Abstract
Post-translational modifications including protein ubiquitination regulate a plethora of cellular processes in distinct manners. RNA N6-methyladenosine is the most abundant post-transcriptional modification on mammalian mRNAs and plays important roles in various physiological and pathological conditions including hematologic malignancies. We previously determined that the RNA N6-methyladenosine eraser ALKBH5 is necessary for the maintenance of acute myeloid leukemia (AML) stem cell function, but the post-translational modifications involved in ALKBH5 regulation remain elusive. Here, we show that deubiquitinase ubiquitin-specific peptidase 9X (USP9X) stabilizes ALKBH5 and promotes AML cell survival. Through the use of mass spectrometry as an unbiased approach, we identify USP9X and confirm that it directly binds to ALKBH5. USP9X stabilizes ALKBH5 by removing the K48-linked polyubiquitin chain at K57. Using human myeloid leukemia cells and a murine AML model, we find that genetic knockdown or pharmaceutical inhibition of USP9X inhibits leukemia cell proliferation, induces apoptosis, and delays AML development. Ectopic expression of ALKBH5 partially mediates the function of USP9X in AML. Overall, this study uncovers deubiquitinase USP9X as a key for stabilizing ALKBH5 expression and reveals the important role of USP9X in AML, which provides a promising therapeutic strategy for AML treatment in the clinic.
Keywords: deubiquitination, RNA m6A, acute myeloid leukemia, ALKBH5, USP9X
N6-methyladenosine (m6A) is the most common modification on mammalian mRNAs (1, 2), which is catalyzed by the METTL3–METTL14 methyltransferase complex associated with other subunits and is removed by either demethylase ALKBH5 or FTO (3, 4). m6A plays an important role in regulating mRNA fates under various physiological and pathological conditions such as hematologic malignancies (5, 6, 7, 8, 9, 10, 11, 12, 13, 14). Given these essential functions of m6A modifiers in various biological processes, it becomes necessary to investigate the mechanisms of how m6A modifiers are regulated at different layers, including the post-translational level. Protein ubiquitination is one of the most powerful post-translational modifications of proteins and regulates a plethora of cellular processes in distinct manners (15). A recent work finds that METTL3 is phosphorylated by extracellular signal–regulated kinase at S43/S50/S525, which decreases the ubiquitination of METTL3 by ubiquitin-specific peptidase 5 (16). The subunit of m6A writer complex WTAP can also be phosphorylated by extracellular signal–regulated kinase signaling (16). Lactylation of METTL3 is found to be essential for capturing target RNA (17). METTL3 is also modified by SUMO1 mainly at lysine residues K177, K211, K212, and K215, and SUMOylation of METTL3 significantly represses its m6A methytransferase activity without its stability, localization, and interaction with METTL14 and WTAP (18). However, post-translational modifications of other m6A modifiers remain largely unknown.
Acute myeloid leukemia (AML) is an aggressive hematological malignancy, which is characterized by blocked differentiation and uncontrolled proliferation of myeloid progenitors (19, 20, 21). Despite great progress achieved about the molecular basis of AML pathogenesis, the overall survival of adult AML patients has not significantly improved in the past 3 decades. Thus, it is critical to further explore the molecular mechanisms of AML development, which might bring hope for developing new effective therapeutic strategies. Recently, we found that ALKBH5 is specifically required for maintaining the function of AML stem cells but not normal hematopoietic stem cells. We also revealed that KDM4C regulates ALKBH5 expression via increasing chromatin accessibility of ALKBH5 locus, reducing H3K9me3 levels, and promoting recruitment of MYB and pol II. These findings lay the foundation for targeting ALKBH5 for AML therapy (7, 13). Therefore, fully understanding the mechanisms of how ALKBH5 expression level is regulated is of great significance.
Deubiquitinating enzymes remove ubiquitin from protein substrates and impact protein activity, localization, or stability. Among 100 human deubiquitinating enzymes, ubiquitin-specific peptidase 9X (USP9X) is implicated in development and various cellular functions including protein trafficking and apoptosis (22). Here, using mass spectrometry (MS), we identify that USP9X interacts with ALKBH5 to deubiquitinate it, which is essential for maintaining AML cell survival. This finding uncovers USP9X as a key deubiquitinase for ALKBH5 protein stabilization and a potential target for AML therapy.
Results
Deubiquitinase USP9X interacts with ALKBH5
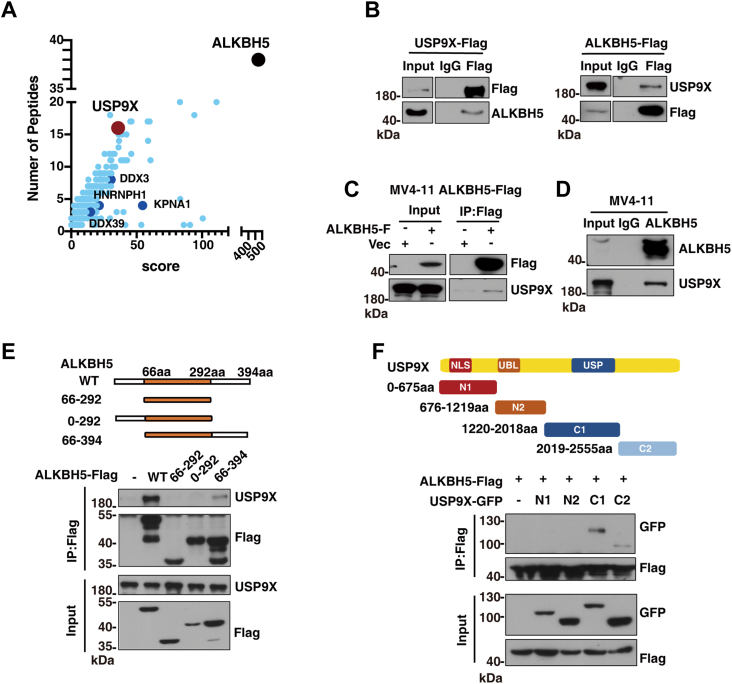
To identify potential regulators of ALKBH5, we performed MS using human myeloid leukemia cell MV4-11 and human embryo kidney human embryonic kidney 293T (HEK293T) cell, which ectopically expressed FLAG-tagged ALKBH5 (FLAG-ALKBH5). Total cellular lysates were treated with benzonase nuclease to remove nucleic acids before immunoprecipitation (IP), and immunoprecipitates pulled down with FLAG from these lysates were decomposed by SDS-PAGE and analyzed by LC–MS. As shown in Figure 1A, some proteins (e.g., DDX3 (23), HNRNPH1 (24), KPNA1, and DDX39 (25)) that are known to interact with ALKBH5 were detected, which validates our LC–MS data. Interestingly, deubiquitinase USP9X was observed among the top candidates (Fig. 1A), suggesting that USP9X might interact with and regulate ALKBH5 ubiquitination. We further examined the physical interaction of USP9X with ALKBH5. We first overexpressed either FLAG-ALKBH5 or FLAG-tagged USP9X in HEK293T cells and conducted co-IP assay. As expected, endogenous ALKBH5 or USP9X was pulled down by either FLAG-tagged USP9X or FLAG-ALKBH5 (Fig. 1B), indicating the interaction between the two proteins. In addition, in human myeloid leukemia cell, endogenous USP9X was also immunoprecipitated by ectopic-expressed FLAG-ALKBH5 (Fig. 1C). Moreover, interaction between endogenous USP9X and ALKBH5 was also observed in human myeloid leukemia cells (Fig. 1D). Thus, these data indicate that deubiquitinase USP9X interacts with ALKBH5, which promotes us to seek the precise region of ALKBH5 responsible for this interaction. As the highly conserved domain in the catalytic core of ALKBH5 is necessary for its activity, we kept the domain intact and generated ALKBH5 truncations by deletion at either N terminus (0–66 aa) or C terminus (292–394 aa), or both. Interestingly, we found that deletion of the C terminus but not N terminus interrupted the interaction between ALKBH5 and USP9X (Fig. 1E), suggesting that ALKBH5 C terminus mediates its interaction with USP9X. Similarly, we tried to define the region of USP9X that mediates its interaction with ALKBH5. Because of the extreme difficulty to obtain full-length USP9X, we generated four GFP-tagged USP9X segments (N1, N2, C1, and C2) in line with previous study (26, 27). We coexpressed these segments with FLAG-ALKBH5 into HEK293T cells for co-IP assay. We found that USP9X-C1 segment was pulled down by ALKBH5, suggesting that this region of USP9X is responsible for its interaction with ALKBH5 (Fig. 1F). Therefore, these data indicate that deubiquitinase USP9X directly binds to ALKBH5.
