Abstract
The systemic delivery of exogenous proteins to cells within the brain and central nervous system (CNS) is challenging due to the selective impermeability of the blood-brain barrier (BBB). Herein, we hypothesized that protein delivery to the brain could be improved via functionalization with DNA aptamers designed to bind transferrin (TfR) receptors present on the endothelial cells that line the BBB. Using β-Galactosidase (β-Gal) as a model protein, we synthesized protein spherical nucleic acids (ProSNAs) comprised of β-Gal decorated with TfR aptamers (Transferrin-ProSNAs). The TfR aptamer motif significantly increases the accumulation of β-Gal in brain tissue in vivo following intravenous injection over both the native protein and ProSNAs containing nontargeting DNA sequences. Furthermore, the widespread distribution of β-Gal throughout the brain is only observed for Transferrin-ProSNAs. Together, this work shows that the SNA architecture can be used to selectively deliver protein cargo to the brain and CNS if the appropriate aptamer sequence is employed as the DNA shell. Moreover, this highlights the importance of DNA sequence design and provides a potential new avenue for designing highly targeted protein delivery systems by combining the power of DNA aptamers together with the SNA platform.
Keywords: Spherical Nucleic Acids, Protein Delivery, Blood-Brain Barrier, Cellular Uptake
Graphical Abstract
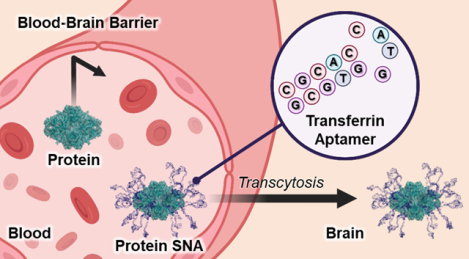
INTRODUCTION
The targeted delivery of proteins to the brain and central nervous system (CNS) is inherently challenging1–4 because of the many natural mechanisms and barriers that living systems have evolved to maintain homeostasis.5 Firstly, unmodified proteins are rapidly degraded by serum proteases6, which inherently limits their effective delivery to target sites. Moreover, the blood-brain barrier (BBB) is a highly selective barrier that prevents circulating materials from reaching the CNS and is regulated by endothelial cells whose tight junctions form the impermeable walls of blood vessels7, 8. Finally, even if the issues with proteolysis and the BBB are overcome, the cell membrane itself prevents the passive movement of exogenous materials into cells within the brain. As such, strategies to improve the intracellular delivery of proteins to the brain and CNS are warranted.
The BBB is a highly regulated barrier, where endothelial cells control the influx of nutrients and molecules into the brain via both passive and active pathways. Lipid-soluble substances (CO2 and O2) freely cross the BBB9, while larger solutes gain access to the brain through active transport via receptors abundant in the endothelium10, 11. Accordingly, transport receptors, such as those for insulin12, 13, glucose14, or transferrin15–22 (TfR), provide potential avenues for the transport of proteins across the BBB through chemical modifications with the respective ligands. Indeed, lysosomal enzymes modified with antibodies specific to both the insulin12, 13 and Tfr receptor16, 21, 22 have shown significantly increased BBB penetration in vivo. Likewise, aptamers—short single-stranded DNA or RNA oligonucleotides that can assume secondary structures with high affinity and specificity to a target—have been evolved to transport oligonucleotides across the BBB via TfR receptors20.
Spherical nucleic acids (SNAs) comprise a class of nanomaterials where oligonucleotides are densely functionalized around a nanoparticle core in a highly oriented manner.23, 24 This spherical arrangement imparts biological properties to the composite oligonucleotides that are distinct from their linear counterparts, including rapid cellular uptake through scavenger receptor-mediated endocytosis and enhanced stability to degrading enzymes.25–29 Importantly, proteins that are normally cell impermeable and highly susceptible to protease degradation are rendered permeable and protease-resistant when functionalized with oligonucleotides as protein spherical nucleic acids (ProSNAs).29–31 Previous work has found that the DNA shell not only protects the protein, but also alters its biodistribution following systemic administration.29
Herein, we hypothesized that the intracellular delivery of proteins to cells within the CNS could be maximized using the ProSNA platform via covalent functionalization with DNA aptamers specific to the TfR receptor. DNA shell design is a critical parameter influencing the cellular uptake and biodistribution of SNAs29, 32–37, and we reasoned that a ProSNA’s distribution could be specifically directed by introducing a targeting sequence. Therefore, we synthesized ProSNAs bearing TfR aptamers using β-Galactosidase (β-Gal) as a model protein (Transferrin-ProSNAs, Figure 1) and assessed their uptake in an in vitro BBB model and their accumulation in the brain following intravenous injection into healthy mice.
Figure 1. Receptor-Mediated Transcytosis of ProSNAs Across the Blood-Brain Barrier.
