Abstract
The year 2023 marks the 25th anniversary of the discovery of RNAi. RNAi-based therapeutics enable sequence-specific gene knockdown by eliminating target RNA molecules through complementary base-pairing. A systematic review of published and ongoing clinical trials was performed. Web of Science, PubMed, and Embase were searched from January 1, 1998, to December 30, 2022 for clinical trials using RNAi. Following inclusion, data from the articles were extracted according to a predefined protocol. A total of 90 trials published in 81 articles were included. In addition, ongoing clinical trials were retrieved from ClinicalTrials.gov, resulting in the inclusion of 48 trials. We investigated how maturation of RNAi-based therapeutics and developments in delivery platforms, administration routes, and potential targets shape the current landscape of clinically applied RNAi. Notably, most contemporary clinical trials used either N-acetylgalactosamine delivery and subcutaneous administration or lipid nanoparticle delivery and intravenous administration. In conclusion, RNAi therapeutics have gained great momentum during the past decade, resulting in five approved therapeutics targeting the liver for treatment of severe diseases, and the trajectory depicted by the ongoing trials emphasizes that even more RNAi-based medicines also targeting extra-hepatic tissues are likely to be available in the years to come.
Keywords: MT: Oligonucleotides: Therapies and Applications, systematic review, RNA interference, clinical trials, RNAi, siRNA, shRNA, miR-shRNA, LNP, N-acetylgalactosamine
Graphical abstract
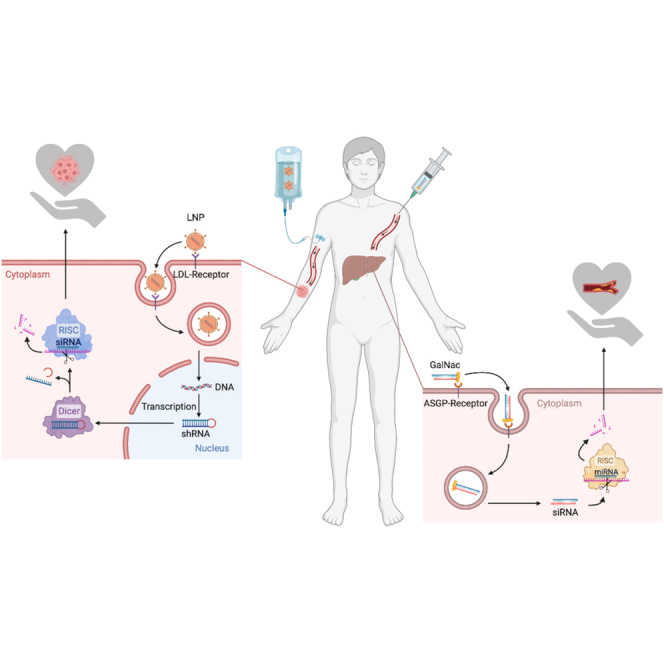
Corydon and colleagues investigate how 25 years of maturation shape the current and future landscape of RNAi-based therapeutics on the basis of published and ongoing clinical trials. RNAi therapy is making headway in the clinic, implying that RNAi therapeutics also targeting extra-hepatic tissues will be available in a few years.
Introduction
In 1998, Andrew Fire and Craig Mello described the pathway of RNAi in the worm Caenorhabditis elegans.1 RNAi induces post-transcriptional gene silencing (PTGS). This conserved and potent gene silencing mechanism may be harnessed for gene therapy and clinical application. PTGS can be induced by short-lived RNAi-based molecules such as small interfering RNA (siRNA), the cells own effectors, microRNAs (miRNAs), or promotor-dependent RNA species such as short hairpin RNA (shRNA).2 Importantly, siRNA-based PTGS is mediated by cytosolic cleavage of target mRNA and involves the RNA-inducing silencing complex (RISC), including argonaut protein 2 (Ago2) and the target mRNA-specific siRNA guide strand (Figure 1).3 Ago2 has slicer activity which is used to cleave target mRNA, when the siRNAs are bound to the target mRNA and associated with RISC.4 siRNAs originate from long double-stranded RNA (dsRNA) sequences, which are cleaved in the cytosol by Dicer.5 With regard to the outcome of silencing miRNAs induce translational repression, whereas siRNAs, which are fully complementary to target sequences, induce Ago2-mediated degradation.6
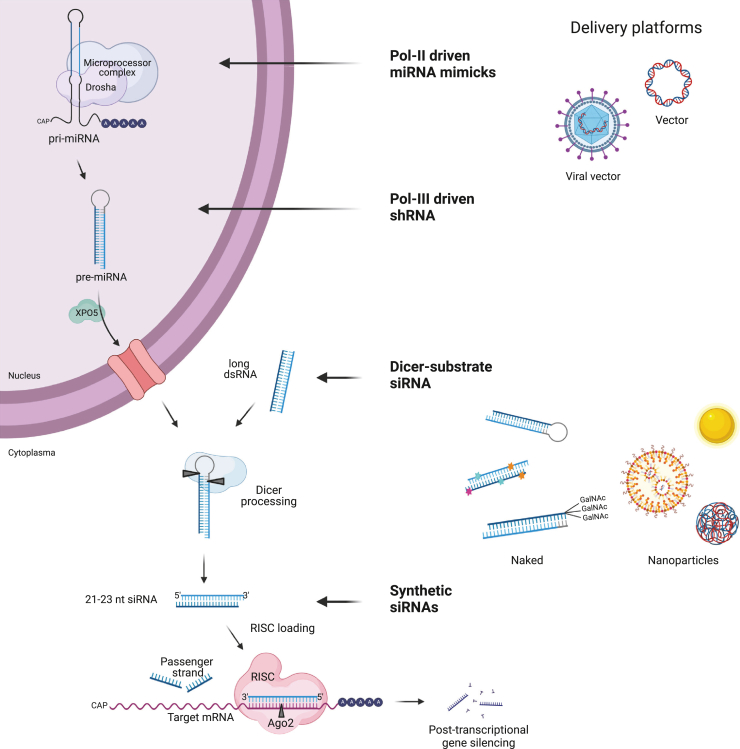
Figure 1.
The RNAi mechanism and entry points of RNAi therapeutics
The left-hand side shows the RNAi pathway and formation of siRNAs. The right-hand side shows different entry points for DNA-based or RNA-based RNAi therapeutics that enters the cell and are loaded into RISC to mediate homology-dependent degradation of target mRNA. Delivery platforms for DNA-based therapeutics include viral and non-viral vectors encoding Pol II driven miRNA mimics or Pol III-driven shRNA. RNAi-based therapeutics are delivered naked as GalNAc-conjugated, modified, or unmodified Dicer-substrate siRNA or synthetic siRNAs or complexed in nanoparticles (gold, lipid, or polymer based). Created using BioRender.com.