Figure 1.
ALKBH5 interacts with USP9X.A, LC–tandem mass spectrometry analysis of ALKBH5 interacting protein. The top candidates were highlighted. B, 293T cells were transfected with FLAG-USP9X or FLAG-ALKBH5, and cell lysates were coimmunoprecipitated with Anti-FLAG Affinity Gel by immunoblotting analysis using the indicated antibodies. C, lysates of MV4-11 cells overexpressing FLAG-ALKBH5 were subjected to coimmunoprecipitation with Anti-FLAG Affinity Gel and subjected to immunoblot analysis with antibodies against indicated proteins. D, MV4-11 cell lysates were subjected to coimmunoprecipitation with anti-ALKBH5 antibody, and the immunoprecipitates were analyzed by indicated antibodies. E, schematic presentation of various human ALKBH5 truncations (up). 293T cells were transfected with ALKBH5-FLAG or truncations, and cell lysates were coimmunoprecipitated with Anti-FLAG Affinity Gel and subjected to immunoblotting analysis with indicated antibodies (down). F, schematic diagram of USP9X including the domain containing nuclear localization signal (NLS), ubiquitin-like domain (UBL), and ubiquitin-specific protease domain (USP). GFP-USP9X-truncation as shown in the top schematics were cotransfected with FLAG-ALKBH5 in 293T cells. Western blot analysis with anti-FLAG or anti-GFP antibody in the input and immunoprecipitates (down). USP9X, ubiquitin-specific peptidase 9X.
USP9X deubiquitinates and stabilizes ALKBH5
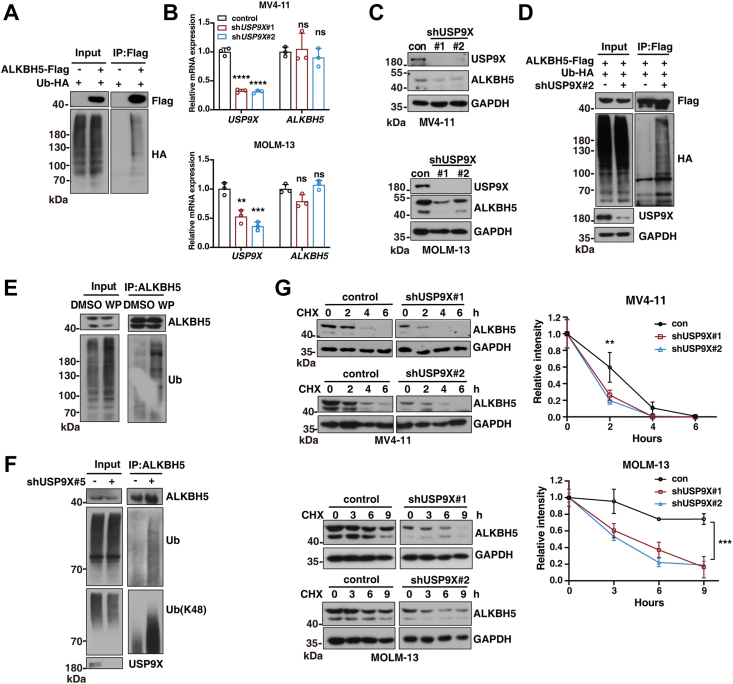
To investigate the molecular mechanisms on how USP9X regulates ALKBH5, we first examined whether ALKBH5 is modified by ubiquitination and conducted ubiquitination assay by cotransfecting hemagglutinin (HA)-tagged Ub (HA-Ub) with FLAG-ALKBH5 into HEK293T cells. As expected, ubiquitination of ALKBH5 was detected (Fig. 2A). Similarly, endogenous ubiquitination of ALKBH5 was also observed in different human leukemia cells (MV4-11 and MOLM-13) and HEK293T cells (Fig. S1A). Interestingly, we found that, while ALKBH5 displayed stable in HEK293T cells, it exhibited obvious less stable in human leukemia cells, especially MV4-11 and MOLM-13 cells (Fig. S1, B and C). This difference may reflect the different biological roles of ALKBH5 and related USP9X in distinct cells. Next, we assessed the potential role of USP9X in regulating ALKBH5 stability. We knocked down USP9X in human leukemia cells (MV4-11, MOLM-13, and Kasumi cells) using shRNAs targeting USP9X (shUSP9X#1/#2). The knockdown efficiency of USP9X was confirmed at the mRNA and protein levels (Figs. 2, B and C and S1D). We found that USP9X knockdown obviously decrease the protein level of ALKBH5 but did not affect its mRNA level (Fig. 2, B and C). We next determined whether USP9X deubiquitinates ALKBH5. As expected, we found that USP9X knockdown increased polyubiquitination of endogenous ALKBH5 (Fig. 2D). WP1130 is an inhibitor of USP9X, and treatment with WP1130 also markedly enhanced polyubiquitination of ALKBH5 (Fig. 2E). Thus, these data indicate that USP9X is responsible for ALKBH5 ubiquitination, which drive us to determine which types of polyubiquitination of ALKBH5 mediated by USP9X. Different ubiquitin mutants (lysine [K] is mutated to arginine [R]) were used and cotransfected with FLAG-ALKBH5 into HEK293T cells. We found that USP9X knockdown led to the increases of polyubiquitin levels of ALKBH5 with K6R, K11R, K27R, K29R, and K33R but not ALKBH5 with K48R that normally mark proteins for proteasomal degradation. Furthermore, ubiquitin mutants (only a single lysine residue) were also used, and knockdown of USP9X significantly increased K48-linked polyubiquitination of ALKBH5 (Fig. S1E). We also observed an obvious increase of endogenous K48-linked polyubiquitination of ALKBH5 upon knocking down USP9X in MV4-11 leukemia cells (Fig. 2F). Moreover, cycloheximide (CHX) chase assays showed that USP9X depletion accelerated the decay of ALKBH5 protein, showing decreased half-lives of ALKBH5 in leukemia cells (Figs. 2G and S1F). WP1130 treatment also increased ALKBH5 decay (Fig. S1G). Taken together, these results indicate that USP9X regulates ALKBH5 stability by mediating its K48-linked polyubiquitination.
