Abstract
Oncolytic viruses, modified for tumor-restricted infection, are a promising cancer immunotherapeutic, yet much remains to be understood about factors driving their activity and outcome in the tumor microenvironment. Here, we report that oncolytic herpes simplex virus C134, previously found to exert T cell-dependent efficacy in mouse models of glioblastoma, exerts T cell-independent efficacy in mouse models of medulloblastoma, indicating this oncolytic virus uses different mechanisms in different tumors. We investigated C134’s behavior in mouse medulloblastomas, using single cell RNA sequencing to map C134-induced gene expression changes across cell types, timepoints, and medulloblastoma subgroup models at whole-transcriptome resolution. Our work details substantial oncolytic virus-induced transcriptional remodeling of medulloblastoma-infiltrating immune cells, 10 subpopulations of monocytes and macrophages collectively demonstrating M1-like responses to C134, and suggests C134 be investigated as a potential new therapy for medulloblastoma.
Keywords: oncolytic virus, medulloblastoma, brain tumors, cancer immunotherapy, pediatric cancer
Graphical abstract
Miller et al. show that OV C134 exhibits different mechanisms of efficacy in different brain tumors, prolonging median survival of medulloblastoma-bearing mice regardless of T cell presence, and inducing significant alterations in the tumor immune microenvironment, particularly through induction of an M1-like macrophage response.
Introduction
Medulloblastoma represents ∼20% of pediatric brain tumors and has an overall mortality rate of 30%.1,2,3,4,5,6 Four major molecular subgroups of medulloblastoma have been defined—WNT, sonic hedgehog (SHH), group 3, and group 4, with characteristic somatic alterations and prognoses. Sequencing studies have shown medulloblastomas to be “immune cold” tumors and although several immunotherapy modalities for medulloblastoma including cancer vaccines, CAR T cell therapy, and immune checkpoint inhibitors are being investigated, continued exploration of new therapies is desperately needed.7,8,9 One such class of immunotherapy emerging for CNS tumors and other malignancies is oncolytic virotherapy. Premised on the concept that the aberrant cellular signaling driving oncogenesis enables increased susceptibility to viral replication, oncolytic viruses (OVs) have been modified to improve tumor specificity and reduce off-target virulence.10 Herpes simplex 1-based OVs, which are neurotropic and thus primed for use in CNS malignancies, have been investigated for three decades.11,12 In 2015, a modified oncolytic herpes simplex 1 virus (oHSV), Talimogene laherparepvec (TVEC) was the first OV approved by the U.S. Food and Drug Administration (FDA) for the treatment of advanced melanoma.13,14 At present, more than 50 studies of OVs are active and/or recruiting in the United States (clinicaltrials.gov searched “oncolytic virus” on June 22, 2023, filtered for “recruiting” and “active, not recruiting”).
C134 is a second-generation chimeric oHSV with both copies of its principal virulence gene, γ134.5, knocked out to improve safety and decrease neurovirulence. Additionally, C134 contains the human cytomegalovirus IRS1 gene knocked in to selectively improve the viral protein synthesis deficits that otherwise accompany γ134.5 disruption.15 C134 was proven to be safe in pre-clinical assessments and is currently undergoing a phase I clinical trial for recurrent glioblastoma (NCT03657576).16,17 Additionally, in a phase Ib trial of patients with recurrent glioblastoma treated with a similar oHSV, G207, expression of approximately 500 genes, one-half of which were immune-related, correlated with patient survival.18,19,20 These early clinical studies are establishing the safety of oHSV and indicate that focus should be given to understanding their intratumoral immune-modulating activities.
Here, we evaluated the therapeutic potential of C134 in two syngeneic mouse medulloblastoma models, one that aligns with the gene expression profile associated with the SHH subgroup (hereafter, MYCN) medulloblastoma, and another that aligns with group 3 subgroup (hereafter, CMYC).21,22 We used single cell RNA-sequencing (scRNA-seq), which revealed that C134 remodels the transcriptome of the entire medulloblastoma immune compartment, particularly altering genes related to antigen presentation, interferon signaling, and cytokine signaling throughout these tumors. Furthermore, C134 treatment caused statistically significant increases in M1 macrophages, dendritic cells, CD4+ and CD8+ T lymphocytes, and natural killer T (NKT) cells. Additionally, we demonstrate that C134 strongly induced an M1 gene expression signature in macrophages, a finding consistent with continued C134 efficacy in our athymic nude mice.
Results
Oncolytic HSV therapy increases survival in syngeneic models of medulloblastoma
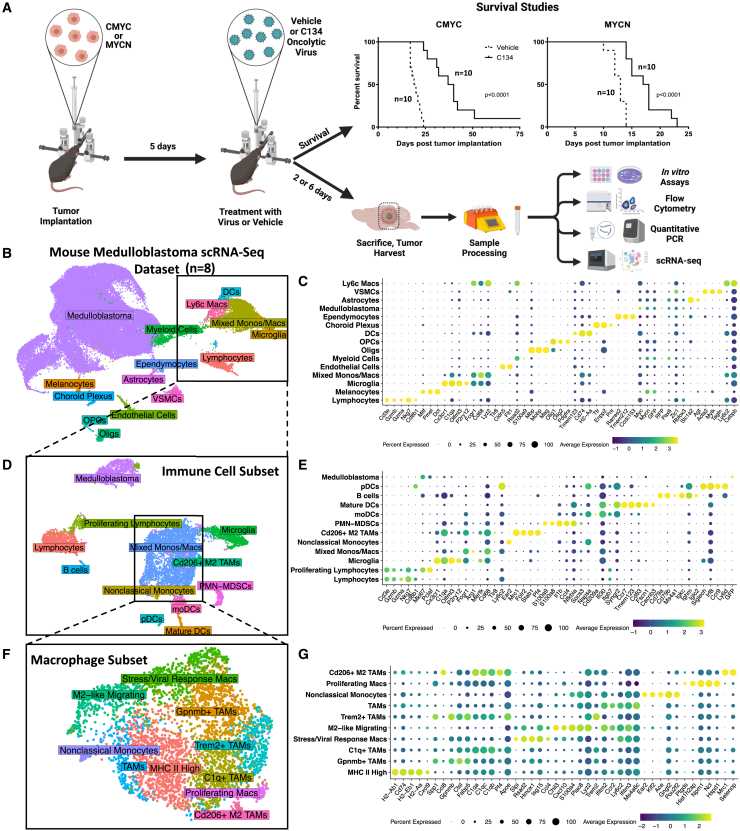
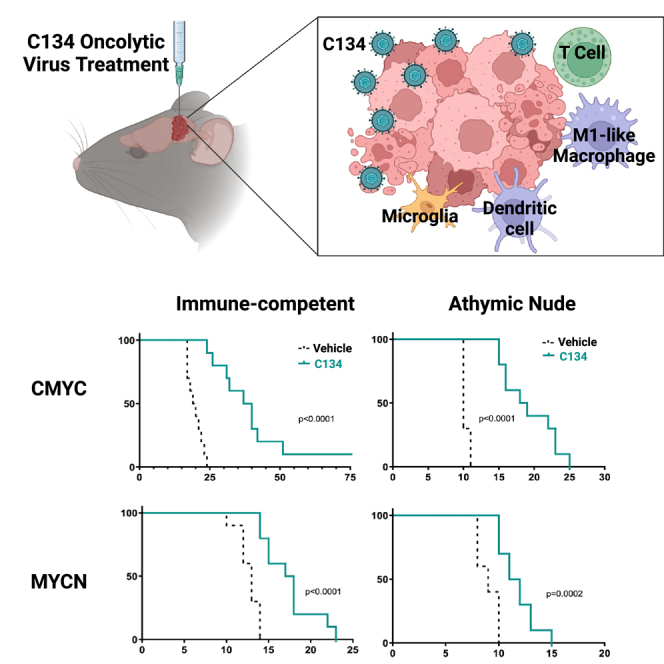
We evaluated the efficacy of C134 (distinct from first-generation Δγ134.5 oHSVs) in syngeneic orthotopic medulloblastoma models representing two subgroups of the human disease. Both models showed statistically significant improvement in overall survival when treated with C134 compared with vehicle (PBS) (Figure 1A). The group 3 medulloblastoma model (CMYC) showed the most significant response (p < 0.0001), with a median survival in C134-treated animals of 38.5 days (range 24–75 days, n = 10) vs. 19.5 days (range 17–24 days, n = 10) in vehicle-treated animals. Median survival for C134-treated MYCN tumor-bearing mice was 17.5 days (range 14–23 days, n = 10), a statistically significant (p < 0.0001) increase vs. 13 days (range, 10–14 days; n = 10) for vehicle-treated mice.
Figure 1.
Single cell analysis of C134-treated mouse medulloblastomas
(A) Summary of orthotopic medulloblastoma mouse model experiments and survival data. Medulloblastoma tumors were established in the right cerebral cortices of immune-competent C57BL/6 mice. Five days after tumor implantation, the mice received 1 × 107 pfu of C134 or vehicle. Mice were then either followed for survival studies, or on days 2 or 6 post-treatment, sacrificed, and brain tumors were harvested and processed for downstream sample characterization experiments. Kaplan-Meier curves depict differences in survival and statistical differences determined using the log rank test. UMAP plots show annotated cell types from single cell RNA sequencing of mouse medulloblastomas for the overall dataset (B), immune cell subset (D), and macrophage subset (F). Accompanying dot plots show normalized expression of marker genes by cell types in the overall dataset (C), the immune cell subset (E), and macrophage subset (G).
To determine whether the therapeutic efficacy observed in immunocompetent C57BL/6 animals was T cell dependent, we also tested virotherapy in athymic nude mice. In the absence of functional T cells, C134 treatment continued to demonstrate statistically significant efficacy in the CMYC (p < 0.0001) and MYCN (p = 0.0002) syngeneic models (Figure S1). Median survival for C134-treated CMYC tumor-bearing mice was 18.5 days (range, 15–25 days; n = 10) vs. 10 days (range, 10–11 days; n = 10) in vehicle-treated mice, while median survival of C134-treated MYCN tumor-bearing mice was 11.5 days (range, 11–15 days; n = 10) vs. 9 days (range, 8–10 days; n = 10) in vehicle-treated mice.
