Abstract
The CRISPR/Cas13b system has been demonstrated as a robust tool for versatile RNA studies and relevant applications. New strategies enabling precise control of Cas13b/dCas13b activities and minimal interference with native RNA activities will further facilitate the understanding and regulation of RNA functions. Here, we engineered a split Cas13b system that can be conditionally activated and deactivated under the induction of abscisic acid (ABA), which achieved the downregulation of endogenous RNAs in dosage- and time-dependent manners. Furthermore, an ABA inducible split dCas13b system was generated to achieve temporally-controlled deposition of m6A at specific sites on cellular RNAs through conditional assembly and disassembly of split dCas13b fusion proteins. We also showed that the activities of split Cas13b/dCas13b systems can be modulated by light via using a photoactivatable ABA derivative. Overall, these split Cas13b/dCas13b platforms expand the existing repertoire of the CRISPR and RNA regulation toolkit to achieve targeted manipulation of RNAs in native cellular environments with minimal functional disruption to these endogenous RNAs.
Graphical Abstract
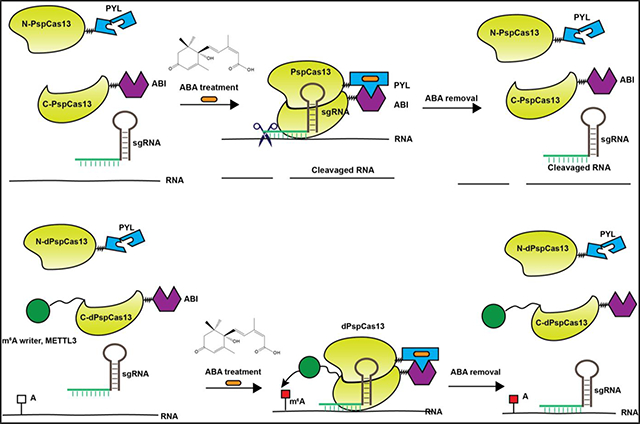
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR), originated from the adaptive immunity systems in the bacteria and archaea,1–2 have been reprogrammed for various biological applications with different CRISPR associated (Cas) proteins and the corresponding single guide RNA (sgRNA).3–10 For example, Cas9 and Cas12a systems have been extensively utilized for genome editing, while their nuclease-inactive version (dCas9 and dCas12a) have been used to recruit various effectors to specific genome loci for desired purposes.5–6, 11–13 RNA-targeting CRISPR systems have also been discovered, including the Cas13 subfamily, which have been used for RNA silencing and detection.14–17 Moreover, nuclease-inactive Cas13 (dCas13) have been employed in different applications, including RNA imaging, splicing control and the editing of RNA bases or posttranscriptional modifications (e.g., N6-methyladenosine, m6A).8–10, 16, 18–22 To offer precise spatiotemporal controls of CRISPR activities, which can be crucial for studying the associated biological activities within the dynamically changing cellular contexts, conditional CRISPR methods regulated by small molecules, light or magnet have been reported.12, 23–29
One concern regarding the CRISPR systems is the bulky size of Cas proteins. Besides the challenges for their in vivo delivery, it has been shown that the bulky Cas proteins can impede endogenous cellular processes through steric interference.30–31 The steric disruptions can be a critical concern for RNA studies, as bulky Cas fusion proteins may interfere with the native functions of the bound RNAs in unexpected ways. For example, it can alter their 3-dimensional structures/conformations, or disrupt their interactions with various endogenous RNA-binding proteins (RBPs) and heterogeneous nuclear ribonucleoproteins (hnRNPs), which are vital for carrying out their biological activities.32–35 This drawback limits the wider applications of RNA-targeting CRISPR systems for purposes other than RNA suppression or detection. To overcome the size-related limitations of CRISPR-based RNA targeting system, a modular CRISPR-Cas-inspired RNA-targeting system (CIRTS) has been reported, which uses a much smaller RNA-targeting protein component engineered from the human protein parts.36 In addition, the small molecule inducible CIRTS system has been developed to provide temporal control of the targeted RNA manipulation.37 Although the CIRTS system reduces the steric interference by employing a protein that is much smaller than the Cas proteins, the constitutive binding of CIRTS fusion proteins on target RNAs can still be an issue in certain applications.
To introduce a new temporally-controlled CRISPR-Cas13b technology and address the critical issue of potential steric interference caused by dCas13b fusion proteins with target RNAs, we engineered two inducible split Cas13b/dCas13b platforms, which allow conditional assembly and disassembly of Cas13b/dCas13b complex under the control of the abscisic acid (ABA)-based chemical induced proximity (CIP) system. We showed that the split Cas13b-mediated RNA silencing can be rapidly induced upon ABA addition and reversed after ABA removal. Moreover, we demonstrated that the same splitting strategy can be applied to the nuclease-inactive Cas13b (dCas13b) to generate a split dCas13b-based m6A editing platform. This m6A editing platform (i.e., split dCas13b-M3), incorporating a m6A writer, methyltransferase-like 3 (METTL3),38–39 successfully achieved site-specific m6A writing on target RNA transcripts in a time-dependent manner. Through studying the kinetics of the inducible assembly and disassembly of the split dCas13b-M3 fragments, we confirmed that the split dCas13b-M3 complex was only transiently reconstituted at the targeted RNA locus for site-specific m6A writing in the presence of ABA and was rapidly released from the target RNA after ABA removal. We observed that the artificially deposited m6A persisted for an extended period after the split dCas13b-M3 fusion proteins were released, which offers an ideal temporal widow to investigate m6A functions in its native cellular environment without disruptions from the dCas13b complex. In addition, we demonstrated that light can be introduced as an alternative trigger to control the activities of split Cas13b/dCas13b system using a synthetic photoactivatable ABA derivative (ABA-DMNB).40 Taken together, we developed novel inducible split Cas13b/dCas13b systems, regulated by both small molecules and light, which expanded the CRISPR toolkit for RNA studies. The inducible and traceless m6A editing platform developed based on this strategy provides a unique tool for unveiling m6A’s biological roles at specific RNA loci, while maintaining minimal interference with the native functions of target RNAs.
Results and Discussions
Design of chemically inducible split PspCas13b
The split protein strategy offers a unique way to conditionally control the activities and functions of chosen proteins with specific triggering signals.12, 41–44 Previous studies have shown that Cas9 and Cas12a proteins can be split into two fragments and reconstituted upon rapamycin-induced dimerization for targeted gene silencing.43–44 We hypothesize that the inducible assembly of split Cas13b can be achieved similarly. This inducible system was designed by splitting PspCas13b protein into the N- and C-terminal fragments, which were fused separately with PYL and ABI, two inducer-binding protein domains that can be heterodimerized by ABA (Fig. 1). A suitable split PspCas13b pair should fulfill the following requirements. First, neither of the two split protein fragments can bind the sgRNA by itself and target the RNA of interest. Second, these two fragments cannot self-assemble effectively in the absence of ABA. Third, the intact and functional Cas13b can be efficiently assembled from these split fragments upon ABA induction. Last, the reconstituted Cas13b should be disassembled rapidly and released from the target RNA after ABA removal.
