Abstract
Background
The balance of the intestinal commensal microbiome of fish and other animals plays an important role in the physiological processes of healthy animals, contributes to the defense against pathogens, stimulates the immune system and facilitates nutrient metabolism. In the last decade, the interest in the application of the insects in fish nutrition increased, although little is known regarding the effects of insect meals on the gastrointenstinal tract microbiome of the sea trout fingerlings. The aim of this study was to evaluate the effect of two diets containing mealworm (MW) and superworm (SW) on the microbiome of the digesta of sea trout fingerlings and the relative abundances of different taxa among communities under controlled conditions.
Results
The insect meals produced a similar weight gain and survival rate to sea trout fed fishmeal. The most abundant bacterial phylum in all the treatment groups was Firmicutes followed by Proteobacteria and Actinobacteria, and significant differences in the amount of Cyanobacteria were observed in the SW group.
Conclusions
The insect meals did not produce differences in the three most abundant phyla in the sea trout digesta. However, the effect of each type of meal on the lower taxonomic levels was evident, particularly in the case of the superworm meal. These microbiome differences indicated that mealworm meal was more related to fishmeal than superworm meal. Our results highlight the potential effects of insect meals, such as mealworm and superworm meals, on the microbiota of sea trout.
Keywords: Sea trout, Next generation sequencing, NGS, Microbiome, Metagenome, Fish, Mealworm, Superworm
Background
Fish, as well as other animals, must maintain the microbiome (bacteria, archaea, fungi, and viruses) in their intestinal tract in a balanced state, preserving the mutualistic relationship along their life cycles. The microbiome contributes to the defense against pathogens, stimulates the immune system and assists with nutrient metabolism [1–3]. In the last decade, the interest of the application of insects in fish nutrition has increased, although little is known about the effects of insect meals on the microbiome of the digesta of sea trout fingerlings.
The impact of insect products on gut health and microbiota has been analyzed with Rainbow trout (O. mykiss), Atlantic salmon (Salmo salar), Sea trout (Salmo trutta m. trutta), European sea bass (Dicentrarchus labrax), Gilthead seabream (Sparus aurata), Siberian sturgeon (Acipenser baerii), [4–14]. In the microbiota studies fish were fed mainly on Hermetia illucens (HI), Tenebrio molitor (TM), and Musca domestica (MD), and some studies investigated the effect of Gryllus sigillatus (GS), Blatta lateralis (BL) and Zophobas morio (ZM) [15].
In general, insect meal does not adversely affect the gut morphology of fish. However, have been found that the inclusion of insects in fish diet can induce some mild gut histological changes. The mucosal and muscular layer thickening of the GIT in sturgeon fed both TM and HI [4]. Moreover, the dietary inclusion of insects increased submuscosa cellularity and production of neutral mucin in the proximal intestine of rainbow trout [8, 16].
Several studies have analyzed the fish microbiota using 16S rRNA gene amplicon sequencing. The microbial communities are affected by certain factors such as the species, the stage of development, the type of food consumed, and the intestinal morphology [4, 5]; environmental and physiological factors also modify the gut microbiota of fish [6]. The type of microbiome will also be conditioned by the feeding habit of the species; in salmonids such as rainbow trout, the predominant phyla are Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria and Actinobacteria [7, 8]. However, Rimoldi et al. (2018) found that rainbow trout fed higher levels of plant meals and rendered animal meals, and lower levels of fishmeal had higher quantities of Fusobacteria and Bacteroidetes in the intestine, and this difference was related to the lower growth performance [9]. Furthermore, Huyben et al. (2019) observed a variation in the abundance of a group of bacteria present in the gut of the rainbow trout according to the stage of development of the insect meal (larvae, prepupae, and pupae) [10]. The Proteobacteria and Firmicutes have been detected as dominated phyla in all gut regions of brown trout (Salmo trutta L.) [11]. Moreover, Michl et al. (2019) observed significantly increased abundances of Proteobacteria and Fusobacteria following the consumption of fishmeal, whereas plant-derived proteins increased the abundance of Firmicutes and Bacterioidetes [7].
In general, plant-based protein meals markedly modify the microbiome, as Kononova et al. (2019) showed the effects of soybean protein and carbohydrates, which are associated with some anti-nutritional factors, on the autochthonous microbiota, provoking inflammatory processes in the intestine of salmonids [12]. Regarding insect meals, Antonopoulou et al. (2019) found that rainbow trout fed 0 and 60% mealworm meal did not differ in the bacterial species or their amounts of relative abundance [13]. A possible explanation for this finding is that insects are part of the natural diet of this species.
Because insects are also part of the natural diet of sea trout, at least in the first stages of development, the aim of this study was to evaluate the effects of two insect meal diets on the microbiome of the digesta of sea trout fingerlings and the abundances of different taxa among communities.
Results
Growth performance
At the end of the experimental period, no significant differences in body weight gain and survival rates were observed among groups fed the different experimental diets, as shown in Fig. 1.
Fig. 1.
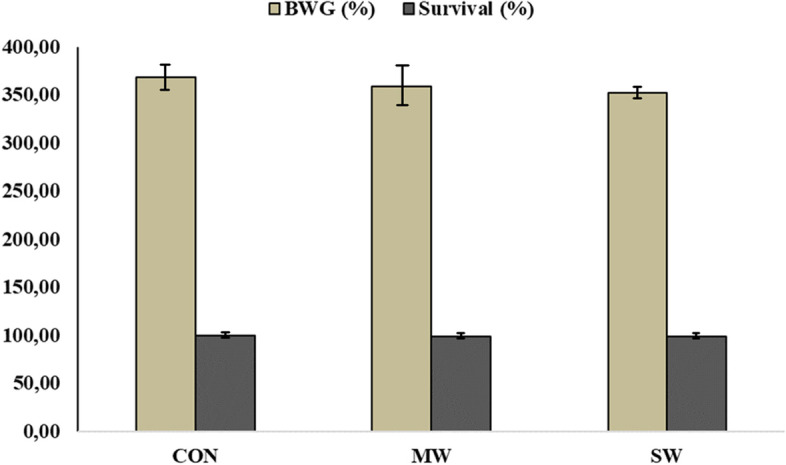
Bodyweight gain (BWG) and survival rate of sea trout at the end of the experiment. Experimental diets: fishmeal diet (CON), mealworm diet (MW), and superworm diet (SW)
Microbiota diversity
In general, 99.95% of the gut microbiota was constituted by bacteria, 0.03% by archaea and 0.01% by viruses, and 0.02% of the microorganisms were not identified. Considering bacteria, 24 phyla were identified, and 17 were represented with an abundance less than 1%, whereas the most predominant bacteria among the remaining phyla were Firmicutes (Fig. 2). In general, significant differences in the abundances of Actinobacteria, Proteobacteria, and Firmicutes were not observed, although the abundance of Cyanobacteria exhibited significant differences, as the SW group presented the lowest content of this phylum as well as the combined data for the 20 phyla (p < 0.05).