The cell has its own endogenous PTGS system on the basis of miRNA effectors. Several pathways exist, and the canonical pathway starts in the nucleus (Figure 1).7 RNA polymerase II (Pol II) or polymerase III (Pol III) transcribes the primary miRNA transcript, which initially is cleaved by Drosha in the nucleus to release the ∼70-nt-long stem-loop structure. This pre-miRNA is then transported to the cytosol by exporting 5 (XPO5), and finally cleaved by Dicer.5,8,9 Next, one of the miRNA duplex strands is incorporated in Ago1–4 inside RISC and here specifies mRNA targets silencing via translation inhibition and by inducing mRNA decay (Figure 1).10,11 Non-canonical miRNA pathways also exist. One example is the Dicer-independent, Ago2-dependent miRNA, called miR451, where no passenger strand is produced.12,13,14 Another example is miRtrons, which bypass Drosha.15 An alternative to siRNAs are shRNAs, used to express siRNA in the cell.16 shRNAs are transcribed from DNA in vivo by a Pol III promotor and these vector encoded molecules were described in 2002. shRNA bypass Drosha, and XPO5 mediates the nuclear export of the pre-miRNA-like molecules.16 shRNA can be embedded in a pri-miRNA scaffold, which allows the cell to use the tissue specific Pol II for transcription called miR-shRNA (Figure 1).16
RNAi represents an entirely new way to treat disease with an unprecedented potential for specific gene targeting through the endogenous miRNA machinery. Several potential disease targets for RNAi-based therapy currently have unattractive treatment options or no treatment options at all. Hence, the mechanism of RNAi and the emergence of RNAi therapeutics have been studied extensively over the years, and recently reviewed in several prominent papers.17,18,19,20 Selected highlights during this journey include in 2001, three years after Fire and Mello’s discovery, the finding that precision duplex silencers consisting of 21 and 22 nt dsRNA sequences with terminal 2 nt 3′ overhangs could elicit RNAi-based PTGS in mammalian cells (Figure 2).21,22 Another important finding, the demonstration that RNAi holds therapeutic promise to prevent liver injury, was reported in 2003.23 In this study Song et al. showed that RNAi targeting Fas protects mice from hepatitis (Figure 2). In 2004 the first clinical trial using a siRNA-based treatment called siRNA-027, was initiated for the treatment of age-related macular degeneration (AMD).24,25 One year later, in 2005, potent nuclear RNAi-mediated knockdown was shown in the nucleus of human cells.26 In 2006, Fire and Mello were awarded the Nobel Prize in Physiology or Medicine for their groundbreaking discovery (Figure 2).27 Evidence for RNAi-based nanotherapeutics from systemically administered siRNA via targeted nanoparticles (NPs) for cancer treatment in humans was provided in 2008.28,29
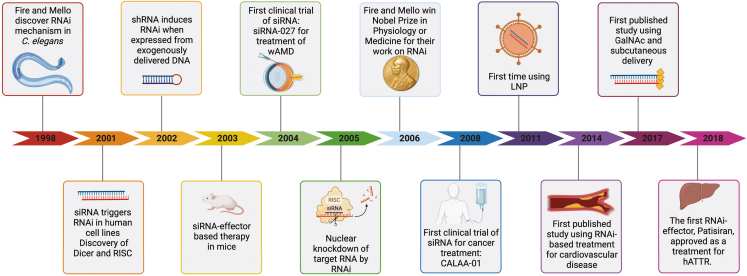
Figure 2.
Milestones in the development of RNAi-based therapeutics
Timeline of important milestones since the discovery of the RNAi mechanism in 1998: 1998,1 2001,5,22 2002,16 2003,23 2004,∗24,25 2005,26 2006,27 2008,∗28,29 2011,∗30 2014,∗31 2017,∗32 2018.∗33,34,35 siRNA, short-interfering RNA; RISC, RNA-inducing silencing complex; shRNA, short hairpin RNA; wAMD, wet age-related macular degeneration; LNP, lipid nanoparticle; GalNAc, N-acetylgalactosamine. The asterisks before or after numbers indicate that the study mentioned is included in the published clinical trials analyzed in this systematic review. Created using BioRender.com.
A few years later, in 2011, the first study demonstrating delivery of RNAi therapeutics using lipid NPs (LNPs) and its potential applications to treatment was published.30 Interestingly, Gish and co-workers used LNPs composed of a plasmid encoding RNAi-based molecules complexed with cholesteryl spermine.30 In 2014, the first RNAi-based treatment for cardiovascular disease using a potent inhibitor of PCSK9 was published.31 N-acetylgalactosamine (GalNAc)-conjugated delivery was introduced in 2017 (Figure 2).32 It all culminated in 2018, when the first RNAi-based drug, patisiran, was approved by the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) as treatment for hereditary transthyretin amyloidosis (hATTR) (Figure 2).33,34,35 Since then, four other RNAi therapeutics (givosiran, lumasiran, inclisiran, and vutrisiran) have been approved for medical use.34,35
By its 25th anniversary, RNAi-based therapy has established itself as an important treatment strategy that may benefit patients with otherwise incurable diseases. This systematic review investigates how the maturation of RNAi therapeutics and developments in delivery platforms, administration routes, and potential targets shape the current landscape of clinically applied RNAi. The most remarkable finding was that the approved RNAi drugs successfully but solely target the liver and targeting of extra-hepatic tissues seems to be the next frontier.
Results
During the search period, which was restricted to the publication year of Fire and Mello’s seminal paper, 1998, and forward, we retrieved 2,723 articles: 1,016 articles from Web of Science, 1,060 from PubMed, and 647 from Embase; 1,234 duplicates were removed. The two reviewers screened 1,489 titles/abstracts in Covidence; 1,382 articles were found to be irrelevant. The kappa coefficient for agreement between reviewers was 0.92. On the basis of full text screening 118 clinical trials were extracted; 28 not original or duplicate trials were excluded. In total, 90 clinical trials published in 81 articles were included for data extraction (Figure S1A). Four hundred fifty ongoing clinical trials were retrieved from ClinicalTrials.gov. One reviewer screened the entries in the database; 402 were found to be irrelevant, and 48 ongoing clinical trials were included for data extraction (Figure S1B). Ongoing clinical trials were listed by the year of trial initiation. An overview of the published and ongoing clinical trials is shown in Tables S1 and S2. Seven articles presented two or more clinical trials with individual National Clinical Trial (NCT) numbers. A list of these articles is provided as Table S3. Design, inclusion/exclusion criteria, and search strategy are described in materials and methods.
Types of RNAi therapeutics
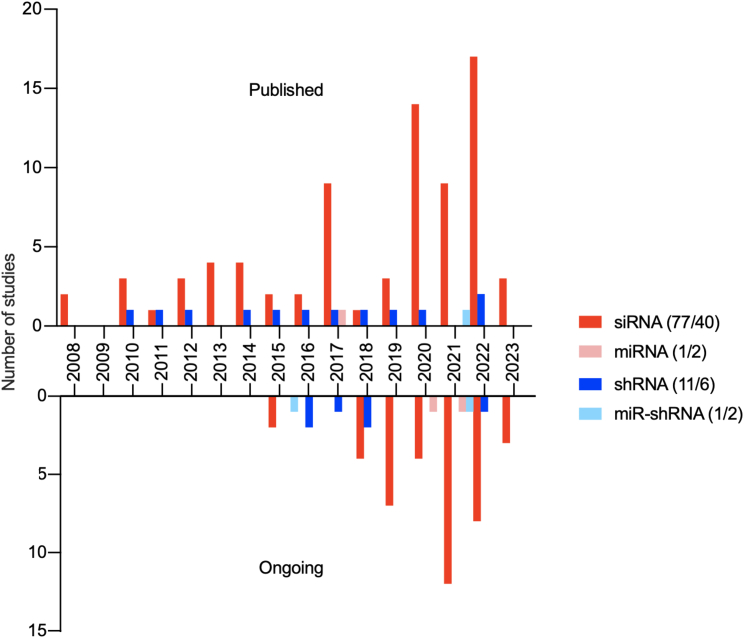
The first papers describing findings from clinical trials using RNAi therapeutics were published in 2008, ten years after the discovery of the RNAi mechanism. From 2008 to 2022, the annual number of published clinical trials using RNAi-based drugs increased almost 10-fold (Figure 3). Likewise, the number of ongoing clinical trials initiated from 2015 to 2021 increased approximately 7-fold. However, in 2022 the number of ongoing trials using siRNA-based drugs decreased by almost 40%. In each of the investigated years, siRNA was by far the most frequently used type of RNAi-inducing molecule in both the published and ongoing clinical trials, respectively. In 2008 and 2019 siRNA was used in all of the published and ongoing clinical trials. In 2022 siRNAs still dominated; it was used in ∼90% of both published and ongoing trials, and in 2023, so far all of the published and ongoing clinical trials have been siRNA-based. Delivery of expressed shRNAs as a treatment option has a low but almost constant frequency in the published trials through the years. A similar frequency of shRNA-based drugs was observed in the ongoing trials. Only few miRNAs and miR-shRNAs have been investigated in the published studies or are under investigation in the ongoing clinical trials (Figure 3).
