Abstract
Acidocalcisomes are membrane-bounded, electron-dense, acidic organelles, rich in calcium and polyphosphate. These organelles were first described in trypanosomatids and later found from bacteria to human cells. Some of the functions of the acidocalcisome are the storage of cations and phosphorus, participation in pyrophosphate (PPi) and polyphosphate (polyP) metabolism, calcium signaling, maintenance of intracellular pH homeostasis, autophagy, and osmoregulation. Isolation of acidocalcisomes is an important technique for understanding their composition and function. Here, we provide detailed subcellular fractionation protocols using iodixanol gradient centrifugations to isolate high-quality acidocalcisomes from Trypanosoma brucei, which are subsequently validated by electron microscopy, and enzymatic and immunoblot assays with organellar markers.
Keywords: Trypanosomatids, Trypanosoma brucei, Acidocalcisome, Subcellular fractionation, Organellar markers
1. Introduction
Acidocalcisomes are lysosome-related organelles rich in cations and polyphosphate, which were originally identified in trypanosomatids and have been conserved from prokaryotes to eukaryotes (1,2). Trypanosoma brucei, the causative agent of sleeping sickness or African trypanosomiasis, is an excellent model to study the acidocalcisome because it is the trypanosome that is most amenable to a number of molecular and genetic tools, easy to cultivate, and possesses acidocalcisomes with up to molar levels of PPi and polyP in the two best-studied life-cycle stages: procyclic and bloodstream forms (PCF and BSF) (3). Studies with this parasite have provided fundamental insights into the acidocalcisome biology and its roles in cation and phosphorus storage, calcium signaling, pH and osmotic regulation, response to stress, autophagy, and growth and infectivity (1–4).
Subcellular fractionation, a practical approach for obtaining purified organelles, is accomplished by cell membrane lysis and density gradient centrifugation to separate the organelles from other contaminating subcellular structures. Several gradients, such as Nycodenz®, sucrose and Percoll®, have been applied for the isolation of mitochondria, lysosomes, Golgi, glycosomes or endoplasmic reticulum from parasitic protozoa (5,6). Percoll® was initially used to isolate acidocalcisomes from trypanosomes (7), but was effectively replaced by iodixanol (OptiPrep™) in our laboratory (8,9). Iodixanol