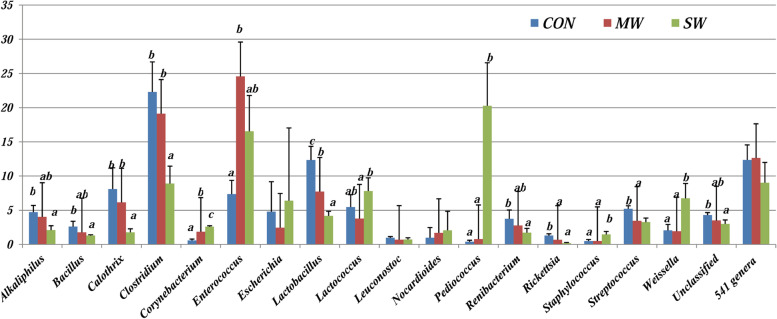
Fig. 2.
The relation between the genera proportions of sea trouts’ gut digesta in the different treatments: control (CON), mealworm meal (MW), and superworm meal (SW)
Regarding the species relations (Fig. 3), 40.26% of the species were shared among treatments and 11.96 to 13.17% of the species are unique to each treatment; a lower number of shared species were observed between two treatments. Additionally, the amount of lactic acid bacteria (LAB) belonging to Enterococcus, Pediococcus, Lactococcus, and Weisella increased significantly in the fish fed the insect meals, namely, 36.16% and 47.98% in the MW and SW groups, respectively, compared to fish fed the control diet at 27.29% (p < 0.05).
Fig. 3.
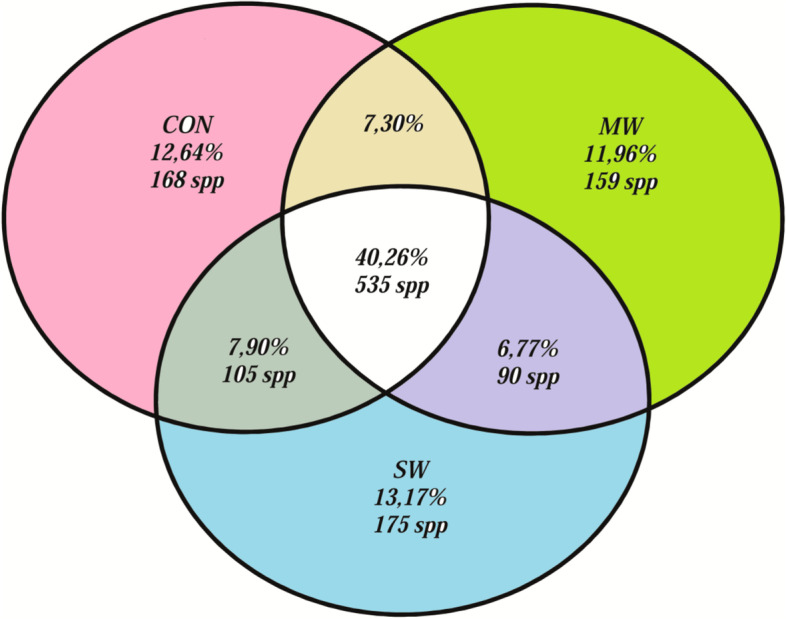
Relation of the number of species per treatment as well as those shared among treatments: Control diet (CON), mealworm diet (MW), and superworm diet (SW). The values are expressed in percentage and amount of species identified
Regarding species richness, 1328 species were identified, and the species richness was 905 species for the CON group, 879 species for the MW group, and 905 species for the SW group. At the same time, the Margalef index (DMg) showed no significant differences among treatments. The alpha diversity indexes, such as Simpson Diversity Index (D), Menhinick index, and Shannon index (H), revealed that insect meals did not affect the species richness. Meanwhile, the Evenness index (e^H/S), and equitability Brillouin index also showed no significant differences among treatments. The dominance index (Berger-Parker) and Dominance D displayed similar values among meal-fed groups, and the dominance was low among groups. Regarding the abundance estimator, the Chao1 calculation showed no significant differences among treatments as well (Table 1).
Table 1.
Alpha diversity indexes in sea trout gut microbiota species, comparison among treatments
| CON | MW | SW | SEM | P-value | |
|---|---|---|---|---|---|
| Individuals | 73.50 | 74.00 | 74.50 | 0.78 | 0.6760 |
| Dominance (D) | 0.15 | 0.12 | 0.12 | 0.01 | 0.3782 |
| Simpson 1-D + | 0.85 | 0.88 | 0.88 | 0.01 | 0.3781 |
| Shannon (H) | 3.05 | 3.15 | 3.16 | 0.06 | 0.3632 |
| Evenness e^H/S | 0.04 | 0.05 | 0.05 | 2.1–3 | 0.1354 |
| Brillouin | 1.17 | 1.30 | 1.36 | 0.05 | 0.0966 |
| Menhinick | 51.05 | 49.28 | 50.80 | 1.69 | 0.7314 |
| Margalef | 118.58 | 114.28 | 117.63 | 4.09 | 0.7441 |
| Equitability (J) | 0.49 | 0.51 | 0.51 | 0.01 | 0.2154 |
| Berger Parker | 0.32 | 0.27 | 0.30 | 0.03 | 0.4398 |
| Chao-1 | 510.50 | 492.75 | 508.00 | 16.88 | 0.7314 |
Values in the same row having different superscript letters are significantly different at p < 0.05, (n = 4)
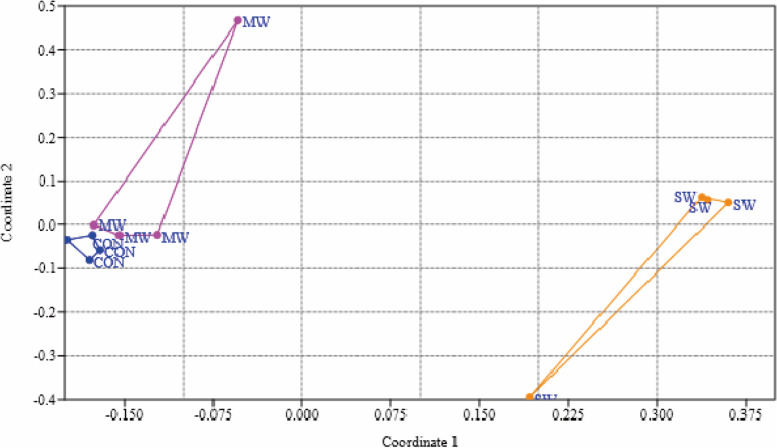
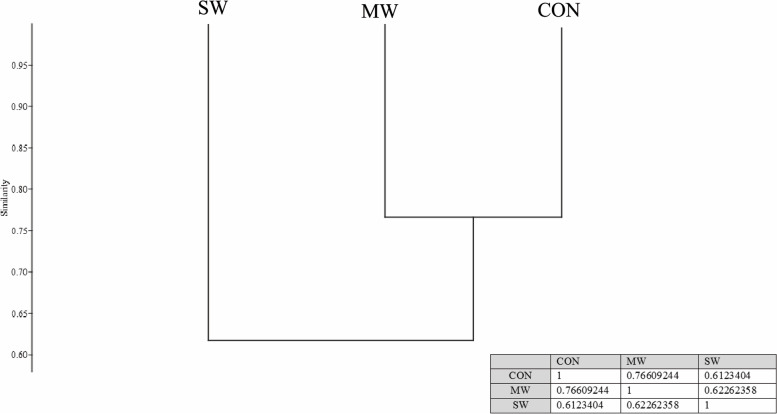
The Bray–Curtis analysis of beta diversity is presented in Table 2. Additionally, the nonmetric multidimensional scaling (NMDS) analysis (Fig. 4, and Fig. 5) and the clusters showed that CON and MW groups were much more related, with 76.6% similarity, than the SW group (61.2%) (Fig. 6).
