Abstract
Background:
Exposure to diesel exhaust particles (DEP) has been linked to a variety of adverse health effects, including increased morbidity and mortality from cardiovascular diseases, chronic obstructive pulmonary disease (COPD), metabolic syndrome, and lung cancer. The epigenetic changes caused by air pollution have been associated with increased health risks. However, the exact molecular mechanisms underlying the lncRNA-mediated pathogenesis induced by DEP exposure have not been revealed.
Methods:
Through RNA-sequencing and integrative analysis of both mRNA and lncRNA profiles, this study investigated the role of lncRNAs in altered gene expression in healthy and diseased human primary epithelial cells (NHBE and DHBE-COPD) exposed to DEP at a dose of 30 μg/cm2.
Results:
We identified 503 and 563 differentially expressed (DE) mRNAs and a total of 10 and 14 DE lncRNAs in NHBE and DHBE-COPD cells exposed to DEP, respectively. In both NHBE and DHBE-COPD cells, enriched cancer-related pathways were identified at mRNA level, and 3 common lncRNAs OLMALINC, AC069234.2, and LINC00665 were found to be associated with cancer progression. In addition, we identified two cis-acting (TMEM51-AS1 and TTN-AS1) and several trans-acting lncRNAs (e.g., LINC01278, SNHG29, AC006064.4, TMEM51-AS1) only differentially expressed in COPD cells, which could potentially play a role in carcinogenesis and determine their susceptibility to DEP exposure.
Conclusions:
Overall, our work highlights the potential importance of lncRNAs in regulating DEP-induced gene expression changes associated with carcinogenesis, and individuals suffering from COPD are likely to be more vulnerable to these environmental triggers.
Keywords: diesel exhaust particle, COPD, lncRNAs, lncRNA-mRNA interaction, carcinogenesis, susceptibility
1. Introduction
As the usage of diesel-powered engines in private transportation has increased, diesel exhaust particles (DEP) have become one of the most prominent anthropogenic pollutants globally, particularly in densely populated metropolitan areas. DEP is one major class of traffic-related particles comprised of numerous toxic constituents, including polycyclic aromatic hydrocarbons (PAHs). Several molecular mechanisms linking excessive exposure to PAHs and the development of lung cancer have been identified. For example, it is known that the metabolism of PAHs in the biological system by cytochrome P450 (CYP) and other metabolic enzymes results in the formation of reactive metabolites such as diol-epoxides and o-quinones, which then contribute to the formation of DNA adducts, changes in gene expression, DNA mutations, and, eventually, carcinogenesis (Moorthy et al. 2015). DNA adducts caused by PAHs have been widely used as a marker of possible cancer risk and have been linked to cancer risk in both experimental and epidemiologic studies (Kriek et al. 1993). In addition, DEP exposure has been reported to induce pulmonary oxidative stress and inflammation, both of which have been implicated in the onset or exacerbation of respiratory diseases such as COPD, and genotoxicity is a significant outcome that is associated with oxidative stress and inflammation (Ahmed et al. 2018; Schwarze et al. 2013; Steiner et al. 2016). Furthermore, DEP exposure may contribute to the pathogenesis of COPD by releasing inflammatory mediators, possibly via NF-κB, MAPK and PI3K signaling pathways (Wang et al. 2020). While multiple metabolic pathways, cellular signaling, and genetic susceptibility all contribute to the development of lung cancer, the causal relationship and underlying molecular processes linking DEP and lung carcinogenesis remain to be elucidated.
A number of long noncoding RNAs (lncRNAs) have been implicated in the etiology of lung cancer (Jiang et al. 2019). LncRNAs are noncoding transcripts that exceed 200 nucleotides in length, and they have recently been identified as one of the biggest and most diverse RNA families. LncRNAs are categorized as intergenic (between genes), intragenic/intronic (within genes), or antisense depending on their proximity to protein-coding genes (Derrien et al. 2012). While the biological functions of most lncRNAs are unclear, several lncRNAs have been identified as regulators of cancer initiation and progression at the transcriptional and post-transcriptional levels, including cell proliferation, apoptosis, metastasis, and differentiation (Sun et al. 2018). The expression of both lncRNA and mRNA profiles has been shown to be significantly affected by traffic-related PM exposure, which is thought to be linked to a variety of disorders. In particular, lncRNAs have a vital regulatory function in the metabolic reprogramming associated with human cancer (Sellitto et al. 2021). The expression levels of lncRNAs are tightly controlled in the healthy state and can be disrupted by a variety of mechanisms during the development of disease.
Increasing evidence suggests that COPD and lung cancer may be distinct manifestations of the same disease (Durham and Adcock 2015). Furthermore, COPD has also been considered as a significant risk factor for lung cancer, with COPD patients having a twofold increased risk of developing lung cancer (Papi et al. 2004). This high prevalence of lung cancer in COPD patients suggests that there may be common mechanisms or common pathogenic factors for either disease (e.g., genetic susceptibility, activation of intracellular pathways, or epigenetics) (Barnes and Adcock 2011). As an epigenetic factor, lncRNAs are critical for regulating gene expression at the transcriptional and post-transcriptional levels (Dykes and Emanueli 2017), ultimately contributing to lung carcinogenesis and associated consequences (He et al. 2017; Reddy et al. 2015; Schones et al. 2015). However, the lncRNA-mediated pathogenic changes after DEP exposure, as well as their potential roles in susceptibility and disease progression, are not fully understood.
In this study, we investigated the role of lncRNAs in lung carcinogenesis induced by DEP using primary human bronchial epithelial cells from healthy (NHBE) and diseased (DHBE-COPD) donors. We hypothesize that through activation of oncogenes or loss-of-function of tumor suppressor genes, lncRNAs mediate the regulatory pathways in lung cancer initiation. Due to the fact that lung cancer does not exhibit obvious symptoms in its early stages, the majority of lung cancers are identified in their late stages, complicating therapy, and considerably lowering the overall lung cancer survival rate (Knight et al. 2017). Therefore, identification of lncRNAs as novel biomarkers involved in lung cancer development due to DEP exposure and elucidation of lncRNAs regulatory networks will contribute to the current scientific understanding of source-specific adverse outcome pathways, which is critical for protecting vulnerable populations from increased risks of traffic-related PM induced adverse health outcomes.
2. Materials and Methods
2.1. Diesel Exhaust Particles (DEP) and Particle Extraction.
The DEP standard reference material (SRM 1650b, with certified 26 PAHs and 7 nitro-PAHs) was purchased from the National Institute of Standards and Technology (NIST, Gaithersburg, MD, USA). The DEP extracts were dissolved in cell media and used for subsequent cell exposures.