Figure 1.
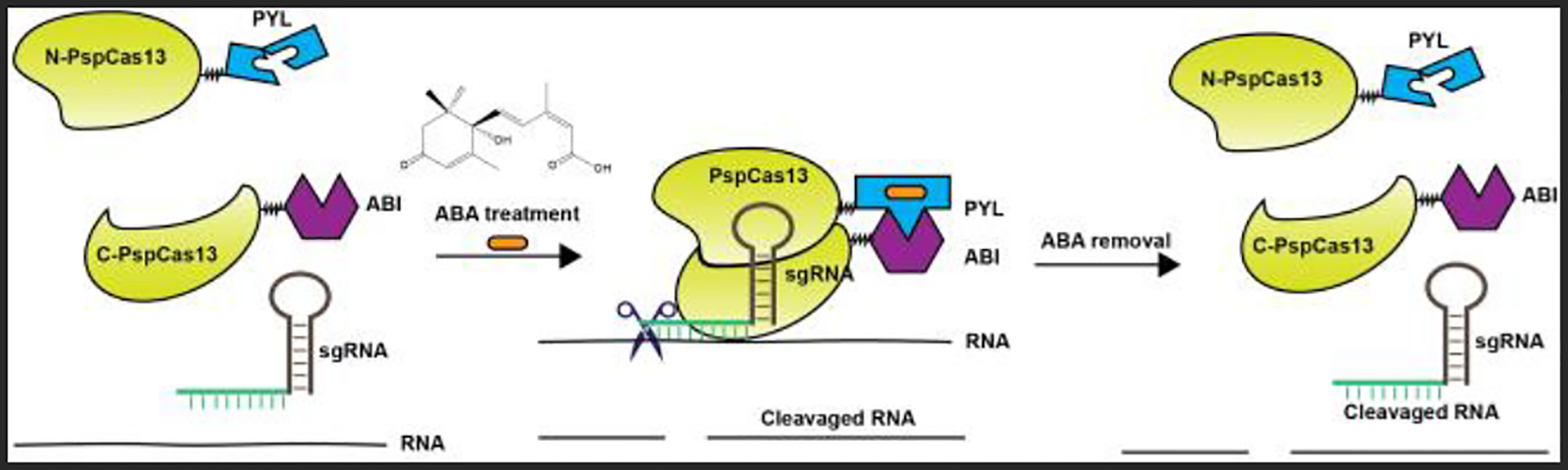
The chemically inducible split PspCas13b platform. The N- and C-terminal of PspCas13b are fused to PYL and ABI separately. The addition of ABA initiates the heterodimerization of PYL and ABI, which induces the reconstitution of the functional PspCas13b protein from its two fragments for RNA cleavage. When ABA is removed, the reconstituted PspCas13b will be dissembled and released from the target RNA.
Screening and testing the chemically inducible split PspCas13b pair
To identify the optimal split site on PspCas13b to generate the inducible split Cas13b pair, different split sites will be screened on the wild type PspCas13b based on its ribonuclease activity. The reconstituted nuclease active Cas13b from the inactive halves under ABA induction was expected to cleave the target RNA in the presence of sgRNA; in contrast, no assembly of the functional Cas13b should occur without ABA trigger, therefore, no RNA cleavage should be observed (Fig. 1).
Because the crystal structure of PspCas13b is unknown, an alignment analysis using the Phyre2 web portal45 was performed to predict PspCas13b structure by comparing its amino acid sequences to those of PbuCas13b whose structure has been reported.46 Based on the predicted structure of PspCas13b (or Cas13b hereafter), we chose 10 potential split sites including the ones within the catalytic or sgRNA binding domains as well as some random sites in the unstructured regions (Fig. S1). Accordingly, 10 pairs of plasmids expressing the N-Cas13b-PYL and ABI-C-Cas13b tagged with HA and Flag separately were successfully cloned (Fig. 2a).
Figure 2.
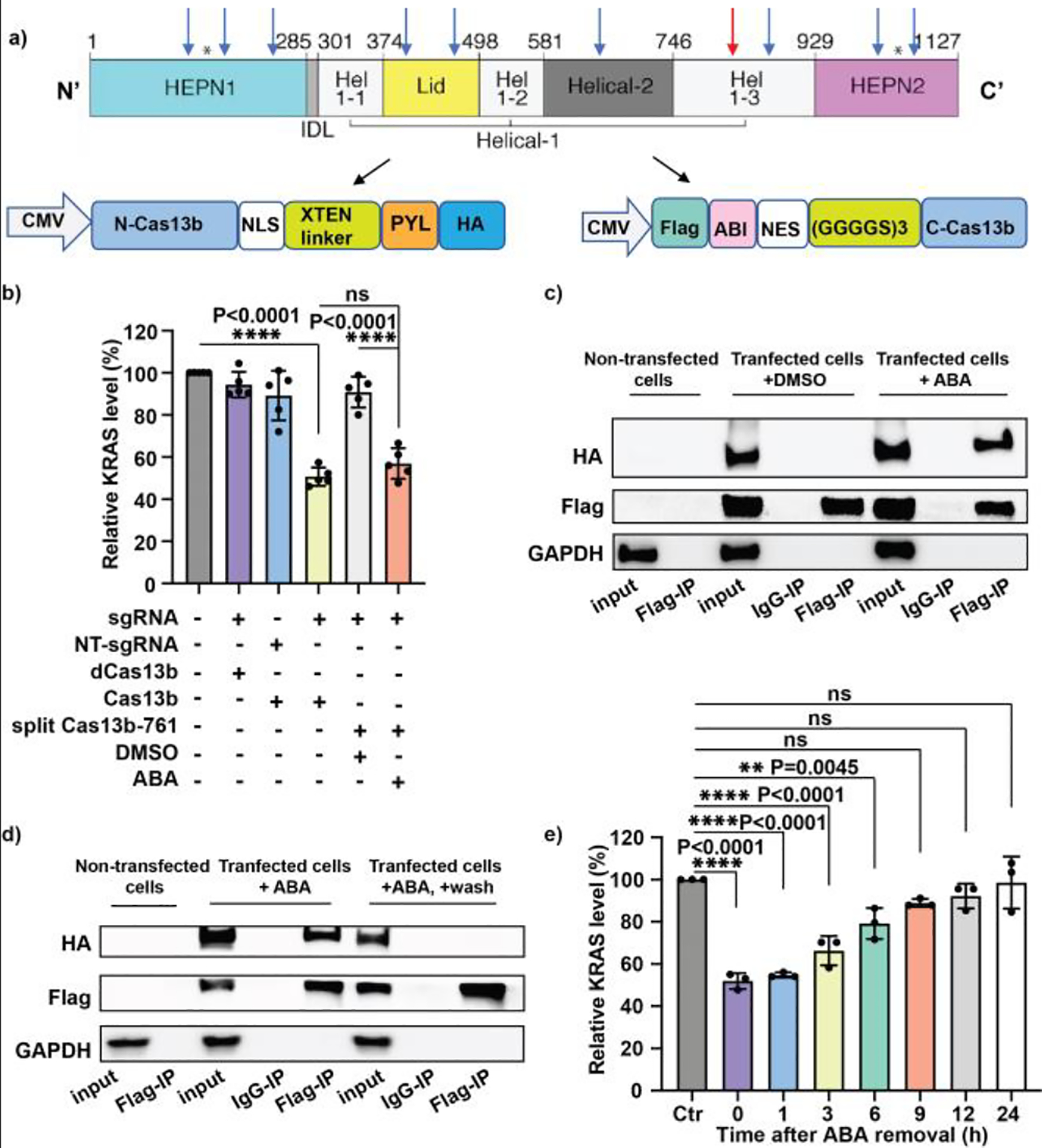
The development and validation of the ABA-inducible split Cas13b system. (a) The protein domains of Cas13b and DNA constructs of the split Cas13b fusion protein. The arrows indicate the 10 split sites to give each N- and C-terminal pair. The red arrow indicates the amino acid 761 site. The N- and C-terminal fragments of Cas13b were sub-cloned into the constructs as shown. (b) Downregulation of KRAS mRNA using the split Cas13b-761 pair with 100 μM ABA. (c) The ABA-dependent interaction of split Cas13b-761 fusion proteins determined by the co-IP assay. (d) The interaction between the split Cas13b-761 pair after ABA removal determined by the co-IP assay. (e) The time-course of KRAS mRNA silencing mediated by the split Cas13b-761 upon ABA withdrawn. The Flag-tagged C-terminus of Cas13b fusion protein was pulled down and analyzed to detect its physical interaction with the HA-tagged N-terminus of Cas13b fusion protein by western blotting in (c) and (d). Values and error bars in (b) and (e) represent the mean and s.e.m. of 5 or 3 independent biological experiments. The KRAS mRNA percentage changes were calculated by normalizing to the results in the non-transfected and non-treated cells (Ctr). P values were determined by one-way ANOVA.
