Highlights
-
•
We summarize the detection frequency of BLV in samples of human origin.
-
•
Nested PCR and tax gene are the main technique and gene targets reported for detecting the bovine leukemia virus.
-
•
We calculated an overall bovine leukemia virus detection frequency of 27%.
Keywords: Bovine leukemia virus, Humans, Epidemiology, Risk factors
Abstract
To review the available studies on the frequency of detection of the bovine leukemia virus in human samples, a systematic review with meta-analysis of the scientific literature was carried out, including papers published in English, Spanish, and Portuguese in 5 multidisciplinary databases. We collected information from different populations following a detailed and reproducible search protocol in which two researchers verified the inclusion and exclusion criteria. We identified 759 articles, of which only 33 met the inclusion criteria. Analyzed studies reported that the presence of the virus was measured in human samples, such as paraffin-embedded breast tissue and peripheral blood from 10,398 individuals, through serological and molecular techniques. An overall virus frequency of 27% (Ranging between 17 and 37%) was observed, with a high-frequency data heterogeneity between studies. The presence of this virus in different human biological samples suggests the need to investigate further its transmission route to humans and its potential role in developing and progressing diseases.
1. Introduction
Bovine Leukemia Virus (BLV) belongs to the Deltaretrovirus genus and Retroviridae family; it is closely related to human T-cell lymphotropic virus 1 (HTLV-1) (Pluta et al., 2020). Morphologically, the viral particle has a diameter that ranges between 60 and 125 nm. Like all retroviruses, BLV possesses the genes gag, pro, pol, and env, which encode the internal structural proteins, viral protease, reverse transcriptase, and envelope glycoproteins of the virion, respectively, which are essential to produce infectious viral particles. Two identical long terminal repeats (LTRs) are flanking these genes (Chameettachal et al., 2023). Moreover, BLV and HTLV-1 have a pX sequence between the env gene and the 3′LTR region that encodes the regulatory proteins Tax and Rex. In BLV and HTLV-1, the Tax protein acts as an activator of transcription with oncogenic potential, while Rex interferes with the exportation of messenger RNA of both viruses from the nucleus (Derse, 1987; Felber et al., 1989; Willems et al., 1987; Willems et al., 1990).
BLV can naturally infect cattle, capybaras, and sheep. Nevertheless, in vitro, BLV can infect different cell types from various species, including humans, such as human lung embryonic cells (WI-38), human tumor (ARH77 and K562), and neural-origin human cells are also highly susceptible to BLV infection (Altaner et al., 1989; Camargos et al., 2004; Delarmelina et al., 2020). For many years, it was believed that humans were not exposed; however, various reports have emerged in the last decades about the presence of BLV in humans for the reason that the identification of different biological markers and gene fragments in the proviral stage (Giovanna et al., 2013; Buehring et al., 2014; Khalilian et al., 2019), proteins (Uribe et al., 2006) and antibodies reactive against the virus as a sample of exposure to this virus (Buehring et al., 2003). It is now known that BLV may be present in the breast (Buehring et al., 2014; Khalilian et al., 2019; Schwingel et al., 2019; Baltzell et al., 2018; Lendez et al., 2018; Buehring et al., 2017; Buehring et al., 2015), lung (Robinson et al., 2016), and blood cells (Buehring et al., 2019) in humans.
Studies published since 1976 demonstrated the ability of BLV to infect various human cell lines. Cell cultures from humans, primates (Chimpanzees and Rhesus monkeys), canines, sheep, goats, and bats were infected with BLV-infected cell lines and by inoculation with cell-free virus preparations in all cell cultures, the production of the complete virus was observed (Graves and Ferrer, 1976). Several studies report the persistence of BLV in various types of cells (Doménech et al., 2000; Heeney et al., 1992; Rovnak et al., 1991; Aida et al., 1989). However, it has been defined that the primary cellular target of the virus is B lymphocyte that expresses surface immunoglobulin M. In addition to B lymphocytes, BLV also persists in cells of the monocyte/macrophage lineage (Aida et al., 1993; Mirsky et al., 1993). Until now, two proteins involved in cellular transport have been proposed as potential cellular receptors for BLV, AP3D1, and CAT1/SLC7; these proteins are common in mammals, sharing high percentages of identity between species (Corredor et al., 2018; Bai et al., 2019). In the natural host, the infection is transmitted horizontally by transferring infected cells by direct contact, iatrogenic route, and through hematophagous insects (Ferrer, 1978; Gillet et al., 2007). Another transmission route is vertical transmission (cow to calf) across the placenta and colostrum and milk alimentation from infected animals (Ferrer, 1979).
With the evolution of diagnostic methods, various reports have emerged in the last decades about the presence of BLV in humans. Several studies have explored the possible link between BLV and human disease, especially breast cancer, but the data remains controversial (Buehring et al., 2003; Burridge, 1981; Burny et al., 1985). There are a few papers linking a related mouse retrovirus (Mouse mammary tumor virus [MMTV]) as well as other viruses to human breast cancer and other diseases (Lawson and Glenn, 2017). MMTV has convincingly been shown to infect and replicate in human cells (Indik et al., 2007), and even sites of integration of the MMTV provirus in infected human cells have been analyzed (Faschinger et al., 2008).
The exact route of BLV transmission for humans has yet to be discovered; some hypotheses about the BLV infection routes for humans are consuming raw milk and undercooked meat, close contact between humans and bovines, production of vaccines from cell cultures that use bovine fetal serum, and exposition to contaminated meat from butchers and slaughterhouses. However, all these forms of transmission are yet to be confirmed (Schwingel et al., 2019).
Regardless of the lack of conclusive evidence about the role of BLV on human health, this study aims to review the BLV detection frequency in human biological samples to explore the available knowledge about this oncogenic retrovirus that affects the health of cattle herds could affect human health and potentially become a public health problem.
2. Methodology
2.1. Study design
Firstly, we systematically reviewed the literature based on the PRISMA 2020 (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines. Then we performed a statistical synthesis of the data.
2.2. Identification
We searched the information in the following databases: PubMed, Science Direct, Scielo, Scopus, Google Scholar (Including grey literature), and LILACS. The Mesh-indexed keywords, including "Leukemia Virus, Bovine", "prevalence", "risk factors", and "humans", and their synonyms in Spanish and Portuguese. The keywords were validated in the Thesauri Descriptors in Health Sciences (DeCS) and Medical Subject Headings. The combination gave rise to 9 different search strategies in the six databases. Additional related papers were collected by reviewing the references of the selected documents.