Figure 2.
USP9X deubiquitinates and stabilizes ALKBH5.A, 293T cells were transfected with ALKBH5-FLAG and HA-Ub for 24 h before ubiquitination assays. Cell lysates were immunoprecipitated with Anti-FLAG Affinity Gel and then analyzed by immunoblotting. B and C, MV4-11 and MOLM-13 cells were transducted with lentiviruses for control shRNA or different sets of USP9X shRNAs. Cellular extracts and total RNA were prepared and analyzed by quantitative qRT–PCR (B) and Western blotting (C). D, USP9X-knockdown 293T cells were cotransfected with the ALKBH5-FLAG and HA-Ub for 24 h before ubiquitination assays. Cell lysates were immunoprecipitated with Anti-FLAG Affinity Gel and then analyzed by immunoblotting using the indicated antibodies. E, MV4-11 cells were treated with 2 μM WP1130 (WP) for 4 h before endogenous ubiquitination assays. Cell lysates were immunoprecipitated with Anti-ALKBH5 antibody and then analyzed by immunoblotting using the indicated antibodies. F, endogenous ubiquitination assay was analyzed in USP9X-knockdown MV4-11 cells. Cell lysates were immunoprecipitated with Anti-ALKBH5 antibody and then analyzed by immunoblotting using the indicated antibodies. G, MV4-11 and MOLM-13 cells with or without USP9X by shRNA were treated with 50 μg/ml CHX or 100 μg/ml CHX and harvested at the indicated time followed by Western blotting analysis. Intensity of each band from biological triplicate experiments was quantified by densitometry with the ImageJ software with GAPDH as a normalizer. The data in (B and G) were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. The error bars represent mean ± SD; ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. CHX, cycloheximide; HA-Ub, hemagglutinin-tagged Ub; USP9X, ubiquitin-specific peptidase 9X.
USP9X stabilizes ALKBH5 by mediating K48-linked polyubiquitination at K57
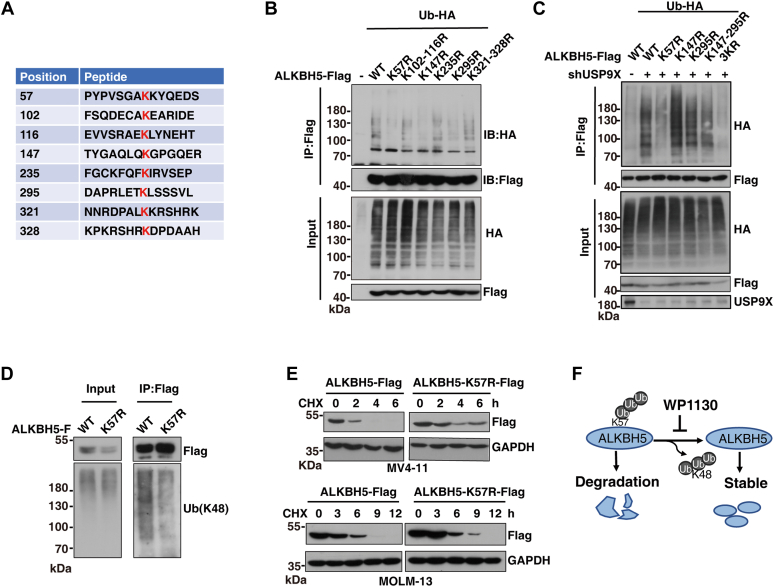
Next, we sought to figure out the residues in ALKBH5 that are deubiquitinated by USP9X. Since ubiquitination of substrates frequently occurs at a lysine residue, eight potential ubiquitination sites in ALKBH5 were predicted using GPS-PLMD (28) and CPLM software (29) (Biocuckoo), including K57, K102, K116, K147, K235, K295, K321, and K328 (Fig. 3A). We then mutated these lysine residues to arginine and investigated whether these ALKBH5 mutants could be modified by polyubiquitination. Using these ALKBH5 ubiquitination-deficient mutants, we performed ubiquitination co-IP assay and found that ALKBH5 mutants with K57R, K147R, and K295R showed reduction of ALKBH5 ubiquitination (Fig. 3B). To investigate which of these three lysine residues could be targeted by USP9X, we knocked down USP9X and found that USP9X knockdown did not affect the ubiquitination level of ALKBH5 with K57R rather than K147R and K295R (Fig. 3C), suggesting that ALKBH5 K57 is the major ubiquitination site that is affected by USP9X. We further investigated whether K57 is modified by K48-linked polyubiquitination. Blotting with the specific Ub (K48) antibody, we found that ALKBH5 K57R markedly reduced its K48-linked polyubiquitination when compared with WT ALKBH5 (Fig. 3D). Thus, these data indicate that USP9X removes K48-linked polyubiquitination at K57 of ALKBH5.
Figure 3.
USP9X mediates K48-linked polyubiquitination of ALKBH5 at K57.A, the table showed the predicted ALKBH5 ubiquitination candidate sites by the GPS-PLMD and CPLM software. B, ubiquitination assays showed that polyubiquitination of ALKBH5 and its mutants. FLAG-tagged WT ALKBH5 or ALKBH5 lysine (K) to arginine (R) mutants were coexpressed with HA-tagged Ub in 293T cells. Cell lysates were immunoprecipitated with Anti-FLAG Affinity Gel and then analyzed by immunoblotting using the indicated antibodies. C, USP9X-knockdown 293T cells were cotransfected with the HA-Ub and ALKBH5-FLAG WT or mutants before ubiquitination assays. The final immunoprecipitates and cell lysates were analyzed by immunoblots for detection of the indicated proteins. D, endogenous ubiquitination assay in MV4-11 cells that were transducted by lentivirus of ALKBH5 or ALKBH5 K57R. Cell lysates were immunoprecipitated with Anti-FLAG Affinity Gel and then analyzed by immunoblotting using antibody against ubiquitin (K48). E, MV4-11 or MOLM-13 stably expressing ALKBH5 or ALKBH5 K57R mutant were treated with 50 μg/ml CHX or 100 μg/ml CHX and harvested at the indicated time followed by Western blotting analysis. F, working model showed that USP9X mediates K48-linked polyubiquitination of ALKBH5 at K57. HA-Ub, hemagglutinin-tagged ubiquitin; USP9X, ubiquitin-specific peptidase 9X.
