Abstract
Alpha interferon (IFN-α) treatment is effective on a long-term basis in only 15 to 25% of patients with chronic hepatitis C. The results of recent trials indicate that response rates can be significantly increased when IFN-α is given in combination with ribavirin. However, a large number of patients do not respond even to combination therapy. Nonresponsiveness to IFN is characterized by evolution of the hepatitis C virus (HCV) quasispecies. Little is known about the changes occurring within the HCV genomes when nonresponder patients are retreated with IFN or with IFN plus ribavirin. In the present study we have examined the genetic divergence of HCV quasispecies during unsuccessful retreatment with IFN or IFN plus ribavirin. Fifteen nonresponder patients with HCV-1 (4 patients with HCV-1a and 11 patients with HCV-1b) infection were studied while being retreated for 2 months (phase 1) with IFN-α (6 MU given three times a week), followed by IFN plus ribavirin or IFN alone for an additional 6 months (phase 2). HCV quasispecies diversification in the E2 hypervariable region-1 (HVR1) and in the putative NS5A IFN sensitivity determining region (ISDR) were analyzed for phase 1 and phase 2 by using the heteroduplex tracking assay and clonal frequency analysis techniques. A major finding of this study was the relatively rapid evolution of the HCV quasispecies observed in both treatment groups during the early phase 1 compared to the late phase 2 of treatment. The rate of quasispecies diversification in HVR1 was significantly higher during phase 1 versus phase 2 both in patients who received IFN plus ribavirin (P = 0.017) and in patients who received IFN alone (P = 0.05). A trend toward higher rates of quasispecies evolution in the ISDR was also observed during phase 1 in both groups, although the results did not reach statistical significance. However, the NS5A quasispecies appeared to be rather homogeneous and stable in most nonresponder patients, suggesting the presence of a single well-fit major variant, resistant to antiviral treatment, in agreement with published data which have identified an IFN sensitivity determinant region within the NS5A. During the entire 8 months of retreatment, there was no difference in the rate of fixation of mutation between patients who received combination therapy and patients who were treated with IFN alone, suggesting that ribavirin had no major effects on the evolution of the HCV quasispecies after the initial 2 months of IFN therapy.
Hepatitis C virus (HCV) is a positive-strand, RNA virus classified within the Flaviviridae family (7). HCV infects about 2% of the human population and is currently recognized as one of the main causes of chronic liver disease, cirrhosis, and hepatocellular carcinoma worldwide (1). In vivo, HCV is present as a pool of different though closely related genetic variants referred to as a quasispecies (21). HCV quasispecies change composition over time in the individual infected host as a consequence of the low fidelity of the viral-RNA-dependent RNA polymerase and of selective pressure on the viral proteins by host factors such as the immune response (10, 18, 20, 30). Several studies have been conducted to analyze the clinical implications of HCV heterogeneity, and data have been provided suggesting that the evolution of HCV quasispecies may be related to disease outcome and to response to interferon (IFN) therapy (3, 15).
Standard treatment with 3 to 5 MU of IFN-α given three times per week for 6 to 12 months in chronic hepatitis C leads to sustained response in only 15 to 25% of treated patients (17). Nonresponse has been associated with an increased rate of HCV quasispecies diversification occurring in nearly all patients when the envelope 2 gene hypervariable region 1 (HVR1) is analyzed and in about one-third when the putative IFN sensitivity determining region (ISDR) within the nonstructural 5A gene (NS5A) is considered (13, 19, 23–26). Patients who fail to respond to a first cycle of IFN-α are often retreated with IFN alone or with IFN-ribavirin combination therapy (4, 8, 27). To date, no studies have been reported on the behavior of HCV quasispecies during retreatment with these schedules. Therefore, in the present study we examined HCV quasispecies diversification within the HVR1 and the ISDR regions in a group of patients who had failed to respond to a first cycle of IFN and were again nonresponders when retreated with IFN alone or with IFN plus ribavirin. These patients were among those included in a recently conducted clinical trial in which patients were retreated for 2 months with IFN alone followed by randomization to continue with IFN monotherapy or to add ribavirin in combination with IFN. This type of design allowed us to assess sequentially the effect of monotherapy and of addition of ribavirin on the circulating HCV quasispecies in patients with virological nonresponsiveness to those treatments.
MATERIALS AND METHODS
Patients.
