Abstract
Background
The genomes of present-day non-Africans are composed of 1–3% of Neandertal-derived DNA as a consequence of admixture events between Neandertals and anatomically modern humans about 50–60 thousand years ago. Neandertal-introgressed single nucleotide polymorphisms (aSNPs) have been associated with modern human disease-related traits, which are risk factors for pancreatic ductal adenocarcinoma (PDAC), such as obesity, type 2 diabetes, and inflammation. In this study, we aimed at investigating the role of aSNPs in PDAC in three Eurasian populations.
Results
The high-coverage Vindija Neandertal genome was used to select aSNPs in non-African populations from 1000 Genomes project phase 3 data. Then, the association between aSNPs and PDAC risk was tested independently in Europeans and East Asians, using existing GWAS data on more than 200 000 individuals. We did not find any significant associations between aSNPs and PDAC in samples of European descent, whereas, in East Asians, we observed that the Chr10p12.1-rs117585753-T allele (MAF = 10%) increased the risk to develop PDAC (OR = 1.35, 95%CI 1.19–1.54, P = 3.59 × 10–6), with a P-value close to a threshold that takes into account multiple testing.
Conclusions
Our results show only a minimal contribution of Neandertal SNPs to PDAC risk.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40659-023-00457-y.
Keywords: Neandertal, Pancreatic cancer, Association study, Introgression, Eurasians, Admixture
Introduction
By comparing the genome sequences of Neandertal and modern genomes it has been shown that ~ 1–3% of the genomes of present-day non-Africans are of Neandertal ancestry [1–3] with 8–20% higher levels of Neandertal ancestry in East Asians compared to Europeans [4–6]. Through phenotypic information from genome-wide association studies it has been shown that introgressed Neandertal DNA still significantly influences the phenotypic variability of anatomically modern humans (AMHs) today. Neandertal-introgressed Single Nucleotide Polymorphisms (aSNPs) have for example been associated with several human traits, such as the genetic susceptibility of type 2 diabetes (T2D), obesity, age of menopause, neurological traits, morning preference, skin and hair morphology, immune response, and inflammation [7–17]. Among these traits are several factors, such as overweight, obesity, T2D, deregulation of the immune system, and chronic inflammation that play a key role in pancreatic ductal adenocarcinoma (PDAC) onset and progression [18–21].
Alongside a small number of environmental risk factors [22–24], PDAC susceptibility has a strong genetic component. Rare high penetrance variants involved in hereditary syndromes (reviewed in Gentiluomo et al.) and frequent low and moderate penetrance variants, discovered through candidate gene and genome-wide association studies (GWAS), have been identified as playing a role in PDAC onset [25–39]. However, the common risk loci discovered so far explain only a small proportion of the overall heritability of the disease [40]. Furthermore, PDAC is a late onset disease [29, 41, 42], thus loci associated with PDAC susceptibility tend to persist in the AMH gene pool, eluding purifying selection.
Considering that aSNPs are associated with several PDAC risk factors and that the genetic contribution to PDAC etiology still needs to be elucidated, we aimed at investigating the Neandertal legacy of PDAC genetic risk. We analysed PDAC GWAS cohorts from different Eurasian populations for significant associations with aSNPs to study the role of Neandertal admixture and PDAC risk in different ancestry groups. This study is the first attempt to investigate the role of archaic admixture on PDAC development.
Results
In this study, 389 144 aSNPs were identified among the non-African populations of the 1000 Genomes project [43]. The association between aSNPs and the risk of developing PDAC was tested in three ancestry groups: non-Finnish Europeans, Finns, and East Asians.
For non-Finnish Europeans, 161 283 aSNPs were available to be analysed in the discovery phase, using the genotypes of PanScan + PanC4 studies. Considering a P < 0.05, 263 aSNPs resulted associated with PDAC risk in the combined PanScan + PanC4 dataset. All 263 of these aSNPs also passed the P < 0.05 threshold when only PanScan or PanC4 were considered separately. Among them, 212 showed residual LD (r2 > 0.5). After pruning, 51 independent aSNPs associations spanning across 51 loci were observed (Fig. 1, Additional File 1). None of the 51 aSNPs remained associated with PDAC after correction for multiple testing (pj = 2.30 × 10–6). The SNP with the lowest P-value was Chr2p14-rs12998719, (OR = 1.11, 95%CI 1.05–1.16, P = 5.51 × 10–5) (Table 1). This variant has been already reported to be associated with PDAC risk [32] and was genotyped in the context of the PANDoRA consortium (replication phase). The results of the replication phase did not show a statistically significant association (OR = 1.46, 95%CI 0.95–1.15, P = 0.38) (Table 1).
Fig. 1.
aSNPs filtering and analysis workflow for each ancestry group. The figure displays aSNPs analysis workflow for non-Finnish Europeans, Finns, and East Asians. The 389 144 aSNPs identified in all non-African populations from 1000 Genomes project phase 3, were filtered and analysed for each ancestry group. aSNP Neandertal introgressed SNP. 1aSNPs that showed an association P-value < 0.05 in PanScan, PanC4 and in the two datasets combined. aSNPs with a P < 0.05 in PanScan (7850), PanC4 (8141), and the combined datasets (8718). 2aSNPs with a P-value < 0.05 and an identical direction of the effect in all the three GWASs included in JaPAN
Table 1.