Moreover, CHX chase assay showed that the half-life of ALKBH5 K57R was higher than that of WT ALKBH5 (Fig. 3E), indicating that mutant of ubiquitination site at K57 increases ALKBH5 stability. Next, we analyzed whether ALKBH5 ubiquitination influences its demethylase activity of RNA m6A. Dot blot analysis showed that ALKBH5 knockdown significantly increased the global levels of RNA m6A, which were rescued by restoration of both WT ALKBH5 and ALKBH5 K57R, the ubiquitination-deficient mutant (Fig. S2A). We further purified ALKBH5 and ALKBH5 K57R proteins. In vitro demethylation assays also showed that both ALKBH5 and ALKBH5 K57R efficiently decreased m6A levels (Fig. S2, B and C). Thus, these data suggest that ALKBH5 ubiquitination at K57 does not affect its demethylase activity. Taken together, these data demonstrated that ubiquitination of ALKBH5 at K57 affects its stability but not the m6A demethylase activity.
USP9X is required for maintaining leukemia cell survival and AML development
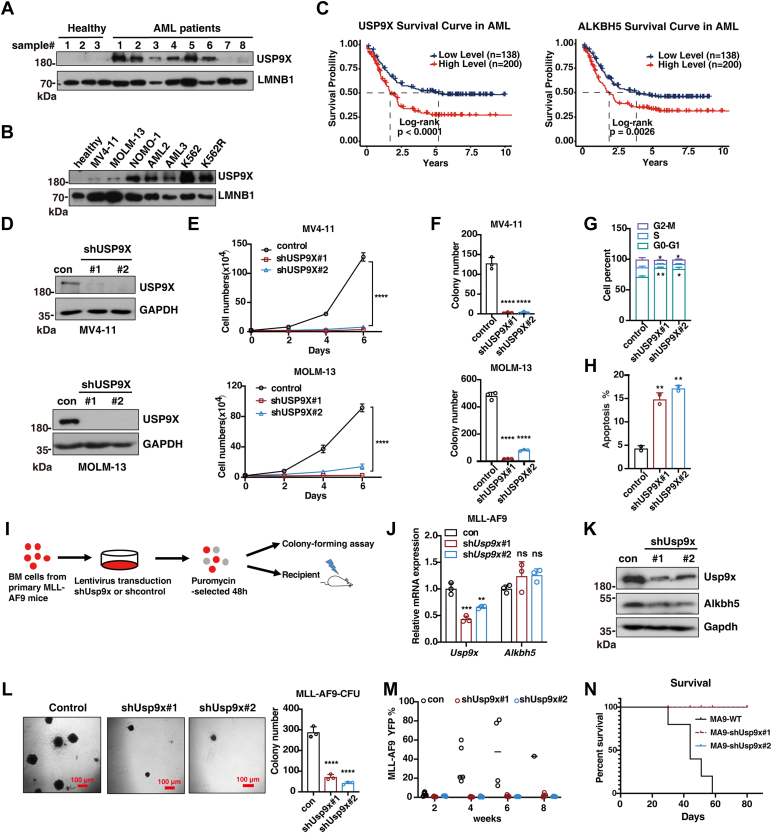
Our recent study has demonstrated that ALKBH5 is required for AML development. However, the role of USP9X in AML remains still elusive. Interestingly, we found that USP9X was aberrantly elevated at the protein level in AML patient–derived leukemia cells, when compared with those of healthy control cells (Fig. 4A). Similarly, higher USP9X protein levels were detected in various human leukemia cell lines (Fig. 4B). These data suggest that abnormal expression of USP9X might be associated with AML progression. To support this idea, we further interrogated two AML patient cohorts and found that elevated expression of USP9X positively correlated with overall shorter survival of AML patients, similar to ALKBH5 (Fig. 4C). To test the function of USP9X in leukemia cells, we knocked down USP9X in MV4-11 and MOLM-13 cell lines (Figs. 4D and S3A). As expected, USP9X knockdown significantly reduced cellular growth and clonogenic ability of leukemia cells (Fig. 4, E and F). In addition, USP9X knockdown resulted in cell cycle arrest in G0–G1 phase (Figs. 4G and S3B) and significantly promoted apoptosis of leukemia cells, showing that the percentage of Annexin V+ cells was higher after USP9X deletion (Figs. 4H and S3C). Furthermore, treatment with WP1130 also induced apoptosis of leukemia cells in a dose-dependent manner (Fig. S3D). These data suggest that USP9X is required for maintaining human AML cell survival.
Figure 4.
USP9X is required for maintaining leukemia cell survival and AML development.A, immunoblot showing USP9X expression in bulk BM mononuclear cells from cord blood (CD34+) (n = 3) and primary patients with AML (n = 8). LMNB1 served as a loading control. B, Western blotting analysis of the expression of USP9X in multiple cell lines. CD34+ cells derived from cord blood were used as a normal control, and LMNB1 served as the loading control. C, Kaplan–Meier plots of overall survival in TCGA and TARGET cohorts for AML patients, stratified on the basis of USP9X or ALKBH5 expression above (high) or below (low) the median. The log-rank test was used to compare survival curves. D, MV4-11 and MOLM-13 cells were transducted with lentiviruses for control shRNA or different sets of USP9X shRNAs. Cellular extracts were prepared and analyzed by Western blotting. E, growth curves of MV4-11 and MOLM-13 cells after transduction with indicated lentiviruses. F, colony-forming assay of MV4-11 and MOLM-13 cells after transduction with indicated lentiviruses. G, flow cytometry was used to quantitatively analyze cell cycle. MV4-11 cells were labeled with Hoechst 33342 at day 3 post-transmission and evaluated by flow cytometry. H, flow cytometry analysis of apoptosis after transduction with indicated lentiviruses at day 3 in MV4-11. I, experimental scheme for (J–N). YFP+ cells were isolated from MLL-AF9-YFP AML mice and used in the experiment. J, quantitative RT–PCR showed the efficiency of Usp9x knockdown and Alkbh5 mRNA level. K, Western blotting showed the efficiency of Usp9x knockdown and Alkbh5 level in MLL-AF9-YFP cells after transfection with shRNA lentivirus targeting Usp9x. L, colony-forming assay of murine leukemia cells after transduction with the indicated lentiviruses. Scale bars represent 100 μm. M, percentages of YFP+ cells in peripheral blood (PB) at the indicated time after transplantation (n = 5). N, Kaplan–Meier survival plot of recipients transplanted with MLL-AF9-YFP+ AML cells transduced with indicated lentiviruses (n = 5). The log-rank test was used to compare survival curves. The data in (E, F, G, H, J, and L) were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. The error bars represent mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. AML, acute myeloid leukemia; BM, bone marrow; TCGA, The Cancer Genome Atlas; USP9X, ubiquitin-specific peptidase 9X.
