Abstract
Colorectal cancer (CRC) is one of the most frequent gastrointestinal malignancies that are considered as a global health challenge. Despite many progresses in therapeutic methods, there is still a high rate of mortality rate among CRC patients that is associated with poor prognosis and distant metastasis. Therefore, investigating the molecular mechanisms involved in CRC metastasis can improve the prognosis. Epithelial-mesenchymal transition (EMT) process is considered as one of the main molecular mechanisms involved in CRC metastasis, which can be regulated by various signaling pathways. PI3K/AKT signaling pathway has a key role in CRC cell proliferation and migration. In the present review, we discussed the role of PI3K/AKT pathway CRC metastasis through the regulation of the EMT process. It has been shown that PI3K/AKT pathway can induce the EMT process by down regulation of epithelial markers, while up regulation of mesenchymal markers and EMT-specific transcription factors that promote CRC metastasis. This review can be an effective step toward introducing the PI3K/AKT/EMT axis to predict prognosis as well as a therapeutic target among CRC patients.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-023-01225-x.
Keywords: Colorectal cancer, PI3K/AKT, EMT, Metastasis, Prognosis
Background
Colorectal cancer (CRC) is the third most common cancer globally and the second cause of cancer-related deaths, with approximately 935,000 deaths in the year 2020 [1]. Since, there is not any significant clinical symptoms in the early stages of CRC, majority of the patients are diagnosed in advanced tumorstages with a poor prognosis [2]. The standard CRC treatment is primarily surgery, while combined adjuvant chemotherapy has increased the overall survival of patients with advanced cancer. Nevertheless, the metastasis and proliferation of tumor cells are the major causes of CRC-related death [3, 4]. Approximately 22% of CRC patients have metastatic CRC with a poor prognosis [5, 6]. Although early-stage CRC patients have a 5-year OS of 80–90%, the survival rates reduce substantially to 40–60% for those with late-stage disease and further decrease to 5–10% among metastatic patients [7]. Therefore, it is required to assess the molecular mechanisms of CRC metastasis to reduce the tumor relapse and distant metastasis among these patients. Epithelial-to-mesenchymal transition (EMT) is a biological process that plays a crucial role in various physiological and pathological conditions such as tissue homeostasis and tumorigenesis. It is characterized by transforming epithelial cells into mesenchymal cells, which decreases their capacity for adhesion and apoptosis while increasing their ability for migration and invasion [8, 9]. EMT process attenuates cell–cell adhesion and downregulates epithelial markers (E-cadherin), whereas it induces cell mobility and the expression of mesenchymal markers (fibronectin, vimentin, and N-cadherin) [10]. It is also implicated in tumor progression, metastasis, and drug resistance. EMT enables tumor cells at the primary tumor site to acquire migratory and invasive capabilities, facilitating their dissemination to distant organs and eventually metastasis [11, 12]. Upregulation of EMT stimulator can induce this cellular process that results in CRC metastasis. These stimulators down regulate the CDH1 while up regulate the mesenchymal factors such as CDH2 and VIM via regulation of EMT-mediated signaling pathways and EMT transcription factors. MicroRNAs (miRNAs) also target mRNAs of EMT-transcription factors. Therefore, down regulation of these miRNAs can be involved in CRC metastasis [13]. A network of signaling pathways, including WNT, NOTCH, and PI3K/AKT modulate the the molecular mechanisms involved in EMT process. The WNT signaling pathway induces EMT by modulating the expression of EMT-associated transcription factors, such as Snail, Slug, and Twist [14, 15]. NOTCH signaling affects EMT by promoting the expression of mesenchymal markers (Vimentin and CDH2) and suppressing epithelial markers (CDH1 and β-catenin) [16]. PI3K/AKT pathway has also a vital role in EMT by activating downstream effectors that regulate cellular processes, including cell survival, migration, and invasion [17, 18]. PI3K/AKT pathway is a crucial signaling cascade involved in regulation of cell growth, survival, and metabolism [19]. It can be activated via the binding of growth factors to receptor tyrosine kinases (RTKs) [20, 21]. RTKs activation recruits and activates the phosphatidylinositol 3-kinase (PI3K) to generate PIP3 that activates the AKT kinase [22]. Additionally, the activated AKT kinase phosphorylates and regulates a wide range of downstream targets such as transcription factors (FOXOs), cell cycle regulators (p21 and p27), and components of the mTOR pathway (mTOR and p70S6K) [23, 24]. AKT can induce EMT by upregulating transcription factors such as Snail, Slug, and Twist, or directly through repressing expression of epithelial markers and promoting mesenchymal markers [25, 26]. For example, PI3K/AKT-induced WDR5 overexpression provoked CRC metastasis via modifying EMT-related markers and enhancing ZNF407 transcription [27]. The CCL20/CXCL8 axis also triggered the PI3K/AKT pathway to promote EMT [28]. Understanding the molecular mechanisms of EMT and its crosstalk with signaling pathways paves the way for the development of targeted therapies to prevent metastasis and improve prognosis among CRC patients. Therefore, in the present review we discussed the role of PI3K/AKT in regulation of EMT process during CRC progression and metastasis (Table 1).
Table 1.
Role of PI3K/AKT in regulation of EMT process during CRC progression
| Study | Year | Axis | Effect on the EMT | Samples | Clinical application |
|---|---|---|---|---|---|
| Wei [17] | 2019 | FAT4/PI3K/AKT/GSK-3B | Induced EMT |
100 T 100Na HCT116, LOVO and SW480 cell lines Xenograft model |
Therapeutic |
| Tan [27] | 2017 | PI3K/AKT/WDR5 | Induced EMT |
161 T 161N HT-29, SW620, HCT-15, HCT116, and COLO205 cell lines Xenograft model |
Therapeutic and prognostic |
| Pan [29] | 2020 | miR-328–3p/ Girdin/AKT | Suppressed EMT |
HCT116 and SW620 cell lines Xenograft model |
Therapeutic |
| Gao [30] | 2022 | MOR/AKT | Induced EMT |
180 T 180N HCT116, Caco-2, SW480, and LoVo cell lines |
Therapeutic and prognostic |
| Liu [31] | 2022 | CENPO/AKT | Induced EMT |
100 T 100N HCT116 and RKO cell lines Xenograft model |
Therapeutic and prognostic |
| Yang [32] | 2018 | GLI1/PI3K/AKT/NF-KB | Induced EMT |
109 T 35N HT29 and HCT116 cell lines |
Therapeutic |
| Zhang [33] | 2021 | PI3K/AKT/AGR2 | Induced EMT |
LoVo, SW480, HT-29, DLD-1, SW48 and HCT116 cell lines Xenograft model |
Therapeutic |
| Li [34] | 2020 | UCHL3/AKT/SOX12 | Induced EMT |
8 T 8N Lim1215, DLD1, SW48, HCT116, SW620, and SW480 cell lines Xenograft model |
Therapeutic and prognostic |
| Chen [35] | 2021 | MYSM1/PI3K/AKT | Suppressed EMT |
41 T 41N SW620 and LOVO cell lines Xenograft model |
Therapeutic and prognostic |
| Golhati [36] | 2011 | mTORC/Rac1/RhoA | Suppressed EMT |
18 T 18 M 18N HCT116, SW480 and KM20 cell lines Xenograft model |
Therapeutic |
| Cui [37] | 2019 | TTN-AS1/ miR-497 | Induced EMT |
95 T 95N SW480, SW620, HT29, HCT116 cell lines Xenograft model |
Therapeutic and prognostic |
| Liao [38] | 2022 | KIFC3/PI3K/AKT/mTOR | Induced EMT |
HT29, HCT116, SW480, DLD-1 cell lines Xenograft model |
Therapeutic |
| Duan [39] | 2018 | IMPDH2/AKT/mTOR/FOXO1 | Induced EMT |
248 T 248N HCT116, SW620, M5, SW480, HT29, DLD-1 and LoVo cell lines Xenograft model |
Therapeutic and prognostic |
| Xu [40] | 2020 | LACTB/AKT/mTOR | Induced EMT |
80 T 80N LOVO, SW480 and HCT116 cell lines Xenograft model |
