Abstract
Retroviral particles contain two positive-strand genomic RNAs linked together by noncovalent bonds that can be dissociated under mild conditions. We studied genomic RNAs of wild-type and mutant avian leukosis viruses (ALVs) in an attempt to (i) better understand the site(s) of RNA dimerization, (ii) examine whether the primer binding site (PBS) and tRNA primer are involved in dimerization, and (iii) determine the structure of genomic RNA in protease-deficient (PR−) mutants. We showed that extensively nicked wild-type ALV genomic RNAs melt cooperatively. This implies a complex secondary and/or tertiary structure for these RNAs that extends well beyond the 5′ dimerization site. To investigate the role of the PBS-tRNA complex in dimerization, we analyzed genomic RNAs from mutant viruses in which the tRNATrp PBS had been replaced with sequences homologous to the 3′ end of six other chicken tRNAs. We found the genomic RNAs of these viruses are dimers that dissociate at the same temperature as wild-type viral RNA, which suggests that the identity of the PBS and the tRNA primer do not affect dimer stability. We studied two ALV PR− mutants: one containing a large (>1.9-kb) inversion spanning the 3′ end of gag and much of pol, rendering it deficient in PR, reverse transcriptase, and integrase, and another with a point mutation in PR. In both of these mutant viruses, the genomic RNA appears to be either primarily or exclusively monomeric. These data suggest that ALV can package its RNA as monomers that subsequently dimerize.
The genomes of retroviruses consist of two identical sense-strand RNA monomers that individually sediment at 35S but are present in mature virions as a 70S dimer (48). One electron microscopic (EM) study that used bacteriophage T4 gene 32 protein to denature Rous sarcoma virus (RSV) genomic RNA showed complex structures which were interpreted as being linked at multiple sites (31). However, most EM studies revealed a relatively stable site of interaction near the 5′ end of viral genomic RNAs which has become known as the dimer linkage site (DLS) (2, 3, 27, 35; for reviews, see references 10 and 48). The DLS was easily visualized in RNA from mammalian type C retroviruses but proved more difficult to visualize in RNAs from avian retroviruses (2, 27, 35). Kung et al. (27) suggested the RSV RNA dimer may be less stable than other retroviral dimers, making it more difficult to visualize by EM. Another possible explanation is that the DLS of RSV is not much more resistant to denaturation than other sites of RNA-RNA interaction within the genome (10, 35).
Based on EM measurements, which are relatively inaccurate, the DLS of Moloney murine leukemia virus (MoMuLV) and RSV were estimated to be less than 50 bases long and located 300 to 500 nucleotides from the 5′ end of the genomic RNAs (3, 10, 35). Attempts have been made to determine the location of the DLS by studying spontaneous dimerization of partial RNA transcripts in vitro. These studies have been done with RNA from several retroviruses and have suggested that the DLS is relatively large and/or that multiple regions are involved in dimerization (RSV [4], Harvey sarcoma virus [15], and MoMuLV [41]). In the case of human immunodeficiency virus type 1 (HIV-1), in vitro studies point to a distinct region that seems to be directly involved in the dimer structure and have led to the “kissing-loop” model for initiation of RNA dimerization 8, 22, 28, 29, 34, 43; for a review, see reference 38). However, it is unclear whether the dimer structures seen in vitro are equivalent to those present in vivo. To test the extent of the interactions of the 35S subunits, we measured the melting behavior of intact and extensively nicked wild-type avian leukosis virus (ALV) genomic RNA.
Retroviral RNA dimers can be dissociated under mild conditions by heating or treatment with denaturing agents such as dimethyl sulfoxide, glyoxal, formamide, or urea (2, 3, 27, 35). This suggests the dimers are held together by some type of weak, noncovalent interactions such as hydrogen bonds. Since dimeric RNA is resistant to phenol extraction, sodium dodecyl sulfate (SDS), and digestion with proteases, either the dimers do not involve proteins or any proteins present are somehow shielded from these treatments (27, 46). Dimer linkages could involve base pairing directly between the subunits or could involve some sort of short RNA linkers (possibly tRNAs) as has been suggested by several groups (6, 27, 46). Based on the 5′ sequence of RSV strain Prague-C (RSV Pr-C), Haseltine and coworkers (23) proposed a complex model of the DLS that involves an antiparallel alignment of the genomic RNAs and base pairing between them and two tRNA primer molecules. An analogous model has been proposed for MoMuLV (10). To test this model, we measured the melting temperature of ALV RNAs containing different tRNA primers and primer binding sites (PBS) and of viral RNAs which do not have tRNA bound at the PBS.
Rapid-harvest virions collected at very short intervals (every 3 to 5 min) consist primarily of immature particles. Early work showed that in freshly budded RSV (6, 7) and visna virus (5), genomic RNA is monomeric but incubation of the virus results in dimerization. This implies that for RSV and visna virus, the viral genome is packaged as a 35S monomer and dimerization to 70S RNA occurs after budding. However, Stoltzfus and Snyder (46) reported that genomic RNA isolated from rapid-harvest RSV (strain B77) virions is a mixture of monomeric and dimeric RNAs. They also showed that extraction at a higher salt concentration or lower temperature resulted in a greater proportion of the RNA in dimeric form and that using higher-ionic-strength buffers also increased the melting temperature of the dimers.
Dimeric RNA from rapid-harvest RSV and MoMuLV has a lower Tm (temperature at which half of the RNA has dissociated) than RNA from mature virus (RSV [46] and MoMuLV [17]), and immature MoMuLV dimers migrate more slowly than mature dimers in nondenaturing agarose gels (17). Pure populations of immature viral particles can be prepared from protease-deficient (PR−) mutants; transient interactions that occur during virus assembly may be preserved in such mutant particles. Therefore, analysis of PR− mutants has been useful for studying viral morphogenesis. Dimeric RNAs from MoMuLV and HIV-1 PR− mutants are similar to those isolated from rapid-harvest virus in that these dimers exhibit lower melting temperatures and altered migration in agarose gels compared to wild-type RNAs (17, 18). This suggests that the dimeric RNA in immature MoMuLV and HIV-1 particles may be in a conformation different from that present in mature virions. This led to the hypothesis that MoMuLV and HIV-1 RNAs are initially packaged as immature dimers and subsequently undergo a structural change termed maturation to become stable, mature dimers (17, 18). However, in the case of HIV-1, mutations in the region of the RNA genome believed to be associated with the initiation of dimerization can result in the production of virions that contain what appears to be a mixture of monomeric and dimeric RNAs (9, 22). Although RNA maturation is a different phenomenon than the maturation of the viral proteins, the two processes may be inextricably linked since protein maturation seems to be a prerequisite for RNA maturation. It should be emphasized that RNA maturation refers to a conformational change that occurs after RNA dimerization.