Candidate aSNPs for each ancestry group
| Panel A. Association studies results | ||||||||
|---|---|---|---|---|---|---|---|---|
| Population | aSNP | Position (GRCh37) | Alleles (M/m) | Genea | MAFb | OR (95%C.I.) | P | |
| PanScan + PanC4c | rs12998719 | 2:67583252 | G/A | – | 29% | 1.11 (1.05–1.16) | 5.51 × 10–5 | |
| PANDoRAc | rs12998719 | 2:67583252 | G/A | – | 28% | 1.05 (0.95–1.15) | 0.38 | |
| FinnGen | rs113955626 | 3:23107748 | C/T | – | 7% | 1.35 (1.17–1.55) | 4.79 × 10–5 | |
| JaPAN | rs117585753 | 10:25190572 | C/T | PRTFDC1 | 10% | 1.35 (1.19–1.54) | 3.59 × 10–6 | |
| Panel B. Top SNP population-based frequency | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | aSNP | Position (GRCh37) | Alleles (M/m) | 1000G MAF | gnomAD MAF | HGDPd | |||||
| EUR | FIN | EAS (%) | EUR (%) | FIN | EAS (%) | EUR (%) | EAS | ||||
| PanScan + PanC4c | rs12998719 | 2:67583252 | G/A | 27% | 30% | 43 | 36 | 29% | 42 | 25 | 35% |
| FinnGen | rs113955626 | 3:23107748 | C/T | 4% | 4% | 13 | 5 | 8% | 14 | 8 | 18% |
| JaPAN | rs117585753 | 10:25190572 | C/T | 8‰ | 5‰ | 10 | 1 | 3‰ | 9 | 1 | 8% |
Panel A shows the summary statistics of aSNPs with the lowest association P-value, panel B shows the minor allele frequency of SNPs with the lowest P-value of associations across several datasets
aSNP Neandertal introgressed SNP; M major allele; m, minor allele; MAF minor allele frequency; OR Odds Ratio; 95%C.I. 95% Confidence Interval; PRTFDC1 phosphoribosyl transferase domain containing 1; 1000G 1000 Genomes project phase 3 [43]; gnomAD Genome Aggregation Database [44]; HGDP Human Genome Diversity Project [45] EUR Europeans; FIN Finns; EAS East Asians
aGene in which aSNP lies
bMAF of the SNP of interest in the analysed datasets. Since allele frequencies are not freely available in JaPAN dataset, the MAF in East Asians from 1000G is reported
cPanScan + PanC4 (discovery phase), PANDoRA (replication phase)
dIn HGDP, SNP frequency in Finns is not available. Frequency in all European populations is displayed
In FinnGen, 251 090 aSNPs were found, and after LD-pruning (r2 > 0.5), 1154 independent aSNPs with a P < 0.05 were observed (Fig. 1). The aSNP with the lowest P-value in FinnGen was Chr3p24.3-rs113955626 (OR = 1.35, 95%CI 1.17–1.55, P = 4.79 × 10–5) (Table 1); this aSNP did not reach the Bonferroni adjusted significance threshold (pj = 2.30 × 10–6).
In the JaPAN dataset, which includes data of the meta-analysis of three GWAS conducted on individuals of Asian descent, 158 393 aSNPs were analysed. The association analysis showed 656 independent aSNPs with a P < 0.05 in all the three GWASs (Fig. 1). The best candidate was Chr10p12.1-rs117585753 (OR = 1.35, 95%CI 1.19–1.54, P = 3.59 × 10–6), whose P-value was very close to the Bonferroni-adjusted threshold (pj = 2.28 × 10–6) (Table 1).
Discussion
We tested the effects of Neandertal introgression on PDAC susceptibility in three ancestry groups. In non-Finnish Europeans and Finns, no novel significant associations between aSNPs and PDAC were observed.
In JaPAN, we found that the T allele of Chr10p12.1-rs117585753 increased the risk to develop PDAC (P = 3.59 × 10–6). This association was not statistically significant when considering multiple testing. However, it is very close to the Bonferroni corrected threshold (pj = 2.28 × 10–6). The functional implications of this aSNP have not been clarified yet: according to GWAS catalog, it is not associated with any complex human trait.
Interestingly, the T allele of Chr10p12.1-rs117585753 is present in EAS (MAF = 10%), whereas it is almost absent in the other populations represented in 1000 Genomes project (e.g., MAF < 0.01 in Europeans from 1000 Genomes project). Since Chr10p12.1-rs117585753 is polymorphic only in Asians, it is possible that the role of this aSNP in complex traits has not been elucidated yet because most of the association studies have been conducted in cohorts with participants of European descent [46]. The lower number of studies with Asian individuals implies that the associations between SNPs, which are rare in Europeans but common in Asians, still need further investigation to be understood entirely.