Therapeutic and prognostic |
| Li [41] | 2022 | GREM1/PI3K/AKT/mTOR | Induced EMT |
55 T 55N SW480 and HCT116 cell lines Xenograft model |
Therapeutic and prognostic |
| Long [42] | 2022 | ECM1/PI3K/AKT/GSK3B/SNAIL | Induced EMT |
75 T 70N NCM460, HCT116, HCT15, HT29, SW480, 293 T, and SW620 cell lines Xenograft model |
Therapeutic and prognostic |
| Zhang [43] | 2021 | P2X7R/AKT/GSK3B | Induced EMT |
SW620 and HCT116 cell lines Xenograft model |
Therapeutic |
| Zhao [44] | 2019 | CAPS1/PI3K/AKT/GSK3B | Induced EMT |
126 T 126N HT29, SW480, DLD1, and SW620 cell lines |
Therapeutic and prognostic |
| Yu [45] | 2021 | CDX2PTEN/PI3K | Suppressed EMT |
161 T 161N RKO, Caco-2, HT-29, SW480 and Lovo cell lines Xenograft model |
Therapeutic and prognostic |
| Shen [46] | 2017 | CXCL8/PI3K/AKT/NF-KB | Induced EMT |
LoVo cell line Xenograft model |
Therapeutic |
| Cheng [47] | 2014 | CXCL8/ PI3K/AKT | Induced EMT |
213 T 213N SW480 and Caco-2 cell lines |
Therapeutic |
| Gao [48] | 2019 | CCR7/PI3K/AKT | Induced EMT |
190 T 190N HCT116, Caco-2, DLD-1, SW620, SW480, HT-29 and LoVo cell lines |
Therapeutic and prognostic |
| Wang [49] | 2020 | miR425-5p, miR-130b-3p, and miR-25-3p/ PTEN | Induced EMT |
17 T 12N HCT116 and SW620 cell lines Xenograft model |
Therapeutic |
| Wei [50] | 2019 | CCL22/PI3K/AKT | Induced EMT |
68 T 68N DLD1 and HT29 cell lines |
Therapeutic and prognostic |
| Chen [51] | 2021 | Id4/PI3K/AKT | Induced EMT |
HCT116 cell line Xenograft model |
Therapeutic |
| Yu [52] | 2019 | GATA1/AKT | Suppressed EMT |
74 T 74N HCT-116 and HCT-8 cell lines Xenograft model |
Therapeutic and prognostic |
| Zhang [53] | 2020 | miR-758/ PAX6 | Suppressed EMT |
84 T 84N HCT-116 and SW620 cell lines |
Therapeutic and prognostic |
| Cong [54] | 2019 | miR-760/ FOXA1 | Suppressed EMT |
54 T 54N SW620, HT29, DLD1 and HCT116 cell lines |
Therapeutic and prognostic |
| Miller [55] | 2020 | LSD1/AKT | Induced EMT | HT29, SW480, HCT116, LoVo and RKO cell lines | Therapeutic |
| Zhao [56] | 2016 | SPOCK1/PI3K/AKT | Suppressed EMT |
HCT116, HT29, SW480, and Lovo cell lines Xenograft model |
Therapeutic |
| Chen [57] | 2016 | STC2/PI3K/AKT | Induced EMT |
77 T 77N HT29 cell line Xenograft model |
Therapeutic and prognostic |
| Chen [58] | 2019 | CLCA4/PI3K/AKT | Induced EMT |
64 T 64N SW620 and LoVo cell lines |
Therapeutic and prognostic |
aTumor (T) tissues, Normal (N) margins
PI3K/AKT axis
PI3K/AKT axis has a pivotal role in regulation of EMT process during CRC progression (Fig. 1). Girdin is an actin-binding protein that is activated by AKT to regulate cytoskeletal remodeling and cell motility. It also promotes AKT signaling by RTKs and G protein-coupled receptors. The p85α subunit of PI3K and phosphorylated Girdin produce the Girdin-p85α complex that stimulates the PI3K/AKT signaling pathway [59]. AKT-dependent Girdin phosphorylation provokes the biological role of Girdin [60]. MiR-328–3p suppressed the EMT in CRC cells through CDH1 up regulation and negative regulation of Snail, Vimentin, and CDH2. Additionally, miR-328–3p repressed the PI3K/AKT pathway by reducing p-AKT and p-Girdin in metastatic liver specimens. Therefore, miR-328–3p stimulated the PI3K/AKT signaling axis via regulating Girdin [29]. Mu-opioid receptor (MOR) belongs to the G protein-coupled receptor family that binds to opioids, including heroin, fentanyl, and morphine [61]. MOR is extensively expressed in various normal tissues as well as human cancers, and its upregulation has been associated with a poor prognosis in cancer patients [62–64]. MOR depletion dramatically inhibited EMT and migration of CRC cells and downregulated p-AKT, which can be restored by SC79 as an activator of PI3K/AKT [30]. Collagen as a key extracellular matrix protein facilitates tumor CRC progression by stimulating the PI3K/AKT signaling axis through the integrin α2β1. PI3K/AKT also promoted EMT by snail up regulation and CDH1 down regulation that resulted in CRC metastasis [65]. Matrix metalloproteinases (MMPs) belong to the zinc-containing endopeptidases family that is implicated in cancer progression via remodelling of the extracellular matrix [66–68]. There was MMP1 up regulation in CRC samples that was correlated with poor prognosis. MMP1 down regulation inhibited AKT pathway and affected the levels of CDH1, CDH2, vimentin, and Twist1 expressions [69]. CENPO modulates the recovery of spindle injury and cell apoptosis by preventing premature separation of chromatids [70]. The centromere has a pivotal role in chromosome separation during cell prolifaration [71]. Dysregulation of the centromere protein CENPO results in chromosomal aneuploidy and abormal cell division [72]. A significant upregulation of CENPO was detected in CRC patients that were associated with decreased survival time. CENPO inhibition also reduced colorectal tumor cell growth and migration. Loss of the CENPO up regulated E-cadherin while down regulated Vimentin and N-cadherin. The suppression of CENPO negatively regulated AKT phosphorylation, CCND1, PIK3CA, and enhanced MAPK9 expression [31]. HH/GLI signaling pathway is a critical modulator of cancer progression as it is involved in cancer stem cell differentiation, metastasis, survival, and growth [73–77]. There was upregulation of GLI1 that was associated with an invasive phenotype and poor prognosis in CRC patients. GLI1 targeted the PI3K/AKT/NFκB pathways to regulate the metastatic features of CRC cells, which reduced the survival rate of CRC patients. The loss of GLI1 also led to CDH1 upregulation while Snail and vimentin down regulations in CRC cells. Additionally, AKT inhibition significantly repressed GLI1. Therefore, PI3K/AKT axis was an upstream effector of GLI1 and induced CRC characteristics by activating GLI1. PI3K/AKT/NF-κB signaling pathway ameliorated the growth and metastasis and attenuated the survival time of CRC cells via GLI1 up regulation [32].
Fig. 1.
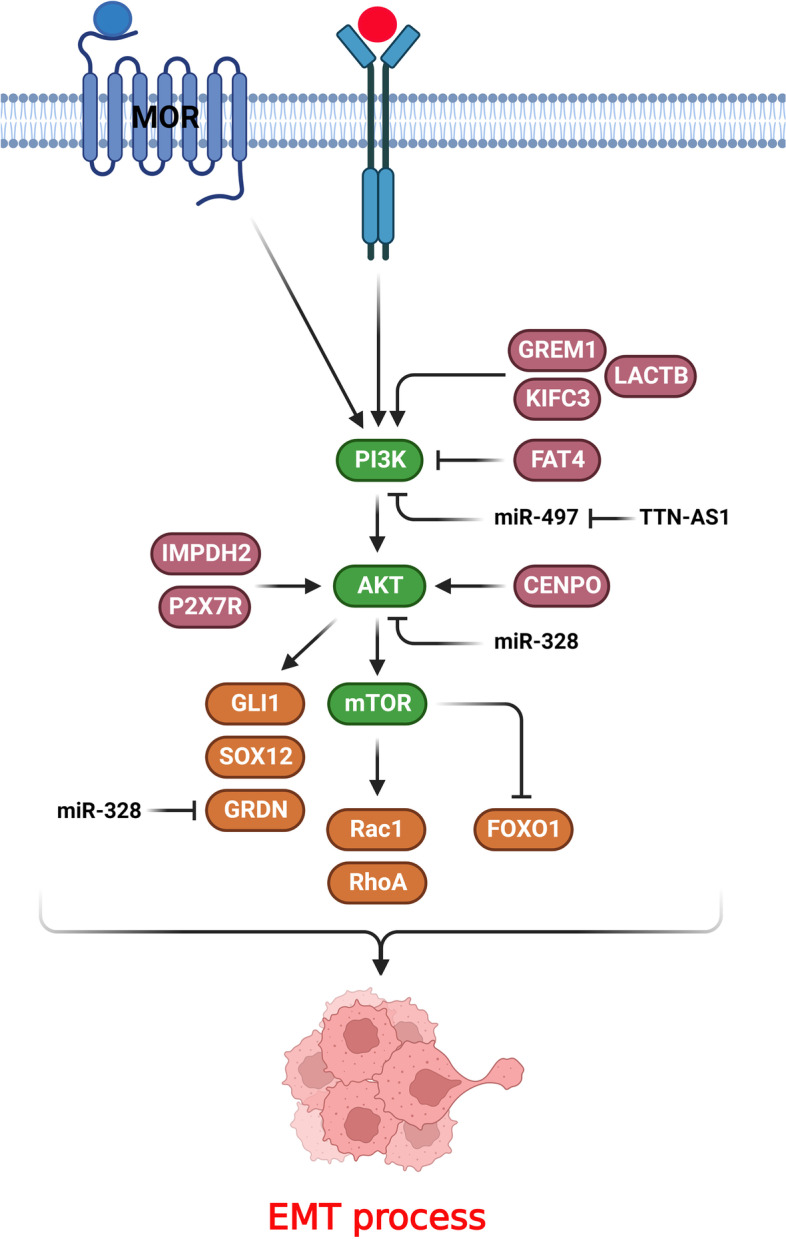
PI3K/AKT/mTOR axis has a pivotal role in regulation of EMT process during CRC progression. (Created with BioRender.com)
Anterior gradient-2 (AGR2) belongs to disulfide isomerase protein family that has a pivotal role in cancer progression. It encompasses a KTEL motif that is a carboxy-terminal endoplasmic reticulum (ER) retention pattern [78–82]. KTEL facilitates the AGR2 attachment to KDEL receptors on the Golgi for retrograde transport and localization in the ER, where it promotes appropriate protein folding [83, 84]. It has been reported that intracellular AGR2 increased CRC metastasis via inducing EMT, resulting in SLUG and SNAIL up regulations. There was AGR2 upregulation following PGE2 stimulus through the EP4-PI3K-AKT axis, indicating its critical role in the regulation of PGE2-mediated EMT and the crosstalk between TAMs and tumor cells [33]. Ubiquitination is a critical mechanism that regulates protein functions posttranslationally [85, 86]. Deubiquitinating enzymes (DUBs), including UCHL3 are pivotal modulators of the ubiquitination [87–89]. There was UCHL3 up regulation in CRC tissues that was correlated with poor prognosis. UCHL3 induced the growth, invasion, and malignancy of CRC cells. Moreover, suppression of UCHL3 led to E-cadherin upregulation, whereas vimentin, CDH2, Slug, Snail, and ZEB1 down regulations. Therefore, UCHL3 triggerd the proliferation of CRC cells via AKT-induced SOX12 over expression [34]. MYSM1 excludes monoubiquitin from H2AK119ub1 and activates transcription in cooperation with histone acetylation [90]. Dysregulation of MYSM1 results in immune system disorders, anemia, and infammatory reactions, as well as various tissue dysfunctions [91–95]. MYSM1 inhibited the CRC progression via miR-200/CDH1 and inhibition of PI3K/AKT axis [35].
