Abstract
Micrococcus luteus, also known as M. luteus, is a bacterium that inhabits mucous membranes, human skin, and various environmental sources. It is commonly linked to infections, especially among individuals who have compromised immune systems. M. luteus is capable of synthesizing the enzyme superoxide dismutase (SOD) as a component of its protective response to reactive oxygen species (ROS). This enzyme serves as a promising target for drug development in various diseases. The current study utilized a subtractive genomics approach to identify potential therapeutic targets from M. luteus. Additionally, genome mining was employed to identify and characterize the biosynthetic gene clusters (BGCs) responsible for the production of secondary metabolites in Bacillus licheniformis (B. licheniformis), a bacterium known for its production of therapeutically relevant secondary metabolites. Subtractive genomics resulted in identification of important extracellular protein SOD as a drug target that plays a crucial role in shielding cells from damage caused by ROS. Genome mining resulted in identification of five potential ligands (secondary metabolites) from B. licheniformis such as, Bacillibactin (BAC), Paenibactin (PAE), Fengycin (FEN), Surfactin (SUR) and Lichenysin (LIC). Molecular docking was used to predict and analyze the binding interactions between these five ligands and target protein SOD. The resulting protein–ligand complexes were further analyzed for their motions and interactions of atoms and molecules over 250 ns using molecular dynamics (MD) simulation analysis. The analysis of MD simulations suggests, Bacillibactin as the probable candidate to arrest the activities of SOD. All the five compounds reported in this study were found to act by directly/indirectly interacting with ROS molecules, such as superoxide radicals (O2–) and hydrogen peroxide (H2O2), and transforming them into less reactive species. This antioxidant activity contributes to its protective effects against oxidative stress-induced damage in cells making them likely candidate for various applications, including in the development of antioxidant-based therapies, nutraceuticals, and functional foods.
Keywords: Micrococcus luteus, Bacillus licheniformis, Genome mining, Molecular dynamics, Simulation
1. Introduction
Micrococci are a group of Gram-positive bacteria that belong to the genus Micrococcus (Becker et al., 2015). They are spherical or slightly elongated in shape and typically occur in clusters or pairs. Micrococci are known for their ability to adapt to various environmental conditions and can be found in diverse habitats such as soil, water, air, and human skin (Aly, 1991, Kooken et al., 2012). Micrococci have diverse metabolic capabilities and can consume a broad range of carbon sources (Sumarsih et al., 2018). They are also identified for their prospective capability to produce various enzymes and metabolites of industrial and biotechnological interest (Kirk et al., 2002, Prakash et al., 2017, Gupta et al., 2019). Micrococcus luteus (M. luteus) is a species of Gram-positive bacterium belonging to the genus Micrococcus. It is commonly found to be originating in various environments, such as soil, dust, water, and human skin. M. luteus is usually considered a non-pathogenic bacterium and is part of the normal microbial flora on human skin. It is not typically associated with causing diseases in healthy individuals (Seifert et al., 1995, Zhu et al., 2021). However, under certain circumstances, M. luteus can become an opportunistic pathogen and cause infections with indwelling medical devices (Zhu et al., 2021). It has been identified as a causative agent of hospital-acquired infections, bloodstream infections, surgical site infections and the infections associated with indwelling catheters or prosthetic devices. M. luteus infections have been reported in patients with conditions such as endocarditis, osteomyelitis, peritonitis, and bacteremia (Peces et al., 1997, Ianniello et al., 2019). It is reported that M. luteus infect individuals with weakened immune systems, such as those undergoing chemotherapy, organ transplant recipients, or patients with HIV/AIDS (Dürst et al., 1991, Zhu et al., 2021). The treatment of M. luteus infections typically involves the use of antibiotics (Szczerba 2003). However, it's important to note that M. luteus is generally resistant to numerous frequently used antibiotics, such as penicillin and erythromycin (Eady et al., 2000, Zhu et al., 2021). This intrinsic resistance is primarily attributed to the thick peptidoglycan cell wall of M. luteus which makes it less susceptible to the action of antibiotics that target cell wall synthesis or disrupt bacterial membranes (Jensen and Campbell, 1976, Vollmer et al., 2008, Zdorovenko et al., 2021). To the best of our knowledge, there are no specific drugs or therapies that have been developed specifically for M. luteus infections. However, the field of drug discovery is constantly evolving, and new treatments may become available in the future. In this study we used subtractive genomics approach which is a computational method used to identify unique genetic elements or features of an organism by comparing the genomes of closely related organisms. It involves the systematic comparison of genomic sequences to identify regions that are present in one organism but absent or significantly different in another.
Bacillus licheniformis (B. licheniformis) is a Gram-positive, spore-forming bacterium belonging to the genus Bacillus. It is generally found in soil, water and various natural environments (Parkinson 2019). B. licheniformis has gained substantial attention due to its versatile metabolic capabilities and potential applications in industry and biotechnology (Muras et al., 2021, Ramirez-Olea et al., 2022). It is recognized to produce an array of secondary metabolites including certain antibiotics with antimicrobial activity (Mondol et al., 2013, Muras et al., 2021). B. licheniformis itself is not used as a direct source of drugs in the context of traditional pharmaceuticals. However, certain products including antibiotics, enzymes and antimicrobial peptides derived from B. licheniformis have been explored for potential therapeutic applications (Muras et al., 2021, Romo-Barrera et al., 2021). In this study, we employed subtractive genomics and genome mining approach to explore and analyze the genomic data of M. luteus and B. licheniformis to identify target protein and potential drug candidates/ligands. The central role of genome mining consists in finding new biosynthetic gene clusters (BGCs) within the genomes of sequenced organisms (Albarano et al., 2020). The genomics era has also enabled genome mining tools to find the candidate biosynthetic gene clusters (Medema et al., 2015). AntiSMASH is one of the algorithm that have been developed to search for secondary metabolite gene clusters (Medema et al., 2011, Blin et al., 2019). Subtractive genomics approach is a well-known and extensively used Insilico approach where host cell sequences and proteomes of pathogens are subtracted to obtain pathogen specific protein sequences which are present only in pathogen but are absent in host cells. This approach involves removal of paralogous sequences, identification of essential genes in pathogen which are non-homologous to host proteome and druggability analysis of essential proteins (Ashraf et al., 2022). And Genome mining refers to the systematic exploration and analysis of genomic data to identify and characterize genes, gene clusters, and biosynthetic pathways involved in the production of natural products or biologically active compounds. It involves searching through various genomic databases and using bioinformatics machineries to extract valuable information regarding the potential drug candidates, enzymes, secondary metabolites, and other important biological molecules. Further, molecular docking method was carried out to predict the binding mode and affinity among ligands and a target protein. Then, the molecular dynamics (MD) simulation technique was explored to assess the behavior and interactions of atoms and molecules over time to get insights into the dynamic behavior of the selected protein–ligand complexes. The MD simulations of five complexes were carried out for 250 ns and the results were analyzed using GROMAS utility. The findings of the study reports as Superoxide dismutase (SOD), a group of enzymes that play a crucial role in the antioxidant defense system. SOD is an enzyme which plays a crucial role in shielding the cells from the damaging effects caused by reactive oxygen species (ROS). SOD works by catalyzing the conversion of superoxide radicals (O2–) into hydrogen peroxide (H2O2) and molecular oxygen (O2). This process helps to neutralize the harmful effects of superoxide radicals, which can damage DNA, proteins, and other cellular components. This can be beneficial in some contexts, such as cancer therapy, where the goal is to surge oxidative stress in cancer cells to induce apoptosis or cell death. SOD is the primary and the strongest detoxification enzyme in the cell (Yasui and Baba, 2006, Kaur et al., 2017). Several studies have investigated the SOD activity in micrococci, and it has been shown that different species of micrococci have varying levels of SOD activity. For example, M. luteus has been found to have high levels of SOD activity, while other species such as M. luteus and Micrococcus variants have lower levels. Understanding the role of SOD in micrococci is important for developing strategies to control the growth and persistence of these bacteria. Additionally, the use of SOD inhibitors may offer a potential therapeutic approach for treating bacterial infections caused by micrococci and other bacteria that produce SOD.