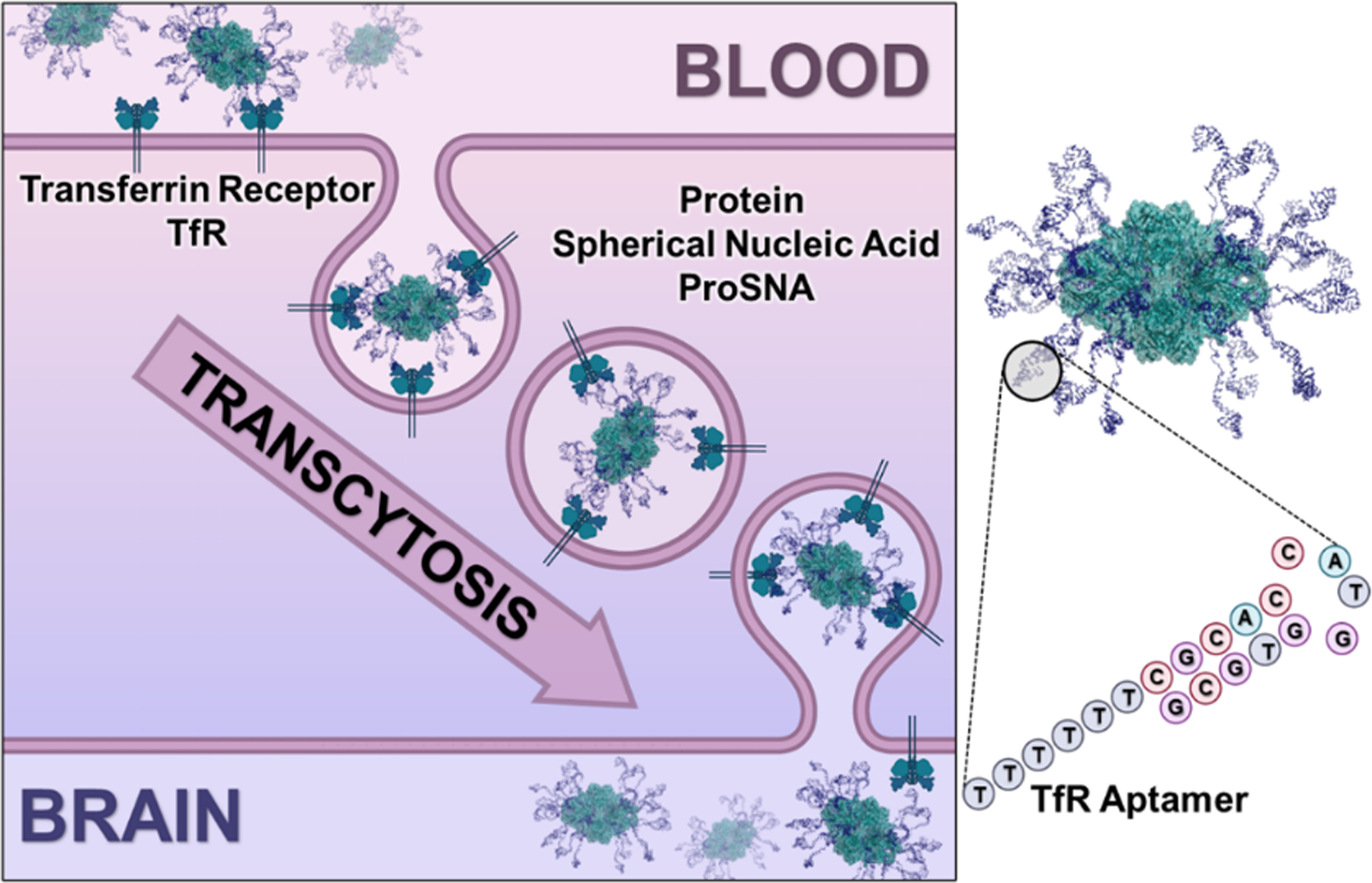
Cartoon representation of β-Gal ProSNAs being shuttled across an endothelial cell to the brain via the transferrin receptor. The surface of β-Gal is decorated with a shell of transferrin-specific aptamers covalently conjugated to surface lysine residues, resulting in a protein spherical nucleic acid (ProSNA). The representation was adapted from PDB ID 1PX3.
RESULTS & DISCUSSION
Transferrin-ProSNAs Are Successfully Synthesized through the Functionalization of β-Galactosidase with Transferrin Aptamer DNA. The transferrin (TfR) aptamer sequence was selected based on prior research using SELEX to evolve and truncate aptamers, resulting in a 14-mer sequence with a strong binding affinity to the extracellular domain of the mouse transferrin receptor.38–40 A scrambled DNA sequence, which differs from TfR by four bases in the loop region and has no receptor binding affinity, was synthesized as a control for aptamer-mediated uptake (Table S1.1). Protein SNAs bearing either TfR aptamers (Transferrin-ProSNAs) or the scrambled DNA sequence (Scr-ProSNAs) were synthesized using published protocols29, 30. β-Gal was chosen as a model system to assess brain delivery efficacy because it is a large (464 kDa) protein, easily amenable to DNA conjugation, and does not naturally traverse cell membranes. To track the distribution of β-Gal in vivo, an AlexaFluor647 (AF647) fluorophore tag was conjugated to surface-accessible cysteines on the protein. Following successful fluorophore labeling, the lysine residues were modified with azide-functionalized-PEG4, which was subsequently reacted with DBCO-dT-terminated aptamer DNA strands through a strained-promoted alkyne azide cycloaddition (Figure S1, Table S1.2)41. A thymidine DNA spacer (T4) was appended to the 3’ end of each sequence to distance the aptamer from the protein surface and promote sequence folding. UV-vis spectroscopy showed that the extent of oligonucleo-tide loading were nearly identical (39 Transferrin DNA per protein and 37 Scr DNA per protein) regardless of sequence (Figure S2, Table S1.3). Covalent DNA conjugation to the protein core was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which showed mobility shifts commensurate with the increased mass following DNA addition (Figure S3). Moreover, dynamic light scattering (DLS) showed an increase in hydrodynamic diameter (Figure S4) following DNA functionalization, confirming the successful synthesis of ProSNAs (β-Gal AF647: 18 ± 2 nm, Transferrin-ProSNA: 24 ± 3 nm). Finally, circular dichroism (CD) spectroscopy confirmed that the secondary structure of β-Gal was not altered following fluorophore and DNA conjugation (Figure S5).
Transferrin-ProSNAs Penetrate Cell Monolayer Faster in an In Vitro BBB Model.
With Transferrin-ProSNAs in hand, we sought to determine the rate at which the aptamer is able to promote receptor-mediate transcytosis.11 For these experiments, bEnd.3 mouse brain endothelial cells, known to highly express the TfR receptor15, 42, were cultured to form a monolayer and simulate the tight endothelial junctions that comprise the BBB. To confirm that the bEnd.3 cell barrier was not compromised by the treatment, transendothelial resistance (TEER) measurements of the monolayer was assessed immediately before beginning the experiment, as well as 24 h post-treatment. TEER values for the bEnd.3 cells did not significantly change over time in the presence of the protein (Figure S6) indicating a restrictive barrier and match the reported finding for a stable cell layer43. Once the monolayer was established, Transferrin-ProSNAs, Scr-ProSNAs, or β-Gal AF647 were applied to the apical side (Figure 2A). The fluorescence of the media on the basolateral side was measured over time using a plate reader fluorimeter as a proxy for protein transcytosis, as a means to determine the rates of transport. The fluorescence intensity of the media in wells treated with both ProSNA constructs increased steadily as a function of time and at a much faster rate than the AF647 modified protein or TMR dextran 70 kDa (Figure 2B). It is to be expected that the Scr-ProSNA would exhibit some degree of transcytosis capability, as it has been previously shown in a clinical trial that SNAs synthesized with gold cores cross the BBB and accumulate in glioblastoma brain tumors following systemic injection, presumably facilitated by scavenger-mediated movement.44, 45
Figure 2. In vitro BBB Analysis Demonstrates Specificity of Transferrin Aptamer for Receptor.