Because of data showing the presence of both monomeric and dimeric RNAs in rapid-harvest RSV (46), it is not clear whether ALV packages monomeric RNAs or, like MoMuLV, immature dimers. The analysis of RNA isolated from immature particles is also ambiguous. One laboratory reported that RNA from an ALV PR active-site mutant is a mixture of monomeric and dimeric RNAs whose ratio varies between experiments (45), while another laboratory found only monomeric RNA (36). In an attempt to resolve these discrepancies, we have revisited the question of the status of genomic RNA in RSV pol and PR− mutants by examining their migration in nondenaturing agarose gels.
MATERIALS AND METHODS
Viruses.
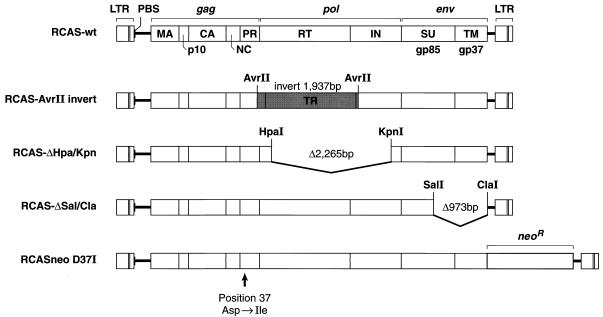
RCAS(A) and RCASBP(A) are replication-competent wild-type ALV vectors derived from the Schmidt-Ruppin (SR-A) strain of RSV (39). Mutant viruses were derived from RCAS(A) or RCASBP(A) by standard molecular biology techniques. Mutants RCASBP(tRNAMet), RCASBP(tRNAPro), RCASBP(tRNALys), RCASBP(tRNAPhe), RCASBP(tRNAIle), and RCASBP(tRNASer) were previously described by Whitcomb and coworkers (49) and are shown in Fig. 3. RCAS-AvrII, RCAS-ΔHpa/Kpn, and RCAS-ΔSal/Cla were described by Fu et al. (19), and their proviral structures are shown in Fig. 5. RCASneoD37I (45), containing a point mutation at the PR active site and the neomycin phosphotransferase gene (neo), was kindly supplied by Volker Vogt (Cornell University, Ithaca, N.Y.) and is depicted in Fig. 5.
FIG. 3.
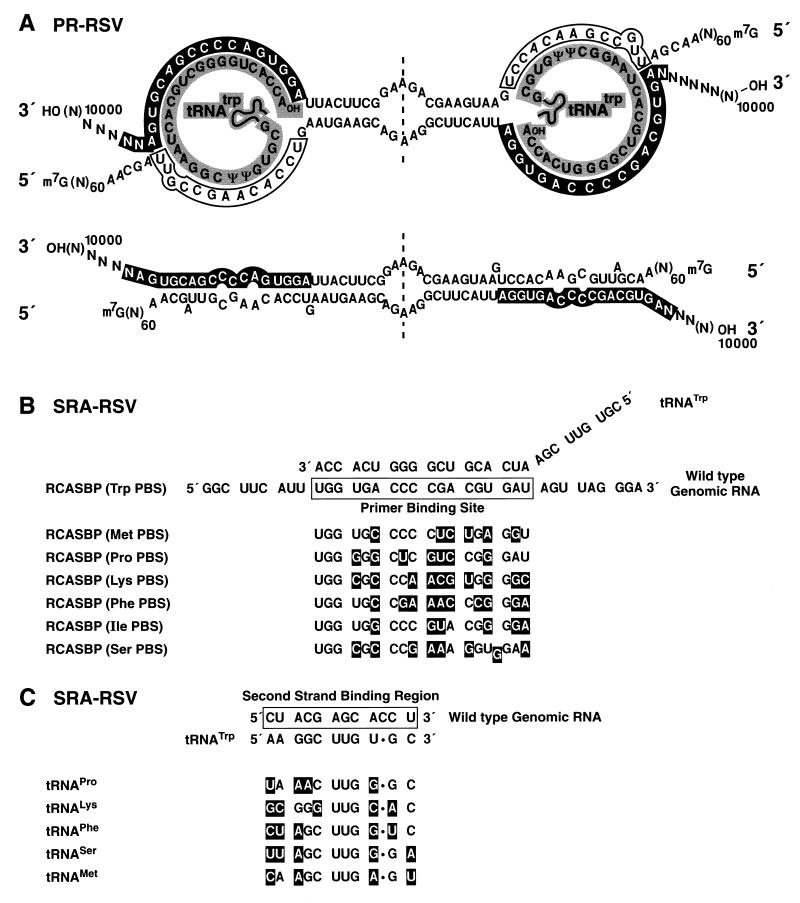
(A) Hypothetical model of RSV dimer linkage structure originally proposed by Haseltine et al. (23), based on the nucleotide sequence of RSV Pr-C strong-stop DNA. The diagram at the top shows potential base pairing between two antiparallel genomic RNA monomers and includes two tRNATrp primer molecules as part of the linkage. The tRNA primers are shaded in gray, PBSs are shown as white letters in black boxes, and the second-strand binding sites are enclosed by boxes. The scheme below shows base pairing between the U5 and PBS regions of the monomers in the absence of tRNA primers (redrawn, with permission, from reference 10). (B and C) Schematics of RSV SR-A derived wild-type RCASBP(A) and PBS mutants showing base pairing between a tRNA primer and each strand of genomic RNA. (B) Sequence of RSV SR-A derived RCASBP(A) wild-type PBS with the 3′ end of the tRNATrp annealed to it and also the altered PBS sequences in the mutant viruses. The white letters in black boxes signify the differences between the wild-type and mutant PBS sequences. The tRNA anticodon specificities are tRNAMet(CAU), tRNAPro(AGG), tRNALys(CUU), tRNAPhe(GAA), tRNAIle(AAU), and tRNASer(UCA). (These mutants were previously described in and the figure is redrawn from reference 49.) (C) Proposed base pairing between RSV SR-A-derived wild-type RCASBP(A) genomic RNA and the tRNATrp primer molecule of the second genomic RNA strand showing mispairing between alternatively specified tRNAs of the mutant viruses and the putative secondary binding site in genomic RNA. The white letters in black boxes signify the differences between the wild-type and mutant tRNA sequences.
FIG. 5.