The molecular docking and MD simulations suggested the top five compounds Bacillibactin (BAC), Paenibactin (PAE), Fengycin (FEN), Surfactin (SUR) and Lichenysin (LIC) for further studies. Among them, Bacillibactin has shown very significant interactions. Bacillibactin functions as a chelator for iron ions. It forms a stable complex with iron, called the ferric-bacillibactin complex, through its specific structure and coordination chemistry (Lee et al., 2011). This complex is recognized by specific receptors present on the bacterial cell surface and is internalized into the bacterial cytoplasm through the transport systems (Dertz et al., 2006). Inside the bacterial cell, the ferric-bacillibactin complex is processed, and iron is released. The released iron is then utilized by the bacteria for various cellular processes, including the synthesis of essential enzymes and proteins involved in metabolism, respiration, and DNA replication. Inhibiting the biosynthesis or utilization of siderophores can potentially deprive bacteria of essential iron, making them more vulnerable to the host immune response or antimicrobial treatments. Hence, through this study we propose Bacillibactin as a candidate ligand to arrest the activities of SOD. The connection between SOD and Bacillibactin lies in their roles in bacterial defense mechanisms (Lee et al., 2011, Wang et al., 2011). SOD protects bacteria from oxidative stress by converting superoxide radicals into less harmful compounds. Oxidative stress can arise when bacteria encounter high levels of ROS, such as during host immune responses. Bacillibactin, on the other hand, helps bacteria scavenge iron, which is crucial for various cellular processes (Miethke and Marahiel, 2007, Page, 2019). Iron limitation can occur in the host environment as a defense strategy against bacterial pathogens.
2. Materials and methods
2.1. Retrieval of Micrococcus luteus and human proteome
The present study employed the subtractive genomics approach to discover the novel and potential drug targets against the M.luteus, which has been identified to cause plethora of infections including native valve endocarditis, septic arthritis, bacteremia, hepatic and brain abscess in immunosuppressive patients (Zhu et al., 2021). Micrococcus luteus and Human proteomes were acquired from the Uniprot database (https://www.uniprot.org/) as reported previously (Bagewadi et al., 2023). The complete set of the data was considered as set (0).
2.2. Selection of paralogous proteins
A web server CD-HIT suit (Huang et al., 2010) was used for clustering and comparing biological sequences. This tool suggests pre-calculated clusters for numerous public sequence databases that are updated on a regular basis. We subjected the Set (0) dataset to CD-HIT suite with a cut-off of 0.6 (60% sequence identity). Other parameters considered includes the removal of paralogous proteins with more than 60% sequence identity were removed (Fatoba et al., 2021, Ashraf et al., 2022). This set of non-paralogous proteins generated using CD-HIT was considered as Set (1).
2.3. Identification and retrieval of non-homologous sequences
The human proteins that are alike to pathogen proteins might interfere in binding of therapeutic compounds or ligands obtained, to the active site of pathogen proteins (Khan et al., 2020). Hence, the homologous sequences that are functionally comparable between the M. luteus and Human proteomes were screened and only non-homologous sequences were considered for further analysis. This dataset was subjected for Galaxy tool (https://usegalaxy.org/) (Afgan et al., 2018) for sequence similarity searching using the Blastp database (Camacho et al., 2009) against human proteome as reported previously with the threshold expectation value cut-off (e-value) 0.0001 (Khan et al., 2020, Bagewadi et al., 2023). The resulting data set of proteins was listed as non-homologous proteins (set2).
2.4. Prediction of sub-cellular location of proteins
The subcellular localization of proteins can provide valuable information related to its activity. The subcellular localization of proteins in a cell is critical for revealing the processes engaged at the cellular level. The subcellular localization of protein analysis was used to classify the proteins from the study into various category including inner membrane, extracellular space, cytoplasm, outer membrane, and periplasm. The subcellular localization approaches are used in identifying a drug target proteins. In this study, we used PSORTdb (https://db.psort.org/) (Lau et al., 2021) among the available subcellular localization prediction tool. PSORTdb consists two databases: ePSORTdb, database of experimentally validated protein subcellular localizations which is manually curated, and cPSORTdb, database of PSORTb-predicted subcellular localizations for the NCBI's RefSeq-derived archaeal and bacterial proteomes which is pre-computed. The search concentrated on extracellular proteins in order to improve the likelihood that it may be used as a drug target. The resulting protein sequences were retrieved in the.FASTA format and listed as Set (3).
2.5. Metabolic pathways analysis
Understanding the biological pathway of the pathogens involves information on the original kinetics governing the molecules and enzymes associated with the pathway. This understanding of specific enzymes that may be crucial targets in the control of M. luteus is essential. In this study, we used KEGG (Kyoto Encyclopedia of Genes and Genomes) via the KAAS server (https://www.genome.jp/kaas-bin/kaas_main) (Moriya et al., 2007). The server operates by rapidly assigning K values to genes in the genome, allowing reconstruction of KEGG pathways based on certain heuristics, sequence similarities and bi-directional best hit information. The data set from the previous analysis (set 3) in the FASTA format was subjected to KEGG analysis and the protein with assigned KEGG Orthology (KO) were considered (set4) for further studies as reported earlier (Bagewadi et al., 2023).
2.6. Genome mining of B. licheniformis
Genome mining refers to the systematic exploration and analysis of genomic data to identify and characterize genes, gene clusters, and biosynthetic pathways involved in the production of natural products or biologically active compounds. It involves searching through various genomic databases and using bioinformatics machineries to extract valuable information regarding the potential drug candidates, enzymes, secondary metabolites, and other important biological molecules. Genome mining was performed to explore SMs (Specialized metabolites) produced and BGCs (Biosynthetic Gene Clusters). BGC prediction and annotation tool antiSMASH was used to automate Biosynthetic Gene Clusters finding in genome sequencing (Kenshole et al., 2021). Uploading the GCF file of B. licheniformis, which can be extracted by downloading the complete genome assembly in NCBI database (https://www.ncbi.nlm.nih.gov/), secondary metabolites producing regions were generated. Most similar known clusters were screened and few ligands were selected for further studies. The vast majority of antibiotics presently in clinical use are derived from microorganisms' naturally occurring small compounds. Many microorganisms' metabolisms have been reported to contain bioactive compounds which are novel in nature, including that of B. licheniformis, which is found to be a rich source of such compounds which can have potential pharmaceutical applications as antibiotics and other class of drugs (Harwood et al., 2018). To screen the secondary metabolites of B. licheniformis, we employed the secondary metabolite analysis shell (antiSMASH) (Blin et al., 2021) tool to find the microbial genomes for secondary/specialized metabolite (SM) biosynthetic gene clusters (BGCs). This was achieved by first downloading the complete genome assembly of B. licheniformis from NCBI database (https://www.ncbi.nlm.nih.gov/) and then subjecting it to antiSMASH as reported previously (Bagewadi et al., 2023). Most similar known clusters were screened and few ligands were selected for further studies.
2.7. Molecular docking of novel inhibitors from B. licheniformis with superoxide dismutase protein
Retrieval and preparation of the ligands and target structure: The 3D structures of the five ligands namely, Bacillibactin, Lichenysin, Paenibactin, Fengycin, and Surfactin selected from Genome mining approach was considered for molecular docking against the superoxide dismutase protein selected from the subtractive genome approach. The 3D structures of these ligands were not readily available in PubChem. Hence, canonical SMILES from PUBCHEM database were used to generate 3D chemical structures of ligands using the CACTUS tool. Later the generated ligands were energy minimized with Avogadro to obtain the least energy conformational state and subsequently converted to pdbqt format file using open babel. Supplementary Fig. 1 shows the 2D structure of all five ligands considered for protein–ligand docking. Additionally, we employed an in-silico pharmacoinformatics approach to screen the identified possibly active candidate compounds from B. licheniformis utilizing ADME/drug-likeness features in order to validate potential compounds for further research (Lipinski et al., 2001). Similarly, the superoxide dismutase proteins’ 3 Dimensional structure was modeled using SWISS MODEL software (Waterhouse et al., 2018), which is automated protein structure homology-modelling server. The structure of SOD obtained from Swiss model is represented in Supplementary Fig. 2. The modelled structure was validated using PROCHECK software (Laskowski et al., 1993) which checks the stereo chemical quality of protein structures based on Ramachandran plot. Then the, active site of the target protein SOD was recorded using the Computed Atlas of Surface Topography of proteins (CASTp) server which give information about surface features and functional regions of protein (Binkowski et al., 2003). The catalytic/functional residues of SOD included 67ALA, 70LEU, 71GLY, 74THR, 75ASN, 142PHE, 145GLN, 146GLN, 149VAL, 150PRO, 151VAL, 152ALA, 153THR, 155PRO and 158GLN. Molecular docking of the ligands and the target was achieved using the Autodock Vina tool (Valdés-Tresanco et al., 2020). Grid box was optimized to include the entire binding site of protein target with a box parameter of 36.882 * 13.805 * 39.34 (x, y, and z coordinates) and 50 * 55 * 35 as grid size for ×, y, and z respectively. PyMOL (https://pymol.org/) was used for visualization of the docking interactions and LigPlot + for generating the interaction diagram.