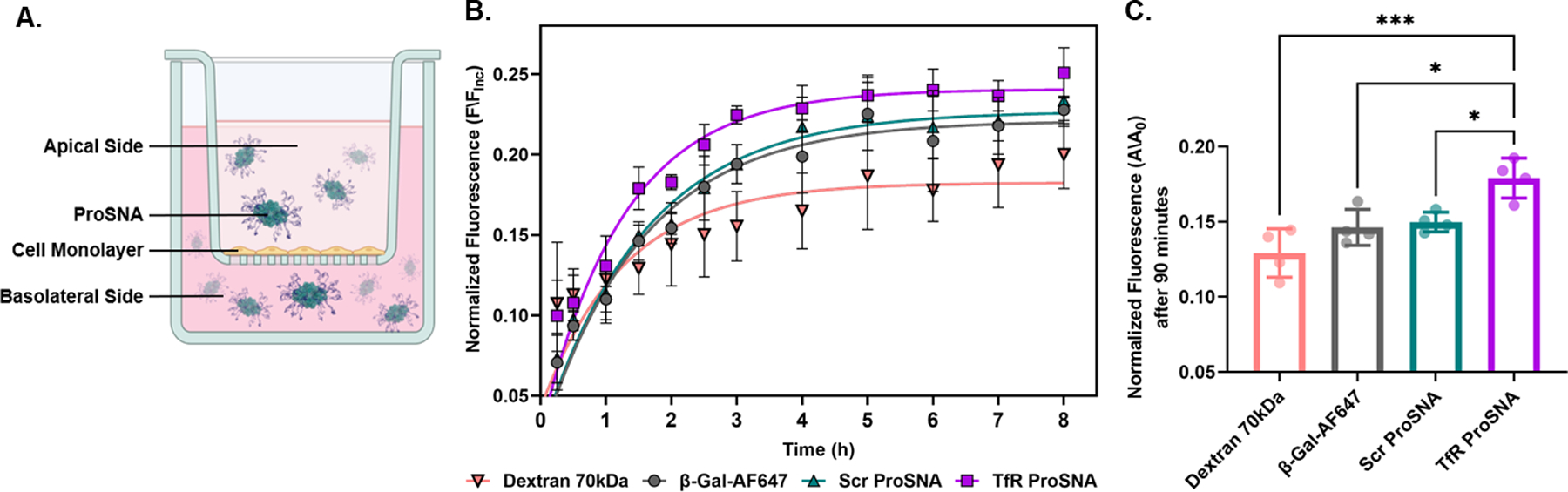
(A) Representation of ProSNAs crossing an in vitro BBB model. Cells were treated with fluorescently labeled proteins on the apical side of a transwell insert. (B) Normalized fluorescence readings of the basolateral side were taken periodically and plotted to measure the protein’s transcytosis efficiency. Each point represents the mean of n =4 measurements, the error bars show SD, and the line is a nonlinear regression fit graphed using GraphPad Prism. (C) Explicit fluorescence significance comparison after 90 minutes incubation of either Dextran 70 kDa, β-Gal-AF647, TfR ProSNA or Scr ProSNA. The statistical significance was analyzed using ordinary one-way ANOVA, followed by tukey multiple comparison. **** p-value < 0.0001, *** p < 0.001, ** p < 0.01, and * p < 0.05.
From these analyses, the apparent permeability coefficient (Papp) was calculated to precisely analyze the kinetics of monolayer transcytosis. This index is widely used as part of the screening process of the flux of a molecule across a barrier, to measure the integrity of the in vitro BBB model46, 47. A significant increase in fluorescence on the basolateral side was observed at 90 min in cells incubated with Transferrin-ProSNAs (Figure 2C). In addition, Transferrin-ProSNAs showed a higher Papp value (7.571·10−6 cm/s) relative to both β-galactosidase (6.628·10−6 cm/s) and Scr-ProSNAs (Papp = 6.941·10−6 cm/s), presumably due to their increased affinity for the transferrin receptor. A table for all the calculated values is included in the SI (Table S2.1).
Transferrin-ProSNAs Significantly Increase the in vivo Accumulation of Proteins in the Brain.
By employing TfR aptamers as the DNA shell, we hypothesized that Transferrin-ProSNAs can target the BBB. Therefore, to study the brain targeting efficiency of Transferrin-ProSNAs, CD-1 mice (n = 6) were injected intravenously (IV) with either Transferrin-ProSNAs, Scr-ProSNAs, or native β-Gal at a dosage of 6.5 mg/kg with respect to protein concentration. At 1-h post-injection, the biodistribution of each construct was assessed using an in vivo imaging system (IVIS) to measure the fluorescence signal in each organ (Figure 3) following sacrifice and perfusion. Importantly, Transferrin-ProSNAs show a significant increase in brain accumulation (p = 0.0001, Figure 3A, C) as compared to the Scr-ProSNAs, which only differs in sequence by four bases. Moreover, the targeting by Transferrin-ProSNAs is specific to the brain, as there was no significant increase in accumulation in other filtration organs when comparing mice treated with Transferrin-ProSNAs to those administered Scr-ProSNAs (Figure 3B, D). Together, this indicates that the DNA aptamers remain folded and recognize their target receptor in a living system.
Figure 3. Biodistribution of β-Galactosidase Following Systemic Injection in Mice.