Next, we tested the protein expression of these split Cas13b pairs in human embryonic kidney 293T (HEK293T) cells. We found that all split protein fragments can be expressed in cells, although at varied levels (Fig. S2). Then, we tested the RNA silencing efficiency of all 10 split Cas13b pairs with the sgRNA targeting KRAS mRNA in HEK293T cells. We observed that some split pairs reduced KRAS mRNA levels in cells treated with ABA or DMSO (Fig. S3), indicating the spontaneous self-assembly of functional Cas13b from these pairs. Other pairs failed to decrease KRAS mRNA levels even in the presence of ABA, suggesting that these pairs cannot be assembled into a functional Cas13b upon ABA induction. Out of the 10 pairs tested, the pair generated by splitting at amino acid 761 of Cas13b resulted in the downregulation of KRAS mRNA only in the presence of ABA with an efficiency comparable to that of the intact Cas13b, while ABA itself did not impact the level of KRAS mRNA (Fig. 2b and Fig. S3). These results suggested that a chemically inducible split Cas13b platform was successfully established by splitting at amino acid 761 of Cas13b (i.e., split Cas13b-761).
To verify the physical interaction of two fragments of the split Cas13b-761 pair in the presence of ABA, we transfected cells with N-Cas13b-PYL-HA and Flag-ABI-C-Cas13b plasmids, followed by ABA or DMSO treatment. Cell lysates were then collected and subjected to co-immunoprecipitation (co-IP) experiments. We found that when pulling down the Flag-tagged C-terminal fragment, the HA-tagged N-terminal fragment can be co-immunoprecipitated only when cells were treated with ABA (Fig. 2c), indicating that these two fragments can physically associate only in the presence of ABA. Taken together, the above mRNA silencing studies and co-IP experiments confirmed that the split Cas13b-761 pair can be effectively assembled into a functional Cas13b to cleave the target RNA under ABA induction.
Dosage and temporal controls of split Cas13b assembly and RNA cleavage induced by ABA
We further investigated the dosage dependence and the time course of ABA-induced RNA cleavage mediated by the split Cas13b-761 system. We observed a clear ABA dosage-dependent downregulation of KRAS mRNA in cells treated with ABA at different concentrations (Fig. S4). Moreover, the time-course study of the KRAS mRNA silencing revealed that KRAS mRNA started to reduce as early as 2 h post ABA treatment and continued to decrease during the 48 h-observation period (Fig. S5). These results demonstrated that the activity of the split Cas13b system can be precisely controlled by ABA in dosage- and time-dependent manners.
Disassembly of split Cas13b after ABA removal
The ABA-induced binding between ABI and PYL fusion proteins are readily reversible upon ABA removal.11, 47 To investigate if the ABA-induced assembly of the Cas13b-761 pair can be reversed after ABA removal, co-IP experiments were performed. Cells transfected with the split Cas13b-761 pair were then treated with 100 μM of ABA for 24 h, followed by a washing step (replacing with fresh media) to remove ABA. After another 24 h incubation, cells were harvested for the co-IP study. We observed that while the HA-tagged N-terminal fragment can be co-immunoprecipitated with the Flag-tagged C-terminal fragment in the presence of ABA, this interaction was lost after ABA was removed (Fig. 2d).
Next, we investigated if the ABA removal can also lead to the reversal of RNA silencing. After KRAS mRNA was knocked down by split Cas13–761 pair upon ABA induction, ABA was removed and cells were harvested at different time points to determine KRAS mRNA levels. We observed that the KRAS mRNA de-repressed over time after ABA withdrawal and its level was fully restored after 9 h (Fig. 2e). These results showed that the RNA silencing mediated by the inducible split Cas13b system can be reversed after removing ABA. Overall, the findings from the co-IP and the RNA knock-down experiments suggested that the reconstituted Cas13b fusion protein can be disassembled and detached from the targeted RNA upon ABA removal.
Light-induced activation of the split Cas13b system
Besides small molecules, light has been shown as another triggering signal to manipulate the activity of Cas protein.23, 28, 48 To enable the optical control of the split Cas13b platform, a synthetic photo-caged ABA (ABA-DMNB) was used, which can be uncaged under light irradiation and generate active ABA.40 To test the light control of the split Cas13b-761 activity, cells were transfected with the split Cas13b-761 pair and sgRNA for 24 h, followed by treatment of ABA, pre-cleaved ABA-DMNB and ABA-DMNB with or without light exposure (365 nm light for 2 min). After another 24 h, cells were harvested to determine the KRAS mRNA levels. In cells treated with ABA-DMNB, the downregulation of KRAS mRNA was only observed when cells were exposed to light (Fig. S6). These results demonstrated that the light inducible split Cas13b platform was successfully developed.
Engineering a split dCas13b-M3 system for inducible m6A writing
Encouraged by the results of the inducible split Cas13b system described above, we further generated a catalytic inactive split dCas13b system. To demonstrate the utility of this system, we applied this platform to achieve site-specific m6A editing. m6A is the most prevalent chemical modifications in eukaryotic RNAs, which is dynamically regulated by various m6A effectors in the cells.49–50. It is known that m6A can impact the RNA fate and activities in a context-dependent manner.10, 51–54 Therefore, tools enabling programmable m6A editing in transcript and site-specific manners will be invaluable for dissecting the function of m6A. So far, several CRISPR dCas13-based methods have been reported, which recruit different m6A effectors to specific RNA transcripts/loci to achieve targeted m6A editing.10, 21–22, 28–29, 55–56 However, in these methods, the bulky dCas13 fusion proteins remain constitutively bound to the target RNAs, which can disrupt the interactions between various endogenous effectors and these RNAs or the installed m6A modification. We hypothesize that a split dCas13b-based m6A editing platform allowing conditional assembly of the dCas13b fusion proteins on the target RNA can address this concern.