Diagram of genomic structures of wild-type ALV/RCAS(A) (RCAS-wt) and mutant proviruses. Mutant RCAS-AvrII has an inversion of the sequence between nucleotides 2435 and 4372, disrupting PR, RT, and IN. RCAS-ΔHpa/Kpn has a deletion of nucleotides 2734 to 4999 disrupting RT and IN. RCAS-ΔSal/Cla has a deletion of nucleotides 6057 to 7030 disrupting SU (surface) and TM (transmembrane) proteins. Nucleotide positions are given with the 5′ end of the genomic RNA taken as position 1 (these mutants were described in and the figure is redrawn from reference 19). Mutant RCASneoD37I is a point mutant in which the PR active-site aspartic acid at position 37 is changed to an isoleucine, rendering it PR−. LTR, long terminal repeat; MA, CA, and NC, matrix, capsid, and nucleocapsid, respectively.
Cell culture.
Viruses were grown in either chicken embryo fibroblasts (CEFs) or DF-1 (42), a CEF-derived cell line. Both the CEFs and the DF-1 cell line were derived from EV-O chick embryos and do not harbor endogenous proviruses closely related to exogenous avian sarcoma-leukosis retroviruses (ASLV) (1). All cells were grown in Dulbecco’s modified Eagle medium (Life Technologies, Inc., Grand Island, N.Y.) supplemented with 10% tryptose-phosphate broth (Life Technologies), 5% heat-inactivated fetal calf serum (HyClone Laboratories, Logan, Utah), 5% heat-inactivated newborn calf serum (Life Technologies), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Quality Biological, Inc., Gaithersburg, Md.). Cells were split 1:3 at confluence and refed with 10 ml of medium per 100-mm-diameter tissue culture dish (Becton Dickinson, Bedford, Mass.), usually every other day.
Transfection, virus preparation, and RNA purification.
DF-1 cells were transfected by the CaPO4 technique (50) including a glycerol shock 4 h after the addition of the precipitate (24). For the replication-competent mutants (all PBS mutants), cells were passaged every other day and supernatant was collected just before passage. For the deletion and inversion mutants (which were replication defective), the cells were not passaged after transfection; instead, viral supernatants were collected at 24, 48, and 72 h after transfection. All supernatants were stored at −80°C and were clarified by sedimentation at 3,000 rpm for 10 min in a Sorvall RC-3 centrifuge and filtration through a 0.45-μm-pore-size polyethersulfone filter (Nalge Nunc International, Rochester, N.Y.). Virions were pelleted through a 15% sucrose cushion by sedimentation at 25,000 rpm for 1 h at 4°C, using a Beckman SW-27 rotor in a Beckman L-8 ultracentrifuge. For all experiments except the high-salt melting curve, RNA was purified as described previously (49) and stored in ethanol at −20°C prior to use. In the case of the high-salt experiment, the lysis buffer contained 50 mM Tris-Cl (pH 7.4), 0.5 M NaCl, 0.01 M EDTA, 0.1% SDS, and 100 μg of proteinase K per ml.
Protein preparation and Western analysis.
Viral supernatants (1 or 10 ml) were clarified by sedimentation at 3,000 rpm for 10 min in a Sorvall RC-3 centrifuge. Virions were pelleted through a 15% sucrose cushion by sedimentation at 35,000 rpm for 1 h at 4°C, using a Beckman SW-40 rotor in a Beckman L-8 ultracentrifuge. The supernatant was removed, and the virus pellet was resuspended in either 15 or 50 μl of protein gel loading buffer (125 mM Tris-Cl [pH 6.8], 1.25% SDS, 10% glycerol, 0.0125% bromophenol blue, 568 mM β-mercaptoethanol) by alternately freezing on dry ice and thawing at 50°C several times. The proteins were fractionated on an SDS–12% polyacrylamide gel with a 4% stacking gel run until the bromophenol blue dye reached the bottom of the gel, and molecular weights were estimated by comparison with prestained protein standards (Life Technologies). The proteins were transferred to nitrocellulose (Schleicher & Schuell, Keene, N.H.), baked at 80°C for 2 h under vacuum, and then detected as described previously (49).
Melting curves, gel electrophoresis, and RNA transfer.
Ethanol-precipitated RNA was collected by sedimentation at 14,000 rpm in an Eppendorf 5415 microcentrifuge (Brinkmann, Westbury, N.Y.) for 15 to 30 min at 4°C. The supernatant was removed by aspiration, and the pellet was dried briefly under vacuum. Viral genomic RNA was usually dissolved in R buffer (10 mM Tris-Cl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1% SDS), but high-salt R buffer (10 mM Tris-Cl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 0.1% SDS) was used for the high-salt experiment. For melting curves, RNA was aliquoted, incubated at various temperatures for 10 min, immediately transferred to ice, and then used for Northern blot analysis (26) essentially as described by Fu and Rein (17). Specifically, RNA was separated by electrophoresis through a 1% nondenaturing agarose gel in TBE buffer (89 mM Tris-Cl [pH 8.3], 89 mM boric acid, 2.5 mM EDTA) and subsequently denatured by incubation at 65°C in 3 gel volumes of 7% formaldehyde for 30 min. The RNA was transferred to GeneScreen Plus nylon membrane (Dupont NEN Research Products, Inc., Boston, Mass.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight and then baked at 80°C for 2 h under vacuum. For RNase nicking experiments, the RNA was resuspended in either R buffer (see above) or H buffer (20 mM HEPES [pH 8.0], 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol), DNA oligonucleotides were annealed to the RNA, and the complex was digested with Escherichia coli RNase H (Boehringer Mannheim, Indianapolis, Ind.) at 37°C for 10 min; then the RNA was aliquoted and a melting curve, gel electrophoresis, and Northern blotting were performed as described above.
Northern blot prehybridization, hybridization, and washes.
The procedure was adapted from that of Fu and Rein (17). Blots were prehybridized at 42°C for at least 3 h in a mixture containing 50% formamide, 10× Denhardt’s solution, 50 mM Tris-Cl (pH 7.8), 1 M NaCl, 1% SDS, 0.1% sodium pyrophosphate, 10% dextran sulfate, and 0.12 mg of salmon sperm DNA per ml. A riboprobe was generated by using pRPPI, a plasmid containing the RCAS(A) XhoI-KpnI fragment (spanning the pol/env junction) cloned into pBluescript KS (Stratagene, La Jolla, Calif.). pRPPI was linearized with Asp718, and T7 RNA polymerase (Promega Corp., Madison, Wis.). was used to synthesize a 32P-labeled antisense transcript homologous to nucleotides 4993 to 5256 of RCAS(A) (taking the 5′ end of the genomic RNA as position 1). Unincorporated nucleotides were removed on an RNase-free Quick-spin G25 column (Boehringer Mannheim). Hybridization was at 42°C for approximately 24 h in a solution containing 50% formamide, 10× Denhardt’s solution, 50 mM Tris-Cl (pH 7.8), 1% SDS, and 0.1% sodium pyrophosphate. Blots were washed in 0.2× SSC containing 0.1% SDS for 5 min at room temperature, then twice for 15 min/wash at 65°C, and finally for 3 to 5 min in room temperature diethyl pyrocarbonate-treated water. The blots were dried and exposed to X-ray film for a minimum of 30 min.