We further studied whether Usp9x is required for murine AML development. We knocked down Usp9x in primary leukemia cells from MLL-AF9–induced AML mice (Fig. 4I). The efficiency of Usp9x knockdown was confirmed at the protein and mRNA levels (Fig. 4, J and K). We also found that knockdown of Usp9x obviously reduced Alkbh5 protein level but not its mRNA level in murine leukemia cells (Fig. 4, J and K). As expected, Usp9x knockdown inhibited the clonogenic ability of leukemia cells (Fig. 4L). We further transplanted leukemia cells upon Usp9x knockdown into sublethally irradiated recipient mice and found that the leukemia burden in recipients of leukemia cells with Usp9x knockdown was significantly lower than that of the control group (Fig. 4M), and knockdown of Usp9x also significantly delayed AML development (Fig. 4N). Together, these data indicate that USP9X is required for maintaining leukemia cell survival and AML development.
ALKBH5 partially mediates the function of USP9X in AML
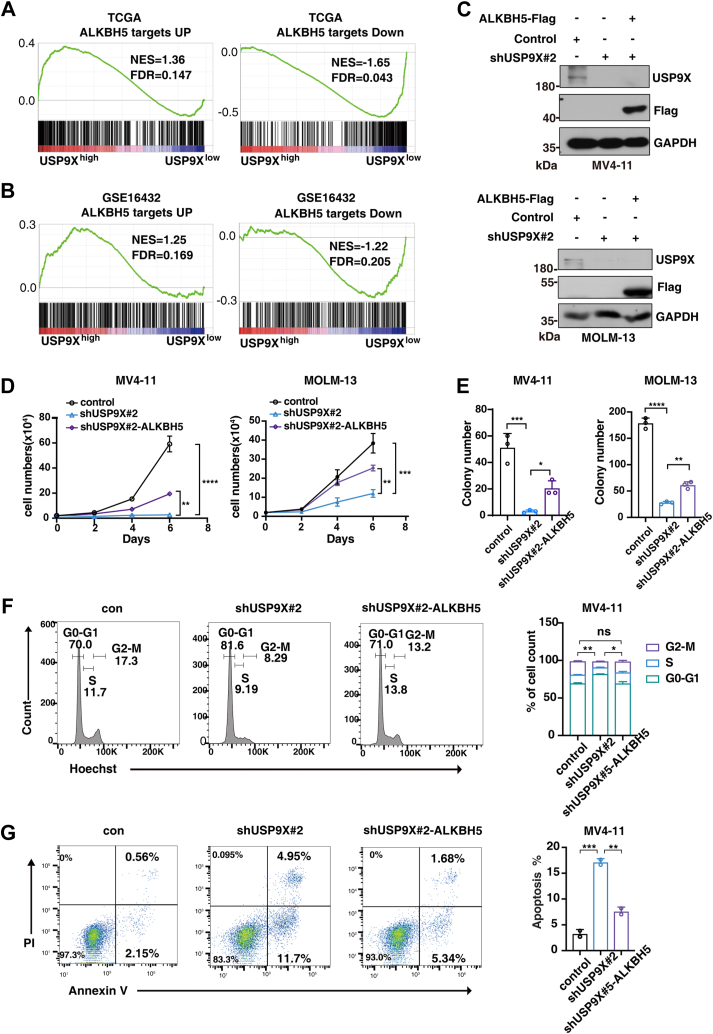
Given that USP9X stabilizes ALKBH5, we asked whether ALKBH5 mediates USP9X function in AML. By interrogating public datasets (GSE14632 and The Cancer Genome Atlas Program for two AML patient cohorts), interestingly, we observed significant enrichment of upregulated ALKBH5 targets in USP9Xhigh AML patients; in contrast, downregulated ALKBH5 targets were enriched in USP9Xlow AML patients (Fig. 5, A and B). Next, we tested whether ALKBH5 mediates the function of USP9X in AML cells. We expressed ALKBH5 in USP9X-knockdown leukemia cells (Fig. 5C) and found that ectopic expression of ALKBH5 partially rescued the clonogenic and proliferation defects caused by USP9X deficiency (Fig. 5, D and E). Moreover, ectopic expression of ALKBH5 also prevented apoptosis and restored cell cycling of USP9X-deficient leukemia cells (Fig. 5, F and G). We further reintroduced K57R-mutant form of ALKBH5 in MOLM-13; as expected, the restoration of ALKBH5-K57R overexpression gave rise to a slightly better rescue effect, indicating that cells expressing ALKBH5-K57R resist USP9X depletion-induced reduction in cellular growth and clonogenic ability (Fig. S4, A–C). Overall, these data indicate that ALKBH5 is one of the functional downstream target genes of USP9X in AML.
Figure 5.
ALKBH5 partially mediates the function of USP9X in AML.A and B, GSEA plot showing enrichment of ALKBH5 targets in USP9Xhighversus USP9Xlow groups from AML patient cohorts TCGA (A), and GSE16432 (B). C, MV4-11 and MOLM-13 cells were transducted with indicated lentiviruses. Cellular extracts were prepared and analyzed by Western blotting. D, growth curve of MV4-11 and MOLM13 after transduction with indicated lentiviruses. E, colony-forming assay of MV4-11 and MOLM13 cells after transduction with indicated lentiviruses. F, flow cytometry analysis of cell cycle after transduction with indicated lentiviruses at day 3 in MV4-11. G, flow cytometry analysis of apoptosis after transduction with indicated lentiviruses at day 3 in MV4-11. The data in (D–G) were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Error bars denote mean ± SD. AML, acute myeloid leukemia; GSEA, gene set enrichment analysis; TCGA, The Cancer Genome Atlas; USP9X, ubiquitin-specific peptidase 9X.
Discussion
ALKBH5 is one of RNA m6A erasers and plays a key role in maintaining RNA m6A homeostasis. Our previous experiments have confirmed the abnormally high expression of ALKBH5 in AML and the role of ALKBH5 in AML promotion. Therefore, exploring the underlying mechanisms of how ALKBH5 expression is regulated is a key scientific question. In here, we identify USP9X as a key deubiquitinase for ALKBH5 and find that USP9X stabilizes ALKBH5 protein by removing K48-linked polyubiquitin chains of ALKBH5 at K57. We also find that USP9X is required for maintaining leukemia cell survival and AML development, which is partially mediated by ALKBH5. Therefore, our findings uncover a new mechanism for regulating ALKBH5 expression and reveal a key role of USP9X in AML, which might provide a promising therapeutic strategy for AML treatment in clinic.