Mouse and human medulloblastoma cells display differential C134-mediated cytotoxicity
We next sought to quantify C134’s replicative potential and cytotoxic effects in medulloblastoma cells, as they have not been previously investigated in this histology. This important first step predicts the potential safety and efficacy in humans, namely, whether the oncolytic activity is high enough to generate antitumor activity, but low enough to avoid causing autoimmune sequelae that jeopardize safety. To determine whether the efficacy observed in the syngeneic models was associated with oncolysis, we tested the viability of the two murine medulloblastoma cell lines after infection with C134 in vitro (Figure S2A). The CMYC- and MYCN-derived cell lines displayed sensitivity to the virus in dose- and time-dependent manners. CMYC showed the highest amount of cytotoxicity at 48 h after infection at a multiplicity of infection (MOI) of 1 infectious virion per cell, with 45% of cells viable, compared with 80% for MYCN. After 96 h at an MOI of 1, 25% (CMYC) and 45% (MYCN) of the cells remained viable. In contrast, human-derived medulloblastoma cell lines D283med (derived from peritoneal metastatic medulloblastoma cells23) and D425med (derived from a group 3 medulloblastoma24) were more sensitive than the mouse model-derived lines to C134-mediated killing (Figure S2B). At 48 h after virus infection, 25% of the D283med and D425med cells remained viable at an MOI of 0.1, and by 96 h, less than 10% of cells remained viable at an MOI of 0.01.
Different amounts of C134 recovered from mouse vs. human medulloblastoma cells
We next examined the amount of virus that could be recovered in vitro from cell lines derived from resected murine medulloblastoma tumors using standard plaque assays on Vero cells (Figure S3A). The MYCN tumor model-derived cell line was the only one from which amounts of recovered virus increased over time. However, there was a less than 25% increase in virus yield over input at any of the time points evaluated for MCYN. To test whether CMYC and MYCN models supported C134 replication in vivo, we quantified virus recoverable from CMYC and MYCN tumors using the same implantation and treatment regimen as performed in the survival study (Figure 1A). After resection of brain tumor quadrants at 3, 24, and 96 h after treatment with C134, we quantified the amount of virus by standard plaque assays on Vero cells. As demonstrated at the 24- and 96-h timepoints, none of the tumor models supported virus replication (Figure S3B), as we detected less than 10% of the input virus (at 3 h). Conversely, when viral recovery was examined from D283med and D425med cells, substantial increases were observed, with 2- and 4-fold increases in virus yield, respectively (Figure S3C).
Single cell RNA-seq reveals diverse tumor-infiltrating immune cells in response to C134 treatment
To characterize post-virotherapy tumors, we performed scRNA-seq of freshly harvested mouse medulloblastomas. We sequenced a total of eight samples, consisting of medulloblastoma subgroup models CMYC or MYCN, vehicle or C134 treatment, and timepoints at day 2 or day 6 after treatment (timepoints relevant to the innate and early adaptive immune response in this model, respectively, based on our prior experience) (Figure 1A and Table S1).25 The resulting dataset from 55,703 high quality cells yielded 49 clusters corresponding with 15 cell types (Figure 1B–C and Table S1). We identified 32 of these clusters as medulloblastoma cells based on high expression of genes characteristic of the SHH and group 3 subgroups, as well as transcripts for GFP and RFP genes engineered into these models (Figure 1C and Table S1). Immune cell types we detected in these samples included lymphocytes (encompassing T cells, B cells, and NK cells), dendritic cells (DCs), microglia, myeloid cells, and a heterogeneous population of monocytes and macrophages. A variety of non-immune stromal cells formed distinct clusters in our analysis, including vascular smooth muscle cells, oligodendrocytes, and melanocytes.
Our immune subset included a total of 7,767 cells, which resulted in 17 clusters by UMAP projection representing more than 12 cell types (Figures 1D and 1E). The resulting increased resolution revealed cell types that included a mature DC population highly expressing Ccr7, monocyte-derived DCs, plasmacytoid DCs, B cells, activated M2 tumor-associated macrophages, Ear2+ nonclassical monocytes, and myeloid-derived suppressor cells. Additionally, a group of proliferating lymphocytes expressing Hist1h2ap, Hmgb2, Hist1h2ae, and Hist1h2ae clustered away from the primary lymphocyte cell cluster, despite regressing out cell cycle genes during normalization of gene counts in this dataset. We also identified a population of cells originating from our medulloblastoma models that expressed GFP, as well as genes negatively regulating apoptosis including Nme2, Ybx1, Ybx3, Mif, Ptma, and Npm1 (Table S1).
Ten distinct populations of macrophages in C134-treated medulloblastomas
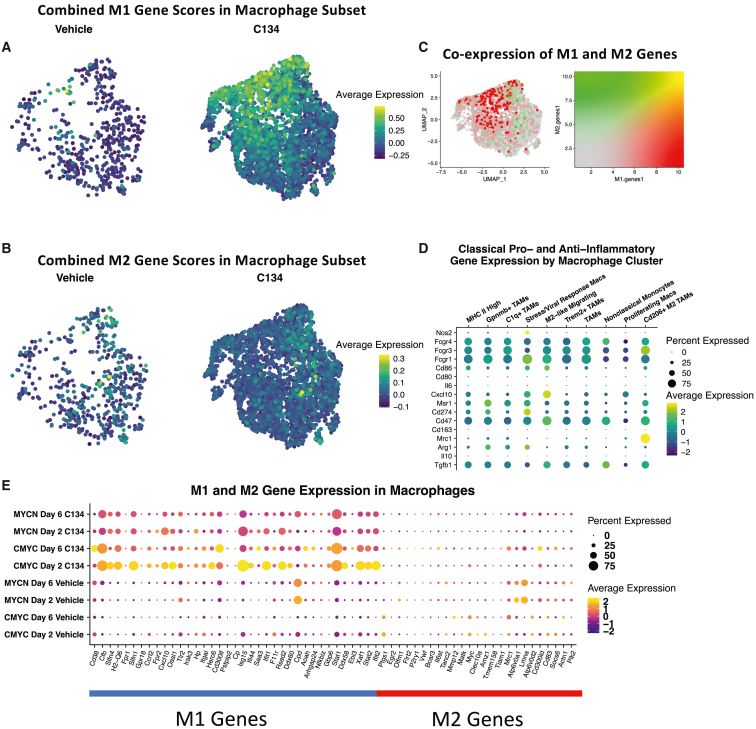
We sought to further refine subtypes of monocytes and macrophages present in the immune cell subset by computationally subsetting these cells into their own dataset, yielding 10 distinct populations of macrophages and monocytes (Figures 1F and 1G). Mapping gene signature scores for 53 murine M1 and 32 M2 genes identified in a landmark study26 demonstrated a strong cluster of M1 cells (Figure 2A), as well as a sparse presence of M2 cells (Figure 2B). Simultaneously mapping both gene signatures demonstrated that distinct cells were responsible for high signals in M1 vs. M2 macrophage-scoring genes (Figure 2C). Gene expression of a variety of tumor-relevant macrophage genes showed potentially major functional differences in these macrophage subtypes, including a population highly expressing Cd206 (Mrc1), a population highly expressing Nos2, and another population highly expressing MHC II genes (Figures 2D and 1G). Visualizing expression of specific M1 and M2 marker genes across samples in the macrophage subset demonstrated that C134-treated samples had a selective increase in M1 gene expression (Figure 2E).
Figure 2.
Macrophage subset analysis
Visually weighted expression of a murine M1 (A) and M2 (B) macrophage gene signature score across the macrophage subset split by treatment groups. (C) Simultaneous visual weighting of both M1 and M2 gene signatures to demonstrate individual cells that have high signal for both gene signatures (yellow), M1 genes (green), M2 genes (red), or neither (gray). (D) Expression of selected tumor-relevant immunomodulatory genes in macrophage subset by cell type. (E) Expression of M1 and M2 genes by scRNA-seq sample.
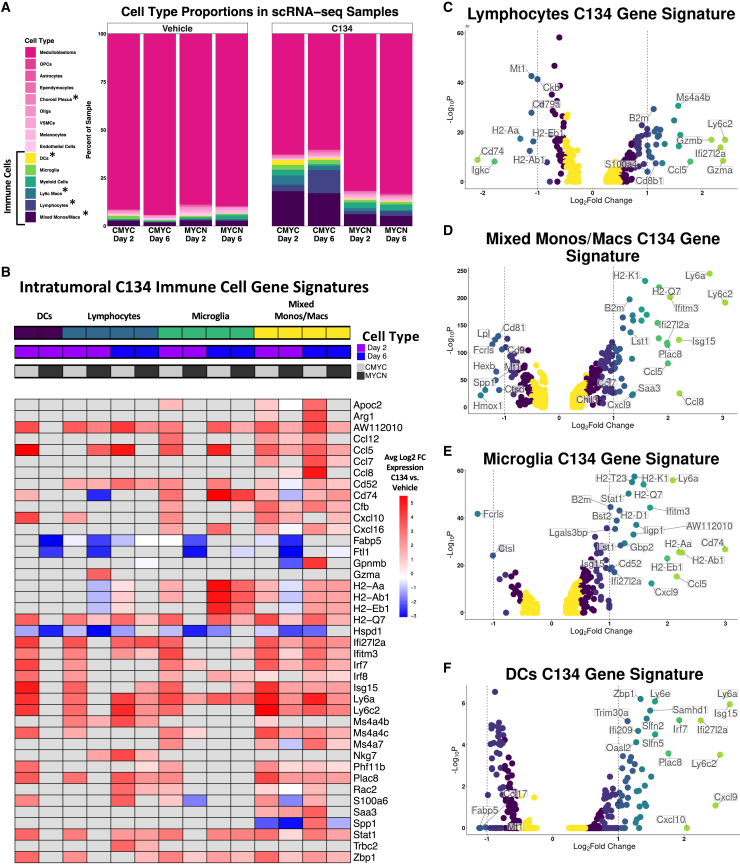
C134 induces differences in cell type proportions
We observed statistically significant (false discovery rate of <0.05) decreases in scRNA-seq sample proportions of medulloblastoma cells, and increases in mixed monocytes/macrophages, lymphocytes, Ly6c+ macrophages, DCs, and choroid plexus cells between C134- and vehicle-treated tumors (Figure 3A). Cell type proportions in the immune cell subset also showed increasing trends in several immune cell types, including monocytes/macrophages, lymphocytes (primarily T cells), microglia, and polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) (Figure S4).
Figure 3.