RESULTS
Melting profile of wild-type ALV RNA.
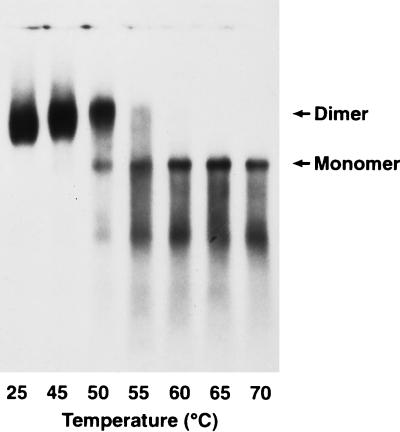
To study the conformation of wild-type ALV RNA, we transfected DNA for a molecular clone, RCASBP(A), into chicken cells, collected virus every 48 h, and used it for protein and RNA analyses. For all experiments, an aliquot of viral supernatant was used for Western blot analysis (as described in Materials and Methods) to check viral production and determine the amount of supernatant needed for the RNA experiments (data not shown). The remainder was subsequently used to purify genomic RNA, perform melting curves, separate the RNAs on nondenaturing agarose gels, and visualize the RNAs on Northern blots. Unless otherwise stated, all melting curves were done with the RNA resuspended in R buffer. This buffer was used in studies of the stability of MoMuLV RNA dimers in which unambiguous results were obtained (17). It should be noted that the stability of nucleic acid base pairs is, in general, highly dependent on the ionic strength of the buffer. As expected, Fu and coworkers have shown that increasing concentrations of Na+ stabilize dimeric HIV-1 RNA (18). Figure 1 shows the dimeric 70S genomic RNA present in mature wild-type RCASBP(A) virions; the dimeric 70S RNA can be dissociated into the more rapidly migrating 35S monomer upon heating. The monomeric RNA migrates as a much more discrete band, a characteristic of retroviral 35S molecules (17, 18). In contrast, the dimeric RNA migrates as a rather diffuse band that becomes slightly more compact and migrates slightly more slowly when the RNA is heated to 50°C and begins melting (Fig. 1, lane 3). This characteristic shift is also observed in genomic RNAs from MoMuLV and HIV-1 (17, 18). This suggests that retroviral RNAs undergo some sort of slight conformational change, perhaps a preliminary unfolding just before or as they begin to dissociate. Upon heating at 50°C (lane 3), some of the RNA dissociates into monomers, at 55°C (lane 4), a greater percentage of the dimers dissociate, and at 60°C (lane 5), all of the RNA is converted to monomers. Based on such melting profiles, we estimate the Tm of wild-type RCASBP/ALV in R buffer is about 55°C.
FIG. 1.
Northern blot showing thermal stability of genomic RNA from wild-type ALV/RCASBP(A) virus. Purified viral RNA was aliquoted and incubated at the indicated temperature for 10 min in standard R buffer. After fractionation on a 1% nondenaturing agarose gel, the RNA was transferred to a positively charged nylon membrane and hybridized with a 32P-labeled riboprobe homologous to nucleotides 4993 to 5256 (taking the 5′ end of the genomic RNA as position 1). Arrows indicate the positions of migration of the 70S RNA dimer and 35S RNA monomer.
Multiple sites or a single RNA interaction site.
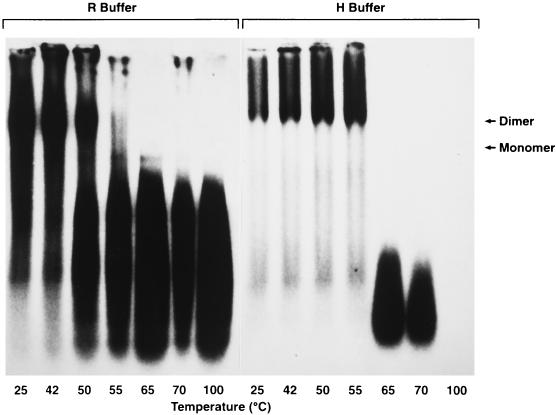
As discussed in the introduction, previous studies have shown that the most stable interactions between two retroviral monomers are in a region near the 5′ end of the RNA, called the DLS (2, 3, 8, 9, 27–29, 34, 35, 43). In an attempt to delineate the extent of the RSV DLS in full-length wild-type genomic RNA, we analyzed the denaturation of nicked genomic RNA. As shown in Fig. 2, in both R buffer and H buffer, the nicked RNAs melt cooperatively, suggesting that RNA-RNA interactions within the 70S dimer are much more extensive and complex than is implied by a small dimerization segment. As seen with the intact wild-type RNA (Fig. 1), heavily nicked wild-type RNA in R buffer begins to dissociate at 50°C (Fig. 2, lane 3) and is completely dissociated by heating at 65°C (lane 5). However, since H buffer contains a divalent cation (10 mM MgCl2) in addition to a monovalent one (50 mM KCl), dimeric RNA in this buffer does not begin to dissociate unless it is heated to a higher temperature, in this case, 65°C (compare lanes 3 and 12). This is presumably due to the stabilizing effect of the Mg2+ in the H buffer on the putative nucleic acid base pairs present in the RNA dimer. In agreement with this result, Fu and coworkers (18) also noted that Mg2+ stabilized wild-type HIV-1 RNA dimers.
FIG. 2.
Northern blot showing thermal stability of nicked genomic RNA from wild-type ALV/RCASBP(A) virus. Purified viral RNAs were resuspended in either standard R buffer or H buffer. Electrophoresis and Northern blot analysis were performed as described in the legend to Fig. 1.
tRNA involvement in RNA dimer structure.
As discussed in the introduction, Haseltine and coworkers (23) proposed a model for the structure of RSV dimeric RNA that includes two tRNATrp primer molecules in addition to the two genomic monomers. This model is shown in Fig. 3A, redrawn from reference 10. We decided to test this model by conducting melting curves on ALV RNAs containing different PBS. These mutants were previously described by Whitcomb et al. (49), and the sequences of their PBSs and putative tRNA second-strand-binding regions are shown in Fig. 3B and C, respectively. The mismatches between the wild-type tRNATrp primer and the alternative PBS are highlighted in Fig. 3B; similarly, the mismatches between the wild-type genomic RNA second-strand-binding region and the alternatively specified tRNAs are shown in Fig. 3C.