2.2. NHBE and DHBE-COPD cell culture and differentiation.
Primary normal human bronchial epithelial (NHBE) cells and diseased human bronchial epithelial (DHBE) cells from donors with COPD were purchased from Lonza (NHBE, Catalog # CC-2540s; DHBE-COPD, Catalog # 00195275; Walkersville, MD). BEGM bronchial epithelial cell growth media (Lonza, Catalog # CC-3170) and Human Bronchial/Tracheal Epithelial Cells (HBTEC) Air-liquid Interface (ALI) differentiation media were purchased from LONZA (Catalog # 00193514, Walkersville, MD) and Lifeline Cell Technology (Frederick, MD), respectively. A type I human atelocollagen solution VitroCol® (Advanced BioMatrix, Catalog #5007, San Diego, CA) was used for coating the culture flasks and plates. Briefly, the BEGM growth medium was used to revive NHBE and DHBE-COPD cells from cryopreservation, seeded as passage one (P1) into T75 cell culture flasks, and incubated at 37°C, 5% CO2. Upon 70–80% confluency, cells were then sub-cultured at ALI to allow differentiation on 24 mm Transwell® inserts with 0.4 μm pore size (Corning; Fisher Scientific) at a density of 1 × 105 cells/insert housed in 6 well cell culture plates. The cells were cultured at ALI for 28 days to facilitate differentiation into ciliated, mucus-producing cells (Ghio et al. 2013). 2.0 mL of fresh medium was supplied in the basal chamber every 48 hours.
2.3. Particle Exposure.
After 4 weeks, the apical chambers of the grown cells were rinsed with 1 mL of Hanks’ Balanced Salt Solution (HBSS) buffer before being exposed to particles, and the buffer was immediately removed. DEP particles were suspended in 250 μL of BEGM medium to prepare the desired dosages (30 μg/cm2, 20 μg/cm2, and 10 μg/cm2) before being applied on the cells in the apical chamber of the Transwell® inserts and gently stirred for at least 2 minutes. All treatments were performed in triplicate. A schematic diagram for the timeline of cell culture, ALI differentiation, and the subsequent DEP exposure is shown in Figure 1.
Figure 1:
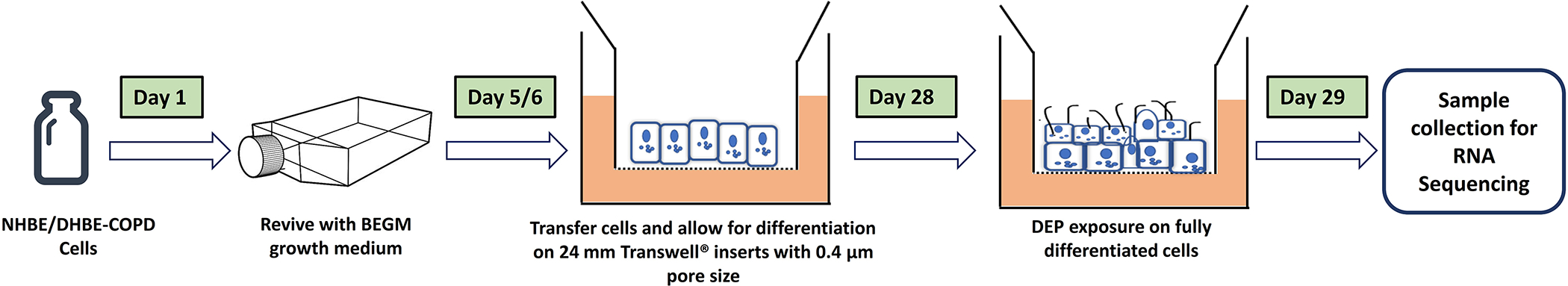
A schematic diagram for the timeline of cell culture, ALI differentiation, and the subsequent DEP exposure.
2.4. Cytotoxicity Assay.
As a prescreen for the downstream transcriptome analysis, the lactate dehydrogenase (LDH) test was performed according to the manufacturer’s protocol (Roche) to detect the cytotoxicity of cells exposed to DEP at dosages of 10 μg/cm2, 20 μg/cm2, and 30 μg/cm2. The supernatants were collected 24 hours after the exposure. To induce 100% cell death, Triton X-100 (0.1%) was used as a positive control. Absorbance was measured at 490 nm with a reference wavelength of 620 nm (TECAN SpectraFluor Plus).
2.5. RNA isolation, library construction, and RNA sequencing.
NHBE and DHBE (COPD) cells exposed to 30 μg/cm2 of DEP were selected for transcriptomic analysis. The TRI Reagent and the spin column-based Direct-zol RNA MiniPrep kit were used to extract and purify total RNA (Zymo Research). All RNA integrity numbers (RINs) exceeded 8.0. The RNA-Seq libraries for both mRNA and lncRNA were prepared with 150–300 ng of total RNA and the NEBNext ultra II Directional RNA Library Prep Kit. Then, the rRNA was removed using the NEBNext rRNA Depletion Kit. RNA-Seq analysis (Illumina NextSeq 500 high throughput 75bp pair end) was carried out at the University of California, Riverside’s Institute for Integrative Genome Biology (IIGB) (UCR).
2.6. RNA-Seq Data Analysis for both mRNA and lncRNA.
FastQC (version 0.11.7) (Andrews 2010) was used to assess read quality after RNA sequencing. Trimming was accomplished using the Trimmomatic (version 0.35) program (Bolger et al. 2014). MINLEN:50 was used to keep reads that were at least 50 bases long. After that, we used kallisto (version 0.46.1) to quantify the abundances of transcripts from our RNA-Seq data (Bray et al. 2016) and annotated transcripts with GENCODE version 37 (Frankish et al. 2021). Kallisto is based on pseudoalignment, which allows rapid determination of read compatibility with targets without alignment (Bray et al. 2016). DESeq2 (version 1.18.1) (Love et al. 2014) was used for normalization and differential mRNA and lncRNA expression analyses in R (version 3.6.3). For mRNA and lncRNAs between exposed and unexposed samples, significance was defined as the adjusted p value (i.e., false discovery rate (FDR)) ≤ 0.05 and the absolute log 2-fold change (log2FC) ≥ |±1|.
2.7. Prediction of lncRNA Target Genes.
To investigate how DE lncRNAs interact with nearby or distant target genes to regulate gene expression, we divided target genes of DE lncRNAs into two groups: cis and trans targets. Based on prior research, cis target genes were initially defined as genes located within 10 kb upstream or downstream of the differentially expressed lncRNA (Ahmed et al. 2021). However, using bedtools (version 2.29.2) (Quinlan 2014), we were unable to locate any nearby target genes within 10 kb upstream and downstream. Thus, we performed further analysis to identify cis-targeted genes at longer distances (up to 50 kb upstream and downstream) to locate potential long-range cis-regulatory elements (Laverré et al. 2022). The trans-regulated genes of DE lncRNAs were then predicted using rtools (http://rtools.cbrc.jp/cgi-bin/RNARNA/index.pl) (Iwakiri et al. 2017). To construct the interaction network for NHBE and DHBE-COPD cells based on the minimum energy of lncRNA and mRNA interaction, the top 200 trans-targeted genes were chosen from a total of 1,000 genes in NHBE and 1,400 genes in DHBE-COPD cells, respectively, using the network (version 1.16.0) and ggnet2 (version 0.1.0) packages in R (version 3.6.3).