Analysis of C134-induced changes in immune cell infiltrate and gene expression in medulloblastomas
(A) Cell type proportions in C134 vs. vehicle-treated medulloblastomas. Differences in each respective cell type’s proportions in C134 vs. vehicle-treated samples were statistically evaluated using t-tests after logit-transformation and using an empirical Bayes framework. ∗Denotes a false discovery rate (FDR) of less than 0.05 (B) Inclusion criteria for genes were an average log2-fold change (Log2FC) of greater than 3 or less than −3 and an adjusted p value of less than 0.05 in any of the six immune cell types shown. The expression of these genes is shown for all immune cell types on the heatmap regardless of Log2FC, however, values with an adjusted p value of greater than 0.05 are colored in gray. Volcano plots show results of differential gene expression analysis performed on a cell type basis from pooling C134-treated and vehicle-treated samples for lymphocytes (C), mixed monocytes/macrophages (D), microglia (E), and DCs (F). Differentially expressed genes in (B–F) were evaluated using a Wilcoxon rank-sum test.
To compare scRNA-seq findings with protein-level information, we performed flow cytometry using the same experimental models, treatments, and time points as for scRNA-seq. This experiment demonstrated increased (p < 0.05) MHCIIhi/CD206lo M1-like macrophages in C134-treated tumors at day 6 (Figure S5A). MHCIIlo/CD206hi M2-like macrophages significantly decreased (p < 0.05) in MYCN medulloblastomas after C134 treatment, on both days 2 and 6 (Figure S5A). No such changes were observed in CMYC medulloblastomas, which appeared to have very few M2-like macrophages regardless of treatment or time point. However, C134-treated CMYC medulloblastomas, which had demonstrated the greatest increase in median survival, exhibited a significant (p < 0.01) increase in their ratio of M1 to M2 macrophages on post-treatment day 6 as compared with vehicle treatment on day 6 or C134 treatment on day 2 (Figure S5B). Abundances of tumor associated macrophages (F4/80+) and neutrophils (Ly6Ghi) were similar in C134- and vehicle-treated tumors at days 2 and 6 (Figure S5C). MDSCs (Ly6Chi) were more abundant in the C134-treated tumors at days 2 and 6. Evaluation of additional cell types of the innate immune response revealed a non-statistically significant increase in NK cells after C134 treatment on days 2 and 6, but a statistically significant (p < 0.05) increase in NKT cells (B220−CD49b+CD3e+) after C134 treatment at day 6 (Figure S5D).
We observed adaptive immune responses at day 6 within the tumor microenvironment of both CMYC and MYCN tumors after C134 treatment (Figure S6A). This included a statistically significant (p < 0.05) increase in the percentage of CD4+ and CD8+ T cells, as well as antigen-experienced (CD44+) T cells (Figures S6A and S6B). Evaluation of immune checkpoint gene expression showed an increase (p < 0.01) in programmed cell death protein 1 (PD-1) expression on CD4+ and CD8+ T cells isolated from both tumor models at day 6 after C134 treatment, but not in controls (Figure S6B). Concomitant with C134 treatment was an increase in the mean fluorescence intensity of cells within the tumor microenvironment expressing the immune checkpoint molecule PD-L1 (Figure S6C). The percentage of Lag3+ CD4+ and Lag3+CD8+ T cells also increased in both models on day 6 only after C134 treatment (Figure S7).
scRNA-seq differential gene expression
We next investigated differences in gene expression induced by C134 in each scRNA-seq-identified cell type. This revealed significantly (adjusted p < 0.05) up- and down-regulated genes throughout many cell types in C134-treated medulloblastomas as compared with vehicle, including DCs, lymphocytes, microglia, and mixed monocytes/macrophages, among many others (Figure 3B and Table S1). Lymphocytes increased expression of Gzma, Gzmb, Nkg7, and decreased expression of major histocompatibility complex (MHC) class II presentation genes H2-Aa, H2-Eb1, H2-Ab1, and Cd74 (Figure 3C). Monocytes, macrophages, and microglia increased expression of multiple MHC class I genes, cytokines, and chemokines (Figures 3D and 3E). DCs increased expression of multiple interferon-response genes and chemokines including Cxcl9 and Cxcl10 (Figure 3F). We observed similar gene expression signatures when C134 vs. vehicle samples were analyzed for each medulloblastoma subgroup model independently (Table S1).
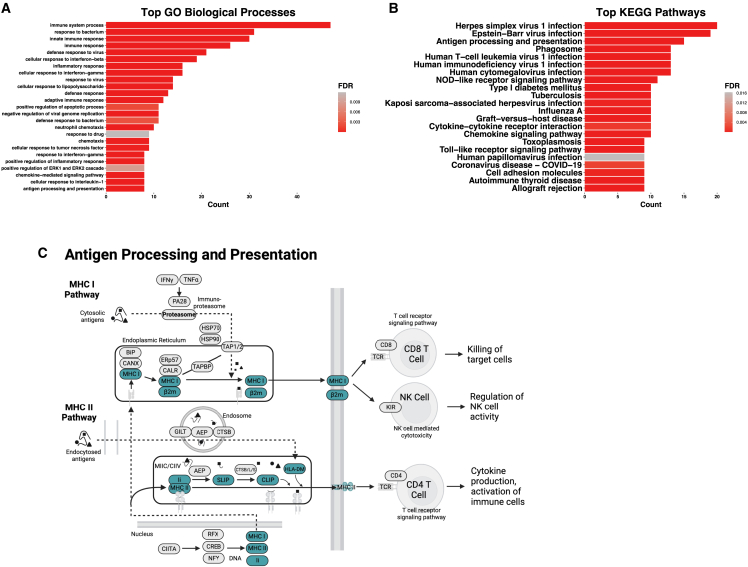
We then performed a Gene Ontology (GO) analysis of the 33 genes with significant (adjusted p < 0.05) differential expression in C134-treated cell types and an average log2-fold change of more than 2 (gene list in Table S1). These genes mapped to cellular pathways involving viral infection, interferon-gamma signaling, and antigen presentation (Figure 4A). Mapping the same genes to KEGG pathways produced a top pathway of herpes simplex 1 virus infection, various other viral infection response pathways, chemokine signaling, and antigen presentation (Figures 4B and 4C).
Figure 4.
GO analysis of C134 gene signatures
Top GO biological processes (A) and KEGG pathways (B) for 33 unique genes with an average log2-fold change (Log2FC) of greater than 2 and an adjusted p value of less than 0.05 in C134-treated samples for one or more cell types. (C) Reconstruction of KEGG pathway denoting (blue highlighted) C134 signature genes involved in antigen processing and presentation.
C134 treatment induces a pro-inflammatory tumor microenvironment
We next used qPCR to measure the expression of cytokines and immune-relevant genes to supplement our scRNA-seq findings. C134-treated tumors had increased expression of the inflammatory markers tumor necrosis factor alpha (Tnfa) and interferon gamma (Ifng) (Figure S8). Ifng expression was significantly (p < 0.01) increased in both models on day 2, but not day 6. Interestingly, CMYC tumors exhibited a significant (p < 0.01) increase in Tnfa expression on day 2, whereas MYCN tumors exhibited a significant (p < 0.05) increase on day 6 after C134 treatment. Expression of chemokines Cxcl10 and Ccl5 were both increased in CMYC and MYCN tumors treated with C134 compared with vehicle-treated tumors (Figure S8), wherein Ccl5 expression was significantly (p < 0.05) increased on days 2 and 6. Similarly, Cxcl10 expression significantly (p < 0.01) increased on days 2 and 6 in C134-treated MYCN tumors, but only on day 2 (p < 0.001) in CMYC tumors. We also observed significantly (p < 0.01) increased expression of the serine protease granzyme B (Gzmb) in C134-treated tumors on day 6 (Figure S8). Flow cytometry evaluation of tumor-infiltrating CD8+ T cells demonstrated a significant (p < 0.05) increase in the percentage specific for HSV glycoprotein B (GB), as well as the immunodominant tumor antigen gp70 (p < 0.01), which is highly expressed in many mouse tumors (Figure S9).27
Direct detection of C134 transcripts within medulloblastomas
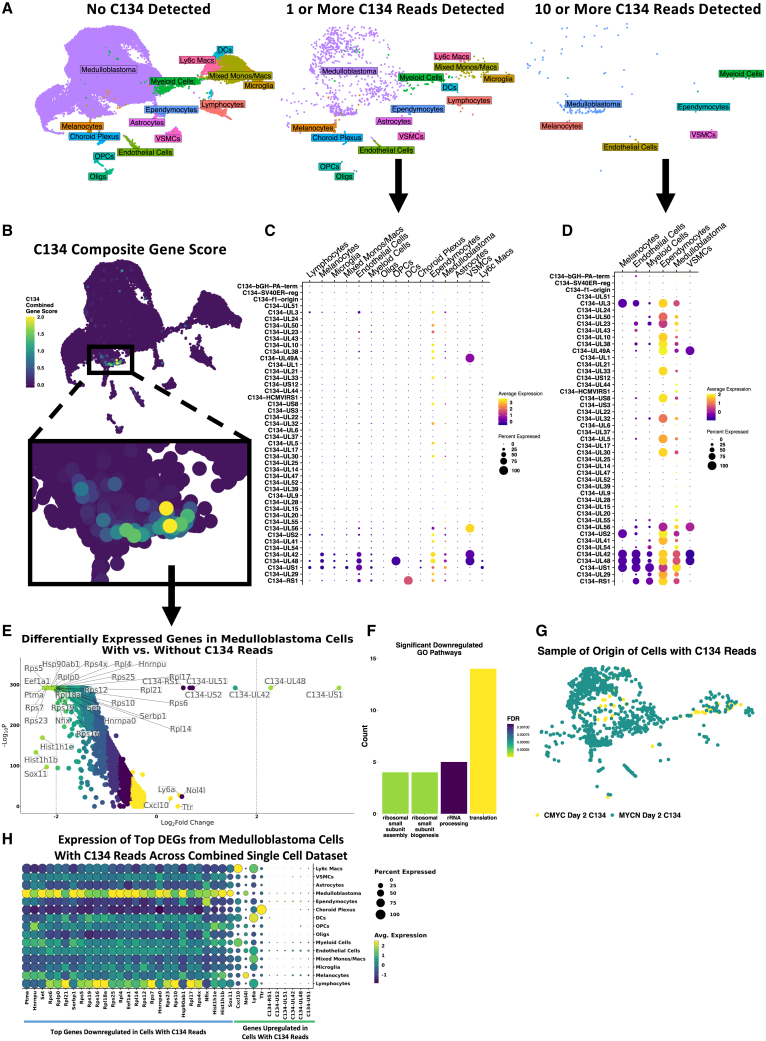
We evaluated our scRNA-seq data for evidence of C134 gene expression, identifying a total of 973 cells with at least 1 sequence read that mapped to the C134 genome and 126 cells with 10 or more such reads (Figure 5A and Figures S10A–S10D). When we applied a combined gene signature of all C134 genes to the scRNA-seq data and visualized this in dimensionally reduced space (Figure 5B), one cluster of medulloblastoma cells demonstrated high gene expression. Three C134 genes, UL42, UL48, and US1, were expressed across a diverse range of cell types, whereas small populations of medulloblastoma cells and ependymocytes demonstrated high expression of many C134 genes (Figures 5C and 5D). Medulloblastoma cells containing at least one C134 read, compared with those without any C134 reads, exhibited a genome-wide decrease in the levels of most transcripts. These cells exhibited with upregulation of just 10 genes, 6 viral and 4 murine (Figure 5E and Table S1). When we conducted a similar analysis on non-medulloblastoma cell types with at least 30 cells (mixed monos/macs and myeloid cells), we observed a similar global downregulation of gene expression. The genes with the greatest decreases corresponded with ribosomal processes, based on GO analysis (Figure 5F).28,29 In both CMYC and MYCN models, C134 reads were detected at day 2 but not day 6 after C134 treatment (Figure 5G). We identified that 0.72% of cells in the C134-treated CMYC day 2 sample, and 9.38% of cells in the C134-treated MYCN day 2 sample contained at least one C134 read. More than 80% of cells containing at least one C134 read were medulloblastoma cells. There were also nonmalignant cells in which C134 transcript was detected, albeit a minority. No vehicle-treated samples contained any reads mapping to C134 genes (all vehicle samples = 0 reads; MYCN day 2 C134 = 31,016 reads; CMYC day 2 C134 = 1,278 reads; MYCN day 6 C134 = 2 reads; and CMYC day 6 C134 = 13 reads). qPCR of the HSV polymerase (pol) gene from harvested intracranial tumor RNA extracts additionally demonstrated the presence of C134 transcripts in both C134-treated medulloblastoma models on day 2, as well as on day 6 in MYCN (Figure S8). Visualization of top differentially expressed genes in medulloblastoma cells with C134 reads from Figure 5E across the combined single cell dataset demonstrated that genes found to be most downregulated in cells with C134 reads displayed high baseline expression in medulloblastoma cells overall, whereas genes found to be upregulated in cells with C134 reads did not exhibit such behavior (Figure 5H).