2.8. MD simulations
The atomic mobility of a simulated system over time under the influence of a force field is predicted by MD simulations. This MD feature helps to study native and protein–ligand complexes to computationally infer the dynamics of a protein molecule by mimicking the physiological condition of a protein molecule. System inputs for MD were generated with Charmm-gui and GROMACS 2020.6 was employed to achieve SOD protein and protein–ligand complex MD simulations under the influence of the chosen CHARMM36m force field. In total, MD simulation of five ligands in complex with protein SOD were carried out for 250 ns time as reported previously (Gangadharappa et al., 2020a, Bagewadi et al., 2023). The complexes include (1) SOD-Bacillibactin (SOD-BAC), (2) SOD-Paenibactin (SOD-PAE), (3) SOD-Fengycin (SOD-FEN), (4) SOD-Surfactin (SOD-SUR) and (5) SOD-Lichenysin (SOD-LIC).
2.9. Analysis of simulated trajectories
Following the production MD run for 250 ns, the resultant trajectory files were analyzed using gromacs utilities such as ‘gmx rms’, ‘gmx rmsf’, ‘gmx gyrate’ and ‘gmx sasa’ for Root Mean Square Deviation (RMSD), Root Mean Square Fluctuations (RMSF), Solvent Accessible Surface Area (SASA) and Radius of Gyration (Rg) respectively (Gangadharappa et al., 2020a). The data was examined for a minimum steady trajectory of 60 ns in a window size of 15 ns. VMD (Humphrey et al., 1996) and PyMol (Schrodinger, 2010, Yuan et al., 2017) software were used to visualize the structural figures. Xmgrace (Turner 2005) tool was used to generate the graphs and plots. These data analysis was performed to check for the rigidity, compactness, fluctuations and stability of ligand–protein complex (Yaraguppi et al, 2021). To study the binding free energy (G binding) of the ligand with protein over simulation time, the Molecular Mechanics Poisson-Boltzmann surface area (MM-PBSA) (Miller et al., 2012) approach was used. The binding free energy was calculated using the GROMACS function g_mmpbsa (Kumari et al., 2014).
3. Results
This study employed Subtractive Genomics strategy combined with molecular docking and MD simulations approach to identify and characterize the drug target from the proteome of M. luteus, and screen the secondary metabolites of B. licheniformis as a therapeutic target. The results of the analysis are explained in the following subsections.
3.1. Paralogous sequence exclusion
To screen out M. luteus paralogous sequences, the CD-hit server was used. In total, 41,924 proteins out of 50,703 were revealed to be paralogous. When the cut off was set to 60% sequence identity on the CD-hit suite server, only 8779 proteins were identified to be non-paralogous. This step rejected 41,924 proteins which are paralogous in nature. As an outcome of this analysis, 8779 proteins were listed as Set1.
3.2. Human non-homologous protein selection
Proteins that function in similar cellular systems between humans and bacteria evolved as homologs over time. As a result, treatments designed to bind pathogen target proteins should not cross-react with host homologous proteins (Khan et al., 2020). BLASTp was used in web galaxy to select non-homologous proteins to the Human proteome. This analysis resulted in removal of 1,909 proteins with a cut off (threshold E-value) of 10-4. These pathogen specific proteins that are homologous to human proteins were further excluded from analysis. The remaining, 6,870 proteins found to be non-homologous proteins (have no hits) and were listed in as set2.
3.3. Sub-cellular localization analysis reveals 15 extracellular proteins
Subcellular localization of non-homologous proteins was carried out-using PSORTdb. The analysis was focused on listing the extracellular proteins because of their probability as drug targets. This is based on the reference to the published papers who used similar strategy to consider the membrane proteins (Uddin et al., 2018, Khan et al., 2020). It is evident that the extracellular or membrane bound proteins are mostly selected for drug targets or vaccine targets because they have the favorable regions to be considered as drug targets (Bakheet and Doig 2009). Extracellular proteins are an important class of drug targets due to their involvement in various disease processes and their accessibility from outside the cells. Targeting these proteins with drugs can modulate their activity, interfere with disease progression, and potentially lead to therapeutic benefits. Hence, in this study we focused on screening the extracellular proteins. Out of 2,215 proteins, according to the results, 55.6% proteins were localized in cytoplasm, 26.7% proteins were found in cytoplasmic membrane, 16.6% proteins location was unknown, 9 proteins in cell wall and 15 proteins in extracellular space. All the 15 extracellular proteins were considered for further analysis and protein sequences of these 15 were noted and listed as set3. The numbers of proteins located in different regions of cell are as shown in Table 1.
Table 1.
Proteins count in different locations of the cell using PSORTdb version 3.0.
| Sl. No. | Predicted localization | Count |
|---|---|---|
| 1 | Cell Wall | 9 |
| 2 | Cytoplasm | 1229 |
| 3 | Cytoplasmic Membrane | 590 |
| 4 | Extracellular | 15 |
| 5 | Unknown | 367 |
| Total | 2215 |
3.4. Identification of drug targets
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a comprehensive database that assimilates information on genes, proteins, pathways, and diseases. It provides a wealth of knowledge on the molecular interactions and biological functions of various molecules, including drug targets. In KEGG, drug targets are often associated with specific diseases or pathways. The database contains information on the relationship between drugs and their targets, including the target protein or enzyme, the specific drug that interacts with the target, and the associated disease or pathway (Gangadharappa et al., 2020b, Kanehisa and Sato, 2020). Metabolic pathway analysis in KEGG involves the systematic study of the interconnected metabolic reactions and pathways within an organism. It aims to understand the overall metabolism, identify key metabolic pathways, and investigate the impact of genetic or environmental factors on metabolic processes. The pathway analysis of the FASTA sequence file of targets (set3) using KEGG through KAAS server (https://www.genome.jp/kaas-bin/kaas_main) resulted in identifying 10 pathways which are specific to Micrococcus luteus. the list includes the MAPK signaling pathway – fly, Nicotinate and nicotinamide metabolism, FoxO signaling pathway, Lipid and atherosclerosis pathway, Longevity regulating pathway, Peroxisome, Chemical carcinogenesis - reactive oxygen species, Longevity regulating pathway for Huntington disease, worm and multiple species. The parameters considered for this analysis includes the selection of ‘hsa’ for Homo sapiens in GENES data set section of KEGG and (single-directional best hit (SBH) as assignment method. Interestingly, 4 proteins out of 15 extracellular proteins considered for the analysis were associated with KO (KEGG Orthology). In most of the pathways listed above, Superoxide Dismutase (SOD) was prominently available, indicates the completeness of the pathway and the genes presence in the genome. Superoxide dismutase (SOD) is a group of enzymes that play a crucial role in the antioxidant defense system of cells. Their primary function is to catalyze the dismutation of superoxide radicals (O2–) into hydrogen peroxide (H2O2) and molecular oxygen (O2). This enzymatic reaction helps protect cells from oxidative damage caused by reactive oxygen species (ROS). These ROS are oxygen-containing chemically reactive molecules that are produced as natural byproducts of cellular metabolism. They include molecules such as superoxide radicals (O2–), hydrogen peroxide (H2O2) and hydroxyl radicals (OH–) (Maurya et al., 2021). While ROS are produced as part of normal cellular processes, their levels are tightly regulated to maintain cellular homeostasis. ROS are implicated in various pathological conditions and diseases, including cancer, neurodegenerative disorders, cardiovascular diseases, and inflammatory conditions. Therefore, targeting ROS and their associated pathways has emerged as a potential therapeutic strategy.
Superoxide dismutase (SOD) being an enzyme that catalyzes the conversion of superoxide radicals (O2–) into molecular oxygen (O2) and hydrogen peroxide (H2O2), plays a critical role in antioxidant defense. While SOD itself is not a direct drug target, modulating SOD activity or expression can have therapeutic implications. Table 2 shows list of enzymes associated with KEGG Orthology (KO). The list of proteins which are not associated with KO assignment are provided in Supplementary Table 1.