Table 2.
ANOSIM and Bray–Curtis results
| ANOSIM | Bray–Curtis |
|---|---|
| Permutation N: | 9999 |
| Mean rank within: | 14.67 |
| Mean rank between: | 40.56 |
| R2: | 0.787 |
| p (same): | 0.003 |
Fig. 4.
Non-metric multidimensional scaling (NMDS) analysis plot of gut microbiota of sea trouts fed with two experimental diets: CON (control diet), MW (mealworm diet), and SW (superworm diet)
Fig. 5.
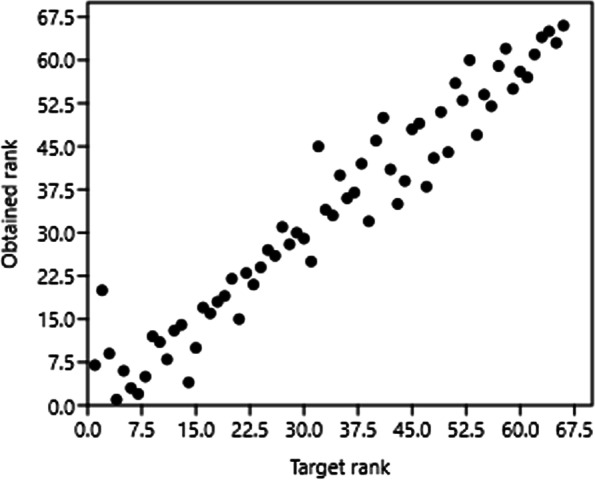
Shepard analysis plot and stress value of sea trouts’ microbiota fed with three experimental diets: CON (control diet), MW (mealworm diet), and SW (superworm diet)
Fig. 6.
Sea trouts’ microbiota clusters similarities and dissimilarities between treatments; CON (control diet), MW (mealworm diet), and SW (superworm diet)
After observing the class distribution in sea trout digesta, Bacilli and Clostridia were the most predominant classes in the groups treated with the three experimental diets, although the SW group exhibited the highest percentage of Bacilli, but the lowest percentages of Clostridia, Nostocophycideae and other classes compared to groups fed the CON and MW diets (Table 3). A similar trend was observed for the class distribution, where Bacilli and Clostridia classes were the most predominant and the SW group exhibited the highest percentage of Bacilli and the lowest percentage of Clostridia among all three treatment groups. Zophobas morio meal also reduced the amount of Nostocophycideae and the grouped orders (Table 3). In terms of the family distribution, fish fed the SW diet exhibited lower percentages of Bacillaceae, Clostridiaceae, Lachnospiraceae, Rickettsiaceae, and Rivulariaceae than fish fed the CON and MW diets. In contrast, higher abundances of Corynebacteriaceae, Lactobacillaceae, and Leuconostocaceae were observed in the SW group than in fish fed the CON and MW diets. However, the percentage of Enterococcaceae was significantly lower in fish fed the CON diet those fed the SW and MW diets (p < 0.05).
Table 3.
Relative abundance of the most dominant classes, orders, and families present in the digesta samples of sea trouts fed with the three experimental diets: control diet (CON), mealworm diet (MW), and superworm diet (SW)
| Identifications | CON | MW | SW | SEM | P-value |
|---|---|---|---|---|---|
| Classes | |||||
| Actinobacteria | 7.99 | 9.12 | 10.32 | 1.71 | 0.6415 |
| Alphaproteobacteria | 2.05 | 3.37 | 0.41 | 0.81 | 0.0826 |
| Bacilli | 41.57a | 50.27ab | 65.69b | 5.47 | 0.0350 |
| Clostridia | 32.92b | 26.77b | 13.53a | 2.75 | 0.0022 |
| Gammaproteobacteria | 5.43 | 2.97 | 7.43 | 3.74 | 0.7054 |
| Nostocophycideae | 8.24b | 6.24b | 1.80a | 1.07 | 0.0060 |
| 40 classes* | 1.79c | 1.26b | 0.78a | 0.10 | 0.0003 |
| Order | |||||
| Actinobacteria | 7.99 | 9.12 | 10.32 | 1.71 | 0.6415 |
| Alphaproteobacteria | 2.05 | 3.37 | 0.41 | 0.81 | 0.0826 |
| Bacilli | 41.57a | 50.27ab | 65.69b | 5.47 | 0.0350 |
| Clostridia | 32.92b | 26.77b | 13.53a | 2.75 | 0.0022 |
| Gammaproteobacteria | 5.43 | 2.97 | 7.43 | 3.74 | 0.7054 |
| Nostocophycideae | 8.24b | 6.24b | 1.80a | 1.07 | 0.0060 |
| 87 orders* | 1.79c | 1.26b | 0.78a | 0.10 | 0.0003 |
| Families | |||||
| Bacillaceae | 3.05b | 2.00a | 1.74a | 0.25 | 0.0106 |
| Clostridiaceae | 27.70b | 23.39b | 11.22a | 2.74 | 0.0056 |
| Corynebacteriaceae | 0.58a | 1.85b | 2.58c | 0.13 | 0.0001 |
| Enterobacteriaceae | 5.26 | 2.77 | 7.31 | 3.73 | 0.6993 |
| Enterococcaceae | 8.02a | 25.84b | 17.07ab | 4.10 | 0.0395 |
| Lachnospiraceae | 1.87b | 1.00ab | 0.71a | 0.28 | 0.0391 |
| Lactobacillaceae | 12.76a | 8.52a | 24.62b | 1.90 | 0.0006 |
| Leuconostocaceae | 3.09a | 2.63a | 7.55b | 0.74 | 0.0020 |
| Microbacteriaceae | 0.25 | 1.46 | 1.05 | 0.47 | 0.2370 |
| Micrococcaceae | 4.10 | 3.09 | 2.75 | 0.47 | 0.1608 |
| Nocardioidaceae | 1.13 | 1.82 | 2.21 | 1.07 | 0.7790 |
| Peptostreptococcaceae | 1.61 | 1.29 | 0.87 | 0.22 | 0.1073 |
| Rickettsiaceae | 1.30b | 0.68a | 0.20a | 0.12 | 0.0005 |
| Rivulariaceae | 8.10b | 6.16b | 1.77a | 1.05 | 0.0060 |
| Ruminococcaceae | 1.11 | 0.64 | 0.42 | 0.21 | 0.1072 |
| Streptococcaceae | 10.70 | 7.23 | 11.07 | 0.99 | 0.0431 |
| 203 families* | 9.27 | 9.52 | 6.80 | 0.96 | 0.1435 |
Values in the same row having different superscript letters are significantly different at p < 0.05, (n = 4)
*Grouped data of the rest of classes, orders and families of bacteria with values lower than 0.5% plus the unidentified groups
Five hundred forty-one genera were identified, which represented the 85.10 ± 3.51% of the total samples. After comparing the most predominant genera present in fish digesta (Fig. 3), sea trout fed the CON diet presented higher amounts of Clostridium and Lactobacillus, and the highest content was observed for Enterococcus followed by Clostridium in the MW group, but the most representative genera in the SW group were Pediococcus and Enterococcus.