Figure 5.
C134 Viral Detection and Gene Expression in Mouse Medulloblastomas
(A) UMAP plots showing the positions and cell types of cells with no C134 reads, 1 or more C134 reads, and 10 or more C134 reads, across the entire single cell dataset. (B) Visually weighted expression of a C134 genome signature score across the dataset, showing a specific region of medulloblastoma cells with high scores. Normalized expression by cell type of C134 genes in cells with 1 or more C134 reads (C) and cells with 10 or more C134 reads (D). (E) Volcano plot showing differentially expressed genes in medulloblastoma cells containing one or more C134 reads vs. medulloblastoma cells with no C134 reads, evaluated using a Wilcoxon rank-sum test. Significantly downregulated genes (adjusted p < 0.05, avg log2-fold change < −2) were subjected to GO analysis, and significant (false discovery rate [FDR] of <0.05) biological processes are shown (F). (G) Sample of origin of cells with one or more reads mapping to the C134 genome. (H) Dot plot quantifying normalized gene expression values across combined single cell dataset for top downregulated and upregulated genes differentially expressed in medulloblastoma cells containing C134 reads shown in (E).
Temporal changes in gene expression in C134-treated medulloblastomas
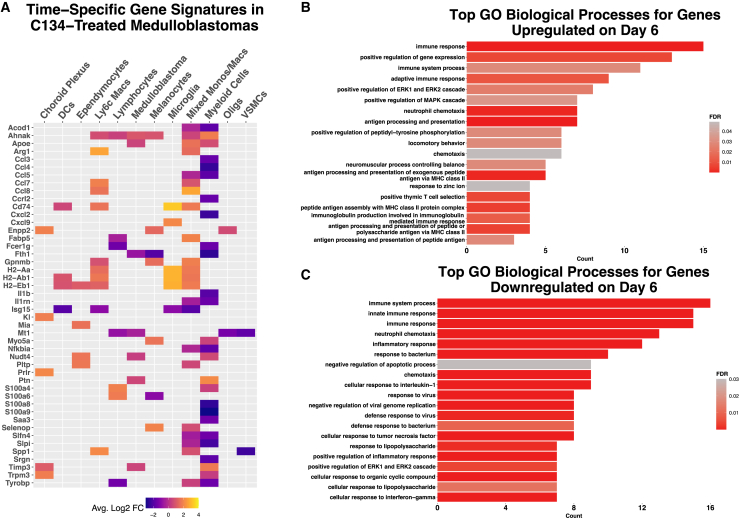
We conducted differential gene expression analyses to evaluate changes throughout C134 infection and clearance. Notably, myeloid and macrophage populations significantly (adjusted p < 0.05) increased expression of Arg1, Ccl7, Ccl8, Cd74, Fabp5, H2-Aa, H2-Ab1, and H2-Eb1 on day 6, whereas expression of Ccl4, Cxcl2, S100a8, and S100a9 significantly decreased (Figure 6A). Top GO biological processes enriched in genes upregulated on day 6 included adaptive immune response, antigen processing and presentation, and immunoglobulin production (Figure 6B), whereas top GO biological processes enriched for genes downregulated on day 6 included innate immune response, cellular response to interferon gamma, and defense response to virus (Figure 6C).
Figure 6.
Temporal scRNA-seq gene expression signatures in medulloblastomas
(A) Heatmap showing differentially expressed genes in C134-treated day 6 medulloblastomas compared with C134-treated day 2, with an average log2 fold change of greater than 1.5 or less than −1.5. Top GO biological processes for greater than 1 average Log2 FC upregulated (B) and less than –1 average Log2 fold change downregulated (C) differentially expressed genes on day 6 vs. day 2 C134-treated medulloblastomas.
Discussion
In this study, we investigated the effect of C134 OV treatment in community-accepted models of medulloblastoma subgroups SHH (MYCN), a subgroup with intermediate outcomes, and Group 3 (CMYC), a subgroup with poor outcomes in the human disease setting.21,22 Gene expression patterns of these models are highly similar to their human medulloblastoma subgroup counterparts.22 C134 is currently under study in a clinical trial of adult relapsed glioblastoma, and its tropism for nectin-expressing brain tumors, as well as the FDA approval of a related modified herpesvirus (TVEC) for use as an anti-cancer therapeutic, emphasizes their possible usefulness in treating human malignancies.
The increase in median survival observed with C134 treatment in C57BL/6 mice for both medulloblastoma models was complemented by the presence of one long-term survivor in the CMYC group (Figure 1A). This long-term survivor led us to hypothesize that T cell-related function may be important for C134 efficacy. To test this hypothesis, we performed a second survival study in athymic nude mice. C134 treatment also prolonged median survival in these mice, although no long-term survivors were observed in the CMYC model in this cohort (Figure S1). We also recognized that variation in response to C134 seen in all of our mouse groups may stem from biological heterogeneity of our tumor models and their interactions in each mouse, as well as heterogeneity in the murine response to C134. Future efforts should seek to understand the mechanisms responsible for the impressive survival observed in some C134-treated mice as opposed to the shorter survival in others. Our result of C134 prolonging survival in athymic nude mice suggested that a non-T cell mechanism may be operating to prolong median survival, yet T cells may not be dispensable for long-term survival. We, therefore, examined the immune response to C134 treatment in tumor-bearing C57BL/6 mice using flow cytometry, and overall these data revealed highly similar immune phenotypes between the two medulloblastoma subgroups. Only subtle differences were appreciated between the subgroups, such as the ratio of M1:M2 macrophages increasing on day 6 in CMYC but not MYCN (Figure S5B).
We used scRNA-seq to characterize the observed immune cell recruitment, using the same post-treatment day 2 and day 6 timepoints to capture potential innate or early adaptive immune changes. Previous medulloblastoma studies employing single cell techniques have focused on subgroup cells of origin and developmental trajectories of malignant cells, and responses to targeted therapies.7,30,31 Other studies have investigated the tumor immune microenvironment of medulloblastoma in the context of radiation therapy response, and identified the landscape of human medulloblastoma immune cells, providing a framework for comparisons with other models.8,32 Our data revealed 29 unique cell populations, and, despite limitations inherent to sample size across scRNA-seq experiments, we observed statistically significant increases in the proportions of DCs, Ly6c+ macrophages, lymphocytes, and a mixed population of monocytes and macrophages in virus-treated vs. vehicle-treated tumors. It should also be considered that cell type-specific rates of lysis or dropout during sample processing and single cell droplet encapsulation, as well as computational quality control excluding low-quality and dead cells, could impact these results. This dataset allowed us to identify both conserved and context-specific gene expression changes elicited by C134 OV treatment in different cell types, at different times, in two medulloblastoma models.
By scoring macrophages with highly specific M1 and M2 gene modules characterized in C57BL/6 mice, we observed C134 induces broad increases in expression of macrophage M1 genes at all time points and in all models. By contrast, there was a negligible effect on the expression of M2 genes (Figure 2E). Although there seem to be beneficial effects induced by C134 in recruiting M1 macrophages, there also seem to be immunosuppressive effects including recruitment of MDSCs, as evidenced by flow cytometry at both day 2 and 6 in C134-treated vs. vehicle-treated medulloblastomas (Figure S5A). This result correlates with an increased proportion of PMN-MDSCs in C134-treated samples from scRNA-seq analysis (Figure S4). Although these cells displayed expression of some neutrophil markers such as S100a9 and Retnlg as used in other studies,32 they shared more marker genes with PMN-MDSCs as identified in other single cell RNA sequencing studies in mice, including S100a8, Nfkbia, Ccl3, Ccl4, Socs3, Atf3, and Klf6,27,33 and our flow studies had confirmed the presence of MDSCs outside of single cell sequencing (Figure S5A). Oncolytic herpesviruses have been shown to recruit MDSCs to tumor microenvironments,34,35 and one future avenue for investigation will be assessing whether combination therapies capable of antagonizing these immunosuppressive cell types are capable of further improving C134’s effects.