is ideal for density gradient isolation of intact subcellular organelles from tissues or cells (10), since it is a true solute that forms a self-generated gradient capable of maintaining the environment of different organelles close to isoosmoticity, that does not cause co-sedimentation of colloidal silica particles with high-density organelles (such as acidocalcisomes), and that does not interfere with most downstream enzymatic assays and electro-blotting analyses without removal. In order to minimize or eliminate other membrane and organelle contaminations, we have recently developed a new iodixanol-based protocol for subcellular fractionation to isolate high-quality acidocalcisomes from PCF trypanosomes through the modification of our previous method (8,9) using additional gradient centrifugation with higher density iodixanol (Fig. 1). After fractionation, the yield and purity of isolated acidocalcisomes were analyzed by enzymatic and immunoblotting assays using organelle markers for acidocalcisomes (vacuolar proton pyrophosphatase-TbVP1), mitochondria (succinate cytochrome c reductase, voltage-dependent anion channel-TbVDAC), glycosomes (hexokinase, pyruvate phosphate dikinase-TbPPDK) and lysosomes (α-mannosidase, Tbp67). The results have validated this method that can efficiently separate acidocalcisomes from other organelles, significantly enhancing their purification (Table 1) by eliminating or decreasing mitochondrial, glycosomal and lysosomal contaminations (Figs. 2 and 3). Our protocols, described here, have successfully been used for isolation and characterization of acidocalcisomes from T. brucei, but can be easily adapted to other trypanosomatids.
Fig. 1.
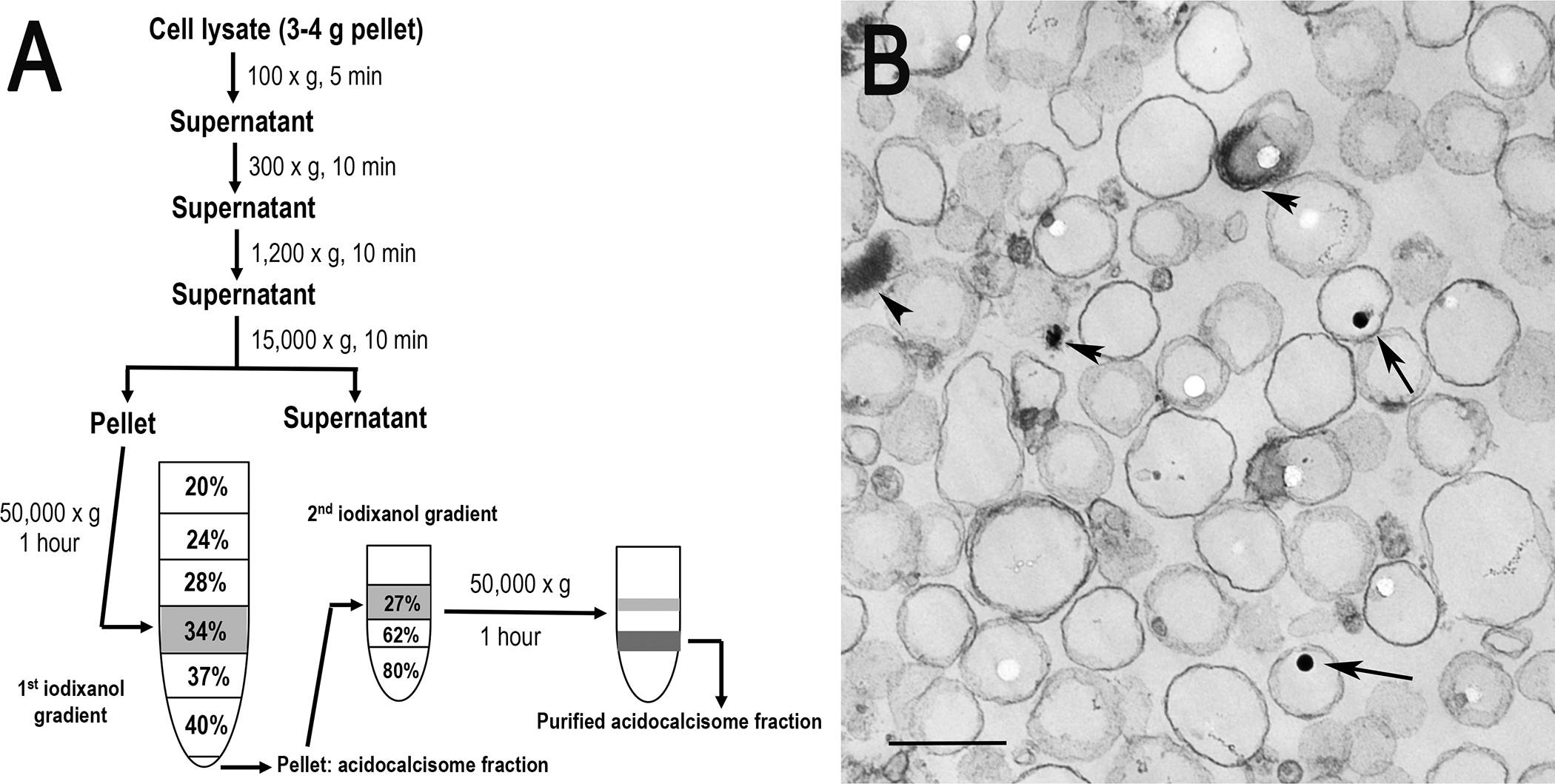
Subcellular fractionation of acidocalcisomes. (a) Trypanosome lysates are obtained by grinding with silicon carbide, decanting by low speed centrifugation to eliminate debris and silicon carbide, and centrifuging at 15,000 × g for 10 min to isolate the organellar fraction that is applied to the 34% step of a discontinuous iodixanol gradient. After centrifugation at 50,000 × g for 1 h, the pellet is resuspended and applied to the 27% step of a second iodixanol gradient and centrifuged at 50,000 × g for 1 h. Aliquots from each fraction are used for enzymatic assays and Western blot analyses. (b) Electron microscopy of acidocalcisome fraction prepared by the iodixanol procedure (fraction 5). Scale bar = 0.2 μm. Arrows and arrowheads show electron-dense material inside acidocalcisomes (reproduced from Huang et al. (2014) (ref. 3) with permission from the Public Library of Science).