Some search strategies were the following: in PubMed (leukemia virus, bovine) AND (prevalence) AND (humans); in Scopus (leukemia virus, bovine) AND (risk factors) AND (humans); in LILACS (leukemia Virus, Bovine) AND (epidemiology) AND (humans). The temporality was established according to the first article published, which was found in the 70 s, until November 2022. The database obtained was imported into EndNote 20 for the elimination of duplicates.
2.3. Screening
Once the duplicates were eliminated, the generated database was imported into Rayyan—a web and mobile app for systematic reviews. Studies with the search terms in the title, abstract and keywords, published in English, Spanish, and Portuguese, available in full text, original, in humans, and that estimated the frequency of BLV or related risk factors were included. All remaining articles were excluded. The full texts of the preselected papers, including the bibliography, were reviewed for articles not retrieved during the search in the identification phase, and disagreements between the two reviewers were resolved.
The following criteria were applied for the selection of qualified studies in this research:
-
(a)
Original articles that estimate the frequency of BLV in humans or the probable risk factors associated with its detection in humans.
-
(b)
Studies that specify the technique used for the detection of BLV.
While those studies focused on the detection and risk factors associated with BLV transmission in animal species were excluded.
2.4. Inclusion
the papers that met the criteria described above were included for the qualitative synthesis of the variables title, authors, year, place of production and publication, study population, identification method, target and sample used, number of people evaluated, and number of positive people. The data obtained were recorded in a spreadsheet format (Microsoft Excel) table.
The process of identification, screening and analysis of the files was carried out by two researchers independently following the PRISMA 2020 statement checklist. The methodological quality was evaluated with the criteria of the STROBE guide (Strengthening the Reporting of Observational Studies in Epidemiology). Though this is an editorial guide, it contains standards that allow evaluation of the methodological quality of the descriptive studies.
2.5. Analysis
Data tables summarize the information extracted from the papers qualitatively, registering for each one the variables title, author(s), year of publication, year of development of the study, journal, URL, country of publication, country of development of the study, language, type of study, sample, study population, the technique and target of diagnosis, number of people studied, number of positive people and reported prevalence. To consolidate the information reported in the studies identified from the systematic review (n = 33), a meta-analysis was performed to estimate the global proportion of positives. Statistical synthesis was carried out to assess the global frequency of BLV (data available from 31 studies) and the Odds Ratio with 95% confidence intervals (data available from 12 studies) (Fig. 1). Two global measures were estimated, positivity frequency and log odds of positivity, considering people with cancer as exposed. The included studies presented a lot of variabilities I2 >90%, so the global effect was estimated with a random-effects model. We performed the statistical analysis using Jamovi version 2.3.18.
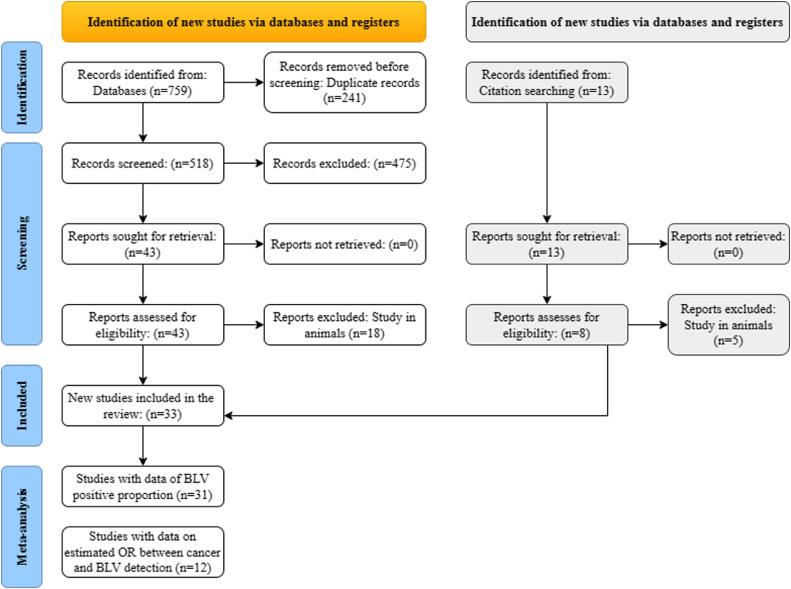
Fig. 1.
The flowchart describing the screening process of 33 articles analyzed in this study.
3. Results
In the initial search, we identified 759 articles by combining the keywords in 9 search strategies in the six databases, PubMed (n = 135), Science Direct (n = 38), Scielo (n = 1), Scopus (n = 113), Google Scholar (n = 187) and LILACS (n = 285). Of these, we removed 241 papers because they were duplicating. Then, two researchers independently read and analyzed the title, keywords, and abstracts of 518 articles. Reports that focused on general aspects of the virus were research review articles or written in a language other than English, Spanish, or Portuguese were also excluded. We selected 43 papers that met the inclusion criteria for reading the full text. Of these, 18 articles that detected the presence of the virus or reported risk factors associated with the virus in animal samples or farm products were excluded resulting in 25 preselected articles. In addition, eight papers identified by screening the bibliography after reading the full text of the 25 preselected articles were included in the present study. Finally, in this study, we analyzed 33 papers (Table 1).
Table 1.
Information summary about the 33 articles analyzed.