PI3K/AKT/mTOR axis
The mTOR as one of the main effectors of PI3K/AKT axis has a pivotal role in regulation of EMT process during CRC progression (Fig. 1). Mammalian target of rapamycin (mTOR) is one of the main effectors of PI3K/AKT pathway that regulates cell proliferation, apoptosis, and metabolism. Rac1 and RhoA stimulation regulate the primary steps of the tumor metastasis via actin rearrangement and cell migration [96]. Rac1 triggers lamellipodia production, while RhoA forms cell–cell adhesions and actin stress fibers. mTORC1/2 modulates F-actin reorganization and lamellipodia formation in tumor cells [97]. There was an association between Raptor, Rictor, and mTOR expression and higher stages of CRC. There were Raptor, Rictor, and mTOR up regulations in matastatic CRC samples. mTORC1/2 suppression mitigated the invasion and migration of CRCs, probably through regulating the rearrangement of the cytoskeleton and deactivating Rac1 and RhoA. Moreover, hindering mTORC1/2 enhanced E-cadherin, cell–cell adhesion, and oxaliplatin-induced apoptosis while inhibited lamellipodia formation, fibronectin, SMA, vimentin, and MMP-9 [36]. There was significant up regulation of TTN-AS1 in CRC tissues that was associated with lymph node involvement, TNM stage, and poor prognosis. TTN-AS1 promoted CRC cell proliferation and invasion. TTN-AS1 stimulated PI3K/AKT/mTOR axis partly via targeting miR-497 in CRC cells [37]. Kinesins (KIFs) are motor proteins that have a critical role in intracellular transportation of mRNAs, protein complexes, and organelles through ATP molecules along microtubules. They also modulate the spindle and chromosomal dynamics throughout meiosis and mitosis [98, 99]. KIF2A inhibition promotes apoptosis by PI3K/AKT suppression in tumor cells [100]. There was KIFC3 up regulation in CRC tissues and cell lines, and its suppression decreased invasion of CRC cells. KIFC3 induced the expression of MMP2/9 and mesenchymal-related markers. KIFC3 also phosphorylated mTOR, AKT, and PI3K. Moreover, there was a positive association between KIFC3 and aggressiveness of CRC cells through EMT and the PI3K/AKT/mTOR pathway [38]. Inosine 5′-monophosphate dehydrogenase (IMPDH) facilitates the generation of xanthosine monophosphate that is a vital process in the guanine synthesis [101]. IMPDH modulates the guanine nucleotide levels and play an importat role in synthesis of RNA and DNA. IMPDH2 induced the EMT, invasion, and growth in CRC cells via up regulating Ki-67 and cyclin D1 and down regulating p27Kip1 and p21Cip1. IMPDH2 promoted the G1/S transition via activating AKT and mTOR and FOXO1 down regulation [39].
Beta-lactamase-like (LACTB) modulates the membrane organization in mitochondria that affects the lipid metabolism and oxidative phosphorylation. Autophagy is a critical homeostatic process that degrades cellular proteins, organelles, and cytoplasmic constituents to balance the consumption and supply of energy under stress conditions [102]. Inhibition of LACTB mitigated the autophagy and promoted the EMT and invasion in CRC cells. LACTB regulated the growth of tumor cells via 4E-BP1, C-Myc, and CCND1 by the PI3K/AKT/mTOR axis. LACTB promoted epithelial polarity and amplified cell–cell junctions to suppress EMT via autophagy. LACTB provoked autophagy via modulating PIK3R3 expression and repressing EMT and cell growth via the regulation of PI3K/AKT/mTOR axis [40]. FAT4 facilitated autophagy by suppressing PI3K, p-AKT, and mTOR. Inhibition of FAT4 also suppressed autophagy while enhanced invasion and EMT in CRC cells. FAT4 regulated the EMT via targeting Twist1 and E-cadherin. Therefore, FAT4 ameliorated autophagy and attenuated the EMT through the PI3K/AKT/GSK-3β and mTOR pathways [17]. Gremlin-1 (GREM1) is a glycoprotein that is classified as a member of the DAN/Cerberus protein family that includes various proteins such as VEGF and TGF-β [103]. GREM1 stimulates organ fibrosis as a key step within the EMT process [104, 105]. Endoplasmic reticulum (ER) participates in protein folding, protein transport, calcium storage, and lipid biosynthesis [106, 107]. Stress stimuli disturb the ER proteostasis, which is followed by unfolded protein response (UPR) stimulation. Three ER membrane receptors, including ATF6, PERK, and IRE1α rescue ER proteostasis, induce cell death, and initiate UPR signaling [108–112]. GREM1 ameliorated the UPR-induced EMT in CRC cells through ATF6 up regulation while ATF4 down regulation. GREM1 also regulated the ATF6 and ATF4 expressions via the VEGF and BMP pathways. There was GREM1 up regulation in advanced stage CRC patients that was correlated with an unfavorable prognosis. Additionally, GREM1 stimulated the PI3K/AKT/mTOR pathway as a target of the VEGF-VEGFR2 axis, which up regulated the ATF6 [41].
PI3K/AKT/GSK-3β axis
GSK-3β is a serine-threonine kinase that inhibits glucose homeostasis to regulate ER-stress and apoptotic pathways. GSK-3β as one of the main effectors of PI3K/AKT axis has a pivotal role in regulation of EMT process during CRC progression (Fig. 2). Extracellular matrix protein 1 (ECM1), as a secretory glycoprotein, regulates numerous cellular mechanisms such as angiogenesis, epithelial cell growth, and tumor progression [113]. There was ECM1 up regulation in CRC cancer tissues that was correlated with tumor size, lymph node metastasis, and TNM staging. It also promoted the invasion and migration of CRC tumor cells. ECM1 inhibition down regulated Snail, pGSK3β, and p-AKT in CRC cells. Additionally, ECM1 induced CRC metastasis by promoting EMT via modulating the PI3K/AKT/GSK3β/Snail pathway [42]. P2X purine receptors are ATP-dependent cation channel receptors that regulate potassium ions outflow and sodium and calcium ions influx [114–116]. ATP and its analogs, BzATP, induced the growth and EMT of CRC cells via activating the P2X7R that stimulated the PI3K/AKT/GSK-3β/β-catenin axis. ATP and BzATP triggered the P2X7R to down regulate CDH1, while up regulated the fibronectin, Snail, and Vimentin. P2X7R also phosphorylated GSK-3β and AKT and increased the growth of CRC cells [43]. Snail is a critical suppressor of CDH1 that can be modulated by ERK, TGFβ, and AKT/GSK-3β signaling pathways [117–121]. Exocytosis is a mechanism that delivers various substances between the plasma and intercellular membrane [122]. Calcium-dependent secretion activator 1 (CAPS1) is involved in exocytosis that induced metastasis in CRC cells via PI3K/AKT/GSK3β/Snail axis-induced EMT. CAPS1 interacted with p85 to activate the PI3K/AKT/GSK3β pathway and then increased the expression of Snail, which followed by promoting EMT process and CRC metastasis [44]. Histone deacetylase inhibitors (HDACIs) such as VPA regulate the EMT in numerous cancer cells [123–126]. It has been demonstrated that VPA dramatically increased the EMT in CRC cells. Snail was stabilized by acetylation, and GSK-3β suppression was involved in the VPA-induced EMT of CRC cells. Furtheremore, VPA stabilized and upregulated the Snail via activating the AKT/GSK-3β axis [127]. Caudal-related homoeobox transcription factor 2 (CDX2) is an intestine-related transcription factor involved in the maintenance and growth of intestinal tissue [128]. Loss of CDX2 increased the levels of MMP-9, vimentin, and fibronectin expressions while down regulated the CDH1 and ZO-1. CDX2 knockdown induced EMT-related markers, and PTEN reduced tumor invasion and phosphorylation of AKT and GSK-3β. CDX2 stimulated PTEN expression and subsequent inhibition of the PI3K/AKT/GSK-3β pathway, which led to negative regulation of Snail and β-catenin. β-catenin down regulation reduced the levels of ZEB1, Slug, and Snail expressions while up regulated CDH1 which restrained cell invasion and migration [45].
Fig. 2.
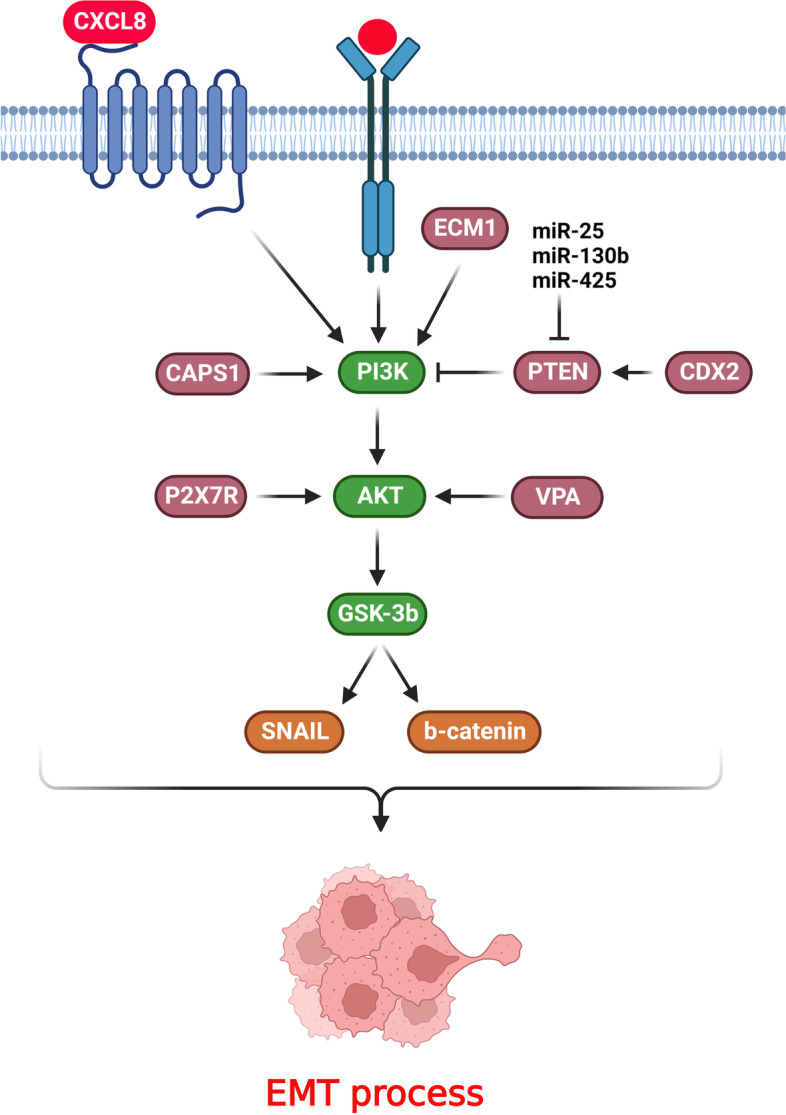
PI3K/AKT/GSK-3β axis has a pivotal role in regulation of EMT process during CRC progression. (Created with BioRender.com)
Chemokine mediated PI3K/AKT activation
Chemokines are the key regulators of the leukocytes migration that are highly expressed in numerous cancers [129, 130]. They stimulate the PI3K/AKT axis, thereby inhibiting the CDH1/β-catenin complex to facilitate invasion and migration of CRC cells [131–133]. Chemokines are involved in regulation of EMT process via PI3K/AKT axis during CRC progression (Fig. 2). The CCL20 and CXCL8 co-activation restrained invasion, growth, and CDH1 levels while upregulating snail, vimentin, and CDH2. Moreover, the CCL20/CXCL8 axis triggered the PI3K/AKT/ERK pathway to promote EMT [28]. CXCL8 is an autocrine growth factor that induces tumor proliferation, drug resistance, angiogenesis, and aggressiveness [134, 135]. PI3K/AKT pathway also activates the NF-κB signaling through κBα (IκBα) protein phosphorylation [136]. CXCL8 increased the growth and invasion of CRC cells. CXCL8 induced EMT through the PI3K/AKT/NF-κB pathway by PI3K and NF-κB phosphorylations. Moreover, CXCL8 triggered EMT by downregulating E-cadherin and upregulating α-SMA, vimentin, and N-cadherin through PI3K/AKT/NF-κB pathway [46]. CXCL8 and CCL20 promoted EMT in human CRC cells to preserve cell invasion, migration, and proliferation through inducing the PI3K/AKT-ERK1/2 axis. CXCL8 and CCL20 coexpression was associated with liver metastases and a poor prognosis in CRC patients [47]. EGFR is correlated with poor prognosis, and its downstream pathways, including MAPK and PI3K/AKT, induce tumor development [137, 138]. SLC/CCR7 is a pivotal modulator of the EMT in tumor cells [139]. A remarkable correlation has been indicated between CCR7 up regulation, regional lymph node metastasis, and tumor infiltration. SLC activated CCR7, which resulted in PI3K/AKT stimulation in CRC cells. CCR7 induced cetuximab resistance in CRC cells under the regulation of the EMT process [48].