To develop an inducible and reversible “traceless” m6A editing system, a split dCas13b protein pair was engineered by splitting the full length dCas13b (the H133A/H1058A mutant of Cas13b) at amino acid 761. We cloned plasmids with the N-terminal fragment (1–761 aa) of dCas13b fused to PYL and an HA tag, while the C-terminal fragment (762–1090 aa) was fused to Flag-tagged ABI and a truncated METTL3 without the RNA-binding motif (Fig. 3a). It is expected that the assembly and disassembly of the split dCas13b-METTL3 (or split dCas13b-M3 hereafter) on the target RNA can be controlled by the addition and removal of ABA to achieve inducible and reversible site-specific m6A writing (Fig. S7). The expression of these split dCas13b-M3 fusion proteins was confirmed by western blotting (Fig. S8). The ABA inducible m6A writing activity of the split dCas13b-M3 system was tested at the A1216 site of ACTB mRNA in HEK293T cells, which was shown to be amenable to m6A deposition.10, 29 We transfected cells with ACTB-targeting sgRNA and the split dCas13b-M3. The non-split version of dCas13b-M3 and dCas13-PYL were used as positive and negative controls. Cells were then treated with 100 μM ABA or DMSO for another 24 h before harvested for subsequent methylated RNA immunoprecipitation with anti-m6A antibody (m6A-MeRIP) and qPCR assays. We observed that m6A levels on ACTB mRNA increased only when ABA was added to cells transfected with the split dCas13b-M3 system, and the elevated m6A level was comparable to that caused by intact dCas13b-M3 (Fig. 3b). To confirm that the observed m6A writing occurred selectively on the targeted ACTB mRNA, the m6A levels at other RNA loci (e.g., GAPDH A690, FOXM1 A3488/A3504, and SOX2 A1398/A1405 sites) not targeted by the ACTB sgRNA were also quantified.29 As expected, we did not find the increase of m6A level at these sites (Fig. S9). Taken together, these results suggested that the split dCas13b-M3 can be reconstituted into a fully functional m6A writer for site-specific m6A writing under ABA induction.
Figure 3.
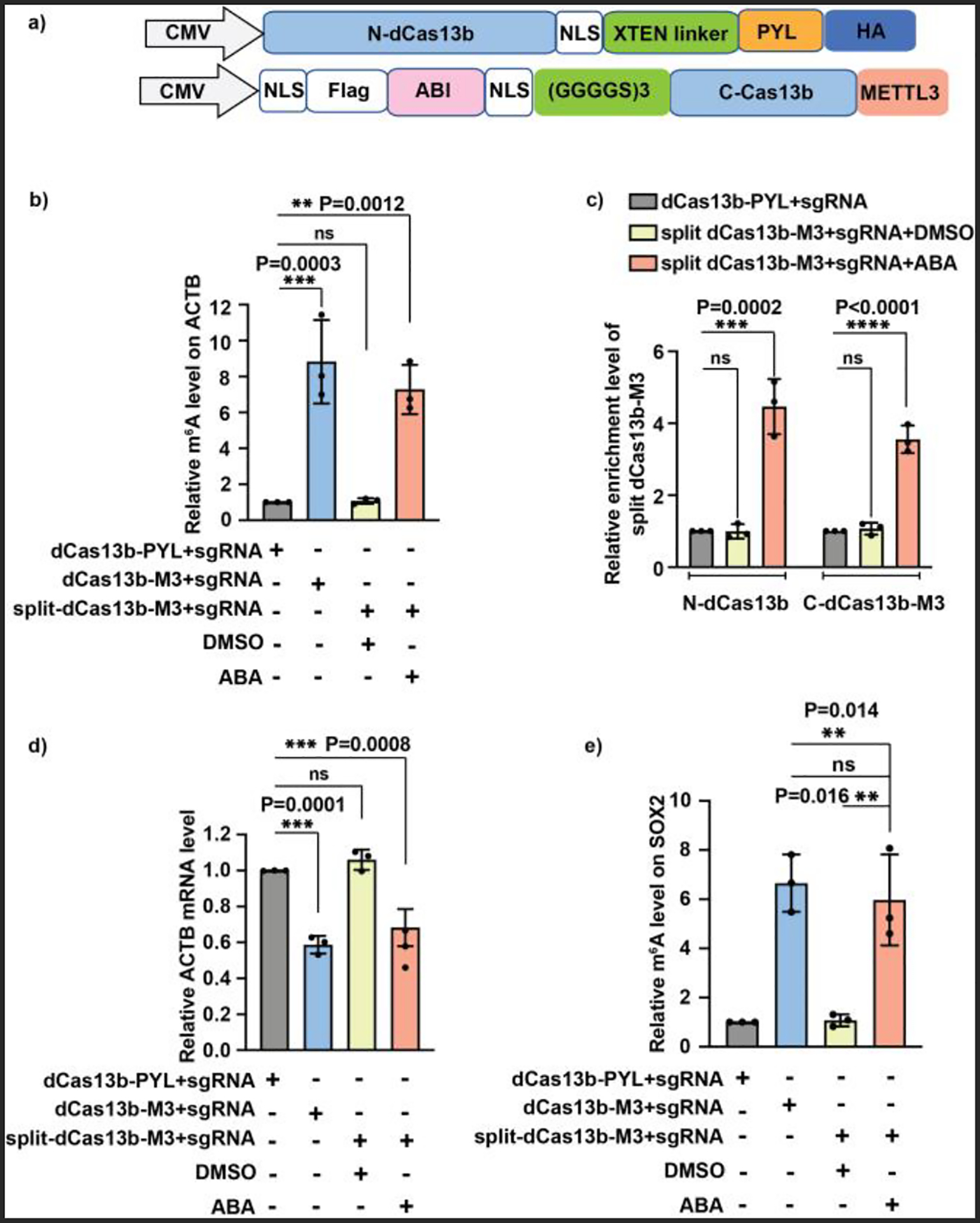
Site-specific m6A writing by the ABA-inducible split dCas13b-M3 platform. (a) The DNA constructs of split dCas13b-M3. (b) ABA-induced m6A writing on A1216 of ACTB mRNA by the split dCas13b-M3 platform. (c) ABA-dependent recruitment of the N- and C-terminal of split dCas13b-M3 to the ACTB mRNA A1216 site. (d) Impact of the induced m6A writing on the stability of ACTB mRNA. (e) Targeted m6A writing at A1398/A1405 site of SOX2 mRNA by the split dCas13b-M3 platform. All enrichment fold changes in (b) to (e) were calculated by normalizing to the results from the condition using dCas13b-PYL with sgRNA. Values and error bars represent the mean and s.e.m. of 3 independent biological experiments. P values were determined by one-way ANOVA.