Table 1.
Purification of acidocalcisomes on iodixanol step gradients.
| Yield (%) | Purification-fold | |||
|---|---|---|---|---|
| 1st | 2nd | 1st | 2nd | |
| Protein (mg) | 0.14 (3) | 0.05 (3) | ||
| Pyrophosphatase* | 9.81 (3) | 4.96 (3) | 70 | 99 |
| Succinate cytochrome c reductase | 0.12 (3) | 0.02 (3) | 1 | 0.2 |
| Hexokinase | 0.46 (3) | 0 (3) | 3 | 0 |
| α-mannosidase | 0.27 (3) | 0.08 (3) | 2 | 1.6 |
Yield values are percentages relative to the 15,000 × g pellet fraction and represent averages from number of preparations in parentheses.
Pyrophosphatase activities in the 15,000 × g pellet, and the 1st and 2nd gradient acidocalcisome preparations were 0.22 ± 0.09, 15.6 ± 3.2, and 22.6 ± 2.1 μmol min−1 mg−1 protein, respectively (mean ± SD) (reproduced from Huang et al. (2014) (ref. 3) with permission from the Public Library of Science).
Fig. 2.
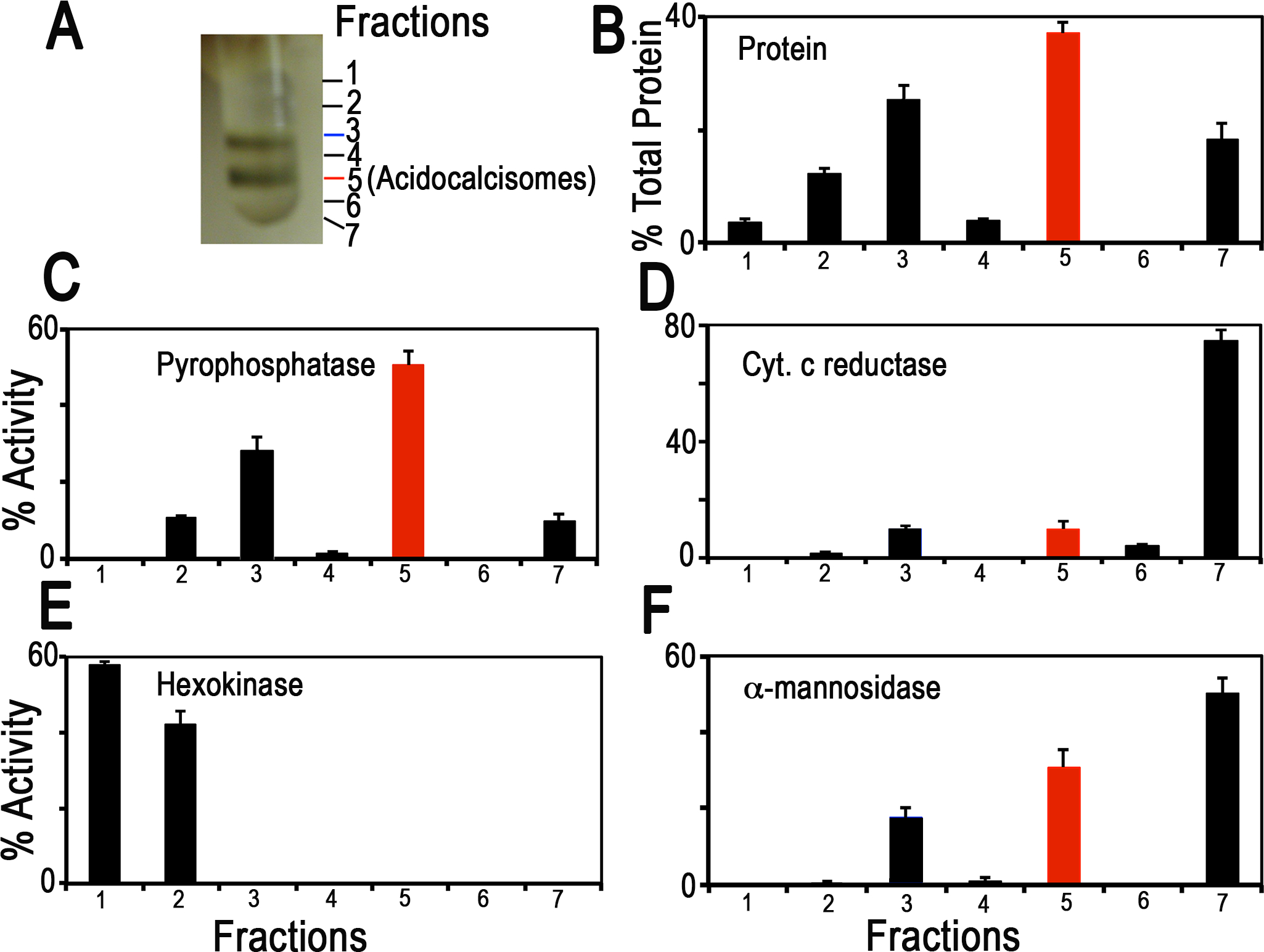
Distribution on iodixanol gradients of organellar markers from PCF trypanosomes. (a) Photograph showing bands obtained after the second iodixanol gradient centrifugation. Fraction 5 corresponds to the purified acidocalcisomes. (b) Protein distribution. (c) TbVP1 activity (measured as the AMDP-sensitive Pi release) is concentrated in fractions 3 and 5. (d) Mitochondrial marker distribution, succinate cytochrome c reductase. (e) Glycosomal marker distribution, hexokinase. (f) Lysosomal marker distribution, α-mannosidase. In (b-f) the y-axis indicates relative distribution; the x-axis indicates fraction number; bars show means ± SD (as a percentage of the total recovered activity) from two or three independent experiments (reproduced from Huang et al. (2014) (ref. 3) with permission from the Public Library of Science).
Fig. 3.
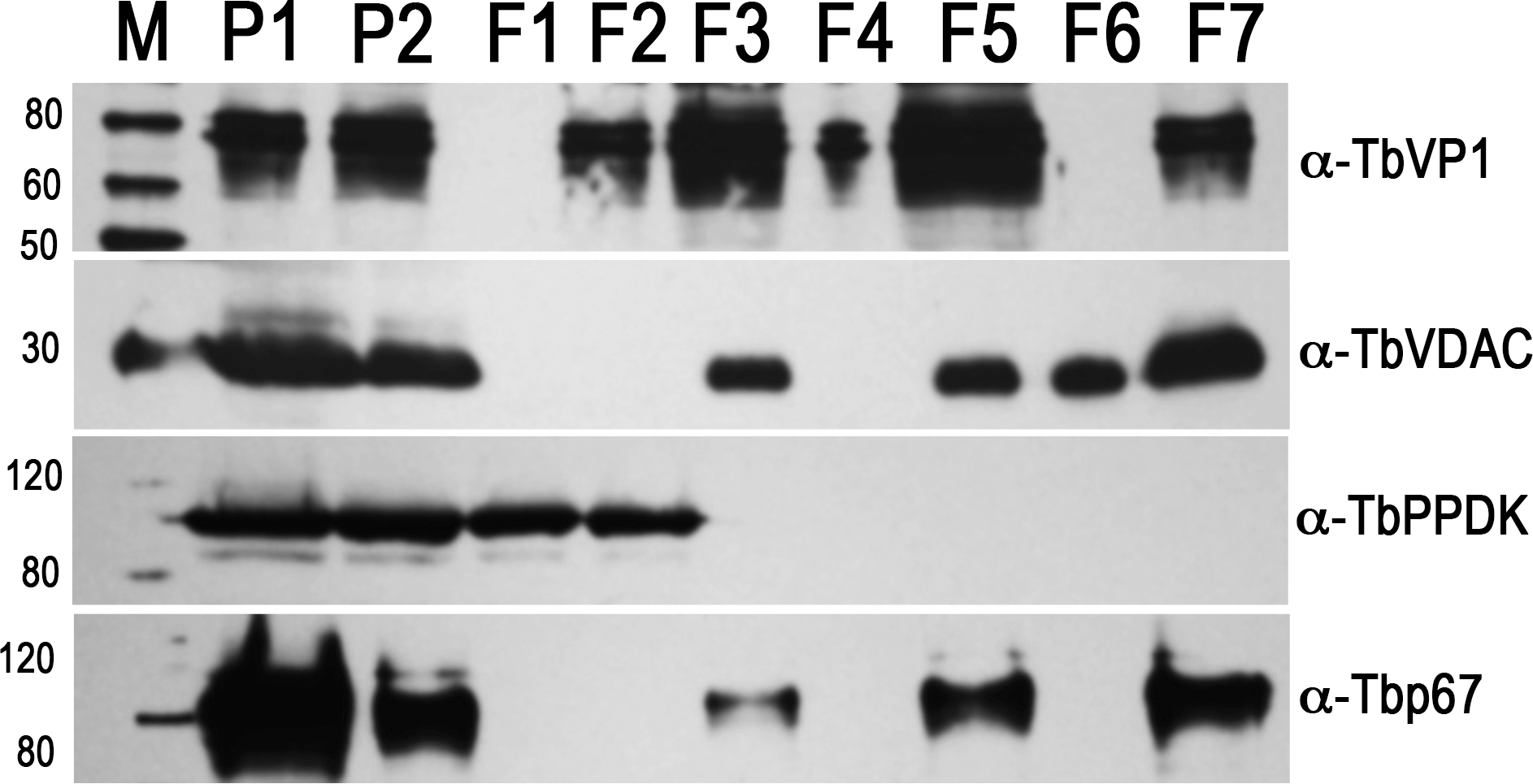
Western blot analyses of subcellular fractions using antibodies against acidocalcisome marker TbVP1, mitochondrial marker voltage-dependent anion channel (TbVDAC), glycosomal marker pyruvate, phosphate dikinase (TbPPDK), and lysosome marker Tbp67. P1, the 15,000 x g pellet (30 μg); P2, the first gradient pellet (2 μg); F1 to F7, the second gradient fractions (2 μg each). M, Magic Marker protein standards (reproduced from Huang et al. (2014) (ref. 3) with permission from the Public Library of Science).