| Publication information | |||||
|---|---|---|---|---|---|
| # I.D. | Title | Year | Journal | Site* | Refs. |
| 1 | Seroepidemiologic testing in men for evidence of antibodies to the feline leukemia virus and bovine leukemia virus | 1976 | Bibliotheca Haematologica | USA |
Caldwell et al. (1976) |
| 2 | Seroepidemiologic studies on the possible relationships of human and bovine leukemia: Brief communication | 1977 | Journal of the National Cancer Institute | USA | Donham et al. (1977) |
| 3 | No Involvement of Bovine Leukemia Virus in Childhood Acute Lymphoblastic Leukemia and Non-Hodgkin's Lymphoma | 1988 | Cancer Research | USA | Bender et al. (1988) |
| 4 | Lack of evidence for infection with known human and animal retroviruses in patients with chronic fatigue syndrome | 1994 | Clinical Infectious Diseases | USA | Heneine et al. (1994) |
| 5 | Evaluation of HIV type 1 western blot-indeterminate blood donors for the presence of human or bovine retroviruses | 1995 | AIDS Res Hum Retroviruses | USA | Sherman et al. (1995) |
| 6 | Humans have antibodies reactive with Bovine leukemia virus | 2003 | AIDS Res Hum Retroviruses | USA | Buehring et al. (2003) |
| 7 | Investigation of the bovine leukemia virus proviral DNA in human leukemias and lung cancers in Korea | 2005 | Journal of Korean Medical Science | South Korea | Lee et al. (2005) |
| 8 | Estudio del potencial zoonotico del virus de la leucosis bovina y su presencia en casos de cancer de seno | 2006 | Universitas Scientiarum | Colombia | Uribe et al. (2006) |
| 9 | Serological and genomic detection of bovine leukemia virus in human and cattle samples | 2010 | Iranian Journal of Veterinary Medicine | Iran | غن et al. (2010) |
| 10 | Serological detection of bovine leukemia virus in slaughterhouse workers from San Nicols de los Garza, Nuevo Len, Mexico | 2013 | African Journal of Microbiology Research | Mexico | Zamora-Avila et al. (2013) |
| 11 | Bovine leukemia virus gene segment detected in human breast tissue | 2013 | Scientific research | Colombia | Giovanna et al. (2013) |
| 12 | Bovine leukemia virus DNA in human breast tissue | 2014 | Emerg Infect Dis | USA | Buehring et al. (2014) |
| 13 | Exposure to Bovine Leukemia Virus Is Associated with Breast Cancer: A Case-Control Study | 2015 | PLoS One | USA | Buehring et al. (2015) |
| 14 | Whole genome sequencing of 51 breast cancers reveals that tumors are devoid of bovine leukemia virus DNA | 2016 | Retrovirology | Mexico / USA | Gillet and Willems (2016) |
| 15 | Multiple oncogenic viruses are present in human breast tissues before development of virus associated breast cancer | 2017 | Infectious Agents and Cancer | Australia | Lawson and Glenn (2017) |
| 16 | Bovine leukemia virus linked to breast cancer in Australian women and identified before breast cancer development | 2017 | PLoS One | Australia | Buehring et al. (2017) |
| 17 | Bovine leukemia virus linked to breast cancer but not coinfection with human papillomavirus: Case-control study of women in Texas | 2018 | Cancer | USA | Baltzell et al. (2018) |
| 18 | Bovine leukemia virus presence in breast tissue of Argentinian females and its association with cell proliferation and prognosis markers | 2018 | Multidisciplinary Cancer Investigation | Argentina | Lendez et al. (2018) |
| 19 | Molecular Detection of Bovine Leukemia Virus (BLV) in Patients with Breast Cancer in Khartoum State, Sudan | 2019 | Scholarena Journal of Cancer Science | Sudan | Ahmed et al. (2020) |
| 20 | Presencia de anticuerpos contra el Virus de la Leucosis Bovina (VLB) en mujeres colombianas | 2019 | Undergraduate dissertation | Colombia | Trujillo Piñeros (2019) |
| 21 | Bovine leukemia virus detected in the breast tissue and blood of Iranian women | 2019 | Microbial Pathogenesis | Iran | Khalilian et al. (2019) |
| 22 | Bovine leukemia virus DNA associated with breast cancer in women from South Brazil | 2019 | Scientific Reports | Brazil | Schwingel et al. (2019) |
| 23 | Bovine leukemia virus discovered in human blood | 2019 | BMC infectious diseases | USA | Buehring et al. (2019) |
| 24 | Detecção de DNA do vírus da leucose bovina (BLV) em tecidos mamários humanos | 2020 | Master's Thesis | Brazil | Nogueira APMdS (2020) |
| 25 | High positivity values for bovine leukemia virus in human breast cancer cases from Minas Gerais, Brazil | 2020 | PLoS One | Brazil | Delarmelina et al. (2020) |
| 26 | Absence of bovine leukemia virus in the buffy coats of breast cancer cases from Alabama, USA | 2021 | Microbial Pathogenesis | USA | Adekanmbi et al. (2021) |
| 27 | Prevalence of bovine leukosis virus in water buffaloes in West-central Colombia | 2021 | Revista Mexicana De Ciencias Pecuarias | Colombia | Flórez et al. (2021) |
| 28 | Risk factor for breast cancer development under exposure to bovine leukemia virus in Colombian women: A case-control study | 2021 | PLoS One | Colombia | Olaya-Galan et al. (2021) |
| 29 | Virus de la leucosis bovina (VLB) y evidencias de su potencial zoonótico | 2021 | Doctoral thesis | Colombia | Olaya-Galán (2021) |
| 30 | Virus de la leucosis bovina: evaluación de la presencia del provirus en sangre humana y analisis in silico de los receptores celulares involucrados en su interacción | 2021 | Master's Thesis | Colombia | Velandia Álvarez (2021) |
| 31 | Bovine leukemia viral DNA found on human breast tissue is genetically related to the cattle virus | 2021 | One Health | Brazil | Canova et al. (2021) |
| 32 | Co-Circulation of Bovine Leukemia Virus Haplotypes among Humans, Animals, and Food Products: New Insights of Its Zoonotic Potential | 2021 | Int J Environ Res Public Health | Colombia | Corredor-Figueroa et al. (2021) |
| 33 | Molecular investigation of possible relationships concerning bovine leukemia virus and breast cancer | 2022 | Scientific reports | Pakistan | Khan et al. (2022) |
Country of origin for the human population studied.
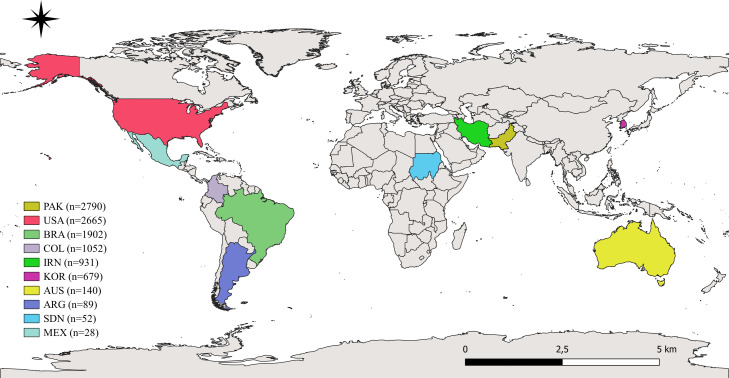
The selected scientific articles were published mainly in the United States (n = 13), the United Kingdom (n = 5), and Colombia (n = 4) and were published between the years 1976 and 2022. The type of studies consisted mainly of descriptive studies (67%) and cases and controls (33%) (Table 2). In total, in the 33 papers evaluated, samples from 10,398 humans were analyzed, of which about 63% were women. Analyzed studies were carried out in countries such as Pakistan (n = 2790; 26.8%), the USA (n = 2665; 25.6%), Brazil (n = 1902; 18.3%), Colombia (n = 1052; 10.1%), Iran (n = 931; 9%), South Korea (n = 679; 6.5%), Australia (n = 140; 1.3%), Argentina (n = 89; 0.9%), Sudan (n = 52; 0.5%), Mexico (n = 28; 0.3%), and there were 70 participants from no specified origin (0.7%) (Fig. 2).