The total amount of bacterial species identified varied among treatment groups. In the CON diet group, only 67.75 ± 4.63% of the bacteria were identified, whereas the MW diet group exhibited the highest percentage of identified species at 73.25 ± 3.36%, followed by the SW diet group at 70.07 ± 7.30%. When observing the most predominant species in each treatment group (Table 4), the fish fed the CON diet presented the highest percentage of Clostridium cadaveris and Calothrix parietina. In the case of the MW group, the most abundant species were also C. cadaveris and C. parietina along with Enterococcus silesiacus. Moreover, the highest abundances were observed for Pediococcus pentosaceus, C. cadaveris, and Enterococcus durans. In addition, significant differences were detected in the SW group, with the lowest levels observed for Alkaliphilus crotonatoxidans, C. parietina, C. cadaveris, Lactobacillus antri, L. delbrueckii, and Streptococcus gallinaceus, and the highest values observed for E. durans, E. gallinarum, E. gilvus, Lactococcus garvieae, P. pentosaceus, P. acidilactici, P. stilesii and Weisella cibaria (p < 0.05).
Table 4.
ANOVA results of the most representative bacterial phylotypes (%) isolated from the gut digesta of sea trout fed with the three experimental diets: control diet (CON), mealworm diet (MW), and superworm diet (SW)
| Phyla | Species | CON | MW | SW | SEM | P-value |
|---|---|---|---|---|---|---|
| Firmicutes | Alkaliphilus crotonatoxidans | 4.04b | 3.38b | 1.75a | 0.47 | 0.0186 |
| Cyanobacteria | Calothrix parietina | 8.10b | 6.15b | 1.77a | 1.05 | 0.0060 |
| Firmicutes | Clostridium cadaveris | 15.47b | 12.96b | 5.94a | 1.69 | 0.0085 |
| Firmicutes | Enterococcus avium | 4.49 | 5.58 | 2.90 | 0.87 | 0.1455 |
| Firmicutes | Enterococcus casseliflavus | 0.66 | 0.69 | 0.51 | 0.10 | 0.4619 |
| Firmicutes | Enterococcus durans | 0.30a | 1.16a | 5.78b | 0.63 | 0.0004 |
| Firmicutes | Enterococcus gallinarum | 0.05a | 0.25a | 1.08b | 0.10 | 0.0001 |
| Firmicutes | Enterococcus gilvus | 0.51a | 0.64a | 2.20b | 0.23 | 0.0010 |
| Firmicutes | Enterococcus lactis | 0.18a | 2.69b | 2.60b | 0.50 | 0.0095 |
| Firmicutes | Enterococcus mundtii | 0.004 | 2.10 | 0.04 | 0.90 | 0.2225 |
| Firmicutes | Enterococcus silesiacus | 0.01 | 7.39 | 0.07 | 3.00 | 0.1909 |
| Proteobacteria | Escherichia albertii | 1.53 | 0.77 | 2.10 | 1.08 | 0.6909 |
| Firmicutes | Lactobacillus antri | 1.35b | 1.14b | 0.52a | 0.14 | 0.0064 |
| Firmicutes | Lactobacillus delbrueckii | 2.41b | 0.11a | 0.30a | 0.41 | 0.0054 |
| Firmicutes | Lactobacillus oris | 2.32b | 1.94ab | 0.80a | 0.30 | 0.0156 |
| Firmicutes | Lactococcus garvieae | 1.36a | 1.23a | 3.93b | 0.48 | 0.0049 |
| Firmicutes | Pediococcus acidilactici | 0.06a | 0.12a | 2.24b | 0.21 | 0.0001 |
| Firmicutes | Pediococcus pentosaceus | 0.15a | 0.32a | 8.80b | 0.84 | 0.0001 |
| Firmicutes | Pediococcus stilesii | 0.04a | 0.08a | 1.89b | 0.16 | 0.0001 |
| Firmicutes | Streptococcus gallinaceus | 2.80b | 1.83ab | 1.56a | 0.30 | 0.0380 |
| Firmicutes | Weissella cibaria | 1.52a | 1.42a | 5.39b | 0.55 | 0.0009 |
| Firmicutes, Cyanobacteria, Proteobacteria, and Actinobacteria | 1307 spp | 20.39 | 20.33 | 18.48 | 1.51 | 0.6123 |
Values in the same row having different superscript letters are significantly different at p < 0.05, (n = 4)
Discussion
In the last several decades, the importance of the gut microbiota has been documented in numerous studies showing that growth performance and fish health are closely related to the microbiota. As Butt and Volkoff (2019) commented, feeding habits influence the structure and composition of the gut microbiota [2]. Additionally, plant-based proteins change the content and structure of the autochthonous microbiota of carnivorous species such as sea trout; in contrast, the use of a natural source of protein such as insect meals may play a role in maintaining the amount of these types of microorganisms that are part of the gut environment of the fish and enhance fish health [12]. Furthermore, insect meals would be able to modulate the microbiota of these animals due to the chitin and antimicrobial peptide contents [14, 15].
In this trial, more than 40% of FM (fish meal) was replaced with insect meals in the two diets, although the insect meals produced similar growth and survival rates when observing the weight gain. When analyzing the bacteria present in the digesta, the dominant phylum in all the treatment groups was Firmicutes, in contrast to brown trout fed a commercial diet, in which the dominant phylum was Proteobacteria, ranging from 88.4 to 92.6% [11]. However, Michl et al. (2019) reported a reduction in the amount of Proteobacteria and an increase in the amounts of Firmicutes and Bacteriodetes in the same species fed diets with more plant-based meals [7]. In contrast, Rimoldi et al. (2019) detected a gradual increase in the abundance of Tenericutes and a reduction in the abundances of Proteobacteria and Firmicutes in rainbow trout fed different amounts of black soldier fly meals [16]. Moreover, Kononova et al. (2019) affirmed that Proteobacteria is more abundant in carnivorous species and Firmicutes is more abundant in herbivorous species [12]. The results from the present trial showed that the abundance of Firmicutes would be conditioned by the amount of plant meal in the three diets, which was approximately 47% of the total, but not the inclusion of insect meals.
After performing a detailed analysis of the classes present in the digesta, Bacilli was the most abundant in all treatment groups, followed by Clostridia, both of which belong to the Firmicutes phylum, but the sum of Alphaproteobacteria and Gammaproteobacteria, which belong to the Proteobacteria phylum, presented similar amounts in all treatment groups, indicating that MW and SW meals exerted the same effect as FM on the digesta of sea trouts. In addition, the phylum Firmicutes and class Clostridia have been repeatedly identified in the digestive tracts of herbivorous fish, and as described above, the higher abundance of this class would be related to the higher amount of plant meal present in the diets [5].