2.8. Pathway Enrichment Analysis for Susceptibility.
DE mRNAs from both NHBE and DHBE-COPD cells were used for pathway enrichment analysis using the ConsensusPathDB database (Kamburov et al. 2009) to compare and contrast the gene expression changes in healthy and diseased cellular models. Overrepresentation analysis was performed. The significance (p-value) of the observed overlap between the DE mRNAs and members of established biological pathways was calculated using the hypergeometric distribution (Zhavoronkov et al. 2014). The results were obtained using the following criteria: (1) the input list and pathways must share at least 5 genes, and (2) a p-value cut-off of 0.01 is required.
2.9. Code availability.
Data analysis and codes are available at https://github.com/biplabua/DEP_lncRNA_mRNA_Co-expression_2021.
3. Results
3.1. Cytotoxicity Assay
Following a 24-h exposure to DEP, the cytotoxicity of NHBE and DHBE-COPD was assessed using the LDH assay. As shown in Figure 2, there was no significant cytotoxicity (≤30%) observed in cells exposed to DEP at levels of 30 μg/cm2, 20 μg/cm2, and 10 μg/cm2, indicating that the cells were stressed during exposure, but the exposure dosage was not too toxic to preclude further transcriptomic evaluation (ISO 2009). Thus, we used a dose of 30 μg/cm2 for the subsequent transcriptomic analysis in this study. Notably, doses of 30 μg/cm2 and 20 μg/cm2 induced relatively higher cytotoxicity in DHBE-COPD cells than in the healthy NHBE cells, which might be connected to the pre-existing COPD condition.
Figure 2: Cytotoxicity in NHBE and DHBE-COPD cells following DEP exposure.

Cells were exposed to DEP at concentrations of 10 μg/cm2, 20 μg/cm2, and 30 μg/cm2 for 24 h. LDH release was used to calculate the percentage of cytotoxicity compared to negative controls of unexposed cells maintained in cell media and positive controls treated with Triton X-100 (0.1% v/v). To determine statistical significance in comparison to the negative controls, two-way ANOVA was used; ns: not significant (p > 0.05), *** (p < 0.001), and **** (p < 0.0001).
3.2. DE mRNAs
DESeq2 identified 503 and 563 DE mRNAs in NHBE and DHBE-COPD cells, respectively, using the of log2FC > |±1| and FDR < 0.05 criteria (Figure 3). A total of 142 mRNAs were common between NHBE and DHBE-COPD cells exposed to DEP (Figure 3c). As the exposed cells did not exhibit substantial cytotoxicity (Figure 2), these results reflected the real transcriptional changes in NHBE and DHBE-COPD cells under the given exposure condition (i.e., a dosage of 30 μg/cm2 following 24 hr exposure).
Figure 3: Differential expression of mRNAs (DE mRNAs) in NHBE and DHBE-COPD cells.

DE mRNA volcano plots in (a) NHBE and (b) DHBE-COPD cells after DEP exposure. The X-axis represents the log 2-fold change, while the Y-axis represents the adjusted p values: −log 10 (padj). The blue dots represent significantly upregulated mRNAs, while red dots represent significantly downregulated mRNAs. Non-differentially expressed mRNAs are represented by black dots. (c) A Venn diagram depicts the number of distinct and common (overlapping) DE mRNAs in NHBE and DHBE-COPD cells.
3.3. DE lncRNAs
DESeq2 identified 10 and 14 DE lncRNAs in NHBE and DHBE-COPD cells, respectively, using the log2FC > |±1| and the FDR < 0.05 criteria (Figure 4). In addition, we found a total of 4 lncRNAs were common between NHBE and DHBE (COPD) cells exposed to DEP (Figure 4c). Some lncRNAs may co-express with mRNAs and regulate gene expression. Integrative investigation of DE lncRNAs and DE mRNAs may uncover epigenetic regulation of gene expression via lncRNAs.
Figure 4: Differential expression of lncRNAs (DE lncRNAs) in NHBE and DHBE-COPD cells.

DE lncRNA volcano plots of in (a) NHBE and (b) DHBE-COPD cells after DEP exposure. The X-axis represents log 2-fold change, while the Y-axis represents the adjusted p values: −log 10 (padj). Blue dots represent significantly upregulated lncRNAs, while red dots represent significantly downregulated lncRNAs. Non-differentially expressed lncRNAs are represented by black dots. (c) A Venn diagram shows the number of distinct and common (overlapping) DE lncRNAs in NHBE and DHBE-COPD cells.
3.4. Prediction of cis-targeted Genes of the DE lncRNAs
Within the 50 kb window, we did not find any differentially expressed cis-targeted mRNA in NHBE cells under the given exposure condition. However, two cis-targeted mRNAs were identified in DHBE-COPD cells, including KAZN (kazrin, periplakin interacting protein), and TTN (titin). KAZN is targeted by lncRNA TMEM51-AS1, and TTN is targeted by lncRNA TTN-AS1, respectively. Notably, TMEM51-AS1 is also identified as one of the trans-acting lncRNAs only found in DHBE-COPD cells (discussed in detail in Section 4.2.3). Additionally, TTN-AS1 has been reported as a potential diagnostic and prognostic biomarker for multiple cancers, including lung cancer (Qi and Li 2020; Zheng et al. 2021b).
3.5. Prediction of trans-targeted Genes of the DE lncRNAs
A total of 5 out of 10 DE lncRNAs in NHBE cells and a total 7 out of 14 DE lncRNAs in DHBE-COPD cells were identified to construct regulatory networks with trans-target genes from rtools. Among the 1,000 predicted trans-acting genes in NHBE cells, 8 target genes were found to be differentially expressed (log2FC > |±1| and FDR < 0.05) in our analysis and to have the same expression pattern (for up and down regulation), whereas 13 DE target genes had the opposite expression trend as the trans-acting lncRNAs (Figure 5). In contrast, among the 1,400 trans-acting genes in DHBE-COPD cells, 28 DE target genes (log2FC > |±1| and FDR < 0.05) showed the same expression trend (for up and down regulation) as the trans-acting lncRNAs, while 16 genes showed the opposite expression trend (Figure 6). Most coding genes are regulated by different lncRNAs, but some, like BMP1 in NHBE cells and PEX5, TMEM63B, and IGF2R in DHBE-COPD cells, are controlled by more than one lncRNA (Figures 5–6).