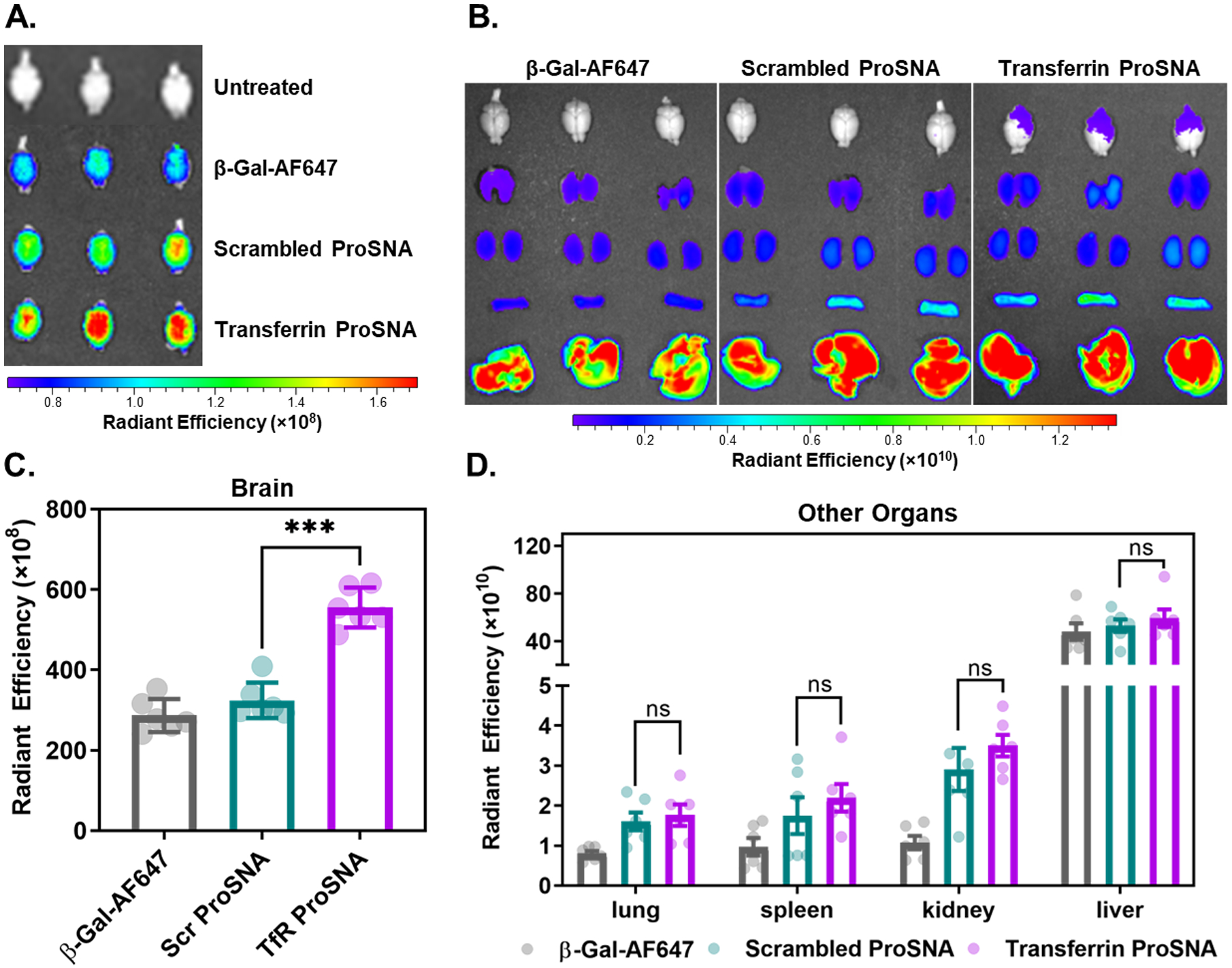
After a 1-h treatment with 6.5 mg of β-Gal/kg mouse via a tail-vein injection; mice (n = 6) were sacrificed, perfused, and their organs dissected and fixed. Using IVIS, the fluorescence from the protein’s AF647 tag was measured in the (A) brain and (B) other filtration organs. A region of interest was drawn around each organ, and the fluorescence radiant efficiency were quantified (C-D). Each dot represents an individual mouse, the bars represent the mean, and the error bars show SD. A paired t-test was performed in GraphPad Prism. *** p ≤ 0.0001, ns = not significant
Transferrin-ProSNAs Visualized Throughout the Brain via Confocal Microscopy.
Encouraged by the increased brain accumulation of Transferrin-ProSNAs on a whole-organ level, we evaluated the tissue and cellular distribution of all materials using immunofluorescent histochemistry (IFC) of brain tissue slices. CD-1 mice (n = 3) were injected intravenously with either Transferrin-ProSNAs, Scr-ProSNAs, or β-Gal AF647 at a dosage of 6.5 mg/kg with respect to protein concentration. At 1-h post-injection, animals were sacrificed and perfused, then brain tissues were sectioned and incubated with a DAPI nucleus stain. Strikingly, the AF647 signal originating from the protein conjugated fluorophore was present throughout the entirety of the brain and not sequestered in a single region. Intense, punctate red fluorescence signals present in the same z-plane as DAPI signals, indicating that Transferrin-ProSNAs are taken up by cells (Figure 4, Figure S7). Conversely, there was no detectable signal in brain tissue slices of animals administered the Scr-ProSNA (Figure S8). This is in agreement with the whole-organ-level data, where minimal brain accumulation was observed via IVIS for these two systems. Together, this shows that not only to Transferrin-ProSNAs increase the accumulation of protein in the brain via enhanced BBB penetration, but also enable the efficient cellular entry in cells present in the brain.
Figure 4. Brain Distribution of Transferrin-ProSNAs.
