Abstract
Most simple retroviruses induce tumors of a single cell type when infected into susceptible hosts. The SRS 19-6 murine leukemia virus (MuLV), which originated in mainland China, induces leukemias of multiple cellular origins. Indeed, infected mice often harbor more than one tumor type. Since the enhancers of many MuLVs are major determinants of tumor specificity, we tested the role of the SRS 19-6 MuLV enhancers in its broad disease specificity. The enhancer elements of the Moloney MuLV (M-MuLV) were replaced by the 170-bp enhancers of SRS 19-6 MuLV, yielding the recombinants ΔMo+SRS+ and ΔMo+SRS− M-MuLV. M-MuLV normally induces T-lymphoid tumors in all infected mice. Surprisingly, when neonatal mice were inoculated with ΔMo+SRS+ or ΔMo+SRS− M-MuLV, all tumors were of T-lymphoid origin, typical of M-MuLV rather than SRS 19-6 MuLV. Thus, the SRS 19-6 MuLV enhancers did not confer the broad disease specificity of SRS 19-6 MuLV to M-MuLV. However, all tumors contained ΔMo+SRS M-MuLV proviruses with common enhancer alterations. These alterations consisted of tandem multimerization of a subregion of the SRS 19-6 enhancers, encompassing the conserved LVb and core sites and adjacent sequences. Moreover, when tumors induced by the parental SRS 19-6 MuLV were analyzed, most of the T-lymphoid tumors had similar enhancer alterations in the same region whereas tumors of other lineages retained the parental SRS 19-6 MuLV enhancers. These results emphasize the importance of a subregion of the SRS 19-6 MuLV enhancer in induction of T-cell lymphoma. The relevant sequences were consistent with crucial sequences for T-cell lymphomagenesis identified for other MuLVs such as M-MuLV and SL3-3 MuLV. These results also suggest that other regions of the SRS 19-6 MuLV genome contribute to its broad leukemogenic spectrum.
Tumor induction by nonacute murine retroviruses involves multiple interactions with the infected host (17). Extensive mutagenesis experiments have shown that transcriptional enhancers within the long terminal repeats (LTRs) are major determinants of retroviral disease specificity and latency (10, 13, 16, 29). Strong LTR enhancer activity is required for the efficient activation of cellular proto-oncogenes by proviral insertion, a common step in viral leukemogenesis (25). High enhancer activity is in turn dependent on the binding of cellular transcription factors that are often expressed in a tissue-specific manner. Thus, cells that support a high level of viral transcription are likely targets for tumor development for a given retrovirus. Indeed, prior studies have shown that enhancers of various murine leukemia viruses (MuLVs) that induce T-cell lymphomas stimulate transcription to a much greater extent in T cells than in other infectable cell types (9, 40, 43, 46).
We have previously described the molecular cloning and characterization of a unique murine retrovirus called the solid-type reticulum sarcoma virus (SRS 19-6), which was originally isolated in mainland China (7). SRS 19-6 MuLV is a replication-competent MuLV that induces leukemias with a mean latency of 6 months. However, unlike most other MuLV, SRS 19-6 MuLV induces tumors derived from multiple cell lineages. SRS 19-6 MuLV induced 35% myeloid, 9% erythroid, 28% B-lymphoid, 14% T-lymphoid, and 2% non-T non-B lymphoid leukemias when inoculated into neonatal NIH Swiss mice (7). Infected animals often had more than one tumor type.
The U3 region of the SRS 19-6 MuLV LTR contains a putative enhancer element approximately 170 bp upstream from the start site of transcription. Typical of many viral and cellular enhancer sequences, this region is composed of multiple motifs for sequence-specific DNA binding proteins (6). Unlike many retroviral enhancers, however, the SRS enhancer consists of a single copy of this array. Binding sites of note are a central LVb/ets motif and an immediately adjacent imperfect core/AML1 motif that are highly conserved among the LTRs of most members of the C-type retrovirus family (21). Prior mutagenesis studies have shown that this pair of binding sites is crucial for the T-cell specificity of Moloney MuLV (M-MuLV) (42). On the other hand, the LVb and core binding sites are also shared with Friend MuLV (F-MuLV), which induces exclusively erythroid leukemia. It has been speculated that regions adjacent to the LVb and core sites also influence the type of leukemia induced by both M-MuLV and F-MuLV (20). The SRS 19-6 MuLV enhancer has a mixture of motifs in common with both M-MuLV and F-MuLV in the regions flanking the LVb and core sites; thus, it is plausible that the SRS 19-6 enhancer element could determine the broad disease specificity of SRS 19-6 MuLV.
In this study we tested the hypothesis that the putative SRS 19-6 MuLV enhancer sequences determine the disease specificity of SRS 19-6 MuLV. This was done by inserting these sequences into an enhancerless plasmid clone of M-MuLV. Infectious virus was recovered by transfection of NIH 3T3 cells, and its pathogenic potential was assessed by injecting it into neonatal NIH Swiss mice. Somewhat surprisingly, the SRS 19-6 MuLV enhancers did not confer the broad disease spectrum characteristic of wild-type SRS 19-6 MuLV onto M-MuLV. Additional detailed analysis of LTRs in tumors revealed consistent enhancer alterations associated with the development of T-cell lymphoma for both the chimeric virus and the parental wild-type SRS 19-6 MuLV.
MATERIALS AND METHODS
Construction of recombinant provirus and production of an infected cell line.
The 170-bp SRS 19-6 MuLV enhancer fragment was generated by PCR amplification with the forward primer: 5′-CAAGATCTAGAATAGGGAAGTTCAGA-3′ and the reverse primer 5′-AATCTAGAAACATCTGATGGGTCTCT-3′ which were complementary to the SRS LTR sequences from positions −300 to −275 and from positions −123 to −148 respectively. PCR amplification (1 cycle of 94°C for 4 min, 56°C for 1 min, and 72°C for 2 min; 33 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and 1 cycle of 94°C for 1 min, 56°C for 1 min, and 72°C for 10 min) in a Perkin-Elmer thermal cycler was carried out on a plasmid template containing the SRS 19-6 MuLV LTR (7). The resulting fragment was resolved and purified from a 1% agarose gel by using the Gene Clean Plus DNA extraction kit (Bio 101) and was digested with XbaI to generate cohesive ends. The digested fragment was ligated into the enhancerless M-MuLV LTR construct ΔMo (23) at the XbaI site at position −150 in the M-MuLV LTR. The resulting LTRs were designated ΔMo+SRS+ and ΔMo+SRS− M-MuLV depending on the orientation of the SRS fragment. Full-length proviral constructs were generated by replacing the downstream LTR of the full-length M-MuLV molecular clone (p63-2) (1) with the ΔMo+SRS+ or ΔMo+SRS− M-MuLV LTR fragment from ClaI to EcoRI as previously described (34). NIH 3T3 fibroblasts were transfected with these constructs, whereupon the U3 regions of the recombinant downstream LTRs are copied to the upstream LTRs during viral DNA synthesis. Following three passages in medium containing 2 μg of Polybrene per ml (to aid the spread of viral infection), an initial screen by the UV-XC plaque assay (38) was used to verify confluent infection with ecotropic virus. Single cell clones were then isolated and screened for the presence of recombinant proviral sequences by Southern blot analysis with the SRS 19-6 MuLV enhancer fragment to probe PstI-digested DNA by standard methods (41). The presence of a ΔMo+SRS M-MuLV provirus in the transfected cells was indicated by the appearance of an 839-bp hybridizing fragment for ΔMo+SRS+ M-MuLV and a 757-bp hybridizing fragment for ΔMo+SRS− M-MuLV.