ABA-dependent assembly of the split dCas13b-M3 on the target RNA
To further confirm that the assembly of the split dCas13b-M3 did occur at the targeted RNA site upon ABA addition, we quantified the enrichments of the HA-tagged N-terminal and Flag-tagged C-terminal of the split dCas13b-M3 at the targeted RNA locus by cross-linking immunoprecipitation (CLIP) and qPCR analyses. We found that neither the N- nor C-terminal fragment of the split dCas13b-M3 was enriched at the target ACTB site in the absence of ABA (Fig. 3c). However, both of them were significantly enriched at the target RNA site upon ABA addition. These results confirmed that the split dCas13b-M3 were assembled and localized at the targeted locus in an ABA-dependent manner.
m6A-dependent RNA destabilization via the split dCas13b-M3 controlled by ABA
The m6A at A1216 locus of ACTB mRNA has been reported to destabilize the mRNA.10, 29 To confirm that the artificially installed m6A by the split dCas13b-M3 can lead to the same biological outcome, cells were transfected with the split dCas13b-M3 and sgRNA, followed by ABA or DMSO treatment. Then cells were treated with 5 μg/ml actinomycin D to inhibit RNA synthesis before harvested to determine the mRNA level. As expected, the ACTB mRNA level was significantly reduced only when cells were treated with ABA (Fig. 3d), and the ABA-induced mRNA decrease was comparable to the destabilization caused by the intact dCas13b-M3. These results confirmed that the m6A deposited by the inducible split dCas13b-M3 platform was biologically relevant and can regulate the fate of target RNA under ABA control.
Customizable m6A writing for different RNAs using the inducible split dCas13b-M3 platform
To demonstrate that this inducible split dCas13b-based m6A writing platform can be applied to edit m6A on other RNAs, we designed a new sgRNA to target the A1398/A1405 area of SOX2 mRNA by switching the spacer sequence in ACTB sgRNA.29 We observed that the m6A level at the A1398/A1405 sites on SOX2 mRNA increased only when ABA was added to cells transfected with the split dCas13b-M3 and SOX2 sgRNA (Fig. 3e). This increased level was comparable to the m6A increase caused by intact dCas13b-M3. To further confirm that the m6A writing occurred selectively at SOX2 mRNA, we examined the m6A levels on other RNA loci not targeted by the SOX2 sgRNA and found no obvious changes of m6A levels at those loci (Fig. S10). These results demonstrated that the inducible split dCas13b-M3 platform can be easily tailored to achieve site-specific m6A modification on different RNAs by customizing the sgRNA spacer sequences.
Temporal control on the assembly of split dCas13b-M3 and m6A writing
To characterize the kinetics of split dCas13b-M3 assembly and the m6A writing as well as its biological impact in response to ABA induction, we investigated the recruitment of each half of the split dCas13b-M3 and the m6A level at the A1216 locus on ACTB mRNA as well as the m6A-mediated RNA destabilization in time-course experiments after ABA addition. We found that both fragments of the split dCas13b-M3 were significantly enriched at the targeted site as early as 1 h post ABA treatment. Such enrichment continued to increase gradually within 24 h (Fig. 4a and 4b). Correspondingly, we observed a significant increase of the m6A level within 1 h after ABA addition and its level increased in a time-dependent manner (Fig. 4c), which correlated well with the kinetics of the split dCas13b-M3 assembly. Moreover, we found that the destabilization of ACTB mRNA mediated by m6A occurred rapidly after 3 h post ABA treatment, and the mRNA level continued to decrease over time (Fig. 4d). Taken together, the inducible split dCas13b-M3 platform enabled rapid and temporal control of the site-specific m6A deposition and the induced biological impact on target RNA.
Figure 4.
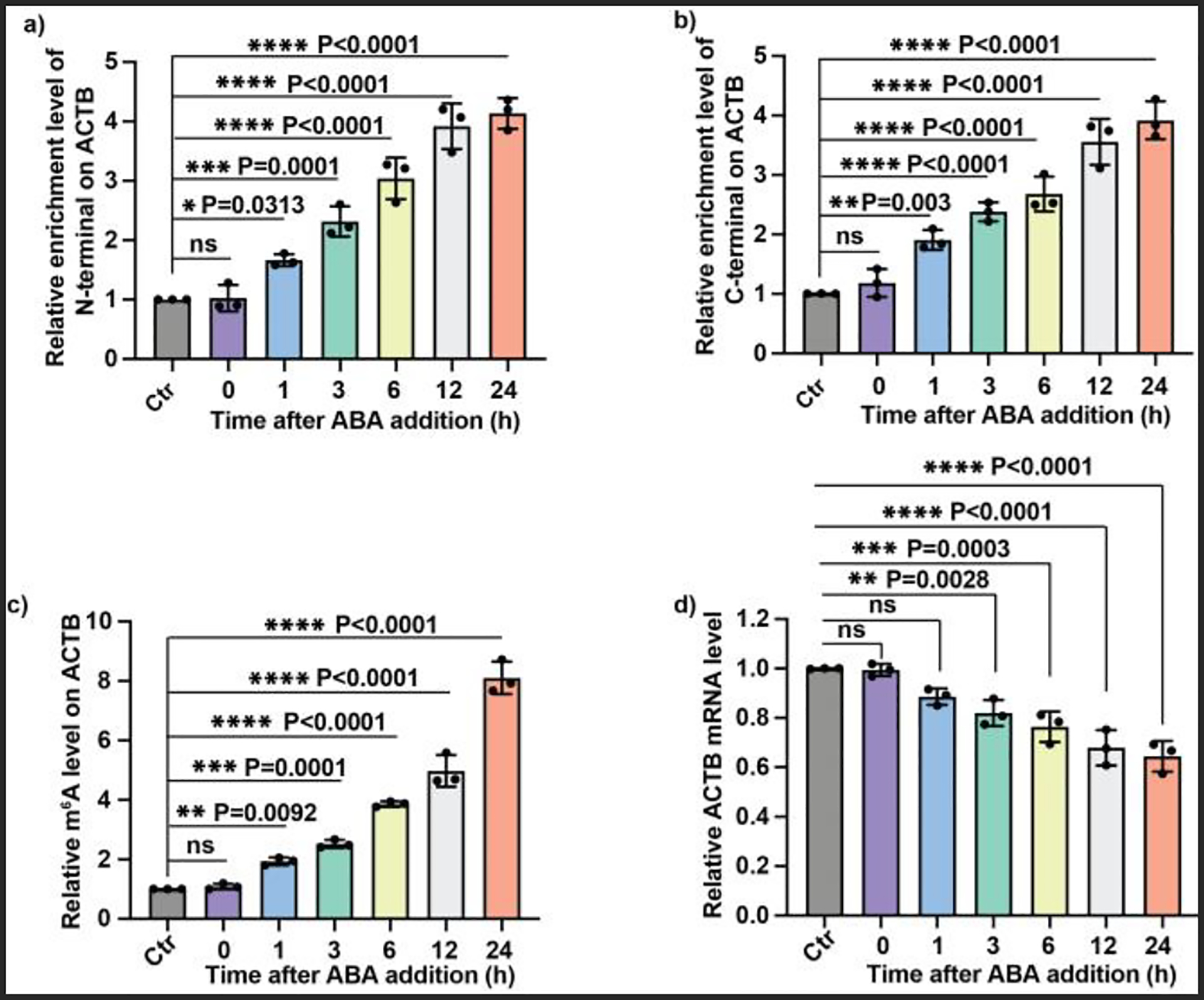
Kinetics of split dCas13b-M3 assembly-mediated m6A writing and the impact on RNA stability induced by ABA. Enrichment of the HA-tagged N-terminal fragment (a) or the Flag-tagged C-terminal fragment (b) of the split dCas13b-M3 on ACTB mRNA at different time points after ABA addition. (c) The relative m6A level at A1216 of ACTB mRNA at different time points after the ABA addition. (d) The time-course impact of m6A on the stability of ACTB mRNA. All relative enrichment levels were calculated by normalizing to the results from the group of dCas13b-PYL with sgRNA (Ctr). Values and error bars represent the mean and s.e.m. of 3 independent biological experiments. P values were determined by one-way ANOVA.