The order and family distribution followed a similar trend as the class distribution. Although the bacterial genera exhibited changes based on the type of protein meal source, the most representative genera in the CON group were Clostridium and Lactobacillus and those in the MW group were Enterococcus and Clostridium, but the most representative genera in the SW group were Pediococcus and Enterococcus. The bacterial genera exhibited increased in that study are used as probiotics in aquaculture, increasing bacterial diversity [2], which probably occurred in fish fed these insect meals. The type of meals exerted a direct effect on the abundance of different genera in the intestinal digesta.
An analysis of the species abundance showed that fish fed the diet with SM exhibited a decrease in the relative abundance of C. cadaveris compared to the CON and MW groups; this species is known as a component of the normal fecal flora of humans and animals, which affects people with a poor overall condition of immunosuppression [17]. On the other hand, C. cadaveris is one of the most prominent bacterium present during the decay of dead bodies [17]. Moreover, C. cadaveris might trigger bacteremia that is related to a high mortality rate in humans. In the present study, the relative abundance of the commensal species C. cadaveris was decreased in the SW group, but significant differences were not observed between the CON and MW groups. Therefore, the reduction in the relative abundance C. cadaveris in the gastrointestinal tract of fish induced by the diet containing SW may be considered as a positive effect on public health.
C. parietina belongs to phylum Cyanobacterium and was previously detected in alkaline and oxygenated freshwaters [18]. The growth of Cyanobacteria is stimulated by the hypoxia of water reservoirs. Moreover, the contamination of dry food and feed with Cyanobacterium is considered a risk of toxin prevalence. Moreover, C. parietina is the bacteria with a higher potential for endotoxin production. The diet containing Zoophobas morio caused a decrease in the abundance of the bacterial species C. parietina in the fish GIT, which may reduce possible cyanobacterial toxin reservoirs in the fish GIT.
The diet containing SW improved the commensal probiotic microbiome in intestinal digesta of Salmo trutta vr. trutta. The SW diet increased the abundance of some bacterial genera, such as Pediococcus, which is considered a bacteria that can positively affect GIT of fish. Pediococcus is a genus of gram-positive lactic acid-producing bacteria belonging to the family Lactobacillaceae. Pediococcus pentosaceus genus is considered as a promising strain for both the food industry and biological applications [19]. The mechanism of action includes bacteriocins (pediocins) or bacteriocin-like substances (BLISs). It has been shown that P. pentosaceus can inhibit the growth of bacteria such as Staphylococcus aureus, Enterococcus faecalis, Clostridium perfringens, Shigella flexneri, Salmonella enteritidis, Listeria monocytogenes, and Escherichia coli [19–24]. The inhibiting effect was also observed against fungi especially Aspergillus, Penicillium, Fusarium, and Candida species [25–28].
In the SW group, an increase in the abundance of pediococci was observed, with the most abundant species identified as P. pentosaceus in the SW group. The bacterial species P. pentosaceus exerts bacteriocynogenic effects against some pathogenic bacteria [29]. Moreover P. pentosaceus regulate environmental homeostasis enforcing systemic immunity and enhancing anti-inflammatory ability [19].
Enterococcus is a key component of the intestinal flora of humans and is widespread in the intestines of most animals, including fish. Some species belonging to the Enterococcus genus, such as Enterococcus faecalis from fish intestine, are used as aquatic probiotics [30]. The SM diet increased the abundance of some Enterococcus species in the fecal digesta, among which E. durans may be considered a possible probiotic, because it potentially produces bacteriocins, namely, durancins [31]. Another species with probiotic potential that have been isolated from fish is Enterococcus gallinarum that regulates the innate immune response [32]. An increase in the abundance of Enterococcus gilvus was also observed in the fecal digesta of the analyzed SW group. The analysis of gene expression in Enterococcus gilvus has identified novel carotenoid biosynthesis genes that improve the multistress tolerance of Lactococcus lactis and promotes their activity toward methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE) [33]. Additionally, W. cibaria, which was more abundant in the SW group, has shown to be an effective probiotic in hybrid surubim [34]. Although significant differences among certain groups of bacteria were observed, the composition of the most representative species shows that they are part of the digestive tract flora, the environment, or part of the protein sources with probiotic properties that help the fish to thrive and achieve target growth and survival rates. In addition, Gajardo et al. (2016) commented that LAB are more abundant in salmon fed a plant-based diet than in fish fed a fishmeal-based diet [35], although, Ringø and Gatesoupe (1998) commented that LAB, such as the Lactobacillus, Carnobacterium, and Streptococcus genera, are also commonly detected in healthy fish microbiota of different fish species, including salmonids [36]. However, insect meals also increase the amount of LAB, as observed in the present study.
Furthermore, when comparing alpha diversity parameters, the inclusion of insect meal in the diet did not modify the different parameters measured, such as richness, evenness, and dominance. The Shannon H values were similar to those obtained in rainbow trout fed only FM and greater than 60% of Tenebrio molitor meal [13]. Additionally, the Chao1 values obtained in the present study were similar to those observed in the digesta of the proximal intestine of salmon fed 45% FM and 38% plant meals [35]. These authors obtained a higher Shannon H index than observed in our results. Moreover, in brown trout fries fed three experimental diets, 100% FM, 50% and 90% plant-based diets followed by a crossover feeding design, plant-based diets produced higher Chao1 and Shannon indexes than the FM diet, although the Chao1 values were lower than the values reported for sea trout in this experiment [7]. In general, the diversity among treatments was similar. Additionally, the NMDS analysis and the similarities of the clusters showed that the microbiome of the Tenebrio molitor group is more similar to the FM group than the Zoophobas morio group, which would be more useful for salmonid nutrition, as described by Antonopoulou et al. (2019) [13].
As mentioned above, the two insect meals exerted a similar effect to FM on maintaining the alpha diversity, and the values of dominance, equitability, and evenness were similar between all treatment groups, showing a balanced microbiome population that varied in abundance among bacterial classes, orders, genus and species as a natural consequence of the type of protein sources used. Nevertheless, the different meals that the fish consumed exerted positive effects on the microbiome, growth and survival performance of the sea trout, although the predominance of phylum Firmicutes in all treatment groups would be a consequence of the amount of plant meals, which were higher (47.17%) than animal meals (33.5%) in the diets, particularly for soybean meal, as highlighted by Kononova et al. (2019) and Michl et al. (2019) [7, 12]. Nevertheless, we cannot forget that plant meals are part of all commercial diets because of their availability and lower prices than fishmeal, and they are used to study the effects of alternative meals, such as insect meals.
Conclusion
Insects are part of the sea trout diet in nature, at least in the first stages of development, before these fish feed on more diverse prey, including other fish. To conclude, this finding may explain why the main phyla present in the digesta were similar in all the treatment groups. However, the effect of each type of meal on the lower taxonomic levels was evident, particularly in the case of superworm meal. These differences were highlighted through the NMDS and the clusters, where fish fed mealworm meal were more related to fish fed fishmeal than fish fed superworm meal. Nevertheless, further studies are necessary to corroborate the finding that insect meals are one of the best alternatives to replace fishmeal in the diets of carnivorous fish.
