Abstract
Advanced three-dimensional structure variations of chromatin in large genome fragments, such as conversion of A/B compartment, topologically associated domains (TADs) and chromatin loops are related closely to occurrence of malignant tumors. However, the structural characteristics of lung cancer still remain uncovered. In this study, we used high-throughput chromosome (Hi-C) conformation capture technology to detect the advanced structural variations in chromatin of two non-smoking lung adenocarcinoma (LUAD) tumor and paired normal tissues. The results indicate that significant chromatin variations are detected in tumor tissues compared with normal tissues. At compartment scale, the main conversion type of compartment is A → B in tumor tissues, which are concentrated mainly on chromosome 3 (Chr3) (33.6%). A total of 216 tumor-specific TADs are identified in tumor tissues, which are distributed mainly in Chr1 (19), Chr2 (15) and Chr3 (17). Forty-one distinct enhancer-promoter loops are observed in tumor tissue, which are associated closely to tumor-related pathways including mitogen-activated protein kinase (MAPK), Phosphatidylinositol-3-kinase-Protein kinase B (PI3K-AKT), Ras, Wnt and Ras1. The most important observation in this study is that we identify five important genes (SYT16, NCEH1, NXPE3, MB21D2, and DZIP1L), which are detected in both A → B compartment, TADs and chromatin loops in tumor samples, and four of these genes (NCEH1, NXPE3, MB21D2, and DZIP1L) locate on q arm of Chr3. Further gene expression and invasion experiment analysis show that NCEH1, MB21D2 and SYT16 are involved in the tumor development. Thus, we provide a comprehensive overview of advanced structures in LUAD for the first time and provide a basis for further research on the genetic variation of this tumor.
Keywords: High-throughput chromosome, Lung cancer, Advanced structural variations, Chromosome 3, Tumor-related genes
Introduction
Lung cancer is the malignant tumor with the highest morbidity and mortality, and non-small cell lung cancer (NSCLC) accounts for 80% in all lung cancer patients (Bray et al. 2018). Smoking is considered an important factor leading to lung cancer, but over 50% of female lung cancer patients have no history of smoking (Molina et al. 2008). The occurrence of lung cancer involves mutations of multiple tumor-related genes, including Tumor Protein P53 (TP53), Epidermal Growth Factor Receptor (EGFR), KRAS Proto-Oncogene (KRAS), etc., which can dramatically promote tumor development. Besides mutations in onco- and onco-suppressive genes, structure variations caused by chromatin instability, such as deletion, reversion, translocation, insertion and fusion, also act as critical roles in lung cancer development (Edwards 2010). In recent years, advanced conformational variations in large genome fragments, including A/B compartment, topologically associated domains (TADs) and chromatin loops, are also considered important in tumoregenesis (Allen et al. 2016; Valton and Dekker 2016; Zheng and Xie 2019).
High-throughput chromosome (Hi-C) conformation capture is a novel technology that combines high-throughput sequencing and chromatin conformation capture for quantifying long-range physical interactions, and detecting large fragments and advanced conformational variations, including A/B compartment, TADs and chromatin loops at the genome scale (Spielmann et al. 2018). A/B compartment is that each chromosome occupies a separate space (chromosome territory) in the interphase nucleus and is partitioned into distinct neighborhoods or compartments (Lieberman-Aiden et al. 2009). And interphase chromosomes can partitioned into megabase-sized topological domains and smaller sub-domains (known as topologically associated domains or TADs), form the basis for higher-level structures referred to as the ‘A’ and ‘B’ compartments (Barutcu et al. 2018; Chang et al. 2020). TAD has been reported to be relatively stable in different cells or tissues, and CCCTC-binding factor (CTCF) and housekeeping genes are enriched in the TAD boundary, while H3K4me3, H3K36me3, transcription start site (TSS) and short interspersed nuclear elements (SINE) repeating elements, etc. are enriched in TAD. The chromatin loop is formed by chromatin interactions between distal cis-regulatory elements to allow modulation of promoter activity by enhancers (Grubert et al. 2020; Rao et al. 2014).
Hi-C is now applied in multiple tumor cell lines to detect specific advanced chromatin variations. In multiple myeloma (MM) cell lines (RPMI-8226 and U266), more than 100 chromosome interactions and the A/B compartment switch regions closely related to mitogen-activated protein kinase (MAPK) signaling and cytokine-cytokine receptor interaction pathways were detected by combining ChIP-seq and whole genome sequence (WGS) (Wu et al. 2017). In prostate cancer, Hi-C and WGS were performed in normal prostate epithelial cell line PrEC and two prostate cancer cell lines (PC3 and LNCaP), which revealed that TADs were more numerous in tumor cells and copy number variations associate with formation of new TADs (Taberlay et al. 2016). In another study about prostate cancer, it also identified specific enhancer-promoter loops in prostate cancer cell line (C42B and 22Rv1) (Rhie et al. 2019). All these results suggest that advanced structure variations (A/B compartment, TADs and chromatin loop) may play important roles in tumor development.
However, our knowledge of advanced structure variations in malignant tumors is still scarce. Variation of chromatin interaction patterns among diverse tumor tissue types remains poorly defined, and its functional relationship with gene regulation remains to be characterized. Previous studies on structure variations have almost used tumor cell lines, rather than tumor tissues from clinical patients, while the molecular events in tumor cell lines cannot reflect the real situation in tumor development.
In this study, we performed Hi-C sequence on tumor and adjacent normal tissues from two paired non-smoking lung adnocarcinoma (LUAD) patients. Further analysis about the difference of A/B compartment, TADs and chromatin loops between tumor and normal tissues identified tumor-specific A/B compartment, TADs and chromatin loops. Our results also suggested that the genomic structural variation of LUAD is mainly derived from Chr3 and five genes (SYT16: Synaptotagmin 16, NCEH1: Neutral Cholesterol Ester Hydrolase 1, NXPE3: Neurexophilin And PC-Esterase Domain Family Member 3, MB21D2: Mab-21 Domain Containing 2, DZIP1L: DAZ Interacting Zinc Finger Protein 1 Like) are identified, which may play critical functions in the development of LUAD.
Materials and Methods
Sample Collection
Two pairs of fresh tumor (C1 and C2) and adjacent normal tissues (N1 and N2) were collected from non-smoking female patients with primary lung cancer undergoing surgical resection without neoadjuvant therapy before surgery and smoking history at West China Hospital (WCH) of Sichuan University (Chengdu China), then store samples at − 80 °C until use. This study was approved by the Ethics Committee on Biomedical Research of West China Hospital of Sichuan University (Chengdu China), and all patients signed informed written consent.
Hi-C Experiments
We added the phosphate buffer saline (PBS) and formaldehyde to the freshly mixed sample and incubated 20% SDS at 37 °C for 15 min before starting the experiment. Disrupted the frozen tissue (20–40 mg) by grinding it with a mortar and pestle in liquid nitrogen. Then transfered the disrupted tissue samples to a 1.5 mL microcentrifuge tube which contain 1 mL 1X PBS and 40.5 μL 37% formaldehyde, further vortexed the tube to mix for 30 s and rotated the tube at room temperature for 20 min. Pipetted off the supernatant and washed the pellet with 300 μL wash buffer, added the 99 μL Wash Buffer; 1 μL 40 mM Calcium Chloride; 25 μL 1 mg/mL Collagenase to the pellet, then incubated it for 1 h at 37 °C in an agitating thermal mixer (1,250 rpm). After incubation, added 6.3 μL 20% SDS into the sample, then broken up the large clumps, after briefly vortexed to resuspend, transfered the supernatant to a new 1.5 mL microcentrifuge tube. Finally, performed the Hi-C experiment following the in situ Hi-C protocol (Rao et al. 2014). Briefly, the cross-linked cells were lysed and digested with DpnII, filled with biotin-14-dATP, proximately ligated with T4 DNA ligase and reverse crosslinked with 5 M sodium chloride. Then the genome DNA was purified, sheared and size-selected. Biotin pull-down was performed to enrich target DNA fragments, followed by standard Illumina library construction, lastly using the Bioanalyzer (Agilent 2100) to verify the size distribution of the size-selected library.
Quantification and Statistical Analysis
Library and Sequencing Data Quality Control
Detecting the libraries concentration and insert size using Qubit2.0 and Agilent 2100, respectively, and the effective concentration of the libraries were accurately quantified using Q-PCR method. Then, the libraries were sequenced on the Illumina NovaSeq 6000 platform at Novogene, Beijing, China, and generated 313.12 Gb raw data averagely of each sample. For the sequencing data, we filtered the joints and low-quality reads, then counted the total number of clean data bases and evaluated the GC content, Q20 and Q30 for each sample, finally reflected base quality value using Phred quality evaluation formula (Qphred = − 10log10(e)).