Chr10p12.1-rs117585753 lies in an intron of the protein-coding PRTFDC1 gene, in which, according to the GWAS catalog, there are SNPs associated with blood cell count [47–49]. Several white and red blood cell count parameters have been used to predict immune response and inflammation in various diseases, including PDAC [50, 51]. One SNP in PRTFDC1 (Chr10p12.1-rs7905553) is in weak LD (r2 = 0.14, D’ = 0.96) with Chr10p12.1-rs117585753 in EAS, and according to GWAS catalog it is associated with red blood cell distribution width (RDW) [52], which is a parameter of erythrocyte variation. RDW has been proposed as a biomarker of the inflammatory state that could predict progression/prognosis in PDAC [53], suggesting a potential contribution of the PRTFDC1 genomic region and Chr10p12.1-rs117585753 in PDAC and immunity.
Several Neandertal-derived haplotypes involved in immunity have been reported to be under selection after Neandertal-AMH introgression. In fact, the positive selection of aSNPs that lead to adaptation (adaptive introgression) has been observed to be driven by the immune response to pathogens [8, 9, 54–57].
Possible limitations of our approach are represented by the fact that we could have underestimated the role of rare variants (MAF < 1%) because we did not have enough statistical power to detect associations between rare aSNPs and PDAC, although we used the largest PDAC datasets currently available, which included more than 200 000 individuals of three different ancestries. An additional potential limitation of this work is that 93 695 out of 389 144 aSNPs identified in Eurasian genomes could not be found in PanScan + PanC4, FinnGen, and JaPAN. Therefore, the role of these aSNPs in PDAC susceptibility was not explored. In future analyses, larger reference panels for imputation could be used to maximize the investigated Neandertal-derived genetic variability.
Conclusions
In conclusion, we observed that the Neandertal introgressed DNA does not influence PDAC susceptibility in populations of European descent. Interestingly, we observed a potential association between Chr10p12.1-rs117585753-T and an increased risk of developing PDAC in populations of Asian descent, although not formally significant after correction for multiple testing. This aSNP is polymorphic only in East Asians and is situated in a genomic region involved in immunity. Further investigations are needed to elucidate the evolutionary processes that lead to these aSNPs in the AMH gene pool and the role of aSNPs in PDAC risk, and more broadly, to explore the Neandertal legacy in the susceptibility to other cancer types.
Methods
Neandertal SNPs identification
The method to select aSNPs was previously described [12]. Briefly, to define a potential introgressed allele, we used four criteria that needed to be fulfilled: (a) the allele is shared between the Vindija Neandertal [5] and at least one non-African population from 1000 Genomes project phase 3 [43]; (b) the allele is not present in Yoruba from sub-Saharan Africa; (c) the allele is carried in homozygous state by Vindija Neandertal; (d) based on the haplotype length, the allele is more likely derived from Neandertal-AMH admixture than incomplete lineage sorting (ILS). To apply the fourth criterion, an approach, that was previously described by Huerta-Sánchez et al., and Dannemann et al. was used [54, 58]. Briefly, it allows the identification of putative Neandertal introgressed regions in all non-African 1000 Genomes project populations. Two recombination maps [59, 60] were used to calculate the expected ILS segments length based on the local recombination rate. Then, the probability that a segment length was consistent with ILS was computed and the resulting P-values were corrected through Benjamini–Hochberg method. Haplotypes that showed an adjusted P-value < 0.05 were considered as introgressed from Neandertal. The aSNPs used in the following analyses lay on one of these Neandertal-derived haplotypes.
All the analyses were based on human genome assembly GRCh37, and only biallelic loci were considered, excluding indels.
Study populations
The association between aSNPs and PDAC risk was tested in three ancestry groups: non-Finnish Europeans, Finns and East Asians. A two-phase association study (discovery and replication) was performed to examine if aSNPs identified in non-Finnish Europeans affected PDAC susceptibility. On the other hand, a validation set was not available for Finns and East Asians, and the association between aSNPs and PDAC was tested by searching for aSNPs in FinnGen and JaPAN datasets, respectively (see below).
For non-Finnish European analyses, the discovery set included data of the Pancreatic Cancer Cohort Consortium (PanScan) and the Pancreatic Cancer Case–Control Consortium (PanC4). The data were downloaded from the database of Genotypes and Phenotypes (dbGaP, https://www.ncbi.nlm.nih.gov/gap/). The dbGaP study accession numbers were: phs000206.v5.p3 and phs000648.v1.p1.; the project reference number was #12644. Details about data collection, genotyping methods and analyses are described in the original publications [26, 31, 32, 61].
Genotype data were imputed separately, for each dataset, using the Michigan Imputation Server (https://imputationserver.sph.umich.edu) [62] and the Haplotype Reference Consortium (HRC, V.r1.1) as reference panel [63]. Prior to the imputation, the following quality controls were applied: genotypes missingness (call rate < 0.9), heterozygosity (> 3 SD from the mean), relatedness (PI_HAT > 0.2), PCA outliers (using PCA), and Hardy–Weinberg equilibrium (P < 1 × 10−6). After imputation, SNPs with low imputation quality (INFO score r2 < 0.7) were excluded. Finally, the imputed datasets were merged. A total of 7 543 430 SNPs passed the quality controls on the autosomal genome, and 8738 PDAC cases and 7034 controls were used in the analysis (Table 2).