A major takeaway from our study was the substantial diversity of myeloid cells in medulloblastomas. The immune and macrophage subset cell populations we identified here show striking similarities with those identified in a study of 28 human medulloblastomas performed by Riemondy et al.36 (Figures S11A and S11B). The presence of immune subpopulations in our syngeneic models such as Mrc1+ M2 macrophages and complement-expressing macrophages that are independently identified in human tumors suggests that our syngeneic models are capable of reflecting immunophenotypic nuances observed in patients. Another study employing single cell sequencing on SHH murine medulloblastomas performed by Dang et al.32 identified transcriptional markers Siglech and Clec12a as distinguishing of microglia and microglial-like tumor macrophages (Siglech+Clec12a–) from peripherally recruited monocytes (Clec12a+Siglech–). In our own immune cell subset, these populations are identifiable in our microglia and mixed monos/macs clusters, and C134 treatment seems to invoke alterations in the Clec12a+ population that are particularly apparent in the CMYC model (Figure S11C). Additionally, both of the aforementioned studies described substantial heterogeneity in myeloid and macrophage populations. This is consistent with our findings of macrophage subpopulations that, based on their transcriptional profiles (Figures 2D and 2E), may contain a diversity of tumor-supportive and tumor-destructive populations, both of which may be modified by C134 treatment (Figure 3A).
In examining the DC cluster, we noted an increased population of Ccr7+ DCs in C134-treated tumors (Figures 1E and S4), which indicates mature, migrating DCs that have taken up antigen and are in transit to present to T cells in nearby lymph nodes.37 Furthermore, plasmacytoid DCs (pDCs) are known to produce large amounts of type I interferon in response to viral infection,38 and an increase in pDC cell number is appreciated in our C134-treated medulloblastomas (Figure S4). The significance of these DCs in the antitumor response and increased survival of C134-treated mice is yet to be understood, but they might address the results we obtained in athymic mice. Lymphocytes upregulated markers of activation and cytotoxicity in C134-treated tumors (Figure 3C), suggesting the adaptive immune response plays a functional role in the antitumor effect. The significance of intratumoral T cells in medulloblastoma is unclear39,40 and has not been explored in the context of OV replication, but our discovery of increased markers of antigen recognition and specificity to both viral and tumor antigens in T cells suggests a beneficial role (Figures S6B and S9). Collectively, our data indicate that C134 evokes a complex immune response across all immune cell types in the tumor microenvironment. In comparing our results with RNA sequencing data from a phase IB trial testing the G207 oncolytic herpesvirus in patients with recurrent glioblastoma, both G207 and C134 treatment led to differentially expressed genes in their respective tumors that demonstrated similar enrichment of biological processes, including immune response, positive regulation of the inflammatory response, and antigen processing and presentation (Figure 4A).18 Additionally, G207-treated tumors demonstrated significant increases in a wide breadth of immune cells, including tumor-infiltrating lymphocytes, macrophages, and DCs, concordant with our results in medulloblastoma (Figure 3A). Additionally, among a set of 502 genes correlated with patient survival in the G207 study was Cxcl10, a gene that in our models consistently demonstrated significant increases in C134-treated medulloblastomas across several immune cell types and timepoints (Figure 3B). Collectively, these results are consistent with the idea that different oncolytic herpesviruses operating in different brain tumor types possess some degree of similarity in the immune responses they elicit. Understanding not only the breadth of such immune responses elicited by different OVs across tumor types, but also the functional components responsible for driving outcomes and clinically relevant effects is a crucial area for investigation in the OV field. The gene expression signatures obtained here for C134 lay the groundwork for such comparisons.
The direct detection of C134 gene expression at single cell resolution was another unique outcome of our work. High expression of C134 genes in medulloblastoma cells and ependymocytes demonstrates increased viral activity in these cells (Figure 5D). The finding of genome-wide decreased expression of mouse genes within medulloblastoma cells that contained reads mapping to C134 is consistent with the known ability of herpes simplex virus to suppress host transcription.41 The genes that seem to be most downregulated in medulloblastoma cells containing C134 reads (Figure 5E) display high levels of expression in the combined dataset; thus, it is not unexpected that such genes experience the most impressive fold changes upon infection with a virus that degrades host cell RNAs.42 While we are hesitant to draw further conclusions about the biological significance of the differentially expressed genes in medulloblastoma cells with C134 reads, we do not discount that these genes and their corresponding pathways, including ribosomal biogenesis (Figure 5F), may hold phenotypically relevant roles as cellular functions co-opted by C134 or predisposing a particular subset of medulloblastoma cells to C134 infection. Cellular factors participating in ribosomal biogenesis have indeed been connected to medulloblastoma in terms of its oncogenesis and subgroup biomarkers, and are being explored as therapeutic targets in a number of cancers.43,44,45,46 Overall, these data are consistent with the known biology of herpes simplex virus 1 (HSV1) and C134, revealing improved replication within the target medulloblastoma cells compared with all other cell types within the short viral replication period (Figure 5G).
The mouse models used in this study allowed us to assess tumor-immune interactions in a syngeneic setting; however, the only natural host for HSV1 is humans, and it is known that C57BL/6 mice in particular are resistant to this virus.47,48,49 Consistent with this, we observed more limited C134-induced direct cytolytic effects and viral recovery of C134 from mouse tumor cells in vitro (Figures S2A and S3A). For this reason, we generated initial data on the behavior of C134 in the community-used human medulloblastoma cell lines D283med and D425med, derived from a medulloblastoma metastasis and a group 3 medulloblastoma isolated from 6-year-old patients, respectively, with the former demonstrating characteristics of cancer stem-like cells.50 It was not unexpected that C134 demonstrated increased cytolytic effects (Figure S2B) and yielded higher viral recovery (Figure S3C) in these human cells as compared with our mouse cells. This may be a positive indicator that C134 is capable of exhibiting therapeutic effects in human tissue; however, these data are intended to inform additional studies comprehensively investigating C134’s behavior in human models. Furthermore, although a substantial body of literature documents that Δγ134.5 oHSVs like C134 are safe for intracranial injection as they are unable to conduct efficient replication in post-mitotic cells, we did not test C134 in non-tumor human cells. Thus, these in vitro data should be interpreted with caution, and future work focused on thoroughly characterizing C134’s behavior in human cells should include both tumor and non-tumor comparators. For the purpose of this work, however, the goal was to establish whether C134 could exert cytolytic activity and replicate in human medulloblastoma cells.
Ongoing clinical trials of oncolytic herpesviruses are an additional source of valuable information regarding the tumor specificity and replicative potential of oncolytic herpesviruses in the CNS. For example, the oHSV G207 is derived from the same first-generation oHSV as C134, and G207 has completed a first-in-human study demonstrating its safety.20 As C134 continues in a phase I clinical trial in adults with recurrent glioblastoma, there are three clinical trials currently investigating G207 in the context of pediatric brain tumors, including two phase I studies and one phase II study (clinicaltrials.gov searched “oncolytic herpes virus” for ‘intervention/treatment’ and “pediatric” for ‘other terms’ on June 26, 2023, filtered for “recruiting,” “not yet recruiting,” and “active, not recruiting”). The results of these trials will likely carry relevance for the expected behavior of C134 in pediatric patients; however, consideration must be given to the differences between G207 and C134 when making these comparisons.
We have longitudinally profiled the immune landscape of response to an OV and the corresponding immune cell gene signatures across two post-treatment timepoints in two syngeneic medulloblastoma mouse models, establishing a rationale for further exploring C134 as a potential therapy in humans.
Material and methods
Study design
All experiments and analyses of medulloblastoma-bearing mice stem from an identical methodology illustrated in Figure 1A. Mice intracranially implanted with medulloblastomas were treated in the same site 5 days after implantation with C134 OV or vehicle, then for survival studies, mice were followed until endpoint, and for all characterization experiments, mice were sacrificed 2 and 6 days after treatment for brain harvesting, sample collection, and downstream experiments. Please refer to specific methods below for more detailed information.
Cell culture
The Vero African green monkey kidney cell line was obtained from the American Type Culture Collection (Manassas, VA). Mouse medulloblastoma cell lines were derived from fresh surgical resections from tumor bearing animals. Briefly, resected tumor tissue was mechanically dissociated and passed through a 100-micron cell strainer. Mouse cell lines (CMYC, MYCN, Ptch) were maintained in Neurobasal-A medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 2 μM L-glutamine, 1% N-2 supplement, 2% B-27 supplement (Gibco, Thermo Fisher Scientific), 20 ng/mL human epidermal growth factor, 20 ng/mL human basic fibroblast growth factor (Miltenyi Biotec, Auburn, CA), and 20 ng/mL Leukemia inhibitory Factor (Millipore Sigma, St. Louis, MO). Human medulloblastoma cell lines D283med and D425med were acquired from the ATCC and Dr. Darrell Bigner (Duke University, Durham, NC), respectively. All cell lines were maintained in DMEM supplemented with 1% penicillin/streptomycin and 10–20% fetal bovine serum (FBS; 10% serum concentration for Vero cells, 20% serum concentration for all other human cells). All cell lines were verified to be free of Mycoplasma contamination by the MycoAlert Mycoplasma Detection Kit (Lonza Inc., Allendale, NJ) before use.
Virus replication assays
Human and mouse medulloblastoma cells were plated in 12-well plates at a density of 5.0 × 105 cells per well. Cells were infected 24 h later with C134 at an MOI of 1 (CMYC, MYCN) or 0.01 (D283med, D425med) in 100 μL of Opti-MEM (Gibco), MOIs reflective of the known resistance of C57BL/6 mice to HSV1 and similar to that previously used for C134 infection of murine cells.47,51 Following a 2-h incubation at 37°C with gentle shaking every 20 min, the medium containing unabsorbed virus was removed and replaced with fresh growth medium. Both cells and supernatants were collected at 3, 24, 48, and 72 h after initial infection. These samples were then freeze-thawed three times, centrifuged to pellet debris, and serially diluted onto previously plated Vero cells. Virus titers were determined by standard plaque assays. Results represent three biological replicates evaluated in triplicate and are representative of a second independent experiment.
For in vivo virus replication studies, mice were treated as described in the material and methods section, “orthotopic injections.” At 3, 48, and 96 h after C134 injection, brain tumor quadrants were harvested and mechanically homogenized. The samples were then freeze-thawed on dry ice three times and centrifuged at 1,500×g for 10 min to pellet debris. The supernatants were collected for titration on Vero cells per established protocol.40 An n = 4 for each sample and time point was collected and titrated in triplicate. Error bars represent the SD.
In vitro assessment of cytotoxicity
Cells were plated in 96-well plates at a density of 2.5 × 104 cells per well. Twenty-four hours after seeding, the cells were infected for 2 h with various MOIs of C134 in 0.1 mL of Opti-MEM at 37°C. Each MOI was assessed in eight replicates. At the end of the incubation period, the virus was removed, and the cells were maintained in their appropriate media as described in the “cell culture” section. Uninfected cells were used as controls. Cell viability was quantified by the CellTiter 96 Aqueous One Solution Cell Proliferation MTS assay (Promega, Madison, WI). The percentage of surviving cells was calculated by dividing the absorbance at 490 nm recorded from the infected wells by the absorbance from the uninfected wells corresponding with the same time point. Results are representative of a second independent experiment.
