García-Nafría J., Leslie A.G.W., Shukla A.K., et al. Molecular basis of β-arrestin coupling to formoterol-bound β1-adrenoceptor. Nature. 2020;583:862–866. doi: 10.1038/s41586-020-2419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first high resolution structure of an arrestin-coupled GPCR bound to a biased ligand, which allowed a direct comparison with the same receptor bound to the same ligand coupled to a G protein mimetic nanobody. Surprisingly, there were discernible differences in the orthosteric binding site that explains the lower affinity of agonist binding in the arrestin-coupled state compared to the G-protin-coupled state.
- 38•.Staus D.P., Hu H., Robertson M.J., Kleinhenz A.L.W., Wingler L.M., Capel W.D., Latorraca N.R., Lefkowitz R.J., Skiniotis G. Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc. Nature. 2020;579:297–302. doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; This structure was the first where the receptor was in a lipid nanodisc and shows a much less tilted arrestin than observed where the receptor is in a detergent micelle(e.g. [35•]). The structure also highlights differences in the structure of the arrestin finger loop inserted into the receptor compared to other arrestin-GPCR structures.
- 39•.Bous J., Fouillen A., Orcel H., Trapani S., Cong X., Fontanel S., Saint-Paul J., Lai-Kee-Him J., Urbach S., Sibille N., et al. Biochemistry; 2022. Structure of the vasopressin hormone-V2 receptor-β-arrestin1 ternary complex. [DOI] [PMC free article] [PubMed] [Google Scholar]; The low resolution structures and accompanying analysis indicate that V2R exists in at least two different states when coupled to arrestin. How these affect receptor biochemistry is unclear.
- 40••.Cao C., Barros-Álvarez X., Zhang S., Kim K., Dämgen M.A., Panova O., Suomivuori C.-M., Fay J.F., Zhong X., Krumm B.E., et al. Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD. Neuron. 2022;110:3154–3167. doi: 10.1016/j.neuron.2022.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a beautiful paper that sets the standard for studying receptor activation, as they compared structures of the same receptor (5HT2BR) bound to the same ligand (LSD) and in three different states, either coupled to arrestin-2, coupled to the G protein Gq or in the transducer-free state. Having the same ligand present in all the receptor states means that any structural differences observed are related to the receptor and its transducer, not the ligand.
- 41.Pin J.P., Kniazeff J., Prezeau L., Liu J.F., Rondard P. GPCR interaction as a possible way for allosteric control between receptors. Mol Cell Endocrinol. 2019;486:89–95. doi: 10.1016/j.mce.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Dumont M.E., Konopka J.B. Comparison of experimental approaches used to determine the structure and function of the class D G protein-coupled yeast alpha-factor receptor. Biomolecules. 2022;12:761. doi: 10.3390/biom12060761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milligan G., Ward R.J., Marsango S. GPCR homo-oligomerization. Curr Opin Cell Biol. 2019;57:40–47. doi: 10.1016/j.ceb.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferre S., Casado V., Devi L.A., Filizola M., Jockers R., Lohse M.J., Milligan G., Pin J.P., Guitart X. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenkamp R.E. Identifying G protein-coupled receptor dimers from crystal packings. Acta Crystallographica Section D Structural Biology. 2018;74:655–670. doi: 10.1107/S2059798318008136. [DOI] [PubMed] [Google Scholar]
- 46••.Mao C., Shen C., Li C., Shen D.-D., Xu C., Zhang S., Zhou R., Shen Q., Chen L.-N., Jiang Z., et al. Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 2020;30:564–573. doi: 10.1038/s41422-020-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structure of a Class C receptor coupled to a G protein. The real surprise was that the G protein coupling site did not conform to the paradigm for Class A, B and F receptors and was formed predominantly by the intracellular loops. However, the position of the G protein binding site and the fact that there was only one G protein coupled per dimer was consistent with all the biochemical data. The higher resolution structure of this complex with additional details was published the following year [47•].
- 47•.Shen C., Mao C., Xu C., Jin N., Zhang H., Shen D.-D., Shen Q., Wang X., Hou T., Chen Z., et al. Structural basis of GABAB receptor–Gi protein coupling. Nature. 2021;594:594–598. doi: 10.1038/s41586-021-03507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; See comments for [46••].
- 48•.Lin S., Han S., Cai X., Tan Q., Zhou K., Wang D., Wang X., Du J., Yi C., Chu X., et al. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021;594:583–588. doi: 10.1038/s41586-021-03495-2. [DOI] [PubMed] [Google Scholar]; CryoEM structures of mGlu2 and mGlu4 were determined coupled to Gi, highlightng the asymmetric activation process of these receptors.