Table 2.
Frequency of different types of variables from the 33 studies included in this systematic review.
| Variable | n | % |
|---|---|---|
| Study | ||
| Descriptive | 22 | 67 |
| Case-control | 11 | 33 |
| Population group | ||
| Female | 6595 | 63 |
| Male | 318 | 3 |
| Kids | 293 | 3 |
| Not specified | 3192 | 31 |
| Sample | ||
| B.M. | 1 | 2.6 |
| BM & PB | 1 | 2.6 |
| BTFFP | 19 | 50 |
| Fresh breast tissue | 1 | 2.6 |
| P.B./ Cell fraction | 8 | 21 |
| P.B./ Serum | 5 | 13.2 |
| P.B./ Cell fraction & serum | 3 | 8 |
| Technique | ||
| AGID assay | 3 | 6.8 |
| Complement fixation test | 1 | 2.3 |
| ELISA | 4 | 9 |
| IHC | 3 | 6.8 |
| Immunoblotting | 1 | 2.3 |
| Immunoperoxidase | 1 | 2.3 |
| Nested PCR | 13 | 29.5 |
| Nested PCR & Nested In situ PCR | 1 | 2.3 |
| PCR | 6 | 13.6 |
| PCR In situ | 7 | 15.9 |
| qPCR | 1 | 2.3 |
| Southern blot | 1 | 2.3 |
| WSG | 2 | 4.5 |
| Target | ||
| env | 8 | 13 |
| gag | 12 | 19 |
| tax | 16 | 26 |
| pol | 5 | 8 |
| px | 1 | 2 |
| LTR | 4 | 6 |
| WSG | 1 | 2 |
| gp51 | 4 | 6 |
| p24 | 6 | 10 |
| Not specified | 5 | 8 |
P.B.: Peripherally blood, BTFFP: Breast tissue specimen fixed in neutral formalin and embedded in paraffin, B.M.: Bone marrow, WSG: Whole genome sequencing, IHC: Immunohistochemistry, AGID: Agar gel immunodiffusion, ELISA: Enzyme-Linked ImmunoSorbent Assay, PCR: Polymerase Chain Reaction.
Fig. 2.
Geographic location of the origin of the populations included in the 33 articles analyzed in this study. PAK: Pakistan; USA: United States of America; BRA: Brazil; COL: Colombia; IRN: Iran; KOR: South Korea; AUS: Australia; ARG: Argentina; SDN: Sudan; MEX: Mexico, n: Number of people analyzed by country.
Human samples analyzed for BLV presence were peripheral blood (42%), neutral formalin-fixed paraffin-embedded breast tissue specimen (50%), fresh breast tissue (2.6%), and others. One study analyzed two samples from the same person, seeking to correlate the results of BLV detection in peripheral blood and fresh breast tissue. The authors found that BLV was detected both in blood and breast tissues with a correlation of 94% in the positive samples of the study (Olaya-Galan et al., 2021; Corredor-Figueroa et al., 2021) (Table 3).
Table 3.
Frequency of BLV detection by sample type, population analyzed, and technique used.
| # I.D. | Sample | Population | Population characteristics | Technique | n | Frequency of positive samples |
|---|---|---|---|---|---|---|
| 1 | P.B./ Serum | Volunteer men and women | Volunteer men and women | Complement fixation test | 192 | 0% |
| 2 | P.B./ Serum | Volunteer men and women | Leukemia patients | AGID assay | 358 | 0% |
| Veterinarians | ||||||
| Dairy farmers | ||||||
| Control group | ||||||
| 3 | B.M. and P.B./ Serum | Kids with ALL or NHL | Cases | Southern blot | 293 | 0% |
| Healthy kids | Control group | |||||
| 4 | P.B./ Cell fraction | Chronic fatigue syndrome case-patient | Health control | PCR | 42 | 0% |
| Chronic fatigue syndrome case-patient | ||||||
| 5 | P.B./ Cell fraction | Blood donors | Blood donors | PCR | 20 | 0% |
| 6 | P.B./ Serum | Volunteer men and women | Volunteer men and women | Immunoblotting | 257 | 74.3% |
| AGID assay | 25 | 0% | ||||
| 7 | B.M. | Leukemia patients | Acute myeloid leukemia patients | Nested PCR | 517 | 0% |
| Chronic myelogenous leukemia cases patients | ||||||
| BTFFP | Lung cancer cases | Lung cancer cases | 162 | |||
| 8 | BTFFP | Women with breast surgery | Malignant breast cancer | Immunoperoxidase | 56 | 7% |
| 9 | P.B./ Cell fraction & serum | Volunteer men and women | Volunteer men and women | Nested PCR | 77 | 9% |
| ELISA | 454 | 13% | ||||
| 10 | P.B./ Cell fraction & serum | Slaughterhouse workers | Slaughterhouse workers | AGID assay | 28 | 7% |
| PCR | 0% | |||||
| 11 | BTFFP | Women with breast surgery | Benign (controls) | PCR | 106 | 45% |
| Malignant (cases) | 35% | |||||
| 12 | BTFFP | Women with breast surgery | Benign and malignant breast surgery | Nested PCR/ Nested-In situ PCR | 219 | 44% |
| IHC | 215 | 16% | ||||
| 13 | BTFFP | Women with breast surgery | Benign (controls) | PCR in situ | 239 | 29% |
| Malignant (cases) | 59% | |||||
| Premalignant | 38% | |||||
| NA | IHC | 236 | 5% | |||
| 14 | BTFFP | Women with breast surgery | DNA sequences from whole genomes of normal breast tissues | WSG | 70 | 0% |
| DNA sequences from whole genomes of breast tumors | ||||||
| 15 | BTFFP | Women with breast surgery | Benign (controls) | PCR in situ | 44 | 35% |
| Malignant (cases) | 74% | |||||
| 16 | BTFFP | Women with breast surgery | Benign (controls) | PCR in situ | 96 | 41% |
| Malignant (cases) | 80% | |||||
| 17 | BTFFP | Women with breast surgery | Benign (controls) | PCR in situ | 216 | 20% |
| Malignant (cases) | 57% | |||||
| Premalignant | 34% | |||||
| 18 | BTFFP | Women with breast surgery | Benign (controls) | PCR in situ | 89 | 25% |
| Malignant (cases) | 22.4% | |||||
| 19 | BTFFP | Women with breast surgery | Breast cancer patients | PCR | 52 | 3.8% |
| 20 | P.B./ Serum | Volunteer women | Volunteer women | ELISA | 226 | 0% |
| 21 | BTFFP | Women with breast surgery | Malignant (cases) | Nested PCR | 200 | 24% |
| Non-malign | 2% | |||||
| Non-tum | 4% | |||||
| Malignant (cases) | 6% | |||||
| Non-malign | 0% | |||||
| Non-tum | 2% | |||||
| P.B./ Cell fraction | Volunteer women | Volunteer women | 200 | 17% | ||
| 22 | BTFFP | Women with breast surgery | Benign (controls) | Nested PCR | 144 | 14% |
| Malignant (cases) | 31% | |||||
| P.B./ Serum | Blood donors | Blood donors | ELISA | 1500 | 0.13% | |
| 23 | P.B./ Cell fraction & serum | Volunteer women | Volunteer women | Nested PCR | 95 | 38% |
| ELISA | 58% | |||||
| 24 | BTFFP | Women with breast surgery | Benign (controls) | Nested PCR | 111 | 73% |