Liver metastasis is a major challenge that accounts for approximately 70% of CRC deaths [140]. Tumor-associated macrophages (TAMs) are the most common type of immune-related cells that infiltrate into the tumor microenvironment to promote metastasis [141]. The classically activated (M1) and alternatively activated (M2) phenotypes are two polarized subtypes of TAMs [142]. M1 macrophages classically activate immune cells that release type I pro-inflammatory cytokines. M2 macrophages enhance tumor progression via immune suppression, angiogenesis, and metastasis [143, 144]. CRC cells delivered miR-425-5p, miR-130b-3p, and miR-25-3p to TAMs via exosomes following the activation of CXCL12/CXCR4 axis. These miRNAs induced the M2 macrophages via targeting PTEN by the PI3K/Akt pathway, leading to increased metastasis and angiogenesis of CRC cells via promoting EMT and releasing VEGF [49]. There was a positive association between CD163 + M2 macrophages presence and CCL22 expression in CRC tissues. M2 macrophages induced 5-FU resistance in CRC cells by releasing CCL22. M2 macrophages also conveyed CCL22 to cancer cells, leading to the promotion of EMT and 5-FU resistance in CRC cells. Furtheremore, 5-FU inhibited the proliferation of CRC cells with PI3K and AKT dephosphorylation. M2 macrophages impaired the inhibitory effect of 5-FU by activating the PI3K/AKT pathway. Therefore, M2 macrophage-secreted CCL22 counteracted the impact of 5-FU on tumor cells by triggering PI3K/AKT [50].
Transcription factors and chromatin remodelers
It has been shown that transcription factors and chromatin remodelers have a key role in EMT process via the regulation of PI3K/AKT pathway during CRC progression (Fig. 3). DNA-binding (Id) inhibitors are members of the basic helix-loop-helix (bHLH) transcription factor family that have lost their DNA-binding domain [145]. The Id family critically regulates cell apoptosis, growth, and differentiation [146, 147]. Id4 dramatically suppressed tumor proliferation and metastasis in the xenograft model. The Id4-transfected cells exerted p21 and p27 up regulation, which was followed by cell cycle arrest at the G0/G1. Id4 inactivated AKT and PI3K, suppressing CRC cell proliferation through modulating PI3K/AKT signaling. Id4 mitigated the EMT process as it up regulated TIMP1/2 and down regulated the snail, slug, twist, β-catenin, MMP2, and MMP7 [51]. GATA1 is a critical modulator of erythroid cell apoptosis, proliferation, and differentiation. It has been indicated that GATA1 was up regulated in breast cancer and increased VEGF-induced tumor angiogenesis and proliferation via interaction with the SET7 histone methyltransferase [148, 149]. GATA1 promotes EMT in cancer cells by CDH1 down regulation [150]. There was GATA1 up regulation in CRC tissues, which was associated with poor prognosis. GATA1 provoked CRC cell invasion and growth through AKT phosphorylation [52]. PAX6 as a highly preserved transcription factor is a pivotal regulator of human tumor progression [151]. There was miR-758 downregulation in CRC tissues that was correlated with malignant features and an unfavorable prognosis. MiR-758 suppressed the cell survival and metastasis in CRC cells. It also repressed the EMT and PI3K/AKT axis and promoted apoptosis via Bcl-2 and Bax in CRC. Therefore, miR-758 inhibited cell metastasis and EMT in CRC by targeting PAX6 and inhibition of PI3K/AKT pathway [53]. FOXA1 exhibits oncogenic function in numerous malignancies by modulating cell cycle, growth, and death [152, 153]. There was a significant miR-760 down regulation in CRC, which was correlated with a poor prognosis. MiR-760 targeted the FOXA1 to inhibit the growth, migration, and aggressiveness of CRC cells via modulating EMT and PI3K/AKT [54].
Fig. 3.
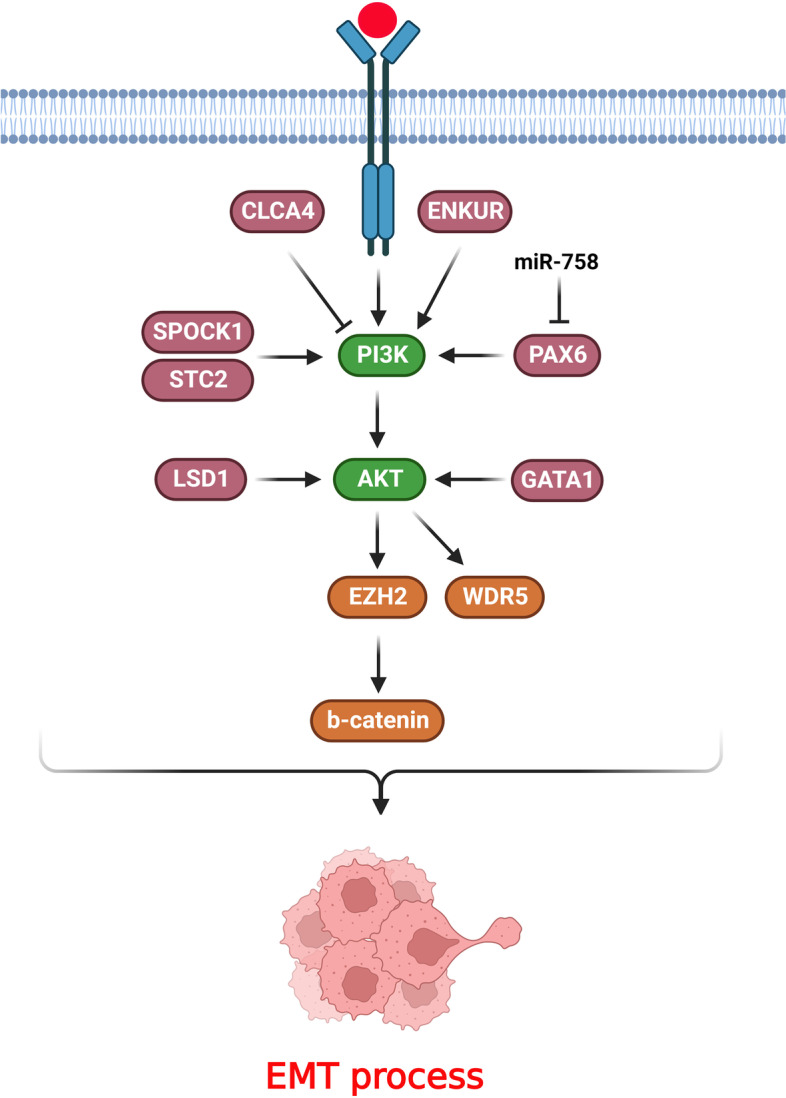
Transcription factors, chromatin remodelers, and ion binding proteins have key roles in EMT process via the regulation of PI3K/AKT pathway during CRC progression. (Created with BioRender.com)
Polycomb repressive complex 2 (PRC2) is involved in tumor progression and tissue homeostasis by modulating the chromatin remodeling. Enhancer of zeste homolog 2 (EZH2) is one of the principal constituents of the PRC2 complex. EZH2 regulates the trimethylation of histone 3 at lysine 27 (H3K27me3), thus repressing transcription. AKT phosphorylated EZH2 following PI3K/AKT axis activation, which is necessary for the cross talk between methylate β-catenin and EZH2. Phosphorylated EZH2 enhanced the β-catenin function, followed by regulating genes involved in metabolic processes and cell migration. Additionally, PI3K/AKT as a critical activated axis in CRC patients led to EZH2 phosphorylation at S21 (pS21-EZH2) [154]. LSD1 belongs to the RE1 silencing transcription factor corepressor (CoREST) complex that includes the RCOR1 as a scaffolding protein and histone deacetylase 1 and 2 (HDAC1/2) as chromatin-modifying subunits [155–157]. CoREST regulates the acetylation of active chromatin to sustain a restrained chromatin phase. LSD1 is implicated in demethylation of H3K4me2 in the promoter of epithelial genes to promote CRC [158–160]. There was significant upregulation of LSD1 in patients who carried PIK3CA mutation in gastrointestinal tumor tissues. LSD1 decreased the proliferation of PIK3CA-mutant colorectal and stomach cancer cells. LSD1 also modulated the phosphorylation of AKT and EMT via CoREST complex. Therefore, LSD1 was up regulated upon PIK3CA mutation that resulted in tumor invasion and EMT features [55]. WDR5 is a highly preserved subunit of COMPASS-related complexes that are involved in H3K4me3 [161, 162]. WDR5 plays an important role in embryonic cell self-renewal and the reprogramming of somatic cells [163, 164]. WDR5 up regulation was prominently associated with an unfavorable prognosis in non-metastatic CRC tissues. Overexpression of WDR5 has also been indicated to trigger CRC metastasis. PI3K/AKT-induced WDR5 expression provoked CRC metastasis via modifying EMT-related markers and up regulating ZNF407 [27].