Figure 5: Predicted trans-targeted genes (log2FC>|±1|) and regulatory network of DE lncRNAs in NHBE cells.

The DE lncRNAs regulatory network was built for the NHBE cells using the R package (version 3.6.3). The colors represent the different types of RNAs; orange: mRNA, and green: lncRNA. The triangles represent upregulation, while the dots represent downregulation.
Figure 6: Predicted trans-targeted genes (log2FC>|±1|) and regulatory network of DE lncRNAs in DHBE-COPD cells.

The DE lncRNA regulatory network was constructed for DHBE-COPD cells using the R package (version 3.6.3). The colors represent the different types of RNAs; blue: mRNA, and red: lncRNA. The triangles represent up-regulation, while the dots represent downregulation.
4. Discussion
4.1. DEP exposure and biological pathway perturbation at mRNA levels
The airway epithelium is the initial defensive barrier that protects the airway from external stimuli. When epithelial functions are compromised, cellular signaling pathways involved in homeostasis maintenance, such as inflammation, repair, and differentiation, may be disturbed. The reduced airway epithelial function is a common pathological alteration in COPD, suggesting that populations with such pre-existing health issues may be more vulnerable to pollutant exposure.
In this study, the most significantly altered pathways from unique DEGs only found in NHBE cells are associated with control mechanisms of cell cycles to maintain the genomic stability, including RND GTPase cycles (FDR value 1.96E-02) and G2/M transition (FDR value 2.36E-02). As the DEP consists of PAHs and their derivatives, DEP-induced ROS generation, oxidative stress and DNA damage may occur via the xenobiotic metabolism (Nemmar et al. 2013). Following pathway analysis, we found that pathways associated with chromatin modifying enzymes (FDR value 3.27E-04) and direct p53 effectors (FDR value 8.79E-03) were enriched in both NHBE and DHBE-COPD cells. Furthermore, we found several pathways such as TNF alpha signaling pathway (FDR value 4.12E-03), p53 signaling pathway (FDR value 9.50E-03), and PIP3 activates AKT signaling pathway (FDR value 5.91E-03) enriched only in DHBE-COPD cells. All these pathways are linked to carcinogenesis (Altomare and Testa 2005; Muller and Vousden 2013; Wajant 2009).
4.2. Trans-acting gene regulation by lncRNAs in DEP exposed NHBE and DHBE-COPD cells
At the transcriptional and post-transcriptional levels, lncRNAs play a major role in pathogenesis by regulating gene expression (Ahadi 2021; Jandura and Krause 2017). The expression of these non-coding lncRNA transcripts has been found to be associated with the expression of target genes via cis and trans mechanisms (Ahmed et al. 2021; Li et al. 2019). Due to the fact that most lncRNAs in the human genome are still uncharacterized, functional annotations of lncRNAs remain challenging and are currently under extensive research (Li et al. 2019; Ramakrishnaiah et al. 2020; Zhang et al. 2021). In this study, as we identified very few differentially expressed cis-targeted genes within the 50 kb window in our search (discussed in detail in Section 3.4), we focused on the discussion of potential regulatory function of DE lncRNAs by the trans mechanism.
4.2.1. Common lncRNAs differentially expressed in both NHBE and DHBE-COPD cells
In this study, 3 common lncRNAs were found differentially expressed in both NHBE and DHBE-COPD cells, including LINC00665, OLMALINC (Oligodendrocyte Maturation-Associated Long Intergenic Non-Coding RNA) and AC069234.2 (Figures 5–6). Among identified DE lncRNAs, prior research has revealed that LINC00665 is involved in tumor progression, DNA damage repair, and is upregulated in a variety of cancers (Dai et al. 2021; Ding et al. 2020), such as the lung adenocarcinoma (LUAD) (Cong et al. 2019). Interesting correlations were found for these 3 common lncRNAs in both NHBE and DHBE-COPD cells: (1) LINC00665 was upregulated in both NHBE (log2FC = 21.98, FDR value =1.23E-06) and DHBE-COPD (log2FC = 20.00, FDR value =2.51E-05) cells; (2) OLMALINC was upregulated in NHBE cells (log2FC = 24.2, FDR value =6.36E-07), whereas downregulated in DHBE-COPD cells (log2FC = −22.12, FDR value =1.02E-05); (3) AC069234.2 were downregulated in NHBE cells (log2FC = −30.92, FDR value = 2.74E-12), whereas upregulated in DHBE-COPD cells (log2FC =19.94, FDR value = 5.67E-05). The different expression patterns in NHBE and DHBE-COPD cells underscore the function of lncRNAs in regulating gene expression.
Through integrative analysis of our lncRNA and mRNA profiling data, we found that lncRNAs OLMALINC and AC069234.2 are associated with ZMIZ2 (Zinc Finger MIZ- Type Containing 2) and ATXN2L (Ataxin 2 like) genes, respectively (Figures 5–6). It has been reported that the depletion of ZMIZ2 gene plays a significant role in attenuation of the colorectal tumorigenesis, which is also considered as a potential therapeutic target (Zhu et al. 2020). Our study shows that the ZMIZ2 gene was downregulated in both NHBE and DHBE-COPD cells, whereas expression pattern was opposite for the lncRNA OLMALINC in NHBE (upregulated) and DHBE-COPD cells (downregulated), respectively (Figures 5–6). In addition, the ATXN2L genes has been suggested to promote cell invasiveness and oxaliplatin resistance via EGF via PI3K/Akt signaling (Lin et al. 2019a). In our study, the ATXN2L gene was shown to be elevated in both NHBE and DHBE-COPD cells, but the lncRNA AC069234.2 was found to be downregulated in NHBE cells but upregulated in DHBE-COPD cells (Figures 5–6). These findings suggest that altered epithelial functions in DHBE-COPD cells may result in lung disease aggravation in response to DEP exposure, while identified lncRNAs may act as early indicators for disease development.