Figure 3.
Types of RNAi therapeutics
Frequency distribution of RNAi-based drugs (siRNA, miRNA, shRNA, and miR-shRNA) used in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. Color-coded annotations for the distribution are included.
A clear tendency from the trials reported during the past 25 years is that synthetic siRNA dominates the field of RNAi therapeutics used in the clinic.
Delivery platforms of RNAi therapeutics
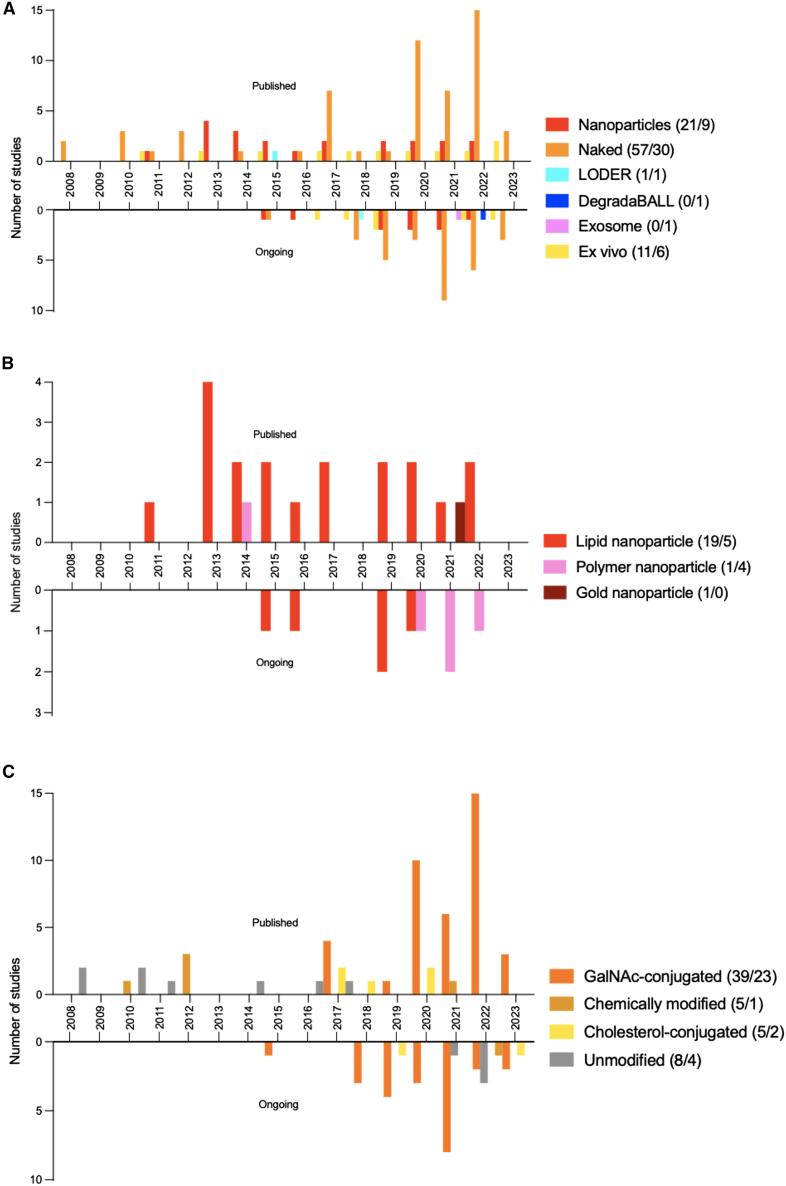
The delivery platform, that is the way to modify or package the RNAi-based molecule, is key to the effectiveness of the RNAi drug. Several factors, including oligonucleotide charge and size, RNase susceptibility, and immunogenicity, as well as interaction with reticuloendothelial system and endocytosis play important roles of the applied delivery platform. From 2008 to 2016 several different delivery platforms were used in the published and ongoing clinical trials, e.g., ex vivo methods (see Table S4), LODER technology (Local Deliver EluteR; a cancer drug delivery platform developed by Silenceed to enable insertion of the RNAi therapeutic directly into the core of solid tumors), and NPs (LNPs, polymer NPs, and gold NPs), with NPs being the most frequently used (Figure 4A). From 2017 onward, naked delivery of RNAi-based drugs is the most common method, and in 2022 79% and 63% of the published and ongoing trials used naked delivery. In comparison, in total 11% and 19% of the published and ongoing trials describes NP delivery of RNAi-based molecules, and 11% and 13% of the published and ongoing trial were delivered by ex vivo methods (Figure 4A). In 2023 naked delivery is used in all of the published and ongoing trials. LODER technology was used in 1% and 2% of all published and ongoing trials. DegradaBALL technology (a porous NP drug delivery platform developed by Lemonex) and exosomes were both used in 2% of all ongoing clinical trials.
Figure 4.
Delivery platforms of RNAi therapeutics
(A) Frequency distribution of methods used to deliver RNAi therapeutics to the target tissue used in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. See Table S4 for explanation of the “Ex vivo” category. (B) Frequency distribution of the types of NP types delivered in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. (C) Frequency distribution of the type of naked RNAi molecules used in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. Color-coded annotations are included for every distribution.
LNPs are most frequently used in the trials for NP delivery. LNPs were used in 91% and 56% of all published and ongoing clinical trials (Figure 4B). Other types of NPs delivery include polymer NPs used in 5% and 44% of all the published and ongoing clinical trials. Gold NP delivery was used in 5% of all the published clinical trials. Notably, the last clinical trial based on LNP delivery was initiated in 2020. From this time forward, polymer NPs seem be the preferred type of NP in the ongoing trials.
GalNAc-conjugated delivery of RNAi-based drugs is the most common form of naked delivery with 68% and 77% in total of the published and ongoing clinical trials (Figure 4C). GalNAc conjugates were not used before 2017, and after its introduction a significant shift in the delivery method unfolded, from LNP based to naked based (Figures 4B and 4C). Other types of naked delivery include chemical modifications (9% and 3%), cholesterol conjugation (9% and 7%), and unmodified delivery (14% and 13%), in the published and ongoing clinical trials, respectively (Figure 4C).
The most remarkable finding in delivery technology since the discovery of RNAi was the replacement of LNP-based delivery by GalNAc conjugation as the predominant method in 2017.
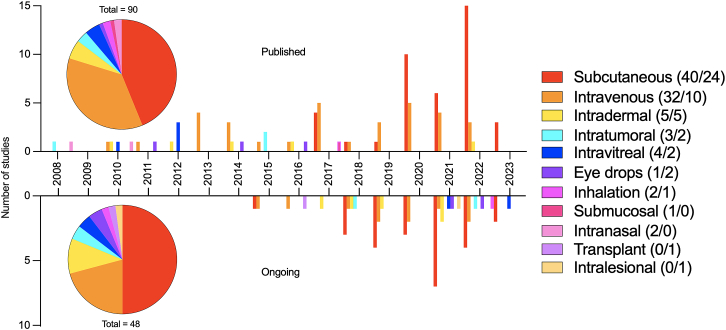
Administration of RNAi therapeutics
A similar shift in administration route was observed after 2017. Hence, before this year various deliver routes had been applied, whereas subcutaneous (s.c.) administration of RNAi therapeutics predominated after 2017. This finding is further substantiated by assessing the accumulated numbers as depicted in the pie charts in Figure 5: 44% and 50% of the published and ongoing trials use s.c. administration, and 36% and 21% use intravenous (i.v.) administration.
Figure 5.
Administration of RNAi therapeutics
Frequency distribution of the types of administration routes used in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. Pie charts showing the overall distribution of the types of administration routes used in the published and ongoing clinical trials. Color-coded annotations are included for every distribution.
In 2017, i.v. administration was used more frequently than s.c. administration (50% and 40%), and before 2017, s.c. administration was not used at all (Figure 5). However, from 2020 to 2022 s.c. administration was the predominant type of administration, with 79% of the published clinical trials using this type of administration in 2022 alone, compared with 16% of the trials using i.v. administration (Figure 5). In 2023 so far, every published clinical trial used s.c. administration. In the ongoing clinical trials, s.c. administration has in general been the most used form of administration, with 44% of trials being based on this form in 2022.