2. Materials
Prepare all solutions using ultrapure water (prepared by purifying deionized water, to attain a sensitivity of 18 MΩ-cm at room temperature (RT) and analytical grade reagents. Prepare and store all reagents at RT (unless indicated otherwise).
2.1. Cell Culture and Standard Equipment
T. brucei procyclic form (PCF) trypanosomes (wild type strain 427) grown in SDM-79 medium (11) supplemented with hemin (7.5 μg/mL) and 10% heat-inactivated fetal bovine serum.
Pipettes (20, 200, and 1000 μl, multichannel pipette).
Pipet-Aid.
Serological pipets (5 ml, 10 ml and 25 ml).
Pasteur glass pipettes with cotton.
Eppendorf tubes (1.5 ml and 1.8 ml).
Glass tubes (disposable culture tubes, 4 ml and 10 ml).
Refrigerated centrifuge for microcentrifuge tubes.
Centrifuge with fixed angle rotor.
Ultracentrifuge with swinging–bucket (SW) rotor.
Centrifuge tubes (polycarbonate thick-wall centrifuge tubes, 50 ml).
Ultracentrifuge tubes (polycarbonate thick-wall centrifuge tubes, 38 ml; ultra-clear or polyallomer tubes, 5 ml).
Stainless steel lab spoon spatula. Precooled at −20°C.
Mortar and pestle. Precooled at −20°C.
Glass tissue homogenizer with tight-fitting pestle (2 ml and 5 ml). Precooled at 4°C.
Tissue homogenizer mixer with stand.
96-well microplate.
No. 5 filter paper.
Microplate Reader.
Microplate incubator shaker.
Nitrocellulose membranes.
Mini-Gel electrophoresis system.
Trans-Blot apparatus.
2.2. Reagents
2.2.1. Gradient Centrifugations
Complete, EDTA-free, protease inhibitor cocktail tablet. Store at 4°C.
Buffer A with glucose (BAG): 116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 5.5 mM glucose, 50 mM Hepes (free acid), pH 7.2. Store at 4°C (see Note 1).
Isolation buffer (IB) (see Note 2): 125 mM sucrose, 50 mM KCl, 4 mM MgCl2, 0.5 mM EDTA, 5 mM dithiothreitol (DTT), 20 mM Hepes-KOH, pH 7.2, and 1 x protease inhibitor cocktails (see Note 3). Store at 4°C.
OptiPrep™ solution: 125 mM sucrose, 300 mM KCl, 24 mM MgCl2, 3.0 mM EDTA, 30 mM dithiothreitol (DTT) (see Note 3), 120 mM Hepes-KOH, pH 7.2. Store at 4°C.
OptiPrep™ (60% w/v iodixanol, 1.32 g/ml). Store at 4°C.
Working solution: 50% w/v iodixanol, 5 Vol of 60% w/v iodixanol plus 1 vol of OptiPrep™ solution (see Note 4). Freshly prepared. Store at 4°C.
OptiPrep™: 90% w/v iodixanol. Dry 60% w/v iodixanol solution completely at 70°C and then resuspend in IB. Store at 4°C.
Silicon carbide, ~400 mesh. Store at −20°C
Sucrose solutions: 10% w/v sucrose (5 ml) and 1 M sucrose (5 ml) in IB.
RIPA buffer: 150 mM NaCl, 20 mM Tris/HCl, pH 7.5, 1 mM EDTA, 1% SDS, 0.1% Triton X-100, and 1 x protein inhibitor cocktails. Freshly prepared.
Bovine serum albumin (BSA) standards: 1 mg/ml, dilute to a series of final concentrations of 0, 50, 200, 400, 600, and 800 (μg/ml). Store at 4°C.
2.2.2. Electron Microscopy
50% Glutaraldehyde solution. Store at 4°C.
16% Paraformaldehyde, methanol free.
0.2 M Sodium cacodylate buffer, pH 7.4. Store at 4°C.
Fixation solution: 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. Freshly prepared (see Note 5).
2.2.3. Marker Enzyme Assays
10 mM KH2PO4 for phosphate standards. Store at 4°C.
100 mM Potassium pyrophosphate (PPi). Store at −20°C.
10 mM Aminomethylenediphosphonate (AMDP): synthesized by Michael Martin (University of Illinois at Urbana-Champaign). Store at −20°C
PPase reaction buffer: 130 mM KCl, 2 mM MgCl2, 10 mM Hepes-KOH, pH 7.2.
0.045% (w/v) Malachite green (MG): dissolve 45 mg malachite green oxalate salt in 100 ml H2O and stir for at least 30 min. Store in dark bottle.
4.2% (w/v) Ammonium molybdate (AM) in 4 N HCl: dissolve 4.2 g ammonium molybdate in 100 ml 4 N HCl.
0.2 M Succinate (pH 7.2): dissolve 2.36 g succinic acid in 100 ml H2O, and then adjust pH to 7.2 with 5 M NaOH.
1 M Hepes-KOH, pH 7.5.
10 mM KCN: dissolve 6.5 mg of KCN in 10 ml H2O under a fume hood (see Note 6). Freshly prepared.
5 mM Cytochrome c from equine heart. Store at −20°C.
Succinate-cytochrome c reductase (SCR) reaction medium: 0.1 mM cytochrome c, 0.3 mM KCN, and 40 mM Hepes-KOH pH 7.5. Freshly prepared. Store at 4°C.