- 49.Kim Y., Jeong E., Jeong J.-H., Kim Y., Cho Y. Structural basis for activation of the heterodimeric GABAB receptor. J Mol Biol. 2020;432:5966–5984. doi: 10.1016/j.jmb.2020.09.023. [DOI] [PubMed] [Google Scholar]
- 50•.Shaye H., Ishchenko A., Lam J.H., Han G.W., Xue L., Rondard P., Pin J.-P., Katritch V., Gati C., Cherezov V. Structural basis of the activation of a metabotropic GABA receptor. Nature. 2020;584:298–303. doi: 10.1038/s41586-020-2408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Four cryo-EM structures of the GABAB receptor heterodimer were determined either in a ligand-free state, or bound to an agonist with or without a PAM. The paper describes in detail the complex series of events from the closure of the VFT upon agonist binding through to an active state that is stabilised by the PAM.
- 51•.Seven A.B., Barros-Álvarez X., de Lapeyrière M., Papasergi-Scott M.M., Robertson M.J., Zhang C., Nwokonko R.M., Gao Y., Meyerowitz J.G., Rocher J.-P., et al. G-protein activation by a metabotropic glutamate receptor. Nature. 2021;595:450–454. doi: 10.1038/s41586-021-03680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structures of mGluR2 were determined in different conformational states and also coupled to Gi. The activation process required re-orientation of the heterodimer transmembrane bundles in relation to one another to form an asymmetric interface.
- 52••.Yue Y., Liu L., Wu L.-J., Wu Y., Wang L., Li F., Liu J., Han G.-W., Chen B., Lin X., et al. Structural insight into apelin receptor-G protein stoichiometry. Nat Struct Mol Biol. 2022;29:688–697. doi: 10.1038/s41594-022-00797-5. [DOI] [PubMed] [Google Scholar]; First high-resolution structure of a Class A GPCR dimer coupled to a G protein. Functional data support the assignment of the dimer interface and show that the wild type dimer has lower basal activity and lower Emax upon agonist binding compared to a dimer interface mutant that is predominantly monomeric.
- 53••.Velazhahan V., Ma N., Pándy-Szekeres G., Kooistra A.J., Lee Y., Gloriam D.E., Vaidehi N., Tate C.G. Structure of the class D GPCR Ste2 dimer coupled to two G proteins. Nature. 2021;589:148–153. doi: 10.1038/s41586-020-2994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structure of Class D GPCR that had many surprises. The native ligand α-factor bound in an unusual conformation with the main determinant for agonism at the periphery of the receptor. The receptor existed as a dimer with an extensive interface and two G proteins coupled simultaneously.
- 54••.Velazhahan V., Ma N., Vaidehi N., Tate C.G. Activation mechanism of the class D fungal GPCR dimer Ste2. Nature. 2022;603:743–748. doi: 10.1038/s41586-022-04498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; A series of structures showing the Ste2 dimer in the ligand-free state, antagonist-bound state and in two states bound to the agonist α-factor (an inactive-like state and an active-like state) in equilibrium with each other. The mechanism of receptor activation was shown to be different from any other class of GPCR.
- 55.Park J., Fu Z., Frangaj A., Liu J., Mosyak L., Shen T., Slavkovich V.N., Ray K.M., Taura J., Cao B., et al. Structure of human GABAB receptor in an inactive state. Nature. 2020;584:304–309. doi: 10.1038/s41586-020-2452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papasergi-Scott M.M., Robertson M.J., Seven A.B., Panova O., Mathiesen J.M., Skiniotis G. Structures of metabotropic GABAB receptor. Nature. 2020;584:310–314. doi: 10.1038/s41586-020-2469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J., Qu L., Wu L., Tang X., Luo F., Xu W., Xu Y., Liu Z.-J., Hua T. Structural insights into the activation initiation of full-length mGlu1. Protein & Cell. 2021;12:662–667. doi: 10.1007/s13238-020-00808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Du J., Wang D., Fan H., Xu C., Tai L., Lin S., Han S., Tan Q., Wang X., Xu T., et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature. 2021;594:589–593. doi: 10.1038/s41586-021-03641-w. [DOI] [PubMed] [Google Scholar]; First structure of a metabotropic glutamate receptor heterodimer describing differences with the homodimer.