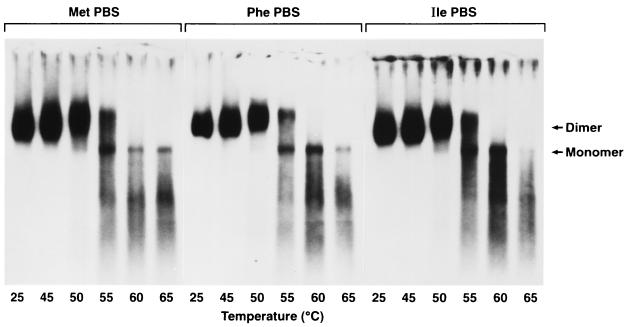
These PBS mutant viruses were transfected into CEFs and allowed to spread throughout the culture. Viral supernatants were collected every 48 h just prior to cell passage. We have previously shown these PBS mutants can use the alternatively specified tRNA for replication, but the PBS sequence does revert to wild type if the viruses are passaged several times from one cell culture to another (49). Previous studies showed that the virus never reverted during the time it takes the virus to spread throughout the cell culture following transfection. To ensure that the PBS had not reverted, virus was obtained from cells that had been directly transfected. As seen in Fig. 4, genomic RNAs from viruses with PBS sequences specifying tRNAMet, tRNAPhe, and tRNAIle all exhibit melting curves similar to those of RNA from wild-type virus (compare with Fig. 1). These dimeric RNAs dissociate partially at 55°C (Fig. 4, lanes 4, 10, and 16) and completely at 60°C (lanes 5, 11, and 17). Similar results were obtained with three other PBS mutants, RCASBP(ProPBS), RCASBP(LysPBS), and RCASBP(SerPBS) (data not shown). Therefore, all of the PBS mutant viruses contain dimeric RNAs that dissociate at essentially the same temperature as the wild type, showing that the identity of the PBS and primer tRNA does not measurably affect the formation or maintenance of the dimeric structure of these mutant viral RNAs.
FIG. 4.
Northern blot showing thermal stability of genomic RNAs from ALV/RCASBP(A) PBS mutants RCASBP(A)(MetPBS), RCASBP(A)(PhePBS), and RCASBP(A)(IlePBS). Arrows indicate the positions of migration of the dimeric and monomeric RNAs. Melting was done in standard R buffer at the indicated temperatures as described in the legend to Fig. 1. PBS mutant constructs are described in Fig. 3.
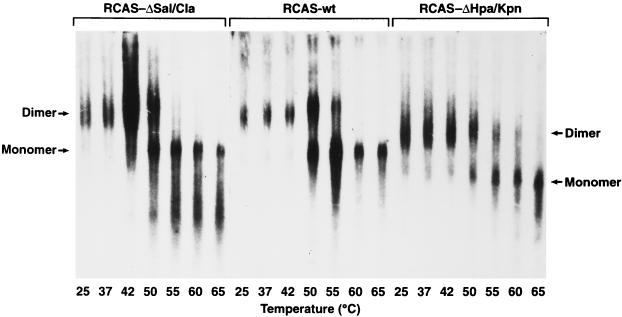
We also studied the melting profile of the pol mutant RCAS-ΔHpa/Kpn. Genomic RNA from this pol mutant has little or no primer tRNATrp annealed at the PBS (19). If the primer is involved in the formation and/or maintenance of the dimeric RNA structure, then genomic RNA from this mutant would be expected to have an altered structure that would be reflected in a difference in its melting profile. Because the large deletion in RCAS-ΔHpa/Kpn may have altered the overall secondary/tertiary structure of the RNA, which could affect the dimeric structure; we also studied the melting profile of RCAS-ΔSal/Cla, a mutant with a large deletion in env, as a control to check for nonspecific effects of large deletions on the melting of the genomic RNA dimers.
The mutant viral genomes are shown in Fig. 5 and have been previously described (19). Specifically, mutant RCAS-ΔHpa/Kpn has a 2.27-kb deletion spanning reverse transcriptase (RT) and integrase (IN) that removes all but the first 77 amino acids of RT. The other mutant, RCAS-ΔSal/Cla, has a 973-bp deletion in env, making it Env−. Since both of these viruses are defective, we used transient transfections of CEFs, collected viral supernatants at 24, 48, and 72 h, extracted the viral RNA, and analyzed the RNAs on Northern blots. Figure 6 shows the results of this RNA melting curve experiment. Both the pol mutant (RCAS-ΔHpa/Kpn) and the env (RCAS-ΔSal/Cla) mutant contain dimeric RNAs with melting curves similar to that of wild-type RNA (Fig. 6; compare lanes 1 to 7 and 15 to 21 with lanes 8 to 14). RNAs from all three viruses dissociate between 50 and 60°C (lanes 4 to 6, 11 to 13, and 18 to 20). Since RCAS-ΔHpa/Kpn has a very large deletion (2.27 kb), its monomers are significantly shorter than those of the wild type. Therefore, dimeric and monomeric RNAs from this mutant migrate more rapidly (lanes 15 to 21) than the corresponding wild-type RNAs (lanes 8 to 14). Taken together with the PBS mutant data, these data suggest the tRNA primer does not play a critical role either in the formation or stability of the dimer linkage.
FIG. 6.
Northern blot showing thermal stability of genomic RNAs from wild-type ALV/RCAS(A) and two deletion mutants. Virions were isolated from transiently transfected CEFs, and viral RNAs were purified. Melting was done in standard R buffer at the indicated temperatures as described in the legend to Fig. 1. RCAS-ΔHpa/Kpn is RT− and IN−, while RCAS-ΔSal/Cla is SU− and TM−. Mutants are further described in Fig. 5.
Genomic RNA structure in ALV PR− mutants.
Previous work with MoMuLV and HIV-1 has shown that dimeric RNA is present in PR− mutant particles but that the conformation of these RNA dimers differs from that of RNA from wild-type particles (17, 18). For ALV, one group reported that a PR− (active-site point) mutant contained a mixture of monomeric and dimeric RNAs (45), while another group found only monomeric RNA in their PR− mutant (36). To resolve this apparent discrepancy, we analyzed the status of genomic RNA in two ALV PR− mutants in an attempt to determine (i) whether monomers and/or dimers are present and (ii) if dimers are present, whether they are in an immature conformation.