Other administration routes have also been investigated in the published clinical trials, especially before the emergence of s.c. administration in 2017. These include, e.g., intradermal (6% and 10%), intravitreal (5% and 4%), eye drops (1% and 4%), and inhalation (2% and 2%) in total in the published and ongoing clinical trials (Figure 5). The ongoing clinical trials in 2021–2023 re-investigated a number of administration routes used before 2017, and they also explore transplant and intralesional administration as new administration options, which have not been investigated in the published clinical trials.
Development of the GalNac technology has been the reason for the radical shift toward naked delivery in 2017 and the switch to s.c. administration. As shown in Figure S2A almost all (97% and 96%) of the published and ongoing clinical trials combining naked delivery with s.c. administration, used GalNAc-conjugated RNAi-based drugs. Ninety percent and 83% of the published and ongoing trials used i.v. administration in combination with LNPs (Figure S2B).
In summary, 2017 indicated a shift from the i.v. to the s.c. administration route in the clinical trials, and most clinical trials have been based on GalNAc conjugates combined with s.c. administration, or LNP combined with i.v. administration.
Diseases targeted by RNAi
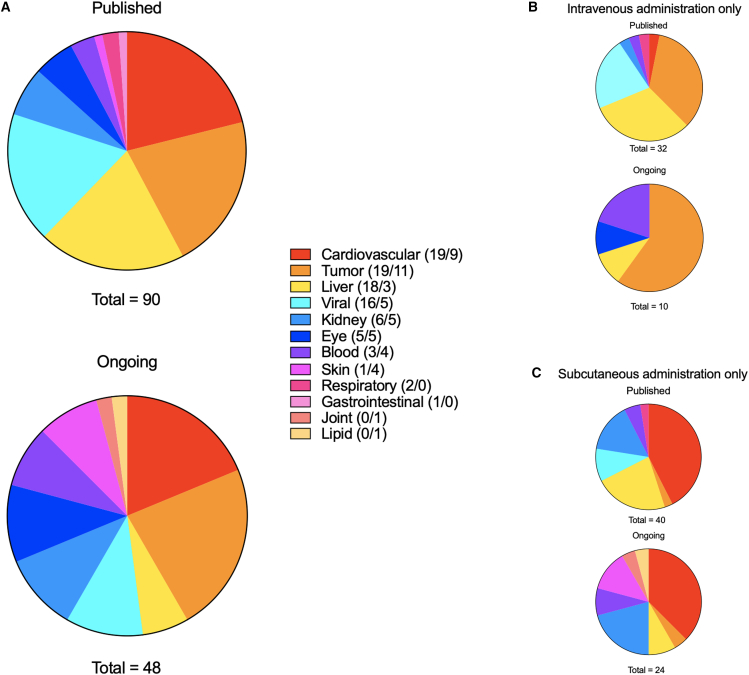
Diseases were grouped on the basis of etiology (Figure 6) or the tissue targeted by the RNAi therapeutics (Figure 7). Types of disease etiology included broad categories such as cardiovascular, tumor, liver, and viral (Figure 6).
Figure 6.
Disease etiology
(A) Overall distribution of the types of RNAi-treated diseases in the published and ongoing clinical trials, from 2008 to 2023 and 2015 to 2023, respectively. (B) Overall distribution of the types of RNAi-treated diseases using i.v. administration in the published and ongoing clinical trials, from 2008 to 2023 and 2015 to 2023, respectively. (C) Overall distribution of the types of RNAi-treated diseases using s.c. administration in the published and ongoing clinical trials from 2008 to 2023 and 2015 to 2023, respectively. Diseases are grouped by etiology. Color-coded annotations are included for every distribution.
Figure 7.
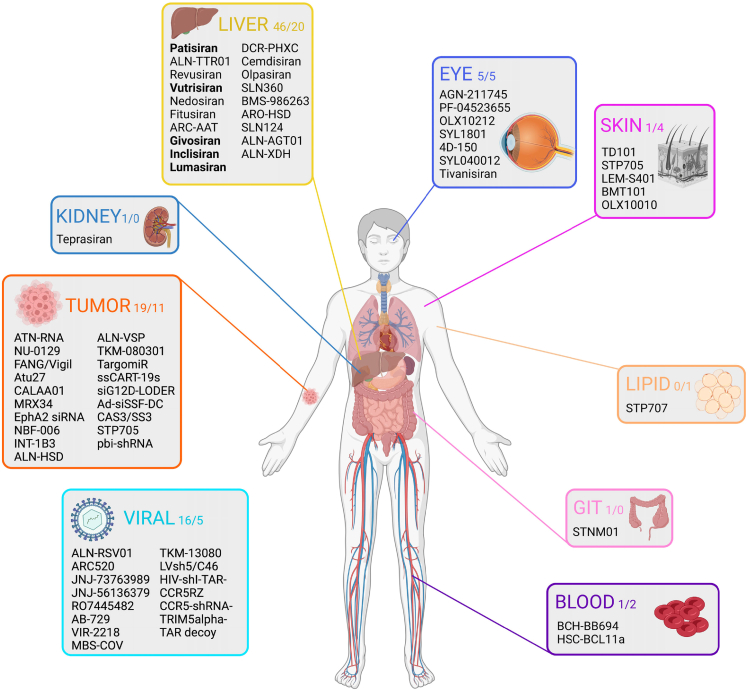
Tissues targeted by the RNAi therapeutics
Overall distribution of tissues targeted by the RNAi therapeutics in the published and ongoing clinical trials, from 2008 to 2023 and 2015 to 2023, respectively. The names of the RNAi therapeutics are included in the organ box together with the number of RNAi therapeutics found to target the specific organ in the published and ongoing clinical trials. The color of the border of the organ boxes correlates with the color-coded annotations in Figure 6. The five FDA- and EMA-approved therapeutical RNAi-based drugs are highlighted in bold. Inspired by Hu et al.20 Created using BioRender.com.
Etiology of the disease
RNAi therapy has been evaluated for several disease categories (Figure 6A). Cardiovascular, liver diseases, and tumor were targeted in about one-fifth of the published and ongoing clinical trials. Viral disease (e.g., HIV, respiratory-syncytial virus [RSV], and Ebola virus) was targeted in 18% and 10% of the published and ongoing clinical trials. Other diseases targeted in the published and ongoing trials include kidney (7% and 10%), eye (6% and 10%), and blood-manifested (3% and 8%) diseases.
Intravenous administration was primarily used to treat tumors (34% and 60%) and liver disease (31% and 10%) in the published and ongoing clinical trials (Figure 6B). The LNP delivery platform was used in more than half (56%) of the published trials with i.v. administration (Figure S2B). On the other hand, 43% and 38% of all s.c. administered RNAi therapeutics in the published and ongoing trials were used to treat cardiovascular-related diseases (Figure 6C).
Tissues and genes targeted by RNAi therapeutics
The number of RNAi therapeutics targeting specific tissues were assessed in the published and ongoing trials (Figure 7). Most RNAi-based drugs from the published and ongoing clinical trials target the liver (52% and 42%), although this may influence disease etiology in other organ systems. Tumors (21% and 23%) and viral infections are the second and third most frequent target, respectively (18% and 10%) of the published and ongoing clinical trials (Figure 7).