Viral stocks and mouse inoculation.
Viral stocks were harvested as cell culture supernatant from infected confluent NIH 3T3 fibroblasts to which fresh growth medium had been added 24 h prior to harvest. The supernatants were passed through a 0.45-pore-size filter and stored in aliquots at −70°C. Titers of viral stocks were determined by the UV-XC plaque assay (38). Neonatal mice (1 to 2 days old) were injected intraperitoneally with 0.2 ml of viral supernatant containing ΔMo+SRS M-MuLVs (7 × 104 XC PFU for ΔMo+SRS+ and 1 × 104 XC PFU for ΔMo+SRS−M-MuLV).
Detection of proviral DNA in tumors and molecular characterization.
Genomic DNAs were harvested from the spleen, thymus, and lymph nodes of moribund leukemic mice as previously described (19). Restriction endonuclease digestion of genomic DNA (5 μg), agarose gel electrophoresis (0.8% agarose), and transfer to GeneScreen Plus (New England Nuclear) were performed as described previously (19). Hybridization probes included the random primed SRS enhancer PCR fragment for the detection of input ΔMo+SRS+ and ΔMo+SRS− M-MuLVs, as well as DNA fragments containing c-myc, Pim-1, Pvt-1, T-cell receptor beta (TCR-β), immunoglobulin heavy-chain (IgH), and Igκ gene sequences for the molecular characterization of each tumor type as described previously (18).
PCR analysis of tumor DNAs.
PCR amplification of genomic tumor DNAs was performed in a 100-μl reaction mixture consisting of Taq polymerase buffer (Perkin-Elmer) containing 0.25 mM deoxynucleoside triphosphates, 20 pmol of each primer, 6 mM MgCl2, 2.5 U of Taq polymerase (Perkin-Elmer), and 1 μg of genomic tumor DNA. Each reaction product was amplified by the following PCR schedule: 1 cycle of 94°C for 4 min, 56°C for 1 min, and 72°C for 2 min; 33 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and 1 cycle of 94°C for 1 min, 56°C for 1 min, and 72°C for 10 min in a Perkin-Elmer thermal cycler. Each reaction product was analyzed by agarose gel electrophoresis (1.5% agarose).
Cloning and sequencing of PCR fragments.
DNA fragments from PCR analysis were cloned into the plasmid pCR2.1 TA cloning system (Invitrogen), and standard α-complementation with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates was used to screen for recombinants. White colonies were isolated, and the Qiagen Mini-Prep purification system was used to purify plasmid DNA that was sequenced by the Sanger et al. chain termination method (39) with [α-35S]dATP and the USB Sequenase kit. DNA sequences were resolved by polyacrylamide sequencing gel electrophoresis (6% polyacrylamide) followed by autoradiography.
RESULTS
Generation of ΔMo+SRS M-MuLV.
To study the role of the SRS 19-6 MuLV enhancer sequences in the broad tumor specificity of SRS 19-6 MuLV, a recombinant M-MuLV, in which the putative SRS 19-6 MuLV enhancers were substituted for the M-MuLV enhancers, was generated (Fig. 1). The SRS 19-6 MuLV enhancer is present in a single copy, as opposed to the tandemly repeated M-MuLV enhancers. The SRS 19-6 MuLV enhancer was inserted into an M-MuLV LTR lacking the enhancer sequences, ΔMo (31). A 170-bp SRS 19-6 MuLV enhancer-containing fragment (from positions −296 to −127) was generated by PCR amplification of SRS 19-6 MuLV plasmid DNA with the oligonucleotide primers specified in Materials and Methods. The primers contained terminal XbaI sites; following cleavage with XbaI, the PCR product was ligated into the XbaI site at position −150 of a plasmid containing the ΔMo LTR to generate the recombinant ΔMo+SRS+ and ΔMo+SRS− M-MuLV LTRs (Fig. 1). These two LTRs were then used to generate M-MuLV provirus clones containing the recombinant LTRs at the 3′ ends as described previously (34). To generate infectious ΔMo+SRS+ and ΔMo+SRS− M-MuLVs, NIH 3T3 fibroblasts were transfected with the recombinant plasmids. During transfection and reverse transcription, the recombinant 3′ LTRs were copied to both ends of the resulting proviruses. To prevent the possible outgrowth of wild-type M-MuLV recombinants that might arise during transfection, single-cell producer clones from the transfected NIH 3T3 cells were isolated and Southern blot hybridization was used to verify that they were infected with the recombinant M-MuLV proviruses only (data not shown). These clones were used as sources of infectious recombinant M-MuLV.