HCV genomes were analyzed in serial serum specimens obtained from 15 selected Italian patients (13 male and 2 female; mean age, 40.1 years; range, 26 to 59 years) with chronic hepatitis C. All 15 patients had been nonresponders during a first cycle of IFN monotherapy and were again nonresponders to a second cycle of IFN alone or of a combination of IFN plus ribavirin. Retreatment was initiated in all patients with 6 MU t.i.w. of IFN-α for 2 months, followed by randomization to add ribavirin (1,000 to 1,200 mg daily) for 6 months in combination with an identical schedule of IFN-α (group 1, including nine patients) or to continue with IFN alone (group 2, including six patients). These patients were selected because they were infected by HCV genotype 1 (either 1a or 1b) and had not developed a sustained response to retreatment. All patients in group 2 remained viremic during the entire treatment period, while in group 1 patients 4 and 9 became transiently HCV RNA negative during therapy by both b-DNA and reverse transcription-PCR (RT-PCR) at month 2 (patient 4) and at the end of treatment (patient 9). Blood samples were obtained from each patient before therapy, at the end of month 2 (before the addition of ribavirin in group 1), and at the end of retreatment. Changes in serum HCV RNA were monitored by b-DNA version 2.0 (Chiron Corporation, Emeryville, Calif.) or by quantitative PCR as described elsewhere (14).
PCR and cloning.
Total RNA was extracted starting from 100 μl of serum by the single-step guanidinium method (6) and resuspended in 10 μl of diethylpyrocarbonate-treated distilled water. RT was performed as described previously (31). The RNA was incubated at 70°C for 5 min and then reverse transcribed in a 25-μl reaction mixture containing 50 pmol of the external antisense primer, 3 mM MgCl2, 1 mmol of each deoxynucleoside triphosphate (dNTP), 1 mM dithiothreitol, 75 mM KCl, 5 mM Tris-HCl (pH 8.3), 20 U of RNase inhibitor (Pharmacia LKB, Piscataway, N.J.), and 130 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL, Gaithersburg, Md.). The mixture was incubated at 37°C for 60 min and then at 95°C for 5 min. The HVR1 and the ISDR in the NS5A gene were amplified after RT by nested PCR. For both of the regions the first round PCR was performed as follows. First, 10 μl of cDNA was added to 40 μl of mixture containing 50 pmol of the external, sense primer, 1.5 mM MgCl2, 23.5 mM Tris-HCl (pH 8.3), 35.5 mM KCl, and 1.5 U of Taq polymerase (Perkin-Elmer, Norwalk, Conn.). Nested “hot-start” PCR was performed as follows. The bottom mixture contained 2.0 mM MgCl2, 0.2 mmol of each dNTP, 10 mM Tris-HCl (pH 8.3), 15 mM KCl, and 50 pmol of each internal primer. A wax layer was used to separate the lower mixture from the top reaction mixture containing 40 mM Tris-HCl (pH 8.3), 60 mM KCl, 1.5 U of Taq polymerase, and 1 μl of the first-round PCR product. For the amplification of a 196-nucleotide fragment of the E2-HVR1, the two following sets of primers were used: outer primers AS (5′-CATTGCAGTTCAGGGCCGTGCTA-3′) and S (5′-GGTGCTCACTGGGGAGTCCT-3′) and inner primers AS (5′-TGCCAACTGCCATTGGTGTT-3′) and S (5′-TCCATGGTGGGGAACTGGGC-3′). Subtype-1a/1b-specific primers were used for the amplification of a 219-nucleotide fragment of the NS5A containing the putative ISDR. For genotype 1a, the outer primer set 5A-1a-2 (AS, 5′-GAGACTTCCGCAGGATTTCT-3′; S, 5′-TGACGTCCATGCTCACTGAT-3′) and inner primer set 5A-1a-1 (AS, 5′-CGAAGGAGTCCAGAATCACC-3′; S, 5′-CCTCCCATATAACAGCAGAG-3′) were used; for genotype 1b, the outer primer set 5A-1b-2 (AS, 5′-CTGGATTTCCGCAGGATCTC-3′; S, 5′-CAGAGACGGCTAAGCGTAGG-3′) and inner primer set 5A-1b-1 (AS, 5′-TCCCTCTCATCCTCCTCCGC-3′; S, 5′-TCCTTGGCCAGCTCTTCAGC-3′) were used. For first- and second-round HVR1 PCR 30 cycles with the following conditions were used: 30 s at 94°C, 25 s at 55°C, and 30 s at 72°C. Cycling parameters for first-round and nested PCR of the NS5A were as follows: 30 s at 94°C, 25 s at 65°C, and 30 s at 72°C for 30 cycles. PCR products were purified (QIAQuik columns; Qiagen, Chatsworth, Calif.) and ligated into TA cloning vectors (Invitrogen, San Diego, Calif.) according to the manufacturer.
HTA.