This work reveals the underlying mechanism of how ALKBH5 expression is regulated at the protein level. Using MS, the unbiased approach, we identified USP9X as key deubiquitinase for ALKBH5. Given the important function of ALKBH5 in multiple malignancies, such as breast cancer (30, 31), glioma (32, 33), and AML (7, 13), ALKBH5 is an attractive therapeutic target for the cancer treatment. Our findings in here indicate that inhibiting USP9X might be an alternative approach for targeting ALKBH5. Previous study indicates that the SUMO E2-conjugating enzyme UBC9 and E3 ligase, PIAS4, mediate ALKBH5 SUMOylation, which significantly inhibits its demethylase activity (34). Interestingly, we found that, unlike SUMOylation, ubiquitination of ALKBH5 at K57 residue does not affect its demethylase activity. In addition, we also observed that USP9X may affect K63-linked polyubiquitination of ALKBH5, its function needs to be further investigated. Moreover, although we identify USP9X as a key deubiquitinase for ALKBH5, currently, we do not find the effective E3 ubiquitin ligases for ALKBH5, which definitely needs to be explored in the future. Overall, our work indicates that fully understanding the regulatory mechanism of ALKBH5 expression at post-translational level would make it possible to target ALKBH5.
Our findings also reveal the important role of USP9X in promoting AML development. USP9X was found to stabilize MCL1 and promotes lymphoma cell survival (22). USP9X also binds to the amino-terminal extensions of phosphatase and tensin homolog (PTEN)α/β and deubiquitinate lysines 235 and 239 in PTENα to regulate PTENα/β stability and promote tumorigenesis (26). In here, our work clarifies the role of USP9X in AML. We also confirmed that ALKBH5 is one of the functional downstream targets of USP9X in leukemia. As expression of ALKBH5 can only partially rescue the phenotypes of USP9X-deficient leukemia cells, the other targets of USP9X in leukemia should be further investigated. Overall, our findings uncover USP9X as a key deubiquitinase for stabilizing ALKBH5 and maintaining AML cell survival.
Experimental procedures
Reagents and antibodies
USP9X (Abcam; catalog no.: ab180191), GAPDH (Proteintech; catalog no.: 60004-1-Ig), ALKBH5 (Sigma; catalog no.: HPA007196), Ubiquitin (P4D1) (catalog no.: sc-8017), K48-linkage specific polyubiquitin (CST; catalog no.: 8081), m6A (Abcam; catalog no.: ab151230), FLAG (Sigma; catalog no.: F2922), HA (Sigma; catalog no.: B9183), GFP (Proteintech; catalog no.: 66002-1-Ig), horseradish peroxidase (HRP)-mouse (Proteintech; catalog no.: 00014-1-Ig), HRP-rabbit (CST; catalog no.: 7074), HRP-mouse (light-chain specific) (ROCKLAND; catalog no.: 38282), HRP-rabbit (light-chain specific) (CST; catalog no.: 93702), goat anti-Rabbit IgG antibody (Sigma; catalog no.: AP132). WP1130 (MedChemExpress; catalog no.: HY-13246), CHX (Sigma; catalog no.: 01810), benzonase nuclease (Sigma; catalog no.: E8263), and 3× FLAG peptide (Sigma; catalog no.: F4799).
Plasmid constructions
Lentivirus pLKO.1, pCDH-CMV-puro, and pHKO-23 vectors were used. Expression plasmids for FLAG-tagged ALKBH5 WT, 66 to 292, 66 to 394, and 0 to 292 complementary DNA (cDNA) were cloned by PCR using specific primers and inserted into pCDH vector. USP9X segments were cloned by standard molecular cloning methods in pEGFP-N1 vector. For the ALKBH5 rescue experiment, human ALKBH5 WT or K57R cDNA was cloned into vector following shALKBH5-3′, and ALKBH5 cDNA was cloned into vector following shUSP9X#2. HA-Ub (WT, K6 only, K11 only, K27 only, K29 only, K33 only, K48 only, and K63 only; K6R, K11R, K27R, K29R, K33R, K48R, and K63R) were generously provided by Dr Bo Zhong (Wuhan University).
The shRNAs of USP9X, ALKBH5, and Usp9x were cloned into pLKO.1 according to standard molecular biology techniques. The following sequences of mRNA were targeted: human USP9X#1: 5′-GGTCGTTACAGCTAGTATTTA-3′;
Human USP9X#2: 5′-GAGAGTTTATTCACTGTCTTA-3′;
Human USP9X#3: 5′-CGATTCTTCAAAGCTGTGAAT-3′;
Human ALKBH5#3′UTR: 5′-GAAAGGCTGTTGGCATCAATA-3′;
Mouse shUsp9x shRNA#1: 5′-GATAATTGCAGCCCTTATTAA-3′;
Mouse shUsp9x shRNA#2: 5′-TCGTAATGTATGCCAATTTAG-3′.
Cell culture and lentivirus production
The human AML cell lines (MOLM-13, Kasumi, and MV4-11) were maintained in RPMI1640 or Iscove’s modified Dulbecco’s medium with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. 293T were cultured in Dulbecco's modified Eagle's medium (Hyclone), supplemented with 10% FBS, and 1% penicillin and streptomycin.
293T cells plated on 100 mm dishes were transfected with the indicated lentiviral plasmids together with the pSPAX2 and pMD2.G plasmids. The culture medium was replaced with new medium without antibiotics at 12 h after transfection. After additional 48 h, the viruses were harvested to infect MV4-11 cells or MOLM-13 cells in the presence of 8 μg/ml polybrene (Sigma; catalog no.: H9268). The infected MV4-11 cells or MOLM-13 cells were selected with 2 μg/ml puromycin (Merck; catalog no.: 540411) at least 2 days.
Co-IP and immunoblot analysis
For transient transfection and co-IP experiments, cells were harvested and lysed with IP buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol, 1 mM DTT, 1 mM PMSF, and 1× cocktail). After brief sonication, the lysates were centrifuged at 12,000 rpm for 10 min at 4 °C. For IP of FLAG-tagged proteins, whole cell lysates were incubated with anti-FLAG M2 Affinity Gel (Bimake; catalog no.: B23102) for 2 h at 4 °C. Otherwise, supernatants were incubated with indicated antibodies and protein G-agarose beads (Thermo; catalog no.: 10004D) at 4 °C overnight. The beads were then washed for three times with 1 ml lysis buffer containing 0.5 M NaCl, boiled in sample buffer, and subjected to immunoblot assay. The cell lysates or pull-downed proteins were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk for 30 min at room temperature and incubated with the specified antibodies at 4 °C overnight. Membranes were subsequently incubated with HRP-conjugated secondary antibodies (Cell Signaling Technology) at room temperature for 1 h. The signals were detected by the luminescent image analyzer LAS-4000 mini (Fujifilm).