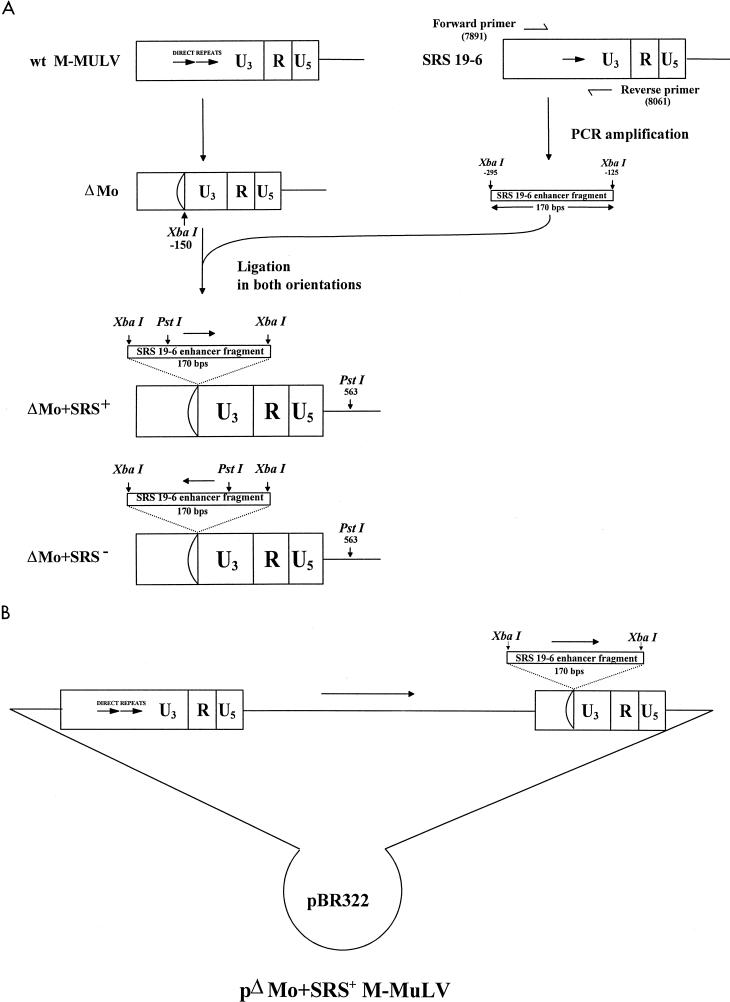
FIG. 1.
Generation of the ΔMo+SRS LTRs. (A) The DNA sequences deleted from positions −150 to −357 in the M-MuLV LTR to give the ΔMo LTR are shown (24). The 170-bp DNA fragment containing the SRS enhancer sequences (positions −298 to −125) was PCR amplified and cloned into the ΔMo LTR in either orientation to give the ΔMo+SRS+ and ΔMo+SRS− LTRs. The PstI restriction endonuclease sites used to discriminate enhancer orientation are shown. The downstream PstI site is in the 5′ M-MuLV sequences. wt, wild type. (B) The organization of the pΔMo+SRS+ M-MuLV plasmid is shown, with the chimeric LTR only in the downstream position. The internal viral sequences are all derived from M-MuLV.
Pathogenicity of ΔMo+SRS M-MuLV in NIH Swiss mice.
To investigate the leukemogenic potential of ΔMo+SRS+ and ΔMo+SRS− M-MuLVs, 2-day-old neonatal mice were inoculated with the viruses and monitored for appearance of disease. Moribund animals were sacrificed, and samples from diseased organs and blood were analyzed as described previously (4). The time course of ΔMo+SRS M-MuLV-induced disease relative to that of wild-type M-MuLV and SRS 19-6 MuLV-induced disease is shown in Fig. 2. Infections with ΔMo+SRS+ and ΔMo+SRS− M-MuLVs had mean latencies of approximately 5 months, intermediate between those with M-MuLV and SRS19-6 MuLV. As would be expected of enhancer elements, the orientation of the SRS enhancers had little influence on the time of disease onset. Confirming their role as enhancers, the inserted putative SRS enhancer sequences functionally substituted for the M-MuLV enhancers by supporting the replication of the virus in tissue culture and the efficient induction of disease in mice.
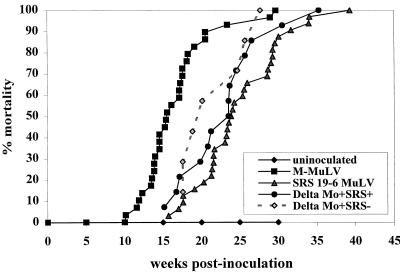
FIG. 2.
Pathogenicity of ΔMo+SRS M-MuLVs. Neonatal NIH Swiss mice were inoculated intraperitoneally with ΔMo+SRS+ (14 animals) and ΔMo+SRS− M-MuLVs (7 animals) as described in Materials and Methods. The time course to death is shown. For comparison, mortality plots for animals inoculated with wild-type M-MuLV and SRS 19-6 MuLV are shown.
While we initially predicted that the disease spectra induced by ΔMo+SRS+ and ΔMo+SRS− M-MuLVs might reflect the SRS 19-6 MuLV enhancer origin, the gross pathology of the resulting tumors more closely resembled that of tumors induced by M-MuLV than of tumors induced by SRS 19-6 MuLV. Characteristic of M-MuLV-induced T-lymphoid leukemia, all ΔMo+SRS M-MuLV-infected moribund mice had enlarged spleens, thymuses and lymph nodes but showed no signs of anemia. In contrast, the majority (∼80%) of SRS 19-6 MuLV-infected mice showed anemia and regressed thymuses and 15% of the mice showed hindlimb paralysis. Indeed, anemia was diagnostic for the presence of myeloid or erythroid leukemias (7).
Molecular characterization of ΔMo+SRS M-MuLV tumors.
To further evaluate the disease specificity of the ΔMo+SRS M-MuLVs, splenic and thymic tumor DNAs were extracted from moribund mice and characterized by Southern blot hybridization for rearrangements of TCR-β and Ig genes. Consistent with the gross pathology, the molecular analysis indicated that ΔMo+SRS M-MuLV-induced tumors were similar to wild-type M-MuLV-induced T-cell lymphomas. All of the tumors showed rearrangements at the TCR-β locus (diagnostic for T-cell lymphomas) (Table 1). As reported previously, some T-cell lymphomas showed TCR-β and IgH but not Igκ rearrangements (4). None of the tumors showed proviral insertions near the evi-1 proto-oncogene, which is characteristic of many SRS 19-6 MuLV-induced myeloid tumors (7). Thus, the combination of the histopathological and molecular analyses indicated that the tumors induced by ΔMo+SRS M-MuLV were T-cell lymphomas.
TABLE 1.
Molecular characterization of tumors induced by ΔMo+SRS MuLVs
| Virus | Animal | Genomic rearrangementsa
|
Insertion near cellular proto-oncogenesa
|
|||||
|---|---|---|---|---|---|---|---|---|
| TCR-β | IgH | Igκ | c-myc | Pim-1 | pvt-1 | evi-1 | ||
| ΔMo+SRS+M-MuLV | 498-1 | R | G | G | − | − | − | − |
| 498-2 | R | G | G | − | − | − | − | |
| 498-3 | R | G | G | + | − | − | − | |
| 498-4 | R | R | G | − | − | − | − | |
| 498-5 | R | R | G | − | − | − | − | |
| 498-6 | R | R | G | − | − | − | ND | |
| 498-7 | R | R | G | + | − | − | ND | |
| 527-12 | R | R | G | − | − | − | ND | |
| 527-13 | R | R | G | − | − | − | ND | |
| Total | 9/9 | 6/9 | 0/9 | 2/9 | 0/9 | 0/9 | 0/5 | |
| ΔMo+SRS−M-MuLV | 501-1 | R | R | G | − | − | − | − |
| 501-2 | R | R | G | − | − | − | − | |
| 501-3 | R | R | G | + | + | − | − | |
| Total | 3/3 | 3/3 | 0/3 | 1/3 | 1/3 | 0/3 | 0/3 | |
G, germ line; R, rearranged; ND, not done. Ratios indicate number of rearrangements per animal for each viral inoculum. Each animal had an enlarged thymus, spleen and lymph nodes.