Table 2.
KO assigned proteins or enzymes list along with its Name, EC number, Molecular function, family and domains.
| Sl. No. | ID | KO List | Name | EC Number | Molecular function | Family and domains |
|---|---|---|---|---|---|---|
| 1 | WP_010079427.1 | K08672 | Extracellular basic protease | 3.4.21 | serine-type endopeptidase activity (Hydrolase, Protease, Serine protease) | Peptidase_S8 (182 – 461) |
| 2 | WP_010078719.1 | K04564 | Superoxide dismutase (SOD) | 1.15.1.1 | metal ion binding superoxide dismutase activity (Oxidoreductase) | Sod_Fe_N (3–84)Sod_Fe_C (91–193) |
| 3 | WP_012751022.1 | K17382 | D-alanyl-D-alanine carboxypeptidase | 3.4.16.4 | (Carboxypeptidase, Hydrolase, Protease) | Beta-lactamase (25–346) |
| 4 | WP_010080124.1 | K00323 | Alanine dehydrogenase | 1.4.1.1 | alanine dehydrogenase activity nucleotide binding (Oxidoreductase) | AlaDh_PNT_N (4–136)AlaDh_PNT_C (148–296) |
3.5. Analysis of genome mining of B. licheniformis for ligands
The antiSMASH server was used to mine the B. licheniformis genome. AntiSMASH enables annotation and study of secondary metabolite biosynthesis gene clusters (BGCs) in fungal and bacterial genomes, as well as quick genome-wide identification. According to the findings, a total of 11 areas were generated, along with the most similar gene clusters, from which 5 ligands were chosen for future investigation. Supplementary Table 2 lists the names of the ligands as well as their PubChem ID (CID).
3.6. Analysis of molecular docking
We employed homology modeling and molecular docking to predict and analyze the interactions among ligand obtained from B. licheniformis and a target protein SOD from M. luteus at the atomic level. The structure of the SOD generated through SWISS Modeller was validated using PROCHECK for its stereo-chemical quality and confirmed the overall protein structure and residue-by-residue geometry. Results of the Ramachandran plot analysis revealed that 91.3% of the residues were evidenced to be in the most favored regions. A good model is expected to have more than 90% of its residues in the most favored areas. Ramachandran plot of Superoxide dismutase for stereo-chemical quality checking is shown in Supplementary Fig. 4. Ramachandran plot analysis of residues in favored and allowed regions is given in Supplementary Table 3. The binding sites of the SOD was predicated using Computed Atlas of Surface Topography of proteins (CASTp) which is a web-based tool and database that provides information about the surface topography and structural features of proteins (Binkowski et al., 2003). The predicted binding sites of the protein SOD are shown (red dots) in the Supplementary Fig. 5. Then, we performed the ADME analysis for the shortlisted ligands as per the Lipinski rule of five. The top five ligands were considered for molecular docking. Molecular docking studies were performed to predict the binding affinities and interaction profiles of the selected 5 ligands with SOD protein residues. Supplementary Table 4 shows the binding energy values for each complex, and Supplementary Fig. 3 shows the binding interaction profile for all complexes. The docking results revealed that the SOD_Bacillibactin and SOD_Paenibactin were the most promising candidates among the other five complexes. Bacillibactin, the ligand, interacts with the target protein complex with the lowest binding energy of −9.3 kcal/mol. The Bacillibactin ligand interacts with the target molecule by creating hydrogen bonds with the amino acids 5THR, 73HIS, 75ASN, 77SER, 152ALA, 153THR and 193ARG.
The binding energy of the Paenibactin ligand to the target SOD was found to be − 7.8 kcal/mol and 777SER and 152ALA are the important amino acid residues implicated in hydrogen bonding between the ligand and receptor. Lichenysin has the third-lowest binding energy, with a value of − 6.7 kcal/mol. It forms hydrogen bonds with 74THR, 152ALA and 153THR to interact with the receptor molecule. Supplementary Fig. 3 shows the interaction between amino acids of target molecule with that of ligand. Praveen et al., has conducted molecular docking studies against the Pseudomonas aeruginosa superoxide dismutase (SOD) and reported the interaction of SOD’s THR residue with ligand propiconazole (Satapute et al., 2019). Molecular interaction of the triazole fungicide propiconazole with homology modelled superoxide dismutase and catalase. Hatai et al., work on interaction of phytochemicals with target SOD has shown energetically favourable results and the binding interactions has ALA as one of the SOD’s interacting residue (Hatai and Banerjee 2019). “Molecular docking interaction between superoxide dismutase (receptor) and phytochemicals (ligand) from Heliotropium indicum Linn for detection of potential phytoconstituents: New drug design for releasing oxidative stress condition/inflammation of osteoarthritis patients. Supplementary Table 4 displays the results of Fengycin (-5.8 kcal/mol.) and Surfactin (-5.6 kcal/mol.) docking investigations, as well as hydrophobic and hydrogen bond interactions. The generated protein–ligand complexes stability was further analyzed using MD simulations approach.
3.7. MD simulation analysis
MD simulation is a computational approach commonly employed to illustrate the behavior and interactions of atoms and molecules over the time. MD simulations can be employed to study the binding of small drug-like molecules to their target proteins. By simulating the complex system over time, MD allows for the exploration of different binding modes, conformational changes, and energetics of protein–ligand interactions. The top three molecules (Bacillibactin, Lichenysin and Paenibactin) picked for MDS have been found to play role in antibacterial activities.
Bacillibactin is a siderophore produced by certain strains of Bacillus subtilis, a Gram-positive bacterium. Siderophores are small molecules that are secreted by bacteria to scavenge and bind iron from the environment, as iron is a vital nutrient for the survival and growth of bacteria. Bacillibactin is a catecholate-type siderophore, which means it contains a catechol moiety in its structure. The catechol groups in Bacillibactin have a high affinity for iron ions, allowing them to form stable complexes with iron. These complexes are then recognized and taken up by specific transport systems on the surface of Bacillus subtilis cells.
Paenibactin is a siderophore, which is a type of small molecule produced by bacteria to scavenge and chelate iron from the environment. It is primarily produced by bacteria of the Paenibacillus genus, including Paenibacillus polymyxa and Paenibacillus larvae.
Lichenysin is a cyclic lipopeptide produced by certain strains of Bacillus licheniformis, a Gram-positive bacterium. It belongs to the family of surfactant lipopeptides and is known for its antimicrobial and surface-active properties. Lichenysin is produced as a mixture of isoforms, with varying lengths and amino acid compositions. The primary structure of lichenysin consists of a cyclic peptide ring connected to a fatty acid chain. The peptide ring is composed of 7 to 10 amino acids, including leucine, isoleucine, valine, and aspartic acid. The fatty acid chain, typically a β-hydroxy fatty acid, is attached to one of the amino acids within the peptide ring. MD simulation analysis of top three complexes namely SOD-BACILLIBACTIN (SOD-BAC), SOD-PAENIBACTIN (SOD-PAE) and SOD_LICHENYSIN (SOD-LIC) along with APO is provided in the following subsection. Gromacs software for 250 ns and the simulation trajectories were analyzed using GROMAS utility.