The heteroduplex tracking assay (HTA) was previously reported as a sensitive and rapid technique for monitoring the evolution of HCV quasispecies within the same patient at different time points. The technique is described in detail elsewhere (15, 25, 29, 31). Briefly, to generate a probe, the insert of one clone (either HVR1 or ISDR) was amplified by PCR and, after column purification, the PCR product was end radiolabelled with T4 polynucleotide kinase (Gibco-BRL) plus [γ-32P]ATP. The probe, which represents the quasispecies major variant at baseline, was hybridized to the heterogeneous PCR product from serum samples obtained at the three different time points, and the different viral sequences were then resolved by nondenaturing polyacrylamide gel electrophoresis. Nucleotide changes between the probe and the target DNA lead to the formation of heteroduplex bands that were characterized by retarded mobility during gel electrophoresis. Probe hybridized with its own unlabelled sequence was used to indicate the homoduplex control. As previously demonstrated (for both the HVR1 and the NS5A portion containing the putative ISDR), the degree of shift of the heteroduplex bands compared to the homoduplex band is directly proportional to the number of nucleotide changes between the probe and the target DNA, and thus the genetic distance between variants can be estimated by calculating the heteroduplex mobility ratio (HMR) (25, 31). The HMR is calculated by measuring the distance in millimeters of the heteroduplex from the origin of the gel and dividing that by the distance of the homoduplex band from the origin. The total HMR of the quasispecies at one time point is calculated as the average of the HMRs of all the variants indicated by the different bands.
CFA.
To characterize individual clones within the circulating quasispecies at different time points, clonal frequency analysis (CFA) was performed by using the same patient-specific probe obtained for the HTA as previously described (26, 31). For CFA, at least 20 independent clones were analyzed per specimen for both the HVR1 and the putative ISDR. In each case, the quasispecies complexity (the total number of variants) was determined by counting the number of unique gel shift patterns. To determine the frequency of each variant, the number of identical gel shift variants was divided by the number of clones analyzed (21), and this value was multiplied by 100 to obtain the percentage.
Reproducibility of quasispecies sampling technique was assessed by performing replicate experiments on undiluted or serially diluted specimens, as previously described (16).
The total HMR of the quasispecies at one time point is obtained by averaging the distances (in millimeters) from the origin of the gel of all the bands representing different clones and by dividing the obtained average distance of the clones by the distance (in millimeters) from the origin of the gel of the homoduplex control.
The changes in quasispecies diversity within the HVR1 and the putative ISDR were measured during the first phase of treatment by using the following formula: (HMRt2 − HMRt1)/HMRt1, where t1 and t2 indicate the pretreatment time point and the month-2 time point, respectively. Similarly, the change in HMR was measured during the second phase of therapy (phase 2), from month 2 to the end of treatment by using the following formula: (HMRt3 − HMRt2)/HMRt2, where t3 indicates the end-of-treatment time point.
Statistical analysis.
A two-sample paired Student’s t test was used to compare mean age, mean serum alanine aminotransferase level, viral RNA titers, and mean HMR value at baseline between the two patients groups. A P value of <0.05 was considered significant. The nonparametric Wilcoxon test was used to compare the rates of change in HMRs between phase 1 and phase 2 in each patient group. The correlation between the HMR values estimated by HTA and CFA was determined by linear regression. The rate of quasispecies diversification over the entire period of observation was assessed and compared in the two groups by the analysis-of-variance (ANOVA) test.
RESULTS
Nine nonresponder patients had been treated for 2 months with IFN monotherapy followed by 6 months of IFN-ribavirin combination therapy, while 6 patients had received 8 months of IFN monotherapy, according to the protocol described in Materials and Methods. HCV quasispecies diversification was assessed in all patients during the 2 phases of treatment, with phase 1 being the same for all 15 patients, while phase 2 differed for patients in group 1 (IFN-ribavirin combination therapy) versus group 2 (IFN monotherapy). Thus, patients in group 1 allowed an internally controlled comparison of HCV quasispecies changes during sequential IFN monotherapy followed by IFN plus ribavirin combination therapy. Group 2 patients represented an external control group for comparison of phase 2 results with group 1 patients.
Baseline virological and clinical characteristics of the 15 patients are summarized in Table 1. No difference was observed between the two groups with regard to the HCV viral load, genotype, and liver histology. HCV quasispecies diversity and complexity were determined in baseline specimens from all 15 cases. Overall, no significant differences in pretreatment HCV quasispecies diversity and complexity between patients of group 1 versus those of group 2 were observed (Table 2).
TABLE 1.
Baseline features of patients included in the two schemes of treatmenta
| Treatment | No. of cases | Sex (M/F) | Mean age (yr) ± SD | Mean serum ALT ± SD | Histological severity of chronic hepatitis
|
HCV genotype (1a/1b) | Serum HCV RNA (MEq/ml) | ||
|---|---|---|---|---|---|---|---|---|---|
| Mild | Moderate | Severe | |||||||
| Group 1 (IFN → IFN-ribavirin) | 9 | 8/1 | 39.8 ± 9.7 | 239 ± 145 | 1 | 5 | 2 | 3/6 | 5.2 ± 7.4 |
| Group 2 (IFN alone) | 6 | 5/1 | 40.5 ± 12 | 130 ± 25.2 | 2 | 2 | 3 | 1/5 | 4.7 ± 6 |
ALT, alanine aminotransferase. P values were not significant for age, serum ALT, and serum HCV RNA values.