Methods
Fish rearing conditions and experimental diets
Live insects were provided by HiProMine S.A (Robakowo, Poland). The larvae were euthanized by freezing at -20 °C for 24 h, after which the insects were oven-dried at 50 °C for 24 h and finely ground. Then, two commercial proteases were used to hydrolyze the dried larvae meals in two subsequent steps. The full-fat larvae meals were ground and mixed using distilled water at a ratio of 4:1 (w:v) to achieve a consistency suitable for enzyme hydrolysis. Initially, the diluted bacterial (Bacillus amyloliquefaciens) endopeptidase enzyme Corolase® 7090 (AB Enzymes GmbH, Darmstadt, Germany) was added to the meals at a concentration of 1.5 g·kg − 1 of protein, and the mixture was heated for five hours at 50 °C, according to the manufacturer’s instructions. Next, 0.75 g·kg − 1 of the fungal protease enzyme Flavourzyme® (endopeptidase and exopeptidase from Aspergillus oryzae; supplied by Novozymes A/S, Denmark) was added to the mixture, which was homogenized and hydrolyzed for three hours. The hydrolyzed meals were kept at 4 °C until diet preparation.
A control diet (CON) and two experimental diets were formulated. A fixed 10% of hydrolyzed insect meal was included in both experimental diets, corresponding to 42% and 44% of fishmeal replacement by hydrolyzed mealworm (MW) and superworm (SW) diets, respectively.
The diets were manufactured at the Aquaculture Experimental Station in Muchocin, Poland using a semi-industrial single-screw extruder (Metalchem S-60, Gliwice, Poland) at 110 °C. Each feed was produced in two pellet sizes with 1.5-mm diameter used from 1st to 30th day of the experiment and 2.5-mm used from 31st to 60th day.
After extrusion, pellets were dried in an oven for 48 h at 40 °C, and then fish oil was added to the mildly heated pellets. The nutritional values are shown in Table 5.
Table 5.
Chemical composition of the three experimental diets: fishmeal diet, enzyme hydrolysed mealworm diet and enzyme hydrolysed superworm diet
| Ingredients (g kg−1) | Diets | ||
|---|---|---|---|
| CONa | MWb (42%) | SWc (44%) | |
| Fish meal | 250 | 145 | 140 |
| Mealworm meala | - | 100 | - |
| Superworm mealb | - | - | 100 |
| Soybean meal | 100 | 100 | 100 |
| Wheat flour | 219 | 220 | 226 |
| Corn gluten | 150 | 150 | 150 |
| Blood meal | 70 | 100 | 100 |
| Brewer yeast | 35 | 35 | 35 |
| Fish oil | 164 | 143 | 140 |
| Dicalcium phosphate | 7.2 | 0.8 | 2.1 |
| Premixc | 1.5 | 1.5 | 1.5 |
| DL-Methionine | 1.2 | 2.2 | 2.4 |
| L-Lysine HCL | 1.1 | 1.8 | 2.0 |
| L-Threonine | 0.6 | 0.6 | 0.7 |
| Proximate analysis (% DM) | |||
| Dry matter | 93.0 | 93.7 | 93.5 |
| Crude protein | 48.0 | 51.1 | 49.8 |
| Crude lipid | 16.3 | 14.6 | 15.3 |
| Ash | 6.5 | 5.4 | 5.1 |
| Crude fibre | 1.7 | 1.7 | 1.7 |
| Chitind | 0 | 9.3 | 4.8 |
| NFEe | 35.0 | 33.8 | 35.0 |
| Gross energy (MJ kg−1) | 22.18 | 22.77 | 22.55 |
Fishmeal diet (CON)
Enzyme hydrolysed mealworm diet (MW)
Enzyme hydrolysed superworm diet (SW)
aMealworm meal (dry matter: 95.58%, crude protein: 47.0%, crude lipid: 29.6%)
bSuperworm meal (dry matter: 96.32%, crude protein: 49.3%, crude lipid: 33.6%)
cPolfamix BASF Poland Ltd. (Kutno, Poland) (g kg−1): vitamin A, 1 000 000 IU; vitamin D3, 200 000 IU; vitamin E, 1.5 g; vitamin K, 0.2 g; vitamin B1, 0.05 g; vitamin B2, 0.4 g; vitamin B12, 0.001 g; nicotinic acid, 2.5 g; D-calcium pantothenate, 1.0 g; choline chloride, 7.5 g; folic acid, 0.1 g; methionine, 150.0 g; lysine, 150.0 g; Fe, 2.5 g; Mn, 6.5 g; Cu, 0.8 g; Co, 0.04 g; Zn, 4.0 g; J, 0.008 g; carrier > 1000.0 g
dCalculated based on chitin content of insect meals
eNitrogen-free extract = 1,000 – (crude protein + ether extract + crude fibre + ash)
Sea trout were transported from the Feed Production Technology and Aquaculture Experimental Station in Muchocin, Poland, to the Division of Inland Fisheries and Aquaculture laboratory where the experiment was conducted.
The diets were manufactured at the Aquaculture Experimental Station in Muchocin, Poland using a semi-industrial single-screw extruder (Metalchem S-60, Gliwice, Poland) at 110 °C. Each feed was produced in two pellet sizes with 1.5-mm diameter used from 1st to 30th day of the experiment and 2.5-mm used from 31st to 60th day.
At the beginning of the experimental period, a total of 225 sea trout fingerlings (5.08 ± 0.9 g) were distributed into nine tanks. The fiberglass tanks with a 60-L capacity were supplied with water from the reservoir in an open-flow system at a rate of 2 L min−1. Water parameters were recorded daily. The temperature was 14.7 ± 0.6 °C, the dissolved oxygen content was maintained at a constant value of 7.5 ± 0.3 mg L−1 and the photoperiod was maintained at 16:8 (light:dark) during the entire experiment. The fish were weighed individually at the beginning and the end of study to measure the body weight gain (BWG) using the following formula: BWG (g) = final body weight (g) − initial body weight (g). The survival rate (SR) was calculated using the formula: SR (%) = (final number of live fish/initial number of live fish) × 100.
During the experiment, the animals were weighed and counted every two weeks to adjust the feed intake ratio, and the growth and feed efficiency parameters were recorded. The feed ratio was based on a feeding chart designed for Atlantic salmon, taking into consideration the average body weight of the fingerlings and the water temperature. Animals were fed with automatic band feeders for 12 h per day, 7 days a week. The mean feed ratios were in the range from 1.39% to 1,48% of fish biomass daily depending on feed consumption.
The fish were housed in the experimental tanks for 60 days, after which the animals were sacrificed by an overdose of clove oil, according to the EU (no 2010/63/EU) regulation for experimental animals. Under sterile conditions, the animals were dissected and the digesta from the distal part of the intestine were collected. The samples were pooled by 4 fish and immediately packed, sealed in sterilized plastic bags, and stored at − 80 °C for analyses of the microbial populations by next-generation sequencing (NGS).
DNA extraction
The research was conducted in accordance with the methodology of the Authors' previous research [37, 38]. DNA was extracted with a commercial kit (Sherlock AX, A&A Biotechnology, Poland) according to the manufacturer’s instructions.