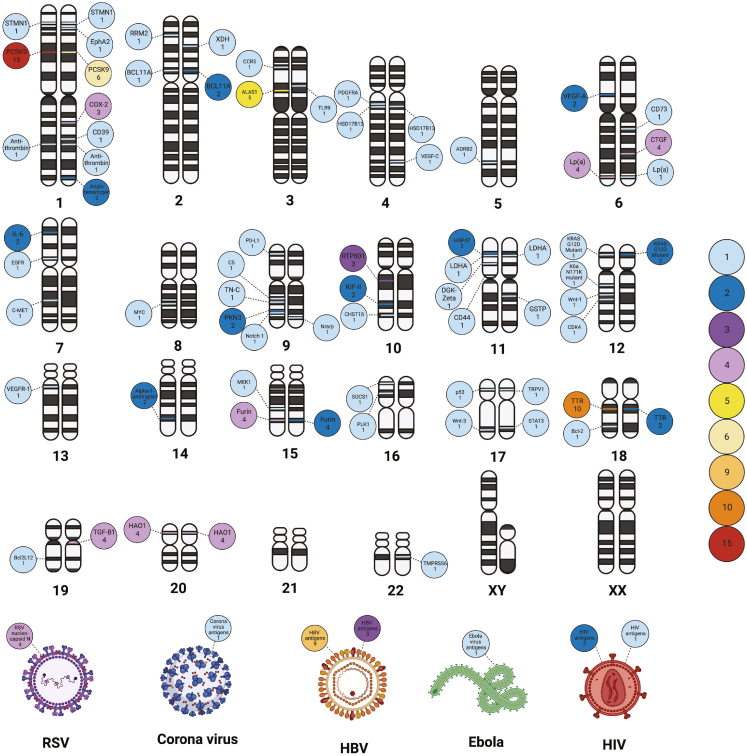
Comparison of the results categorized by disease etiology with target of the therapeutics shows that RNAi-based medicines applied in treatment of cardiovascular diseases all target the liver (Tables S5 and S6). These medicines include inclisiran, olpasiran, SLN360, and ALN-AGT01. In principle, the potency of RNAi enables the targeting of every disease-causing gene in the human genome. The gene loci and resulting proteins targeted by RNAi-based drugs are summarized in Figure 8. PCSK9, located on chromosome 1, is a prominent example of a targeted gene. The encoded protein, PCSK9, is involved in hypercholesterolemia by regulating hepatic lipoprotein metabolism. The mRNA transcribed by PCSK9 is the target in 15 and 6 published and ongoing clinical trials for the treatment of atherosclerotic cardiovascular disease (ASCVD) (hypercholesterolemia). Another example is TTR, located on chromosome 18. It also encodes a liver-expressed protein, and the involved mRNA is targeted 10 and 2 times in the published and ongoing clinical trials for the treatment of hATTR. Other frequent targets in the published and ongoing trials include VEGFA (2 and 0), HBV-antigens (9 and 3), and ALAS1 (5 and 0). A total of 64 genes (including viral genes) have been targeted in the published and ongoing clinical trials.
Figure 8.
Genome distribution of target loci
Heatmap showing the chromosomal location of genes targeted by RNAi therapeutics in the published (circles on the left chromosome) and ongoing clinical trials (circles on the right chromosome). Viral targets are shown below (circles on the left and right side of viruses depict the target in the published and ongoing clinical trials, respectively). Color-coded annotations are included on the right hand. Red refers to the highest number of studies targeting the mRNA of the given protein, whereas light blue refers to the lowest number of studies targeting the mRNA of the given protein (e.g., PCSK9 located on chromosome 1 encodes the PCSK9 protein. It is targeted 15 and 6 times in the published and ongoing clinical trials). Created using BioRender.com.
Approved RNAi-based drugs
At present, five RNAi-based drugs (patisiran, givosiran, lumasiran, inclisiran, and vutrisiran) have been approved by the FDA and the EMA. Selected characteristics are listed in Table S7. All have been identified in the published clinical trials, and all except for givosiran are applied in the ongoing clinical trials. They are all siRNA-based and target the liver. Except for patisiran, which is delivered i.v. in LNPs, they are administrated s.c. as GalNAc conjugates. In total, 68 different experimental RNAi medicines were identified from the published and ongoing clinical trials (Figure 7). Many of the genes, including TTR, LDHA, LPA, CCR5, STMN1, and a number of oncogenes, are targeted by more than one RNAi therapeutics (Figures 7 and 8).
The world of clinical RNAi
RNAi-based treatment options are investigated all around the world and patients are recruited from 60 different countries, with the vast majority of the published and ongoing clinical trials being recruited from the United States (55 and 40) (Figure S3). A high number of studies were likewise recruited from countries include Germany (27 and 11), United Kingdom (28 and 16), France (19 and 13), the Netherlands (18 and 12), and Italy (10 and 6). Fifty-three percent and 46% of the published and ongoing clinical trials recruited participants from more than one country. Cyprus, Iceland, Serbia, and Sierra Leone published trials with available published data, but for the present there are no ongoing trials. On the other hand, even though ongoing clinical trials are recruited from Chile, Croatia, Estonia, Ireland, Latvia, Lithuania, Mauritius, Puerto Rico, Romania, and Thailand, no studies have yet been published (Figure S3).
Discussion
Since its groundbreaking discovery, RNAi has been used extensively in basic research and clinical trials. The purpose of this systematic review was to provide a detailed overview of RNAi agents applied in the clinic, on the basis of published and ongoing clinical trials from the discovery 25 years ago until present day.
Use of RNAi-based molecules comes with inherent challenges because these molecules are vulnerable to nuclease-mediated degradation, have unfavorable physicochemical features that preclude simple transfer into cells, and may contribute to activation of the immune system. Safe and effective RNAi therapeutics therefore require sophisticated delivery technologies.36 The success of RNAi drugs critically depends on chemical modifications and/or technologies designed to protect the RNAi effector from degradation and to warrant their systemic stability, to facilitate localization to the target tissue, and finally, to safeguard successful intracellular delivery. On the other hand, RNAi represents an entirely new way to treat disease applicable to a plethora of targets. Indeed, this systematic review clearly demonstrate that the field of clinically applied RNAi is rapidly evolving. Cardiovascular diseases are currently the dominant group of diseases targeted by RNAi. However, treatment for a wide range of tissues from eyes to joints, and genetic to acquired diseases have been and are being investigated.
Recent inclusive narrative reviews, with detailed description of the mechanisms of RNAi18 and development of novel RNAi therapeutics6,17,19,37,38,39 including the chemistry of the RNAi-based drugs,20 the delivery mechanisms, the cellular uptake mechanisms of antisense oligonucleotides (ASOs) and their endocytotic pathways, should be acknowledged.40,41
The current review includes both published and ongoing clinical trials. Thus, the review not only summarizes the historic and contemporary field of clinical RNAi, but also provides a strong indication of its further direction. Notably, most of both published and ongoing clinical trials have recruited or are recruiting patients from the North America, Europe, South America, India, China, Russia, and South Africa, with the United States being the leading country in terms of number of trials. A clear tendency seems to be that regions or countries from which patients have been recruited continue their efforts by participating in the ongoing studies. Importantly, several “newcomers” from Africa, Central and South America, Europe, and Asia actively contribute to the ongoing trials, thereby adding more efforts, capacity, additive partnership, and knowledge into developing RNAi therapeutics. The continuous and increasing interest in RNAi-based treatment, reflected by the increase in the number of published and ongoing clinical trials clearly has been fueled by the successful development of Patisiran for treatment of hATTR spearheaded by Anylam Pharmaceuticals.33 The approval of the first RNAi therapeutics was a major scientific milestone for the entire field of RNAi and the atmosphere of RNAi optimism has sparked a new flood of interest not only from the scientific society, but importantly also from investors and big pharma. In celebration of the 25th anniversary of RNAi, this important outcome should be highly recognized.
The most remarkable finding was the high number of published clinical trials, amounting to 90 since the discovery of RNAi in 1998. These have paved the way for the five FDA- and EMA-approved RNAi therapeutics for the treatment of severe diseases for which there previously were unattractive or no treatment options at all. The drugs all target the liver which reflects the high accessibility of this organ through hepatic receptors, the specific anatomy of the liver, and the effective transfer of siRNAs by i.v. administrated LNPs or s.c. delivered GalNAc-conjugated siRNAs (see below). A remarkable decrease in the number of ongoing clinical trials targeting the liver was observed, most likely reflecting that the field awaits selection of new targets following the successes obtained by s.c. administration of GalNAc-conjugated siRNAs to hepatic tissue.