Ubiquitination assays
Ubiquitination assays were done following a denaturing IP protocol. FLAG-tagged ALKBH5 or HA-Ub were cotransfected into 293T cells with or without depletion of USP9X for 24 h. Then cells were lysed in lysis buffer containing 1% SDS and denatured by 95 °C heating for 10 min. The samples were centrifuged at 12,000g for 10 min, and then the supernatants were diluted with lysis buffer until SDS concentration was decreased to 0.1% for IP with anti-FLAG M2 affinity gel (Bimake; catalog no.: B23102). The immunoprecipitates were analyzed by immunoblots with the indicated antibodies.
Quantitative RT–PCR
Total RNA was isolated from cells using TRIzol (Takara Bio; catalog no.: 9109), reverse-transcribed (TOYOBO; catalog no.: FSQ-101), and subjected to quantitative PCR analysis to measure mRNA levels of the tested genes with SYBR Green Mix (Bio-Rad; catalog no.: 172-5274). Data shown are the relative abundance of the indicated mRNA normalized to that of human GAPDH and mouse GAPDH, respectively. Quantitative PCR was performed using the following primers:
Human USP9X: 5′-TCGGAGGGAATGACAACCAG-3′ (forward) and 5′-GGAGTTGCCGGGGAATTTTCG-3′ (reverse);
Human ALKBH5: 5′-ATCCTCAGGAAGACAAGATTAG-3′ (forward) and 5′-TTCTCTTCCTTGTCCATCTC-3′ (reverse);
Human GAPDH: 5′-GAGTCAACGGATTTGGTCGT-3′ (forward) and 5′-GACAAGCTTCCCGTTCTCAG-3′ (reverse);
Mouse Usp9x: 5′-TCCAACAGAATCAGACTTCATCG-3′ (forward) and 5′-TGGAAATGCAGGTTCCTCATCT-3′ (reverse);
Mouse Alkbh5: 5′-CGCGGTCATCAACGACTACC-3′ (forward) and 5′-ATGGGCTTGAACTGGAACTTG-3′ (reverse);
Mouse Gapdh: 5′-CATCACTGCCACCCAGAAGACTG-3′ (forward) and 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′ (reverse).
Cell proliferation and in vitro colony-forming assay
For leukemia cell proliferation assays, human leukemia cells (MV4-11 and MOLM-13) were transduced with lentivirus and selected with 2 μg/ml puromycin (Merck; catalog no.: 540411) for 2 days. After selection, cells were seeded into 24-well plates at the concentration of 20,000 cells per well in triplicates. Cell proliferation was assessed by counting cell numbers every 2 days. For colony-forming assay, mouse bone marrow (BM) cells were cultured in MethoCult M3434 (STEMCELL Technologies; catalog no.: 03434) methylcellulose medium. For cell lines, transduced cells were plated in triplicate in 1% methylcellulose medium supplemented with 100 IU/ml penicillin and 100 μg/ml streptomycin as well as 10% FBS. Colonies were evaluated and scored after 5 to 7 days of incubation.
m6A dot blot
For m6A dot blot, total RNA was extracted from cell aliquots using TRIzol. RNA samples were quantified by Nanodrop (Thermo Fisher Scientific) and UV crosslinked to the membrane, and membrane was blocked with 5% nonfat dry milk (in 1× PBS) for 1 h and incubated with a specific anti-m6A antibody (1:1000 dilution; Abcam, catalog no.: ab180191) for 2 h at room temperature. HRP-conjugated secondary antibodies were added to the blots for 1 h at room temperature, and the membrane was developed with ECL Western Blotting Substrate (Bio-Rad) and exposure with X-Ray Super RX Films (Fujifilm).
Flow cytometry analysis
Leukemia cells were stained with Hoechst 33342 (Thermo Fisher; catalog no.: H3570) to analyze cell cycle using flow cytometry. To analyze apoptosis, cells were stained with Annexin V, and propidium iodide (BD Biosciences; catalog no.: 556547) was added before flow cytometric analysis. All flow cytometric analyses and cell sorting were performed on the BD Fortessa X-20 or BD FACS Aria III in the Core Facility of Medical Research Institute, Wuhan University, and data were analyzed with FlowJo software (BD Biosciences).
MS
FLAG-tagged ALKBH5 stably expressed in HEK293T and MV4-11 cells were harvested and lysed with IP buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol, 1 mM DTT, 1 mM PMSF, and 1× cocktail). After brief sonication, the lysates were centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants were treated with a mix of 25 U/ml benzonase (Sigma; catalog no.: E8263) and 2 mM Mg2+ at for 30 min 37 °C that inducing RNA and DNA digested into single nucleosides. And then the supernatants were immunoprecipitated with anti-FLAG affinity gel (Bimake; catalog no.: B23102) and boiled in sample buffer. Eluted proteins were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. Gel slices including all proteins were excised. LC–MS/MS analysis was performed in Novogene Co, Ltd. Briefly, proteins extracted from gels were digested with trypsin in 50 mM ammonium bicarbonate at 37 °C overnight. After treatment with 5 mM DTT and 11 mM iodoacetamide, the resulting peptides were separated by silica capillary column and eluted at a flow rate of 0.3 μl/min with the UltiMate 3000 HPLC system (Thermo Fisher Scientific) coupled with the Q Exactive mass spectrometer (Thermo Fisher Scientific), which was set in the data-dependent acquisition mode using Xcalibur 2.2 software (Thermo Fisher Scientific).
Protein expression and purification
The protocol for protein purification was previously described (35). FLAG-tagged ALKBH5 WT and K57R were expressed in HEK293T cells. For each protein, four 10 cm dishes of cells were prepared and lysed in 2 ml lysis buffer (50 mM Tris–HCl [pH 7.5], 300 mM KCl, 0.5% NP-40, 5% glycerol, 5 μg/ml DNase I, 1% RNase T1/A, 1% protease inhibitor, and 1 mM DTT) at 4 °C for 1 h and sonicated (5 s on, 5 s off, three cycles). The proteins were affinity purified using 30 μl Anti-FLAG Affinity Gel (Bimake; catalog no.: B23102) at 4 °C for 2 h. After extensive wash with wash buffer (50 mM Tris–HCl, pH 7.5. 300 mM KCl, 5% glycerol, and 1 mM DTT), proteins were eluted in 200 μl 1× FLAG elution solution (0.5 μg/ml FLAG peptide in wash buffer) (Sigma; catalog no.: F4799) at 4 °C for 1 h. Protein purity was verified with SDS-PAGE followed by Coomassie staining.
In vitro demethylation assays
The activity assay was performed as reported (3). ssRNA with m6A was transcribed by HiScribe T7 Quick High Yield RNA Synthesis Kit (Abcam; catalog no.: E2050S) and m6A (TriLink; catalog no.: N-1013-1), according to the manufacturer's instructions. Briefly, ssRNA with m6A and FLAG-tagged human ALKBH5 were purified from HEK293T cells were incubated with a mix of l-ascorbic acid (100 μM), (NH4)2Fe(SO4)2·6H2O (50 μM), α-ketoglutarate (100 μM), Tris(2-carboxyethyl)phosphine (1 mM), RNasin (0.4 U/μl), and 50 mM of Hepes buffer (pH 7.0) for 2 h at 37 °C. Then the samples were analyzed by Dot Blot.