Hi-C Library Quality Assessment
We used Hicup_truncater tool of the HICUP software (Babraham Institute) to estimate the proportion of Truncated reads in the all reads, and the ratio should more than 10%. Then the BWA software (Li and Durbin 2009) (version: 0.7.10-r789, align type: aln) was used to align the double-end data of clean reads to the sequence of the reference genome (ftp://ftp.ensembl.org/pub/release-95/fasta/homo_sapiens/dna), and evaluate the efficiency of the alignment. Then, we used the HIC-Pro v2.10.0 (Servant et al. 2015) to analyze the alignment results, identify the valid Interaction Pairs and Invalid Interaction Pairs in the Hi-C sequencing results, to evaluate the quality of the Hi-C library. The standard of library quality is: the proportion of Invalid Interaction Pairs cannot exceed 80% (Belton et al. 2012). And we also counted the number of interactive fragments Cis/Trans after double-end reads aligned. Finally, the Pearson correlation coefficient was calculated between each biological replicate to observe the correlation of the biological replicate library.
Chromatin Interaction Analysis
We calculated the ratio value by the number of observed interaction Hi-C reads versus the number of interaction reads expected to occur between any two chromosomes, which was used as the frequency of interactions between chromosomes to predict the strength of interactions between chromosomes in the genome (Zhang et al. 2012). Interaction decay exponents (IDE) is used to describe the relationship between chromosome interaction frequency and interaction distance. In this paper, HIC-Pro v2.10.0 software is used to divide the Hi-C data according to “bin = 100 kb” to calculate the interaction intensity and normalize the data. Then use HiCdat (Schmid et al. 2015) to calculate the relationship between the interaction frequency and the linear distance of the genome. The slope of the resulting model is the corresponding IDE value.
A/B Compartment Analysis
We used iterative correction and eigenvector decomposition (ICE)-normalized interaction matrices at 100-kb resolution to detect chromatin compartment types by R-package HiTC (v 1.24.0) (Servant et al. 2012). Positive or negative values of the first principal component (PC1) separate chromatin regions into two spatially segregated compartments. The compartment with higher gene density was assigned as A compartments, and the other compartment was assigned as B compartment (Servant et al. 2012). Then we counted the number of genes on the A/B compartment in each sample.
The different conversion of compartments in tumor compared with normal tissues were identified as the different A/B compartment. We also calculated the fold change (FC) value of A/B compartment based on PC1 value between tumor and normal tissues, to evaluate the degree of conversion of the compartments.
TAD Analysis
We used ICE-normalized interaction matrices at 40-kb resolution to call TAD by TadLib software (Zhang et al. 2012) at bin = 20 kb. At the same time, inclustion ratio (IR) was calculated by HOMER software (Wang et al. 2017), which indicates the ratio of the interaction between the internal TAD and the interaction of the same length area upstream and downstream from the external TAD, to filter the TAD. The TADs with IR > 1 and the length is more than five bins were retained.
We firstly calculated the directionality index score (DI) value of the whole genome range of the sample, which is the directional score, then merge the TAD boundaries of all samples, and finally calculated the DI delta value of each TAD (that is, the average difference of DI in each four bins of TAD boundary upstream and downstream). The limma package is used to calculate the different TAD boundaries between tumor and normal tissue. Different TAD boundary was defined as: the difference is significant false discovery rate (FDR) value < 0.01, and the DI delta score value of the two groups of samples is not greater than 200.
We downloaded the CTCF ChIP-seq data from upper lobe of left lung of normal lung tissue (seven samples) in Encyclopedia of DNA Elements (ENCODE) Project Consortium (2012), which also could access from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105949. Then the CTCF distribution curve line plot of C1 and C2 samples were obtained based on this CTCF peak values.
Loop Analysis
We used improved HICCUPS method (Rao et al. 2014) to identify the loop structures at 10 kb resolution, and the size of bins is 10 kb, FDR ≤ 0.01.
To avoid possible deviations in the anchor sites at both ends of the loop, we extended the length of the bin to the left and right of the anchor site by a bin, and intersected the gene transcription start site (upstream 2 kb as the promoter region) to obtain each loop annotation information of the anchor site. The anchor site on one side of the loop is located in the promoter region, and the anchor site on the other side is located in the non-promoter region (potential enhancer region, enhancer-like) is called a promoter–enhancer related loop.
Using the method reported by Barutcu et al. (2015), firstly, we merged the loop results of each sample and de-redundant, then analyzed different loops between tumor and normal tissues based on the original interaction value of each sample, which was calculated for the non-redundant loop, by the edgeR v3.8.6. The different loops were defined as: FDR < 0.01, p-value < 0.05, and FC > 1.5. For the different loops, we calculated the normalized interaction frequency between the two anchors of each loop, and used R language Visualize.
GO and KEGG Analysis
Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using enrichGO and enrichKEGG from clusterProfiler (Yu et al. 2012), and the results were visualized with the R language.
Gene Annotation
Firstly, we merged the intervals corresponding to the two samples (for example: merge the IR intervals of C1 and N1), if the sample has no value in this interval, filled it with 0. Then the difference of PCA, IR, and ICF was represented by the corresponding difference between tumor and normal tissues, the difference of RNAPII and H3K27ac between lung cancer and normal cell from public ChIP-seq data GSE58740 and GSE131604 of NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). Lastly, we visualized these genes using IGV software.
Gene Expression and Survival Analyses
For the genes with chromatin structure variations at A/B compartment, TAD and loop levels, we predicted the difference signification between tumor and normal tissues, non-smoking female patients and normal tissues, as well as the non-smoking female patients, LUAD and normal tissues as the transcript level based on the TCGA-LUAD dataset (https://portal.gdc.cancer.gov/). For the one-by-one paired comparison between two groups, we used the wilcox.test, while other comparisons used the t-test to evaluate the significant difference between any two groups, and p-value < 0.05 represents the significant difference.
For survival analysis, we selected the non-smoking female patients from all patients of TCGA-LUAD dataset, which were divided into two groups (High expression and Low expression) based on the median value of the specific gene in these samples, then were modeled by survival package and visualization.
Assay of Cell Invasion
Human lung cancer cell lines H1299 and H1975 were obtained from the American Type Culture Collection, and were genotyped and authenticated before experiments. We took the cells to be measured in the log-growth phase, washed them with PBS, and then suspended the cells in serum-free medium to adjust the cell density to 1–10 * 105 / mL. SiRNA were synthesized by Hanheng Biotechnology Co., Ltd. and matrigel gel was diluted in serum-free cell culture medium or PBS buffer by 1:8, took 100 μL evenly applied on the polycarbonate membrane surface of the upper chamber, 37 °C for 0.5-1 h, to polymerize into gel. Then the 500–650 μL of medium containing 5–10% fetal bovine serum (FBS) or chemokines was generally added to the 24-well plate lower chamber, the transwell chamber was placed in the 24-well plate with tweezers, then the 100-200 μL cell suspension was added to the upper chamber, and finally placed in the incubator for 48 h. Removing the small chamber, absorbing the culture medium, and gently wiping the cells in the matrigel gel and the upper chamber with a cotton swab. A new 24-well plate was added with 4% paraformaldehyde at 600 μL, and the chamber was placed and postfixed for 20–30 min. The siRNA were provided below:
NCEH1-si-1: CCUGAGCAAAUUCAUGAUGUU TT AACAUCAUGAAUUUGCUCAGG TT.
NCEH1-si-2: CAUCUUAUCAGCAAAAUGU TT ACAUUUUGCUGAUAAGAUG TT.
NCEH1-si-3: CAUCAAGUGGCUAGAUCAA TT UUGAUCUAGCCACUUGAUG TT.
MB21D2-si-1: GGGUGGAGGAAUUGAACAA TT UUGUUCAAUUCCUCCACCC TT.
MB21D2-si-2: AGGUAGAAAAGGUGGAAAA TT UUUUCCACCUUUUCUACCU TT.
MB21D2-si-3: CCUGGACUUAGAUGAGCUUAA TT UUAAGCUCAUCUAAGUCCAGG TT.
SYT16-si-1: GGGAUUUGAAGAUUCCUAU TT AUAGGAAUCUUCAAAUCCC TT.
SYT16-si-2: GAGAAACUAUUCUAUCUCA TT UGAGAUAGAAUAGUUUCUC TT.
SYT16-si-3: CGUUGAUGAUUUCCGUUUA TT UAAACGGAAAUCAUCAACG TT.