60 mM ATP, disodium salt solution. Freshly prepared. Store at −20°C
60 mM NADP+. Freshly prepared. Store at −20°C
Glucose-6-phosphate dehydrogenase, type VII, from baker’s yeast (250 units). Store at −20°C.
Hexokinase (HK) enzyme solution: 0.5–1 unit/μl of hexokinase in cold H2O. Prepared immediately before use. Store at −4°C.
Hexokinase (HK) assay medium: 10 mM D-glucose, 0.6 mM ATP, 0.6 mM NADP+, 10 mM MgCl2, 2.5 units/ml glucose-6-phosphate dehydrogenase, and 50 mM Hepes-KOH, pH 7.8. Freshly prepared. Store at 4°C.
0.4 M Sodium acetate buffer, pH 4.6: dissolve 5.44 g sodium acetate trihydrate in 98 ml H2O, add 1.2 ml glacial acetic acid, and then adjust pH to 4.6 with NaOH.
12.5 mM p-Nitrophenol. Freshly prepared. Store at −20°C.
60 mM p-Nitrophenyl-α-D-mannopyranoside (pNP-Man). Store at −20°C.
1 M Na2CO3.
α-Mannosidase (AMA) reaction medium: 200 mM sodium acetate buffer, pH 4.6, and 0.6 mM pNP-Man. Freshly prepared. Store at 4°C.
2.2.4. Immunoblotting
10% SDS-PAGE gel.
SDS-PAGE running buffer: 25 mM Tris-HCl, pH 8.3, 192 mM glycine, 0.1% SDS.
MagicMark protein ladder. Store at −20°C.
2× Laemmli sample buffer.
PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4.
PBS-T: PBS containing 0.1% Tween-20.
Blocking buffer: PBS-T containing 10% non-fat milk.
Western blot transfer buffer: 25 mM Tris base, 192 mM glycine, 0.01% SDS (see Note 7), 20% methanol.
Horseradish peroxidase conjugated anti-mouse or anti-rabbit IgG (H+L) antibody. Store at −20°C.
Pierce ECL Western blotting substrate. Store at 4°C.
3. Methods
Isolate and purify acidocalcisomes from trypanosomes using two iodixanol gradient centrifugations (Fig. 1). Carry out all procedures at 4°C unless otherwise specified.
3.1. First Gradient Centrifugation
Collect late mid-log PCF trypanosomes (1 L, approximately 3–4 g wet weight) by centrifugation at 1000 × g for 7 min at room temperature (RT).
Wash the trypanosomes twice with BAG at RT.
Wash the trypanosomes once in 25 ml cold isolation buffer (IB) supplied with Complete, EDTA-free, protease inhibitor cocktail.
Transfer the pellet into a pre-cooled mortar using a pre-cooled spoon spatula.
Lyse the washed cells with silicon carbide in IB in a pestle and mortar by grinding for approximately 60 seconds (see Note 8).
Transfer the lysed cells with silicon carbide into a 38 ml thick-wall centrifuge tube using the spoon spatula.
Rinse the mortar, pestle and spoon spatula with 10 ml IB and transfer the lysate-silicon mixture into the 38 ml centrifuge tube (see Note 9).
Resuspend the lysate-silicon mixture (approximately 13 ml) by adding IB to a final volume of 25 ml, incubate it for 20 min to settle some particles out of the mixture, and transfer the supernatant into a new centrifuge tube for centrifugation (see Note 9).
Centrifuge the lysed cells in IB at 100 × g for 5 min.
Decant the supernatant and centrifuge this at 300 × g for 10 min.
Decant the supernatant and centrifuge this at 1,200 × g for 10 min.
Centrifuge the supernatant at 15,000 × g for 10 min and retain the pellet.
Re-suspend the pellet in 1 ml IB with a Wheaton homogenizer (see Note 10) and apply it to the 34% step of a discontinuous gradient.
Prepare density gradient solutions (4 ml each) containing 20, 24, 28, 34, 37 and 40% (w/v) iodixanol in 10 ml glass tubes using 50% (w/v) iodixanol working solution and 1 M sucrose with IB (see Note 11).
Load gradient solutions in 38 ml polycarbonate (thick-wall) ultracentrifuge tubes, starting from the bottom with the less dense iodixanol, using a glass pipette with Pipet-Aid (see Note 12).
Centrifuge at 50,000 × g for 1 hour in a SW Ti rotor in the ultracentrifuge (see Note 13).
Acidocalcisomes form a pellet at the bottom of the gradient (Fig. 1). Aspirate the gradient and resuspend the pellet in IB for the second gradient purification.
3.2. Second Gradient Centrifugation
Re-suspend the pellet containing crude acidocalcisomes from the first gradient in 700 μl IB with a Wheaton homogenizer (see Note 10) and apply it to the 27% step of another discontinuous gradient (Fig. 1).
Prepare density gradient solutions (1 ml each) containing 27, 62 and 80% (w/v) iodixanol in 4 ml glass tubes by diluting 90% (w/v) iodixanol (see Note 11) and 1.4 ml 10% (w/v) sucrose with IB.
Load gradient solutions in 5 ml polyallomer centrifuge tube, starting from the bottom with 10% sucrose and the less dense iodixanol, using a glass pipette with Pipet-Aid (see Note 12).
Centrifuge at 50,000 × g for 1 hour in a SW 55 Ti rotor in the ultracentrifuge (see Note 13).
Collect the separated fractions carefully from the top of the gradient in seven Eppendorf tubes (see Note 14). Fraction 5 corresponds to the purified acidocalcisomes (Fig. 1).
Wash the fractions twice with 3–5 volumes of isolation buffer by centrifugation at 20,000 × g for 15 min to remove the iodixanol (see Note 15).
Quantify the protein concentration by Bradford assay (see Note 16) using a Microplate Reader.
Analyze fractions by various organelle marker enzyme assays (see Note 17) and Western blots (Figs. 2 and 3).
3.3. Electron Microscopy
Observe the morphology and structures of acidocalcisomes (Fig. 1) from fraction 5 of the second gradient by transmission electron microscopy (TEM).