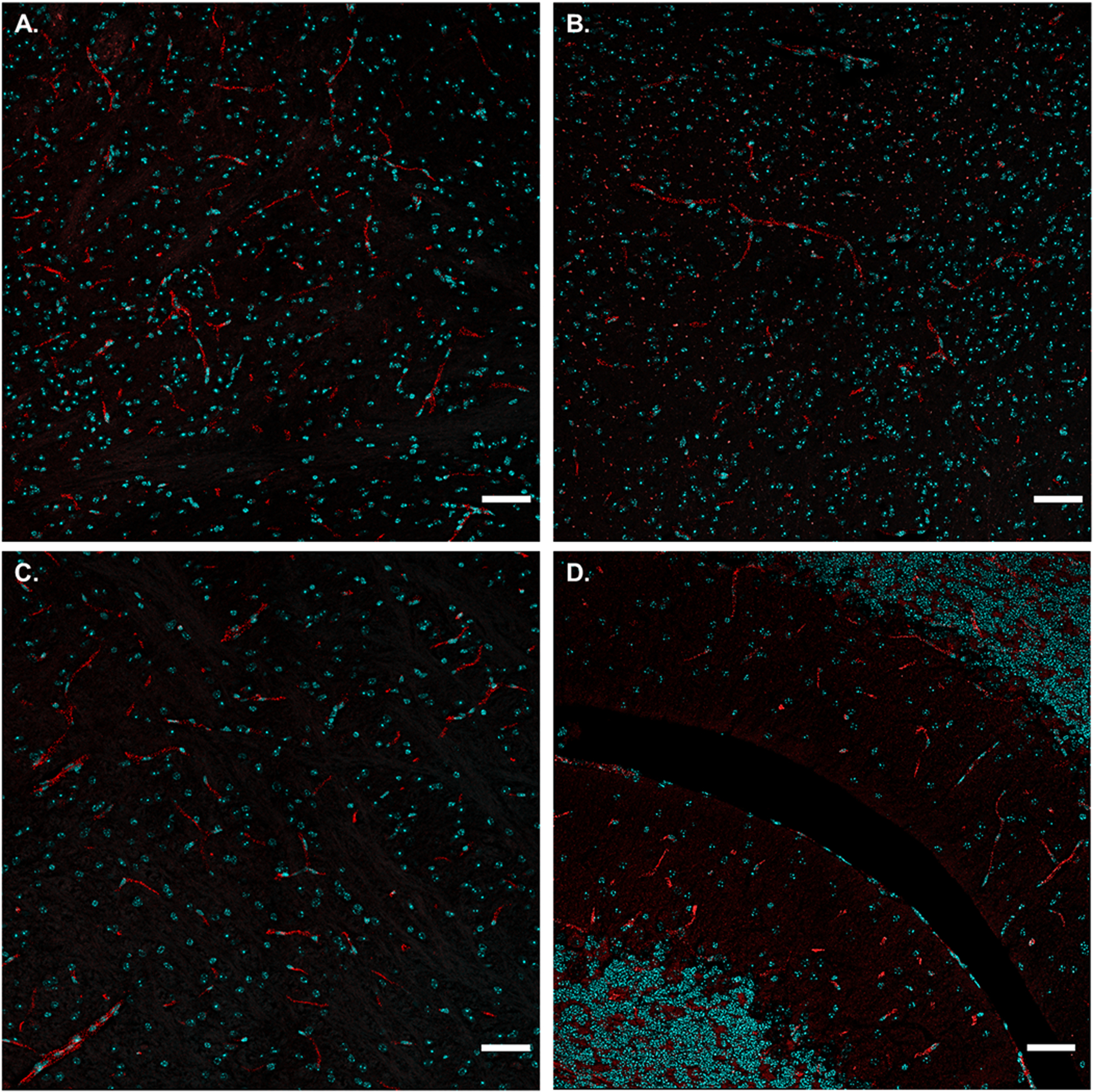
Representative confocal images of brain histology slides from mice 1-h after intravenous injection of 6.5 mg of β-Gal/kg mouse. Punctate red fluorescence signal (ProSNA) is present throughout the brain, in regions such as: (A) thalamus, (B) hypothalamus, (C) pons, and (D) cerebellum. Nuclei are stained with DAPI (cyan) to approximate the location within the cell. Scale bar = 50 μm. Further imaging in other brain regions is represented in Figure S7.
CONCLUSIONS
Herein, we show that the delivery of proteins to the brain is enhanced through their transformation into ProSNAs comprised of DNA aptamers that bind the TfR receptor. This is significant, because although it has been previously shown that adding a DNA shell changes the biodistribution of proteins29, this is a demonstration of using the oligonucleotide sequence as a tissue targeting moiety. This is especially exciting when one considers the vast number of targets and disease sites that aptamers can be easily designed against48–50. Moreover, DNA aptamers have been previously successfully employed in ProSNAs as nucleic acid recognition elements to probe pH or glucose levels in living cells.51 As such, it may be possible to design systems that simultaneously target disease sites (using a targeting aptamer) and probe intracellular conditions upon localization (using a recognition aptamer). Taken together, this work illustrates that DNA shell design can have a profound impact on the resulting distribution of SNAs and can be used to target hard-to-reach organs through intelligent sequence design.
MATERIALS AND METHODS
Synthesis β-Galactosidase Protein Spherical Nucleic Acids.
Oligonucleotides (TfR is 5’-GCG TGG TAC CAC GCT TTT T DBCO-dT-3’ and Scr is 5’-GCG TGT GCT CAC GCT TTT T DBCO-dT-3’) were synthesized using standard phosphoramidite chemistry protocols using universal CPG solid supports and phosphoramidites, as well as coupling reagents purchased from Glen Research. Synthesized strands were purified by HPLC and analyzed using MALDI-MS. ProSNA synthesis was performed based on literature precedence29, 30. β-Gal from an E. coli overproducer was first dissolved in PBS. Next, ten equivalents of AF647-C2-maleimide were added, and the reaction was shaken for 2 hours at 25 °C. Unconjugated AF647 was removed by repeated rounds of centrifugation using a 100 kDa MWCO filter, and the number of fluorophore modifications per protein was calculated using UV-Vis spectroscopy. Then 350 equivalents of NHS-PEG4-azide crosslinker were added to β-Gal-AF647, and the reaction was shaken overnight at 25°C. Unconjugated linker was removed by ten rounds of centrifugation using a 100 kDa filter, and the number of azide modifications was assessed by MALDI-MS. Finally, 350 equivalents of DBCO-dT terminated DNA strands were mixed with β-Gal-AF647-azide and allowed to incubate for 72 h at 25 °C with shaking. Unreacted DNA strands were removed by successive rounds of centrifugation in a 100 kDa filter until the filtrate did not have a detectable absorbance at 260 nm. The number of DNA strands per protein was calculated based on UV-Vis spectroscopy. Characterization of successful covalent conjugation was assessed by SDS PAGE gel.