3.7.1. RMSD and RMSF analysis reveals overall stability at the structure and residue level
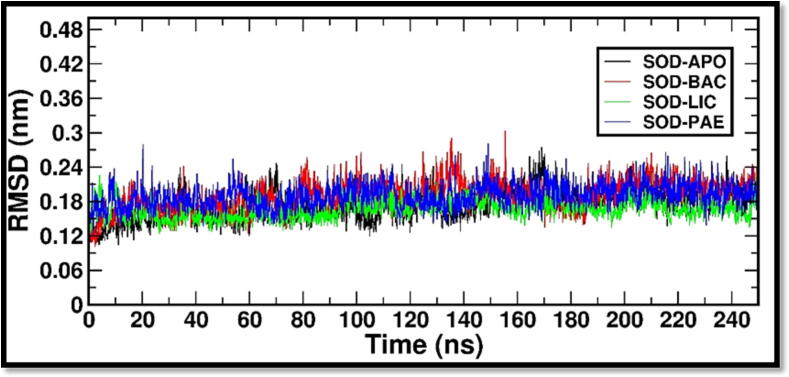
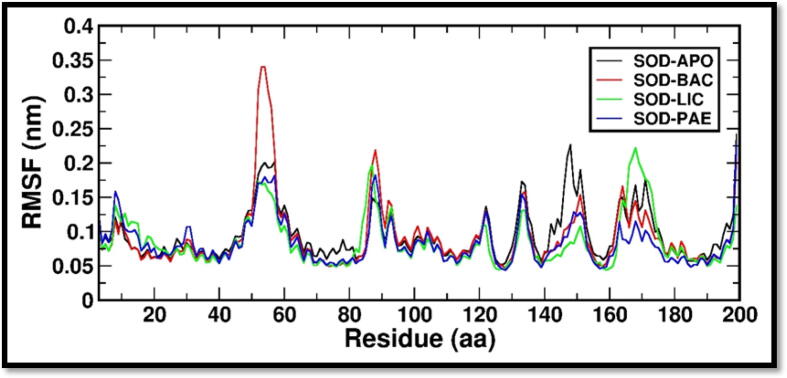
To assess the structural stability of the complexes, RMSD of the simulated system along time trajectory was analyzed by measuring the deviation of SOD with regard to its crystal structure (APO). Within 40 ns of the simulation time the RMSD of all systems converged complying stable well-equilibrated system, as indicated in Fig. 1. Then, the root mean square fluctuations (RMSF) were estimated to assess the region wise loss of stability in all the five systems. RMSF plot analysis of all the SOD bound systems with ligands is shown in Fig. 2. A region with a high RMSF is expected to be more flexible, whereas a region with a low RMSF would possibly have more constrained dynamics. This demonstrated that ligands Bacillibactin, Lichenysin and Paenibactin binding to protein resulted in a stable association and did not cause significant changes in protein structure. RMSF of SOD systems were monitored to examine the loss of stability region wise and it is known to calculate the deviation in fluctuations around averaged position of each atom. The RMSF is a measure of an amino-acid's structural displacement from its average position during the simulation. The RMSF method is effective for assessing local flexibility in protein structures and identifying flexible and rigid area. Pereira et al., has used the computational approach including RMSF to predict the protein stability of SOD and the results have indicated the stable regions with minimal variations (Pereira et al., 2021).
Fig. 1.
RMSD (Root Mean Square Deviation) is a measure of the average distance between the atoms of two superimposed molecular structures. It is commonly used in molecular dynamics simulations to assess the accuracy of the simulation and the stability of the protein structure over time. The comparative RMSD values were estimated with “gmx rms”. SOD-BACILLIBACTIN (SOD-BAC), SOD_LICHENYSIN (SOD-LIC) and SOD-PAENIBACTIN (SOD-PAE).
Fig. 2.
RMSF (Root Mean Square Fluctuation) is a measure of the average deviation of atomic positions from their mean positions over time in molecular dynamic simulations. It provides information about the flexibility and dynamics of a protein structure. Regions with high RMSF values are typically more flexible, while regions with low RMSF values are typically more rigid. The figure indicates a comparative RMSF plot of the complexes with the active site residues encircled in red generated with “gmx rmsf”.
3.7.2. Inhibitors bound SOD found to minimal change in conformational state
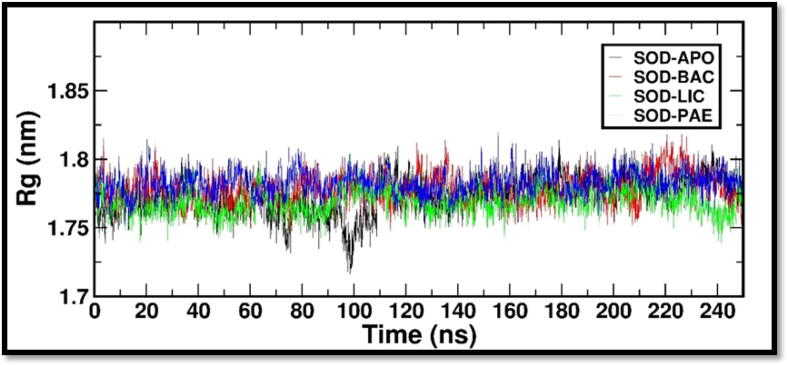
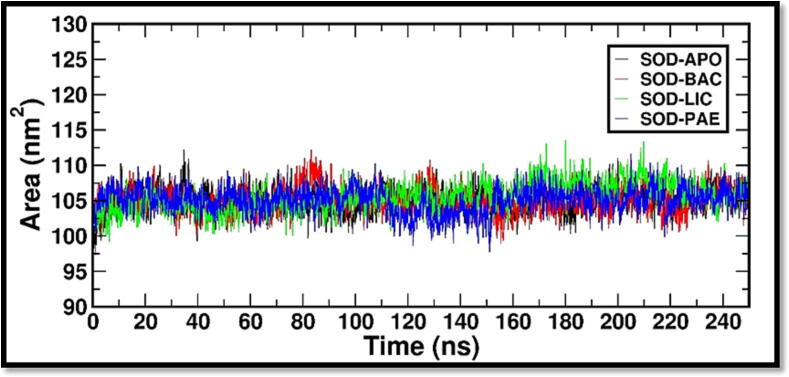
Further, we carried out the radius of gyration (Rg) analysis to evaluate the compactness or overall size of a protein molecule. Rg quantifies the distribution of mass around the center of mass of the protein and provides insights into its structural properties and dynamics. The radius of gyration was used to study the conformational changes and flexibility of a protein over time. By calculating the Rg at different time points during a molecular dynamic simulation, one can assess the expansion or contraction of the protein structure, as well as fluctuations in its shape. Fig. 3 depicts Rg plot for all the three complexes with reference to SOD crystal structure. The Rg analysis determined the compactness of a structure during the simulation time. The Rg plot clearly shows that all the systems Rg-time fluctuations were maintained between 1.75 and 1.8 nm. Then, we calculated Solvent Accessible Surface Area (SASA), which refers to the surface area of a protein that is available to solvent molecules such as water. SASA is a crucial parameter in protein structure analysis as it provides insights into the protein's folding, stability, and interactions with other molecules. We have observed the surface area in the range of 100 – 110 nm2 for all the complexes (Fig. 4).
Fig. 3.
RG (Radius of Gyration) is a measure of the compactness of a protein structure, calculated as the root mean square distance of each atom in the protein from the center of mass of the protein. It provides a quantitative measure of the overall size and shape of a protein structure. Protein structures with a smaller RG value are typically more compact and globular, while structures with a larger RG value are typically more extended and flexible. The “gmx gyrate” module was used to estimate the RG values for the comparative graphs plotted above.
Fig. 4.
SASA (Solvent Accessible Surface Area) is a measure of the surface area of a protein structure that is accessible to solvent molecules. It can provide insights into conformational changes, protein–ligand interactions, and protein–protein interactions. The surface area values over the simulation were predicted with “gmx sasa” and the comparative graphs are plotted.
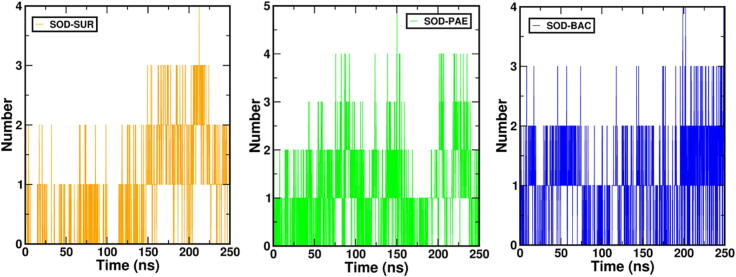
Hydrogen (H) bonds are considered to be an important parameter in stabilization of intermolecular interactions between ligand molecule and protein target (Suganya et al, 2017). Fig. 5 represents H-bonds between the ligands and SOD. In addition, we performed the Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) analysis. MMPBSA is a computational technique employed to estimate the binding free energy of biomolecular complexes. It combines molecular mechanics (MM) calculations to describe the molecular interactions within the complex and Poisson-Boltzmann (PB) continuum electrostatics to account for solvation effects. MMPBSA analysis was performed on all three MDS trajectories (Kumari et al., 2014), and it was discovered that the total binding energy of SOD-LIC and SOD-PAE was relatively larger than that of the SOD-BAC complex. Table 3 indicates the binding energy of all the complexes. The binding energy of the SOD-BAC complex system was −30.522+/-28.764 kJ/mol which showed that the ligand was tightly bound to the protein throughout the simulation duration. To summarize the above results, all the systems have negative binding energy where SOD-BAC has the lowest binding energy. Performing a molecular dynamics simulation after molecular docking can provide additional insights into the stability and behavior of a protein–ligand complex. Molecular docking is a computational method used to predict the binding of a small molecule to a target protein, but it provides only a static picture of the complex. MD simulation can reveal the complex's behaviour over time, including its movement, ability to adapt to environmental changes, and interactions with other molecules. This information can be useful in predicting the stability of the complex and its ability to bind to a target. The movements and interaction of molecules over time could be the reason for getting the difference in binding energy for SOD-BAC complex system. However, the MM-PBSA binding affinity of all the complexes obtained indicates the stable binding over the simulation time. The summary of the binding energy of all the complexes is indicated in Table 3.