Historically, one must acknowledge the variety of types of RNAi therapeutics. From our literature study, it is clear that synthetic siRNAs are used more frequently than vector encoded RNAi-based molecules. An obvious advantage of siRNAs is that they are simpler to manufacture and quality assess, whereas shRNAs needs delivery of large DNA-based constructs to the nucleus. Hence, siRNAs require repeated administration, while shRNA-based therapies are typically more persistent. siRNA duplexes may activate the innate immune system via intra- and extracellular RNA sensors.42,43 shRNAs are less likely to elicit this response. On the other hand, shRNAs are primarily delivered by viral vectors that potentially induce immune and cytotoxicity.44,45 Additionally, shRNA-based molecules may saturate the processing pathway used by miRNAs, thereby interfering with the function of miRNAs and the endogenous RNAi machinery.45,46 Repression of off-target mRNAs, primarily mediated by so-called seed site matches47,48 is a general risk of RNAi. Both siRNAs and shRNAs can elicit off-target effects, but chemical modifications of the seed site and the co-delivered passenger strand may alleviate this concern.49
The biggest challenge of RNAi-based therapy is devising technology that allows efficient, specific, and safe delivery to the target cells. Many of the published and ongoing clinical trials take advantage of the efficient endocytosis of GalNAc-conjugated siRNAs and LNPs by liver cells, where both i.v. and s.c. administration are frequently applied. Hence, the liver can effectively endocytose GalNAc-conjugated siRNAs and LNPs. The first approaches to siRNA administration were focused on LNP-based delivery platforms offering a shielded compartment, isolated from immune components and serum nuclease activity, and a drug-biodistribution profile dictated by the packing agent.36 The function of LNPs is to protect the siRNA from degradation and renal clearance, thereby prolonging half-life and facilitating easy endosomal escape.50,51 To deliver siRNAs across barriers, the LNPs also needs to have a neutral surface charge provided by ionizable cationic lipid components, leading to reduced interaction with and binding of non-specific serum proteins.37,51
LNPs can associate with blood-circulating apolipoprotein E (ApoE) leading to cell-uptake via the low-density lipoprotein receptor (LDLR) on hepatocytes.37,41,52 However, dsRNA is recognized as non-self by extracellular and intracellular receptors, thus activating an immune response. An alternative is modifying the siRNA (e.g., by GalNAc conjugation). This receptor-based delivery platform centered on GalNAc conjugation enables the siRNA to bind to the asialoglycoprotein receptor (ASGPR) densely found on hepatocytes, facilitating clathrin-dependent receptor-mediated endocytosis leading to robust RNAi-mediated gene silencing.53 The unmodified siRNA drug ALN-RSV01 administered intranasally for the treatment of RSV did not make it past phase II, underscoring that competent and discriminating ASGPR-targeting ligands in combination with advantageous administration route have been key factors for clinically translation of GalNAc-siRNA-based molecules.17,54
Comparing the published and ongoing trials a clear tendency toward applying RNAi therapeutics in tumor treatment seems to unfold. The enhanced permeability and retention effect, possibly as a consequence of immature blood vessels and reduced lymphatic flow, seems to drive efficient transfer to and accumulation of RNAi drugs in tumor tissue.55
Interestingly, both unmodified and chemically modified RNAi therapeutics are investigated for the treatment of diseases using local administration. A prominent example of an unmodified RNAi-based drug is SYL1801 (phase II) which is administered locally by eye drops for treatment of neovascular AMD (NCT05637255). An example of a chemically modified RNAi drug is teprasiran (phase II) used to treat acute kidney injury.56
Intravenous administration is attractive as it facilitates quick and widespread distribution of the RNAi therapeutic. Another advantage of i.v. administration is the ability to delivery viral particles encoding shRNA or miR-shRNA. This could allow targeted delivery and safer RNAi treatment. However, i.v. administration is invasive and may result in infection, extravasation, and infiltration. These characteristics may also be challenging if the drug is not well-tolerated by the patient, or if the concentration is too high – resulting in an extensive immune response and cytotoxicity.57 To avoid these disadvantages the field has attempted to replace i.v. administration by s.c. administration, as this method is less invasive and allows slower uptake through the lymphatic system and capillaries into the circulation.58 Hence, s.c. administration has dominated the scene since 2017. A similar dominance of s.c. over i.v. administration is also observed in ongoing clinical trials. The fact that i.v. administration is less frequently used for liver-related diseases in the ongoing clinical trials compared with the published trials, further substantiates the prominent efficacy of s.c. administration developed for liver-targeting RNAi therapeutics. Subcutaneous administration has not been used for delivery of LNPs. A plausible explanation for this observation is that LNPs are often retained at the injection site because they are too large to enter capillaries and the lymph system.
Taken together, LNP systems are currently one of the most sophisticated non-viral nucleic acid delivery platforms enabling gene therapies. The recent decades of using lipid-based delivery systems for small molecule therapeutics have driven the efforts of evolving LNP technology for delivery of RNAi-based molecules38 and profoundly affected the success of, e.g., COVID-19 vaccines.59,60
Before 2017 several other administration routes besides s.c. and i.v. were applied. These routes were abandoned because of their unsuccessful application. However, on the basis of the gained knowledge from the success of s.c. and i.v. administrated drugs, the field seems to have shifted toward reimplementation of previously used administration routes, including intradermal, intratumoral, intravitreal, topical eye drops, and inhalations. The idea is to tailor the therapy using local administration of RNAi therapeutics as in the case of intravitreal injections of naked siRNAs to the eye or intertumoral injection of siRNAs. Notably, ex vivo delivery of RNAi-based drugs seems to slightly lag behind the rest of the field, and the few examples are based on gene transfer of shRNA- or miR-shRNA-encoding genes to tumor cells, T helper cells, B cells, and hemopoietic stem cells (Table S2).
A notable observation is the relative few gene targets currently exploited for treatment despite the almost unlimited potential of RNAi-based drugs.61 The trajectory depicted by the ongoing clinical trials shows that future RNAi therapeutics may be based on siRNAs, delivered naked or in a conjugated form (such as GalNAc) using the well-established s.c.- and intravitreal-based administration routes as well as novel, less proven methods. These approaches may hence be based on eye drop, inhalation, submucosal, intranasal, and intralesional administration for the targeting of a wide variety of diseases affecting the eye, CNS, kidney, blood, skin, joints, or lipid tissues (ClinicalTrials.gov; Table S2). This notion is further supported by the massive activity of pharmaceutical corporations including Alnylam, Gradalis, Dicerna, Olix, Novartis, and Sylentis.
Alnylam has pioneered the translation of RNAi medicines for especially liver-related and liver-targeted diseases and has recently also focused their activities on infectious, cardio-metabolic, and CNS disorders. However, the view on RNAi-based drugs looked quite different at the end of 2016 when Alnylam halted their phase III trial of revusiran because of severe adverse events, resulting in several large pharmaceutical companies backing out of the RNAi field. Despite this setback, Alnylam persisted and performed, on the basis of their enhanced stabilization chemistry (ESC), the first clinical trials using GalNAc-siRNA conjugates. The increased stability of ESC-GalNAc-siRNA conjugates resulted in efficacy ten times greater than standard template chemistry because of considerably increased liver exposure and sustained gene silencing.36 In 2018 Alnylam had six GalNAc-siRNA conjugates in clinical trials and by January 2023 the company has increased its activity to ten active RNAi-therapeutic-based clinical trials.
Gradalis develops shRNA for tumors (ex vivo intradermal delivery and i.v. LNP-based delivery, 3 ongoing clinical trials). Olix targets hypertension (s.c. administration of GalNAc-conjugated siRNA, 1 ongoing clinical trial) and AMD (intravitreal administration of cholesterol-conjugated siRNA, 1 ongoing clinical trial). Novartis focuses on treating ASCVD (s.c. administration of GalNAc-conjugated siRNA, 5 ongoing clinical trials). This atmosphere of RNAi optimism has stimulated a new global wave of investment and licensing deals, reinvolving big pharma, such as Novo Nordisk, Eli Lilly, and Janssen.