Since insertional activation of cellular proto-oncogenes is an important step in M-MuLV leukemogenesis, it was of interest to screen ΔMo+SRS M-MuLV-induced tumors for insertions near proto-oncogenes known to be activated by wild-type M-MuLV (c-myc, Pim-1, and Pvt-1). ΔMo+SRS M-MuLV-induced tumors were tested for such insertions by Southern blot analysis as described previously (18), and the results are summarized in Table 1. Proviral integrations adjacent to Pim-1, c-myc, and Pvt-1 in ΔMo+SRS M-MuLV-induced tumors were observed with frequencies somewhat lower than for wild-type-M-MuLV-induced tumors (18).
ΔMo+SRS M-MuLV proviruses in tumor DNAs display rearrangements in the enhancer.
The LTRs in the ΔMo+SRS M-MuLV-induced tumors were examined to determine if they were altered during the leukemogenic process. Tumor DNAs were digested with PstI and subjected to gel electrophoresis and Southern blot hybridization with an SRS enhancer-specific probe. In all tumors analyzed, the SRS DNA sequences were detected and PstI fragments of the expected size for each enhancer orientation were observed (Fig. 3). However, we and others have detected enhancer alterations in end-stage tumors induced by various MuLV and feline leukemia virus (FeLV) recombinants (3, 5, 14, 33, 35, 44, 49). Thus, a more detailed PCR analysis with SRS-specific oligonucleotide primers was used to search for modifications in the SRS sequences (Fig. 4). Relative to the enhancers in the input ΔMo+SRS M-MuLV LTR, enhancer alterations were observed in splenic and thymic tumors from all mice infected with ΔMo+SRS+ or ΔMo+SRS− M-MuLV. The alterations were evident from ethidium bromide staining of gels of the PCR products; Southern blot hybridization of the gels indicated that most of the tumors contained multiple rearrangements, largely due to the presence of multiple copies of tandemly repeated sequences (see below). It should be noted that the multiple rearrangements evident in the PCR amplifications were consistent with the single PstI fragments present in the Southern blot hybridizations of Fig. 3. This was because the tandemly repeated sequences each contained the PstI site contained in the SRS 19-6 MuLV enhancer (see below).
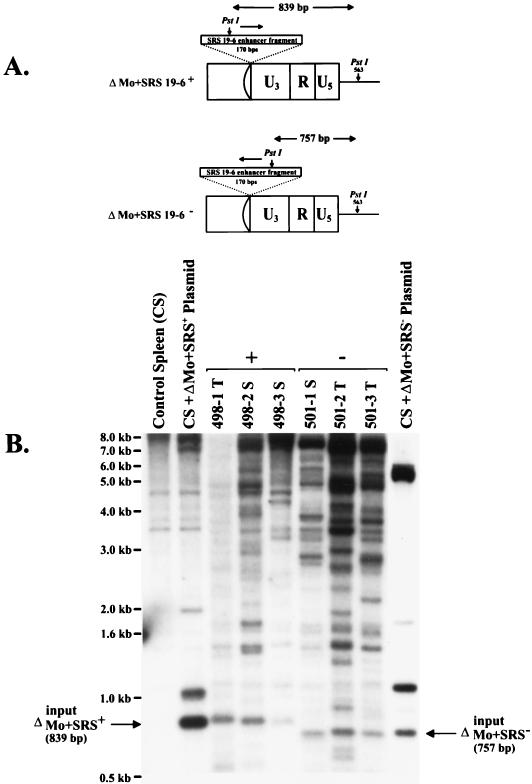
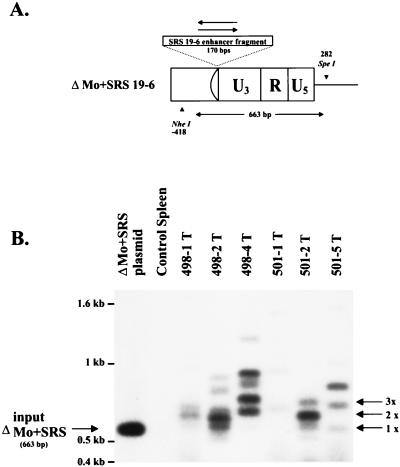
FIG. 3.
Southern blot analysis of ΔMo+SRS M-MuLV-induced tumors. High-molecular-weight DNAs from ΔMo+SRS M-MuLV-induced tumors were digested with PstI, which cleaves asymmetrically within the SRS enhancer fragment. (A) Hybridization with a labeled SRS enhancer-specific probe would yield a diagnostic 839-bp hybridizing fragment for ΔMo+SRS+ M-MuLV and a 757-bp fragment for ΔMo+SRS− M-MuLV. (B) Southern blots of the tumors all show fragments of the expected size when hybridized with an SRS 19-6 MuLV enhancer probe. Larger hybridizing fragments presumably represent endogenous MuLV sequences (also present in control spleen DNA) or host-virus junction fragments from the downstream LTR.
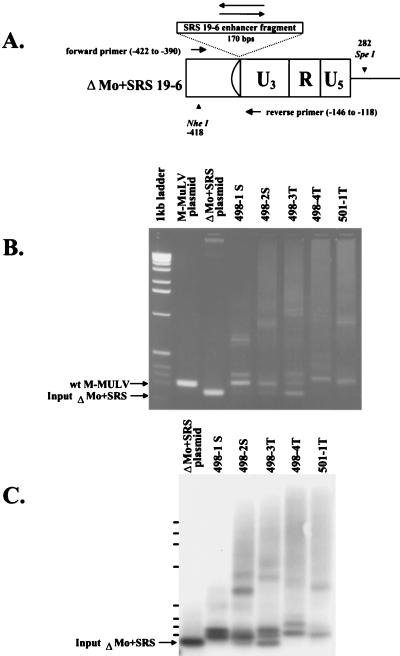
FIG. 4.