Table 2.
Study population description for each ancestry group
| Population | Cases | Controls | Sex | Median age (25–75% percentile) | Total number of subjects | ||
|---|---|---|---|---|---|---|---|
| Male | Female | Cases | Controls | ||||
| non-Finnish Europeans | |||||||
| PanScan + PanC4a | 8738 | 7034 | 54% | 46% | 65 (55–75) | 65 (55–75) | 15 772 |
| PANDoRAa | 1894 | 3089 | 51% | 49% | 67 (59–73) | 59 (49–67) | 4983 |
| Finns | |||||||
| FinnGen | 1249 | 259 583 | – | – | – | – | 260 832 |
| East Asians | |||||||
| JaPANb | 2039 | 32 592 | 49.3–62.6% | – | 62.7–66.3 | 43.6–56.3 | 34 631 |
The table shows the number of cases and controls in PanScan + PanC4, PANDoRA, FinnGen and JaPAN. Male and female count and median age of cases and controls are displayed for each study
aPanScan + PanC4 (discovery phase), PANDoRA (replication phase)
bData for male count and age are displayed as minimum–maximum values of the three GWASs included in JaPAN
Data not available: “- “
The replication of aSNPs with a P-value of association with PDAC risk lower than the Bonferroni-adjusted threshold (see below) was attempted in the Pancreatic Disease Research (PANDoRA) consortium [64, 65]. PANDoRA is a multicentric study on pancreatic cancer based mainly on European countries (Greece, Italy, Germany, Netherlands, Denmark, Czech Republic, Hungary, Poland, Ukraine, Lithuania, UK). In addition, PANDoRA includes a subgroup of Brazilian cases and controls that were excluded from the validation set in this study because PanScan + PanC4 (discovery set) included only Caucasian samples, while Brazilians belong to different ancestries (unlike the other PANDoRA samples). Information on sex, and age (recruitment for controls and diagnosis for the cases) was collected for each participant. The controls were enrolled among the general population, blood donors or hospitalised individuals not affected by cancer, chronic pancreatitis, or diabetes [64]. For this study, 4983 individuals (1894 PDAC cases and 3089 controls) from PANDoRA were included in the analysis (Table 2).
Non-Finnish Europeans and Finns were analysed separately because PanScan + PanC4 and PANDoRA mainly include subjects with Central European ancestry. We used the FinnGen Release 8 (R8) data that consists of GWAS summary statistics of 1249 pancreatic cancer cases and 259 583 controls with Finnish ancestry (Table 2). Subjects affected by other cancer types were excluded from the controls (https://FinnGen.gitbook.io/documentation/) [66].
To examine the association between aSNPs identified in East Asians and PDAC, we downloaded JaPAN consortium dataset that consisted of summary statistics of a meta-analysis of three GWASs (JaPAN, National Cancer Center and BioBank Japan GWASs). Comprehensive information on genotyping and data analysis are given in the original publication [67]. Summary statistics for the GWAS analysis are available on the JaPAN consortium website (http://www.aichi-med-u.ac.jp/JaPAN/current_initiatives-e.html) and include 34 631 individuals of East Asian origin (2039 PDAC cases and 32 592 controls) (Table 2).
Data and statistical analyses
For non-Finnish Europeans, the association between aSNPs and PDAC susceptibility was tested in the PanScan + PanC4 dataset using logistic regression analysis, adjusting for age, sex and the top eight principal components (Fig. 2). To obtain a list of independent aSNPs, all aSNPs in linkage disequilibrium (LD; r2 > 0.5) with each other were excluded, and in each LD block the aSNP with the lowest association P-value was selected. Then, all aSNPs showing an association lower than the threshold for statistical significance corrected for multiple testing in PanScan, PanC4 and in the combined datasets were selected for replication in PANDoRA.
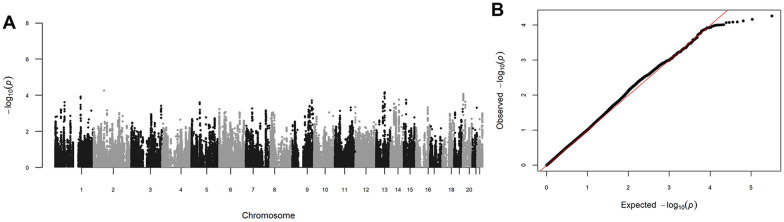
Fig. 2.