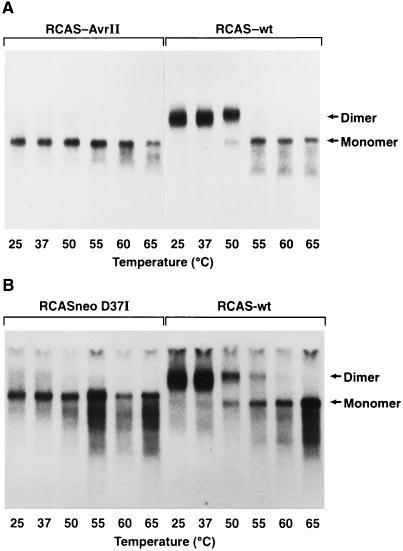
We analyzed the structure of genomic RNA from virions produced by PR− mutant RCAS-AvrII, which contains a large (almost 1.94-kb) inversion that encompasses the 3′ end of gag (p15) and more than half of pol (Fig. 5). Therefore, this mutant is defective in PR, RT, and IN. Wild-type RCAS(A) and RCAS-AvrII plasmids were separately transfected into DF-1 cells, and supernatants containing virus were harvested at 24, 48, and 72 h posttransfection. An aliquot of the supernatant was checked for virus production by Western blotting (data not shown), and the remainder was used for RNA isolation, melting curve, and Northern blotting.
35S RNA is present in RCAS-AvrII virus particles, but no 70S dimer is detected (Fig. 7A, lanes 1 to 6). In contrast, in the same experiment using the same buffer, after RNA incubation at 25°C and 37°C, RNA from wild-type virions is present only as dimers (lanes 7 and 8). The 70S RNA from wild-type virions dissociates into monomers upon heating to at least 50°C (lanes 9 to 12). Since mutant RCAS-AvrII contains such a large (>1.9-kb) inversion, there is the possibility that the overall structure of the RNA molecule could be affected, potentially interfering with dimerization.
FIG. 7.
Northern blot showing thermal stability of genomic RNAs from wild-type ALV/RCAS(A) and two PR− mutants. Virions were isolated from transiently transfected DF-1 cells, and viral RNAs were purified. Melting was done in standard R buffer at the indicated temperatures as described in the legend to Fig. 1. (A) Northern blot melting profiles of genomic RNAs from wild-type ALV/RCAS(A) (RCAS-wt) and mutant RCAS-AvrII. Mutant RCAS-AvrII is PR−, RT−, and IN− and is described further in Fig. 5. (B) Northern blot melting profiles of genomic RNAs from wild-type RSV and mutant RCASneoD37I. Mutant RCASneoD37I has a point mutation at the active site rendering it PR−.
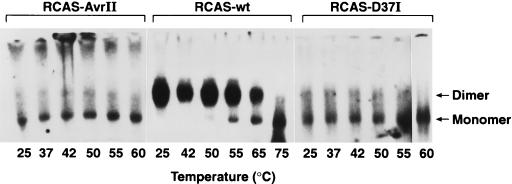
To rule out this possibility, RCASneoD37I, a virus with a point mutation in PR was obtained (45) (Fig. 5). This mutant has the PR active-site aspartic acid at position 37 replaced with an isoleucine, inactivating the PR. It also carries the gene for neomycin resistance cloned into the ClaI site. Genomic RNA from RCASneoD37I (Fig. 7B, lanes 1 to 6), is monomeric, as is the case for RCAS-AvrII. This is consistent with the results obtained by Oertle and Spahr (36), who examined another PR active-site mutant (Asp37Arg). In contrast, Stewart et al. (45) reported a mixture of dimeric and monomeric RNAs in an unheated RNA sample from RCASneoD37I. However, Stewart and coworkers extracted genomic RNA in the presence of 500 mM NaCl, while we extracted RNA in 100 mM NaCl and resuspended it in 50 mM NaCl. To rule out the possibility that the difference in results is due to the difference in the extraction and resuspension of the RNA, the experiments with both PR− mutants were repeated with high-salt conditions. Wild-type RCAS(A), RCAS-AvrII, and RCASneoD37I virions were lysed in the presence of 500 mM NaCl (rather than the usual 100 mM NaCl), and the RNAs were resuspended in high-salt R buffer, containing 500 (rather than 50) mM NaCl. The RNAs were incubated at various temperatures for 10 min and fractionated on a nondenaturing agarose gel that was used for a Northern blot (Fig. 8). Although it is necessary to use higher temperatures to dissociate wild-type RNA dimers (Fig. 8, lanes 7 to 12) under these conditions, RNA obtained from the two PR− mutants, RCAS-AvrII (lanes 1 to 6) and RCASneoD37I (lanes 13 to 18), is still primarily monomeric. The background in this experiment is higher, and it is possible that a small amount of dimeric RNA is present; however, the vast majority of the genomic RNA is clearly monomeric. Since PR− mutants produce immature particles, these data support the theory that RSV packages its genomic RNA as 35S molecules that later dimerize.
FIG. 8.
Northern blot showing thermal stability of genomic RNAs from wild-type ALV/RCAS(A) (RCAS-wt) and mutants RCAS-AvrII and RCASneoD37I. Virions were isolated from transiently transfected DF-1 cells and lysed in high-salt (500 mM NaCl) buffer, and viral RNAs were purified. Melting was done at the indicated temperatures as described in the legend to Fig. 1 except that RNA was suspended in high-salt buffer. Arrows indicate positions of migration of the dimeric and monomeric RNAs. Mutants are further described in Fig. 5.
DISCUSSION
Assembly of type C retroviruses in infected cells begins with the formation of a core structure at the host cell membrane that contains the Gag and Gag-Pol polyprotein precursors, two 35S viral RNAs, and tRNA primer (13; for a review of retroviral assembly, see reference 47). The core buds through the cell membrane, picking up the envelope proteins to become a noninfectious immature particle. The Gag precursor protein is the only viral gene product required for assembly and budding (47). As a late step that occurs either during budding or shortly thereafter, the viral protease cleaves the Gag precursor into the mature matrix, capsid, and nucleocapsid (NC) proteins (48, 51). Concomitant or subsequent protein core maturation results in a mature infectious virion containing dimeric viral RNA tightly associated with NC (10, 48, 51). NC is required for genomic RNA packaging (12, 13, 20, 21) as part of the full-length Gag polyprotein (13, 32, 36, 45) and is believed to interact with psi/E, the encapsidation signal present in the viral genomic RNA. NC has also been suggested to play a role in RNA dimerization (13, 32).