TABLE 2.
HCV quasispecies features at baseline in patients included in the two schemes of treatment
| Treatment group | Mean Qsa:
|
|
|---|---|---|
| Diversity ± SDb | Complexity ± SDc | |
| Group 1 | 0.96 ± 0.028 | 8.0 ± 1.4 |
| Group 2 | 0.93 ± 0.014 | 8.2 ± 7.1 |
Qs, quasispecies.
Relative to HMR.
Number of single shift patterns within 20 clones analyzed by CFA. The P values were not significant.
In the current study both the HTA and the CFA techniques were used to quantify the HCV quasispecies diversity in sequential serum samples in individual patients during treatment, the CFA technique allowing a more detailed assessment of individual clones within a quasispecies population and more accurately defining the quasispecies complexity in specimens previously analyzed by HTA.
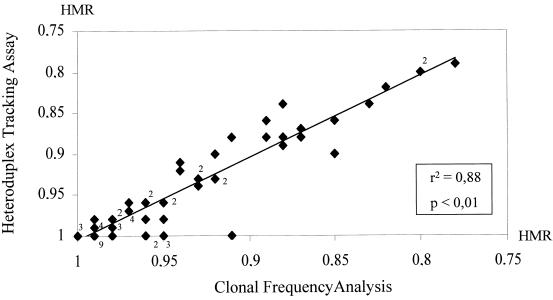
A total of 63 specimens were analyzed by both the HTA and the CFA techniques (for a total of 1,400 independent clones), and data were quantitatively expressed in terms of the HMR, as described in Materials and Methods. Figure 1 depicts the strong correlation between the HMR values of each quasispecies population derived in parallel by the HTA and CFA techniques (r2 = 0.88; P < 0.01) supporting the accurate and reproducible assessment of HCV quasispecies carried out in the present study.
FIG. 1.
Correlation between two methods for HCV quasispecies analysis. Data were derived by using 63 specimens for which the quasispecies diversity was analyzed in the HVR1 or in the NS5A regions by two different methods, HTA and CFA. The y axis depicts HCV quasispecies genetic diversity data expressed as the HMR generated via the HTA technique, while the x axis shows quasispecies genetic diversity data obtained by the CFA technique. A highly significant correlation was observed between the two methods for both genetic regions. The digits close to the solid diamond symbols represent the number of samples with the same value when the value was >1.
Virus load profiles during retreatment.
In 11 patients the HCV RNA serum levels were fairly stable during therapy, with changes not exceeding 1 log. Significant changes in serum HCV RNA levels were observed in four cases during therapy: HCV RNA become undetectable by both b-DNA and PCR at month 2 in patient 4 and at the end of treatment in patient 9, while in patients 5 and 11 viremia decreased by more than 1 log by the end of treatment (from 7.3 to 5.3 log Eq/ml in patient 5 and from 7.3 to 5.9 log Eq/ml in patient 11).
HVR1 quasispecies diversification during IFN-ribavirin therapy.
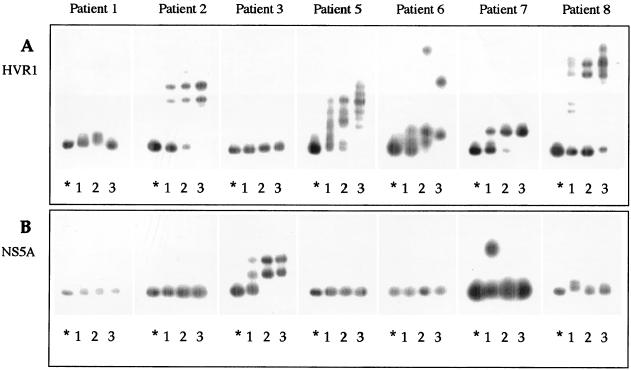
Figure 2 illustrates analysis of HCV quasispecies by HTA during IFN monotherapy followed by IFN-ribavirin combination therapy for 7 patients in treatment group 1. For each patient, HCV quasispecies were analyzed in serum samples obtained before therapy (lane 1), 2 months after the initiation of therapy (end of phase 1) (lane 2), and at the end of therapy (end of phase 2) (lane 3). Both the E2 HVR1 (Fig. 2A) and the putative ISDR (Fig. 2B) were amplified by RT-PCR and hybridized with patient-specific probes derived from the quasispecies major variant of the pretreatment time point. In two cases, patients 4 and 9, the viral RNA could not be detected by RT-PCR in serum samples obtained after 2 months of treatment with IFN or at the end of treatment, respectively. In Fig. 2A, which shows temporal changes in the HVR1 quasispecies, five patients (patients 2, 5, 6, 7, and 8) displayed an expanding gel shift pattern during retreatment, indicating a substantial evolution in genetic diversity between quasispecies variants. Accordingly, in four of these patients, a consistent decrease in HMR values over the entire treatment period (patient 2, from 0.91 to 0.84; patient 5, from 0.96 to 0.88; patient 6, from 0.95 to 0.89; patient 8, from 0.98 to 0.80) was observed. One patient (number 7) showed a minor change in HMR (from 0.96 to 0.94). The increased quasispecies diversity at the end of therapy compared to pretreatment was associated in these five cases with a reduction in mean quasispecies complexity (from 9.5 to 6.7 variants per 20 clones analyzed). In patients 2, 5, 6, and 7, the HTA profiles demonstrated that the major variants detected before treatment were drastically reduced after the first 2 months of IFN treatment and were no longer detected in serum specimens obtained at the end of treatment. The HTA findings were confirmed by the CFA technique, as shown in Fig. 3, where the pretreatment quasispecies major variant of patients 2 and 6, corresponding to clones with mobilities identical to that of the homoduplex control, was represented by only one of 20 clones at month 2 and by none of the 20 clones at the end of treatment.