Precipitate aliquot (25 μl) of fraction 5 (acidocalcisomes) of the second gradient (Fig. 1) by centrifugation at 20,000 × g for 15 min at 4°C.
Fix the pellet in 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at RT for 1 h.
Carefully replace the supernatant with fresh fixative (see Note 5) without disturbing the pellet and then store it at 4°C.
Process samples for transmission electron microscopy according to the standard procedures (12).
3.4. Marker Enzyme Assays
Detect organelle marker enzymatic activities in the 15,000 × g pellet (P1), the first gradient pellet (P2) and the fractions (F1 to F7) of the second gradient (Table 1, Fig. 2).
3.4.1. AMDP-sensitive vacuolar proton pyrophosphatase (V-PPase)
Vacuolar proton (H+) pyrophosphatase (V-PPase) is an acidocalcisome membrane-associated enzyme, which couples the energy of PPi hydrolysis to H+ influx across the organellar membrane. It has been widely used as a specific acidocalcisome marker for subcellular fractionation studies (1–3). The V-PPase activity assay is based on measuring phosphate (Pi) release using the malachite green assay (13).
Aminomethylenediphosphonate (AMDP) is used to distinguish between vacuolar (sensitive) and soluble (insensitive) PPase activities (14).
Prepare phosphate (Pi) standards: 0, 78, 313, 625, and 1250 μM KH2PO4.
Mix three parts of 0.045% malachite green hydrochloride (MG) and one part of 4.2% ammonium molybdate (AM) (3:1) at RT for at least 20 min and then pass through Whatman No. 5 filter paper.
Dilute fraction aliquots to 0.1 μg protein/μl (see Note 18).
Add 5 μl of each Pi standard (duplicated) or each diluted fraction aliquot (in triplicate) into 96-well microplate.
Mix reactions by adding 100 μl of PPase reaction buffer only (blank control) or the reaction buffer containing 100 μM PPi, with or without 40 μM AMDP, using a multichannel pipette.
Incubate at 30°C for 10 min in microplate incubator shaker.
Stop the reaction by adding 95 μl of freshly prepared malachite dye solution (MG-AM).
Immediately measure the absorbance (A) at 660 nm in microplate reader.
Determine the amount of Pi released by comparison with a standard curve, after subtracting blank control values.
Calculate the specific activity of V-PPase as μmol Pi released/min per mg of protein (see Note 19).
3.4.2. Succinate-cytochrome c reductase
Succinate-cytochrome c reductase (SCR) catalyzes the electron transfer reaction from succinate to cytochrome c and involves several oxidation-reduction components in the mitochondrial electron transport chain. The SCR is widely used as a marker of the abundance of mitochondria within a tissue/cell. The enzyme activity of SCR is measured using succinate and cytochrome c as the substrates based on the method of Sottocasa et al. (15).
Wash fraction aliquots twice and re-suspend with IB without DTT (see Note 20).
Add 10 μl of fraction aliquot (in triplicate) into 96-well microplate.
Mix the reactions by adding 90 μl of the SCR reaction medium containing 3 mM succinate (pH 7.2), 0.1 mM cytochrome c, 0.3 mM KCN, and 40 mM Hepes-KOH pH 7.5. The reaction is started by the addition of the substrate (cytochrome c).
Incubate at 30°C for 10 min in microplate incubator shaker.
Monitor the increase in the absorbance (A) at 550 nm at every 2 min intervals in the microplate reader. The amount of enzyme resulting in a change in A550 of 0.001/min is considered as 1 unit of enzyme.
Calculate the activity of SCR as units per mg of protein.
3.4.3. Hexokinase
Hexokinase (HK) is an important glycolytic enzyme in the glycosomes. This enzyme utilizes ATP as phosphyl donor to catalyze the conversion of D-glucose to glucose 6-phosphate, which is then oxidized to 6-phosphogluconate in the presence of oxidized nicotinamide adenine dinucleotide phosphate (NADP+) by glucose-6-phosphate dehydrogenase (G6PDH). The activity of the glycosomal marker HK is measured spectrophotometrically by following the NADPH formation as an increase of absorbance at 340 nm by coupling the reaction to G6PDH as described previously (16).
Prepare HK positive controls for standard curve: 0.1–1.0 units/ml of HK.
Add 10 μl of each HK standard or each fraction aliquot (in triplicate) into 96-well microplate.
Mix the reactions by adding 90 μl of the HK reaction medium containing 10 mM D-glucose, 0.6 mM ATP, 0.6 mM NADP+, 10 mM MgCl2, 2.5 units/ml glucose-6-phosphate dehydrogenase, and 50 mM Hepes-KOH, pH 7.8. The reaction is started by the addition of ATP.
Incubate the reactions at 30°C for 5 min in microplate incubator shaker.
Measure the absorbance (A) at 340 nm in microplate reader.
Determine the HK activities by comparison with the standard curve (HK positive controls). One unit of HK is defined as the phosphorylation of 1 μmol of D-glucose per min at pH 7.8 at 30°C.
Calculate the activity of HK as units per mg of protein.
3.4.4. Alpha-mannosidase
Alpha-mannosidase (AMA) catalyzes the cleavage of the α-form of mannose. This enzyme assists in the breakdown of complex sugars from glycoproteins within the lysosome. The activity of the lysosomal marker AMA is measured spectrophotometrically at 405 nm based on the release of 4-nitrophenol from the synthetic substrate p-nitrophenyl-α-D-mannopyranoside (pNP-Man) as described previously (17).
Prepare p-nitrophenol standards: 0, 25, 75 and 125 μM nitrophenol.
Add 10 μl of each fraction aliquot (in triplicate) or IB (blank control) into 96-well microplate.
Mix the reactions by adding 90 μl of the AMA reaction medium containing 200 mM sodium acetate buffer (pH 4.6) and 0.6 mM pNP-Man.
Incubate the reactions at 30°C for 30 min in microplate incubator shaker.
Stop the reaction by adding 160 μl of 1 M Na2CO3.
Measure the absorbance (A) at 405 nm in microplate reader.