Ion binding proteins
Ion binding proteins have a key role in EMT process via the regulation of PI3K/AKT pathway during CRC progression (Fig. 3). SPOCK1 belongs to the Ca2 + -binding proteoglycan family that has an important regulatory role in metastasis, cell cycle, and DNA repair [165–167]. Decreased expression of SPOCK1 markedly mitigated the migration and aggressiveness of CRC cells by impairing the EMT. The loss of SPOCK1 also down regulated the p-Akt and p-PI3K [56]. The stanniocalcin 1 (STC1) and stanniocalcin 2 (STC2) glycoprotein hormones modulate the secretion of phosphate and calcium [168]. STC2 has a vital role in cell–cell interactions between normal colon epithelia and tumor cells [169]. STC2 expression has been correlated with tumor stage and survival time in CRC patients. It also induced the EMT and migration of CRC cells. STC2 accelerated the tumorigenesis and development of tumor cells via the EMT process by stimulating the PI3K/AKT and ERK/MEK pathways. STC2 up regulated p-ERK, p-MEK, p-AKT, PI3K, and Ras upon altering EMT markers [57]. CaM-binding protein is encoded by ENKUR and interplays with the p85 component of PI3K. p85 induces the PI3K signaling cascade to accelerate tumor cell proliferation, metabolism, and survival while inhibited apoptosis [170–172]. ENKUR exhibited a tumor suppressive role in CRC cells as it modulated tumor growth, migration, and invasion. Down regulation of ENKUR activated the PI3K/Akt signaling pathway in CRC cells. Additionally, inhibition of ENKUR downregulated E-cadherin while upregulated vimentin and N-cadherin in CRC cells [173]. Calcium-activated chloride channel (CLCA) modulators are proteins that have a symmetrical multiple cysteine patterns in the terminal tail [174]. Reduced expression of CLCA4 accelerates cell proliferation and metastasis via modulating EMT [175–177]. CLCA4 inhibited the PI3K-AKT signaling and EMT in CRC cells. CLCA4 suppression was also associated with metastasis to lymph nodes in CRC patients. CLCA4 repressed the invasion and migration of CRC cells by attenuating EMT and PI3K/AKT signaling inactivation [58].
Conclusions
EMT process is considered as one of the main molecular mechanisms involved in tumor metastasis. This process can be directly or indirectly regulated by signaling pathways. It has been reported that the PI3K/AKT pathway plays a key role in promotion of the EMT process during CRC progression by up regulation of mesenchymal markers and EMT specific transcription factors that result in CRC metastasis. Therefore, PI3K/AKT/EMT axis can be used to predict prognosis and as a suitable therapeutic target in metastatic CRC. Since, PI3K/AKT has a key role in promotion of EMT process, it can be expected that the clinical monoclonal antibodies such as Cetuximab and Panitumumab that can target the RTKs as the main triggers of this pathway can be used as the indirect EMT inhibitors to reduce the CRC metastasis and improve prognosis among these patients.
Acknowledgements
None.
Abbreviations
- AGR2
Anterior gradient-2
- bHLH
Basic helix-loop-helix
- LACTB
Beta-lactamase-like
- CLCA
Calcium-activated chloride channel
- CAPS1
Calcium-dependent secretion activator 1
- CDX2
Caudal-related homoeobox transcription factor 2
- CRC
Colorectal cancer
- DUBs
Deubiquitinating enzymes
- ER
Endoplasmic reticulum
- EZH2
Enhancer of zeste homolog 2
- EMT
Epithelial-mesenchymal transition
- GREM1
Gremlin-1
- HDAC1/2
Histone deacetylase 1 and 2
- HDACIs
Histone deacetylase inhibitors
- IMPDH
Inosine 5′-monophosphate dehydrogenase
- KIFs
Kinesins
- mTOR
Mammalian target of rapamycin
- MMPs
Matrix metalloproteinases
- MOR
Mu-opioid receptor
- PI3K
Phosphatidylinositol 3-kinase
- PRC2
Polycomb repressive complex 2
- CoREST
RE1 silencing transcription factor corepressor
- RTKs
Receptor tyrosine kinases
- STC1
Stanniocalcin 1
- TAMs
Tumor-associated macrophages
- UPR
Unfolded protein response
Authors’ contributions
AM were involved in search strategy and drafting. MM designed, revised, structured, and edited the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. 2019;234(10):17127–43. doi: 10.1002/jcp.28473. [DOI] [PubMed] [Google Scholar]
- 3.Huang YJ, Jan YH, Chang YC, Tsai HF, Wu AT, Chen CL, et al. ATP synthase subunit epsilon overexpression promotes metastasis by modulating AMPK Signaling to induce epithelial-to-mesenchymal transition and is a poor prognostic marker in Colorectal Cancer Patients. J Clin Med. 2019;8(7):1070. doi: 10.3390/jcm8071070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbaszadegan MR, Moghbeli M. Genetic and molecular origins of colorectal Cancer among the Iranians: an update. Diagn Pathol. 2018;13(1):97. doi: 10.1186/s13000-018-0774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Oliveira J. Advanced colorectal cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):61–3. doi: 10.1093/annonc/mdp130. [DOI] [PubMed] [Google Scholar]
- 6.Ansa BE, Coughlin SS, Alema-Mensah E, Smith SA. Evaluation of colorectal cancer incidence trends in the United States (2000–2014) J Clin Med. 2018;7(2):22. doi: 10.3390/jcm7020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oki E, Ando K, Nakanishi R, Sugiyama M, Nakashima Y, Kubo N, et al. Recent advances in treatment for colorectal liver metastasis. Ann Gastroenterol Surg. 2018;2(3):167–75. doi: 10.1002/ags3.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, et al. Myosin II isoform switching mediates invasiveness after TGF-β-induced epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2011;108(44):17991–6. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamidi AA, Khalili-Tanha G, Nasrpour Navaei Z, Moghbeli M. Long non-coding RNAs as the critical regulators of epithelial mesenchymal transition in colorectal tumor cells: an overview. Cancer Cell Int. 2022;22(1):71. doi: 10.1186/s12935-022-02501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15(1):129. doi: 10.1186/s13045-022-01347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (London England) 2021;41(3):199–217. doi: 10.1002/cac2.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu T, Datta PK. Regulation of EMT in Colorectal Cancer: a culprit in Metastasis. Cancers. 2017;9(12):171. doi: 10.3390/cancers9120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Lai Q, He C, Fang Y, Yan Q, Zhang Y, et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):334. doi: 10.1186/s13046-019-1330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbaszadegan MR, Taghehchian N, Li L, Aarabi A, Moghbeli M. Contribution of KCTD12 to esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):853. doi: 10.1186/s12885-018-4765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moghbeli M, Mosannen Mozaffari H, Memar B, Forghanifard MM, Gholamin M, Abbaszadegan MR. Role of MAML1 in targeted therapy against the esophageal cancer stem cells. J Transl Med. 2019;17(1):126. doi: 10.1186/s12967-019-1876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38(1):112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navaei ZN, Khalili-Tanha G, Zangouei AS, Abbaszadegan MR, Moghbeli M. PI3K/AKT signaling pathway as a critical regulator of cisplatin response in tumor cells. Oncol Res. 2021;29(4):235–50. doi: 10.32604/or.2022.025323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Therapy. 2021;6(1):425. doi: 10.1038/s41392-021-00828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan K, Mahajan NP. PI3K-independent AKT activation in cancers: a treasure trove for novel therapeutics. J Cell Physiol. 2012;227(9):3178–84. doi: 10.1002/jcp.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moghbeli M, Makhdoumi Y, Soltani Delgosha M, Aarabi A, Dadkhah E, Memar B, et al. ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer. Biol Res. 2019;52(1):2. doi: 10.1186/s40659-018-0208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong D, Yamori T. Advances in development of phosphatidylinositol 3-kinase inhibitors. Curr Med Chem. 2009;16(22):2839–54. doi: 10.2174/092986709788803222. [DOI] [PubMed] [Google Scholar]
- 23.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13(7):815. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210(5):767–76. doi: 10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tania M, Khan MA, Fu J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumor Biol. 2014;35:7335–42. doi: 10.1007/s13277-014-2163-y. [DOI] [PubMed] [Google Scholar]
- 26.Roshan MK, Soltani A, Soleimani A, Kahkhaie KR, Afshari AR, Soukhtanloo M. Role of AKT and mTOR signaling pathways in the induction of epithelial-mesenchymal transition (EMT) process. Biochimie. 2019;165:229–34. doi: 10.1016/j.biochi.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Tan X, Chen S, Wu J, Lin J, Pan C, Ying X, et al. PI3K/AKT-mediated upregulation of WDR5 promotes colorectal cancer metastasis by directly targeting ZNF407. Cell Death Dis. 2017;8(3):e2686. doi: 10.1038/cddis.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen T, Cheng X, Liu X, Xia C, Zhang H, Pan D, et al. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif Cells Nanomed Biotechnol. 2019;47(1):4038–45. doi: 10.1080/21691401.2019.1669620. [DOI] [PubMed] [Google Scholar]