FIG. 2.
HTA of the HCV quasispecies in the HVR1 (A) and NS5A (B) characterized before, during, and at the end of therapy in seven patients who received IFN followed by combination therapy. For each experiment, radiolabeled probes were generated from pretreatment quasispecies major variant and hybridized to itself (homoduplex, indicated by an asterisk) or to heterogeneous target sequences obtained from PCR-amplified products of the same patient before therapy (lane 1), 2 months after initiation of IFN therapy (lane 2), and at the end of therapy (lane 3).
FIG. 3.
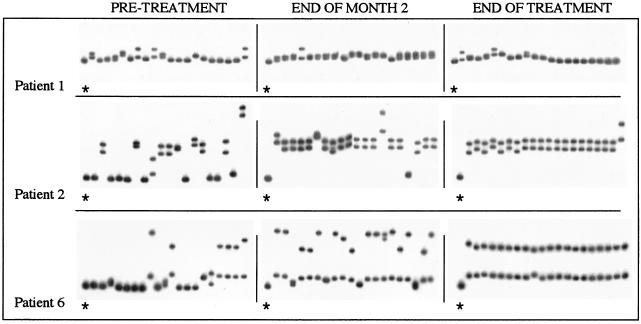
CFA of the HVR1 quasispecies in patients 1, 2, and 3 within patients of group 1. For each patient, a radiolabeled probe was prepared from the pretreatment quasispecies major variant. In each case, 20 independent HVR1 clones were analyzed at three different time points: before treatment, after 2 months of IFN treatment, and at the end of IFN-ribavirin treatment. Each panel depicts the gel shift pattern of the 20 HVR1 clones plus the homoduplex shift control, which is indicated by an asterisk.
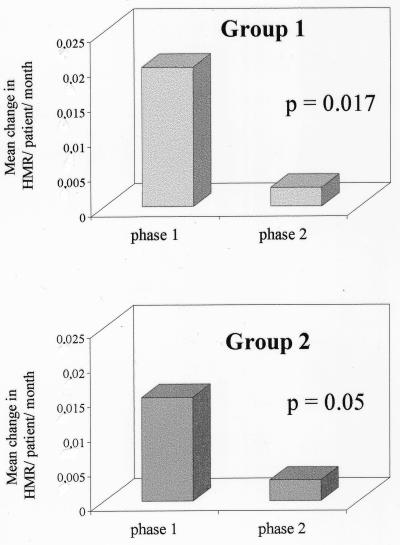
The remaining patients in group 1 (patients 1 and 3) had a low HVR1 genetic diversity as determined by HTA at the pretreatment time point and showed only minor changes in the quasispecies population during the entire treatment period. These observations were confirmed by the corresponding CFA profiles (Fig. 3, patient 1) and by the minor changes in HMR over time (patient 1, from 0.98 to 0.99; patient 3, from 1 to 0.99). In addition, in these cases the quasispecies complexity decreased during retreatment (from 10 to 7 and 4 to 3 clones per 20 clones for patients 1 and 3, respectively). The rate of HCV quasispecies diversification during phase 1 and during phase 2 was assessed by calculating the mean changes in HMR per patient per month. As shown in Fig. 4A, the rate of HVR1 quasispecies diversification was significantly higher during the first 2 months of therapy with IFN alone (phase 1) compared to phase 2, when patients received combination therapy (mean change in HMR/patient/month: phase 1 = 0.02; phase 2 = 0.0027; P = 0.017).
FIG. 4.
Comparison of the rate of HVR1 quasispecies diversification during the first 2 months of treatment (phase 1) versus the second phase of treatment (phase 2) within patients of groups 1 and 2. To estimate the rate of quasispecies genetic divergence during different intervals of treatment, the mean change in HMR per patient/month was derived as described in Materials and Methods.