Results
Hi-C Detection Revealed Multiple Structural Variations in Non-smoking LUAD Tumor Tissues
Genome disorder contributed to occurrence and development of malignant tumors (Flavahan et al. 2016; Hnisz et al. 2016). In this study, we conducted Hi-C analysis on two non-smoking LUAD tumor and adjacent normal tissues to identify the advanced structure variations (Fig. 1a, S1A), their age was 37 years old, and the clinical pathological stages were all stage III. About 54 and 52 million interactions were obtained from the cancer and normal tissues respectively, after filtering by the ratio of invalid interaction pairs, which was analyzed by HiC-Pro software based on unique reads mapped on genome (Table S1).
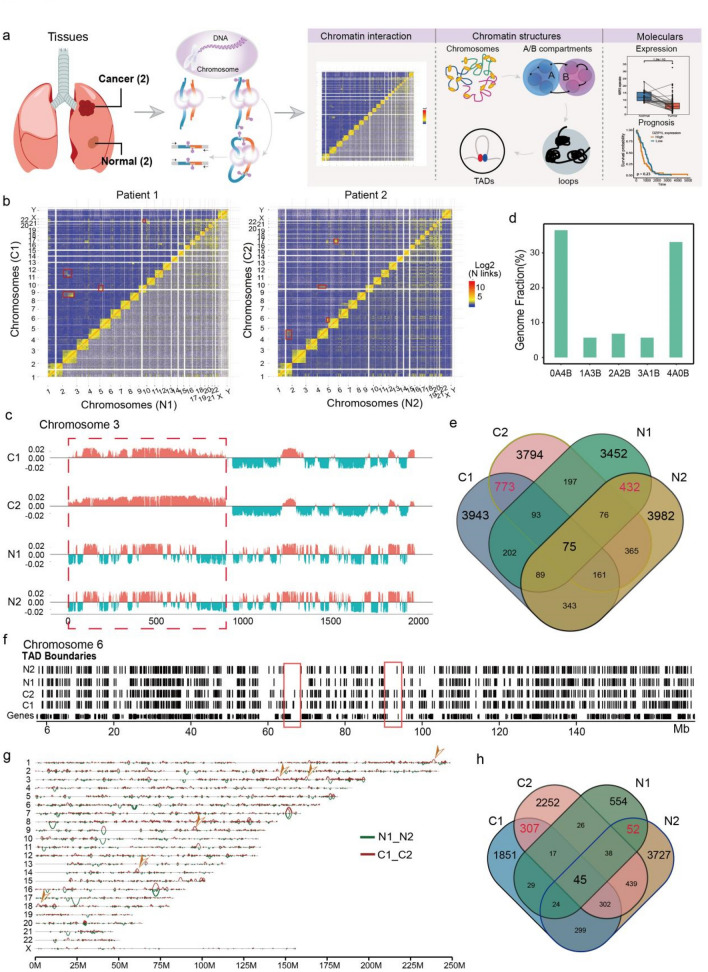
Fig. 1.
Global features of 3D Genome Organization in two LUAD and normal tissues a Sample information and analysis workflow of this study. b Chromatin interaction heatmap of all genome in tumor (C1 and C2) and normal tissues (N1 and N2), and obviously translocation sites were signed by red box. c Genome browser snapshot showing compartment A/B patterns (PC1 value) across Chr3 in four samples. Positive PC1 in red corresponds to compartment A, and negative PC1 in green corresponds to compartment B. Red box representing the significant compartmental transition region on Chr3. d Bar plots showing the degree of conservation of A/B compartment labels of four samples. The y axis is the fraction of the genome conserved by the five possible combinations of compartment A/B designations. The label below each bar represents the composition of the compartment designations. For example, ‘‘1A3B’’ represents the genomic region where one sample exhibits a compartment A label and the other three samples exhibit a compartment B label. e Venn plot showing the overlap of identified TADs in four samples. The red numbers represent the overlapped TADs in tumor and normal tissues. f Genome browser snapshot showing topological domain boundaries across Chr6 in four samples. Boundaries are identified at 40-kb bin resolution, and cancer specific genome regions were signed by red box. g Genome browser snapshot showing chromatin loop across all chromosomes in tumor and normal tissues, and tumor specific loop was marked by yellow arrow. h Venn plot showing the overlap of identified loops in four samples. The red numbers represent the overlapped loops in tumor and normal tissues
Interactions between chromosomal loci might lead to genome rearrangement, which induced spatial structure variation in many tumor-related genes and contributed to occurrence of tumors (Wu et al. 2017). In this study, the ratio of the interaction reads number observed on any two chromosomes to the expected interaction reads was defined as interaction frequency between chromosomes. Compared with normal tissues, the interaction frequency between small gene-rich chromosomes (Chr16-22) was lower than that in tumor tissues (Fig. 1b, S1B), while increased interaction frequency was observed in other chromosomes such as Chr2 and Chr3 in patient 1, ChrY and Chr14 in patient 2 (Fig. 1b).
Chromatin interaction contained two major modes, representing open (type A) and closed (type B) chromatin compartments (Valton and Dekker 2016). Here, we identified multiple A/B compartments based on PCA value of the correlation coefficient matrix from chromatin interaction by HiTC (v1.24.0) software at 100 kb resolution (Servant et al. 2012). The most obvious A/B compartment conversion regions were found in Chr3 in tumor tissues compared with normal tissues (Fig. 1c), followed by Chr4 and 12 (Fig. S1C).
As previously reported for cultured human cells (Zhao et al. 2021), we observed substantial compartment A/B conversions across primary tumor tissues (Fig. 1c, d), finding that 23.67% of the genome is dynamically compartmentalized in different tumor tissues. These data also underscored that as much as 10.1% of the compartment related genomes variant, which was significant (Fig. 1c).
We then analyzed the difference of TADs between tumor and normal tissues, which was the basic unit of gene expression and was important in forming independent regions of gene regulation (Crane et al. 2015). TadLib (Zhang et al. 2012) and HOMER (Wang et al. 2017) were conducted to identify TAD structures at 20-kb bin resolution (Table S2), a total of 17,175 and 15,629 TADs were found in tumor and normal tissues, respectively (Fig. S1D), but with no significant difference in the domain size (Fig. S1E). We further identified 5,071 tumor unique TADs (Fig. 1e), although majority of these TADs were individual specific, we still found some overlapped TADs in the two tumor tissues, which might play important roles in tumorogenesis (Fig. 1f, red boxes).
Finally we analyzed the loop structures in LUAD tumor and normal tissues. Chromatin loops occurred in TADs or sub-TADs and formed enhancer-promoter interactions, which played important roles in tumoregnesis by increasing expression of oncogenes (Ji et al. 2016). Here we performed HICCUPS software at 10-kb resolution to process Hi-C data and identified significant interaction sites, the number and mean length of loops were listed in Table S4, showing that more but smaller loop structures were identified in cancer tissues by comparing with normal tissues (Fig. 1g). A total of 15,848 loops were observed in all samples and 307 loops were identified specific in tumor tissues (Fig. 1h). Three types of loops were identified in our study, including enhancer–enhancer (E–E), enhancer–promoter (E–P) and promoter–promoter (P–P), which increased dramatically in tumor tissues (Fig. S1E–G), suggesting the important roles of these loops in tumor progression.
Our results revealed multiple structure variations, including A/B compartment, TADs and loops in two LUAD tumor tissues, which might play important roles in tumor process.
Conversions of A/B Compartment Concentrated Mainly on Chr3 in Tumor Tissues
In breast cancer, A/B compartment in the chromosomes of tumor contained many genes, which related closely to tumor-related pathways such as Wnt and TCF/LEF (Barutcu et al. 2015). Here, we extensively analyzed converted A/B compartment between LUAD and normal tissue and performed hierarchical clustering of these compartments (Table S3). The results indicated that compartment conversion occurred in every chromosome of all samples (Fig. 2a), while A → B was the main converted type in tumor tissues (Fig. S2A, B), which suggested that multiple genes lost transcriptional activity and prevented the body from performing normal functions and promoted cancer development. In all chromosomes, we noticed that most A/B conversion events were occurred in Chr3 (580/1,728, 33.6%) and the dominant type was B → A conversion (355/580) (Fig. 2a).