Manhattan and Quantile–Quantile (Q-Q) plots of PanScan + PanC4 association study results. The P-values displayed in Manhattan (A) and Q-Q plots (B) are calculated combining PanScan and PanC4 datasets. The plots were done using qqman R package (https://cran.r-project.org/web/packages/qqman/index.html) [68]. The inflation factors (l) did not indicate systematic inflation for PanScan (l = 1.02), PanC4 (l = 1.05), and combined datasets (l = 1.05). The inflation factors were computed using simtrait R package (https://cran.r-project.org/web/packages/simtrait/index.html) [69]
The genomic DNA of the PANDoRA samples was extracted from circulating blood using the QIamp® 96 DNA QIcube® HT Kit (Qiagen, Hilden, Germany). The genotyping was done using TaqMan RealTime PCR assays in 384-well plates. Each plate included cases and controls, duplicated samples for quality controls (QCs) and negative controls. The fluorescent signal detection was detected through a QuantStudioTM 5 Real-Time PCR system (Thermofisher, USA) and genotypes were called using the QuantStudio™ Design and Analysis Software v1.5.1. Samples with a genotyping call rate lower than 75% were excluded from the analysis. Hardy–Weinberg equilibrium test was performed with the Pearson chi-square test. To test the association between aSNPs and PDAC risk in PANDoRA, a logistic regression adjusted for age, sex, and country of origin was used.
For Finns and East Asians, the analyses were carried out in parallel, keeping separated the two ancestry groups. Considering that for FinnGen and JaPAN we used summary statistics, we looked at the P-value for association in these two datasets for the aSNPs selected for the two populations. Since JaPAN is a meta-analysis of three studies, along with P-value, the concordance of the direction of the effect between the three GWASs was considered.
P-value correction for multiple testing was performed using Bonferroni correction and considering the independent (r2 < 0.8) aSNPs. The adjusted significance thresholds were: 0.05/19 623 = 2.55 × 10–6 for PanScan + PanC4 and PANDoRA; 0.05/21 780 = 2.30 × 10–6 for FinnGen; 0.05/21 965 = 2.28 × 10–6 for JaPAN.
Supplementary Information
Additional file 1: Title of data: Independent associations (P<0.05) identified in PanScan+PanC4 association studiesDescription of data: Independent (r2<0.5) aSNPs showing an association P<0.05 in PanScan, PanC4, and combined datasets. The displayed summary statistics and MAF are referred to the analyses with the combined datasets. (Abbreviations: aSNP, Neandertal introgressed Single Nucleotide Polymorphism; m, minor allele; M, major allele, MAF, minor allele frequency).
Acknowledgements
This article is based upon work from COST Action “Identification of biological markers for prevention and translational medicine in pancreatic cancer (TRANSPAN)”, CA21116, supported by COST (European Cooperation in Science and Technology). This research used genotyping data provided to the PANDoRA consortium by the EPIC cohort, for which we would like to thank the contributors from EPIC UK. We want to acknowledge the participants and investigators of the FinnGen study.
Abbreviations
- 95%C.I.
95% Confidence interval
- AMH
Anatomically modern human
- aSNP
Neandertal intogressed single nucleotide polymorphism
- dbGaP
Database of genotypes and phenotypes
- EAS
East Asians from 1000 genomes project phase 3
- EUR
Europeans from 1000 genomes project phase 3
- gnomAD
Genome aggregation database
- GWAS
Genome-wide association study
- HGDP
Human genome diverity project
- HRC
Haplotype Reference Consortium
- ILS
Incomplete lineage sorting
- JaPAN
Japan Pancreatic Cancer Research consortium
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- PanC4
Pancreatic Cancer Case Control consortium
- PANDoRA
Pancreatic Disease Research consortium
- PanScan
Pancreatic Cancer Cohort consortium
- PCA
Principal component analysis
- PDAC
Pancreatic ductal adenocarcinoma
- QC
Quality controls
- Q-Q plot
Quantile–quantile plot
- RDW
Red blood cell distribution width
- T2D
Type 2 diabetes
Author contributions
The study was designed by DC, MD and ST; the experiments and statistical analyses were carried out by MP (Margherita Piccardi), MG (Manuel Gentiluomo), and SB (Stefania Bertoncini). The manuscript was drafted by MP (Margherita Piccardi), MG (Manuel Gentiluomo), MD, FC, ST and DC. The other authors collected the samples, provided the data and contributed in the analysis of the results. All authors critically read, and approved the manuscript.
Funding
This study was supported by Associazione Italiana Ricerca Cancro (AIRC IG n. 26343) [Aldo Scarpa], Italian Ministry of Health (FIMPCUP_J38D19000690001) [Aldo Scarpa], the Ministry of Health of the Czech Republic (Grant number NV19-08–00113) [Pavel Soucek], Cancer Research UK (C7690/A26881) [Eithne Costello], Pancreatic Cancer UK Research Innovation Fund [Eithne Costello], Charles University Research Fund (Cooperatio No. 43 -Surgical Disciplines) [Ludmila Vodickova], Ministry of Health of the Czech Republic (AZV NU21-07–00247) [Ludmila Vodickova], Ministry of Health of the Czech Republic (grant no. NV19-03–00097 to B.M-D.) [Beatrice Mohelníková Duchoňová], Italian Ministry of Health grants to Fondazione “Casa Sollievo della Sofferenza” IRCCS Hospital, San Giovanni Rotondo (FG), Italy [Francesco Perri], and by the “5 × 1000” voluntary contribution [Francesco Perri].