Reversibility and kinetics of the split dCas13b-M3 mediated m6A writing
To investigate the reversibility of the inducible split dCas13b-M3 system, the enrichment of the split dCas13b-M3 fragments and the corresponding m6A level on ACTB mRNA after ABA withdrawal were studied. We found that the abundance of both fragments at the A1216 region of ACTB mRNA decreased rapidly and significantly within 1 h after ABA removal and reached background levels after 3 h (Fig. 5a and 5b). Interestingly, the m6A level reduced at a much slower pace after ABA removal, which returned to the background level after 24 h (Fig. 5c). The observed differential kinetics between the release of split dCas13b-M3 from RNA and the decrease of installed m6A on RNA offers a wide temporal window and an optimal opportunity to investigate m6A functions in a native environment without the potential interference from the bulky dCas13b fusion protein.
Figure 5.
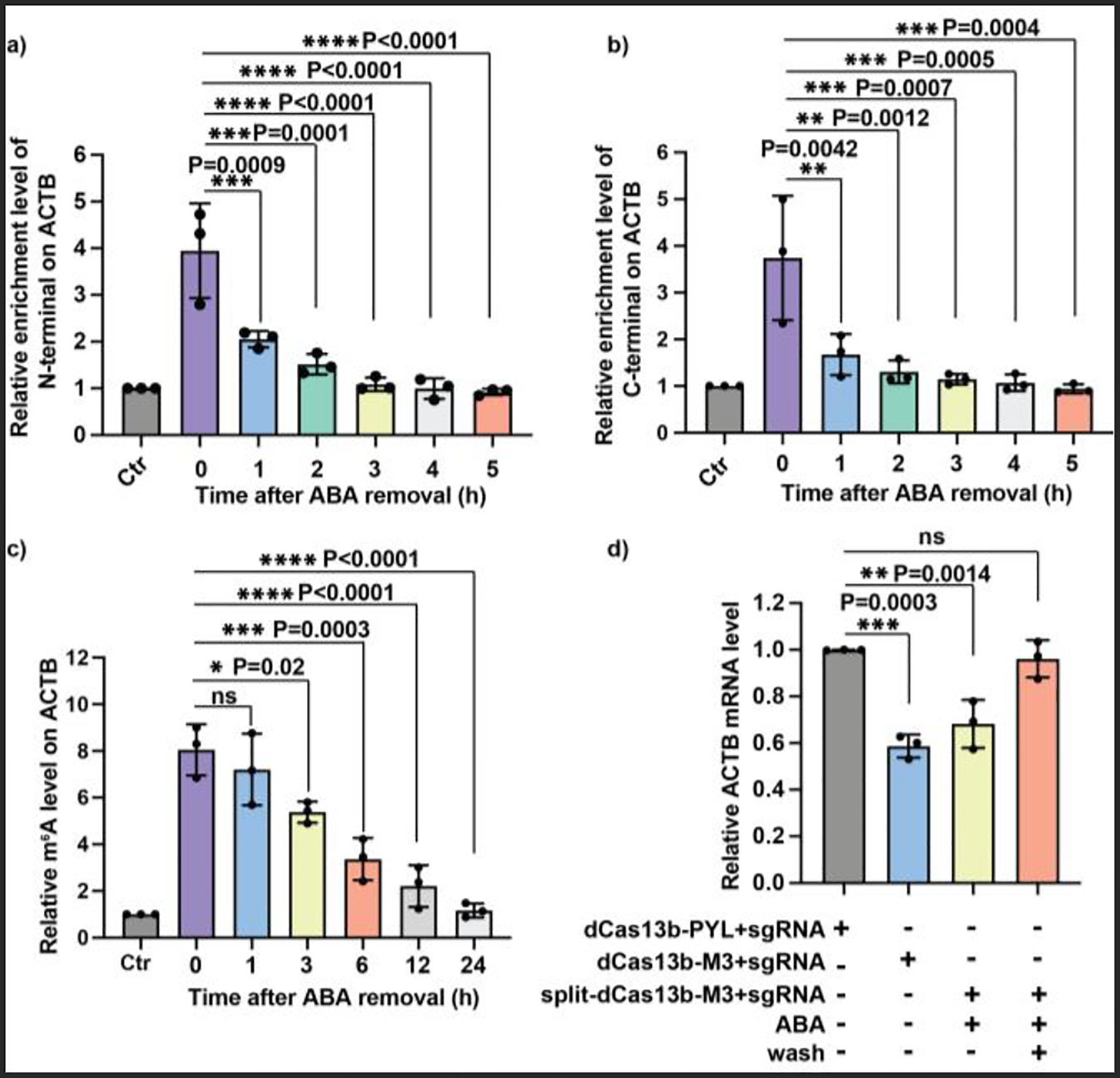
Reversibility of the split dCas13b-M3-mediated m6A writing. The relative enrichment levels of the HA-tagged N-terminal fragment (a) and the Flag-tagged C-terminal fragment (b) of the split dCas13b-M3 on ACTB mRNA at different time points after ABA removal. (c) The m6A level at the A1216 site of ACTB mRNA at different time points after ABA removal. (d) Reversibility of m6A-induced ACTB mRNA destabilization. All the relative enrichment and mRNA levels were calculated by normalizing to results from the group of dCas13b-PYL with sgRNA (Ctr). Values and error bars represent the mean and s.e.m. of 3 independent biological experiments. P values were determined by one-way ANOVA.
Furthermore, the reversibility of the m6A-dependent ACTB mRNA destabilization was investigated. We observed that the decreased ACTB mRNA level was restored after ABA removal (Fig. 5d). These results suggested that the m6A-mediated biological effect regulated by the split dCas13b-M3 was not only inducible but also reversible.