| Malignant (cases) | 81% | |||||
| No diagnosis | 45% | |||||
| 25 | BTFFP | Women with breast surgery | Benign (controls) | Nested PCR | 88 | 59% |
| Malignant (cases) | 96% | |||||
| 26 | P.B./ Cell fraction | Volunteer women | Breast cancer patients | qPCR | 258 | 0% |
| WSG | ||||||
| 27 | P.B./ Cell fraction | Volunteer men and women | Volunteer men and women | Nested PCR | 9 | 0% |
| 28 | P.B./ Cell fraction | Women with breast surgery | Benign (controls) | Nested PCR/PCR in situ | 158 | 48% |
| Malignant (cases) | 62% | |||||
| Fresh breast tissue | Benign (controls) | IHC | 10% | |||
| Malignant (cases) | 12% | |||||
| 29 | BTFFP | Dead women | Benign | Nested PCR | 145 | 71% |
| PCR in situ | ||||||
| 30 | P.B./ Cell fraction | Volunteer men and women | Volunteer men and women | Nested PCR | 352 | 27.3% |
| 31 | BTFFP | Women with breast surgery | Women with breast surgery | PCR | 59 | 86% |
| Nested PCR | ||||||
| 32 | P.B./ Cell fraction | Women with breast surgery | Same population and technique that study #ID 28 | |||
| BTFFP | ||||||
| 33 | BTFFP | Women with breast surgery | Benign (controls) | Nested PCR | 2790 | 10% |
| Malignant (cases) | 27% | |||||
# ID: Corresponds to the same article in Table 1, ALL: Acute Lymphoblastic Leukemia, NHL: Non-Hodgkin's Lymphoma, P.B.: Peripherally blood, BTFFP: Breast tissue specimen fixed in neutral formalin and embedded in paraffin, B.M.: Bone marrow, WSG: Whole genome sequencing, AGID assay: Agar gel immunodiffusion assay, ELISA: Enzyme-Linked Immunosorbent Assay, PCR: Polymerase Chain Reaction, IHC: Immunohistochemistry.
Serological studies based on the detection of antibodies against BLV in humans show conflicting results. Studies based only on classical serological techniques such as complement fixation and agar gel immunodiffusion (AGID), showed no evidence of human antibodies against BLV (Caldwell et al., 1976; Donham et al., 1977). One study compared the sensitivity of AGID test versus the immunoblot, which is a more sensitive technique. It detected anti-BLV antibodies by immunoblot in 39% of serum tested negative by AGID (Buehring et al., 2003). In a study which evaluated the presence of BLV by AGID and PCR in the same blood sample, they found a positivity of 7% with the AGID technique and 0% with the PCR test, for which the authors conclude that the presence of antibodies may be due to previous exposure to virus antigens and does not guarantee an active viral infection (Zamora-Avila et al., 2013). On the other hand, solid phase serological techniques, such as enzyme immunoassay (ELISA) using the virus p24 antigen, showed that humans have IgG, IgM and IgA antibodies against BLV, indicating that antibodies reactive with the BLV antigen capsid may serve as a biomarker for BLV exposure (Buehring et al., 2019; غن et al., 2010).
BLV p24 protein was detected in human breast tissue samples analyzed by Immunohistochemistry (IHC), indicating the presence of viral proteins as evidence of active viral replication (Buehring et al., 2014; Buehring et al., 2015).
The samples were mainly analyzed by molecular techniques such as nested PCR (Polymerase Chain Reaction) using different target genes such as tax, gag, env, pol, and LTR. One study analyzed the whole genome of the BLV. The other studies used molecular and serological virus detection techniques (Table 3).
In general, the results of PCR techniques indicate the presence of the virus in the proviral stage (integrated into the host cell genome). Molecular techniques employed include standard PCR, which is based on DNA extracts from target tissues and cannot be used to identify the location of viral gene sequences in specific cell types, unlike in situ PCR which can identify viral gene sequences in particular cells, studies using in situ techniques were able to confirm that BLV was located within mammary epithelial cells (Buehring et al., 2014; Lendez et al., 2018; Buehring et al., 2017; Buehring et al., 2015; Lawson and Glenn, 2017; Olaya-Galan et al., 2021).
Only one study evaluated the presence of BLV using probe-based qPCR, which is a much more sensitive and specific molecular technique and is widely used for the evaluation of gene expression of specific genes in a relative way. However, neither in this study nor in any of the studies included in this review was gene expression assessed. The authors did not detect the presence of BLV by qPCR or WSG in peripheral blood samples from women with cancer; however, some important aspects in the methodology of this study are not reported, such as the negative controls or the inhibition controls they used to confirm the functionality of their detection assay (Adekanmbi et al., 2021).
Two studies performed massive genome sequencing to identify specific viral gene segments but did not yield positive results (Gillet and Willems, 2016; Adekanmbi et al., 2021). This result may be because whole genome sequencing detects only viral DNA integrated into the human genome, which may be present at too low concentrations (<1% of reads) to be caught without amplification (Buehring et al., 2019).
Nested PCR is a modification of the standard PCR that was designed to improve sensitivity and specificity. Due to this, it was the most used technique in the different studies included in this review (29.5%), reporting different percentages of positivity (Table 3). A possible explanation for the negative results could be due to low viral loads, partial deletion or polymorphism of the target viral gene that could hinder hybridization with viral DNA, accidental loss of virus during cell division which can result in a virally transformed cell with no detectable virus (“hit-and-run virus” phenomenon) (Joshi and Buehring, 2012).