Transendothelial electrical resistance (TEER) measurements.
The transendothelial resistance of bEnd.3 cell monolayers was measured using a Millicell ERS-2 voltammeter equipped with a MERSSTX01 electrode after cells reached confluence in a transwell plate. Once TEER values reached that of standard monolayers with tight junctions, cells were treated and resistances measured again at 24 h post-treatment. The final TEER result is expressed (Ω×cm2) = (TEER total (Ω)-TEER blank (Ω))×cm2.
In Vitro Blood-Brain Barrier Model.
6.5 mm Transwell®-COL collagen-coated 0.4 μm Pore PTFE membrane inserts were placed in a 24-well plate and pre-incubated with full cell culture media overnight at 37°C. The next day, bEnd.3 endothelial cells were seeded on the apical side of the transwell insert and cultured at 37°C in 5% CO2. After nine days, media was removed, fresh media was added to each well, and treatments added to the appropriate insert. The treatment consisted of 60 nM with respect to protein concentration (300 μL) of β-Gal-AF647, Transferrin-ProSNA, or Scr-ProSNA diluted in full cell culture media and added to the apical side. Cell culture medium (1000 μL) was added to the basolateral side of the insert. Cells were incubated at 37°C in 5% CO2 and 75 μL of media was removed from the lower chambers at set timepoints and fluorescence measured via plate reader fluorimeter (λEx= 640 nm, λEm= 681 nm) and transferred back to the basolateral side. Cumulative transport across the membrane was plotted as increase in fluorescence as a function of time. The slopes of the linear regions were used to calculate the permeability coefficients as previously described52. The following equation was used:
Where dQ/dt is the transport rate, defined as the slope obtained from the regression of the transported amount, C0 is the initial concentration on the donor side and A is the surface area of the inserts (0.33 cm2).
Ex Vivo Near-Infrared Fluorescence (NIRF) Imaging.
Female CD1 mice (28–30 g) were administered a single injection via tail vein at a dose of 6.5 mg β-Gal / kg body weight. After 1 h, mice (n = 6) were humanely euthanized by cardiac perfusion with PBS while anesthetized. Tissues were harvested and fixed. Organs were imaged using an IVIS system using 650 nm/700 nm excitation/emission filters, and data was quantified by measuring radiant efficiency with the Living Image software.
Brain Histology and Imaging.
Female CD1 mice (28–30 g) were administered a single injection via tail vein at a dose of 6.5 mg β-Gal / kg body weight. 1 h post-injection, mice (n = 3) were humanely euthanized by cardiac perfusion while anesthetized. Each mouse was first perfused with 20 mL of PBS, then 20 mL of 4% paraformaldehyde (PFA), and the brains were dissected and stored in 4% PFA with shaking at 4 °C for 2 h. Organs were equilibrated at 4°C in 15% sucrose then 30% sucrose until the organs sunk to the bottom of their respective container. Each organ was then embedded and cryo-sectioned in the sagittal plane at the midline at a 10 μm thickness. Slides were stained with DAPI to visualize nuclei. A Leica TCS SP8 Confocal Microscope was used to image different regions of each slide at a 40X magnification.
Supplementary Material
ACKNOWLEDGMENT
This material is based on research sponsored by the Air Force Research Laboratory under agreement FA8650-15-2-5518. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Air Force Research Laboratory or the U.S. Government. Research reported in this publication was also supported by the Lefkofsky Family Foundation, Polsky Urologic Cancer Institute of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University at Northwestern Memorial Hospital, and the National Cancer Institute of the National Institutes of Health under Awards U54CA199091, R01CA208783, and P50CA221747. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.E.C. was supported by a Postdoctoral Fellowship, PF-20-046-01 - LIB, from the American Cancer Society, as well as the Eden and Steven Romick Postdoctoral Fellowship through the American Committee for the Weizmann Institute of Science. This work was supported by the Northwestern University Keck Biophysics Facility and a Cancer Center Support Grant (NCI CA060553). This work made use of the IMSERC MS facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-2025633), the State of Illinois, and the International Institute for Nanotechnology (IIN). Histology services were provided by the Northwestern University Research Histology and Phenotyping Laboratory which is supported by NCI P30-CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center”. Microscopy was performed at the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), graciously supported by the Chemistry for Life Processes Institute, the NU Office for Research, the Department of Molecular Biosciences and the Rice Foundation. Portions of Figure 1 and 2 were created with BioRender.com.
ABBREVIATIONS
- CNS
Central nervous system
- BBB
Blood-brain barrier
- TfR
transferrin
- β-Gal
β-Galactosidase
- SNA
Spherical nucleic acid
- ProSNA
Protein spherical nucleic acid
- Scr
scrambled
- AF647
AlexaFluor647
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- CD
Circular dichroism
- IV
Intravenous
- IVIS
In vivo imaging system
- IFC
Immunofluorescent histochemistry
- HPLC
High-performance liquid chromatography
- MALDI-MS
Matrix-assisted laser desorption ionization-time of flight mass spectroscopy
- PBS
Phosphate-buffer saline
- MWCO
Molecular weight cut off
- PFA
paraformaldehyde
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at DOI ________________:
Experimental procedures, oligonucleotide sequences, UV-Vis traces, SDS PAGE gel of purified ProSNAs, DLS, circular dichroism spectra of protein and ProSNA constructs, TEER measurements of in vitro BBB model, and fluorescence histology images.
Add financial interests here.