Library preparation, sequencing, basic analysis
An analysis of the bacterial population was performed based on the hypervariable region V3-V4 of the 16S rRNA gene. The specific sequences of the 341F and 785R primers with adaptors (341F primer 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; 785R primer 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC) were used to amplify the selected region and prepare the library. The primers contain Illumina adaptor sequence (in italics) and V3-V4 16S rRNA locus specific sequence [39]. All steps including amplification, indexing and library quantification were performed according to the protocol "16S Metagenomic Sequencing Library Preparation" (Illumina, USA). Briefly, the PCR reaction was performed with Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs, UK) under the following reaction conditions: 95 °C for 3 min; 25 cycles of: 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, 72 °C for 5 min, Hold at 4°. The resulting amplicons were then indexed with Nextera XT Index Kit (Illumina, USA). Library size was evaluated on Bioanalyzer 2100 DNA High Sensitivity chip (Agilent). The library was validated to the expected size on a Bioanalyzer trace for the final library of ~ 630 bp. The libraries were quantified using a fluorometric quantification method using dsDNA binding dyes. Individual concentrations of DNA libraries were calculated in nM, based on the size of DNA amplicons, as determined by an Agilent Technologies 2100 Bioanalyzer [40, 41].
Sequencing was performed on the MiSeq, 2 × 250 PE (paired-end) in order to obtain at least 50 000 read pairs per sample. Automatic data analysis was performed on the MiSeq apparatus, using the MiSeq Reporter (MSR) v2.6 software and 16S Metagenomics app. The analysis consisted of three stages: automatic demultiplexing of samples, generating fastq files containing raw reads and reads classification at several taxonomic levels: kingdom, phylum, class, order, family, genus, and species. 16S Metagenomics app provides taxonomic classification to species level based on the SILVA reference sequence database [42, 43].
Statistical analysis
Alpha diversity was analyzed in the QIIME. The beta diversity measure was calculated based on the Bray–Curtis method [44]. Bacterial diversity was assessed by Shepard analyses. Nonmetric multidimensional scaling (NMDS) has been performed to analyze bacterial community composition.
The Kolmogorov–Smirnov test was used to determine the normality of the data distribution and equality of variances. Data are presented as the means ± standard errors of the means (SEM). Statistical significance was declared at p ≤ 0.05. Bioinformatic analysis ensuring the classification of readings by species level was carried out with the free Infostat software was used for the one-way ANOVA, and if significant differences were observed among treatment groups, data were further analyzed using Tukey’s post hoc test.
Acknowledgements
We would like to thank Silvia Nogales-Mérida for helping in the study.
Abbreviations
- NGS
Next generation sequencing
- CON
Control diet
- MW
Mealworm diet
- SW
Superworm diet
- FM
Fish meal
- DM
Dry matter
- NMDS
Non-metric Multi-dimensional Scaling
Authors’ contributions
AJ: preparation of manuscript and data analysis. MR, BK, AJ: running of laboratory and statistical analysis. JM, DJ, MR: conducted and designed the experiment. All authors contributed to the analysis of the data, discussion of results and implications and commented on the manuscript at all stages. All authors read and approved the final manuscript.
Funding
This work was funded two sources: the project TEAM TECH no. POIR.04.04.00–00-204E/16–00, entitled Insects as Novel Protein Sources for Fish and Poultry, financed by the Foundation of Polish Science (POIR 4.4).
Availability of data and materials
All data are included in this published article. The raw datasets are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All animal handling protocols and methods complied with the recommendations of Directive 2010/63/EU of the European Parliament and the council of 22 September 2010 on the protection of animals used for scientific purposes, the Polish law of 15 January 2015 on the protection of animals used for scientific purposes (Dz.U.2015 poz. 266), and the good practices and recommendations of the National Ethics Committee for Animal Experiments and the Local Ethics Committee for Animal Experiments of Poznan University of Life Sciences. The trial was performed in the Division of Inland Fisheries and Aquaculture, a unit of the Faculty of Veterinary Medicine and Animal Science of Poznan University of Life Sciences with certify for animal experiments (approved unit no. 0091) by the National Ethics Committee for Animal Experiments (based on authorization by the Ministry of Science and Higher Education). The study was carried out in compliance with the ARRIVE guidelines. All methods were carried out in accordance with relevant guidelines and regulations.
According to Polish law and the EU directive (no 2010/63/EU), the experiments conducted within this study did not require the approval of the Local Ethical Committee for Experiments on Animals in Poznan.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gagliardi A, Totino V, Cacciotti F, et al. Rebuilding the gut microbiota ecosystem. Int J Environ Res Public Health. 2018;15:1679. doi: 10.3390/ijerph15081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butt RL, Volkoff H. Gut microbiota and energy homeostasis in fish. Front Endocrinol. 2019;10:9. doi: 10.3389/fendo.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semova I, Carten JD, Stombaugh J, et al. Microbiota Regulate Intestinal Absorption and Metabolism of Fatty Acids in the Zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye L, Amberg J, Chapman D, et al. Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 2014;8:541–551. doi: 10.1038/ismej.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egerton S, Culloty S, Whooley J, et al. The gut microbiota of marine fish. Front Microbiol. 2018;9:873. doi: 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dehler CE, Secombes CJ, Martin SAM. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.) Aquaculture. 2017;467:149–157. doi: 10.1016/j.aquaculture.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michl SC, Beyer M, Ratten J-M, et al. A diet-change modulates the previously established bacterial gut community in juvenile brown trout (Salmo trutta) Sci Rep. 2019;9:2339. doi: 10.1038/s41598-019-38800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michl SC, Ratten J-M, Beyer M, et al. The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): Diet-dependent shifts of bacterial community structures. PLoS One. 2017;12:e0177735. doi: 10.1371/journal.pone.0177735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimoldi S, Terova G, Ascione C, et al. Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PLoS One. 2018;13:e0193652. doi: 10.1371/journal.pone.0193652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huyben D, Vidaković A, Werner Hallgren S, Langeland M. High-throughput sequencing of gut microbiota in rainbow trout (Oncorhynchus mykiss) fed larval and pre-pupae stages of black soldier fly (Hermetia illucens) Aquaculture. 2019;500:485–491. doi: 10.1016/j.aquaculture.2018.10.034. [DOI] [Google Scholar]