NS5A (ISDR) quasispecies diversification in patients of group 1.
Figure 2B shows the HTA profile for the NS5A quasispecies in patients of group 1. In four out seven patients (1, 2, 5, and 6) no changes were observed during the treatment period. Of the remaining three patients, the HMR decreased slightly in patient 3 (from 0.95 to 0.93), while it increased in patient 7 (from 0.94 to 0.99); a marginal change in the NS5A quasispecies composition was observed in patient 8 (from 0.98 to 0.99).
HVR1 and ISDR quasispecies diversification in patients of group 2.
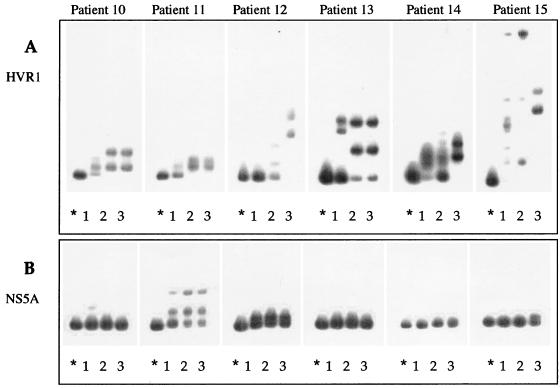
Patients in group 2 were treated with IFN monotherapy throughout the 8-month study period. Figure 5A illustrates the HTA profiles of the HVR1 quasispecies in patients 10, 11, 12, 13, 14, and 15 analyzed at different time points. In all of these patients, the HVR1 quasispecies composition consistently changed during the entire treatment period, since the major variants detected before therapy were not observed at the end of treatment. In particular, in patients 10, 11, and 13 major changes occurred mainly during the first 2 months of therapy and were maintained throughout the treatment period, as indicated by the mobility pattern at month 2 (lane 2) and at the end of therapy (lane 3).
FIG. 5.
HTA of the HCV quasispecies in the HVR1 (A) and NS5A (B) characterized before, during, and at the end of therapy in six patients who received IFN monotherapy. For each experiment, radiolabeled probes were generated from pretreatment quasispecies major variant and hybridized to itself (homoduplex, indicated by an asterisk) or to heterogeneous target sequences obtained from PCR-amplified products of the same patient before therapy (lane 1), 2 months after initiation of IFN therapy (lane 2), and at the end of therapy (lane 3).
In contrast, in patients 12, 14, and 15 different shift patterns were observed at the three time points, indicating that new subsets of quasispecies variants could continuously emerge.
HMR decreased after IFN therapy in patient 11 (from 0.94 to 0.92), in patient 12 (from 0.99 to 0.87), and in patient 14 (from 0.87 to 0.80), indicating an increasing genetic divergence between variants before and at the end of treatment. In contrast, HMR values increased in patients 13 and 15 (from 0.91 to 0.96 and from 0.93 to 0.96, respectively), indicating a reduction in quasispecies genetic diversity.
Figure 5B shows the quasispecies tracking profiles of NS5A sequences in the same patients before treatment, after 2 months of treatment, and at the end of therapy. As observed for patients treated with IFN plus ribavirin, the NS5A quasispecies population in this group of patients appeared not to be affected by IFN, since similar or identical gel shift patterns were observed in most patients at the time of sequential testing. In only one patient (number 15) was a marked change in the major quasispecies variants observed at the end of treatment. The overall stability of the NS5A quasispecies was associated, in most patients, with a simple genetic quasispecies composition. Only patient 11 showed a more complex NS5A quasispecies that remained stable over the period of observation.
Figure 4B illustrates the comparison of the rates of genetic diversification between phase 1 and phase 2 of treatment in patients of group 2. As we observed in group 1, the rate of quasispecies diversification was significantly different in the two phases of treatment: the mean change in HMR/patient/month was approximately fivefold higher during the first 2 months of treatment (0.015) compared to the following 6 months (0.003) (P = 0.05).
In agreement with these observations, there was no difference in the patterns of HCV quasispecies diversification over the entire treatment period in patients of group 1 compared to those of group 2 (P = 0.90 by ANOVA).
DISCUSSION
With respect to the treatment of chronic hepatitis C, it is becoming apparent that response rates with current treatment regimens remain suboptimal (2, 17). Since one goal of successful therapy is the eradication of virus from the infected host, studies examining the dynamic responses of HCV to various treatment regimens are warranted. In particular, the effect of retreatment with IFN alone or with IFN plus ribavirin on the evolution of the HCV quasispecies in nonresponsive patients is unknown.
In the present study, a detailed assessment of HCV quasispecies diversification rates in a group of IFN nonresponders who were randomized to receive retreatment with IFN alone for 8 months or IFN alone for 2 months followed by IFN plus ribavirin was carried out. The two treatment groups were well matched for the virological predictors of IFN response, including pretreatment viral load and infecting HCV genotype (all were genotype 1) (17); in addition, the pretreatment quasispecies diversity was similar in the two groups.