Primary AML patient and cord blood samples
AML patient samples were collected from BM aspirations with informed consent. Mononuclear cells were isolated by density gradient centrifugation with Ficoll (GE Healthcare Life Science). Normal cord blood units, designated for research use, were obtained from Tongji Medical College. All experiments referring to human samples were conducted in compliance with all relevant ethical regulations and were approved by the ethics committees of medical research of the universities. The cells were harvested and boiled in sample buffer. The samples were analyzed by immunoblots with the indicated antibodies.
Murine MLL-AF9 leukemia model
BM cells extracted from MLL-AF9-YFP+ mice were lysed with RBC and infected with shUsp9x lentivirus in the presence of 8 μg/ml Polybrene (Sigma; catalog no.: H9268), 10 ng/ml IL-3 (PeproTech; catalog no.: 213-13), 10 ng/ml IL-6 (PeproTech; catalog no.: 216-16), and 20 ng/ml stem cell factor (PeproTech; catalog no.: 250-03). The infected cells were transplanted into C57BL/6J (CD45.2) (Jackson Laboratory; catalog no.: 000664) mice irradiated with sublethal dose (4 Gy + 4 Gy) via tail vein injection. Male congenic recipient mice (CD45.2) at 8 to 10 weeks old were used for AML transplantation. The YFP+ leukemia cells in peripheral blood were analyzed every week. All mice were bred and maintained in Animal Center of Medical Research Institute at Wuhan University. All animal experiments were performed according to protocols approved by the Animal Care and Use Committee of Medical Research Institute, Wuhan University.
Statistical analysis
Statistical analysis was assessed by ANOVA test using GraphPad Prism 8.3 software (GraphPad Software, Inc). The log-rank test was used to compare survival curves. All experiments were reproduced at least three times. Data are presented as means ± SD. p Values of less than 0.05 were considered statistically significant. In the figures, asterisks indicate ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Data availability
All data are contained within this article and available from the corresponding author on reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the members of our laboratory for helpful discussion. This work is supported by the grants from the National Key R&D Program of China (grant no.: 2022YFA0103200), the National Natural Science Foundation of China (grant no.: 82230007), and the Fundamental Research Funds for the Central Universities (grant nos.: 2042022dx0003 and 2042021kf0225). This work is also supported by the grants from the Hubei Provincial Natural Science Fund for Creative Research Groups (grant no.: 2021CFA003) and the Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (grant no.: TFJC2018005). We also thank all the staff in the core facility of Medical Research Institute at Wuhan University for their technical support.
Author contributions
P. W., J. W., and H. Z. conceptualization; P. W., J. W., and H. Z. methodology; P. W., J. W., and H. Z. formal analysis; P. W., J. W., S. Y., M. C., Y. C., W. L., Z. G., and J. H. investigation; P. W., J. W., H. Z., and J. Z. writing–original draft; J. Z. and H. Z. supervision.
Reviewed by members of the JBC Editorial Board. Edited by Mike Shipston
Supporting information
References
- 1.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millan-Zambrano G., Robson S.C., et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su R., Dong L., Li Y., Gao M., Han L., Wunderlich M., et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Li Y., Wang P., Han G., Zhang T., Chang J., et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell. 2020;27:81–97.e8. doi: 10.1016/j.stem.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Chen Y., Sun B., Wang L., Yang Y., Ma D., et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 12.Paris J., Morgan M., Campos J., Spencer G.J., Shmakova A., Ivanova I., et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen C., Sheng Y., Zhu A.C., Robinson S., Jiang X., Dong L., et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27:64–80.e9. doi: 10.1016/j.stem.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng Y., Wei J., Yu F., Xu H., Yu C., Wu Q., et al. A critical role of nuclear m6A reader YTHDC1 in leukemogenesis by regulating MCM complex-mediated DNA replication. Blood. 2021;138:2838–2852. doi: 10.1182/blood.2021011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mevissen T.E.T., Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 16.Sun H.L., Zhu A.C., Gao Y., Terajima H., Fei Q., Liu S., et al. Stabilization of ERK-phosphorylated METTL3 by USP5 increases m(6)A methylation. Mol. Cell. 2020;80:633–647.e7. doi: 10.1016/j.molcel.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong J., He J., Zhu J., Pan J., Liao W., Ye H., et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell. 2022;82:1660–1677.e10. doi: 10.1016/j.molcel.2022.02.033. [DOI] [PubMed] [Google Scholar]
- 18.Du Y., Hou G., Zhang H., Dou J., He J., Guo Y., et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46:5195–5208. doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelcovits A., Niroula R. Acute myeloid leukemia: a review. R. I. Med. J. (2013) 2020;103:38–40. [PubMed] [Google Scholar]
- 20.Dohner H., Weisdorf D.J., Bloomfield C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwickart M., Huang X., Lill J.R., Liu J., Ferrando R., French D.M., et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 23.Shah A., Rashid F., Awan H.M., Hu S., Wang X., Chen L., et al. The DEAD-box RNA helicase DDX3 interacts with m(6)A RNA demethylase ALKBH5. Stem Cells Int. 2017;2017 doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uren P.J., Bahrami-Samani E., de Araujo P.R., Vogel C., Qiao M., Burns S.C., et al. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol. 2016;13:400–411. doi: 10.1080/15476286.2015.1138030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho N.H., Cheveralls K.C., Brunner A.D., Kim K., Michaelis A.C., Raghavan P., et al. OpenCell: endogenous tagging for the cartography of human cellular organization. Science. 2022;375 doi: 10.1126/science.abi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen S.M., Zhang C., Ge M.K., Dong S.S., Xia L., He P., et al. PTENalpha and PTENbeta promote carcinogenesis through WDR5 and H3K4 trimethylation. Nat. Cell Biol. 2019;21:1436–1448. doi: 10.1038/s41556-019-0409-z. [DOI] [PubMed] [Google Scholar]
- 27.Theard D., Labarrade F., Partisani M., Milanini J., Sakagami H., Fon E.A., et al. USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 2010;29:1499–1509. doi: 10.1038/emboj.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics. 2017;44:243–250. doi: 10.1016/j.jgg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W., Tan X., Lin S., Gou Y., Han C., Zhang C., et al. Cplm 4.0: an updated database with rich annotations for protein lysine modifications. Nucleic Acids Res. 2022;50:D451–D459. doi: 10.1093/nar/gkab849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Chen Y., Wang L., Ji S. ALKBH5 promotes the proliferation of glioma cells via enhancing the mRNA stability of G6PD. Neurochem. Res. 2021;46:3003–3011. doi: 10.1007/s11064-021-03408-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F., Wei J., Cui X., Yu C., Ni W., Bungert J., et al. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021;49:5779–5797. doi: 10.1093/nar/gkab415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article and available from the corresponding author on reasonable request.