Orthotopic injections and medulloblastoma models
Medulloblastoma tumor models were propagated solely in vivo by serial intracranial passage. Specifically, CMYC or MYCN medulloblastomas harvested from endpoint mice were subjected to filtration through a 100-μm cell strainer, washed three times via adding 5 mL of neurobasal media and centrifugation at 300×g for 5 min at 4°C, and frozen down until implantation into downstream mice. At implantation time, 1/20th of an upstream tumor cell suspension stock was injected into each downstream mouse, corresponding to approximately 2 × 105 viable tumor cells. For experiments in which multiple stock tubes were needed, these stock tubes were first mixed into a master stock, which was then used for implanting all mice in the respective medulloblastoma model group, ensuring standardization of the tumors. For each injection of tumor, medulloblastoma cells were suspended in 5 μL Matrigel (BD Biosciences, Franklin Lakes, NJ). Tumor cells were then implanted into the cerebral cortices of 5-week-old B6(Cg)-Tyrc-2J/J (Jackson Laboratory, Bar Harbor, ME) or 5-week-old Hsd: Athymic Nude-Foxn1nu (ENVIGO, Frederick, MD) mice using the small animal stereotactic frame (David Kopf Instruments, Tujunga, CA). The injection coordinates were 1.75 mm caudal from bregma, 1.5 mm to the right of the sagittal suture, and 3 mm ventral from the skull surface. Cells were injected at 2.5 μL/min using a 26G Hamilton syringe under isoflurane gas anesthesia. Treatment with C134 (1 × 107 pfu/dose) was delivered in 5μL total volume or an equivalent volume of vehicle (10% glycerol in PBS) was administered five days after tumor implantation by intratumoral injection using the same coordinates on the stereotactic frame as those used for implantation. The mice were euthanized if they developed neurologic deficits such as hemiparesis or lethargy. All animal experiments were approved by the Nationwide Children’s Hospital Institutional Animal Care and Use Committee (AR17-00039).
Flow cytometry
Two and 6 days after treatment, mice were sacrificed using deep anesthesia with isoflurane, followed by cervical dislocation. The intracranial tumors (n = 4), implanted and treated as described above, were harvested. Single cell tumor suspensions were prepared by mechanical disruption followed by passage through a 100-μM cell strainer in Neurobasal-A medium with a sterile 1 mL syringe plunger. Tumor cell suspensions were subsequently lysed with ACK RBC lysis buffer (Lonza, Inc., Basel, Switzerland) and blocked with 5% mouse Fc blocking reagent (2.4G2, BD Biosciences) in FACS buffer (1% FBS and 1 mM EDTA in PBS). Cells were labeled with one of the following antibody staining panels for analysis of the innate and adaptive immune cells, as previously described.52 For T and NK cell analysis: CD4-fluorescein isothiocyanate (FITC) (GK1.5), NK1.1-phycoerythrin (PE) (PK136), CD8a-PE-Cy7 (53–6.7), CD3e-Violet 421 (145-2C11), B220-PerCP/Cy5.5 (RA3-6B2) and CD44-allophycocyanin (APC) (IM7). For T cell exhaustion markers: CD4-FITC, CD8a-PE-Cy7, CD3e-Violet 421 and B220-PerCP/Cy5.5 with LAG3-PE (C9B7W) and Tim3-APC (RMT3-23) or CTLA4-PE (UC10-4B9) and PD-1-APC (29F.1A12). For tumor associated macrophages: CD206-fluorescein isothiocyanate (C068C2), Ly6C-PE (AL-21), MCH class II-APC (M5/114.15.2), CD11c-PerCP/Cy5.5 (N418), CD11b-Violet 421 (M1/70), F4/80-PE-Cy7 (BM8), and Ly-6G-APC-Cy7 (1A8). HSV-GB-TETRAMER-APC. HSV GB monomer (SSIEFARL) was obtained from the National Institutes of Health (NIH) tetramer core facility (Human B2M H-2Kb). GP70-TETRAMER-APC, mouse MuLV gp70 p15E, monomer (KSPWFTTL) was obtained from the NIH tetramer core facility (Human B2M H-2Kb). Tumor cells were stained with PD-L1-PE (10F.9G2). Single samples were stained with the above staining panels for 30 min on ice and washed one time with FACS buffer. After labeling, cells were fixed in 1% paraformaldehyde and a minimum of 100,000 events were collected and analyzed on a BD FACS LSR II (BD Biosciences). Analysis was carried out using the FlowJo software, version 10.0.3 (Tree Star Inc., Ashland, OR). All staining antibodies were purchased from BioLegend (San Diego, CA) except for anti-MHC II (eBioscience). See flow cytometry gating strategies (Figure S12).
Tissue harvest and single cell dissociation
After confirmation of mouse death, brain tumors were harvested and chopped into small pieces. A 1-mL pre-warmed Papain solution at 2 mg/mL was added, and samples were incubated for 20 min at 37°C on a heating block with gentle shaking. The Papain solution components were then temporarily removed, and wide bore pipette tips were used to pipette the remaining larger tissue pieces up and down several times. The Papain solutions were then re-added, and mixed with 5 mL PBS +1% BSA. The debris was allowed to settle for 1 min before the solution was transferred to a new tube and centrifuged at 300×g for 5 min. Supernatants were discarded, and pellets were resuspended in 5 mL 10× buffer, followed by filtration through a 100-micron and then 70-micron filters. Filtered solutions were then centrifuged at 300×g for 5 min, and a red blood cell lysis protocol was carried out by resuspending pellets in 1 mL chilled 1× Red Blood Cell lysing Buffer Hybri-Max (Sigma #R7757). Solutions were incubated at 4°C for 10 min, then 10 mL chilled RPMI were added, and samples were centrifuged at 300×g for 10 min. Supernatants were discarded, and cells were resuspended in 5 mL chilled PBS +1% BSA for counting. Minimal cell death was noted during processing (1.2%–6.9% death), and final cell suspensions were diluted to thousands of cells per microliter. For samples with significant debris noted, a dead cell removal protocol was performed according to manufacturer instructions (Miltenyi Biotec, 130-090-101).
scRNA-seq
A Chromium Next GEM Single Cell 3′ v3.1 kit from 10× Genomics was used to carry out sequencing library preparation according to the manufacturer’s protocol. Targeted cell recovery was 8,000 cells. Output and quality control metrics of both the raw and processed single cell data are summarized (Table S1).
RNA isolation and qPCR gene expression assays
Intracranial tumors (n = 3), implanted and treated as described above, were harvested two- and six-days following C134 treatment. Resected tumor tissue was stored in RNAlater Solution (Invitrogen by Thermo Fisher Scientific) at 4°C. RNA was isolated from resected tumor tissue using the method described in the PureLink RNA Mini kit (Invitrogen by Thermo Fisher Scientific). Briefly, brains were homogenized in Lysis Buffer using a Kimble Kontes pellet pestle and cordless motor (DWK Life Sciences, Millville, NJ). After binding of RNA to the column, samples were treated with PureLink DNase as described in the PureLink RNA Mini kit protocol. RNA was eluted with RNase-free water and stored at −80°C. Two micrograms of total RNA were used to synthesize cDNA using SuperScript IV Reverse Transcriptase (Invitrogen by Thermo Fisher Scientific) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems by Thermo Fisher Scientific). iTaq Universal SYBR Green Supermix kit (Bio-Rad, Hercules, CA) was used to quantify the gene transcripts in 10 μL reactions according to the manufacturer’s instructions. Cycling conditions included an initial step of 2 min at 50°C and 5 min denaturation at 95°C, followed by 40 thermal cycles of denaturation at 95°C for 15 s, annealing at 58°C for 30 s, and elongation at 72°C for 30 s. The comparative quantitation method was used, and the results are represented as fold gene expression relative to Gapdh: 2− (C t target gene − C t Gapdh). The primers used in this study are as follows: Gapdh (forward 5′-TGCACCACCAACTGCTTAGC-3′, reverse 5′- GGCATGGACTGTGGTCATGAG-3′), Tnfa (forward 5′- TGGCCTCCCTCTCATCAG-3′, reverse 5′- GGCTGGCACCACTAGTTG-3′), Ifng (forward 5′-TGATGGCCTGATTGTCTTTCAA-3′, reverse 5′-GGATATCTGGAGGAACTGGCAA-3′), Gzmb (forward 5′-TGCTGCTAAAGCTGAAGAGTAAG-3′, reverse 5′-CGTGTTTGAGTATTTGCCCATTG-3′), Cxcl10 (forward 5′-GGATGGCTGTCCTAGCTCTGTAC-3′, reverse 5′-TGGGCATGGCACATGGT-3′), Ccl5 (forward 5′-CTGCCGCGGGTACCATGAAG-3′, reverse 5′-TACAGGGTCAGAATCAAG-3′), HSV pol (forward 5′-ACCGCCGAACTGAGCAGAC-3′, reverse 5′-TGAGCTTGTAATACACCGTCAGGT-3′).
Single cell sequencing quality control, sample integration, and dimension reduction
Raw FASTQ files were analyzed by the CellRanger pipeline using a custom reference genome consisting of mouse version mm10 and the C134 genome, and sequences for GFP and RFP present in the medulloblastoma cells. Cellranger outputs were read into an Rstudio environment using version 4.1.0 of the Seurat package.53,54,55,56 Cells were then subsetted to meet a variety of quality control standards including minimum and maximum unique genes detected, percent mitochondrial genes (to remove dead or dying cells), percent of hemoglobin genes (to remove red blood cells). Next, cell cycle phase scores were calculated using a reference expression matrix from Nestorowa et al.57 The v2 version of the Seurat function SCTransform58 was then used to compute normalized gene counts for each cell, regressing out cell cycle phase as well as percent mitochondrial reads. Subsequently, the DoubletFinder package59 was implemented, and likely doublets were removed from the dataset. Integration was then performed, followed by dimension reduction using uniform manifold approximation and projection.60 The Clustree package61 was then used to visualize potential resolutions by which to define clusters on the dimensionally reduced UMAP plots, and resolution 2.0 was chosen as this appeared to encompass cluster divisions that were most biologically accurate.