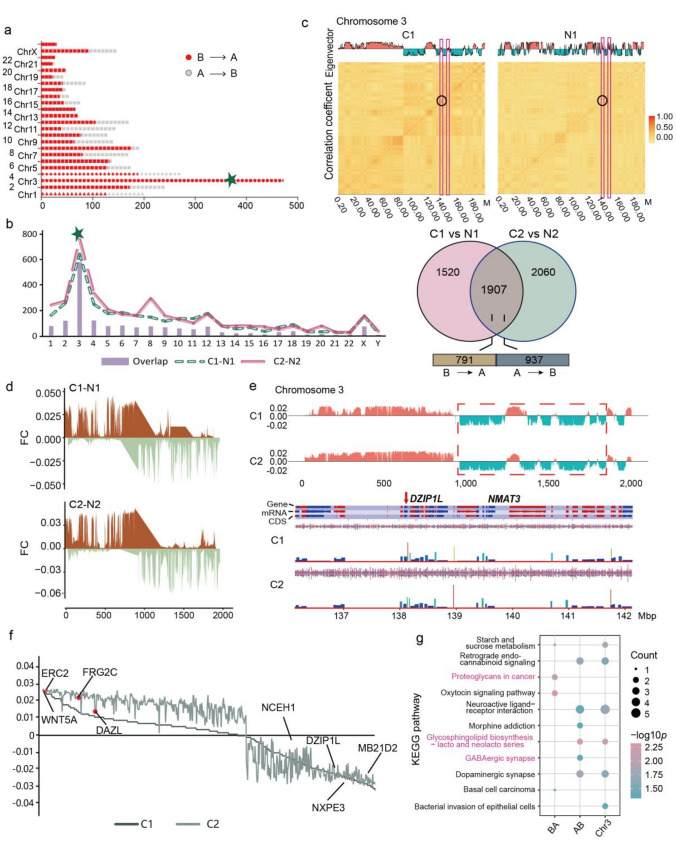
Fig. 2.
Conversions of A/B Compartment concentrated mainly on Chr3 in tumor tissues a Distribution map of the number in two compartment conversion types (A → B and B → A) of each chromosome in tumor tissues. The green five-pointed star signing the Chr3, which contain the most different compartments. b Statistics distribution chart of different and overlapped compartment in each chromosome and the of two tumor tissues. Venn plot showing the number of overlapped and sample specific compartments and the number of different conversion type in the overlapped compartments. c Heatmap of A/B compartment interaction in tumor and normal tissue. The horizontal and vertical coordinates of the figure below are the chromosome positions, and the darker the color, the higher the correlation. d Distribution map of difference between tumor and normal tissue in each patient based on PC1 value across Chr 3. e Gene annotation on the regions with compartment conversion of Chr3 by the SeqMonk Course Manual solfware. f Distribution map of different compartment-related genes in two tumor samples based on the PC1 value, and some important genes were marked on the plot. g Bar plot showing the KEGG pathways enriched in different compartment conversions on Chr3 of tumor samples
We then analyzed the compartment switch in each tumor tissue, and the results also showed that Chr3 exhibited the most A/B compartment switch events. Specifically, a total of 1,907 A/B compartment variations were found in tumor tissues while 580 (30.4%) located on Chr3 (Fig. 2b, S2C). Combining the A/B compartment distribution and chromosome genome correlation on Chr3 indicated that the correlation intensity of A → B converted type in the tumor tissue was also higher than that in normal tissue (Fig. 2c), which meant the formation of new chromatin structures in the genome region and promoted tumorigenesis.
Next, we conducted subtraction between the compartments of paired samples from the same patient based on PC1 value on whole genome and Chr3 to assess the degree of compartment structural variation in tumor tissue, which showed that the compartment related region of Chr3 with the most frequent and highest degree variation was clearly divided into two regions, the first 12.5 Mb tends to be converted from A → B, and the retained tend to be converted from B → A (Fig. 2d, S2D), and these regions contained some important genes, such as ELKS/RAB6-Interacting/CAST Family Member 2 (ERC2), Wnt Family Member 5A (WNT5A) and Matrin 3 (NMAT3) (Fig. 2e).
Finally we further visualized the 89 converted compartments identified in tumor tissues, which revealed the most important tumor-related genes, including ERC2, FSHD Region Gene 2 Family Member C (FRG2C), Deleted In Azoospermia Like (DAZL), and Mab-21 domain containing 2 (MB21D2), et al. (Fig. 2f). KEGG pathway analysis indicated that the A → B converted type related genes were enriched in glycosphingolipid biosynthesis−lacto and neolacto series and dopaminergic synapse pathways (Fig. 2g). We also annotated all genes on converted A/B compartment regions between tumor and normal tissues, which revealed some tumor associated signaling pathways, including PI3K−Akt, MAPK, Ras and Rap1 (Fig. S2E). In general, we demonstrated that A → B was the major compartment conversion type in LUAD and related closely to important signal pathways in tumor process.
Distinct TAD Boundaries Variations were Identified in LUAD
Occurrence of TAD was a common characteristic and more than 85% TADs were conserved in cells, however, tumor cells still harbored distinct specific TADs, involving tumor-related genes such as GATA Binding Protein 1 (GATA1), SRY-Box Transcription Factor 9 (SOX9), c-myc (MYC), which might contribute to cancer development (Spielmann et al. 2018). Therefore, we analyzed the TAD structures between tumor and normal tissues at 40-kb resolution based on DI delta value to observe the specific TADs in LUAD and the related functional genes (Wang et al. 2017). A total of 1,500 different TADs were detected in this study, and tumor and normal tissues harbored 650 and 850 TADs respectively (Fig. 3a). Distributing of these TADs on chromosomes, we found that Chr2 contained the highest number of TADs (149), and Chr1 (109), Chr3 (107), Chr4 (96) and Chr8 (104) also hold more TADs than other chromosomes (Fig. 3a). We then analyzed the TADs in each patient and identified 446 and 420 specific TADs in C1 and C2 tumor tissues respectively, and 216 TADs were overlapped in both two tumor tissues (Fig. 3b). Furthermore, the majority of overlapped TADs were concentrated on Chr1 (19), Chr3 (17), and Chr2 (15) (Fig. 3c), which revealed the importance of the three chromosomes in TAD formation.
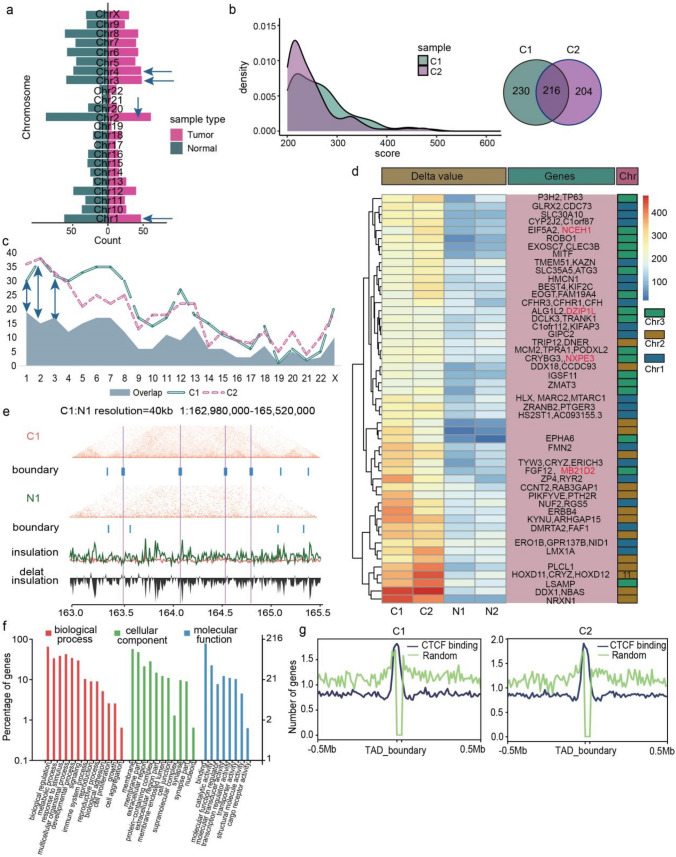
Fig. 3.