4.2.2. DE lncRNAs only found in NHBE cells
In NHBE cells, lncRNAs NUTM2B-AS1 (NUTM2B antisense RNA 1) (log2FC = 6.15, FDR value = 3.38E-02) and OTUD6B-AS1 (OTUD6B antisense RNA 1) were both found to be upregulated (log2FC = 5.66, FDR value = 1.53E-03). In our analyses, lncRNA NUTM2B-AS1 was found to be associated with four genes, including ASH1L (ASH1 Like Histone Lysine Methyltransferase), IFT172 (Intraflagellar Transport 172), BMP1 (Bone Morphogenetic Protein 1), PBXIP1 (PBX Homeobox Interacting Protein 1) (Figure 5). Downregulation of IFT172 has been linked to dysfunction of intraflagellar transport machinery and maintenance of cilia length seen in COPD (Hessel et al. 2014), suggesting that DEP exposure may be a risk factor for COPD development. In addition, lncRNA AC069234.2 has the opposite interaction with MYO5A (Myosin VA), MAP4K4, and MLEC (Malectin) genes (Figure 5). Previous studies reported that downregulation of MAP4K4 results in induction of apoptosis (Liu et al. 2011). MYO5A has been reported to be remarkably upregulated in esophageal squamous cell carcinoma tissues and cells (Liang et al. 2020), while upregulation of MLEC (Malectin) has been linked to colorectal cancer (Mao et al. 2020). On the other hand, the ACVR1 (Activin A Receptor Type 1) gene was downregulated together with lncRNA AC069234.2 in our study (Figure 5). Increased expression of ACVR1 has been linked to decreased survival in a large cohort of 227 hepatocellular carcinoma cases (Li et al. 2015). Furthermore, CANX (Calnexin) is detected in high abundance in breast cancer patients at both primary and metastatic stages (Moradpoor et al. 2020), and high CANX expression has been linked to a worse survival rate for breast cancer patients (Geng et al. 2019). Our study found that lncRNA OTUD6B-AS1 was upregulated whereas CANX gene was downregulated (Figure 5). Thus, overexpression of lncRNA OTUD6B-AS1 in NHBE cells may provide protective functions for maintaining cellular homeostasis.
4.2.3. DE lncRNAs only found in DHBE-COPD cells
Previous studies suggested that pre-exiting COPD conditions may play as an independent risk factor for cancer progression (Durham and Adcock 2015; Papi et al. 2004). Mounting evidence suggest that altered expression of lncRNAs can lead to carcinogenesis by acting like the oncogene and interacting with the enhancer of a gene (Ahadi 2021; Martínez-Terroba and Dimitrova 2020; Zhang et al. 2013). Therefore, the associations between lncRNAs and mRNAs and their expression patterns are important to decipher their molecular mechanisms. For example, lncRNA FGD5-AS1 that was found to be upregulated in DHBE-COPD cells in this study (log2FC = 19.43, FDR value = 2.08E-04) has been reported as an oncogene and promoted tumorigenesis (Fan et al. 2020). Elevated expression of FGD5-AS1 upregulates the FGFRL1 (fibroblast growth factor receptor like 1) gene by sponging miRNA has-miR-107 and induces non-small cell lung carcinoma cell proliferation (Fan et al. 2020). Another upregulated lncRNA LINC01278 (log2FC = 17.24, FDR value = 1.01E-03) was reported to accelerate cancer progression via miRNAs (e.g., miR-134-5p/Lysine demethylase 2A (KDM2A) axis) (Liu et al. 2021) and upregulate the KDM2A expression (associated with the cyclin D1 expression) to induce cancer progression and proliferation. By forming axis with miR-559/TCF12, miR-376c-3p/DNM3, miR-133a-3p/PTHR1, LINC01278 is known to induce hepatocellular carcinoma (Song et al. 2020), papillary thyroid carcinoma (Lin et al. 2019b), and osteosarcoma (Qu and Li 2020), respectively. We found that lncRNA LINC01278 regulates 6 target genes with same direction (all upregulated) (Figure 6). Upregulation of DYNC1H1 (Dynein Cytoplasmic 1 Heavy Chain 1) and ANP32E (Acidic Nuclear Phosphoprotein 32 Family Member E) genes is known to promote cell proliferation and migration (Gong et al. 2019; Huang et al. 2020).
On the other hand, lncRNA SNHG29 (also named as LRRC75A-AS1) that activates the p53/p21 signaling and results in cell senescence (Jiang et al. 2021) was found downregulated in DHBE-COPD cells (log2FC = −6.42, FDR value = 1.82E-05), while its trans-targeted gene COL5A2 (Collagen Type V Alpha 2 Chain) was found to be upregulated (Figure 6). Recent studies reported that the gene expression of COL5A2 was significantly upregulated in colorectal cancer (Wang et al. 2021b), gastric cancer (Tan et al. 2021), bladder cancer (Zeng et al. 2018), and prostate cancer (Ren et al. 2021). The expression pattern indicates the potential risk of DHBE-COPD cells in cancer development (Figure 6).
Another lncRNA TMEM51-AS1 was found to be upregulated in our study (log2FC = 20.30, FDR value = 1.64E-07), which induced the cell proliferation by sponging miRNA response elements in laryngeal squamous cell carcinoma (Lyu et al. 2020). Furthermore, lncRNA TMEM51-AS1 is considered as a competitive endogenous RNA (ceRNA) of ARID4A (AT-Rich Interaction Domain 4A) gene, which targets hsa-miR-1254. In addition, TRAPPC10 (Trafficking Protein Particle Complex Subunit 10) gene targets hsa-miR-106B (Hui et al. 2019) to induce cell proliferation in laryngeal squamous cell carcinoma (Lyu et al. 2020). Downregulation of TMEM51-AS1 (strong relevance with p53-R273H) was found preventing the tumorigenic capacity of colorectal cancer stem cells in vitro and in vivo (Zhao et al. 2019). This result indicates the upregulation of lncRNA TMEM51-AS1 in DHBE-COPD may be involved in carcinogenesis.
Furthermore, we identified downregulated lncRNA AC006064.4 (log2FC = −10.85, FDR value = 2.5E-02) coexpressed with upregulated RAC2 and MECOM (MDS1 And EVI1 Complex Locus) genes (Figure 6). Elevated expression of the RAC2 gene has been associated with the cell migration and cell proliferation of osteosarcoma by regulating the Wnt signaling pathway (Xia et al. 2019). It has been reported that the mRNA level of MECOM gene in glioblastoma multiforme (GBM) tissues was significantly higher than that in the adjacent tissues, which suggests that MECOM may play a driving role in GBM oncogenesis (Hou et al. 2016). In addition, lncRNA AC006064.4 has recently been linked to SARS-Cov-2 infection (Vishnubalaji et al. 2020), indicating that it might be an important modulator of several biological pathways in severe lung damage.