Fig. 5.
Hydrogen bonds (H-bonds) are non-covalent interactions that play an essential role in the stabilization of protein–ligand interactions. The H-bond interactions were estimated with “hbond” function of gromacs. Figures indicate the H-bond interactions between the protein and its respective ligands over the simulation time.
Table 3.
Energy values of different Complexes obtained during MD simulations.
| Sl. No. | Complex | Binding free energy (Kcal/mol) |
|---|---|---|
| 1 | SOD_BACILLIBACTIN | −30.522+/-28.764 kJ/mol |
| 2 | SOD_PAENIBACTIN | −53.739+/-39.348 kJ/mol |
| 3 | SOD_LICHENYSIN | −128.070+/-13.610 kJ/mol |
4. Discussion
The combined analysis of subtractive genomics and genome mining approaches resulted in analysing the genomic data of M. luteus and B. licheniformis and reporting the target protein and potential drug candidates/ligands. The set of proteins screened in the present study was compared to the previously published studies (Khan et al., 2020) on Bartonella bacilliformis, where only 100 proteins sequences out of 1097 proteins were excluded using the same parameters as mentioned above. Khan et al., used the Subtractive genomics approach towards the identification of novel therapeutic targets against human Bartonella bacilliformis. The primary goal of this study was to find new treatment targets for B. bacilliformis subsp., and a subtractive genomic strategy was used for the entire proteome by utilising multiple online databases and computational tools. ‘ABC transporter permease’ and ‘Flagellar biosynthesis protein FlhA’ were found to be novel drug targets of this study. Whereas, in another study (Uddin et al, 2018) applied similar approach for drug resistant Mycobacterium tuberculosis (XDR KZN 605) and reported the identification of 41 out of 3993 protein sequences as paralogous sequences at a threshold of 80%. The cut-off or threshold may vary in range from 40% and go till 100% for clustering the sequences. However, there is no specified - ideal cut off value. The choice of cut-off value depends on many factors such as selection of set of data, number of sequences and the purpose of application(Chen et al., 2016). The genome sequence of Streptomyces sp. T676 was recently obtained using antiSMASH, and 2 type I modular PKS gene clusters were found (Lee et al., 2020).
Based on BLASTp results an inference can be made by observing the results and number of protein sequences that the number of non-homologous proteins solely depends on the diversity in protein sequences of screened or target organism. Fewer non-homologous target proteins against human proteome can result in choosing a novel protein or enzyme target in pathogens. Focused analysis of subcellular proteins of extracellular which play crucial roles in various biological processes and can serve as potential drug targets. We report 15 proteins in extracellular space, out of which, 4 proteins associated with KO were considered. Reports from (Khan et al., 2020) showed that 7 out of 114 essential proteins were originated to be unique to pathogen pathways and those proteins were used further in the identification of novel drug targets. Uddin et al., reported that out of 129, only two proteins were successfully passed in the KAAS server (Uddin et al., 2018). Only few pathogen specific pathways, which are unique and play a crucial role in their life cycle, can be identified among a pool of metabolic pathways. This in turn, it can be employed in search of essential enzymes or proteins involved in those pathways.
The molecular docking technique was effectively used to screen the ligand molecules. Identified top three molecules Bacillibactin (Chakraborty et al., 2022), Lichenysin (Rodrigues et al., 2006) and Paenibactin (SUGANYA 2020) picked for MDS have been found to play role in antibacterial activities. The role of Bacillibactin in the antibacterial activity of Bacillus subtilis is indirect. By efficiently sequestering iron from the environment, Bacillibactin can limit the availability of iron for pathogenic bacteria, hindering their growth and virulence (Lee et al., 2011). Iron limitation can have detrimental effects on bacterial physiology, including the disruption of crucial cellular processes that require iron as a cofactor, such as DNA synthesis and energy production (Consentino 2019). From a therapeutic perspective, the study of paenibactin and other siderophores has provided insights into potential strategies for combating bacterial infections. Disrupting the iron acquisition systems of pathogenic bacteria, including their siderophore production or iron uptake mechanisms, has been proposed as an important target for the development of novel antimicrobial agents (Benitez et al., 2012). By inhibiting the ability of bacteria to scavenge iron, it may be possible to limit their growth and reduce their virulence. Lichenysin exhibits a range of biological activities, including antimicrobial, hemolytic, and surfactant properties (Díaz et al., 2022). Its antimicrobial activity is particularly notable, as it has been shown to inhibit the growth of various Gram-positive bacteria and fungi. Lichenysin disrupts the integrity of microbial cell membranes, leading to cell lysis and death (Ruiz et al., 2017). Its diverse range of activities makes it an interesting compound with potential applications in various fields, including medicine, industry, and biotechnology.
RMSD values show that they converged within a simulation time of 40 ns further conforming stable and well-equilibrated system. It further demonstrated the complexes' stability. The validation investigation adds to the evidence that ligands have a high affinity for their respective protein molecules. RMSF values of SOD_BAC, SOD_LIC & SOD_PAE complexes were found to deviate in the residue ranges between 48 and 55 and 160–180 with reference to SOD crystal structure. However, none of the active site residues (60–160 residue range) has relatively higher fluctuation than the SOD crystal structure. RMSF analysis demonstrates that ligands Bacillibactin, Lichenysin and Paenibactin binding to protein resulted in a stable association and did not cause any significant changes in protein structure. The findings therefore provide a clear proof of minimal changes in the protein’s conformational state. However, the type of bound ligand will determine the extent of deviation.
Rg-time fluctuations were maintained between 1.75 and 1.8 nm and surface area in the range of 100 – 110 nm2 for all the complexes. The findings therefore provide a clear proof of minimal changes in the protein’s conformational state. Hydrogen bonds play a crucial role in molecular simulations as they are essential for understanding the structure, dynamics, and interactions of biomolecules. In molecular simulations, hydrogen bonds are modeled and analyzed based on the principles of molecular mechanics and quantum mechanics. Total no of hydrogen bond contacts between the ligands and the protein was analyzed for the complete duration and it was observed that at every time frame minimum of 1 hydrogen bond interactions throughout the duration of 250 ns simulation period. The MMPBSA analysis provided insights into the strength and stability of biomolecular interactions of complexes studied and to identify Bacillibactin as the probable candidate molecule to arrest the activities of SOD. Overall, this research may offer insightful information that could be used to design novel SOD inhibitors for future drug discovery studies.
5. Conclusion
In this study, we employed subtractive genomics, a computational approach to identify unique or differentially expressed genes in M. luteus compared to Homo sapiens. This subtractive genomics approach provided valuable insights into the unique genetic features of a M. luteus. The study resulted in the identification of important extracellular protein SOD as a drug target. Then, the genome mining approach was used to identify potential ligands from B. licheniformis genomic data. This analysis resulted in identification of five important lead molecules against the SOD. Further, we used molecular docking technique to predict and analyze the binding interactions between these five ligands and the target protein SOD. These resulting protein–ligand complexes were further subjected to MD simulation analysis to study the motions and interactions of atoms and molecules over 250 ns time. The analysis of MD simulations suggests Bacillibactin as the probable candidate molecule to arrest the activities of SOD. Bacillibactin, being a siderophore, plays a significant role in bacterial iron acquisition and can have implications in antimicrobial strategies. Bacillibactin indirectly influences oxidative stress in bacteria by regulating iron availability and its interaction with ROS. Understanding the interplay between bacillibactin, iron homeostasis, and oxidative stress provides insights into bacterial survival mechanisms and may offer potential targets for antimicrobial strategies aimed at disrupting iron acquisition and oxidative stress management. Future in-vivo studies would aid in the validation of the findings of this study. Subtractive genomics can provide valuable insights into the unique genetic features of an organism and molecular docking and MD simulations techniques provides information that can be validated through experiments.