PCR analysis of proviral enhancers in ΔMo+SRS M-MuLV-induced tumors. (A) A diagram of the ΔMo+SRS+ or ΔMo+SRS− M-MuLV LTRs is shown, along with the locations of the oligonucleotide primers used to amplify the proviral LTRs from tumor DNAs. (B) PCR products from several different ΔMo+SRS M-MuLV-induced tumors were analyzed by agarose gel electrophoresis (2% agarose) and stained with ethidium bromide. Plasmids containing either wild-type (wt) M-MuLV or input ΔMo+SRS M-MuLV DNA provided size marker controls for the PCR amplification products, as shown to the left of the gel. Although some tumor DNAs yielded PCR fragments of the expected size, all tumors gave one or more enhancer-specific fragment of increased size. (C) Southern blot hybridization with an SRS enhancer-specific probe of a gel similar to the one in panel B is shown.
To confirm that the LTR alterations detected by the PCR analysis shown in Fig. 4B and C were present in the ΔMo+SRS M-MuLV-induced tumor DNAs and that they were not artifacts generated by PCR amplification, tumor DNAs were digested with the restriction enzymes NheI and SpeI (diagrammed in Fig. 5A) and Southern blot hybridization was performed with an SRS enhancer-specific probe (Fig. 5B). Although some tumors yielded hybridizing fragments characteristic of the input ΔMo+SRS M-MuLV, all tumors contained additional fragments of lower mobility. The patterns of the bands that resulted from this analysis were consistent with the patterns observed in Fig. 4B and C, confirming that the inserted SRS enhancers had indeed been altered during the process of tumorigenesis.
FIG. 5.
Southern blot analysis of ΔMo+SRS M-MuLV LTRs in tumor DNAs. Tumor DNAs were digested with NheI and SpeI and analyzed by Southern blot hybridization with an SRS enhancer-specific probe. NheI recognizes a site in the U3 region of M-MuLV 5′ of the inserted SRS sequences, and SpeI recognizes a site in the 5′ noncoding region 282 bp downstream from the U3-R junction (Fig. 4A). A diagnostic fragment of 680 bp was indicative of input ΔMo+SRS M-MuLV proviral DNA. The sizes of SRS-hybridizing fragments detected in this analysis corresponded to the fragment sizes detected by PCR amplification (Fig. 4). Fragments corresponding to duplications and triplications of the SRS enhancer sequences are indicated (2× and 3×, respectively). Hybridizing fragments migrating more slowly than the enhancer triplications might have represented virus-host junction fragments originating from the downstream LTRs.
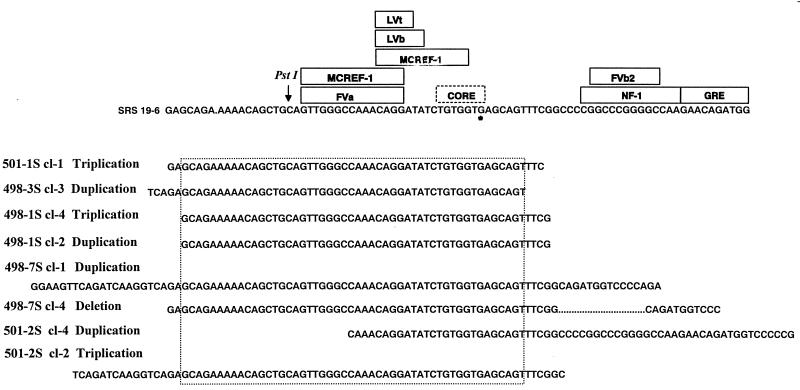
To further analyze these enhancer changes, the prominent DNA fragments resulting from the PCR amplifications in Fig. 4 were cloned and sequenced. In some cases, multiple clones with different-sized amplification products from the same tumor were sequenced. The resulting DNA sequences from each clone were aligned with the input SRS enhancers and are shown in Fig. 6. All tumors showed tandem repetitions of a portion of the SRS 19-6 enhancer sequences; in some tumors (e.g., 501-2S and 498-7S), more than one rearrangement was detected. It was noteworthy that each of the tumors contained ΔMo+SRS M-MuLV proviruses with tandem repetition of a region of the SRS 19-6 MuLV enhancer encompassing the LVb and core motifs and surrounding sites. The common region of repetition is shown in the figure. These results suggested that tandem repetition of these sequences was important for induction of T-cell lymphomas by ΔMo+SRS M-MuLVs.
FIG. 6.
Altered SRS enhancer regions amplified from ΔMo+SRS M-MuLV-induced tumors. PCR products of tumors similar to those shown in Fig. 4 were cloned and sequenced. The sequences were aligned with the input SRS enhancer sequences shown at the top of the figure. Binding sites for sequence-specific DNA binding proteins are included (6). The asterisk indicates a G residue that is not present in the standard core motif; it is an A residue in the M-MuLV enhancers. The sequences shown for the different PCR clones indicate sequences that were tandemly duplicated or triplicated. For 498-7S clone 4, the alteration consisted of a deletion (… …) in the NF-1 sequences. Two clones with different sequences were obtained for tumors 498-1S, 498-7S, and 501-2S. Otherwise, the sequences shown represent the predominant PCR products from the respective tumors. Tumors 498-1S, 498-3S, and 498-7S were induced by M-MuLV+ M-MuLV, and tumors 501-1S and 501-2S were induced by ΔMo+SRS+ M-MuLV. A specific region of the SRS enhancers was amplified in at least one of the proviruses in each tumor. The minimum size of the region of common amplification is indicated by the dotted box. This portion of the enhancers contains the highly conserved NF-1, LVb motifs, and core motif that differs from M-MuLV at the nucleotide indicated in the figure.
Since all ΔMo+SRS M-MuLV-induced tumors contained rearrangements of the inserted SRS enhancers, it was of interest whether these changes occurred at preleukemic times or were associated solely with tumor tissue. The presence of alterations at early times would suggest that these changes might have conferred an in vivo replicative advantage to the virus; on the other hand, the appearance of changes at later times would suggest that the multimerizations were important for late events in leukemogenesis, such as the insertional activation of cellular proto-oncogenes. Indeed, Lenz et al. detected proviruses with duplications of the enhancer region of SL3-3 MuLV inserted into an intron of the c-myc proto-oncogene, which suggested that this alteration is important for insertional activation of c-myc during tumorigenesis (29). We isolated DNA from the spleens, bone marrow, and thymuses of preleukemic ΔMo+SRS+ M-MuLV-inoculated mice at 2, 4, 6, and 8 weeks postinoculation. In addition, two mice from the same litter were allowed to become moribund and DNA was extracted from the tumors. All DNA samples were analyzed for the presence of enhancer alterations by PCR as above. In all cases, the preleukemic animals had no detectable SRS enhancer alterations whereas typical alterations were detected in the tumor DNAs (not shown). Thus, multimerization of the SRS enhancer elements was associated with late events in leukemogenesis by ΔMo+SRS M-MuLV.