It is not definitively known whether retroviral genomic RNAs are packaged as monomers or dimers (9). The apparent overlap of sequences involved in RNA packaging and dimerization suggests a close relationship between these two processes (2, 3, 35). Only dimeric RNA has been isolated from murine rapid-harvest (14) and PR− mutant (17) retrovirus particles and from HIV-1 PR− mutant particles (18). These data led to the hypothesis that RNA dimerization is required for packaging of genomic RNA (17, 18). The model proposes that NC, as part of the Gag polyprotein, is capable of recognizing the packaging signal only in the context of dimeric 70S RNA. This theory is attractive because it explains how the virus ensures that two genomic RNAs are packaged into each virion. However, the data do not exclude the possibility that monomers packaged by MuLV and HIV-1 are converted to dimers so quickly that only dimeric RNA can be recovered from particles.
When carefully examined, the dimeric RNAs from rapid-harvest MoMuLV and PR− mutant MoMuLV and HIV-1 were found to have an altered conformation compared to dimers from mature virions. This immature conformation is characterized by a lower Tm, lower sedimentation rate, and altered migration on nondenaturing agarose gels compared to mature dimers (17, 18). Following protein maturation, the immature dimers undergo some sort of conformational change, also called maturation, which results in a more stable RNA dimer that dissociates at a higher temperature (17, 18). Mature NC protein is thought to mediate the RNA conformational change since in vitro experiments show that HIV-1 NC is capable of converting the transient kissing-loop complex formed by partial HIV-1 transcripts to a more stable interaction (33). HIV-1 NC protein was also shown to have a similar stabilizing effect on dimers formed by a synthetic fragment of Harvey sarcoma virus (16). To summarize the proposed pathway for genomic RNA in MoMuLV and HIV-1: (i) genomic RNA forms immature dimers that are recognized and packaged by the Gag polyprotein, (ii) immature particles bud from the cell membrane, (iii) protein maturation occurs when the viral PR cleaves the Gag precursor to release the mature proteins, and (iv) mature NC mediates maturation (conformational change) of the dimeric RNA resulting in a mature, stable 70S dimer (17, 18).
However, the idea that only RNA dimers are packaged is at odds with data from RSV and visna virus which clearly show the presence of monomeric RNAs in rapid harvest (5–7). Although, as has already been discussed, PR− mutants of HIV-1 contain immature RNA dimers, mutations in the region of the HIV-1 RNA believed to be associated with the initiation of dimerization can lead to the production of virions that contain mixtures of what appears to be monomeric and dimeric RNA (9, 22). Moreover, it has been reported that virions from PR− mutants of RSV contain monomeric RNA (36). Specifically, Oertle and Spahr (36) studied two ALV mutants that were incapable of processing Pr76gag, one with a mutation at the PR active site and the other at the cleavage site between NC and PR. In both mutants, the unprocessed Gag precursor is capable of packaging viral RNA but not dimerizing it. These data agree with results reported herein from experiments with two ALV PR− mutants, RCAS-AvrII (RT−, IN−, and PR−), with a large (1.94-kb) inversion, and RCASneoD37I (PR−), a PR active-site point mutant. We found the genomic RNAs of both of these mutants in monomeric form (Fig. 7). The current data confirm previous findings that RT, IN, and PR are not required for packaging genomic RNA (47).
There are two possible interpretations of the ALV data. The first is that ASLV genomic RNAs are actually packaged as dimers but the immature dimers are so unstable that they dissociate under the conditions used (50 mM NaCl). However, since immature MoMuLV (17) and HIV-1 (18) dimers were visualized in 50 and 100 mM NaCl, respectively, it seems unlikely that immature RSV dimers would not be sufficiently stable at 50 mM NaCl to be visualized. Stewart et al. (45) examined RNA from RCASneoD37I, the same PR− active-site mutant that we studied. They isolated RNA at 500 mM NaCl and found a mixture of monomeric and dimeric RNAs that varied between experiments. To test the possibility that our results differ from those of Stewart et al. (45) because of the different salt concentrations, we repeated our experiments with the two PR− mutants at high salt concentration (500 mM NaCl) and still found primarily monomeric RNA (Fig. 8). We cannot easily explain the differences in our results and those reported by Stewart et al. (45).
The second possible explanation for the presence of largely monomeric RNA in the ALV PR− particles that we examined is that these mutant viruses package monomeric RNA and are unable to dimerize it. We favor this interpretation of the data because it fits with the ALV rapid-harvest data, PR− mutant data, and the fact that dimeric RNA has never been isolated from infected cells. The idea that RNA encapsidation precedes dimerization and that core maturation is required for complete or proper dimerization was initially proposed based on the observation that genomic RNA isolated from RSV rapid-harvest virus is primarily monomeric RNA (6, 7). This idea was also used by Stewart et al. (45) and Oertle and Spahr (36) to explain their PR− mutant data. This implies that dimerization is not required for packaging of genomic RNA and that packaging and dimerization are separable events in ASLV. It may also hold true for visna virus, since 35S RNA has been recovered from rapid-harvest particles of this virus (5).
As stated above, ASLV genomic RNA is thought to be recognized and packaged by NC in the context of the Gag polyprotein (13, 32, 36). This agrees with our data and those of Stewart et al. (45) showing that RNA packaging seems to be unaffected in ASLV PR− mutants. However, since the RNA found in ASLV PR− particles (36) (Fig. 7 and 8) and also an NC/PR cleavage site mutant (36) is primarily monomeric, presumably processing of Pr76gag is required for RNA dimerization. This idea is supported by experiments that show RNA dimerization coincides with polyprotein cleavage and core maturation (7). Mature NC proteins have the ability to bind single-stranded nucleic acids (44) and catalyze the breakage (25) and formation (11, 40) of nucleic acid base pairs. It is tempting to speculate that the protein cleavage events that liberate NC result in RNA dimerization in ASLV. If this is true, then in the case of ASLV, protein maturation occurs before or concomitant with dimerization. To summarize the proposed genomic RNA pathway for ASLV: (i) monomeric RNA is recognized and packaged by the Gag polyprotein, (ii) immature particles bud from the cell membrane, (iii) protein maturation occurs when the viral PR cleaves the precursor proteins to release the mature proteins, and (iv) mature NC mediates the dimerization (and possibly maturation) of ASLV genomic RNA, resulting in a mature, stable 70S dimer.
As discussed above, the data suggest that RSV packages its genome as monomers but MoMuLV packages immature dimers. While it is clear that HIV-1 PR− mutants package immature dimers, some HIV-1 kissing-loop mutants contain mixtures of RNA monomers and dimers. These two disparate pieces of data do not provide a clear answer to the RNA packaging strategy employed by HIV-1. Why different retroviruses would have evolved to employ disparate RNA packaging strategies is not clear; however, evolution relies on chance, not direction. There are precedents for differences in the retroviral life cycle among this group of viruses. For example, ASLV is distinct from other retroviruses in that it encodes protease within the gag gene. Consequently, in ASLV infections, approximately 20 times more PR is produced and incorporated into virions than in cells infected with HIV-1 or MuLV (48). In addition, we have previously shown that a MoMuLV pol mutant is capable of properly annealing tRNA to its primer binding site whereas similar ALV mutants do not (19).