Availability of data and materials
The PanScan and PanC4 genotyping data are available from the dbGaP website (study accession numbers phs000206.v5.p3 and phs000648.v1.p1). The JaPAN data used in this work are available in the JaPAN consortium website (http://www.aichi-med-u.ac.jp/JaPAN/current_initiatives-e.html). FinnGen R8 dataset of PDAC cases and controls (“C3_PANCREAS_EXALLC”) is downloadable from the FinnGen official website (https://finngen.gitbook.io/documentation/). The PANDoRA data for this work will be made available to researchers who submit a reasonable request to the corresponding author, conditional to approval by the PANDoRA Steering Committee and Ethics Commission of the Medical Faculty of the University of Heidelberg. Data will be stripped from all information allowing identification of study participants.
Declarations
Ethics approval and consent to participate
Individual-level genotyping data were used in this work for PanScan, PanC4 and PANDoRA. Each study participating in PanScan and PanC4 obtained approval from the responsible institutional review board (IRB) and IRB certification permitting data sharing in accordance with the NIH Policy for sharing of Data Obtained in NIH-Supported or NIH-Conducted Genome Wide Association Studies. The PANDoRA study protocol was approved by the Ethics Commission of the Medical Faculty of the University of Heidelberg. In accordance with the Declaration of Helsinki, written informed consent was obtained from each participant. FinnGen and JaPAN (meta-analysis of JaPAN consortium, BioBank Japan and National Cancer Center GWASs) summary statistics were used in this study. FinnGen has the permission to use samples from Finnish biobanks and data from national health registers for research. Each sample is coded and individual data will not be released. The project was evaluated by the Coordinating Ethics Committee of the Helsinki and Uusimaa Hospital District. FinnGen adhere to the Biobank Law, the Personal Data Act and EU Data Protection Regulation. JaPAN consortium GWAS study protocol was approved by the Ethical Review Board of Aichi Medical University, the Institutional Ethics Committee of Aichi Cancer Center, the Human Genome and Gene Analysis Research Ethics Committee of Nagoya University, and the ethics committees of the participating hospitals. BioBank Japan GWAS protocol was accepted by the ethics committees of the RIKEN Center for Integrative Medical Sciences. The informed consent for each participant was collected following the corresponding institutional ethics committee guidelines. National Cancer Center GWAS project was approved by the ethics committee of the National Cancer Center.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Margherita Piccardi and Manuel Gentiluomo these authors share the first position.
References
- 1.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514(7523):445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, et al. The complete genome sequence of a Neanderthal from the Altai mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wall JD, Yang MA, Jay F, Kim SK, Durand EY, Stevison LS, et al. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics. 2013;194(1):199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, et al. A high-coverage Neandertal genome from Vindija cave in Croatia. Science. 2017;358(6363):655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Wolf AB, Fu W, Li L, Akey JM. Identifying and interpreting apparent neanderthal ancestry in African individuals. Cell. 2020;180(4):677–687.e16. doi: 10.1016/j.cell.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507(7492):354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 2016;98(1):5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sams AJ, Dumaine A, Nédélec Y, Yotova V, Alfieri C, Tanner JE, et al. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 2016;17(1):246. doi: 10.1186/s13059-016-1098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351(6274):737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCoy RC, Wakefield J, Akey JM. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168(5):916–927.e12. doi: 10.1016/j.cell.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannemann M. The population-specific impact of Neandertal introgression on human disease. Genome Biol Evol. 2021 doi: 10.1093/gbe/evaa250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McArthur E, Rinker DC, Capra JA. Quantifying the contribution of Neanderthal introgression to the heritability of complex traits. Nat Commun. 2021;12(1):4481. doi: 10.1038/s41467-021-24582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reilly PF, Tjahjadi A, Miller SL, Akey JM, Tucci S. The contribution of Neanderthal introgression to modern human traits. Curr Biol. 2022;32(18):R970–R983. doi: 10.1016/j.cub.2022.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McArthur E, Rinker DC, Capra JA. Quantifying the contribution of Neanderthal introgression to the heritability of complex traits. Nat Commun. 2021;12(1):4481. doi: 10.1038/s41467-021-24582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X, Robles CR, Pazokitoroudi A, Ganna A, Gusev A, Durvasula A, et al. The lingering effects of Neanderthal introgression on human complex traits. eLife. 2023;12:e80757. doi: 10.7554/eLife.80757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dannemann M, Kelso J. The contribution of Neanderthals to phenotypic variation in modern humans. Am J Hum Genet. 