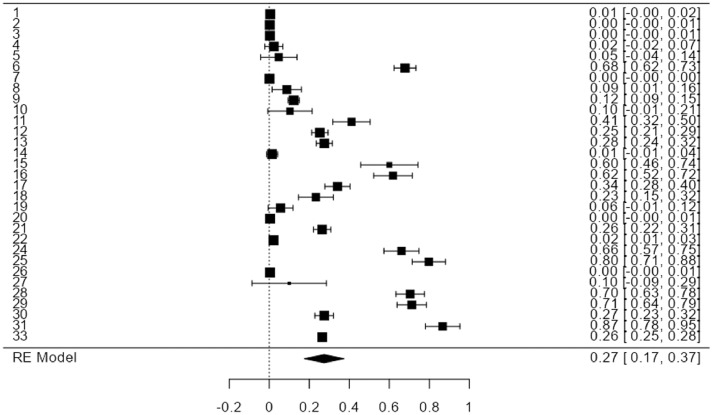
The results of the studies are very heterogeneous with the index I2 =99%, so the global effect was estimated with a random-effects model. Overall, we observed high heterogeneity in the virus detection frequency according to the sample and the technique involved. The analysis was carried out separately according to the design of the studies. In the descriptive ones, a slightly lower frequency was found at 24% (12% to 36%), while in the case-control studies was 34% (16% to 52%). We observed that the frequency of positivity ranged from 0 to 87%. For this reason, the overall estimate resulted in a positivity frequency of 27% with an interval between 17 and 37% (Fig. 3).
Fig. 3.
Forest plot of the individual and global BLV frequencies of positivity detected in the 31 analyzed studies. It shows the estimates of each study, where studies with positivity from 1% to 87% are observed; in eight studies, there were percentages of positivity greater than 50%, eight presented zero positive cases, and 15 studies had positivity between 2 and 41%; the overall estimate of positivity was 27% with a range between 17 and 37%. An analysis excluding data from the four gray literature documents (Trujillo Piñeros, 2019; Nogueira APMdS, 2020; Olaya-Galán, 2021; Velandia Álvarez, 2021) showed the BLV frequencies of positivity decreased by 2%, from 27% to 25%, and the confidence interval ranged from 14% to 35%.
Only the study from Corredor-Figueroa et al. (Corredor-Figueroa et al., 2021) analyzed the potential exposure factors related to the presence of the virus. Through bivariate analysis, they found a correlation between BLV presence and the consumption of dairy products, such as raw milk and homemade yoghurt, and the number of dairy products consumed by the person. Also, they observed a significant statical relationship between virus presence and age (≥50, <50; p = 0.039) and the city of origin of the participants (Bogotá or other, p = 0.036). This study estimated that women with higher consumption of raw milk and homemade yoghurt had a higher risk of the presence of BLV (OR = 2.424, 95% CI: 1.063–5.527, p = 0.035).
Fifteen studies reported Bovine Leukemia Virus gene sequences by PCR in tissue samples from human breast cancers (Table 3). In a PCR-based case-control study, 67 (59%) of 114 US breast cancers tested positive for BLV compared with 30 (29%) of 104 normal breast controls: odds ratio 3 (Buehring et al., 2015). A similar prevalence pattern is observed in the studies included in this review, which had been designed as cases and controls (n = 12) (Table 3). In another study carried out in the USA, an increase in the prevalence of BLV was observed in benign breast tissues (20%), premalignant breast tissues (34%) and malignant breast tissues (57.4%) (Baltzell et al., 2018).
In the papers we reviewed, BLV's prevalence is significantly higher in breast cancer tissues compared to normal or benign control breast tissues. The presence of the infectious agent in normal or benign tissues before cancer development is a key criterion for a causal role in cancer (Hill, 1965). A study using the PCR-in situ technique identified BLV in 23 (74%) of 31 benign breast biopsy tissues 3 to 10 years before the development of BLV-positive breast cancers in the same Australian patients (Buehring et al., 2017).
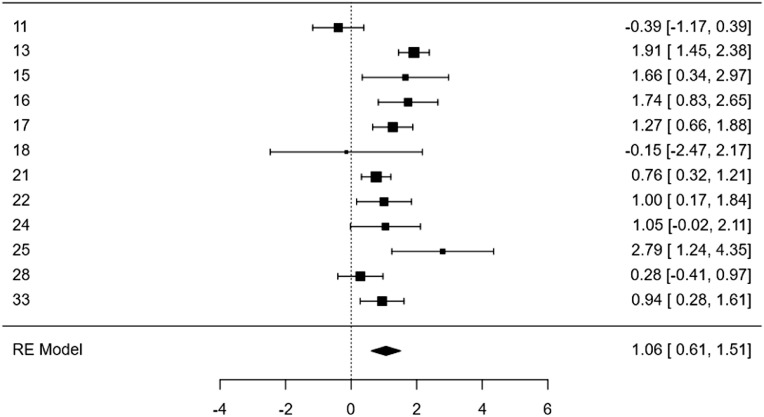
In 12 of the studies, they compared positivity between women with and without breast cancer, for which we evaluated the effect by estimating the log odds and the odds ratio (OR) was assessed. Two studies found a negative point estimate, that is, it decreases the possibility of being positive; the other ten studies showed results indicating that having cancer increases the probability of being positive, and nine showed statistically significant values (intervals do not include zero). The overall estimate was 1.06, which means that the presence of cancer increases the possibility of being positive. The chance of being positive is 2.88 times compared to those without breast cancer, which can vary between 1.84 times and 4.53 (Fig. 4).
Fig. 4.
Log of Odds of the Individual and Global data from 12 analyzed studies. The figure shows how cancer increases the possibility of being positive for BLV in most studies (10/12). Overall, the log odds estimate showed that having cancer increases the chance of being positive for the virus, OR global=e^1.06=2.88 (1.84–4.53), which means that in women with cancer, the possibility of being positive for BLV is 2.88 times more than those without cancer, varying between 1.84 and 4.53 times.
4. Discussion
This review included 10,398 people evaluated using different techniques to detect the presence of BLV. To date, the frequency of detection of BLV by analysing sequences, antigens, or antibodies have been found in 27% (Ranging between 17 and 37%) of people evaluated. The BLV frequencies in the population-assessed groups presented a significant heterogeneity according to the characteristics of the population, the sample and the diagnostic technique used. For example, in a women group diagnosed with a malignancy in the breast tissue, the presence of the virus was evaluated using the nested-PCR method and found a positivity of 95.9%, in contrast in the same study, in the group of women without a diagnosis of malignancy in the breast tissue, the BLV frequency was 59% (Delarmelina et al., 2020).