4.2.4. Trans-acting mRNAs only found in DHBE-COPD cells
A few unique mRNAs that are associated with multiple lncRNAs were identified in DHBE-COPD cells, including TMEM63B (Transmembrane Protein 63B), PEX5 (Peroxisomal Biogenesis Factor 5), and IGF2R (Insulin Like Growth Factor 2 Receptor) (Figure 6). The TMEM63B gene was linked to lncRNAs LINC01278 (log2FC =17.24, FDR value =1.01E-03), and AC069234.2 (log2FC =19.94, FDR value =5.67E-05), while PEX5 gene was connected with lncRNAs LINC00665 (log2FC =20.00, FDR value =2.51E-05) and LINC01278 (Figure 6). Genetic variants of PEX5 in the peroxisome pathway have been linked to cutaneous melanoma, and these variants may serve as novel biomarkers for predicting survival in patients with cutaneous melanoma (Wang et al. 2021a). Furthermore, both OLMALINC (log2FC = −22.12, FDR value =1.02E-05) and AC069234.2 were found to regulate the IGF2R gene. Prior research found that the M6P/IGF2R posttranscriptional dysregulation is a contributing mechanism in breast cancer development and breast cancer response to therapy (Iwamoto and Barber 2007), and IGF2R is also a poor prognostic factor for early-stage cervical cancer (Takeda et al. 2019). In the current study, the upregulated IGF2R is linked to downregulated lncRNA OLMALINC and upregulated lncRNA AC069234.2. For lncRNA AC069234.2 that is connected to multiple genes (Figure 6), a study by Zheng et al. (2021a) reported that upregulation of PIP5K1A promotes the progression of glioma, while downregulation of IFNGR2 (Interferon Gamma Receptor 2) gene may impair the interferon-γ (IFN-γ) signaling that is related to tumor progression (Castro et al. 2018). The same expression patterns observed in DHBE-COPD cells (Figure 6) indicates that these lncRNA-mRNA pairs may function in concert in cancer progression.
4.4. Potential Limitations and Future Research Needs
Some potential limitations of this study should be noted. First, while the Encyclopedia of DNA Elements (ENCODE) project classified over 9,640 human genome loci as lncRNAs, only ~100 have been thoroughly characterized to determine their cellular functions (Bánfai et al. 2012), and since lncRNA expression is tissue specific, we were only able to annotate a limited number of known lncRNAs in human airway epithelial cells in the current study. In addition, the expression of mRNAs and lncRNAs is highly dynamic and is influenced by many internal and external factors. The results shown in the current study only represent a snapshot of the complex system. Lastly, the relationships that have been identified do not prove causality. Further validation studies are necessary to confirm the effects of lncRNA alterations on DEP-induced health outcomes. Experiments using RNA interference (RNAi), antisense oligonucleotides (ASOs), or CRISPR/Cas9 genome editing methods, in particular, will be useful to validate the regulatory function of identified lncRNAs in controlling gene expression.
5. Conclusions
Overall, this study highlights the vulnerability of people with pre-existing lung diseases to DEP exposure. Dysfunctional DHBE-COPD cells are shown to be more sensitive to the same level of DEP, with the altered expression of mRNAs (e.g., IGF2R, IFNGR2, PEX5, TMEM63B) and lncRNAs (e.g., LINC01278, SNHG29, AC006064.4, TMEM51-AS1) clearly linked to carcinogenesis (Figure 7). These findings help understand lncRNA-mRNA interactions and the relevance of air pollution in lung cancer. As a result, the lncRNAs identified in this work could provide an experimental basis for the potential use of lncRNAs as biomarkers for diagnosis and prognosis to assist with the early detection of lung cancer and protect vulnerable populations from increased risks of traffic-related PM induced adverse health outcomes.
Figure 7:

A schematic diagram of DEP exposure-mediated health outcomes in the NHBE and DHBE-COPD cells.
Supplementary Material
Acknowledgements
Ying-Hsuan Lin gratefully acknowledges the support from the University of California, Riverside (UCR) Regents’ Faculty Fellowships for this research. Alexa Canchola was supported by an NRSA T32 training grant (T32 ES18827).
Abbreviations
- COPD
chronic obstructive pulmonary disease
- DE
differentially expressed
- DEP
diesel exhaust particles
- DHBE
diseased human bronchial epithelial cells
- FDR
false discovery rate
- lncRNA
long non-coding RNA
- log2FC
log2 fold change
- NHBE
normal human bronchial epithelial cells
- PAHs
polycyclic aromatic hydrocarbons
Footnotes
Disclosure statement
The authors report no conflict of interest.
Supplementary material
Supplementary material is available for: (1) certified mass fraction values for PAHs in SRM 1650b, (2) list of differentially expressed mRNAs in NHBE cells, (3) list of differentially expressed mRNAs in DHBE-COPD cells, (4) differentially expressed known lncRNAs in NHBE cells, (5) differentially expressed known lncRNAs in DHBE-COPD cells, (6) list of the enriched pathways from unique DEGs only in NHBE cells exposed to diesel exhaust particles, (7) list of the enriched pathways from common DEGs in both NHBE and DHBE-COPD cells exposed to diesel exhaust particles, (8) list of the enriched pathways from unique DEGs only in DHBE-COPD cells exposed to diesel exhaust particles, (9) cis-targeted mRNAs of DE-lncRNAs in DHBE-COPD cells, (10) trans-targeted mRNAs of DE-lncRNAs in NHBE cells, and (11) trans-targeted mRNAs of DE-lncRNAs in DHBE-COPD cells.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
- Ahadi A 2021. Functional roles of lncRNAs in the pathogenesis and progression of cancer. Genes & Diseases 8, 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed C, Jiang H, Chen J, Lin Y-H, Ahmed CMS, Jiang H, Chen JY and Lin Y-H 2018. Traffic-Related Particulate Matter and Cardiometabolic Syndrome: A Review. Atmosphere 9, 336–336. [Google Scholar]
- Ahmed CS, Paul BC, Cui Y, Frie AL, Burr A, Kamath R, Chen JY, Nordgren TM, Bahreini R and Lin Y-H 2021. Integrative Analysis of lncRNA–mRNA Coexpression in Human Lung Epithelial Cells Exposed to Dimethyl Selenide-Derived Secondary Organic Aerosols. Chem. Res. Toxicol. 34, 892–900. [DOI] [PubMed] [Google Scholar]
- Altomare DA and Testa JR 2005. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24, 7455–7464. [DOI] [PubMed] [Google Scholar]
- Andrews S 2010. FastQC: A quality control tool for high throughput sequence data. Available online at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. The Babraham Institute. [Google Scholar]
- Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Gunawardena HP, Yu Y and Xie L 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 22, 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ and Adcock IM 2011. Chronic obstructive pulmonary disease and lung cancer: a lethal association. Am. J. Respir. Crit. Care Med. 184, 866–867. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M and Usadel BJB 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P and Pachter L 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotech. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Castro F, Cardoso AP, Gonçalves RM, Serre K and Oliveira MJ 2018. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Frontiers in Immunology 9, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao C, Ji S, Shen Y, De W and Qiang Y 2019. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Sheng X, Sha R, Peng J, Yang F, Zhou L, Lin Y, Xu Y, Zhang S and Yin W 2021. Linc00665 can predict the response to cisplatin-paclitaxel neoadjuvant chemotherapy for breast cancer patients. Front. Oncol. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A and Knowles DG 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Zhao J, Huan L, Liu Y, Qiao Y, Wang Z, Chen Z, Huang S, Zhao Y and He X 2020. Inflammation‐Induced Long Intergenic Noncoding RNA (LINC00665) Increases Malignancy Through Activating the Double‐Stranded RNA–Activated Protein Kinase/Nuclear Factor Kappa B Pathway in Hepatocellular Carcinoma. Hepatology 72, 1666–1681. [DOI] [PubMed] [Google Scholar]