- 29.Pan S, Ren F, Li L, Liu D, Li Y, Wang A, et al. MiR-328-3p inhibits cell proliferation and metastasis in colorectal cancer by targeting Girdin and inhibiting the PI3K/Akt signaling pathway. Exp Cell Res. 2020;390(1):111939. doi: 10.1016/j.yexcr.2020.111939. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Yang L, He Y, Liu Y, Xu P, Zhang J, et al. MOR promotes epithelial-mesenchymal transition and proliferation via PI3K/AKT signaling pathway in human colorectal cancer. Acta Biochim Biophys Sin. 2022;55(1):72–80. doi: 10.1093/abbs/gmt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Chen C, Yan M, Zeng X, Zhang Y, Lai D. CENPO regulated proliferation and apoptosis of colorectal cancer in a p53-dependent manner. Discover Oncol. 2022;13(1):8. doi: 10.1007/s12672-022-00469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Zhang C, Qi W, Cui Y, Xuan Y. GLI1 promotes cancer stemness through intracellular signaling pathway PI3K/Akt/NFκB in colorectal adenocarcinoma. Exp Cell Res. 2018;373(1–2):145–54. doi: 10.1016/j.yexcr.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Chi J, Hu J, Ji T, Luo Z, Zhou C, et al. Intracellular AGR2 transduces PGE2 stimuli to promote epithelial-mesenchymal transition and metastasis of colorectal cancer. Cancer Lett. 2021;518:180–95. doi: 10.1016/j.canlet.2021.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Zheng Y, Li X, Dong X, Chen W, Guan Z, et al. UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via AKT/mTOR signaling pathway. Am J Transl Res. 2020;12(10):6445–54. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Wang W, Li Y, Huo Y, Zhang H, Feng F, et al. MYSM1 inhibits human colorectal cancer tumorigenesis by activating miR-200 family members/CDH1 and blocking PI3K/AKT signaling. J Expe Clin Cancer Res. 2021;40(1):341. doi: 10.1186/s13046-021-02106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71(9):3246–56. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Z, Han B, Wang X, Li Z, Wang J, Lv Y, Long Non-Coding RNA TTN-AS1 promotes the Proliferation and Invasion of Colorectal Cancer cells by activating miR-497-Mediated PI3K/Akt/mTOR signaling. OncoTargets and therapy. 2019;12:11531–9. doi: 10.2147/OTT.S229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao H, Zhang L, Lu S, Li W, Dong W. KIFC3 promotes proliferation, Migration, and Invasion in Colorectal Cancer via PI3K/AKT/mTOR signaling pathway. Front Genet. 2022;13:848926. doi: 10.3389/fgene.2022.848926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan S, Huang W, Liu X, Liu X, Chen N, Xu Q, et al. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res. 2018;37(1):304. doi: 10.1186/s13046-018-0980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu W, Yu M, Qin J, Luo Y, Zhong M. LACTB regulates PIK3R3 to promote autophagy and inhibit EMT and proliferation through the PI3K/AKT/mTOR signaling pathway in Colorectal Cancer. Cancer Manage Res. 2020;12:5181–200. doi: 10.2147/CMAR.S250661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R, Zhou H, Li M, Mai Q, Fu Z, Jiang Y, et al. Gremlin-1 promotes colorectal Cancer Cell Metastasis by activating ATF6 and inhibiting ATF4 pathways. Cells. 2022;11(14):2136. doi: 10.3390/cells11142136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long S, Wang J, Weng F, Xiang D, Sun G. Extracellular matrix protein 1 regulates colorectal Cancer cell proliferative, migratory, invasive and epithelial-mesenchymal transition activities through the PI3K/AKT/GSK3β/Snail signaling Axis. Front Oncol. 2022;12:889159. doi: 10.3389/fonc.2022.889159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang WJ, Luo C, Huang C, Pu FQ, Zhu JF, Zhu ZM. PI3K/Akt/GSK-3β signal pathway is involved in P2 × 7 receptor-induced proliferation and EMT of colorectal cancer cells. Eur J Pharmacol. 2021;899:174041. doi: 10.1016/j.ejphar.2021.174041. [DOI] [PubMed] [Google Scholar]
- 44.Zhao GX, Xu YY, Weng SQ, Zhang S, Chen Y, Shen XZ, et al. CAPS1 promotes colorectal cancer metastasis via snail mediated epithelial mesenchymal transformation. Oncogene. 2019;38(23):4574–89. doi: 10.1038/s41388-019-0740-7. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, Li S, Xu Z, Guo J, Li X, Wu Y, et al. CDX2 inhibits epithelial-mesenchymal transition in colorectal cancer by modulation of snail expression and β-catenin stabilisation via transactivation of PTEN expression. Br J Cancer. 2021;124(1):270–80. doi: 10.1038/s41416-020-01148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen T, Yang Z, Cheng X, Xiao Y, Yu K, Cai X, et al. CXCL8 induces epithelial-mesenchymal transition in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway. Oncol Rep. 2017;37(4):2095–100. doi: 10.3892/or.2017.5453. [DOI] [PubMed] [Google Scholar]
- 47.Cheng XS, Li YF, Tan J, Sun B, Xiao YC, Fang XB, et al. CCL20 and CXCL8 synergize to promote progression and poor survival outcome in patients with colorectal cancer by collaborative induction of the epithelial-mesenchymal transition. Cancer Lett. 2014;348(1–2):77–87. doi: 10.1016/j.canlet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Gao L, Xu J, He G, Huang J, Xu W, Qin J, et al. CCR7 high expression leads to cetuximab resistance by cross-talking with EGFR pathway in PI3K/AKT signals in colorectal cancer. Am J Cancer Res. 2019;9(11):2531–43. [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Wang X, Si M, Yang J, Sun S, Wu H, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, et al. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. Onco Targets Ther. 2019;12:3051–63. doi: 10.2147/OTT.S198126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen HJ, Yu Y, Sun YX, Huang CZ, Li JY, Liu F, et al. Id4 suppresses the Growth and Invasion of Colorectal Cancer HCT116 cells through CK18-Related inhibition of AKT and EMT signaling. J Oncol. 2021;2021:6660486. doi: 10.1155/2021/6660486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J, Liu M, Liu H, Zhou L. GATA1 promotes colorectal cancer cell proliferation, migration and invasion via activating AKT signaling pathway. Mol Cell Biochem. 2019;457(1–2):191–9. doi: 10.1007/s11010-019-03523-w. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Xu J, Zhang H, Sun J, Li N, Huang X. MicroRNA-758 acts as a tumor inhibitor in colorectal cancer through targeting PAX6 and regulating PI3K/AKT pathway. Oncol Lett. 2020;19(6):3923–30. doi: 10.3892/ol.2020.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cong K, Li CG, Wei YH, Zhang K, Xu HB. MicroRNA-760 inhibits the biological progression of colorectal carcinoma by directly targeting FOXA1 and regulating epithelial-to-mesenchymal transition and PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(13):5730–40. doi: 10.26355/eurrev_201907_18310. [DOI] [PubMed] [Google Scholar]
- 55.Miller SA, Policastro RA, Savant SS, Sriramkumar S, Ding N, Lu X, et al. Lysine-specific demethylase 1 mediates AKT activity and promotes epithelial-to-mesenchymal transition in PIK3CA-Mutant Colorectal Cancer. Mol Cancer Res. 2020;18(2):264–77. doi: 10.1158/1541-7786.MCR-19-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao P, Guan HT, Dai ZJ, Ma YG, Liu XX, Wang XJ. Knockdown of SPOCK1 inhibits the Proliferation and Invasion in Colorectal Cancer cells by suppressing the PI3K/Akt pathway. Oncol Res. 2016;24(6):437–45. doi: 10.3727/096504016X14685034103554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen B, Zeng X, He Y, Wang X, Liang Z, Liu J, et al. STC2 promotes the epithelial-mesenchymal transition of colorectal cancer cells through AKT-ERK signaling pathways. Oncotarget. 2016;7(44):71400–16. doi: 10.18632/oncotarget.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, Liu Y, Jiang CJ, Chen YM, Li H, Liu QA. Calcium-activated Chloride Channel A4 (CLCA4) plays inhibitory Roles in Invasion and Migration through suppressing epithelial-mesenchymal transition via PI3K/AKT signaling in Colorectal Cancer. Med Sci Monit. 2019;25:4176–85. doi: 10.12659/MSM.914195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin C, Ear J, Pavlova Y, Mittal Y, Kufareva I, Ghassemian M, et al. Tyrosine phosphorylation of the Gα-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci Signal. 2011;4(192):ra64. doi: 10.1126/scisignal.2002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, et al. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. 2005;9(3):389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Zadina JE, Kastin AJ, Ge LJ, Hackler L. Mu, delta, and kappa opiate receptor binding of Tyr-MIF-1 and of Tyr-W-MIF-1, its active fragments, and two potent analogs. Life Sci. 1994;55(24):Pl461–6. doi: 10.1016/0024-3205(94)00533-8. [DOI] [PubMed] [Google Scholar]
- 62.Chen DT, Pan JH, Chen YH, Xing W, Yan Y, Yuan YF, et al. The mu-opioid receptor is a molecular marker for poor prognosis in hepatocellular carcinoma and represents a potential therapeutic target. Br J Anaesth. 2019;122(6):e157–e67. doi: 10.1016/j.bja.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 63.Lennon FE, Mirzapoiazova T, Mambetsariev B, Salgia R, Moss J, Singleton PA. Overexpression of the µ-opioid receptor in human non-small cell lung cancer promotes akt and mTOR activation, tumor growth, and metastasis. Anesthesiology. 2012;116(4):857–67. doi: 10.1097/ALN.0b013e31824babe2. [DOI] [PubMed] [Google Scholar]
- 64.Afsharimani B, Cabot P, Parat MO. Morphine and tumor growth and metastasis. Cancer Metastasis Rev. 2011;30(2):225–38. doi: 10.1007/s10555-011-9285-0. [DOI] [PubMed] [Google Scholar]
- 65.Wu X, Cai J, Zuo Z, Li J. Collagen facilitates the colorectal cancer stemness and metastasis through an integrin/PI3K/AKT/Snail signaling pathway. Biomed Pharmacother. 2019;114:108708. doi: 10.1016/j.biopha.2019.108708. [DOI] [PubMed] [Google Scholar]
- 66.Singh D, Srivastava SK, Chaudhuri TK, Upadhyay G. Multifaceted role of matrix metalloproteinases (MMPs) Front Mol Biosci. 2015;2:19. doi: 10.3389/fmolb.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 68.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8(3):221–33. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K, Zheng J, Yu J, Wu Y, Guo J, Xu Z, et al. Knockdown of MMP–1 inhibits the progression of colorectal cancer by suppressing the PI3K/Akt/c–myc signaling pathway and EMT. Oncol Rep. 2020;43(4):1103–12. doi: 10.3892/or.2020.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19(3):843–54. doi: 10.1091/mbc.e07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25(17):4033–49. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, et al. Comprehensive analysis of the ICEN (interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11(6):673–84. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 73.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–31. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 74.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8(10):743–54. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gulino A, Di Marcotullio L, Ferretti E, De Smaele E, Screpanti I. Hedgehog signaling pathway in neural development and disease. Psychoneuroendocrinology. 2007;32(Suppl 1):52–6. doi: 10.1016/j.psyneuen.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 76.Scales SJ, de Sauvage FJ. Mechanisms of hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30(6):303–12. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 77.Teglund S, Toftgård R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805(2):181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Shih LJ, Lu YF, Chen YH, Lin CC, Chen JA, Hwang SP. Characterization of the agr2 gene, a homologue of X. laevis anterior gradient 2, from the zebrafish, Danio rerio. Gene Expr Patterns. 