REFERENCES
- 1.Pardridge WM, Blood-Brain Barrier and Delivery of Protein and Gene Therapeutics to Brain. Frontiers in Aging Neuroscience 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi X; Manickam DS; Brynskikh A; Kabanov AV, Agile delivery of protein therapeutics to CNS. Journal of Controlled Release 2014, 190, 637–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salameh TS; Banks WA, Delivery of Therapeutic Peptides and Proteins to the CNS. Elsevier: 2014; pp 277–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terstappen GC; Meyer AH; Bell RD; Zhang W, Strategies for delivering therapeutics across the blood–brain barrier. Nature Reviews Drug Discovery 2021, 20 (5), 362–383. [DOI] [PubMed] [Google Scholar]
- 5.Fu A; Tang R; Hardie J; Farkas ME; Rotello VM, Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjugate Chemistry 2014, 25 (9), 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster J; Koulov A; Mahler H-C; Detampel P; Huwyler J; Singh S; Mathaes R, In Vivo Stability of Therapeutic Proteins. Pharmaceutical Research 2020, 37 (2). [DOI] [PubMed] [Google Scholar]
- 7.Langen UH; Ayloo S; Gu C, Development and Cell Biology of the Blood-Brain Barrier. Annual Review of Cell and Developmental Biology 2019, 35 (1), 591–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daneman R; Prat A, The Blood–Brain Barrier. Cold Spring Harbor Perspectives in Biology 2015, 7 (1), a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadry H; Noorani B; Cucullo L, A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids and Barriers of the CNS 2020, 17 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W; Liu QY; Haqqani AS; Leclerc S; Liu Z; Fauteux F; Baumann E; Delaney CE; Ly D; Star AT et al. , Differential expression of receptors mediating receptor-mediated transcytosis (RMT) in brain microvessels, brain parenchyma and peripheral tissues of the mouse and the human. Fluids and Barriers of the CNS 2020, 17 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulgar VM, Transcytosis to Cross the Blood Brain Barrier, New Advancements and Challenges. Frontiers in Neuroscience 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boado RJ; Ka-Wai Hui E; Zhiqiang Lu J; Pardridge WM, Insulin receptor antibody-iduronate 2-sulfatase fusion protein: Pharmacokinetics, anti-drug antibody, and safety pharmacology in Rhesus monkeys. Biotechnology and Bioengineering 2014, 111 (11), 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giugliani R; Giugliani L; De Oliveira Poswar F; Donis KC; Corte AD; Schmidt M; Boado RJ; Nestrasil I; Nguyen C; Chen S; et al. Neurocognitive and somatic stabilization in pediatric patients with severe Mucopolysaccharidosis Type I after 52 weeks of intravenous brain-penetrating insulin receptor antibody-iduronidase fusion protein (valanafusp alpha): an open label phase 1–2 trial. Orphanet Journal of Rare Diseases 2018, 13 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anraku Y; Kuwahara H; Fukusato Y; Mizoguchi A; Ishii T; Nitta K; Matsumoto Y; Toh K; Miyata K; Uchida S at al. Glycaemic control boosts glucosylated nanocarrier crossing the BBB into the brain. Nature Communications 2017, 8 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jefferies WA; Brandon MR; Hunt SV; Williams AF; Gatter KC; Mason DY, Transferrin receptor on endothelium of brain capillaries. Nature 1984, 312 (5990), 162–163. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y; Pardridge WM, Delivery of β-Galactosidase to Mouse Brain via the Blood-Brain Barrier Transferrin Receptor. Journal of Pharmacology and Experimental Therapeutics 2005, 313 (3), 1075–1081. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee D; Liu AP; Voss NR; Schmid SL; Finn MG, Multivalent Display and Receptor-Mediated Endocytosis of Transferrin on Virus-Like Particles. ChemBioChem 2010, 11 (9), 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabezón I; Manich G; Martín-Venegas R; Camins A; Pelegrí C; Vilaplana J, Trafficking of Gold Nanoparticles Coated with the 8D3 Anti-Transferrin Receptor Antibody at the Mouse Blood–Brain Barrier. Molecular Pharmaceutics 2015, 12 (11), 4137–4145. [DOI] [PubMed] [Google Scholar]
- 19.Johnsen KB; Burkhart A; Melander F; Kempen PJ; Vejlebo JB; Siupka P; Nielsen MS; Andresen TL; Moos T, Targeting transferrin receptors at the blood-brain barrier improves the uptake of immunoliposomes and subsequent cargo transport into the brain parenchyma. Scientific Reports 2017, 7 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X; Yang Y; Zhao H; Zhu T; Yang Z; Xu H; Fu Y; Lin F; Pan X; Li L et al. W., Enhanced in Vivo Blood–Brain Barrier Penetration by Circular Tau–Transferrin Receptor Bifunctional Aptamer for Tauopathy Therapy. Journal of the American Chemical Society 2020, 142 (8), 3862–3872. [DOI] [PubMed] [Google Scholar]
- 21.Ullman JC; Arguello A; Getz JA; Bhalla A; Mahon CS; Wang J; Giese T; Bedard C; Kim DJ; Blumenfeld JR et al. Brain delivery and activity of a lysosomal enzyme using a blood-brain barrier transport vehicle in mice. Science Translational Medicine 2020, 12 (545), eaay1163. [DOI] [PubMed] [Google Scholar]
- 22.Kariolis MS; Wells RC; Getz JA; Kwan W; Mahon CS; Tong R; Kim DJ; Srivastava A; Bedard C; Henne KR et al. Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Science Translational Medicine 2020, 12 (545), eaay1359. [DOI] [PubMed] [Google Scholar]
- 23.Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ, A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382 (6592), 607–609. [DOI] [PubMed] [Google Scholar]
- 24.Cutler JI; Auyeung E; Mirkin CA, Spherical Nucleic Acids. Journal of the American Chemical Society 2012, 134 (3), 1376–1391. [DOI] [PubMed] [Google Scholar]