2017;101(4):578–589. doi: 10.1016/j.ajhg.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Incio J, Liu H, Suboj P, Chin SM, Chen IX, Pinter M, et al. Obesity-induced inflammation and desmoplasia promote pancreatic cancer progression and resistance to chemotherapy. Cancer Discov. 2016;6(8):852–869. doi: 10.1158/2159-8290.CD-15-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen DK, Korc M, Petersen GM, Eibl G, Li D, Rickels MR, et al. Diabetes, pancreatogenic diabetes, and pancreatic cancer. Diabetes. 2017;66(5):1103–1110. doi: 10.2337/db16-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eibl G, Cruz-Monserrate Z, Korc M, Petrov MS, Goodarzi MO, Fisher WE, et al. Diabetes mellitus and obesity as risk factors for pancreatic cancer. J Acad Nutr Diet. 2018;118(4):555–567. doi: 10.1016/j.jand.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padoan A, Plebani M, Basso D. Inflammation and pancreatic cancer: focus on metabolism, cytokines, and immunity. Int J Mol Sci. 2019;20(3):676. doi: 10.3390/ijms20030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maisonneuve P, Lowenfels AB. Risk factors for pancreatic cancer: a summary review of meta-analytical studies. Int J Epidemiol. 2015;44(1):186–198. doi: 10.1093/ije/dyu240. [DOI] [PubMed] [Google Scholar]
- 23.Barone E, Corrado A, Gemignani F, Landi S. Environmental risk factors for pancreatic cancer: an update. Arch Toxicol. 2016;90(11):2617–2642. doi: 10.1007/s00204-016-1821-9. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Gentiluomo M, Lorenzo-Bermejo J, Morelli L, Obazee O, Campa D, et al. Mendelian randomisation study of the effects of known and putative risk factors on pancreatic cancer. J Med Genet. 2020;57(12):820–828. doi: 10.1136/jmedgenet-2019-106200. [DOI] [PubMed] [Google Scholar]
- 25.Low SK, Kuchiba A, Zembutsu H, Saito A, Takahashi A, Kubo M, et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS ONE. 2010;5(7):e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campa D, Rizzato C, Stolzenberg-Solomon R, Pacetti P, Vodicka P, Cleary SP, et al. The TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int J Cancer J Int Cancer. 2015;137(9):2175–2183. doi: 10.1002/ijc.29590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campa D, Pastore M, Gentiluomo M, Talar-Wojnarowska R, Kupcinskas J, Malecka-Panas E, et al. Functional single nucleotide polymorphisms within the cyclin-dependent kinase inhibitor 2A/2B region affect pancreatic cancer risk. Oncotarget. 2016;7(35):57011–57020. doi: 10.18632/oncotarget.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campa D, Gentiluomo M, Obazee O, Ballerini A, Vodickova L, Hegyi P, et al. Genome-wide association study identifies an early onset pancreatic cancer risk locus. Int J Cancer. 2020;147(8):2065–2074. doi: 10.1002/ijc.33004. [DOI] [PubMed] [Google Scholar]
- 30.Rizzato C, Campa D, Pezzilli R, Soucek P, Greenhalf W, Capurso G, et al. ABO blood groups and pancreatic cancer risk and survival: Results from the PANcreatic disease ReseArch (PANDoRA) consortium. Oncol Rep. 2013;29(4):1637–1644. doi: 10.3892/or.2013.2285. [DOI] [PubMed] [Google Scholar]
- 31.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47(8):911–6. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amundadottir LT. Pancreatic cancer genetics. Int J Biol Sci. 2016;12(3):314–325. doi: 10.7150/ijbs.15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentiluomo M, Peduzzi G, Lu Y, Campa D, Canzian F. Genetic polymorphisms in inflammatory genes and pancreatic cancer risk: a two-phase study on more than 14 000 individuals. Mutagenesis. 2019 doi: 10.1093/mutage/gez040. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Nakatochi M, Hosono Y, Ito H, Kamatani Y, Inoko A, et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020 doi: 10.1038/s41467-020-16711-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corradi C, Gentiluomo M, Gajdán L, Cavestro GM, Kreivenaite E, Franco GD, et al. Genome-wide scan of long noncoding RNA single nucleotide polymorphisms and pancreatic cancer susceptibility. Int J Cancer. 2021 doi: 10.1002/ijc.33475. [DOI] [PubMed] [Google Scholar]
- 37.Galeotti AA, Gentiluomo M, Rizzato C, Obazee O, Neoptolemos JP, Pasquali C, et al. Polygenic and multifactorial scores for pancreatic ductal adenocarcinoma risk prediction. J Med Genet. 2021;58(6):369–377. doi: 10.1136/jmedgenet-2020-106961. [DOI] [PubMed] [Google Scholar]
- 38.Pistoni L, Gentiluomo M, Lu Y, López de Maturana E, Hlavac V, Vanella G, et al. Associations between pancreatic expression quantitative traits and risk of pancreatic ductal adenocarcinoma. Carcinogenesis. 2021;42(8):1037–45. doi: 10.1093/carcin/bgab057. [DOI] [PubMed] [Google Scholar]
- 39.Corradi C, Lencioni G, Gentiluomo M, Felici A, Latiano A, Kiudelis G, et al. Polymorphic variants involved in methylation regulation: a strategy to discover risk loci for pancreatic ductal adenocarcinoma. J Med Genet. 2023 doi: 10.1136/jmg-2022-108910. [DOI] [PubMed] [Google Scholar]
- 40.Chen F, Childs EJ, Mocci E, Bracci P, Gallinger S, Li D, et al. Analysis of heritability and genetic architecture of pancreatic cancer: a PanC4 study. Cancer Epidemiol Prev Biomark. 2019;28(7):1238–1245. doi: 10.1158/1055-9965.EPI-18-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuda Y. Age-related morphological changes in the pancreas and their association with pancreatic carcinogenesis. Pathol Int. 2019;69(8):450–462. doi: 10.1111/pin.12837. [DOI] [PubMed] [Google Scholar]