Distinct TAD boundaries variations were identified in LUAD a Barplot showing the number of tumor and normal tissue specific TADs in each chromosome, and top four chromosomes were marked by green arrow. b Density distribution of the delta score value in two tumor tissues, and venn plot showing the number of sample specific and overlapped TADs in tumor tissues. c Statistics distribution chart showing the different TADs between paired tumor and normal tissues as well as the overlapped TADs between two tumor samples in each chromosome. Green arrows represent the chromosomes with relatively highly overlapped TADs. d Heat map of tumor specific TADs of Chr1, 2, and 3 based on delta score value, and the related genes was annotated in the pink box on the right, NCEH1, DZIP1L, NXPE3 and MB21D2 were signed by red color. e Hi-C interaction matrix of the region (Chr1: 162,980,000–165,520,000) in patient 1 showing the TAD boundaries distribution between tumor and normal tissues. Top: Hi-C interaction matrix and TAD boundaries (vertical bars), middle: insulation scores, bottom: delta insulation score. The pink line labeled the tumor specific TAD boundary. f GO enrichment analysis of tumor specific TADs related genes. g CTCF binding distribution in tumor samples based on the public ChIP-seq data
Next, we analyzed the important genes involved in overlapped TADs in Chr1, 2 and 3, and found multiple tumor-related genes, including Chromosome 1 Open Reading Frame 87 (C1orf87), Complement Factor H Related 3 (CFHR3) and Regulator Of G Protein Signaling 5 (RGS5) in Chr1, Homeobox D11 (HOXD11), DEAD-Box Helicase 1 (DDX1) and Neurexin 1 (NRXN1) in Chr2, Tumor Protein P63 (TP63), Autophagy Related 3 (ATG3), Limbic System Associated Membrane Protein (LSAMP) and FGF12 fibroblast growth factor 12 (FGF12) in Chr3, and the most interesting observation in TADs was that we also found five tumor-related genes, including SYT16, NCEH1, DZIP1L, NXPE3 and MB21D2, which were former identified in A → B compartment (Fig. 3d), suggesting the important role of the five genes in specific structure variation in lung cancer. Extensive analysis of TAD boundary showed that some significant boundaries were detected in tumor tissues which clearly indicated the formation of novel TADs (Fig. 3e, S3A).
Then we performed GO functional and KEGG pathway analysis by the ClusterProfiler R package to investigate the important pathways of tumor specific TADs related genes and found that they involved in the functions, such as biological adhesion, cell junction, transporter activity, cargo receptor activity and reproductive process which played critical roles in tumor process and metastasis (Fig. 3f), and some important tumor-related pathways including cell adhesion molecules, choline metabolism in cancer, as well as hippo, wnt and mTOR signaling pathways (Fig. S3B).
Finally we explored the CTCF in TADs, which played an important role in organization of genome into TADs, and was critical for coordinating the transcriptional regulation, chromatin states, DNA replication and establishing TAD boundaries (Barutcu et al. 2018; Katoh and Katoh 2006; Wang et al. 2020). We used the ChIP-seq from study of ENCODE Project Consortium (2012) to analyze the CTCF distribution of TAD boundary in important genes, which showed that CTCF had a higher reads ratio value in TAD boundaries (Fig. 3g, S3C).
Taken together, we found 216 tumor-specific TADs in LUAD samples and further identified five important genes (SYT16, NCEH1, DZIP1L, NXPE3 and MB21D2), which were consistent with A/B compartment results.
Specific Chromatin Enhancer-Promoter Loops were Identified in LUAD Tumor Tissues
The chromatin loops with increased formation of active chromatin hubs is composed by interactions of multiple enhancers and promoters (Ji et al. 2016), and previous studies indicated that chromatin loops could override endogenous gene expression programme and provide a possible therapeutic approach in disease treatment such as sickle cell anaemia (Deng et al. 2012, 2014; Fudenberg et al. 2011; Zhang et al. 2019; Zheng and Xie 2019). Some strong interaction signal points at off-diagonal positions was found in the whole genome heat map as shown in Fig. 1b, which suggested the existence of loop structures. Here, we explored the loop structures at the 10 kb resolution and FDR ≤ 0.01 by the HICCUPS software.
A total of 2344 different chromatin loops were identified between tumor and normal tissues by the method reported in the published article (Zhang et al. 2019), and 1,357 loops existed specific in tumor tissues (Fig. 4a, Table S4) and the mean length was lower than that in normal tissues (Fig. 4a). We also observed that most of these loops (312) were concentrated on Chr3, followed by Chr1 (230), Chr8 (220) and Chr2 (155) (Fig. 4b, S4A). Then, we further analyzed the chromatin loops in each patient, and found that they harbored 956 and 1,012 tumor specific loops respectively, and a total of 131 loops were overlapped in both two tumors (Fig. 4c, inner Venn), which were mainly distributed on Chr3 (Fig. 4c). Thus, the Chr3 is important in the formation of chromatin loops.
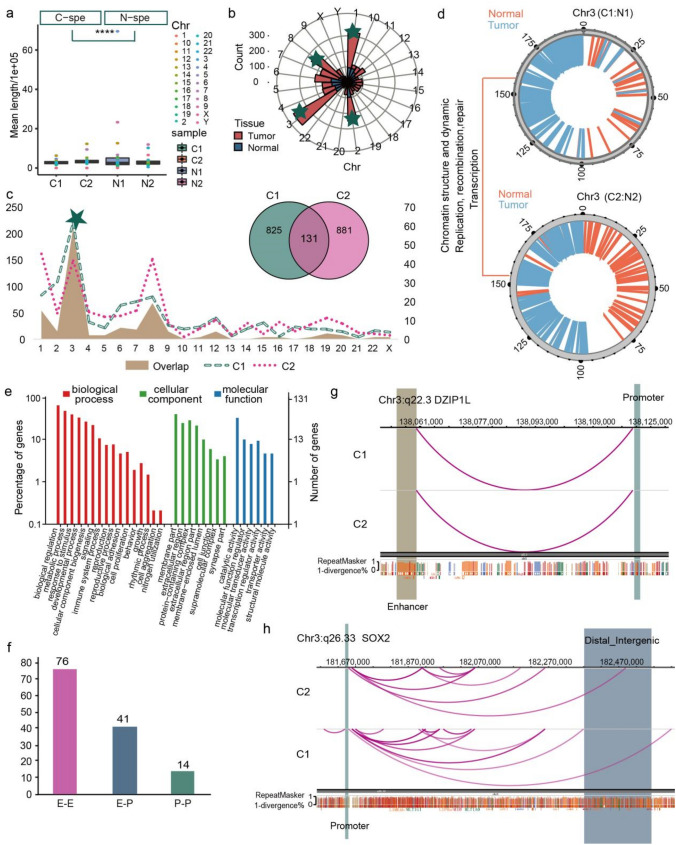
Fig. 4.
Distinct chromatin enhancer–promoter loops were identified in LUAD tissues a Boxplot showing the mean length distribution of tumor and normal specific loops in each sample. T-test was used to calculate the significant difference, ****p < 0.0001. b Polar diagram showing the number of tumor and normal tissue specific loops in each chromosome, and the top four chromosomes were marked by green five-pointed star. c Statistics distribution chart showing the different loops between paired tumor and normal tissues as well as the overlapped loops between two tumor samples in each chromosome, and green five-pointed star signed the Chr3, which with the most number of different and overlapped loops. Venn plot showing the number of overlapped and sample specific loops. d Distribution of tumor and normal specific loops on genome of Chr3, showing the function of these genome regions. e GO enrichment analysis of the tumor specific loops related genes. f Barplot showing the number of different loop type of the 131 overlapped loops in tumor tissues. g Genome browser snapshot of chromatin loop with the annotation of the end anchors at DZIP1L locus, the brown bar represents the enhancer domain of gene, the green bar represents the promoter domain, and the blue bar is the distal enhancers. h The same visualization with g at the SOX2 locus
Next, the important different genome regions in formation of chromatin loops between tumor and normal tissues were determined by HiCPlotter, and we found some tumor process closely related genes, such as SID1 transmembrane family member 1 (SIDT1), 5-Hydroxymethylcytosine Binding (HMCES), Nicotinamide Nucleotide Adenylyltransferase 3 (NMNAT3), and G Protein-Coupled Receptor 87 (GPR87), which all distributed in gene sequence where the tumor specific loops were located (Fig. 4d). We further investigated and annotated the distinct genes on Chr3, and found that they were enriched in immune system process, cell proliferation, biological adhesion, molecular transducer activity, transcription regulator activity and transporter activity (Fig. 4e), and majority of the KEGG pathways (Fig. S4C) were tumor-related.
Enhancer–enhancer (E–E), enhancer–promoter (E–P) and promoter–promoter (P–P) were the three major types of chromatin loops and the enhancer-promoter loops played important regulatory roles in occurrence and development of diseases (Deng et al. 2012). In this study, we obtained 76 E–E, 41 E–P and 14 P–P loops (Fig. 4e), and all enrolled genes are listed in Table S5. Further analysis distinguished that the five genes (SYT16, NCEH1, DZIP1L, NXPE3 and MB21D2) formerly identified in A/B compartment and TADs, which were also found in E–P loops on Chr3. And the anchor site at two ends of the chromatin loop, which is located on q22.3 of Chr3, was formed by the promoter of DZIP1L and enhancer of MB21D2. The known oncogene SRY (sex determining region Y)-box 2 (SOX2), which is also loctaed on Chr3 and stimulated cellular migration and anchorage-independent growth, was observed to have a physical interaction between more than three distal enhancers and the promoter region of itself (Fig. 4f, S4D). Therefore, these results further confirmed the importance of Chr3 in formation of chromatin loops and tumorgenesis.