In general, molecular biology techniques have high sensitivity and specificity. For example, the in-situ PCR technique decreases cross-contamination with the positive control or between samples because DNA is not extracted and fixed within control cells and human tissues and cannot escape and generate cross-contamination. Any contaminating molecule amplified during PCR is eliminated during the rinse steps, while the amplified intracellular DNA remains fixed within the cellular architecture (Baltzell et al., 2018).
In the study in which AGID-positive and PCR-negative results were obtained, the authors conclude that one of the following conditions is likely: (1) Constant exposure may have occurred without infection, eliciting an aroused immune response; however, the virus could not integrate into the human genome. (2) Infected lymphocytes may have become trapped in lymphoid organs and were not in the peripheral circulation. (3) The number of provirus particles may have been deficient, and nested PCR would have been needed to increase sensitivity (Zamora-Avila et al., 2013).
The most common molecular techniques used to detect BLV presence were directed at different targets of the proviral DNA, such as env, gag, pol, tax, and LTR. Most studies use only one of these targets, and some studies use two or more of these, which may increase the likelihood of detecting the virus since partial genome deletions after integration into host cells are considered an essential mechanism of immune response evasion (Rosewick et al., 2017). In a study of the HTLV-1 virus carried out by Kamihira et al., which is closely related to BLV, deletions of the provirus integrated into the host genome began in the gag region, followed by deletions of the pol and env genes; in contrast, tax and LTR regions were the least frequently deleted genes (Kamihira et al., 2005). Analogous results were observed in specimens from cattle infected with BLV; deletions involving parts of gag and env and all of the pol were frequent (Gillet et al., 2007). Therefore, choosing hybridization primers or probes is an important point to consider in designing PCR assays and could be another potential reason for negative results.
Using whole genome sequencing methods, BLV was not identified in 70 breast tissue samples or 20 peripheral blood samples from women with breast cancer (Gillet and Willems, 2016; Adekanmbi et al., 2021). The reason for the negative result based on whole genome sequencing is unclear. One plausible explanation is that complete genome sequencing techniques are not as sensitive as amplification techniques such as PCR, and the authors assumed that BLV would have to be present in all cells in the tissue, which may not be the case. Retroviral genomes rarely exceed 10–12 kb and constitute a minor fraction of the infected host cell genome. The infected cell type may include only a small fraction of the sample, and infected cells may contain a relatively low number of copies of the viral genome (Lawson and Glenn, 2017).
Early studies of BLV in humans based on serologic tests such as AGID and complement fixation tests reported no evidence of the virus in human samples. A possible explanation for some of the negative results could be that sufficiently sensitive reagents and techniques (Buehring et al., 2003). Serological methods based on detecting antibodies directed to virus antigens such as p24 and gp51. This last is an envelope glycoprotein subject to variation within different viral subspecies. Thus, the epitope targeted by the detecting monoclonal antibody could not be present on BLV sub-species that circulate in the region studied and, as such, infected individual's bovine or human (Schwingel et al., 2019). The detection of antibodies against viruses was widely used in different studies; these techniques are useful for diagnosing viral diseases; however, in the case of BLV infections, relying on antibodies to prove the infection has several disadvantages. BLV may not express the p24 capsid protein in blood cells and may not replicate there (Buehring et al., 2019). Studies in cattle indicate that lymphocytes harbouring the BLV provirus rarely produce extracellular virions or express viral proteins, even though cattle have antibodies to BLV (Buehring et al., 2003)
None of the included studies was specifically designed to determine the route of exposure of humans to BLV. Some of the articles mention the consumption of raw milk as a potential transmission route, but this was not a variable under study. BLV transmission from cattle to humans is also plausible, as BLV is widespread in beef and dairy herds. Although pasteurization renders the virus noninfectious, and presumably thorough beef cooking does as well, many people have drunk raw milk and eaten raw or undercooked meat at some point (Buehring et al., 2015).
In the studies included in this review, we observed that the prevalence of BLV is significantly higher in breast cancer tissues compared to normal or benign control breast tissues. In 12 of the studies, they compared positivity between women with and without breast cancer, for which we evaluated the effect by estimating the log odds and the odds ratio (OR) was calculated. The overall estimate was 1.06, which means that the presence of cancer increases the possibility of being positive. The possibility of being positive is 2.88 times compared to those without breast cancer, which can vary between 1.84 times and 4.53. This observation supports that this oncovirus may have a potential role in human breast cancer.
Although many steps are needed to establish causality for any disease, a statistically significant association between the disease and the suspected agent is the first step. Validation of this association by other investigators in different populations is an essential subsequent step (Hill, 1965). Although case-control studies are not conclusive in themselves, this type of study is widely accepted as a valid method of establishing an association between the presence of a causative agent and the outcome of the disease. It would be desirable for prospective studies that show that the viral infection preceded the development of detectable cancer to support the idea of a causal association of BLV with breast cancer. In addition, it is crucial to carry out studies focused on demonstrating that the virus can transform normal cells into malignant cells, understand the oncogenic mechanism of the virus and identify the route of viral transmission.
Depending on the country of origin, all studies have variable percentages of BLV presence, possibly due to different lifestyles, diets, ages, types, and the number of sample sizes used, and techniques applied to detect the virus occurrence. We observed high heterogeneity in the data of frequencies of BLV detection, estimating an overall frequency of 27% (Ranged between 17 and 37%). The percentage of BLV detected in human samples by any study suggests the need to investigate further its transmission route to humans and the role of the virus in developing and progressing diseases (Khalilian et al., 2019).