Assigning cell types in single cell sequencing data
To annotate UMAP clusters with defined cell types, the Seurat function FindAllMarkers was used to determine top upregulated marker genes specific to each cluster, using a Wilcoxon Rank-Sum test. We adjusted the p values using Bonferroni correction. Genes with the greatest average log2-fold change expression within a given cluster were compared with all others, and those with adjusted p values of less than 0.05 were examined for the presence of cell type-defining marker genes. A thorough description of marker genes used with accompanying references can be found in the supplemental methods.
Single cell sequencing subset analysis
To analyze specific cell types in the overall dataset at higher resolution, subset analysis was performed. Cells of interest were computationally extracted from the overall dataset, and all previously transformed gene counts were erased, leaving only the raw RNA counts. An independent round of normalization, sample integration, and dimension reduction was performed on this data as already described in quality control, sample integration, and dimension reduction. This yielded dimensionally reduced plots of the subsetted data (Figures 1B, 1D, and 1F). The same process of assigning cell types as described above was applied to assign cell type identities to these data subsets (Figures 1C, 1E, and 1G).
Single cell sequencing cell type proportion testing
Differences in the proportion of cell types within each sample were assessed using the R package Propeller.62 Briefly, cell type proportions were logit-transformed and subsequently subjected to t-tests moderated with an empirical Bayes framework. A false discovery rate of less than 0.05 was used as a threshold for assigning significant differences in cell type proportions.
Single cell sequencing differential gene expression analysis
Differential gene expression was performed by comparing each respective cell type belonging to a particular model, treatment, or time point, with the same cell type in the other model, treatment, or time point of interest. In the case of Figures 5E and 5F, medulloblastoma cells with one or more reads mapping to the C134 genome were compared with medulloblastoma cells with 0 C134 reads. The Seurat command FindMarkers was then used to perform Wilcoxon Rank Sum tests on each given cell type between relevant conditions. Genes with adjusted p values of less than 0.05 and with the highest average log2-fold changes between the conditions being compared were extracted and used to generate gene signature heatmaps, volcano plots, and for GO and KEGG pathway analysis. The Scpubr package was used to generate plots in Figures 5H and S11C.63
Single cell sequencing pathway analysis
Pathway analysis was conducted by extracting all cell type-specific differentially expressed genes with an average log2-fold change of more than 2 and an adjusted p value of less than 0.05 as determined for the specific comparisons of C134- and vehicle-treated samples. Because some genes came up as highly differentially expressed in multiple different cell types, we then filtered for unique genes, ultimately arriving at a list of 33 differentially expressed genes. This list was input into NIH DAVID (https://david.ncifcrf.gov/), and the top GO biological process and KEGG pathways by number of mapped genes were used to generate plots.64,65
Statistical analysis
Kaplan-Meier survival curves were generated with the GraphPad Prism 8.2.0 software. Statistical significance (p < 0.05) between the vehicle- and C134-treated groups was determined using the log rank test. Statistical analysis was performed on flow cytometry and qPCR results using GraphPad Prism 8.2.0 using Tukey’s multiple comparisons test. Ninety-five percent confidence intervals of difference were used to determine significance. Significant results were denoted with p values: p < 0.05 (∗), p < 0.01, (∗∗), p < 0.001 (∗∗∗).
Data and code availability
The single cell RNA-seq data presented in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through Series accession number GEO: GSE200008. All code used for analysis is available upon request to the corresponding author.
Acknowledgments
Figure 1A, Figure 4C, and the graphical abstract were created with biorender.com. The authors would like to acknowledge the following support: Internal Nationwide Children’s Hospital start-up grant (J.R.L.) and CancerFree KIDS Pediatric Cancer Research Alliance Award (A.W.S.). The authors acknowledge Dr. Martine Roussel and Dr. Frederique Zindy (St. Jude Children’s Research Hospital, Memphis, TN) for the syngeneic mouse medulloblastoma models, as well as Dr. Matthew Cannon for computational support. This work was supported by NIH NCI U54-CA232561, and J.H. was supported by NIH 5T32CA269052-02.
Author contributions
A.S., K.A.C., E.R.M., K.E.M., and J.H. conceptualized this work. A.S., K.E.M., J.H., L.S., J.J.W., M.C., A.G., and R.R. supported the methodology. A.S., J.H., C.Y.C., L.S., I.H.A., A.M., D.K., R.D., and Y.K. were involved in investigation. A.S., K.E.M., and J.H. produced visualization of data. Funding acquisition occurred through J.L., K.A.C., and E.R.M. Supervision was provided by T.P.C., K.A.C., E.R.M., and K.E.M. Writing of the original draft was by A.S., J.H., E.R.M., K.A.C., and K.E.M. Reviewing and editing the manuscript was performed by all authors.
Declaration of interests
The authors in full transparency disclose the following commercial interactions. E.R.M. reports PACT Pharma LLC, scientific advisory board membership, honorarium, and stock options; Scorpion Therapeutics, LLC, scientific advisory board membership, honorarium, and stock options; Qiagen N.V., supervisory board membership, stock; Singular Genomics, Inc., board of directors membership and stock. K.A.C. reports stock in Mustang Bio licensure.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2023.07.006.
Supplemental information
This table contains tabs that contain quality control metrics for single cell RNA sequencing data described in this paper. Additional tabs contain data outputs from differential gene expression analysis identifying cluster markers, cell type markers, and differentially expressed genes for the treatment groups, timepoints, medulloblastoma models, and cells with C134 reads detected, corresponding to the figure panels to which tab titles refer
References
- 1.Northcott P.A., Robinson G.W., Kratz C.P., Mabbott D.J., Pomeroy S.L., Clifford S.C., Rutkowski S., Ellison D.W., Malkin D., Taylor M.D., et al. Medulloblastoma. Nat. Rev. Dis. Primers. 2019;5:11–20. doi: 10.1038/s41572-019-0063-6. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Garancher A., Ramaswamy V., Wechsler-Reya R.J. Medulloblastoma: From Molecular Subgroups to Molecular Targeted Therapies. Annu. Rev. Neurosci. 2018;41:207–232. doi: 10.1146/annurev-neuro-070815-013838. [DOI] [PubMed] [Google Scholar]
- 3.Moxon-Emre I., Taylor M.D., Bouffet E., Hardy K., Campen C.J., Malkin D., Hawkins C., Laperriere N., Ramaswamy V., Bartels U., et al. Intellectual Outcome in Molecular Subgroups of Medulloblastoma. J. Clin. Oncol. 2016;34:4161–4170. doi: 10.1200/JCO.2016.66.9077. [DOI] [PubMed] [Google Scholar]
- 4.Packer R.J., Vezina G. Management of and prognosis with medulloblastoma: therapy at a crossroads. Arch. Neurol. 2008;65:1419–1424. doi: 10.1001/archneur.65.11.1419. [DOI] [PubMed] [Google Scholar]
- 5.Frič R., Due-Tønnessen B.J., Lundar T., Egge A., Kronen Krossnes B., Due-Tønnessen P., Stensvold E., Brandal P. Long-term outcome of posterior fossa medulloblastoma in patients surviving more than 20 years following primary treatment in childhood. Sci. Rep. 2020;10:9371. doi: 10.1038/s41598-020-66328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelstein K., Spiegler B.J., Fung S., Panzarella T., Mabbott D.J., Jewitt N., D’Agostino N.M., Mason W.P., Bouffet E., Tabori U., et al. Early aging in adult survivors of childhood medulloblastoma: long-term neurocognitive, functional, and physical outcomes. Neuro. Oncol. 2011;13:536–545. doi: 10.1093/neuonc/nor015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hovestadt V., Smith K.S., Bihannic L., Filbin M.G., Shaw M.L., Baumgartner A., DeWitt J.C., Groves A., Mayr L., Weisman H.R., et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature. 2019;572:74–79. doi: 10.1038/s41586-019-1434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riemondy K.A., Venkataraman S., Willard N., Nellan A., Sanford B., Griesinger A.M., Amani V., Mitra S., Hankinson T.C., Handler M.H., et al. Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma. Neuro Oncol. 2022;24:273–286. doi: 10.1093/neuonc/noab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griesinger A.M., Birks D.K., Donson A.M., Amani V., Hoffman L.M., Waziri A., Wang M., Handler M.H., Foreman N.K. Characterization of Distinct Immunophenotypes across Pediatric Brain Tumor Types. J.I. 2013;191:4880–4888. doi: 10.4049/jimmunol.1301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Aguirre I., Cassady K.A. Cancer Immunology and Immunotherapy. Elsevier; 2022. Oncolytic viruses in immunotherapy; pp. 375–437. [DOI] [Google Scholar]
- 11.Friedman G.K., Bernstock J.D., Chen D., Nan L., Moore B.P., Kelly V.M., Youngblood S.L., Langford C.P., Han X., Ring E.K., et al. Enhanced Sensitivity of Patient-Derived Pediatric High-Grade Brain Tumor Xenografts to Oncolytic HSV-1 Virotherapy Correlates with Nectin-1 Expression. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martuza R.L., Malick A., Markert J.M., Ruffner K.L., Coen D.M. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science. 1991;252:854–856. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 13.Conry R.M., Westbrook B., McKee S., Norwood T.G. Talimogene laherparepvec: First in class oncolytic virotherapy. Hum. Vaccin. Immunother. 2018;14:839–846. doi: 10.1080/21645515.2017.1412896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehman H., Silk A.W., Kane M.P., Kaufman H.L. Into the clinic: Talimogene laherparepvec (T-VEC), a first-in-class intratumoral oncolytic viral therapy. J. Immunother. Cancer. 2016;4:53. doi: 10.1186/s40425-016-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassady K.A. Human Cytomegalovirus TRS1 and IRS1 Gene Products Block the Double-Stranded-RNA-Activated Host Protein Shutoff Response Induced by Herpes Simplex Virus Type 1 Infection. J. Virol. 2005;79:8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MD J.M. 2022. A Phase I Trial of IRS-1 HSV C134 Administered Intratumorally in Patients with Recurrent Malignant Glioma (clinicaltrials.Gov) [Google Scholar]