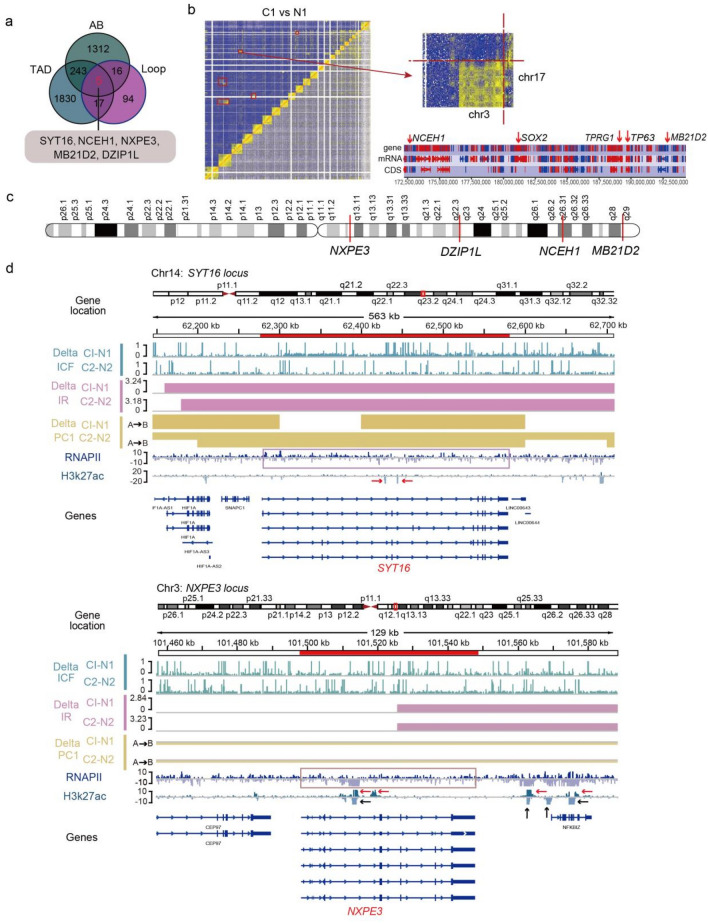
SYT16, NCEH1, NXPE3, MB21D2, and DZIP1L were the Most Important Genes in Advanced Structure Variations in LUAD
The changes in the spatial structure of chromatin could shape the alteration of cancer genome (Zhang et al. 2019), and the chromatin remodeling also induced chromosome instability and led to occurrence of tumors (Barutcu et al. 2015). Our former analyses found the cancer-specific advanced structure variations, which were mainly enriched on Chr3. To further identify the principal cancer-related genes induced by the chromatin structure variation, we attempted to find the overlapped molecules with structure variation in all three chromatin structure levels. The results revealed five cancer-related genes, and four of these genes (NCEH1, DZIP1L, NXPE3 and MB21D2) are located on p arm of Chr3 while SYT16 is located on Chr14 (Fig. 5a–c). In all these genes, SYT16 was related to calcium ion binding and protein heterodimerization activity, which also acted as a prognostic biomarker in LGG (Lower-grade glioma) (Chen et al. 2020). NCEH1 expressed in human atheromatous lesions and played a critical role in the hydrolysis of intracellular cholesterol ester (CE) in human macrophage foam cells, thereby contributing to the initial part of reverse cholesterol transport in human atherosclerosis (Igarashi et al. 2010). NXPE3 was significantly hypermethylated and down regulated in hepatocellular carcinoma (HCC) tumors, and epigenetic silencing of this gene may be associated with the occurrence of HCC (Yamada et al. 2016). Mutated MB21D2 was found in LUAD tumor tissues and suggested to have an oncogenic role and could be a potent neoantigen, just like other passenger mutation (Buisson et al. 2019). DZIP1L functioned as an important molecule in Hedgehog signaling pathway and often activated in gastric cancer (Katoh and Katoh 2006).
Fig. 5.
Five most important genes in advanced structure variations of LUAD a Venn plot showing the related gene number distribution of tumor specific chromatin structures at A/B compartment, TAD, and loop levels. Five genes (marked with red color) were found in each resolution chromatin structure. b Heatmap and partial heatmap of Patient 1 showing a balanced translocation between Chr3 and 17. Heatmaps were coloured by the number of interactions with the colour gradient scaled linearly from five (blue) to 10 (yellow). The bottom plot showing read counts for amplified regions on Chr3. The high peaks show a significantly higher number of reads than in the surrounding regions, and NCEH1, SOX2, TPRG1, TP63 and MB21D2 genes were labeled. c Distribution of the position of five genes on Chr3 genome, and four genes (NXPE3, DZIP1L, NCEH1, MB21D2) are labeled. d Genome browser tracks of ChIP-seq data (RNAPII/H3K27ac), compartments changes (Delta PC1), TAD changes (Delta IR) and loop changes (Delta ICF) in tumor compared with normal tissues at the SYT16 (top) and NXPE3 (bottom) genes locus. Pink box representing the RNAPII peak on gene region, red and black arrows showing the increased or decreased H3K27ac peaks on the gene region
To explore the association between advanced chromatin structure variations and transcriptional regulation of the five genes, we used the public ChIP-seq data (RNAPII and H3k27ac) from previous studies (Kurppa et al. 2020; Sammons et al. 2015) (Fig. 5d, S5). The RNAPII plays an essential role in initiating nascent transcription, and H3k27ac is an active enhancer marker. At the SYT16 locus on Chr14, the regions of gene showed increased decompaction (stronger interaction of chromosomes and higher insulation score) and A/B compartment switching with the presence of RNAPII in tumor tissue, but the deletion of H3k27ac peaks. The similar phenomenon was observed at the DZIP1L and NCEH1 locus on Chr3, but without the H3k27ac peaks at the DZIP1L locus. (Fig. S5A, B). At the NXPE3 locus, expect gene region, the region downstream of gene also showed the similar chromatin structure variations in tumor, and the deletion of RNAPII with the presence or delection of H3k27ac signals. The similar phenomenon was observed at the MB21D2 locus (Fig. S5C). And the results suggested stronger activity of RNAPII while A/B compartment switching may generate new regulation site.
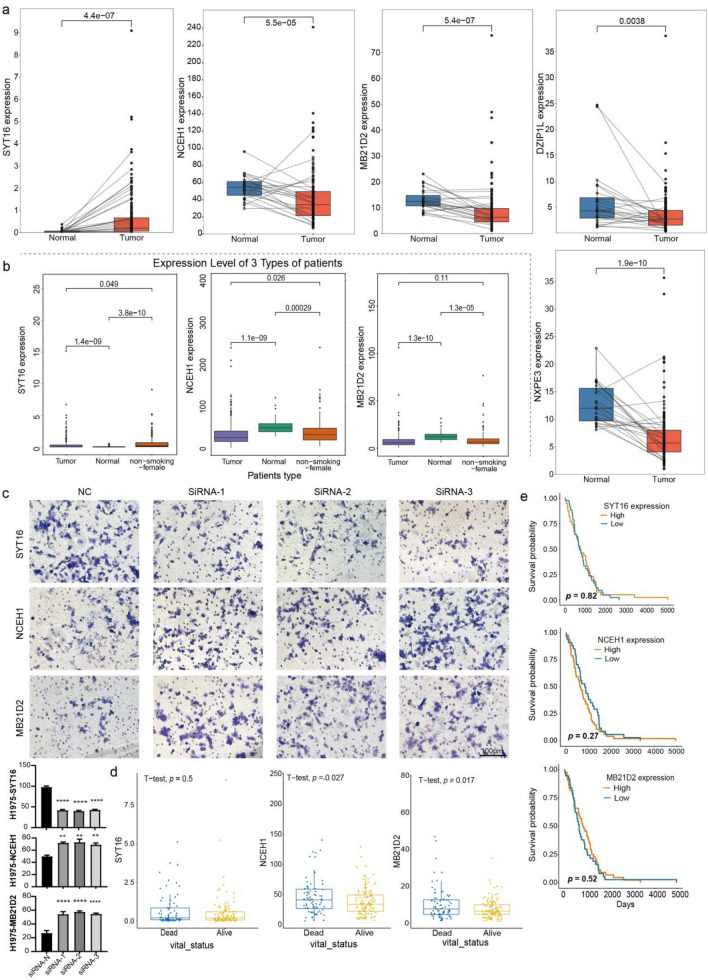
Furthermore, the four genes (NCEH1, DZIP1L, NXPE3 and MB21D2) located on the Chr3 were all down-regulated in the LUAD of non-smoking-female patients based on the TCGA-LUAD dataset, while the SYT16 was up-regulated in tumor samples (Fig. 6a, b and S6A, B). Knocking-down of these genes revealed that the NCEH1 and MB21D2 were markedly increased the invasion of lung cancer H1299 and H1975 cells, while the SYT16 decreased the invasion of cells, suggesting their oncogenic or anti-oncogenic importance (Fig. 6c, S6C). Moreover, despite there was no difference in the prognosis of these genes, they all highly expressed in death group, which may represented the pro-tumor effect of NCEH1 and MB21D2 in non-smoking-female patients (Fig. 6d, e).