Determine the amount of p-nitrophenol released by comparison with a standard curve, after subtracting blank control values. 1 unit of AMA is defined as the amount of enzyme that releases 1 μmol of p-nitrophenol per min at 30°C and pH 4.6.
Calculate the activity of AMA as μmol p-nitrophenol released /min × mg protein.
3.5. Western Blot Analyses
Detect the presence of organelle marker proteins in the 15,000 × g pellet (P1), the first gradient pellet (P2) and the fractions (F1 to F7) of the second gradient by immunoblotting (Fig. 3). Carry out all procedures at RT unless otherwise specified.
Lyse the aliquots of pellets or fractions with RIPA buffer containing protease inhibitor tablet in ice for 1 h.
Mix the lysed fractions with 2× Laemmli sample buffer at 1:1 ratio (v/v) and directly load.
Transfer the separated proteins onto nitrocellulose membranes using a Trans-Blot apparatus.
Block the membranes with 10% non-fat milk in PBS containing 0.5% Tween-20 (PBS-T) at 4°C overnight.
Incubate the blots with antibodies against proteins localized to acidocalcisomes (TbVP1) (18), mitochondria (voltage-dependent anion channel, TbVDAC) (19), glycosomes (pyruvate, phosphate dikinase, TbPPDK) (20), and lysosomes (Tbp67)(21) for 1 h (see Note 21).
After five washings with PBS-T, incubate the blots with horseradish peroxidase conjugated anti-mouse or anti-rabbit IgG (H+L) antibody at a dilution of 1:20,000 for 1 h.
After washing five times with PBS-T, visualize the immunoblots using Pierce ECL Western blotting substrate according to the manufacturer’s instructions.
4. Notes:
Warm BAG to RT before use, because washing trypanosomes with cold BAG could cause cell lysis or cell aggregation/cell clumps formation.
Isolation buffer (IB) is also known as lysis buffer (LB) or homogenization buffer (HB), whose osmolality is approximately 300 mOsm.
Add DTT and 1x protease inhibitor cocktails just before use.
The preparation of the working solution, as described, ensures that the concentrations of KCl, MgCl2, EDTA, DTT and the buffer (Hepes-KOH, pH 7.2) are constant throughout the gradient.
Good fixation requires fresh glutaraldehyde and paraformaldehyde.
KCN is highly toxic. Wear a mask to prepare it under fume hood, and avoid skin contact and inhalation.
For proteins larger than 80 kD or membrane proteins, we recommend that SDS be included at a final concentration of 0.01%.
Mix the cell pellet (approximately 3–4 g wet weight) with 1.5 x wet weight silicon carbide, transfer it to a precooled mortar (−20°C) using a stainless-steel lab spoon spatula, and then grind with a mortar and pestle on ice until lysis is greater than 99% (generally 45–60 s). Always monitor the efficacy of the grinding or lysis by phase contrast microscopy.
Rinse the grinding tools with IB to recover most of the cell-lysates, and also resuspend the lysate-silicon mixtures twice with IB to recover more supernatant, in which acidocalcisome are floating with other organelles.
Suspension and homogenization of the pellet should be carried out on ice as gently as possible to avoid damage not only to the target organelles (acidocalcisomes) but also to any other organelles present, particularly those that may release degradative enzymes. In addition, homogenize by approximately 6 stokes of the pestle (50–70 rpm) using a tissue homogenizer mixer. Aliquots from the homogenizations of the 15,000 × g pellet (P1) and the first gradient pellet (P2) are also taken and used for enzymatic assays and Western blot analyses (Table 1, Fig. 3).
Glass tubes are better for making up density gradient solutions than plastic tubes. Prepare 4 ml of 20, 24, 28, 34, 37 and 40% (w/v) iodixanol gradient solutions in 10 ml glass tubes, by adding 1.60, 1.92, 2.24, 2.72, 2.96, or 3.20 ml of 50% (w/v) iodixanol working solution to each tube, respectively, supplementing with 166, 200, 233, 283, 308 or 333 μl of 1 M sucrose to maintain each gradient solution isosmotic, and finally add isolation buffer to 4 ml in total, except for the 34% iodixanol gradient (which is mixed with the homogenized pellet in IB). Mix the gradient solutions well and store on ice for the gradient loading.
Although overlaying starting from the bottom of an ultracentrifuge tube with the densest layer is the most popular means of creating a discontinuous gradient, underlaying (i.e. starting from the bottom of an ultracentrifuge tube with the least dense layer) using a Pasteur glass pipette with cotton and a pipette aid is easier, more reliable and the recommended method for making discontinuous gradients. The only important requirement is that no air bubbles are introduced which may disturb the lower density layer above, for this reason it requires some practice to maintain a steady liquid flow by pressing the release button slowly.
Label the centrifuge tubes and balance exactly.
Although a Pasteur pipette or syringe can be used to collect fractions from the top of the ultracentrifuge tube, we prefer using the tip of the pipette to aspirate fractions from the top very slowly, moving it across the diameter of the tube and minimizing the aspiration of any liquid from below the fraction or band.
Although iodixanol does not interfere with most downstream analyses (such as SDS-PAGE and enzymatic assays), removal of iodixanol is an absolute requirement for electron microscopy. On the other hand, if it is necessary to concentrate the gradient fractions, TCA precipitation of the proteins is recommended.
Bradford and BCA assays are very commonly used protein estimation methods. Bradford is fast because there is 5 min incubation time whereas for BCA its 30 min. In the case of BCA, it is recommended to use a volume of 10–25 μl, which is quite larger than required for Bradford. However, Bradford is less compatible with detergents (such as 1% SDS) than BCA. Since the aliquots of fractions (containing membrane proteins) are lysed with RIPA buffer, we dilute the lysed fractions with IB to reduce the concentration of the RIPA detergents for determination of the protein concentration by Bradford assay.
The enzyme activities of succinate-cytochrome c reductase (SCR), hexokinase (HK) and α-mannosidase (AMA) are not affected by storage at −80°C for 2 weeks. However, freezing and storage significantly reduce V-PPase activity, so we recommend that the V-PPase activities of the gradient fractions be assayed just right after the subcellular fractionation.