SRS 19-6 MuLV-induced T-cell lymphomas also have rearrangements in the enhancers.
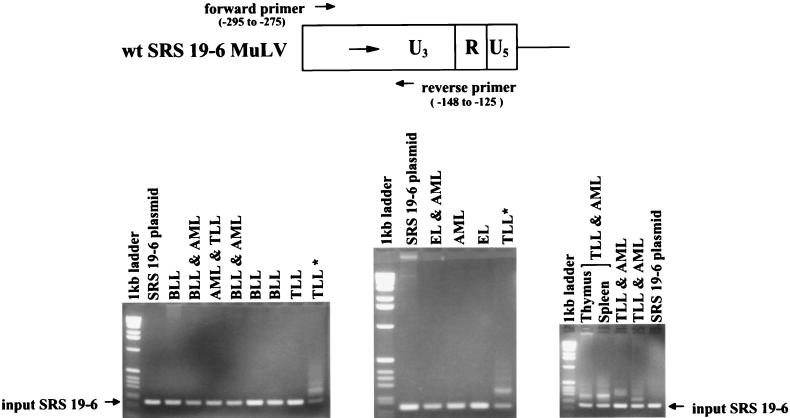
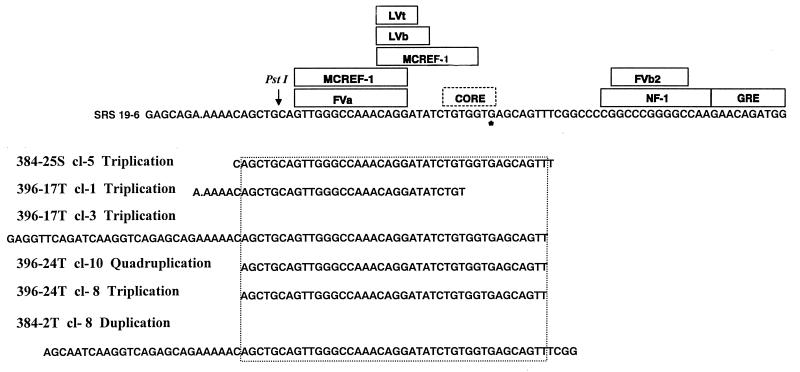
Since the parental SRS 19-6 MuLV also induced T-cell lymphomas in approximately 14% of inoculated mice (7), we investigated whether the lymphomas contained SRS 19-6 MuLV enhancer rearrangements analogous to those detected in ΔMo+SRS M-MuLV-induced tumors. A set of SRS-specific PCR primers that flank the enhancers was used to study enhancer sequences in SRS 19-6 MuLV-induced tumors of B-cell (BLL), myeloid (AML), erythroid (EL), and T-lymphoid (TLL) origin, as shown in Fig. 7. A PCR product corresponding to the size of input SRS 19-6 MuLV was detected in all tumors tested; its identity as an authentic SRS 19-6 MuLV product was verified by Southern blot hybridization with an SRS-specific enhancer probe. However, it was noteworthy that larger PCR products were detected in four of six T-lymphoid tumor DNAs (Fig. 7). In contrast, none of the tumor samples that did not contain T-cell lymphomas showed evidence of enhancer rearrangements. Novel PCR products from each of the T-cell lymphoma-containing tumors were cloned, and their nucleotide sequences were determined. The sequences are displayed in Fig. 8. It was striking that the rearrangements consisted of tandem reiterations of essentially the same SRS enhancer region as was found in the ΔMo+SRS M-MuLV-induced tumors. In one case (396-17T), two rearrangements were detected in the same tumor sample; whereas one contained a triplication that did not include the core motif (clone 1), the other contained a triplication that did include it (clone 3). This provides further support for the importance of the reiteration of the LVb and core-containing regions of the SRS 19-6 enhancer for efficient T-cell lymphomagenesis. It was equally noteworthy that none of the B-lymphoid, myeloid, or erythroid leukemias showed evidence of equivalent LTR rearrangements. This suggests that the single enhancer of SRS 19-6 MuLV can apparently function effectively in the induction of BLL, AML, and EL without the need for rearrangement.
FIG. 7.
Proviral LTRs in tumors induced by parental SRS 19-6 MuLV. The same LTR analysis as in Fig. 5B was applied to tumor DNAs induced by wild-type (wt) SRS 19-6 MuLV. The location of the PCR primers is shown at the top of the figure. Abbreviations: BLL, B-cell lymphoblastic lymphoma; AML, acute myelogenous leukemia; TLL, T-cell lymphoblastic lymphoma; EL, erythroid leukemia; ∗, from the same animal. LTR alterations were detected in four of the six T-lymphoid tumors induced by wild-type SRS 19-6 MuLV, but in none of the tumors that did not involve TLL.
FIG. 8.
SRS enhancer regions amplified in wild-type SRS 19-6 MuLV-induced tumors. The novel sized enhancer-specific PCR products shown in Fig. 7 were cloned and sequenced and are displayed here. The analogous region to that in ΔMo+SRS M-MuLV-induced tumors was amplified in T-lymphoid tumors induced by wild-type SRS 19-6 MuLV.
DISCUSSION
In this study, we investigated the disease specificity of chimeric M-MuLVs driven by the enhancer sequences from SRS 19-6 MuLV. We initially hypothesized that these chimeras would show the broad disease spectrum of SRS 19-6 MuLV, since the enhancer sequences have typically been shown to determine the disease specificity of many nonacute retroviruses and since SRS 19-6 MuLV induces leukemias of multiple hematopoietic lineages. However, contrary to our expectations, the ΔMo+SRS M-MuLVs induced T-cell lymphomas exclusively. Thus, insertion of the SRS 19-6 MuLV enhancers into the M-MuLV LTR was not sufficient to broaden the disease specificity of the chimeric viruses. These results indicate that either M-MuLV has additional determinants of T-cell lymphomagenicity in addition to its enhancers or SRS 19-6 MuLV has additional determinants for the broad disease spectrum in addition to its enhancers or both. Other investigators have shown that replacement of the M-MuLV enhancers with F-MuLV enhancers is sufficient to convert the disease specificity to erythroleukemia (10, 11, 26, 28, 30, 47). Therefore, if other T-cell lymphomagenic determinants exist outside the LTR of M-MuLV, they must be relatively weak. This in turn suggests that SRS 19-6 MuLV has additional determinants involved in its broad disease spectrum outside the LTR enhancer region substituted into the ΔMo+SRS M-MuLV LTRs. Additional determinants could lie within the LTR but outside the main enhancer region or in other regions of the SRS 19-6 MuLV genome. One possibility is that the SRS 19-6 MuLV envelope protein, which is quite distinct from other ecotropic MuLV envelopes (6), allows preferential infection of multipotential hematopoietic cells.