Author contributions
ZB and AK have conceptualized and designed the present study. AK incorporated the designed study and executed all the analysis relating to genome mining. BG and DY were involved in MD and simulation analysis. ZB and AK have written the manuscript. TY and SS were involved in critical manuscript review and provided valuable suggestions. ZB supervised the complete study and guided manuscript preparation. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this research work through small group research project under the grant number R.G. P. 1/123/44.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2023.103753.
Contributor Information
Zabin K. Bagewadi, Email: zabin@kletech.ac.in.
T.M. Yunus Khan, Email: mtatagar@kku.edu.sa.
Bhavya Gangadharappa, Email: bhavyasg@msrit.edu.
Ankita Kamalapurkar, Email: 01fe18bbt012@kletech.ac.in.
Shaik Mohamed Shamsudeen, Email: sshahul@kku.edu.sa.
Deepak A. Yaraguppi, Email: deepak.yaraguppi@kletech.ac.in.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A., Guerler A., Hillman-Jackson J., Hiltemann S., Jalili V., Rasche H., Soranzo N., Goecks J., Taylor J., Nekrutenko A., Blankenberg D. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarano L., Esposito R., Ruocco N., Costantini M. Genome mining as new challenge in natural products discovery. Mar. Drugs. 2020;18:199. doi: 10.3390/md18040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly, R., 1991. Cutaneous microbiology. Dermatology. Los Altos: Appleton & Lange. 22-25.
- Ashraf B., Atiq N., Khan K., Wadood A., Uddin R. Subtractive genomics profiling for potential drug targets identification against moraxella catarrhalis. PLoS One. 2022;17:e0273252. doi: 10.1371/journal.pone.0273252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagewadi Z.K., Aakanksha U.K., Yaraguppi D.A., Yunus Khan T.M., Deshpande S.H., Dammalli M., Revankar A.G., Savalagi A.J., Hiremath S.V. Molecular docking and simulation studies against nucleoside diphosphate kinase (ndk) of pseudomonas aeruginosa with secondary metabolite identified by genome mining from paenibacillusehimensis. J. Biomol. Struct. Dyn. 2023:1–10. doi: 10.1080/07391102.2023.2167118. [DOI] [PubMed] [Google Scholar]
- Bakheet T.M., Doig A.J. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- Becker K., Skov R.L., von Eiff C. Staphylococcus, micrococcus, and other catalase-positive cocci. Manual Clin. Microbiol. 2015:354–382. [Google Scholar]
- Benitez, L.B., Velho, R.V., de Souza da Motta, A., Segalin, J., Brandelli, A., 2012. Antimicrobial factor from bacillus amyloliquefaciens inhibits paenibacillus larvae, the causative agent of american foulbrood. Archives of microbiology. 194: 177-185. [DOI] [PubMed]
- Binkowski T.A., Naghibzadeh S., Liang J. Castp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. Antismash 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. Blast+: Architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K., Kizhakkekalam V.K., Joy M., Chakraborty R.D. Bacillibactin class of siderophore antibiotics from a marine symbiotic bacillus as promising antibacterial agents. Appl. Microbiol. Biotechnol. 2022;106:329–340. doi: 10.1007/s00253-021-11632-0. [DOI] [PubMed] [Google Scholar]
- Chen Q., Wan Y., Lei Y., Zobel J., Verspoor K. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE. 2016. Evaluation of cd-hit for constructing non-redundant databases. [Google Scholar]
- Consentino, L., 2019. Mechanisms of host iron acquisition in bacillus cereus: Role of bacillibactin-feua in iron uptake and expression of genes involved in iron homeostasis in vivo during insect gut infection.
- Dertz E.A., Xu J., Stintzi A., Raymond K.N. Bacillibactin-mediated iron transport in bacillus s ubtilis. J. Am. Chem. Soc. 2006;128:22–23. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
- Díaz P.R., Torres M.J., Petroselli G., Erra-Balsells R., Audisio M.C. Antibacterial activity of bacillus licheniformis b6 against viability and biofilm formation of foodborne pathogens of health importance. World J. Microbiol. Biotechnol. 2022;38:181. doi: 10.1007/s11274-022-03377-3. [DOI] [PubMed] [Google Scholar]
- Dürst U., Bruder E., Egloff L., Wüst J., Schneider J., Hirzel H. Micrococcus luteus: A rare pathogen of valve prosthesis endocarditis. Z. Kardiol. 1991;80:294–298. [PubMed] [Google Scholar]
- Eady E.A., Coates P., Ross J.I., Ratyal A.H., Cove J.H. Antibiotic resistance patterns of aerobic coryneforms and furazolidone-resistant gram-positive cocci from the skin surface of the human axilla and fourth toe cleft. J. Antimicrob. Chemother. 2000;46:205–213. doi: 10.1093/jac/46.2.205. [DOI] [PubMed] [Google Scholar]
- Fatoba A.J., Okpeku M., Adeleke M.A. Subtractive genomics approach for identification of novel therapeutic drug targets in mycoplasma genitalium. Pathogens. 2021;10 doi: 10.3390/pathogens10080921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharappa B.S., Rajashekarappa S., Sathe G. Proteomic profiling of serratia marcescens by high-resolution mass spectrometry. BioImpacts: BI. 2020;10:123. doi: 10.34172/bi.2020.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharappa B.S., Sharath R., Revanasiddappa P.D., Chandramohan V., Balasubramaniam M., Vardhineni T.P. Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors. J. Biomol. Struct. Dyn. 2020;38:3757–3771. doi: 10.1080/07391102.2019.1667265. [DOI] [PubMed] [Google Scholar]
- Gupta V., Chauhan A., Kumar R., Dhyani A., Chakravarty S. Meningitis caused by micrococcus luteus: Case report and review of literature. Int. J. Med. Microbiol. Trop. Dis. 2019;5:63–64. [Google Scholar]
- Harwood C.R., Mouillon J.M., Pohl S., Arnau J. Secondary metabolite production and the safety of industrially important members of the bacillus subtilis group. FEMS Microbiol. Rev. 2018;42:721–738. doi: 10.1093/femsre/fuy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatai B., Banerjee S.K. Molecular docking interaction between superoxide dismutase (receptor) and phytochemicals (ligand) from heliotropium indicum linn for detection of potential phytoconstituents: New drug design for releasing oxidative stress condition/inflammation of osteoarthritis patients. J. Pharmacogn. Phytochem. 2019;8:1700–1706. [Google Scholar]
- Huang Y., Niu B., Gao Y., Fu L., Li W. Cd-hit suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., Schulten, K., 1996. Vmd: Visual molecular dynamics. J Mol Graph. https://doi.org/10.1016/0263-7855(96)00018-5. 14: 33-38, 27-38. [DOI] [PubMed]
- Ianniello N.M., Andrade D.C., Ivancic S., Eckardt P.A., Ramirez J.C.L. Native valve infective endocarditis due to micrococcus luteus in a non-hodgkin’s lymphoma patient. IDCases. 2019;18:e00657. doi: 10.1016/j.idcr.2019.e00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S.E., Campbell J. Peptidoglycan biosynthesis in micrococcus luteus (sodonensis): Transglycosidase and phosphodiesterase activities in membrane preparations. J. Bacteriol. 1976;127:309–318. doi: 10.1128/jb.127.1.309-318.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y. Kegg mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, N., K Sharma, A., Shakeel, A., Kumar, V., Singh, A., Gupta, A., Suhag, D., K Rajput, S., Mukherjee, M., 2017. Therapeutic implications of superoxide dismutase and its importance in kinase drug discovery. Current Topics in Medicinal Chemistry. 17: 2495-2508. [DOI] [PubMed]
- Kenshole E., Herisse M., Michael M., Pidot S.J. Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 2021;60:47–54. doi: 10.1016/j.cbpa.2020.07.010. [DOI] [PubMed] [Google Scholar]
- Khan M.T., Mahmud A., Iqbal A., Hoque S.F., Hasan M. Subtractive genomics approach towards the identification of novel therapeutic targets against human bartonella bacilliformis. Inf. Med. Unlocked. 2020;20 [Google Scholar]
- Kirk O., Borchert T.V., Fuglsang C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002;13:345–351. doi: 10.1016/s0958-1669(02)00328-2. [DOI] [PubMed] [Google Scholar]
- Kooken J.M., Fox K.F., Fox A. Characterization of micrococcus strains isolated from indoor air. Mol. Cell. Probes. 2012;26:1–5. doi: 10.1016/j.mcp.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Lynn A. G_mmpbsa–a gromacs tool for high-throughput mm-pbsa calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Laskowski, R., MacArthur, M., Moss, D., Thornton, J., 1993. Iucr. PROCHECK: a program to check the stereochemical quality of protein structures. urn: issn. 0021-8898.