- 42.Nodari Y, Gentiluomo M, Mohelnikova-Duchonova B, Kreivenaite E, Milanetto AC, Skieceviciene J, et al. Genetic and non-genetic risk factors for early-onset pancreatic cancer. Dig Liver Dis. 2023 doi: 10.1016/j.dld.2023.02.023. [DOI] [PubMed] [Google Scholar]
- 43.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. bioRxiv. 2022 doi: 10.1101/2022.03.20.485034v2. [DOI] [Google Scholar]
- 45.Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367(6484):eaay5012. doi: 10.1126/science.aay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mills MC, Rahal C. A scientometric review of genome-wide association studies. Commun Biol. 2019;2(1):1–11. doi: 10.1038/s42003-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182(5):1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen MH, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182(5):1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167(5):1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng L, Gu S, Wang P, Chen H, Chen Z, Meng Z, et al. White blood cell and granulocyte counts are independent predictive factors for prognosis of advanced pancreatic caner. Gastroenterol Res Pract. 2018;8(2018):8096234. doi: 10.1155/2018/8096234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montagnana M, Danese E. Red cell distribution width and cancer. Ann Transl Med. 2016;4(20):399. doi: 10.21037/atm.2016.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang C, Wang M, Qin T, Qin R. Clinical importance of preoperative red-cell volume distribution width as a prognostic marker in patients undergoing radical surgery for pancreatic cancer. Surg Today. 2021 doi: 10.1007/s00595-021-02374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dannemann M, Andrés AM, Kelso J. Introgression of neandertal- and denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am J Hum Genet. 2016;98(1):22–33. doi: 10.1016/j.ajhg.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quach H, Rotival M, Pothlichet J, Loh YHE, Dannemann M, Zidane N, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167(3):643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nédélec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167(3):657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 57.Jagoda E, Xue JR, Reilly SK, Dannemann M, Racimo F, Huerta-Sanchez E, et al. Detection of Neanderthal adaptively introgressed genetic variants that modulate reporter gene expression in human immune cells. Mol Biol Evol. 2022;39(1):304. doi: 10.1093/molbev/msab304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huerta-Sánchez E, Jin X, Asan BZ, Peter BM, Vinckenbosch N, et al. Altitude adaptation in Tibetans caused by introgression of denisovan-like DNA. Nature. 2014;512(7513):194–7. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 60.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campa D, Rizzato C, Capurso G, Giese N, Funel N, Greenhalf W, et al. Genetic susceptibility to pancreatic cancer and its functional characterisation: The PANcreatic disease ReseArch (PANDoRA) consortium. Dig Liver Dis. 2013;45(2):95–99. doi: 10.1016/j.dld.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Campa D, Gentiluomo M, Stein A, Aoki MN, Oliverius M, Vodičková L, et al. The PANcreatic Disease ReseArch (PANDoRA) consortium: ten years’ experience of association studies to understand the genetic architecture of pancreatic cancer. Crit Rev Oncol Hematol. 2023;1(186):104020. doi: 10.1016/j.critrevonc.2023.104020. [DOI] [PubMed] [Google Scholar]
- 66.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin Y, Nakatochi M, Hosono Y, Ito H, Kamatani Y, Inoko A, et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020;11(1):3175. doi: 10.1038/s41467-020-16711-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. J Open Source Softw. 2018;3(25):731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 69.Yao Y, Ochoa A. Limitations of principal components in quantitative genetic association models for human studies. bioRxiv. 2023 doi: 10.1101/2022.03.25.485885v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Title of data: Independent associations (P<0.05) identified in PanScan+PanC4 association studiesDescription of data: Independent (r2<0.5) aSNPs showing an association P<0.05 in PanScan, PanC4, and combined datasets. The displayed summary statistics and MAF are referred to the analyses with the combined datasets. (Abbreviations: aSNP, Neandertal introgressed Single Nucleotide Polymorphism; m, minor allele; M, major allele, MAF, minor allele frequency).
Data Availability Statement
The PanScan and PanC4 genotyping data are available from the dbGaP website (study accession numbers phs000206.v5.p3 and phs000648.v1.p1). The JaPAN data used in this work are available in the JaPAN consortium website (http://www.aichi-med-u.ac.jp/JaPAN/current_initiatives-e.html). FinnGen R8 dataset of PDAC cases and controls (“C3_PANCREAS_EXALLC”) is downloadable from the FinnGen official website (https://finngen.gitbook.io/documentation/). The PANDoRA data for this work will be made available to researchers who submit a reasonable request to the corresponding author, conditional to approval by the PANDoRA Steering Committee and Ethics Commission of the Medical Faculty of the University of Heidelberg. Data will be stripped from all information allowing identification of study participants.