It was very interesting that at least one of the ΔMo+SRS M-MuLV proviruses in each of the T-cell lymphomas tested showed alterations in the SRS enhancer sequences. These alterations consisted of duplications (or higher multimers) of a portion but not all of the enhancer. This suggests that the multimerized portion might contain elements that favor expression in T-lymphoid cells while the nonmultimerized portion might contain elements that are not active (or are even inhibitory) in these cells. Multimerization of the T-lymphoid-specific portion might improve the ability of the enhancer to function in T-lymphoid cells. This idea was supported by examination of the LTRs in tumors induced by the parental SRS 19-6 MuLV. The only tumors that showed LTR alterations were T-lymphoid tumors (or those that had a T-lymphoid component); B-lymphoid, myeloid, and erythroid tumors induced by the same virus contained proviruses with unrearranged LTRs. Thus, the native SRS 19-6 MuLV enhancer would appear to be optimal for expression in B-lymphoid, myeloid, and erythroid cells but suboptimal for expression in T-lymphoid cells.
Sequence alignment of the alterations observed in both SRS 19-6 MuLV- and ΔMo+SRS M-MuLV-induced T-cell lymphomas enabled the delineation of a minimal T-lymphoid-specific region of the enhancer. In every tumor examined, at least one LTR contained amplified enhancer sequences that included the LVb-core binding sites. This LVb-core region has been strongly implicated in the T-cell-specific expression of multiple cellular genes (8, 27, 36, 37, 48) as well as in the T-lymphoid specificity of M-MuLV and SL3-3 MuLV (20, 32, 42). Sequences flanking the LVb-core sites were also amplified, as diagrammed in Fig. 6 and 8, but they generally did not include the downstream NF-1 and GRE sites. The lack of NF-1 within this amplified region suggests that NF-1 binding does not confer increased transcriptional activity in T-lymphoid cells. We recently carried out in vivo dimethyl sulfate footprinting on the upstream LTR of M-MuLV proviruses in infected cells and found that the NF-1 sites are not occupied in infected T-lymphoid cells or primary thymic tumor cells (22). In addition, FeLV-induced T-lymphoid tumors lack NF-1 binding activity due to posttranscriptional modification of the protein (35). One of the ΔMo+SRS M-MuLV-induced tumors examined here (498-7S) had a rearranged LTR with a deletion of the NF-1 site and one with a duplication of the LVb-core region.
The amplification of the LVb-core region has been documented in proviruses of T-lymphoid tumors induced by several other MuLVs or FeLVs (3, 5, 14, 33, 35, 44, 49). In a prior study by Chen and Yoshimura, proviral insertions adjacent to c-myc that had duplications in the MCF 13 enhancers invariably included the core and LVb binding sites (12). Likewise, when T-lymphoid tumor cell lines derived from FeLV-infected cats were analyzed for enhancer alterations, a portion of the FeLV enhancer that always included the core motifs was duplicated (35). Viral recombinants with multimers of the core sequence appear to have a selective advantage due to an increase in proviral transcriptional efficiency. However, sequences flanking the core element are required for efficient proviral transcriptional activation in T-lymphoid cells, since concatemers of the core sequence alone fail to increase viral transcription in transient-transfection assays (50). Intact binding sites for both LVb and core were found to be necessary for transcriptional activation of promoters of TCR-β as well as M-MuLV (45), and the core sequences and flanking binding sites for ets and myb are necessary for efficient SL3-3 transcription in T cells (50).
The experiments presented here suggest that the altered proviral LTRs in the T-lymphoid tumors induced by parental SRS 19-6 MuLV have higher transcriptional activity than the original SRS 19-6 MuLV enhancer in T-lymphoid cells. It would be interesting to test this directly by generating an SRS 19-6 MuLV containing the altered LTR. It seems likely that such a virus would induce a higher incidence of T lymphomas than the parental virus. Indeed, when T-cell lymphoma arose in a mouse infected with mouse mammary tumor virus, proviral enhancer alterations were observed and a recombinant molecular clone of mouse mammary tumor virus harboring these altered enhancer sequences efficiently induced T-lymphoid disease when introduced into mice (2, 49). Likewise, Ethelberg et al. have shown that an SL3-3 MuLV enhancer variant that arose during the process of pathogenesis exhibited greater potency when molecularly cloned and reintroduced into mice (15).
The experiments here describe a direct test of the hypothesis that the enhancers of SRS 19-6 MuLV are sufficient to confer the broad spectrum of leukemias induced by this virus. Since the ΔMo+SRS M-MuLVs induced only T-cell lymphomas, it will be very interesting to test additional chimeras between SRS 19-6 MuLV and M-MuLV to identify the disease determinants. These chimeras are under construction.
ACKNOWLEDGMENTS
This work was supported by grant CA32455 from the National Cancer Institute. S.W.G. was supported by grant 5 T32 CA09054 from the National Cancer Institute. The support of the UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center is gratefully acknowledged.
We thank Jeff Lander for providing data for the wild-type M-MuLV mortality plot.