2007;7(4):452–60. doi: 10.1016/j.modgep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res. 2013;15(2):204. doi: 10.1186/bcr3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park K, Chung YJ, So H, Kim K, Park J, Oh M, et al. AGR2, a mucinous ovarian cancer marker, promotes cell proliferation and migration. Exp Mol Med. 2011;43(2):91–100. doi: 10.3858/emm.2011.43.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fessart D, Domblides C, Avril T, Eriksson LA, Begueret H, Pineau R, et al. Secretion of protein disulphide isomerase AGR2 confers tumorigenic properties. eLife. 2016;5:e13887. doi: 10.7554/eLife.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dumartin L, Alrawashdeh W, Trabulo SM, Radon TP, Steiger K, Feakins RM, et al. ER stress protein AGR2 precedes and is involved in the regulation of pancreatic cancer initiation. Oncogene. 2017;36(22):3094–103. doi: 10.1038/onc.2016.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higa A, Mulot A, Delom F, Bouchecareilh M, Nguyên DT, Boismenu D, et al. Role of pro-oncogenic protein disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) in the control of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286(52):44855–68. doi: 10.1074/jbc.M111.275529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Persson S, Rosenquist M, Knoblach B, Khosravi-Far R, Sommarin M, Michalak M. Diversity of the protein disulfide isomerase family: identification of breast tumor induced Hag2 and Hag3 as novel members of the protein family. Mol Phylogenet Evol. 2005;36(3):734–40. doi: 10.1016/j.ympev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 85.Morrow JK, Lin HK, Sun SC, Zhang S. Targeting ubiquitination for cancer therapies. Future Med Chem. 2015;7(17):2333–50. doi: 10.4155/fmc.15.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell cycle (Georgetown Tex) 2017;16(7):634–48. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18(2):69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang M, Yu T, Hu L, Cheng Z, Li M. Ubiquitin Carboxy-Terminal HydrolaseL3 correlates with human sperm Count, Motility and Fertilization. PLoS ONE. 2016;11(10):e0165198. doi: 10.1371/journal.pone.0165198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suzuki M, Setsuie R, Wada K. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology. 2009;150(12):5230–9. doi: 10.1210/en.2009-0332. [DOI] [PubMed] [Google Scholar]
- 90.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27(4):609–21. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Panda S, Gekara NO. The deubiquitinase MYSM1 dampens NOD2-mediated inflammation and tissue damage by inactivating the RIP2 complex. Nat Commun. 2018;9(1):4654. doi: 10.1038/s41467-018-07016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Panda S, Nilsson JA, Gekara NO. Deubiquitinase MYSM1 regulates innate immunity through inactivation of TRAF3 and TRAF6 complexes. Immunity. 2015;43(4):647–59. doi: 10.1016/j.immuni.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 93.Wilms C, Krikki I, Hainzl A, Kilo S, Alupei M, Makrantonaki E, et al. 2A-DUB/Mysm1 regulates epidermal development in part by suppressing p53-Mediated programs. Int J Mol Sci. 2018;19(3):687. doi: 10.3390/ijms19030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haffner-Luntzer M, Kovtun A, Fischer V, Prystaz K, Hainzl A, Kroeger CM, et al. Loss of p53 compensates osteopenia in murine Mysm1 deficiency. FASEB J. 2018;32(4):1957–68. doi: 10.1096/fj.201700871R. [DOI] [PubMed] [Google Scholar]
- 95.Bahrami E, Witzel M, Racek T, Puchałka J, Hollizeck S, Greif-Kohistani N, et al. Myb-like, SWIRM, and MPN domains 1 (MYSM1) deficiency: genotoxic stress-associated bone marrow failure and developmental aberrations. J Allergy Clin Immunol. 2017;140(4):1112–9. doi: 10.1016/j.jaci.2016.10.053. [DOI] [PubMed] [Google Scholar]
- 96.Hall A. Rho GTPases and the actin cytoskeleton. Sci (New York NY) 1998;279(5350):509–14. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 97.Liu L, Chen L, Chung J, Huang S. Rapamycin inhibits F-actin reorganization and phosphorylation of focal adhesion proteins. Oncogene. 2008;27(37):4998–5010. doi: 10.1038/onc.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miki H, Setou M, Hirokawa N. Kinesin superfamily proteins (KIFs) in the mouse transcriptome. Genome Res. 2003;13(6b):1455–65. doi: 10.1101/gr.984503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu X, Gong H, Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104(6):651–6. doi: 10.1111/cas.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang K, Lin C, Wang C, Shao Q, Gao W, Song B, et al. Silencing Kif2a induces apoptosis in squamous cell carcinoma of the oral tongue through inhibition of the PI3K/Akt signaling pathway. Mol Med Rep. 2014;9(1):273–8. doi: 10.3892/mmr.2013.1804. [DOI] [PubMed] [Google Scholar]
- 101.Shu Q, Nair V. Inosine monophosphate dehydrogenase (IMPDH) as a target in drug discovery. Med Res Rev. 2008;28(2):219–32. doi: 10.1002/med.20104. [DOI] [PubMed] [Google Scholar]
- 102.Tolue Ghasaban F, Maharati A, Akhlaghipour I, Moghbeli M. MicroRNAs as the critical regulators of autophagy-mediated cisplatin response in tumor cells. Cancer Cell Int. 2023;23(1):80. doi: 10.1186/s12935-023-02925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiang Q, Hong D, Liao Y, Cao Y, Liu M, Pang J, et al. Overexpression of Gremlin1 in mesenchymal stem cells improves Hindlimb Ischemia in mice by enhancing cell survival. J Cell Physiol. 2017;232(5):996–1007. doi: 10.1002/jcp.25578. [DOI] [PubMed] [Google Scholar]
- 104.Rodrigues-Diez R, Rodrigues-Diez RR, Lavoz C, Carvajal G, Droguett A, Garcia-Redondo AB, et al. Gremlin activates the smad pathway linked to epithelial mesenchymal transdifferentiation in cultured tubular epithelial cells. Biomed Res Int. 2014;2014:802841. doi: 10.1155/2014/802841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Investig. 2003;112(12):1776–84. doi: 10.1172/JCI200320530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–97. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 107.Song MJ, Malhi H. The unfolded protein response and hepatic lipid metabolism in non alcoholic fatty liver disease. Pharmacol Ther. 2019;203:107401. doi: 10.1016/j.pharmthera.2019.107401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, et al. The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30(7):1357–75. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu J, Chen YJ, Dobbs N, Sakai T, Liou J, Miner JJ, et al. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J Exp Med. 2019;216(4):867–83. doi: 10.1084/jem.20182192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 111.Zhou K, Zheng Z, Li Y, Han W, Zhang J, Mao Y, et al. TFE3, a potential therapeutic target for spinal cord Injury via augmenting autophagy flux and alleviating ER stress. Theranostics. 2020;10(20):9280–302. doi: 10.7150/thno.46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhardwaj M, Leli NM, Koumenis C, Amaravadi RK. Regulation of autophagy by canonical and non-canonical ER stress responses. Sem Cancer Biol. 2020;66:116–28. doi: 10.1016/j.semcancer.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown AM, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J Biol Chem. 2003;278(19):17491–9. doi: 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- 114.Bergamin LS, Capece M, Salaro E, Sarti AC, Falzoni S, Pereira MSL, et al. Role of the P2 × 7 receptor in in vitro and in vivo glioma tumor growth. Oncotarget. 2019;10(47):4840–56. doi: 10.18632/oncotarget.27106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang WJ, Hu CG, Zhu ZM, Luo HL. Effect of P2X7 receptor on tumorigenesis and its pharmacological properties. Biomed Pharmacother. 2020;125:109844. doi: 10.1016/j.biopha.2020.109844. [DOI] [PubMed] [Google Scholar]
- 116.Zhang WJ, Zhu ZM, Liu ZX. The role and pharmacological properties of the P2 × 7 receptor in neuropathic pain. Brain Res Bull. 2020;155:19–28. doi: 10.1016/j.brainresbull.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 117.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 118.Mali AV, Joshi AA, Hegde MV, Kadam SS. Enterolactone modulates the ERK/NF-κB/Snail signaling pathway in triple-negative breast cancer cell line MDA-MB-231 to revert the TGF-β-induced epithelial-mesenchymal transition. Cancer Biol Med. 2018;15(2):137–56. doi: 10.20892/j.issn.2095-3941.2018.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu LL, Chen XH, Zhang G, Liu ZC, Wu N, Wang H, et al. CCL21 facilitates chemoresistance and cancer stem cell-like properties of colorectal cancer cells through AKT/GSK-3β/Snail Signals. Oxid Med Cell Longev. 2016;2016:5874127. doi: 10.1155/2016/5874127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luo W, Liu X, Sun W, Lu JJ, Wang Y, Chen X. Toosendanin, a natural product, inhibited TGF-β1-induced epithelial-mesenchymal transition through ERK/Snail pathway. Phytother Res. 2018;32(10):2009–20. doi: 10.1002/ptr.6132. [DOI] [PubMed] [Google Scholar]
- 121.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Investig. 2014;124(4):1853–67. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang BL. A unique SNARE machinery for exocytosis of cytotoxic granules and platelets granules. Mol Membr Biol. 2015;32(4):120–6. doi: 10.3109/09687688.2015.1079934. [DOI] [PubMed] [Google Scholar]
- 123.Ji M, Lee EJ, Kim KB, Kim Y, Sung R, Lee SJ, et al. HDAC inhibitors induce epithelial-mesenchymal transition in colon carcinoma cells. Oncol Rep. 2015;33(5):2299–308. doi: 10.3892/or.2015.3879. [DOI] [PubMed] [Google Scholar]
- 124.Jiang GM, Wang HS, Zhang F, Zhang KS, Liu ZC, Fang R, et al. Histone deacetylase inhibitor induction of epithelial-mesenchymal transitions via up-regulation of snail facilitates cancer progression. Biochim Biophys Acta. 2013;1833(3):663–71. doi: 10.1016/j.bbamcr.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 125.Kong D, Ahmad A, Bao B, Li Y, Banerjee S, Sarkar FH. Histone deacetylase inhibitors induce epithelial-to-mesenchymal transition in prostate cancer cells. PLoS ONE. 2012;7(9):e45045. doi: 10.1371/journal.pone.0045045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uchida H, Maruyama T, Nishikawa-Uchida S, Oda H, Miyazaki K, Yamasaki A, et al. Studies using an in vitro model show evidence of involvement of epithelial-mesenchymal transition of human endometrial epithelial cells in human embryo implantation. J Biol Chem. 2012;287(7):4441–50. doi: 10.1074/jbc.M111.286138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feng J, Cen J, Li J, Zhao R, Zhu C, Wang Z, et al. Histone deacetylase inhibitor valproic acid (VPA) promotes the epithelial mesenchymal transition of colorectal cancer cells via up regulation of snail. Cell Adhes Migr. 2015;9(6):495–501. doi: 10.1080/19336918.2015.1112486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grainger S, Savory JG, Lohnes D. Cdx2 regulates patterning of the intestinal epithelium. Dev Biol. 2010;339(1):155–65. doi: 10.1016/j.ydbio.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 129.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150(1):165–78. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zangouei AS, Hamidi AA, Rahimi HR, Saburi E, Mojarrad M, Moghbeli M. Chemokines as the critical factors during bladder cancer progression: an overview. Int Rev Immunol. 2021;40(5):344–58. doi: 10.1080/08830185.2021.1877287. [DOI] [PubMed] [Google Scholar]