Light-controlled m6A writing through the split dCas13b-M3 platform
To validate the ABA-DMNB-mediated light control of the split dCas13b-M3 platform, cells were transfected with split dCas13b-M3 and sgRNA for 24 h before treated with ABA-DMNB and exposed to 365 nm light for 2 min. After another 24 h, cells were harvested to test the light-inducible m6A writing at A1216 of ACTB mRNA. We observed that m6A level was elevated only when cells were exposed to UV light (Fig. S11). These results demonstrated that the light control of the split dCas13b-M3 platform can be successfully achieved. As light has been broadly applied to provide spatiotemporal control of various biological activities, the light-inducible split Cas13b/dCas13b system can potentially offer spatial-specific RNA manipulation in suitable applications.
Conclusions
To expand the CRISPR toolkits and address the critical issue of steric interference with target RNAs from dCas13b fusion proteins, we developed novel small molecule- and light-regulated split Cas13b/dCas13b platforms with inducible and reversible assembly of functional Cas13b/dCas13b complex to achieve conditional RNA silencing and site-specific m6A editing on RNA. Through time course studies, we found that the assembly of functional Cas13b and dCas13b-M3 occurred rapidly upon ABA addition and can be reversed after ABA removal. Most interestingly, the observed release of dCas13b complex was much faster than the diminishment of m6A on the target RNA, which can be essential for studying the native cellular function of m6A modification without perturbation from the bulky dCas13b proteins. Other approaches have also been reported to overcome the size issue of Cas protein for targeted RNA manipulation, specifically the CIRTS system,36–37 which incorporates a much smaller protein component. The CIRTS-based methods have the advantages of small protein size and minimal immunogenicity, which are crucial properties for in vivo applications when compared to current CRISPR methods. However, both CIRTS and existing CRISPR methods will have proteins components constitutively bound on the target RNA. Although the split Cas13b/dCas13b platform shares similar RNA targeting efficiency to the CIRTS and non-split Cas13b systems based on this study and previous reports,36–37 the spatiotemporally controlled assembly and disassembly of the split Cas13b/dCas13b alleviate the concerns of persistent protein binding and perturbation on targeted RNAs. Therefore, this new split Cas13b/dCas13b method provides an alternative strategy to minimize the unwanted interference with target RNAs introduced by current methods in RNA studies. Taken together, we established unique inducible and reversible split Cas13b/dCas13b systems for conditional RNA downregulation and traceless site-specific m6A editing. We expect that these inducible split Cas13b/dCas13b systems will expand the utility of RNA-targeting CRISPR system, which have a significant impact on understanding and manipulating RNA in cells.
Supplementary Material
ACKNOWLEDGMENT
This work is supported by the National Institutes of Health grant R21CA247638.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge on the ACS Publications website.
The details of experimental methods and the supplementary figures are included in the Supporting Information.
The authors declare no competing interest.
REFERENCES
- 1.Barrangou R; Fremaux C; Deveau H; Richards M; Boyaval P; Moineau S; Romero DA; Horvath P, CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315 (5819), 1709–1712. [DOI] [PubMed] [Google Scholar]
- 2.Makarova KS; Wolf YI; Alkhnbashi OS; Costa F; Shah SA; Saunders SJ; Barrangou R; Brouns SJ; Charpentier E; Haft DH; Horvath P; Moineau S; Mojica FJ; Terns RM; Terns MP; White MF; Yakunin AF; Garrett RA; van der Oost J; Backofen R; Koonin EV, An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 2015, 13 (11), 722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickar-Oliver A; Gersbach CA, The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 2019, 20 (8), 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang F; Doudna JA, CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 2017, 46, 505–529. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura M; Gao Y; Dominguez AA; Qi LS, CRISPR technologies for precise epigenome editing. Nat Cell Biol 2021, 23 (1), 11–22. [DOI] [PubMed] [Google Scholar]
- 6.Mali P; Esvelt KM; Church GM, Cas9 as a versatile tool for engineering biology. Nature methods 2013, 10 (10), 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sander JD; Joung JK, CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014, 32 (4), 347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox DBT; Gootenberg JS; Abudayyeh OO; Franklin B; Kellner MJ; Joung J; Zhang F, RNA editing with CRISPR-Cas13. Science 2017, 358 (6366), 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C; Zhou Y; Xiao Q; He B; Geng G; Wang Z; Cao B; Dong X; Bai W; Wang Y; Wang X; Zhou D; Yuan T; Huo X; Lai J; Yang H, Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat Methods 2021, 18 (5), 499–506. [DOI] [PubMed] [Google Scholar]
- 10.Wilson C; Chen PJ; Miao Z; Liu DR, Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol 2020, 38 (12), 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen T; Gao D; Zhang R; Zeng G; Yan H; Lim E; Liang F-S, Chemically controlled epigenome editing through an inducible dCas9 system. Journal of the American Chemical Society 2017, 139 (33), 11337–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X; Dong K; Kong D; Zhou Y; Yin J; Cai F; Wang M; Ye H, A far-red light-inducible CRISPR-Cas12a platform for remote-controlled genome editing and gene activation. Sci Adv 2021, 7 (50), eabh2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breinig M; Schweitzer AY; Herianto AM; Revia S; Schaefer L; Wendler L; Cobos Galvez A; Tschaharganeh DF, Multiplexed orthogonal genome editing and transcriptional activation by Cas12a. Nat Methods 2019, 16 (1), 51–54. [DOI] [PubMed] [Google Scholar]
- 14.Smargon AA; Cox DBT; Pyzocha NK; Zheng K; Slaymaker IM; Gootenberg JS; Abudayyeh OA; Essletzbichler P; Shmakov S; Makarova KS; Koonin EV; Zhang F, Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell 2017, 65 (4), 618–630 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patchsung M; Jantarug K; Pattama A; Aphicho K; Suraritdechachai S; Meesawat P; Sappakhaw K; Leelahakorn N; Ruenkam T; Wongsatit T; Athipanyasilp N; Eiamthong B; Lakkanasirorat B; Phoodokmai T; Niljianskul N; Pakotiprapha D; Chanarat S; Homchan A; Tinikul R; Kamutira P; Phiwkaow K; Soithongcharoen S; Kantiwiriyawanitch C; Pongsupasa V; Trisrivirat D; Jaroensuk J; Wongnate T; Maenpuen S; Chaiyen P; Kamnerdnakta S; Swangsri J; Chuthapisith S; Sirivatanauksorn Y; Chaimayo C; Sutthent R; Kantakamalakul W; Joung J; Ladha A; Jin X; Gootenberg JS; Abudayyeh OO; Zhang F; Horthongkham N; Uttamapinant C, Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng 2020, 4 (12), 1140–1149. [DOI] [PubMed] [Google Scholar]