Only a few studies have focused on diseases of the eye, which otherwise have been at the forefront of gene therapy.62 Notably, the eye is the target tissue in the first siRNA clinical trial and is especially attractive for RNAi-based treatment, primarily because of its small size and the intraocular immune privilege, which both favor local delivery and thus limiting inflammation and off-target effects.63 The only approved RNA-based drug for the eye, Macugen, targeting VEGF in the eye, has been withdrawn by the request of the marketing holder.64 The observed discrepancy most likely reflects that retinal gene therapy has aimed at treating loss-of-function diseases rather than gain-of-function diseases such as hATTR. Another reason is unmet endpoint goals, which lead to discontinuation of a phase III trial of bevasiranib (NCT00499590). However, seven different RNAi-based drugs, fighting various eye diseases including wet AMD (wAMD) (AGN-211745 [phase I], PF-04523655 [phase I/II], OLX10212 [phase I], SYL1801 [phase II], and 4D-150 [phase I/II]), diabetic macular edema (DME; PF-04523655 [phase I]), glaucoma (SYL040012 [phase I]), and eye discomfort, particularly dry eye disease (Sjögren’s syndrome) (tivanisiran (SYL1001) [phase III]) are currently being investigated in clinical trials. As an alternative to using synthetic siRNAs, novel vector encoded shRNAs may offer a more persistent solution.16 A number of recent studies using shRNA-, agshRNA-, and miRtron-based therapy have already been investigate in pre-clinical settings,2,65,66 indicating that such RNAi-based molecules targeting the retina may also find its way into clinical trials.62,67,68
In conclusion, the broad spectrum of diseases identified in the ongoing clinical trials is excellent news not only for patients but also the entire field of RNAi. Twenty-five years ago, RNAi was first described in C. elegans. After two and a half decades of maturation, a handful of approved RNAi treatments targeting the liver exist and treatment for other tissues are dedicatedly investigated. These exiting findings clearly signal that RNAi-based drugs are here to stay and have almost indefinite potential. As RNAi therapy is making headway in the clinic, the progress of the liver therapeutics discussed will hopefully provide the unmet need for treatment of extra-hepatic disease thereby improving the quality of life for a great spectrum of patients.
Materials and methods
Design and inclusion/exclusion criteria
This systematic review was conducted with adherence to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 statement guidelines (the PRISMA 2020 statement checklist provided in Table S8). The review was not registered. Inclusion/exclusion criteria, search strategy, and extraction protocol were defined prior to the literature search. For the identification of published clinical trials, we included all original research articles on clinical trials using RNAi. Non-interventional studies were excluded. The search period was restricted to the publication year of Fire and Mello’s seminal paper,1 1998, and forward. Non-original articles, studies in non-human species, and pure in vitro/ex vivo studies were excluded. For the identification of ongoing clinical trials, we included active entries in ClinicalTrials.gov using RNAi.
Search strategy
We searched Web of Science (All Databases), PubMed, and Embase from January 1, 1998, to December 30, 2022. The search was performed December 30, 2022, using Google Chrome Version 108.0.5359.124 (Official Build) (arm64). The search string was built from synonyms of RNAi together with the above-mentioned inclusion/exclusion criteria. For a limited number of studies, the online publication date was in the end of 2022 with the consequence that the paper first appeared in the journal the following year. In these cases, the studied are figured in 2023.
Articles were imported to Endnote 20.4 (build 18004) with semi-automated removal of duplicates, and with subsequent manual removal of duplicates not found by Endnote. Articles were imported into Covidence systematic review software (Veritas Health Innovation, Melbourne, Australia) and independently screened by titles/abstracts by two reviewers. Included full text articles were screened by two reviewers, who also extracted clinical trials identified in each article. Clinical trials were excluded according to the above-mentioned criteria and limitations. Furthermore, clinical trials sharing NCT number with a trial published earlier, were also excluded. All conflicts between reviewers during the screening process were resolved by consensus. Data from included clinical trials were extracted according to the extraction protocol (Table S9). If data for a given clinical trial (with NCT number) were missing in the corresponding included article, data were, if possible, retrieved from ClinicalTrials.gov.
We searched ClinicalTrials.gov for ongoing clinical trials using the filters “recruiting,” “enrolling by invitation,” and “active, not recruiting.” The search was performed January 18, 2023, using Google Chrome Version 108.0.5359.124 (Official Build) (arm64). The aforementioned extraction protocol was deployed on these studies.
The full search strategy including inclusion/exclusion criteria for identification of published and ongoing clinical trials via databases and ClinicalTrials.gov is provided as in the supplemental information.
Our assessment of LNPs and chemical modifications of RNAi effectors were limited to general categories of LNP-based and chemical modification delivery platforms. Another limitation was that only one reviewer screened/extracted the ongoing clinical trials. We did not assess the risk for bias in the included published and ongoing studies.
Statistics
Descriptive statistics were performed for study characterization and kappa statistical analysis was performed to determine interrater agreement (title/abstract screening). Statistical analyses and accompanying figures were made in Prism 9.5.1 for Mac OS X (GraphPad Software, San Diego, CA).
Acknowledgments
This work was supported by the Faculty of Health Sciences, Aarhus University (PhD scholarships to B.K.F.-J. and T.S.J.), Fight for Sight, Denmark (B.K.F.-J. and T.S.J.), the Synoptik Foundation (T.S.J.), APTaps (T.J.C.), and the Velux Foundation (T.J.C.).
Author contributions
Conceptualization, B.K.F.-J., A.L.A., L.A., and T.J.C.; Methodology, I.J.C., T.S.J., and B.K.F.-J.; Software, B.K.F.-J. and E.G.J.; Article Screening, I.J.C., A.C.J., A.L.A., L.A., and B.K.F.-J.; Data Extraction, I.J.C., B.K.F.-J., and A.C.J.; Writing - Original Draft, I.J.C., L.A., and T.J.C.; Writing - Review and Editing, I.J.C., B.K.F.-J., T.S.J., A.C.J., E.G.J., A.L.A., L.A., and T.J.C.; Visualization, I.J.C., B.K.F.-J., E.G.J., A.L.A., L.A., and T.J.C.; Supervision, A.L.A., L.A., and T.J.C.; Project Administration, L.A. and T.J.C. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.07.018.