There are also unanswered questions about the exact nature of the genomic RNA dimer of retroviruses. Since 70S retroviral RNA can be dissociated under mild conditions and the dimers become more thermostable by increasing salt concentration, the RNAs may be held together by hydrogen bonding between nucleic acid base pairs (18, 27, 46) (compare Fig. 7 and 8). It has been proposed that the RSV tRNA primer plays an important role in the dimer linkage (23). We tested this model by measuring the melting temperature of ALV mutants that have alternate tRNA primers or no tRNA at the PBS. Since RNA from of all these viruses melted at essentially the same temperature as wild-type RNA, we conclude that the tRNA primer does not play a critical role in forming or maintaining the dimeric structure of the RNA.
Although it is often proposed that the dimer linkage structure is a relatively small, discrete portion of the genome, nicked viral RNA migrates as a dimer (Fig. 2), suggesting that it is held together by RNA-RNA interactions involving many sites within the RNA genome. This idea is supported by EM (31) and in vitro dimerization (15) studies. It is not clear from the available data whether the interactions are inter- or intramolecular; however, the highly cooperative nature of the thermal denaturation suggests that the interactions are both extensive and complex. Even though there are, in the mature RNA dimer, such complex interactions, it is possible that the elements near the 5′ end of the RNA are important for the initiation and/or stabilization of the RNA dimer.
Why are retroviral genomes actual physical dimers and what function, if any, does the dimeric RNA structure serve? The 5′ leader region of retroviral RNAs is highly structured, and phylogenetic analyses show that a hairpin structure within the DLS is highly conserved among members of this family. The fact that there is selection pressure to maintain the structure within this region suggests that it is biologically important, and mutagenesis studies support the idea that the DLS is biologically important (9, 30, 37). In addition, the location of the primary site of the DLS near the 5′ end of the virus near elements that function in transcription, translation, encapsidation, and recombination suggest that dimerization may play a role in these functions (38). Since the structure of the RNA dimer has been postulated to regulate several vital steps within the retroviral life cycle, the elucidation of the precise nature and extent of the RNA interactions within the DLS may not only help us to better understand retroviral replication but also lead to the development of novel strategies for combating these pathogens.
ACKNOWLEDGMENTS
We thank William Fu for technical instruction and Alan Rein for helpful discussion and insightful advice and suggestions. We also thank Volker Vogt for providing mutant RCASneoD37I and Hilda Marusiodis for superb secretarial assistance.
This research was sponsored by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Astrin S M, Buss E G, Hayward W S. Endogenous viral genes are non-essential in the chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 2.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 3.Bender W, Chien Y-H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieth E, Gabus C, Darlix J-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–128. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahic M, Vigne R. Properties of visna virus particles harvested at short time intervals: RNA content, infectivity, and ultrastructure. J Virol. 1975;15:1222–1230. doi: 10.1128/jvi.15.5.1222-1230.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canaani E, Helm K V D, Duesberg P. Evidence for the 30-40S RNA as precursor of the 60-70S RNA of Rous sarcoma virus. Proc Natl Acad Sci USA. 1973;70:401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung K S, Smith R E, Stone M P, Joklik W K. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology. 1972;50:851–864. doi: 10.1016/0042-6822(72)90439-4. [DOI] [PubMed] [Google Scholar]
- 8.Clever J L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J. Genome structure. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 261–368. [Google Scholar]
- 11.Dib-Hajj F, Khan R, Giedroc D P. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–243. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupraz P, Oertle S, Meric C, Damay P, Spahr P-F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupraz P, Spahr P-F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.East J L, Allen P T, Knesek J E, Chan J C, Bowen J M, Dmochowski L. Structural rearrangement and subunit composition of RNA from released Soehner-Dmochowski murine sarcoma virions. J Virol. 1973;11:709–720. doi: 10.1128/jvi.11.5.709-720.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y-X, Fu W, Winter A J, Levin J G, Rein A. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J Virol. 1995;69:2486–2490. doi: 10.1128/jvi.69.4.2486-2490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type I nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing-loop structure facilitates genomic RNA dimerisation in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 23.Haseltine W A, Maxam A M, Gilbert W. Rous sarcoma virus genome is terminally redundant: the 5′ sequence. Proc Natl Acad Sci USA. 1977;74:989–993. doi: 10.1073/pnas.74.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes S H, Kosik E. Mutagenesis of the region between env and src of the SR-A strain of Rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology. 1984;236:89–99. doi: 10.1016/0042-6822(84)90250-2. [DOI] [PubMed] [Google Scholar]
- 25.Khan R, Giedroc D P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- 26.Khandjian E W, Meric C. A procedure for Northern blot analysis of native RNA. Anal Biochem. 1986;159:227–232. doi: 10.1016/0003-2697(86)90332-5. [DOI] [PubMed] [Google Scholar]
- 27.Kung H-J, Hu S, Bender W, Bailey J M, Davidson N. RD-114, baboon, and woolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 28.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 29.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 30.Laughrea M, Jette L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangel W F, Delius H, Duesberg P H. Structure and molecular weight of the 60-70S RNA and the 30-40S RNA of the Rous sarcoma virus. Proc Natl Acad Sci USA. 1974;71:4541–4545. doi: 10.1073/pnas.71.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meric C, Spahr P-F. Rous sarcoma virus nucleic acid binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muriaux D, De Rocquigny H, Roques B-P, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 34.Muriaux D, Girard P-M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. J Biol Chem. 1996;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 35.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oertle S, Spahr P-F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paillart J C, Berthoux L, Ottman M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral RNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillart J C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 39.Petropoulos C J, Hughes Replication-competent retroviral vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prats A C, Roy C, Wang P A, Erard M, Housset V, Gabus C, Paoletti C, Darlix J L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaefer-Klein J, Givol I, Barsov E V, Whitcomb J M, VanBrocklin M, Foster D N, Federspiel M J, Hughes S H. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 43.Skripkin E, Paillart J-C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith B J, Bailey J M. The binding of an avian myeloblastosis virus basic 12,000 dalton protein to nucleic acids. Nucleic Acids Res. 1979;7:2055–2972. doi: 10.1093/nar/7.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart L, Schatz G, Vogt V. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoltzfus C M, Snyder P N. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975;16:1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 48.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 49.Whitcomb J M, Ortiz-Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]