- 25.Choi CHJ; Hao L; Narayan SP; Auyeung E; Mirkin CA, Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proceedings of the National Academy of Sciences 2013, 110 (19), 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seferos DS; Prigodich AE; Giljohann DA; Patel PC; Mirkin CA, Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Letters 2009, 9 (1), 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnaby SN; Lee A; Mirkin CA, Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proceedings of the National Academy of Sciences 2014, 111 (27), 9739–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meckes B; Banga RJ; Nguyen ST; Mirkin CA, Enhancing the Stability and Immunomodulatory Activity of Liposomal Spherical Nucleic Acids through Lipid-Tail DNA Modifications. Small 2017, 1702909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusmierz CD; Bujold KE; Callmann CE; Mirkin CA, Defining the Design Parameters for in Vivo Enzyme Delivery Through Protein Spherical Nucleic Acids. ACS Central Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodin JD; Sprangers AJ; McMillan JR; Mirkin CA, DNA-Mediated Cellular Delivery of Functional Enzymes. Journal of the American Chemical Society 2015, 137 (47), 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harguindey A; Culver HR; Sinha J; Bowman CN; Cha JN, Efficient cellular uptake of click nucleic acid modified proteins. Chemical Communications 2020, 56 (35), 4820–4823. [DOI] [PubMed] [Google Scholar]
- 32.Narayan SP; Choi CHJ; Hao L; Calabrese CM; Auyeung E; Zhang C; Goor OJGM; Mirkin CA, The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates. 2015, 11 (33), 4173–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnaby SN; Perelman GA; Kohlstedt KL; Chinen AB; Schatz GC; Mirkin CA, Design Considerations for RNA Spherical Nucleic Acids (SNAs). 2016, 27 (9), 2124–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chinen AB; Ferrer JR; Merkel TJ; Mirkin CA, Relationships between Poly(ethylene glycol) Modifications on RNA–Spherical Nucleic Acid Conjugates and Cellular Uptake and Circulation Time. 2016. [DOI] [PMC free article] [PubMed]
- 35.Chinen AB; Guan CM; Ko CH; Mirkin CA, The Impact of Protein Corona Formation on the Macrophage Cellular Uptake and Biodistribution of Spherical Nucleic Acids. 2017, 1603847. [DOI] [PMC free article] [PubMed]
- 36.Guan CM; Chinen AB; Ferrer JR; Ko CH; Mirkin CA, Impact of Sequence Specificity of Spherical Nucleic Acids on Macrophage Activation in Vitro and in Vivo. Molecular Pharmaceutics 2019, 16 (10), 4223–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrer JR; Sinegra AJ; Ivancic D; Yeap XY; Qiu L; Wang J-J; Zhang ZJ; Wertheim JA; Mirkin CA, Structure-Dependent Biodistribution of Liposomal Spherical Nucleic Acids. ACS Nano 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CHB; Dellamaggiore KR; Ouellette CP; Sedano CD; Lizadjohry M; Chernis GA; Gonzales M; Baltasar FE; Fan AL; Myerowitz R et al. Aptamer-based endocytosis of a lysosomal enzyme. Proceedings of the National Academy of Sciences 2008, 105 (41), 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macdonald J; Houghton P; Xiang D; Duan W; Shigdar S, Truncation and Mutation of a Transferrin Receptor Aptamer Enhances Binding Affinity. Nucleic Acid Therapeutics 2016, 26 (6), 348–354. [DOI] [PubMed] [Google Scholar]
- 40.Macdonald J; Denoyer D; Henri J; Jamieson A; Burvenich IJG; Pouliot N; Shigdar S, Bifunctional Aptamer–Doxorubicin Conjugate Crosses the Blood–Brain Barrier and Selectively Delivers Its Payload to EpCAM-Positive Tumor Cells. Nucleic Acid Therapeutics 2020, 30 (2), 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewett JC; Sletten EM; Bertozzi CR, Rapid Cu-Free Click Chemistry with Readily Synthesized Biarylazacyclooctynones. Journal of the American Chemical Society 2010, 132 (11), 3688–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan AI; Liu J; Dutta P, Iron transport kinetics through blood-brain barrier endothelial cells. Biochimica et Biophysica Acta (BBA) - General Subjects 2018, 1862 (5), 1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan B; Kolli AR; Esch MB; Abaci HE; Shuler ML; Hickman JJ, TEER Measurement Techniques for In Vitro Barrier Model Systems. Journal of Laboratory Automation 2015, 20 (2), 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen SA; Day ES; Ko CH; Hurley LA; Luciano JP; Kouri FM; Merkel TJ; Luthi AJ; Patel PC; Cutler JI et al. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Science Translational Medicine 2013, 5 (209), 209ra152–209ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumthekar P; Ko CH; Paunesku T; Dixit K; Sonabend AM; Bloch O; Tate M; Schwartz M; Zuckerman L; Lezon R et al. A first-in-human phase 0 clinical study of RNA interference–based spherical nucleic acids in patients with recurrent glioblastoma. Science Translational Medicine 2021, 13 (584), eabb3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth R; Kim H, Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab on a Chip 2012, 12 (10), 1784–1792. [DOI] [PubMed] [Google Scholar]
- 47.Palumbo P; Picchini U; Beck B; van Gelder J; Delbar N; DeGaetano A, A general approach to the apparent permeability index. J Pharmacokinet Pharmacodyn 2008, 35 (2), 235–48. [DOI] [PubMed] [Google Scholar]
- 48.Keefe AD; Pai S; Ellington A, Aptamers as therapeutics. Nature Reviews Drug Discovery 2010, 9 (7), 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan W; Wang H; Chen Y; Zhang X; Zhu H; Yang C; Yang R; Liu C, Molecular aptamers for drug delivery. Trends in Biotechnology 2011, 29 (12), 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J; Rossi JJ, Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Molecular Therapy - Nucleic Acids 2014, 3 (6), e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samanta D; Ebrahimi SB; Kusmierz CD; Cheng HF; Mirkin CA, Protein Spherical Nucleic Acids for Live-Cell Chemical Analysis. Journal of the American Chemical Society 2020, 142 (31), 13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa S; Deli MA; Kawaguchi H; Shimizudani T; Shimono T; Kittel A; Tanaka K; Niwa M, A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem Int 2009, 54 (3–4), 253–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