Supplemental information
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Alsing S., Doktor T.K., Askou A.L., Jensen E.G., Ahmadov U., Kristensen L.S., Andresen B.S., Aagaard L., Corydon T.J. VEGFA-targeting miR-agshRNAs combine efficacy with specificity and safety for retinal gene therapy. Mol. Ther. Nucleic Acids. 2022;28:58–76. doi: 10.1016/j.omtn.2022.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Bobbin M.L., Rossi J.J. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 9.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 12.Kaadt E., Alsing S., Cecchi C.R., Damgaard C.K., Corydon T.J., Aagaard L. Efficient Knockdown and Lack of Passenger Strand Activity by Dicer-Independent shRNAs Expressed from Pol II-Driven MicroRNA Scaffolds. Mol. Ther. Nucleic Acids. 2019;14:318–328. doi: 10.1016/j.omtn.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheloufi S., Dos Santos C.O., Chong M.M.W., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 18.Mencia R., Gonzalo L., Tossolini I., Manavella P.A. Keeping up with the miRNAs: current paradigms of the biogenesis pathway. J. Exp. Bot. 2023;74:2213–2227. doi: 10.1093/jxb/erac322. [DOI] [PubMed] [Google Scholar]
- 19.Traber G.M., Yu A.M. RNAi-Based Therapeutics and Novel RNA Bioengineering Technologies. J. Pharmacol. Exp. Therapeut. 2023;384:133–154. doi: 10.1124/jpet.122.001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J. Therapeutic siRNA: state of the art. Signal Transduct. Targeted Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 24.Sirna-027 Study I., Kaiser P.K., Symons R.C.A., Shah S.M., Quinlan E.J., Tabandeh H., Do D.V., Reisen G., Lockridge J.A., Short B., et al. RNAi-Based Treatment for Neovascular Age-Related Macular Degeneration by Sirna-027. Am. J. Ophthalmol. 2010;150:33–39. doi: 10.1016/j.ajo.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Ten Years of siRNA – a Clinical Overview. 2012. https://www.europeanpharmaceuticalreview.com/article/13688/ten-years-of-sirna-a-clinical-overview/
- 26.Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 27.The Nobel Prize in Physiology or Medicine 2006. https://www.nobelprize.org/prizes/medicine/2006/summary/
- 28.Davis M.E., Zuckerman J.E., Choi C.H.J., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hattab D., Gazzali A.M., Bakhtiar A. Clinical Advances of siRNA-Based Nanotherapeutics for Cancer Treatment. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13071009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gish R.G., Satishchandran C., Young M., Pachuk C. RNA interference and its potential applications to chronic HBV treatment: results of a Phase I safety and tolerability study. Antivir. Ther. 2011;16:547–554. doi: 10.3851/IMP1798. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald K., White S., Borodovsky A., Bettencourt B.R., Strahs A., Clausen V., Wijngaard P., Horton J.D., Taubel J., Brooks A., et al. A Highly Durable RNAi Therapeutic Inhibitor of PCSK9. N. Engl. J. Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmermann T.S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B.R., Hutabarat R., Nochur S., Vaishnaw A., Gollob J. Clinical Proof of Concept for a Novel Hepatocyte-Targeting GaINAc-siRNA Conjugate. Mol. Ther. 2017;25:71–78. doi: 10.1016/j.ymthe.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. NEJM. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 34.FDA Approved Drugs. 2023. https://cms.centerwatch.com/directories/1067-fda-approved-
- 35.Table of all EPARs for human and veterinary medicines. 2023. https://www.ema.europa.eu/en/medicines/download-medicine-data#european-public-assessment-reports-
- 36.Kulkarni J.A., Witzigmann D., Thomson S.B., Chen S., Leavitt B.R., Cullis P.R., van der Meel R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021;16:630–643. doi: 10.1038/s41565-021-00898-0. [DOI] [PubMed] [Google Scholar]
- 37.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springer A.D., Dowdy S.F. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Therapeut. 2018;28:109–118. doi: 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 41.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R.G. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 43.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 44.Rao D.D., Vorhies J.S., Senzer N., Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv. Drug Deliv. Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Kenworthy R., Lambert D., Yang F., Wang N., Chen Z., Zhu H., Zhu F., Liu C., Li K., Tang H. Short-hairpin RNAs delivered by lentiviral vector transduction trigger RIG-I-mediated IFN activation. Nucleic Acids Res. 2009;37:6587–6599. doi: 10.1093/nar/gkp714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 47.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 48.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iribe H., Miyamoto K., Takahashi T., Kobayashi Y., Leo J., Aida M., Ui-Tei K. Chemical Modification of the siRNA Seed Region Suppresses Off-Target Effects by Steric Hindrance to Base-Pairing with Targets. ACS Omega. 2017;2:2055–2064. doi: 10.1021/acsomega.7b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 51.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan C., Allen T.M., Cullis P.R. Lipid nanoparticle delivery systems for siRNA-based therapeutics. Drug Deliv. Transl. Res. 2014;4:74–83. doi: 10.1007/s13346-013-0161-z. [DOI] [PubMed] [Google Scholar]
- 53.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel'in A.V., O'Shea J., et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 54.Gottlieb J., Zamora M.R., Hodges T., Musk A.W., Sommerwerk U., Dilling D., Arcasoy S., DeVincenzo J., Karsten V., Shah S., et al. ALN-RSV01 for prevention of bronchiolitis obliterans syndrome after respiratory syncytial virus. infection in lung transplant recipients. J. Heart Lung Transplant. 2016;35:213–221. doi: 10.1016/j.healun.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Wu J. The Enhanced Permeability and Retention (EPR) Effect: The Significance of the Concept and Methods to Enhance Its Application. J. Personalized Med. 2021;11 doi: 10.3390/jpm11080771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thielmann M., Corteville D., Szabo G., Swaminathan M., Lamy A., Lehner L.J., Brown C.D., Noiseux N., Atta M.G., Squiers E.C., et al. Teprasiran, a Small Interfering RNA, for the Prevention of Acute Kidney Injury in High-Risk Patients Undergoing Cardiac Surgery A Randomized Clinical Study. Circulation. 2021;144:1133–1144. doi: 10.1161/CIRCULATIONAHA.120.053029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vijayakumar A., Sharon E.V., Teena J., Nobil S., Nazeer I. A clinical study on drug-related problems associated with intravenous drug administration. J. Basic Clin. Pharm. 2014;5:49–53. doi: 10.4103/0976-0105.134984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McLennan D.N., Porter C.J.H., Charman S.A. Subcutaneous drug delivery and the role of the lymphatics. Drug Discov. Today Technol. 2005;2:89–96. doi: 10.1016/j.ddtec.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 59.Labouta H.I., Langer R., Cullis P.R., Merkel O.M., Prausnitz M.R., Gomaa Y., Nogueira S.S., Kumeria T. Role of drug delivery technologies in the success of COVID-19 vaccines: a perspective. Drug Deliv. Transl. Res. 2022;12:2581–2588. doi: 10.1007/s13346-022-01146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pardridge W.M. Brain gene therapy with Trojan horse lipid nanoparticles. Trends Mol. Med. 2023;29:343–353. doi: 10.1016/j.molmed.2023.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossi J.J., Rossi D.J. siRNA Drugs: Here to Stay. Mol. Ther. 2021;29:431–432. doi: 10.1016/j.ymthe.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Askou A.L., Jakobsen T.S., Corydon T.J. Retinal gene therapy: an eye-opener of the 21st century. Gene Ther. 2021;28:209–216. doi: 10.1038/s41434-020-0168-2. [DOI] [PubMed] [Google Scholar]
- 63.Thakur A., Fitzpatrick S., Zaman A., Kugathasan K., Muirhead B., Hortelano G., Sheardown H. Strategies for ocular siRNA delivery: Potential and limitations of non-viral nanocarriers. J. Biol. Eng. 2012;6:7. doi: 10.1186/1754-1611-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morjaria R., Chong N.V. Pharmacokinetic evaluation of pegaptanib octasodium for the treatment of diabetic edema. Expet Opin. Drug Metabol. Toxicol. 2014;10:1185–1192. doi: 10.1517/17425255.2014.922543. [DOI] [PubMed] [Google Scholar]
- 65.Askou A.L., Alsing S., Benckendorff J.N.E., Holmgaard A., Mikkelsen J.G., Aagaard L., Bek T., Corydon T.J. Suppression of Choroidal Neovascularization by AAV-Based Dual-Acting Antiangiogenic Gene Therapy. Mol. Ther. Nucleic Acids. 2019;16:38–50. doi: 10.1016/j.omtn.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orlans H.O., McClements M.E., Barnard A.R., Martinez-Fernandez de la Camara C., MacLaren R.E. Mirtron-mediated RNA knockdown/replacement therapy for the treatment of dominant retinitis pigmentosa. Nat. Commun. 2021;12:4934. doi: 10.1038/s41467-021-25204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin F.L., Wang P.Y., Chuang Y.F., Wang J.H., Wong V.H.Y., Bui B.V., Liu G.S. Gene Therapy Intervention in Neovascular Eye Disease: A Recent Update. Mol. Ther. 2020;28:2120–2138. doi: 10.1016/j.ymthe.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker J., Fischer N., Grimm D. Right on target: The next class of efficient, safe, and specific RNAi triggers. Mol. Ther. Nucleic Acids. 2022;28:363–365. doi: 10.1016/j.omtn.2022.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.