- 11.Al-Hisnawi A, Ringø E, Davies SJ, et al. First report on the autochthonous gut microbiota of brown trout (Salmo trutta Linnaeus) Aquac Res. 2015;46:2962–2971. doi: 10.1111/are.12451. [DOI] [Google Scholar]
- 12.Kononova SV, Zinchenko DV, Muranova TA, et al. Intestinal microbiota of salmonids and its changes upon introduction of soy proteins to fish feed. Aquacult Int. 2019;27:475–496. doi: 10.1007/s10499-019-00341-1. [DOI] [Google Scholar]
- 13.Antonopoulou E, Nikouli E, Piccolo G, et al. Reshaping gut bacterial communities after dietary Tenebrio molitor larvae meal supplementation in three fish species. Aquaculture. 2019;503:628–635. doi: 10.1016/j.aquaculture.2018.12.013. [DOI] [Google Scholar]
- 14.Gasco L, Finke M, van Huis A. Can diets containing insects promote animal health? J Insects Food Feed. 2018;4:1–4. doi: 10.3920/JIFF2018.x001. [DOI] [Google Scholar]
- 15.Józefiak A, Engberg RM. Insect proteins as a potential source of antimicrobial peptides in livestock production. A review. J Anim Feed Sci. 2017;25:87. doi: 10.22358/jafs/69998/2017. [DOI] [Google Scholar]
- 16.Rimoldi S, Gini E, Iannini F, et al. The Effects of Dietary Insect Meal from Hermetia illucens Prepupae on Autochthonous Gut Microbiota of Rainbow Trout (Oncorhynchus mykiss) Animals. 2019;9:143. doi: 10.3390/ani9040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schade RP, Van Rijn M, Timmers HJLM, et al. Clostridium cadaveris bacteraemia: Two cases and review. Scand J Infect Dis. 2006;38:59–62. doi: 10.1080/00365540500388792. [DOI] [PubMed] [Google Scholar]
- 18.Rinkel BE, Manoylov KM. Calothrix — an evaluation of fresh water species in United States rivers and streams, their distribution and preliminary ecological findings. Proc Acad Natl Sci Phila. 2014;163:43–59. doi: 10.1635/053.163.0108. [DOI] [Google Scholar]
- 19.Jiang S, Cai L, Lv L, Li L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb Cell Fact. 2021;20:45. doi: 10.1186/s12934-021-01537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben Taheur F, Kouidhi B, Fdhila K, et al. Anti-bacterial and anti-biofilm activity of probiotic bacteria against oral pathogens. Micro Pathog. 2016;97:213–220. doi: 10.1016/j.micpath.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Soundharrajan I, Kim D, Kuppusamy P, et al. Probiotic and Triticale Silage Fermentation Potential of Pediococcus pentosaceus and Lactobacillus brevis and Their Impacts on Pathogenic Bacteria. Microorganisms. 2019;7:318. doi: 10.3390/microorganisms7090318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu HH, Tsai CC, Hsih HY, Tsen H-Y. Screening from pickled vegetables the potential probiotic strains of lactic acid bacteria able to inhibit the Salmonella invasion in mice. J Appl Microbiol. 2008;104:605–612. doi: 10.1111/j.1365-2672.2007.03573.x. [DOI] [PubMed] [Google Scholar]
- 23.Shin MS, Han SK, Ji AR, et al. Isolation and characterization of bacteriocin-producing bacteria from the gastrointestinal tract of broiler chickens for probiotic use. J Appl Microbiol. 2008;105:2203–2212. doi: 10.1111/j.1365-2672.2008.03935.x. [DOI] [PubMed] [Google Scholar]
- 24.Cao Z, Pan H, Tong H, et al. In vitro evaluation of probiotic potential of Pediococcus pentosaceus L1 isolated from paocai—a Chinese fermented vegetable. Ann Microbiol. 2016;66:963–971. doi: 10.1007/s13213-015-1182-2. [DOI] [Google Scholar]
- 25.Varsha KK, Priya S, Devendra L, Nampoothiri KM. Control of spoilage fungi by protective lactic acid bacteria displaying probiotic properties. Appl Biochem Biotechnol. 2014;172:3402–3413. doi: 10.1007/s12010-014-0779-4. [DOI] [PubMed] [Google Scholar]
- 26.Bartkiene E, Lele V, Ruzauskas M, et al. Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation. Microorganisms. 2020;8. 10.3390/microorganisms8010064. [DOI] [PMC free article] [PubMed]
- 27.Kim JS, Kim JH, Palaniyandi SA, et al. Yak-Kong Soybean (Glycine max) Fermented by a Novel Pediococcus pentosaceus Inhibits the Oxidative Stress-Induced Monocyte-Endothelial Cell Adhesion. Nutrients. 2019;11:1380. doi: 10.3390/nu11061380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulgasem BY, Lani MN, Hassan Z, et al. Antifungal activity of lactic acid bacteria strains isolated from natural honey against pathogenic candida species. Mycobiology. 2016;44:302–309. doi: 10.5941/MYCO.2016.44.4.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajpai VK, Han J-H, Rather IA, et al. Characterization and antibacterial potential of lactic acid bacterium pediococcus pentosaceus 4i1 isolated from freshwater fish Zacco koreanus. Front Microbiol. 2016;7:2037. doi: 10.3389/fmicb.2016.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao Q, Sun X, Sun J, et al. A candidate probiotic strain of Enterococcus faecium from the intestine of the crucian carp Carassius auratus. AMB Express. 2020;10:40. doi: 10.1186/s13568-020-00973-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanchi H, Mottawea W, Sebei K, Hammami R. The genus enterococcus: between probiotic potential and safety concerns—an update. Front Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Román L, Padilla D, Acosta F, et al. The effect of probiotic Enterococcus gallinarum L-1 on the innate immune parameters of outstanding species to marine aquaculture. J Appl Anim Res. 2015;43:177–183. doi: 10.1080/09712119.2014.928635. [DOI] [Google Scholar]
- 33.Hagi T, Kobayashi M, Kawamoto S, et al. Expression of novel carotenoid biosynthesis genes from Enterococcus gilvus improves the multistress tolerance of Lactococcus lactis. J Appl Microbiol. 2013;114:1763–1771. doi: 10.1111/jam.12182. [DOI] [PubMed] [Google Scholar]
- 34.Mouriño JLP, Pereira G do V, Vieira F do N, et al. Isolation of probiotic bacteria from the hybrid South American catfish Pseudoplatystoma reticulatum×Pseudoplatystoma corruscans (Siluriformes: Pimelodidae): a haematological approach. Aquaculture Rep. 2016;3:166–171. 10.1016/j.aqrep.2016.03.001
- 35.Gajardo K, Rodiles A, Kortner TM, et al. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): A basis for comparative gut microbial research. Sci Rep. 2016;6:30893. doi: 10.1038/srep30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringø E, Gatesoupe FJ. Lactic acid bacteria in fish: a review. Aquaculture. 1998;160:177–203. doi: 10.1016/S0044-8486(97)00299-8. [DOI] [Google Scholar]
- 37.Józefiak A, Benzertiha A, Kierończyk B, et al. Improvement of cecal commensal microbiome following the insect additive into chicken diet. Animals. 2020;10. 10.3390/ani10040577 [DOI] [PMC free article] [PubMed]
- 38.Drażbo AA, Juśkiewicz J, Józefiak A, Konieczka P. The fermentation process improves the nutritional value of rapeseed cake for turkeys—effects on performance, gut bacterial population and its fermentative activity. Animals. 2020;10:1711. doi: 10.3390/ani10091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1–e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo A-TE, Jedlicka JA. Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol Ecol Res. 2014;14:1183–1197. doi: 10.1111/1755-0998.12269. [DOI] [PubMed] [Google Scholar]
- 41.Hess JF, Kohl TA, Kotrová M, et al. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv. 2020;41:107537. doi: 10.1016/j.biotechadv.2020.107537. [DOI] [PubMed] [Google Scholar]
- 42.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray JR, Curtis JT. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in this published article. The raw datasets are available from the corresponding author on reasonable request.