- Lau W.Y.V., Hoad G.R., Jin V., Winsor G.L., Madyan A., Gray K.L., Laird M.R., Lo R., Brinkman F.S.L. Psortdb 4.0: Expanded and redesigned bacterial and archaeal protein subcellular localization database incorporating new secondary localizations. Nucleic Acids Res. 2021;49:D803–D808. doi: 10.1093/nar/gkaa1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hwang S., Kim J., Cho S., Palsson B., Cho B.-K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in streptomyces. Comput. Struct. Biotechnol. J. 2020;18:1548–1556. doi: 10.1016/j.csbj.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Passalacqua K.D., Hanna P.C., Sherman D.H. Regulation of petrobactin and bacillibactin biosynthesis in bacillus anthracis under iron and oxygen variation. PLoS One. 2011;6:e20777. doi: 10.1371/journal.pone.0020777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Maurya R., Namdeo M. Superoxide dismutase: A key enzyme for the survival of intracellular pathogens in host. React. Oxygen Species. 2021 doi: 10.5772/intechopen.94870. [DOI] [Google Scholar]
- Medema M.H., Blin K., Cimermancic P., De Jager V., Zakrzewski P., Fischbach M.A., Weber T., Takano E., Breitling R. Antismash: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., De Bruijn I., Chooi Y.H., Claesen J., Coates R.C. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M., Marahiel M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B.R., 3rd, McGee, T.D., Jr., Swails, J.M., Homeyer, N., Gohlke, H., Roitberg, A.E., 2012. Mmpbsa.Py: An efficient program for end-state free energy calculations. J Chem Theory Comput. https://doi.org/10.1021/ct300418h. 8: 3314-3321. [DOI] [PubMed]
- Mondol M.A.M., Shin H.J., Islam M.T. Diversity of secondary metabolites from marine bacillus species: Chemistry and biological activity. Mar. Drugs. 2013;11:2846–2872. doi: 10.3390/md11082846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. Kaas: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muras A., Romero M., Mayer C., Otero A. Biotechnological applications of bacillus licheniformis. Crit. Rev. Biotechnol. 2021;41:609–627. doi: 10.1080/07388551.2021.1873239. [DOI] [PubMed] [Google Scholar]
- Page M.G. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin. Infect. Dis. 2019;69:S529–S537. doi: 10.1093/cid/ciz825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, T., 2019. Specific infectious diseases causing infertility and subfertility in cattle. In “veterinary reproduction and obstetrics” ten edition, Elsevier BV, Edinburgh, Scotland.
- Peces, R., Gago, E., Tejada, F., Laures, A., Alvarez-Grande, J., 1997. Relapsing bacteraemia due to micrococcus luteus in a haemodialysis patient with a perm-cath catheter. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association-European Renal Association. 12: 2428-2429. [DOI] [PubMed]
- Pereira, G.R.C., Vieira, B.d.A.A., De Mesquita, J.F., 2021. Comprehensive in silico analysis and molecular dynamics of the superoxide dismutase 1 (sod1) variants related to amyotrophic lateral sclerosis. PLoS One. 16: e0247841. [DOI] [PMC free article] [PubMed]
- Prakash O., Nimonkar Y., Chavadar M.S., Bharti N., Pawar S., Sharma A., Shouche Y.S. Optimization of nutrients and culture conditions for alkaline protease production using two endophytic micrococci: Micrococcus aloeverae and micrococcus yunnanensis. Indian J. Microbiol. 2017;57:218–225. doi: 10.1007/s12088-017-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Olea H., Reyes-Ballesteros B., Chavez-Santoscoy R.A. Potential application of the probiotic bacillus licheniformis as an adjuvant in the treatment of diseases in humans and animals: A systematic review. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.993451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L., Banat I.M., Teixeira J., Oliveira R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006;57:609–618. doi: 10.1093/jac/dkl024. [DOI] [PubMed] [Google Scholar]
- Romo-Barrera C.M., Castrillón-Rivera L.E., Palma-Ramos A., Castañeda-Sánchez J.I., Luna-Herrera J. Bacillus licheniformis and bacillus subtilis, probiotics that induce the formation of macrophage extracellular traps. Microorganisms. 2021;9:2027. doi: 10.3390/microorganisms9102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Pinazo A., Pérez L., Manresa A., Marqués A.M. Green catanionic gemini surfactant–lichenysin mixture: Improved surface, antimicrobial, and physiological properties. ACS Appl. Mater. Interfaces. 2017;9:22121–22131. doi: 10.1021/acsami.7b03348. [DOI] [PubMed] [Google Scholar]
- Satapute P., Sanakal R.D., Mulla S.I., Kaliwal B. Molecular interaction of the triazole fungicide propiconazole with homology modelled superoxide dismutase and catalase. Environ. Sustain. 2019;2:429–439. [Google Scholar]
- Schrodinger, L., 2010. The pymol molecular graphics system, version 1.3 r1. (No Title).
- Seifert H., Kaltheuner M., Perdreau-Remington F. Micrococcus luteus endocarditis: Case report and review of the literature. Zentralblatt Bakteriol. 1995;282:431–435. doi: 10.1016/s0934-8840(11)80715-2. [DOI] [PubMed] [Google Scholar]
- SUGANYA, K., 2020. Reg. No. F9172, Madurai Kamaraj University.
- Sumarsih S., Hadi S., Andini D., Nafsihana F. IOP Conference Series: Earth and Environmental Science. IOP Publishing; 2018. Carbon and nitrogen sources for lipase production of micrococcus sp. Isolated from palm oil mill effluent-contaminated soil. [Google Scholar]
- Szczerba I. Susceptibility to antibiotics of bacteria from genera micrococcus, kocuria, nesterenkonia, kytococcus and dermacoccus. Med. Doswiadczalna Mikrobiol. 2003;55:79. [PubMed] [Google Scholar]
- Turner P. Oregon Graduate Institute of Science Technology; 2005. Center for coastal and land-margin research. [Google Scholar]
- Uddin R., Siddiqui Q.N., Azam S.S., Saima B., Wadood A. Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant mycobacterium tuberculosis (xdr kzn 605) through subtractive genomics approach. Eur. J. Pharm. Sci. 2018;114:13–23. doi: 10.1016/j.ejps.2017.11.014. [DOI] [PubMed] [Google Scholar]
- Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. Amdock: A versatile graphical tool for assisting molecular docking with autodock vina and autodock4. Biol. Direct. 2020;15:1–12. doi: 10.1186/s13062-020-00267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W., Blanot D., De Pedro M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Mo X., Zhang L., Wang Q. Four superoxide dismutase (isozymes) genes of bacillus cereus. Ann. Microbiol. 2011;61:355–360. [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. Swiss-model: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K., Baba A. Therapeutic potential of superoxide dismutase (sod) for resolution of inflammation. Inflamm. Res. 2006;55:359–363. doi: 10.1007/s00011-006-5195-y. [DOI] [PubMed] [Google Scholar]
- Yuan S., Chan H.S., Hu Z. Using pymol as a platform for computational drug design. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2017;7:e1298. [Google Scholar]
- Zdorovenko E.L., Gannesen A.V., Dmitrenok A.S., Zhurina M.V., Mart'yanov S.V., Shashkov A.S. Structure of cell-wall glycopolymers of micrococcus luteus c01. Carbohydr. Res. 2021;506 doi: 10.1016/j.carres.2021.108356. [DOI] [PubMed] [Google Scholar]
- Zhu M., Zhu Q., Yang Z., Liang Z. Clinical characteristics of patients with micrococcus luteus bloodstream infection in a chinese tertiary-care hospital. Polish J. Microbiol. 2021;70(3):321. doi: 10.33073/pjm-2021-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.