The diversification of HCV quasispecies was analyzed within the two distinct genomic regions HVR1 and NS5A-ISDR, previously shown to be affected by IFN therapy by using the related techniques HTA and CFA. The systematic application of these well-standardized and highly reproducible methods (26, 29, 31) allowed quasispecies analysis in a relatively large number of serial samples. Although all patients were nonresponders, in some of them the predominant clones have clearly been suppressed during treatment. This was likely due to the antiviral treatment which induced rapid clearance of HCV-sensitive variants, giving minor and resistant variants an opportunity to expand. We cannot exclude, however, that the treatment increased variation within the existing major variants as a result of selective pressure in favor of those mutants that were most fit to replicate. Probably the best approach to address these issues would be to analyze the nucleotide sequences of a representative number of cDNA clones at each time point and their phylogenetic relationship. However, it would be difficult to verify both hypotheses because of the rapid turnover of viral population and the lack of closer time points.
During the first 2 months of IFN retreatment, an increased rate of quasispecies diversification in both HVR1 and NS5A genes was observed. Interestingly, these two genomic regions evolved rather independently, and changes in the quasispecies population occurred more frequently within the HVR1 compared to the NS5A, with rates of diversification that were statistically significant for HVR1 but not for NS5A. In one particular case, however, the behavior of the quasispecies was clearly different, with changes occurring within NS5A but not within HVR1. These results, while confirming that IFN exerts a different selective pressure on the HCV quasispecies in HVR1 compared to the portion of the NS5A which contains the putative ISDR sequence, indicate that independent evolution of these two regions may occur.
The overall lower rate of genetic diversification observed in the NS5A regions compared to the HVR1 may have been a consequence of the selection on an IFN-resistant population in the NS5A quasispecies already during the first cycle of therapy, a notion also in agreement with the rather restricted NS5A quasispecies found before retreatment in most of our patients. However, the data are not significantly different from those previously reported in naive patients (31).
In recent years several studies have associated nonresponsiveness to IFN treatment with the presence of a specific consensus ISDR within the carboxyl-terminal half of the HCV NS5A gene. In particular, it has been shown that patients with an ISDR sequence identical to that of HCV-J (wild type) do not respond, while those with a mutated sequence often do respond to the therapy (5, 9). These findings have not been always confirmed (28, 32). Unlike all previous studies, where the nucleotide sequence of the putative ISDR was determined, in our study we analyzed the NS5A quasispecies by a combination of the HTA and CFA techniques, and we have shown that in nonresponders the NS5A quasispecies is often very homogeneous and highly conserved during treatment. These findings suggest the presence of a single well-fit major variant, one resistant to the antiviral treatment. The presence of such a homogeneous viral population already before the beginning of retreatment might be the result of a previous selection of an IFN-resistant strain that occurred during the first cycle of therapy. Thus, these findings support what has been previously suggested by different authors about the existence of an ISDR motif associated with IFN nonresponsiveness (5, 9, 11).
A major finding of the present study was that the HCV quasispecies changed much more rapidly during the early phase of retreatment than during the late phase, a finding independent of whether patients received IFN alone or IFN plus ribavirin during the second phase. Indeed, mean changes in HMR per patient/month were five- to sevenfold higher for HVR1 during the first 2 months of retreatment than for the following 6 months. This observation is consistent with the concept that IFN resistance occurs early in the course of treatment and indicates the need for assessing other induction schedules for IFN retreatment, such as daily IFN treatment, which may increase the antiviral pressure and reduce the level of virus escape.
The addition of ribavirin to IFN had no major effects on the diversification and evolution of the HCV quasispecies driven by IFN. This appeared to be true for the changes occurring both in the HVR1 and in the NS5A genes. Indeed, the patterns of these quasispecies that had developed during the first 2 months of IFN retreatment remained unchanged in most patients during further treatment, without any differences between monotherapy and combination therapy. These observations are in agreement with previous work indicating that ribavirin alone has no appreciable effect on HCV quasispecies heterogeneity (12). The mechanism by which ribavirin increases the virological response to IFN still remains largely unknown. It has been recently reported that, in cultured peripheral blood mononuclear cells from patients chronically infected with HCV, combination therapy induced increased levels of 2′,5′-oligoadenylate synthetase compared to treatment with IFN alone, suggesting that the enhanced antiviral activity was due to a synergistic effect of ribavirin plus IFN on the expression of this enzyme (22). Since our results were obtained with patients treated sequentially with the two drugs, a similar quasispecies analysis should be done in patients receiving IFN in combination with ribavirin from the beginning of treatment.