- 131.Wang L, Li CL, Wang L, Yu WB, Yin HP, Zhang GY, et al. Influence of CXCR4/SDF-1 axis on E-cadherin/β-catenin complex expression in HT29 colon cancer cells. World J Gastroenterol. 2011;17(5):625–32. doi: 10.3748/wjg.v17.i5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen G, Chen SM, Wang X, Ding XF, Ding J, Meng LH. Inhibition of chemokine (CXC motif) ligand 12/chemokine (CXC motif) receptor 4 axis (CXCL12/CXCR4)-mediated cell migration by targeting mammalian target of rapamycin (mTOR) pathway in human gastric carcinoma cells. J Biol Chem. 2012;287(15):12132–41. doi: 10.1074/jbc.M111.302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang CY, Fong YC, Lee CY, Chen MY, Tsai HC, Hsu HC, et al. CCL5 increases lung cancer migration via PI3K, akt and NF-kappaB pathways. Biochem Pharmacol. 2009;77(5):794–803. doi: 10.1016/j.bcp.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 134.Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neurooncology. 2005;7(2):122–33. doi: 10.1215/S1152851704001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoshimura T, Matsushima K, Oppenheim JJ, Leonard EJ. Neutrophil chemotactic factor produced by lipopolysaccharide (LPS)-stimulated human blood mononuclear leukocytes: partial characterization and separation from interleukin 1 (IL 1) J Immunol (Baltimore, Md: 1950). 1987;139(3):788–93. doi: 10.4049/jimmunol.139.3.788. [DOI] [PubMed] [Google Scholar]
- 136.Patel M, Horgan PG, McMillan DC, Edwards J. NF-κB pathways in the development and progression of colorectal cancer. Transl Res. 2018;197:43–56. doi: 10.1016/j.trsl.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luraghi P, Bigatto V, Cipriano E, Reato G, Orzan F, Sassi F, et al. A molecularly annotated model of patient-derived Colon Cancer Stem-Like cells to assess genetic and nongenetic mechanisms of resistance to Anti-EGFR Therapy. Clin Cancer Res. 2018;24(4):807–20. doi: 10.1158/1078-0432.CCR-17-2151. [DOI] [PubMed] [Google Scholar]
- 139.Pang MF, Georgoudaki AM, Lambut L, Johansson J, Tabor V, Hagikura K, et al. TGF-β1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis. Oncogene. 2016;35(6):748–60. doi: 10.1038/onc.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zarour LR, Anand S, Billingsley KG, Bisson WH, Cercek A, Clarke MF, et al. Colorectal Cancer Liver Metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017;3(2):163–73. doi: 10.1016/j.jcmgh.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Doak GR, Schwertfeger KL, Wood DK. Distant relations: macrophage functions in the metastatic niche. Trends Cancer. 2018;4(6):445–59. doi: 10.1016/j.trecan.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–66. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 143.Ghosh S, Mukherjee S, Choudhury S, Gupta P, Adhikary A, Baral R, et al. Reactive oxygen species in the tumor niche triggers altered activation of macrophages and immunosuppression: role of fluoxetine. Cell Signal. 2015;27(7):1398–412. doi: 10.1016/j.cellsig.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 144.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66(1):157–67. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 145.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 146.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13(8):410–8. doi: 10.1016/S0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 147.Yokota Y, Mori S. Role of id family proteins in growth control. J Cell Physiol. 2002;190(1):21–8. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- 148.Boidot R, Végran F, Jacob D, Chevrier S, Cadouot M, Feron O, et al. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29(17):2577–84. doi: 10.1038/onc.2009.525. [DOI] [PubMed] [Google Scholar]
- 149.Zhang Y, Liu J, Lin J, Zhou L, Song Y, Wei B, et al. The transcription factor GATA1 and the histone methyltransferase SET7 interact to promote VEGF-mediated angiogenesis and tumor growth and predict clinical outcome of breast cancer. Oncotarget. 2016;7(9):9859–75. doi: 10.18632/oncotarget.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li Y, Ke Q, Shao Y, Zhu G, Li Y, Geng N, et al. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget. 2015;6(6):4345–56. doi: 10.18632/oncotarget.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Elso C, Lu X, Weisner PA, Thompson HL, Skinner A, Carver E, et al. A reciprocal translocation dissects roles of Pax6 alternative promoters and upstream regulatory elements in the development of pancreas, brain, and eye. Genesis (New York, NY: 2000). 2013;51(9):630–46. doi: 10.1002/dvg.22409. [DOI] [PubMed] [Google Scholar]
- 152.Zhang S, Liu Q, Zhang Q, Liu L. MicroRNA-30a-5p suppresses proliferation, invasion and tumor growth of hepatocellular cancer cells via targeting FOXA1. Oncol Lett. 2021;22(2):574. doi: 10.3892/ol.2021.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gan HY, Li N, Zhang Q, Feng ZZ. Silencing FOXA1 gene regulates liver cancer cell apoptosis and cell proliferation. Eur Rev Med Pharmacol Sci. 2018;22(2):397–404. doi: 10.26355/eurrev_201801_14187. [DOI] [PubMed] [Google Scholar]
- 154.Ghobashi AH, Vuong TT, Kimani JW, O’Hagan HM. Activation of AKT induces EZH2-mediated β-catenin trimethylation in colorectal cancer. bioRxiv. 2023:2023–01. [DOI] [PMC free article] [PubMed]
- 155.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 156.Iwase S, Januma A, Miyamoto K, Shono N, Honda A, Yanagisawa J, et al. Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun. 2004;322(2):601–8. doi: 10.1016/j.bbrc.2004.07.163. [DOI] [PubMed] [Google Scholar]
- 157.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31(3):353–65. doi: 10.1016/S0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 158.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–16. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109(4):994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S, et al. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 2013;73(1):235–45. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285(43):32967–76. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Sem Cancer Biol. 2012;22(5–6):396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li Y, Chen L, Chan TH, Liu M, Kong KL, Qiu JL, et al. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology. 2013;144(1):179–91e4. doi: 10.1053/j.gastro.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 166.Röll S, Seul J, Paulsson M, Hartmann U. Testican-1 is dispensable for mouse development. Matrix Biol. 2006;25(6):373–81. doi: 10.1016/j.matbio.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 167.Szliter E, McClellan S, Barrett R, Hazlett L. Testican-1/SPOCK1 promotes resistance against P. aeruginosa-induced keratitis via modulation of MMP-driven wound healing and ECM restoration. Investig Ophthalmol Vis Sci. 2009;50(13):1199. [Google Scholar]
- 168.McCudden CR, James KA, Hasilo C, Wagner GF. Characterization of mammalian stanniocalcin receptors. Mitochondrial targeting of ligand and receptor for regulation of cellular metabolism. J Biol Chem. 2002;277(47):45249–58. doi: 10.1074/jbc.M205954200. [DOI] [PubMed] [Google Scholar]
- 169.Zeng X, Yang P, Chen B, Jin X, Liu Y, Zhao X, et al. Quantitative secretome analysis reveals the interactions between epithelia and tumor cells by in vitro modulating colon cancer microenvironment. J Proteom. 2013;89:51–70. doi: 10.1016/j.jprot.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 170.Xie Q, Guo X, Gu J, Zhang L, Jin H, Huang H, et al. p85α promotes nucleolin transcription and subsequently enhances EGFR mRNA stability and EGF-induced malignant cellular transformation. Oncotarget. 2016;7(13):16636–49. doi: 10.18632/oncotarget.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jiang HN, Zeng B, Zhang Y, Daskoulidou N, Fan H, Qu JM, et al. Involvement of TRPC channels in lung cancer cell differentiation and the correlation analysis in human non-small cell lung cancer. PLoS ONE. 2013;8(6):e67637. doi: 10.1371/journal.pone.0067637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ito Y, Hart JR, Ueno L, Vogt PK. The regulatory subunit of PI3K, p85β, induces cellular transformation, enhanced cell proliferation and increased PI3K signaling. Cancer Res. 2014;74(19Supplement):3339. doi: 10.1158/1538-7445.AM2014-3339. [DOI] [Google Scholar]
- 173.Ma Q, Lu Y, Gu Y. ENKUR is involved in the regulation of Cellular Biology in Colorectal Cancer cells via PI3K/Akt signaling pathway. Technol Cancer Res Treat. 2019;18:1533033819841433. doi: 10.1177/1533033819841433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xiao X, Tang YS, Mackins JY, Sun XL, Jayaram HN, Hansen DK, et al. Isolation and characterization of a folate receptor mRNA-binding trans-factor from human placenta. Evidence favoring identity with heterogeneous nuclear ribonucleoprotein E1. J Biol Chem. 2001;276(44):41510–7. doi: 10.1074/jbc.M106824200. [DOI] [PubMed] [Google Scholar]
- 175.Liu Z, Chen M, Xie LK, Liu T, Zou ZW, Li Y, et al. CLCA4 inhibits cell proliferation and invasion of hepatocellular carcinoma by suppressing epithelial-mesenchymal transition via PI3K/AKT signaling. Aging. 2018;10(10):2570–84. doi: 10.18632/aging.101571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hou T, Zhou L, Wang L, Kazobinka G, Zhang X, Chen Z. CLCA4 inhibits bladder cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT pathway. Oncotarget. 2017;8(54):93001–13. doi: 10.18632/oncotarget.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yu Y, Walia V, Elble RC. Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS ONE. 2013;8(12):e83943. doi: 10.1371/journal.pone.0083943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