Fig. 6.
Expression and prognostic value prediction of this five genes based on TCGA-LUAD dataset a One-by-one paired box plots showing the expression of SYT16, NXPE3, NCEH1, DZIP1L and MB21D2 in non-smoking female patients and normal tissues based on the TCGA-LUAD dataset (128 samples). p-value < 0.05 represents significant difference by the wilcox test. b Box plots showing the expression of SYT16, NCEH1 and MB21D2 in tumor, non-smoking female patients and normal samples based on the TCGA-LUAD dataset. p-value < 0.05 represents significant difference by the t-test. c Invasion were determined with a transwell assay of H1975 cell line. Scale bar = 100 μm. The test method was One-way Anova, *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. d Box plots showing the expression of NCEH1, SYT16 and MB21D2 in dead and alive samples based on the non-smoking female patients from TCGA-LUAD dataset. p-value < 0.05 represents significant difference by the t-test. e Survival analysis of NCEH1, SYTY16 and MB21D2 in selected non-smoking female patients based on TCGA database
Taken together, five genes were found have variations in the A/B, TAD and loop structure of genome with the change of RNAPII and H3k27ac signals in tumor, which implicates a potential role for these genes in facilitating the occurrence and development of LUAD.
Discussion
Chromatin instability played important roles in occurrence of malignant tumors. Earlier studies about 3D genome with Hi-C technology in cancer were largely limited to cultured cells or a small collection of primary cancer cell types, such as myeloma and prostate cancer (Rhie et al. 2019; Taberlay et al. 2016; Wu et al. 2017), which ignored the impact of tumor microenvironment and contributed poor manifestation to the true pathological characteristics of malignant tumors, and previous study showed that there was a genomic inconsistency between subcultured cells (U87MG glioma cell line) and primary tumor cells (glioblastoma) (Kaiser and Semple 2017).
In our study, it was the first time to apply Hi-C technology to detect clinical lung adenocarcinoma tissue samples and conducted in-depth analyses about the advanced chromatin structure variations between LUAD and normal tissues. We observed multiple advanced structural variations (A/B compartment, TADs and loops) in tumor tissues. Our results further confirmed the dominant role of Chr3 in the development of lung tumor, which with the most prevalent structural variations at A/B compartment (820/5308, 15.4%), TAD (107/650, 16.5%) and chromatin loop (312/1357, 23%). The previous study about lung cancer revealed that the gain/amplification at q and loss/deletion at p arms of Chr3 were the most frequent and early events in lung cancer, which encoded many candidate driver genes, including Phosphatidylinositol-4,5-bisphosphate 3-kinase Catalytic Subunit Alpha (PIK3CA) and Tumor Protein P63 (TP73L: as defined by the HUGO nomenclature; also known as p63), and they participated in many cancer associated functions, such as cell proliferation, cell adhesion, apoptosis, RAS signaling, and oncogenic transformation (Qian and Massion 2008). Total or part copy deletions in p arm of Chr3 are related closely to the development of clear cell renal carcinoma, and they code many chemokine receptors that aid inflammatory processes, such as C–C Motif Chemokine Receptor 5 (CCR5) and Von Hippel-Lindau (VHL) (Brunelli et al. 2011). Our study also identified many tumor-associated genes, including the ERC2, WNT5A at compartment resolution, TP63, Melanocyte Inducing Transcription Factor (MITF) in TAD structure and SOX2, NMNAT3 at loop scale, are mainly participated in cell proliferation, transporter and regulation functions and MAPK, PI3K-AKT, Ras, WNT, Ras1 pathways.
A → B compartment conversions were frequently observed in malignant tumors cells (Katoh and Katoh 2006), which was also discovered in tumor tissues. But we also observed B → A compartment conversions in tumor tissues with a higher close compartmentalization during the LUAD development, which is related closely to accessibility of transcription factors, such as Zinc Finger DBF-Type Containing 2 (ZDBF2), EGF like repeats and discoidin domains 3 (EDIL3) and Hyaluronan and proteoglycan link protein 1 (HAPLN1) (Zhao et al. 2021). Our result showed that the genes with variation in compartment status were most notably related to extracellular region and participated in cell proliferation, biological adhesion in cancer, as well as the transcription regulator activity.
Our study unveiled multiple chromatin structure variations at TAD and loop scale in LUAD tissues, which were reported to play important roles in tumor development (Deng et al. 2012, 2014; Spielmann et al. 2018; Zheng and Xie 2019). Further interesting observation was that not all changed TADs were functional consequences of genes (Fig. 3d), and non-coding regions in of TADs were also identified in lung tumors, suggesting the regulatory roles of these regions, which still needs further investigation. We also observed some novel TADs (Fig. 3e, S3A, B) in tumor tissue by compared with normal sample, and the formation of these structures may due to the duplications spanning a TAD boundary (Franke et al. 2016).
Furthermore, some tumor associated genes, such as SOX2, Tumor Protein P63 Regulated 1 (TPRG1), were identified at loop anchor points of the tumor specific E-P type loops, which were reported exist somatic variation and were key upregulated factors in lung cancer. Previous study identified enrichment of SNPs for different sets of diseases in each set of group-specific loops by the analysis similar to GO enrichment (Grubert et al. 2020), so intersecting the variants in promoter or enhancer regions of these oncogenes with tumor-specific chromatin loops in our study, may help to explain how such sequence variation leads to LUAD. We also observed obvious individuality (Fig. 4c, S4B) at the loop scale, but the mechanism for this phenomenon remained further exploration.
The most important observation in our study was that three genes (SYT16, NCEH1, and MB21D2) were identified with the variations of both A/B compartment, TADs and chromatin loops in tumor tissues, and also with the pro- or anti-tumor effects. While the other two genes, NXPE3 and DZIP1L, had no differential expression between lung cancer cell lines (H1975, H1299) and normal lung cell line (BEAS) in spite of the obvious changes were observed in the TCGA dataset. Taken together, the SYT16, NCEH1, and MB21D2 genes found in our study may play a key role in the LUAD development.
The disadvantage of our study is that the sample size is small, but the public data verification was performed on the genes we found with conformational changes. In the next step of conformational research, we will expand the sample and combine WGS, RNA-seq, ChIP-seq and other multi-omics data for comprehensive discussion.
Genomics instability in cancer is not a single phenomenon. Instead, many different mutational processes can act to restructure the genome and, in doing so, could generate a notably flexible array of possible structures. With this regard, our results provide a basis for further research on the genomic structure variations of LUAD.
Conclusion
In this study, we did a comprehensive analysis of chromatin structure in LUAD using Hi-C technology firstly, and found the importance of Chr3 and five meaningful genes, which can provide reference basis for future research. Although we have found some important chromatin structural variations and genes in lung cancer tissues, we need to further combine other omics data for joint analysis to interpret the genomic structural variations of tumor tissues from more perspectives.
Acknowledgements
This work was supported by National Guided Science and Technology Development Project of Sichuan Province (No: 2020ZYD009), Clinical Research Incubation Project of West China Hospital of Sichuan University (No: 2018HXFH012), Hospital-enterprise cooperation clinical research innovation project (Sichuan University West China Hospital-Shanghai Pharmaceutical) (No: 2019HXCX04), Sichuan Science and Technology Program (No: 2020ZYD007), Science and Technology Achievement Transformation Fund of Sichuan University West China Hospital (No: CGZH19013), and we sincerely thanks to Biomarker Technologies Corporation (Beijing, China) for the analyses of bioinformation in this study.
Abbreviations
- Hi-C
High-throughput chromosome
- Chr
Chromosome
- TADs
Topologically associated domains
- PBS
Phosphate buffer saline
- FBS
Fetal bovine serum
- SDS
Sodium dodecyl sulfate
- ICE
Iterative correction and eigenvector decomposition
- ChIP-seq
Chromatin immunoprecipitation sequencing
- CTCF
CCCTC-binding factor
- ENCODE
Encyclopedia of DNA elements
- RNAPII
RNA polymerase II
- IGV
Integrative genomics viewer
- NCBI
National Center for Biotechnology Information
- TCGA
The Cancer Genome Atlas
- FDR
False discovery rate
- LUAD
Lung adenocarcinoma
Authors' contributions
LZ and LW designed and funded the research, wrote the paper; TS and MY wrote the paper, prepared the figures; TS designed the research; YY and ZL prepared the figures.