REFERENCES
- 1.Bacheler L T, Fan H. Integrated Moloney murine leukemia virus DNA studied by using complementary DNA which does not recognize endogenous related sequences. J Virol. 1980;33:1074–1082. doi: 10.1128/jvi.33.3.1074-1082.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J K, Diggelmann H, Dekaban G A, Grossi G F, Semmler R, Waight P A, Fletcher R F. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J Virol. 1988;62:2985–2993. doi: 10.1128/jvi.62.8.2985-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belli B, Patel A, Fan H. Recombinant mink cell focus-inducing virus and long terminal repeat alterations accompany the increased leukemogenicity of the Mo+PyF101 variant of Moloney murine leukemia virus after intraperitoneal inoculation. J Virol. 1995;69:1037–1043. doi: 10.1128/jvi.69.2.1037-1043.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brightman B K, Chandy K G, Spencer R H, Gupta S, Pattengale P K, Fan H. Characterization of lymphoid tumors induced by a recombinant murine retrovirus carrying the avian v-myc oncogene. Identification of novel (B-lymphoid) tumors in the thymus. J Immunol. 1988;141:2844–2854. [PubMed] [Google Scholar]
- 5.Brightman B K, Farmer C, Fan H. Escape from in vivo restriction of Moloney mink cell focus-inducing viruses driven by the Mo+PyF101 long terminal repeat (LTR) by LTR alterations. J Virol. 1993;67:7140–7148. doi: 10.1128/jvi.67.12.7140-7148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bundy L, Fan H. Molecular and phylogenetic analysis of SRS 19-6 murine leukemia virus. Virus Genes. 1999;18:65–79. doi: 10.1023/a:1008073419746. [DOI] [PubMed] [Google Scholar]
- 7.Bundy L M, Ru M, Zheng B F, Cheng L, Pattengale P K, Portis J L, Fan H. Biological characterization and molecular cloning of murine C-type retroviruses derived from the TSZ complex from mainland China. Virology. 1995;212:367–382. doi: 10.1006/viro.1995.1494. [DOI] [PubMed] [Google Scholar]
- 8.Cameron S, Taylor D S, TePas E C, Speck N A, Mathey-Prevot B. Identification of a critical regulatory site in the human interleukin-3 promoter by in vivo footprinting. Blood. 1994;83:2851–2859. [PubMed] [Google Scholar]
- 9.Celander D, Haseltine W A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984;312:159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- 10.Chatis P A, Holland C A, Hartley J W, Rowe W P, Hopkins N. Role for the 3′ end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci USA. 1983;80:4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatis P A, Holland C A, Silver J E, Frederickson T N, Hopkins N, Hartley J W. A 3′ end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984;52:248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Yoshimura F K. Identification of a region of a murine leukemia virus long terminal repeat with novel transcriptional regulatory activities. J Virol. 1994;68:3308–3316. doi: 10.1128/jvi.68.5.3308-3316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DesGroseillers L, Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984;52:448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ethelberg S, Hallberg B, Lovmand J, Schmidt J, Luz A, Grudstrom T, Pedersen F S. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997;71:1196–1206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ethelberg S, Sorensen A B, Schmidt J, Luz A, Pedersen F S. An SL3-3 murine leukemia virus enhancer variant more pathogenic than the wild type obtained by assisted molecular evolution in vivo. J Virol. 1997;71:9796–9799. doi: 10.1128/jvi.71.12.9796-9799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan H. Influences of the long terminal repeats on retrovirus pathogenicity. Semin Virol. 1990;1:165–174. [Google Scholar]
- 17.Fan H. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- 18.Fan H, Chute H, Chao E, Pattengale P K. Leukemogenicity of Moloney murine leukemia viruses carrying polyoma enhancer sequences in the long terminal repeat is dependent on the nature of the inserted polyoma sequences. Virology. 1988;166:58–65. doi: 10.1016/0042-6822(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 19.Fan H, Mittal S, Chute H, Chao E, Pattengale P K. Rearrangements and insertions in the Moloney murine leukemia virus long terminal repeat alter biological properties in vivo and in vitro. J Virol. 1986;60:204–214. doi: 10.1128/jvi.60.1.204-214.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golemis E, Li Y, Fredrickson T N, Hartley J W, Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989;63:328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golemis E A, Speck N A, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granger S W, Fan H. In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types. J Virol. 1998;72:8961–8970. doi: 10.1128/jvi.72.11.8961-8970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanecak R, Mittal S, Davis B R, Fan H. Generation of infectious Moloney murine leukemia viruses with deletions in the U3 portion of the long terminal repeat. Mol Cell Biol. 1986;6:4634–4640. doi: 10.1128/mcb.6.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanecak R, Pattengale P K, Fan H. Addition of substitution of simian virus 40 enhancer sequences into the Moloney murine leukemia virus (M-MuLV) long terminal repeat yields infectious M-MuLV with altered biological properties. J Virol. 1988;62:2427–2436. doi: 10.1128/jvi.62.7.2427-2436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward W S, Neel B G, Astrin S M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins N. Genetic basis of disease specificity of nondefective Friend murine leukemia virus. Ann N Y Acad Sci. 1989;567:14–25. doi: 10.1111/j.1749-6632.1989.tb16455.x. [DOI] [PubMed] [Google Scholar]
- 27.Hsiang Y H, Spencer D, Wang S, Speck N A, Raulet D H. The role of viral enhancer “core” motif-related sequences in regulating T cell receptor-gamma and -delta gene expression. J Immunol. 1993;150:3905–3916. [PubMed] [Google Scholar]
- 28.Ishimoto A, Takimoto M, Adachi A, Kakuyama M, Kato S, Kakimi K, Fukuoka K, Ogiu T, Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987;61:1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenz J, Celander D, Crowther R L, Patarca R, Perkins D W, Haseltine W A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Golemis E, Hartley J W, Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987;61:693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linney E, Davis B, Overhauser J, Chao E, Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature. 1984;308:470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- 32.LoSardo J E, Boral A L, Lenz J. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J Virol. 1990;64:1756–1763. doi: 10.1128/jvi.64.4.1756-1763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overhauser J, Fan H. Generation of glucocorticoid-responsive Moloney murine leukemia virus by insertion of regulatory sequences from murine mammary tumor virus into the long terminal repeat. J Virol. 1985;54:133–144. doi: 10.1128/jvi.54.1.133-144.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plumb M, Fulton R, Breimer L, Stewart M, Willison K, Neil J C. Nuclear factor 1 activates the feline leukemia virus long terminal repeat but is posttranscriptionally down-regulated in leukemia cell lines. J Virol. 1991;65:1991–1999. doi: 10.1128/jvi.65.4.1991-1999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser H M, Wotton D, Gegonne A, Ghysdael J, Wang S, Speck N A, Owen M J. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc Natl Acad Sci USA. 1992;89:9934–9938. doi: 10.1073/pnas.89.20.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redondo J M, Pfohl J L, Krangel M S. Identification of an essential site for transcriptional activation within the human T-cell receptor delta enhancer. Mol Cell Biol. 1991;11:5671–5680. doi: 10.1128/mcb.11.11.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Short M K, Okenquist S A, Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987;61:1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 42.Speck N A, Renjifo B, Golemis E, Fredrickson T N, Hartley J W, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 43.Speck N A, Renjifo B, Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starkey C R, Lobelle-Rich P A, Granger S, Brightman B K, Fan H, Levy L S. Tumorigenic potential of a recombinant retrovirus containing sequences from Moloney murine leukemia virus and feline leukemia virus. J Virol. 1998;72:1078–1084. doi: 10.1128/jvi.72.2.1078-1084.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiesen H J, Bosze Z, Henry L, Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988;62:614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt M, Haggblom C, Swift S, Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985;55:184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S W, Speck N A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992;12:89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanagawa S, Kakimi K, Tanaka H, Murakami A, Nakagawa Y, Kubo Y, Yamada Y, Hiai H, Kuribayashi K, Masuda T, et al. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J Virol. 1993;67:112–118. doi: 10.1128/jvi.67.1.112-118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaiman A L, Nieves A, Lenz J. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J Virol. 1998;72:3129–3137. doi: 10.1128/jvi.72.4.3129-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]