If the protein concentration is below 0.1 μg/μl, concentrate the fraction by TCA first or directly add 5 μl of fraction for assays and finally calculate the enzyme activities based on the original protein concentrations.
Calculate the activity of V-PPase as follows: [ΔPi released (μM) × n] / [incubation time (min) × protein (μg/μl)] = ΔPi × 0.04 (μmol/min × mg). The net amount (μM) of Pi released by specific V-PPase (ΔPi) is obtained by subtracting the value of the (PPi + AMDP) reaction from that of the PPi reaction. The incubation time is 10 min. The protein concentration is 0.1 μg/μl, and the dilution factor (n) is 40.
Because DTT interferes with SCR activity, we wash the fraction aliquots with IB without DTT before the SCR assay to minimize the interference.
Incubate the blots with rabbit antibodies against TbVP1 (1:5,000), rabbit antibodies against TbVDAC (1:2,000), mouse antibodies against TbPPDK (1:200), and mouse antibodies against Tbp67 (1:3,000) at RT for 1 h.
Acknowledgements
We thank Norbert Bakalara for the anti-TbVP1 antibody, Minu Chaudhuri for anti-TbVDAC antibody, Frédéric Bringaud for anti-TbPPDK antibody, James Bangs for anti-Tbp67 antibody, Michael Martin for the AMDP, and Mary Ard and Georgia Electron Microscopy (GEM) facility for assistance with TEM sample preparation and imaging. Work was funded by a grant from the US National Institutes of Health (AI-077358 to R.D.)
References
- 1.Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SNJ (2005) Acidocalcisomes-conserved from bacteria to man. Nature Rev Microbiol 3:251–261 [DOI] [PubMed] [Google Scholar]
- 2.Docampo R, Huang G (2016) Acidocalcisomes of eukaryotes. Curr Opin Cell Biol 41:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang G, Ulrich PN, Storey M, Johnson D, Tischer J, Tovar JA, Moreno SNJ, Orlando R, Docampo R (2014) Proteomic analysis of the acidocalcisome, an organelle conserved from bacteria to human cells. PLoS Pathog 10:e1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang G, Bartlett PJ, Thomas AP, Moreno SNJ, Docampo R (2013) Acidocalcisomes of Trypanosoma brucei have an inositol 1,4,5-trisphosphate receptor that is required for growth and infectivity. Proc Natl Acad Sci U S A 110:1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Souza W, Morgado-Diaz JA, Cunha-e-Silva NL (2008) Cell fractionation of parasitic protozoa. In: Posch A. (eds) 2D PAGE: sample preparation and fractionation. Methods in Molecular Biology™, vol 425:313–331. Humana Press; [DOI] [PubMed] [Google Scholar]
- 6.Jardim A, Hardie DB, Boitz J, Borchers CH (2018) Proteomic profiling of Leishmania donovani promastigote subcellular organelles. J Proteome Res 17(3):1194–1215 [DOI] [PubMed] [Google Scholar]
- 7.Scott DA, Docampo R, Dvorak JA, Shi S, Leapman RD (1997) In situ composition analysis of acidocalcisomes Trypanosoma cruzi. J Biol Chem 272:28020–28029 [DOI] [PubMed] [Google Scholar]
- 8.Scott DA, Docampo R (2000) Characterization of isolated acidocalcisomes of Trypanosoma cruzi. J Biol Chem 275:24215–24221 [DOI] [PubMed] [Google Scholar]
- 9.Ferella M, Nilsson D, Darban H, Rodrigues C, Bontempi EJ, Docampo R, Andersson B (2008) Proteomics in Trypanosoma cruzi - localization of novel proteins to various organelles. Proteomics 8:2735–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham J, Ford T, Rickwood D (1994) The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem 220:367–373. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham I (1977) New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool 24:325–329 [DOI] [PubMed] [Google Scholar]
- 12.Kuo J (Ed.), (2007) Electron Microscopy: Methods and Protocols (2nd edition), Methods in Molecular Biology ™, vol 369. Humana Press [Google Scholar]
- 13.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100:95–97 [DOI] [PubMed] [Google Scholar]
- 14.Zhen R, Baykov AA, Bahuleva NP, Rea PA (1994) Aminomethylenediphosphonate: A potent type-specific inhibitor of both plant and phototrophic bacterial H+-pyrophosphatases. Plant Physiol 104:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottocasa GL, Kuylenstierna B, Ernster L, Bergstrand A (1967) An electron-transport system associated with the outer membrane of liver mitochondria. A biochemical and morphological study. J Cell Biol 32:415–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannata JJ, Valle E, Docampo R, Cazzulo JJ (1982) Subcellular localization of phosphoenolpyruvate carboxykinase in the trypanosomatids Trypanosoma cruzi and Crithidia fasciculata. Mol Biochem Parasitol 6:151–160 [DOI] [PubMed] [Google Scholar]
- 17.Liao YF, Lal A, Moremen KW (1996) Cloning, expression, purification, and characterization of the human broad specificity lysosomal acid alpha-mannosidase. J Biol Chem 271:28348–28358 [DOI] [PubMed] [Google Scholar]
- 18.Lemercier G, Dutoya S, Luo S, Ruiz FA, Rodrigues CO, Baltz T, Docampo R, Bakalara N (2002) A vacuolar-type H+-pyrophosphatase governs maintenance of functional acidocalcisomes and growth of the insect and mammalian forms of Trypanosoma brucei. J Biol Chem 277:37369–37376 [DOI] [PubMed] [Google Scholar]
- 19.Singha UK, Sharma S, Chaudhuri M (2009) Downregulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot Cell 8:1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bringaud F, Baltz D, Baltz T (1998) Functional and molecular characterization of a glycosomal PPi-dependent enzyme in trypanosomatids: pyruvate, phosphate dikinase. Proc Natl Acad Sci USA 95:7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander DL, Schwartz KJ, Balber AE, Bangs JD (2002) Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J Cell Sci 115:3253–3263 [DOI] [PubMed] [Google Scholar]