- Durham A and Adcock I 2015. The relationship between COPD and lung cancer. Lung Cancer 90, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM and Emanueli C 2017. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics 15, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Li H, Yu Z, Dong W, Cui X, Ma J and Li S 2020. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci. Rep. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland Jane E., Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, Berry A, Bignell A, Boix C, Carbonell Sala S, Cunningham F, Di Domenico T, Donaldson S, Fiddes Ian T., García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Howe KL, Hunt T, Izuogu OG, Johnson R, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Riera FC, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Wolf MY, Xu J, Yang Yucheng T, Yates A, Zerbino D, Zhang Y, Choudhary Jyoti S, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Tress ML and Flicek P. 2021. GENCODE 2021. Nucleic Acids Research 49, D916–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng N, Zhang W, Li Y and Li F 2019. Aspartyl aminopeptidase suppresses proliferation, invasion, and stemness of breast cancer cells via targeting CD44. Anat 302, 2178–2185. [DOI] [PubMed] [Google Scholar]
- Ghio AJ, Dailey LA, Soukup JM, Stonehuerner J, Richards JH and Devlin RB 2013. Growth of human bronchial epithelial cells at an air-liquid interface alters the response to particle exposure. Particle and Fibre Toxicology 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L-B, Wen T, Li Z, Xin X, Che X-F, Wang J, Liu Y-P and Qu X-J 2019. DYNC1I1 Promotes the Proliferation and Migration of Gastric Cancer by Up-Regulating IL-6 Expression. Frontiers in Oncology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ou C, Xiao Y, Han Q, Li H and Zhou S 2017. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget 8, 71325–71341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessel J, Heldrich J, Fuller J, Staudt MR, Radisch S, Hollmann C, Harvey B-G, Kaner RJ, Salit J and Yee-Levin J 2014. Intraflagellar transport gene expression associated with short cilia in smoking and COPD. PloS one 9, e85453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou A, Zhao L, Zhao F, Wang W, Niu J, Li B, Zhou Z and Zhu D 2016. Expression of MECOM is associated with unfavorable prognosis in glioblastoma multiforme. OncoTargets Ther. 9, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gao W, Liu H, Yin G, Duan H, Huang Z and Zhang Y 2020. Up-regulated ANP32E promotes the thyroid carcinoma cell proliferation and migration via activating AKT/mTOR/HK2-mediated glycolysis. Gene 750, 144681. [DOI] [PubMed] [Google Scholar]
- Hui L, Wang J, Zhang J and Long J 2019. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ 7, e7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO. 2009. 10993–5: 2009 Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- Iwakiri J, Terai G and Hamada M 2017. Computational prediction of lncRNA-mRNA interactions by integrating tissue specificity in human transcriptome. Biol. 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto KS and Barber CL 2007. Radiation‐induced posttranscriptional control of M6P/IGF2r expression in breast cancer cell lines. Molecular Carcinogenesis: Published in cooperation with the University of Texas MD Anderson Cancer Center 46, 497–502. [DOI] [PubMed] [Google Scholar]
- Jandura A and Krause HM 2017. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 33, 665–676. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hu H, Chen Q, Zhang Y, Chen W, Huang Q, Chen X, Li J and Zhong M 2021. Long non-coding RNA SNHG29 regulates cell senescence via p53/p21 signaling in spontaneous preterm birth. Placenta 103, 64–71. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li Z and Wang R 2019. Long non‑coding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications (Review). Int J Oncol 55, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Wierling C, Lehrach H and Herwig R 2009. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Research 37, D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SB, Crosbie PA, Balata H, Chudziak J, Hussell T and Dive C 2017. Progress and prospects of early detection in lung cancer. Open Biology 7, 170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek E, Van Schooten FJ, Hillebrand MJ, Van Leeuwen FE, Den Engelse L, De Looff AJ and Dijkmans AP 1993. DNA adducts as a measure of lung cancer risk in humans exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect 99, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverré A, Tannier E and Necsulea A 2022. Long-range promoter–enhancer contacts are conserved during evolution and contribute to gene expression robustness. Genome Research 32, 280–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Li F, Jiang K, Zhang M, Han R, Jiang R, Li Z, Tian Y, Yan F and Kang X 2019. Integrative analysis of long noncoding RNA and mRNA reveals candidate lncRNAs responsible for meat quality at different physiological stages in Gushi chicken. PLoS One 14, e0215006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, Song F, Zheng H, Yu J and Song T 2015. Regulatory MiR‐148a‐ACVR1/BMP circuit defines a cancer stem cell‐like aggressive subtype of hepatocellular carcinoma. J. Hepatol. 61, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Wu Z, Shen S, Niu Y, Guo Y, Liang J and Guo W 2020. LINC01980 facilitates esophageal squamous cell carcinoma progression via regulation of miR-190a-5p/MYO5A pathway. Arch. Biochem. Biophys. 686, 108371. [DOI] [PubMed] [Google Scholar]
- Lin L, Li X, Pan C, Lin W, Shao R, Liu Y, Zhang J, Luo Y, Qian K and Shi M 2019a. ATXN2L upregulated by epidermal growth factor promotes gastric cancer cell invasiveness and oxaliplatin resistance. Cell Death Dis. 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Tan L, Luo D, Peng X, Zhu Y and Li H 2019b. Linc01278 inhibits the development of papillary thyroid carcinoma by regulating miR-376c-3p/DNM3 axis. Cancer Manag. Res 11, 8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A-W, Cai J, Zhao X-L, Jiang T-H, He T-F, Fu H-Q, Zhu M-H and Zhang SH 2011. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin. Cancer Res. 17, 710–720. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu J and Lin QJE 2021. Histone demethylase KDM2A: Biological functions and clinical values. Exp. Therap. Med. 22, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W and Anders S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu K, Li Y, Xu Y, Yue H, Wen Y, Liu T, Chen S, Liu Q, Yang W and Zhu X 2020. Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma. RNA biology 17, 977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Zhao H, Qin Y, Wei J, Sun J, Zhang W and Kang Y 2020. Post-transcriptional dysregulation of microRNA and alternative polyadenylation in colorectal Cancer. Front. Genet. 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Terroba E and Dimitrova N 2020. Long noncoding RNA amplified in lung cancer rewires cancer pathways. The Journal of cell biology 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy B, Chu C and Carlin DJ 2015. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol Sci 145, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpoor R, Gharebaghian A, Shahi F, Mousavi A, Salari S, Akbari ME, Ajdari S and Salimi M 2020. Identification and Validation of Stage-Associated PBMC Biomarkers in Breast Cancer Using MS-Based Proteomics. Front. Oncol. 10, 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA and Vousden KH 2013. p53 mutations in cancer. Nature cell biology 15, 2–8. [DOI] [PubMed] [Google Scholar]