- 17.Cassady K.A., Bauer D.F., Roth J., Chambers M.R., Shoeb T., Coleman J., Prichard M., Gillespie G.Y., Markert J.M. Pre-clinical Assessment of C134, a Chimeric Oncolytic Herpes Simplex Virus, in Mice and Non-human Primates. Mol. Ther. Oncolytics. 2017;5:1–10. doi: 10.1016/j.omto.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller K.E., Cassady K.A., Roth J.C., Clements J., Schieffer K.M., Leraas K., Miller A.R., Prasad N., Leavenworth J.W., Aban I.B., et al. Immune Activity and Response Differences of Oncolytic Viral Therapy in Recurrent Glioblastoma: Gene Expression Analyses of a Phase IB Study. Clin. Cancer Res. 2022;28:498–506. doi: 10.1158/1078-0432.CCR-21-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markert J.M., Liechty P.G., Wang W., Gaston S., Braz E., Karrasch M., Nabors L.B., Markiewicz M., Lakeman A.D., Palmer C.A., et al. Phase Ib trial of mutant herpes simplex virus G207 inoculated pre-and post-tumor resection for recurrent GBM. Mol. Ther. 2009;17:199–207. doi: 10.1038/mt.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markert J.M., Medlock M.D., Rabkin S.D., Gillespie G.Y., Todo T., Hunter W.D., Palmer C.A., Feigenbaum F., Tornatore C., Tufaro F., Martuza R.L. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 2000;7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 21.Zindy F., Uziel T., Ayrault O., Calabrese C., Valentine M., Rehg J.E., Gilbertson R.J., Sherr C.J., Roussel M.F. Genetic Alterations in Mouse Medulloblastomas and Generation of Tumors De novo from Primary Cerebellar Granule Neuron Precursors. Cancer Res. 2007;67:2676–2684. doi: 10.1158/0008-5472.CAN-06-3418. [DOI] [PubMed] [Google Scholar]
- 22.Kawauchi D., Robinson G., Uziel T., Gibson P., Rehg J., Gao C., Finkelstein D., Qu C., Pounds S., Ellison D.W., et al. A Mouse Model of the Most Aggressive Subgroup of Human Medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman H.S., Burger P.C., Bigner S.H., Trojanowski J.Q., Wikstrand C.J., Halperin E.C., Bigner D.D. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J. Neuropathol. Exp. Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- 24.He X.M., Wikstrand C.J., Friedman H.S., Bigner S.H., Pleasure S., Trojanowski J.Q., Bigner D.D. Differentiation characteristics of newly established medulloblastoma cell lines (D384 Med, D425 Med, and D458 Med) and their transplantable xenografts. Lab. Invest. 1991;64:833–843. [PubMed] [Google Scholar]
- 25.Ghonime M.G., Jackson J., Shah A., Roth J., Li M., Saunders U., Coleman J., Gillespie G.Y., Markert J.M., Cassady K.A. Chimeric HCMV/HSV-1 and Δγ134.5 oncolytic herpes simplex virus elicit immune mediated antigliomal effect and antitumor memory. Transl. Oncol. 2018;11:86–93. doi: 10.1016/j.tranon.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jablonski K.A., Amici S.A., Webb L.M., Ruiz-Rosado J.d.D., Popovich P.G., Partida-Sanchez S., Guerau-de-Arellano M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veglia F., Hashimoto A., Dweep H., Sanseviero E., De Leo A., Tcyganov E., Kossenkov A., Mulligan C., Nam B., Masters G., et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbon S., Douglass E., Dunn N., Good B.M., Unni D.R., Harris N.L., Mungall C.J., Basu S., Chisholm R.L., Dodson R.J., et al. Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., He X., Liu X., Zhang F., Huang L.F., Potter A.S., Xu L., Zhou W., Zheng T., Luo Z., et al. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell. 2019;36:302–318.e7. doi: 10.1016/j.ccell.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ocasio J.K., Babcock B., Malawsky D., Weir S.J., Loo L., Simon J.M., Zylka M.J., Hwang D., Dismuke T., Sokolsky M., et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat. Commun. 2019;10:5829. doi: 10.1038/s41467-019-13657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang M.T., Gonzalez M.V., Gaonkar K.S., Rathi K.S., Young P., Arif S., Zhai L., Alam Z., Devalaraja S., To T.K.J., et al. Macrophages in SHH subgroup medulloblastoma display dynamic heterogeneity that varies with treatment modality. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynn L., Akara J. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. SCIENCE IMMUNOLOGY. 2020;16 doi: 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Currier M.A., Sprague L., Rizvi T.A., Nartker B., Chen C.-Y., Wang P.-Y., Hutzen B.J., Franczek M.R., Patel A.V., Chaney K.E., et al. Aurora A kinase inhibition enhances oncolytic herpes virotherapy through cytotoxic synergy and innate cellular immune modulation. Oncotarget. 2017;8:17412–17427. doi: 10.18632/oncotarget.14885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otani Y., Yoo J.Y., Lewis C.T., Chao S., Swanner J., Shimizu T., Kang J.M., Murphy S.A., Rivera-Caraballo K., Hong B., et al. NOTCH-Induced MDSC Recruitment after oHSV Virotherapy in CNS Cancer Models Modulates Antitumor Immunotherapy. Clin. Cancer Res. 2022;28:1460–1473. doi: 10.1158/1078-0432.CCR-21-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riemondy K.A., Venkataraman S., Willard N., Nellan A., Sanford B., Griesinger A.M., Amani V., Mitra S., Hankinson T.C., Handler M.H., et al. Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma. Neuro. Oncol. 2022;24:273–286. doi: 10.1093/neuonc/noab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riol-Blanco L., Sánchez-Sánchez N., Torres A., Tejedor A., Narumiya S., Corbí A.L., Sánchez-Mateos P., Rodríguez-Fernández J.L. The chemokine receptor CCR7 activates in dendritic cells two signaling modules that independently regulate chemotaxis and migratory speed. J. Immunol. 2005;174:4070–4080. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 38.Cella M., Jarrossay D., Facchetti F., Alebardi O., Nakajima H., Lanzavecchia A., Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 39.Vermeulen J.F., Van Hecke W., Adriaansen E.J.M., Jansen M.K., Bouma R.G., Villacorta Hidalgo J., Fisch P., Broekhuizen R., Spliet W.G.M., Kool M., Bovenschen N. Prognostic relevance of tumor-infiltrating lymphocytes and immune checkpoints in pediatric medulloblastoma. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2017.1398877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murata D., Mineharu Y., Arakawa Y., Liu B., Tanji M., Yamaguchi M., Fujimoto K.-I., Fukui N., Terada Y., Yokogawa R., et al. High programmed cell death 1 ligand-1 expression: association with CD8+ T-cell infiltration and poor prognosis in human medulloblastoma. J. Neurosurg. 2018;128:710–716. doi: 10.3171/2016.11.JNS16991. [DOI] [PubMed] [Google Scholar]
- 41.Knipe D.M., Howley P. Lippincott Williams & Wilkins; 2013. Fields Virology. [Google Scholar]
- 42.Kwong A.D., Kruper J.A., Frenkel N. Herpes simplex virus virion host shutoff function. J. Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vriend J., Rastegar M. Ubiquitin ligases and medulloblastoma: genetic markers of the four consensus subgroups identified through transcriptome datasets. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165839. [DOI] [PubMed] [Google Scholar]
- 44.Korshunov A., Okonechnikov K., Stichel D., Schrimpf D., Delaidelli A., Tonn S., Mynarek M., Sievers P., Sahm F., Jones D.T.W., et al. Gene expression profiling of Group 3 medulloblastomas defines a clinically tractable stratification based on KIRREL2 expression. Acta Neuropathol. 2022;144:339–352. doi: 10.1007/s00401-022-02460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiao L., Liu Y., Yu X.-Y., Pan X., Zhang Y., Tu J., Song Y.-H., Li Y. Ribosome biogenesis in disease: new players and therapeutic targets. Sig Transduct Target Ther. 2023;8:15–22. doi: 10.1038/s41392-022-01285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bish R., Vogel C. RNA Binding Protein-Mediated Post-Transcriptional Gene Regulation in Medulloblastoma. Mol. Cells. 2014;37:357–364. doi: 10.14348/molcells.2014.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez C. Genetics of natural resistance to herpesvirus infections in mice. Nature. 1975;258:152–153. doi: 10.1038/258152a0. [DOI] [PubMed] [Google Scholar]
- 48.Lopez C. Resistance to HSV-1 in the mouse is governed by two major, independently segregating, non-H-2 loci. Immunogenetics. 1980;11:87–92. doi: 10.1007/BF01567772. [DOI] [PubMed] [Google Scholar]
- 49.Lundberg P., Welander P.V., Edwards C.K., van Rooijen N., Cantin E. Tumor Necrosis Factor (TNF) Protects Resistant C57BL/6 Mice against Herpes Simplex Virus-Induced Encephalitis Independently of Signaling via TNF Receptor 1 or 2. J. Virol. 2007;81:1451–1460. doi: 10.1128/jvi.02243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casciati A., Tanori M., Manczak R., Saada S., Tanno B., Giardullo P., Porcù E., Rampazzo E., Persano L., Viola G., et al. Human Medulloblastoma Cell Lines: Investigating on Cancer Stem Cell-Like Phenotype. Cancers (Basel) 2020;12:226. doi: 10.3390/cancers12010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghonime M.G., Saini U., Kelly M.C., Roth J.C., Wang P.-Y., Chen C.-Y., Miller K., Hernandez-Aguirre I., Kim Y., Mo X., et al. Eliciting an immune-mediated antitumor response through oncolytic herpes simplex virus-based shared antigen expression in tumors resistant to viroimmunotherapy. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C.-Y., Wang P.-Y., Hutzen B., Sprague L., Swain H.M., Love J.K., Stanek J.R., Boon L., Conner J., Cripe T.P. Cooperation of Oncolytic Herpes Virotherapy and PD-1 Blockade in Murine Rhabdomyosarcoma Models. Sci. Rep. 2017;7:2396. doi: 10.1038/s41598-017-02503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nestorowa S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., Göttgens B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–e31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McInnes L., Healy J., Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2020 doi: 10.48550/arXiv.1802.03426. Preprint at. [DOI] [Google Scholar]
- 61.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phipson B., Sim C.B., Porrello E., Hewitt A.W., Powell J., Oshlack A. propeller: testing for differences in cell type proportions in single cell data. arXiv. 2021 doi: 10.1101/2021.11.28.470236. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blanco-Carmona E. Generating publication ready visualizations for Single Cell transcriptomics using SCpubr. arXiv. 2022 doi: 10.1101/2022.02.28.482303. Preprint at. [DOI] [Google Scholar]
- 64.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 65.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table contains tabs that contain quality control metrics for single cell RNA sequencing data described in this paper. Additional tabs contain data outputs from differential gene expression analysis identifying cluster markers, cell type markers, and differentially expressed genes for the treatment groups, timepoints, medulloblastoma models, and cells with C134 reads detected, corresponding to the figure panels to which tab titles refer
Data Availability Statement
The single cell RNA-seq data presented in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through Series accession number GEO: GSE200008. All code used for analysis is available upon request to the corresponding author.