- 16.Abudayyeh OO; Gootenberg JS; Essletzbichler P; Han S; Joung J; Belanto JJ; Verdine V; Cox DBT; Kellner MJ; Regev A; Lander ES; Voytas DF; Ting AY; Zhang F, RNA targeting with CRISPR-Cas13. Nature 2017, 550 (7675), 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellner MJ; Koob JG; Gootenberg JS; Abudayyeh OO; Zhang F, SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 2019, 14 (10), 2986–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terns MP, CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol Cell 2018, 72 (3), 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konermann S; Lotfy P; Brideau NJ; Oki J; Shokhirev MN; Hsu PD, Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173 (3), 665–676 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abudayyeh OO; Gootenberg JS; Franklin B; Koob J; Kellner MJ; Ladha A; Joung J; Kirchgatterer P; Cox DBT; Zhang F, A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365 (6451), 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch S; He C; Dickinson BC, Targeted m(6)A Reader Proteins To Study Epitranscriptomic Regulation of Single RNAs. J Am Chem Soc 2018, 140 (38), 11974–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J; Chen Z; Chen F; Xie G; Ling Y; Peng Y; Lin Y; Luo N; Chiang C-M; Wang H, Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic acids research 2020, 48 (10), 5684–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuñez JK; Harrington LB; Doudna JA, Chemical and biophysical modulation of Cas9 for tunable genome engineering. ACS chemical biology 2016, 11 (3), 681–688. [DOI] [PubMed] [Google Scholar]
- 24.Davis KM; Pattanayak V; Thompson DB; Zuris JA; Liu DR, Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nature chemical biology 2015, 11 (5), 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W; Deiters A, Conditional control of CRISPR/Cas9 function. Angewandte Chemie International Edition 2016, 55 (18), 5394–5399. [DOI] [PubMed] [Google Scholar]
- 26.Gangopadhyay SA; Cox KJ; Manna D; Lim D; Maji B; Zhou Q; Choudhary A, Precision control of CRISPR-Cas9 using small molecules and light. Biochemistry 2019, 58 (4), 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X; Chen X; Fang Q; Li J; Bai Z, Inducible CRISPR genome-editing tool: classifications and future trends. Crit Rev Biotechnol 2018, 38 (4), 573–586. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J; Li B; Ma J; Jin W; Ma X, Photoactivatable RNA N6‐Methyladenosine Editing with CRISPR‐Cas13. Small 2020, 16 (30), 1907301. [DOI] [PubMed] [Google Scholar]
- 29.Shi H; Xu Y; Tian N; Yang M; Liang F-S, Inducible and reversible RNA N6-methyladenosine editing. Nature Communications 2022, 13 (1), 1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghavami S; Pandi A, CRISPR interference and its applications. Prog Mol Biol Transl Sci 2021, 180, 123–140. [DOI] [PubMed] [Google Scholar]
- 31.Qi LS; Larson MH; Gilbert LA; Doudna JA; Weissman JS; Arkin AP; Lim WA, Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152 (5), 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis CJ; Pan T; Kalsotra A, RNA modifications and structures cooperate to guide RNA–protein interactions. Nature reviews Molecular cell biology 2017, 18 (3), 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hentze MW; Castello A; Schwarzl T; Preiss T, A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 2018, 19 (5), 327–341. [DOI] [PubMed] [Google Scholar]
- 34.Corley M; Burns MC; Yeo GW, How RNA-binding proteins interact with RNA: molecules and mechanisms. Molecular cell 2020, 78 (1), 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N; Dai Q; Zheng G; He C; Parisien M; Pan T, N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518 (7540), 560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauch S; He E; Srienc M; Zhou H; Zhang Z; Dickinson BC, Programmable RNA-Guided RNA Effector Proteins Built from Human Parts. Cell 2019, 178 (1), 122–134 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch S; Jones KA; Dickinson BC, Small Molecule-Inducible RNA-Targeting Systems for Temporal Control of RNA Regulation. ACS Cent Sci 2020, 6 (11), 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer KD; Jaffrey SR, The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014, 15 (5), 313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J; Yue Y; Han D; Wang X; Fu Y; Zhang L; Jia G; Yu M; Lu Z; Deng X; Dai Q; Chen W; He C, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014, 10 (2), 93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright CW; Guo ZF; Liang FS, Light control of cellular processes by using photocaged abscisic acid. ChemBioChem 2015, 16 (2), 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolberg TB; Meger AT; Boucher JD; Corcoran WK; Schauer EE; Prybutok AN; Raman S; Leonard JN, Computation-guided optimization of split protein systems. Nat Chem Biol 2021, 17 (5), 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shekhawat SS; Ghosh I, Split-protein systems: beyond binary protein-protein interactions. Curr Opin Chem Biol 2011, 15 (6), 789–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nihongaki Y; Otabe T; Ueda Y; Sato M, A split CRISPR-Cpf1 platform for inducible genome editing and gene activation. Nat Chem Biol 2019, 15 (9), 882–888. [DOI] [PubMed] [Google Scholar]
- 44.Zetsche B; Volz SE; Zhang F, A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 2015, 33 (2), 139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley LA; Mezulis S; Yates CM; Wass MN; Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015, 10 (6), 845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slaymaker IM; Mesa P; Kellner MJ; Kannan S; Brignole E; Koob J; Feliciano PR; Stella S; Abudayyeh OO; Gootenberg JS; Strecker J; Montoya G; Zhang F, High-Resolution Structure of Cas13b and Biochemical Characterization of RNA Targeting and Cleavage. Cell Rep 2019, 26 (13), 3741–3751 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanton BZ; Chory EJ; Crabtree GR, Chemically induced proximity in biology and medicine. Science 2018, 359 (6380). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y; Wu X; Guan N; Shao J; Li H; Chen Y; Ping Y; Li D; Ye H, Engineering a far-red light-activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci Adv 2020, 6 (28), eabb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y; Hsu PJ; Chen YS; Yang YG, Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018, 28 (6), 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boccaletto P; Stefaniak F; Ray A; Cappannini A; Mukherjee S; Purta E; Kurkowska M; Shirvanizadeh N; Destefanis E; Groza P; Avsar G; Romitelli A; Pir P; Dassi E; Conticello SG; Aguilo F; Bujnicki JM, MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 2022, 50 (D1), D231–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X; Lu Z; Gomez A; Hon GC; Yue Y; Han D; Fu Y; Parisien M; Dai Q; Jia G; Ren B; Pan T; He C, N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505 (7481), 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viegas IJ; de Macedo JP; Serra L; De Niz M; Temporão A; Silva Pereira S; Mirza AH; Bergstrom E; Rodrigues JA; Aresta-Branco F; Jaffrey SR; Figueiredo LM, N6-methyladenosine in poly(A) tails stabilize VSG transcripts. Nature 2022, 604 (7905), 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alarcon CR; Lee H; Goodarzi H; Halberg N; Tavazoie SF, N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519 (7544), 482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X; Zhao BS; Roundtree IA; Lu Z; Han D; Ma H; Weng X; Chen K; Shi H; He C, N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161 (6), 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia Z; Tang M; Ma J; Zhang H; Gimple RC; Prager BC; Tang H; Sun C; Liu F; Lin P; Mei Y; Du R; Rich JN; Xie Q, Epitranscriptomic editing of the RNA N6-methyladenosine modification by dCasRx conjugated methyltransferase and demethylase. Nucleic Acids Res 2021, 49 (13), 7361–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo J; Chen Z; Qin S; Li S; Liu C; Zhang L; Ran R; Kong Y; Wang F; Liu S; Zhou Y; Zhang X; Weng X; Zhou X, TRADES: Targeted RNA Demethylation by SunTag System. Adv Sci (Weinh) 2020, 7 (19), 2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