- Nemmar A, Holme JA, Rosas I, Schwarze PE and Alfaro-Moreno E.J.B.r.i. 2013. Recent advances in particulate matter and nanoparticle toxicology: a review of the in vivo and in vitro studies. BioMed Res. Int. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi A, Casoni G, Caramori G, Guzzinati I, Boschetto P, Ravenna F, Calia N, Petruzzelli S, Corbetta L and Cavallesco G 2004. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax 59, 679–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G and Li L 2020. LncRNA TTN-AS1 Promotes Progression of Non-Small Cell Lung Cancer via Regulating miR-491–5p/ZNF503 Axis. Onco Targets Ther 13, 6361–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z and Li S 2020. Long noncoding RNA LINC01278 favors the progression of osteosarcoma via modulating miR‐133a‐3p/PTHR1 signaling. J. Cell. Physiol. [DOI] [PubMed] [Google Scholar]
- Quinlan AR 2014. BEDTools: the Swiss‐army tool for genome feature analysis. Current protocols in bioinformatics 47, 11.12. 11–11.12. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnaiah Y, Kuhlmann L and Tyagi S 2020. Towards a comprehensive pipeline to identify and functionally annotate long noncoding RNA (lncRNA). Computers in Biology and Medicine, 104028. [DOI] [PubMed] [Google Scholar]
- Reddy MA, Zhang E and Natarajan R 2015. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 58, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Chen X, Fang K, Zhang X, Wei X, Zhang T, Li G, Lu Z, Song N and Wang S 2021. COL5A2 Promotes Proliferation and Invasion in Prostate Cancer and Is One of Seven Gleason-Related Genes That Predict Recurrence-Free Survival. Front. Oncol. 11, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Leung A and Natarajan R 2015. Chromatin modifications associated with diabetes and obesity. Arterioscler. Thromb. Vasc. Biol. 35, 1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze PE, Totlandsdal AI, Låg M, Refsnes M, Holme JA and Øvrevik J 2013. Inflammation-related effects of diesel engine exhaust particles: studies on lung cells in vitro. BioMed Res. Int. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto A, Pecoraro G, Giurato G, Nassa G, Rizzo F, Saggese P, Martinez CA, Scafoglio C and Tarallo R 2021. Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer. Cancers 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yu Z, Fu A and Zhou D 2020. Proliferation and migration of hepatocellular carcinoma are accelerated by LINC01287 via the miR-559/TCF12 axis. Eur Rev Med Pharmacol Sci 24, 6023–6030. [DOI] [PubMed] [Google Scholar]
- Steiner S, Bisig C, Petri-Fink A and Rothen-Rutishauser B 2016. Diesel exhaust: current knowledge of adverse effects and underlying cellular mechanisms. Arch. Toxicol. 90, 1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Huang Z, Sheng W and Xu M. d. 2018. Emerging roles of long non-coding RNAs in tumor metabolism. J. Hematol. Oncol. 11, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Komatsu M, Chiwaki F, Komatsuzaki R, Nakamura K, Tsuji K, Kobayashi Y, Tominaga E, Ono M and Banno K 2019. Upregulation of IGF2R evades lysosomal dysfunction-induced apoptosis of cervical cancer cells via transport of cathepsins. Cell Death Dis. 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Chen Q, Xing Y, Zhang C, Pan S, An W and Xu H 2021. High expression of COL5A2, a member of COL5 family, indicates the poor survival and facilitates cell migration in gastric cancer. Biosci. Rep. 41, BSR20204293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnubalaji R, Shaath H and Alajez NM 2020. Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS-CoV-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response. Genes 11, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H 2009. The role of TNF in cancer. Death Receptors and Cognate Ligands in Cancer, 1–15. [Google Scholar]
- Wang C, Zhou J, Wang J, Li S, Fukunaga A, Yodoi J and Tian H 2020. Progress in the mechanism and targeted drug therapy for COPD. Signal Transduction and Targeted Therapy 5, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu H, Dai W, Luo S, Amos CI, Lee JE, Li X, Yue Y, Nan H and Wei Q 2021a. Association of genetic variants of TMEM135 and PEX5 in the peroxisome pathway with cutaneous melanoma-specific survival. Annals of translational medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang Y-H, Yang P-Y and Liu F 2021b. Increased Collagen Type V α2 (COL5A2) in Colorectal Cancer is Associated with Poor Prognosis and Tumor Progression. OncoTargets Ther. 14, 2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Gao X, Shao L, Chen Q, Li F, Wu C, Zhang W and Sun Y 2019. Down-regulation of RAC2 by small interfering RNA restrains the progression of osteosarcoma by suppressing the Wnt signaling pathway. Int. J. Biol. Macromol. 137, 1221–1231. [DOI] [PubMed] [Google Scholar]
- Zeng X-T, Liu X-P, Liu T-Z and Wang X-H 2018. The clinical significance of COL5A2 in patients with bladder cancer: A retrospective analysis of bladder cancer gene expression data. Medicine (Baltimore) 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chen Z, Wang X, Huang Z, He Z and Chen Y 2013. Long non-coding RNA: a new player in cancer. J. Hematol. Oncol. 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jia C and Kwoh CK 2021. Predicting the interaction biomolecule types for lncRNA: an ensemble deep learning approach. Briefings in Bioinformatics 22, bbaa228. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li Y, Sheng J, Wu F, Li K, Huang R, Wang X, Jiao T, Guan X and Lu Y 2019. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. Journal of Experimental & Clinical Cancer Research 38, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A, Buzdin AA, Garazha AV, Borisov NM and Moskalev AA 2014. Signaling pathway cloud regulation for in silico screening and ranking of the potential geroprotective drugs. Front. Genet. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xie H, Wu W, Wen X, Zeng Z and Shi Y 2021a. CircRNA PIP5K1A promotes the progression of glioma through upregulation of the TCF12/PI3K/AKT pathway by sponging miR-515–5p. Cancer Cell Int. 21, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q-X, Wang J, Gu X. y., Huang C-H, Chen C, Hong M and Chen Z 2021b. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomedicine & Pharmacotherapy 135, 111169. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Gu L, Lin X, Cui K, Liu C, Lu B, Zhou F, Zhao Q, Shen H and Li Y 2020. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of β-catenin. Cell Death Differ. 27, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.