Funding
National Guided Science and Technology Development Project of Sichuan Province (No: 2020ZYD009), Clinical Research Incubation Project of Sichuan University West China Hospital (No: 2018HXFH012), Hospital-enterprise cooperation clinical research innovation project (Sichuan University West China Hospital -Shanghai Pharmaceutical) (No: 2019HXCX04), Sichuan Science and Technology Program (No: 2020ZYD007), and Science and Technology Achievement Transformation Fund of Sichuan University West China Hospital (No: CGZH19013).
Data availability
Hi-C sequencing data and process data have been deposited to the Gene Expression Omnibus (GEO) database, under the accession GSE161486.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
This study was approved by the Ethics Committee on Biomedical Research of West China Hospital of Sichuan University (Chengdu China). We confirm that all methods of processing human samples and experimental protocols were carried out in accordance with relevant guidelines and regulations in the methods section of the manuscript.
Consent to participate
All patients signed informed written consent.
Consent for publication
Not applicable.
Footnotes
Tingting Song and Menglin Yao contributed equally to this study.
Contributor Information
Li Zhang, Email: zhangli7375@scu.edu.cn.
Weimin Li, Email: weimi003@scu.edu.cn.
References
- Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8(354):354. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- Barutcu AR, Lajoie BR, McCord RP, Tye CE, Hong D, Messier TL, Browne G, van Wijnen AJ, Lian JB, Stein JL, Dekker J, Imbalzano AN, Stein GS. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16:214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Maass PG, Lewandowski JP, Weiner CL, Rinn JL. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat Commun. 2018;9(1):1444. doi: 10.1038/s41467-018-03614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton J-M, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Brunelli M, Fiorentino M, Gobbo S, Sperandio N, Cheng L, Cossu-Rocca P, Segala D, Eble JN, Delahunt B, Novara G, Ficarra V, Martignoni G. Many facets of chromosome 3p cytogenetic findings in clear cell renal carcinoma: the need for agreement in assessment FISH analysis to avoid diagnostic errors. Histol Histopathol. 2011;26(9):1207–1213. doi: 10.14670/HH-26.1207. [DOI] [PubMed] [Google Scholar]
- Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, Lawrence MS. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 2019 doi: 10.1126/science.aaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L-H, Ghosh S, Noordermeer D. TADs and their borders: free movement or building a wall? J Mol Biol. 2020;432(3):643–652. doi: 10.1016/j.jmb.2019.11.025. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang Z, Wang W, Ren S, Xue J, Zhong L, Jiang T, Wei H, Zhang C. SYT16 is a prognostic biomarker and correlated with immune infiltrates in glioma: A study based on TCGA data. Int Immunopharmacol. 2020;84:106490. doi: 10.1016/j.intimp.2020.106490. [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523(7559):240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, Blobel GA. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PAW. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220(2):244–254. doi: 10.1002/path.2632. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan W-L, Spielmann M, Timmermann B, Wittler L, Kurth I, Cambiaso P, Zuffardi O, Houge G, Lambie L, Brancati F, Pombo A, Vingron M, Spitz F, Mundlos S. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538(7624):265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol. 2011;29(12):1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F, Srivas R, Spacek DV, Kasowski M, Ruiz-Velasco M, Sinnott-Armstrong N, Greenside P, Narasimha A, Liu Q, Geller B, Sanghi A, Kulik M, Sa S, Rabinovitch M, Kundaje A, Dalton S, Zaugg JB, Snyder M. Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 2020;583(7818):737–743. doi: 10.1038/s41586-020-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, Reddy J, Borges-Rivera D, Lee TI, Jaenisch R, Porteus MH, Dekker J, Young RA. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M, Osuga J-i, Uozaki H, Sekiya M, Nagashima S, Takahashi M, Takase S, Takanashi M, Li Y, Ohta K, Kumagai M, Nishi M, Hosokawa M, Fledelius C, Jacobsen P, Yagyu H, Fukayama M, Nagai R, Kadowaki T, Ohashi K, Ishibashi S. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ Res. 2010;107(11):1387–1395. doi: 10.1161/CIRCRESAHA.110.226613. [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, Misteli T, Jaenisch R, Young RA. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016;18(2):262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Semple CA. When TADs go bad: chromatin structure and nuclear organisation in human disease. F1000Res. 2017 doi: 10.12688/f1000research.10792.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review) Int J Mol Med. 2006;18(6):1019–1023. [PubMed] [Google Scholar]
- Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, Knelson EH, Xie Y, Lim K, Cejas P, Portell A, Lizotte PH, Ficarro SB, Li S, Chen T, Haikala HM, Wang H, Bahcall M, Gao Y, Shalhout S, Boettcher S, Shin BH, Thai T, Wilkens MK, Tillgren ML, Mushajiang M, Xu M, Choi J, Bertram AA, Ebert BL, Beroukhim R, Bandopadhayay P, Awad MM, Gokhale PC, Kirschmeier PT, Marto JA, Camargo FD, Haq R, Paweletz CP, Wong K-K, Barbie DA, Long HW, Gray NS, Jänne PA. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020 doi: 10.1016/j.ccell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J Thorac Oncol. 2008;3(3):212–215. doi: 10.1097/JTO.0b013e3181663544. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie SK, Perez AA, Lay FD, Schreiner S, Shi J, Polin J, Farnham PJ. A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat Commun. 2019;10(1):4154. doi: 10.1038/s41467-019-12079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015;25(2):179–188. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW, Grob S, Grossniklaus U. HiCdat: a fast and easy-to-use Hi-C data analysis tool. BMC Bioinform. 2015;16(1):277. doi: 10.1186/s12859-015-0678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Lajoie BR, Nora EP, Giorgetti L, Chen C-J, Heard E, Dekker J, Barillot E. HiTC: exploration of high-throughput 'C' experiments. Bioinformatics. 2012;28(21):2843–2844. doi: 10.1093/bioinformatics/bts521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, Heard E, Dekker J, Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19(7):453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- Taberlay PC, Achinger-Kawecka J, Lun ATL, Buske FA, Sabir K, Gould CM, Zotenko E, Bert SA, Giles KA, Bauer DC, Smyth GK, Stirzaker C, O'Donoghue SI, Clark SJ. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016;26(6):719–731. doi: 10.1101/gr.201517.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton A-L, Dekker J. TAD disruption as oncogenic driver. Curr Opin Genet Dev. 2016;36:34–40. doi: 10.1016/j.gde.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-T, Cui W, Peng C. HiTAD: detecting the structural and functional hierarchies of topologically associating domains from chromatin interactions. Nucleic Acids Res. 2017;45(19):e163. doi: 10.1093/nar/gkx735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang L, Chen Y, Li M, Ha M, Li S. Screening and identification of biomarkers associated with the diagnosis and prognosis of lung adenocarcinoma. J Clin Lab Anal. 2020;34(10):e23450. doi: 10.1002/jcla.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Li T, Li R, Jia L, Zhu P, Liu Y, Chen Q, Tang D, Yu Y, Li C. 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat Commun. 2017;8(1):1937. doi: 10.1038/s41467-017-01793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Yasui K, Dohi O, Gen Y, Tomie A, Kitaichi T, Iwai N, Mitsuyoshi H, Sumida Y, Moriguchi M, Yamaguchi K, Nishikawa T, Umemura A, Naito Y, Tanaka S, Arii S, Itoh Y. Genome-wide DNA methylation analysis in hepatocellular carcinoma. Oncol Rep. 2016;35(4):2228–2236. doi: 10.3892/or.2016.4619. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho Y-J, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148(5):908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, Chee S, Ma K, Ye Z, Zhu Q, Huang H, Fang R, Yu L, Izpisua Belmonte JC, Wu J, Evans SM, Chi NC, Ren B. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet. 2019;51(9):1380–1388. doi: 10.1038/s41588-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang M, Gao S, Chen J. Chromatin architecture reorganization during somatic cell reprogramming. Curr Opin Genet Dev. 2021;70:104–114. doi: 10.1016/j.gde.2021.07.006. [DOI] [PubMed] [Google Scholar]
- Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol. 2019;20(9):535–550. doi: 10.1038/s41580-019-0132-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Hi-C sequencing data and process data have been deposited to the Gene Expression Omnibus (GEO) database, under the accession GSE161486.