In summary, the present study describes a detailed analysis of HCV quasispecies diversification in two groups of nonresponder patients, all of whom were infected with HCV genotype 1 and who were then retreated with IFN alone or with IFN plus ribavirin. Retreatment was associated with accelerated quasispecies diversification during the first 2 months of therapy in nearly all patients. After the first 2 months, the level of quasispecies diversification was reduced, regardless of the addition of ribavirin to the treatment schedule. The data argue for clinical trials designed to investigate the efficacy of using more-aggressive drugs with increased antiviral activity on HCV during the early treatment period.
ACKNOWLEDGMENTS
We thank Francesca Dal Pero and Sean Kim for excellent technical support.
This study was supported by an educational grant from Schering-Plough. D.R.G. is partially supported by NIH grants AI40032-02 and AI39049-02.
REFERENCES
- 1.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 2.Brown J L. Efficacy of combined interferon and ribavirin for treatment of hepatitis C. Lancet. 1998;351:78–80. doi: 10.1016/S0140-6736(05)78157-5. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J, Miller R, Purcell R. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 4.Chemello L, Cavalletto L, Bernardinello E, Guido M, Pontisso P, Alberti A. The effect of interferon alpha and ribavirin combination therapy in naive patients with chronic hepatitis C. J Hepatol. 1995;23:8–12. [PubMed] [Google Scholar]
- 5.Chayama K, Tsubota A, Kabayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 8.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxì A, Ling M-H, Alvrecht J. Interferon alpha-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 9.Enamoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 10.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuma T, Enamoto N, Marumo F, Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology. 1998;28:1147–1153. doi: 10.1002/hep.510280433. [DOI] [PubMed] [Google Scholar]
- 12.Gonzales-Peralta R P, Lui W Z, Davis G L, Qian K P, Lau J Y N. Modulation of hepatitis C virus quasispecies heterogeneity by interferon alpha and ribavirin therapy. J Viral Hepatol. 1997;4:99–106. doi: 10.1111/j.1365-2893.1997.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Peralta R P, Qian K, She J Y, Davis G L, Ohno T, Mizokami M, Lau J Y N. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J Med Virol. 1996;49:242–247. doi: 10.1002/(SICI)1096-9071(199607)49:3<242::AID-JMV14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Gretch D R, dela Rosa C, Carithers R L, Willson R A, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann Intern Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 15.Gretch D R, Polyak S J, Wilson J J, Carithers R L, Perkins J D, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gretch D R, Polyak S J Groupe Français d’Etudes Moleculaires des Hepatites (GEMHEP), editors . Proceedings of the Hepatitis C virus GEMHEP Conference. Paris, France: John Libbey Eurotext; 1997. The quasispecies nature of hepatitis C virus: research methods and biological implications; pp. 57–69. [Google Scholar]
- 17.Hoofnagle J H, Di Bisceglie A M. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 18.Holland J J, De La Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 19.Kanazawa Y, Hayashi N, Mita E, Li T, Hagiwara H, Kasahara A, Fusamoto H, Kamada T. Influence of viral quasispecies on effectiveness of interferon therapy in chronic hepatitis C patients. Hepatology. 1994;20:1121–1130. [PubMed] [Google Scholar]
- 20.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martìn J, Navas S, Quiroga J A, Pardo M, Carreno Y. Effects of the ribavirin-interferon α combination on cultured peripheral blood mononuclear cells from chronic hepatitis C patients. Cytokine. 1998;10:635–644. doi: 10.1006/cyto.1997.0333. [DOI] [PubMed] [Google Scholar]
- 23.Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, Kaneshige T, Igimi H, Kamada T, Uchida K. Hepatitis C viral complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology. 1995;108:789–795. doi: 10.1016/0016-5085(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 24.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 25.Polyak S, Faulkner G, Carithers R, Jr, Corey L, Gretch D. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- 26.Polyak S J, McArdle S, Liu S-L, Sullivan D G, Chung M, Hofgärtner W T, Carithers R L, McMahon B J, Mullins J I, Corey L, Gretch D R. Evolution of hepatitis C virus quasispecies in the hypervariable region 1 and the interferon sensitivity determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schalm S W, Hansen B E, Chemello L, Bellobuono A, Brouwer J T, Weiland O. Ribavirin enhances the efficacy but not the adverse effects of interferon in chronic hepatitis C: meta-analysis of individual patient data from European centers. J Hepatol. 1997;26:961–966. doi: 10.1016/s0168-8278(97)80103-1. [DOI] [PubMed] [Google Scholar]
- 28.Squadritto G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alpha. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan D G, Wilson J J, Carithers R L, Perkins J D, Gretch D R. Multigene tracking of hepatitis C virus quasispecies after liver transplantation: correlation of genetic diversification in the envelope region with asymptomatic or mild disease patterns. J Virol. 1998;72:10036–10043. doi: 10.1128/jvi.72.12.10036-10043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- 32.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alpha. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]