- 59.Koehl A., Hu H., Feng D., Sun B., Zhang Y., Robertson M.J., Chu M., Kobilka T.S., Laeremans T., Steyaert J., et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019;566:79–84. doi: 10.1038/s41586-019-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasrallah C., Cannone G., Briot J., Rottier K., Berizzi A.E., Huang C.-Y., Quast R.B., Hoh F., Banères J.-L., Malhaire F., et al. Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang W., Yang F., Xu C., Ling S., Lin L., Zhou Y., Sun W., Wang X., Liu P., Rondard P., et al. Structural basis of the activation of metabotropic glutamate receptor 3. Cell Res. 2022;32:695–698. doi: 10.1038/s41422-022-00623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hino T., Arakawa T., Iwanari H., Yurugi-Kobayashi T., Ikeda-Suno C., Nakada-Nakura Y., Kusano-Arai O., Weyand S., Shimamura T., Nomura N., et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasmussen S.G., Choi H.J., Fung J.J., Pardon E., Casarosa P., Chae P.S., Devree B.T., Rosenbaum D.M., Thian F.S., Kobilka T.S., et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chun E., Thompson A.A., Liu W., Roth C.B., Griffith M.T., Katrich V., Kunken J., Xu F., Cherezov V., Hanson M.A., et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Che T., English J., Krumm B.E., Kim K., Pardon E., Olsen R.H.J., Wang S., Zhang S., Diberto J.F., Sciaky N., et al. Nanobody-enabled monitoring of kappa opioid receptor states. Nat Commun. 2020;11:1145. doi: 10.1038/s41467-020-14889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Che T., Majumdar S., Zaidi S.A., Ondachi P., McCorvy J.D., Wang S., Mosier P.D., Uprety R., Vardy E., Krumm B.E., et al. Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell. 2018;172:55–67.e15. doi: 10.1016/j.cell.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67•.Robertson M.J., Papasergi-Scott M., He F., Seven A.B., Meyerowitz G., Panova O., Peroto M.C., Che T. Structure determination of inactive-state GPCRs with a universal nanobody. Nat Struct Mol Biol. 2022;29:1188–1195. doi: 10.1038/s41594-022-00859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; See text for [68•]
- 68•.Tsutsumi N., Mukherjee S., Waghray D., Janda C.Y., Jude K.M., Miao Y., Burg J.S., Aduri N.G., Kossiakoff A.A., Gati C., et al. Structure of human Frizzled5 by fiducial- assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling. Elife. 2020;9 doi: 10.7554/eLife.58464. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper showed a method of determining a cryo-EM structure of a GPCR in the ligand free state by inceasing its size through antibody binding. Subsequent papers [67•, 69•, 70•] have introduced other methods to determine cryo-EM structures of GPCRs in an inactive state. It is not yet clear which method is most effective to get consistently high-resolution structures of a wide variety of GPCRs, but undoubtedly this important area will see new developments in the future.
- 69•.Zhang K., Wu H., Hoppe N., Manglik A., Cheng Y. Fusion protein strategies for cryo-EM study of G protein-coupled receptors. Nat Commun. 2022;13:4366. doi: 10.1038/s41467-022-32125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; See text for [68•]
- 70•.Xu J., Chen G., Wang H., Cao S., Heng J., Deupi X., Du Y., Kobilka B.K. Calcineurin-fusion facilitates cryo-EM structure determination of a family A GPCR. bioRxiv. 2022 doi: 10.1101/2022.03.27.485993. [DOI] [Google Scholar]; See text for [68•]
- 71.Garcia-Nafria J., Tate C.G. Cryo-electron microscopy: moving beyond X-ray crystal structures for drug receptors and drug development. Annu Rev Pharmacol Toxicol. 2020;60:51–71. doi: 10.1146/annurev-pharmtox-010919-023545. [DOI] [PubMed] [Google Scholar]
- 72.Su M., Zhu L., Zhang Y., Paknejad N., Dey R., Huang J., Lee M.Y., Williams D., Jordan K.D., Eng E.T., et al. Structural basis of the activation of heterotrimeric Gs-protein by isoproterenol-bound beta1-adrenergic receptor. Mol Cell. 2020;80:59–71 e54. doi: 10.1016/j.molcel.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao Y., Hu H., Ramachandran S., Erickson J.W., Cerione R.A., Skiniotis G. Structures of the rhodopsin-transducin complex:insights into G protein activation. Mol Cell. 2019;75:781–790. doi: 10.1016/j.molcel.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou X.E., He Y., de Waal P.W., Gao X., Kang Y., Van Eps N., Yin Y., Pal K., Goswami D., White T.A., et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Isberg V., Vroling B., van der Kant R., Li K., Vriend G., Gloriam D. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2014;42:422–425. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.