Funding
Willington Mendoza was supported as a research assistant in a proyect financed by the “Convocatoria conjunta de proyectos de I + D + i en el marco de la agenda regional de I + D -> i del grupo G8+1″ and the Government of Antioquia. The funders had no role in study design, data collection and analysis, publication decisions, or manuscript preparation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- بروجنی غن, Rabani M., Emam M., Tofighi E. Serological and genomic detection of bovine leukemia virus in human and cattle samples. Iran. J. Vet. Med. 2010;4(4):253–258. doi: 10.22059/ijvm.2010.22101. [DOI] [Google Scholar]
- Adekanmbi F., McNeely I., Omeler S., Kalalah A., Poudel A., Merner N., et al. Absence of bovine leukemia virus in the buffy coats of breast cancer cases from Alabama, USA. Microb. Pathog. 2021:161. doi: 10.1016/j.micpath.2021.105238. [DOI] [PubMed] [Google Scholar]
- Ahmed M., Sami E., Elhasen Abdalla M., Israa M. Molecular detection of bovine leukemia virus (BLV) in patients with breast cancer in Khartoum State, Sudan. SAJ Cancer Sci. 2020;6:105. https://www.scholarena.com/article/Molecular-Detection-of-Bovine-Leukemia-Virus-BLV-in-Patients-with-Breast-Cancer-in-Khartoum-State-Sudan.pdf [Internet][p.]. Available from: [Google Scholar]
- Aida Y., Miyasaka M., Okada K., Onuma M., Kogure S., Suzuki M., et al. Further phenotypic characterization of target cells for bovine leukemia virus experimental infection in sheep. Am. J. Vet. Res. 1989;50(11):1946–1951. PubMed PMID: 2559634. [PubMed] [Google Scholar]
- Aida Y., Okada K., Amanuma H. Phenotype and ontogeny of cells carrying a tumor-associated antigen that is expressed on bovine leukemia virus-induced lymphosarcoma. Cancer Res. 1993;53(2):429–437. PubMed PMID: 8380256. [PubMed] [Google Scholar]
- Altaner C., Altanerová V., Bán J., Niwa O., Yokoro K. Human cells of neural origin are permissive for bovine leukemia virus. Neoplasma. 1989;36(6):691–695. PubMed PMID: 2559338. [PubMed] [Google Scholar]
- Bai L., Sato H., Kubo Y., Wada S., Aida Y. CAT1/SLC7A1 acts as a cellular receptor for bovine leukemia virus infection. FASEB J. 2019;33(12):14516–14527. doi: 10.1096/fj.201901528R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzell K.A., Shen H.M., Krishnamurthy S., Sison J.D., Nuovo G.J., Buehring G.C. Bovine leukemia virus linked to breast cancer but not coinfection with human papillomavirus: case-control study of women in Texas. Cancer. 2018;124(7):1342–1349. doi: 10.1002/cncr.31169. Epub 20171220PubMed PMID: 29266207. [DOI] [PubMed] [Google Scholar]
- Bender A.P., Robison L.L., Kashmiri S.V., McClain K.L., Woods W.G., Smithson W.A., et al. No involvement of bovine leukemia virus in childhood acute lymphoblastic leukemia and non-Hodgkin's lymphoma. Cancer Res. 1988;48(10):2919–2922. PubMed PMID: 2834051. [PubMed] [Google Scholar]
- Buehring G.C., Philpott S.M., Choi K.Y. Humans have antibodies reactive with bovine leukemia virus. AIDS Res. Hum. Retroviruses. 2003;19(12):1105–1113. doi: 10.1089/088922203771881202. [DOI] [PubMed] [Google Scholar]
- Buehring G.C., Shen H.M., Jensen H.M., Choi K.Y., Sun D., Nuovo G. Bovine leukemia virus DNA in human breast tissue. Emerg. Infect. Dis. 2014;20(5):772–782. doi: 10.3201/eid2005.131298. PubMed PMID: 24750974; PubMed Central PMCID: PMC4012802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehring G.C., Shen H.M., Jensen H.M., Jin D.L., Hudes M., Block G. Exposure to bovine leukemia virus is associated with breast cancer: a case-control study. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0134304. e0134304-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehring G.C., Shen H., Schwartz D.A., Lawson J.S. Bovine leukemia virus linked to breast cancer in Australian women and identified before breast cancer development. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179367. Epub 20170622PubMed PMID: 28640828; PubMed Central PMCID: PMC5480893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehring G.C., DeLaney A., Shen H., Chu D.L., Razavian N., Schwartz D.A., et al. Bovine leukemia virus discovered in human blood. BMC Infect. Dis. 2019;19(1):297. doi: 10.1186/s12879-019-3891-9. PubMed PMID: 30940091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burny A., Bruck C.E.M., Cleuter Y., Couez D., Deschamps J., Ghysdael J., et al. Bovine leukemia virus, a versatile agent with various pathogenic effects in various animal species. Cancer Res. 1985;45(9 Suppl) 4578s-82s. PubMed Central PMCID: 2410107. [PubMed] [Google Scholar]
- Burridge M.J. The zoonotic potential of bovine leukemia virus. Vet. Res. Commun. 1981;5(1):117–126. doi: 10.1007/BF02214976. [DOI] [PubMed] [Google Scholar]
- Caldwell G.G., Baumgartener L., Carter C., Cotter S., Currier R., Essex M., et al. Seroepidemiologic testing in man for evidence of antibodies to feline leukemia virus and bovine leukemia virus. Bibl Haematol. 1976 doi: 10.1159/000399139. No. 43:238-41. [DOI] [PubMed] [Google Scholar]
- Camargos M.F., Reis J.K.P.D., Leite R.C. Bovine leukemia virus. Virus Rev. Res. 2004;9(1):44–59. doi: 10.17525/vrrjournal.v9i1.221. [DOI] [Google Scholar]
- Canova R., Weber M.N., Budaszewski R.F., da Silva M.S., Schwingel D., Canal C.W., et al. Bovine leukemia viral DNA found on human breast tissue is genetically related to the cattle virus. One Health. 2021;13 doi: 10.1016/j.onehlt.2021.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chameettachal A., Mustafa F., Rizvi T.A. Understanding retroviral life cycle and its genomic RNA packaging. J. Mol. Biol. 2023;435(3) doi: 10.1016/j.jmb.2022.167924. [DOI] [PubMed] [Google Scholar]
- Corredor A.A.O., González J., Baquero L.A., Curtidor H., Olaya-Galán N.A.O., Patarroyo M.A.O., et al. In silico and in vitro analysis of boAP3d1 protein interaction with bovine leukaemia virus gp51. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0199397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor-Figueroa A.P., Olaya-Galán N.N., Velandia-Álvarez S., Muñoz M., Salas-Cárdenas S.P., Ibáñez-Pinilla M., et al. Co-circulation of bovine leukemia virus haplotypes among humans, animals, and food products: new insights of its zoonotic potential. Int. J. Environ. Res. Public Health. 2021;18(9) doi: 10.3390/ijerph18094883. Epub 20210504PubMed PMID: 34064361; PubMed Central PMCID: PMC8124648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarmelina E., Buzelin M.A., Souza B.S.D., Souto F.M., Bicalho J.M., Câmara R.J.F., et al. High positivity values for bovine leukemia virus in human breast cancer cases from Minas Gerais, Brazil. PLOS One. 2020;15(10) doi: 10.1371/journal.pone.0239745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J. Virol. 1987;61(8):2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. PubMed PMID: 3037109; PubMed Central PMCID: PMC255671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doménech A., Goyache J., Llames L., Jesús Payá M., Suárez G., Gómez-Lucía E. In vitro infection of cells of the monocytic/macrophage lineage with bovine leukaemia virus. J. Gen. Virol. 2000;81(Pt 1):109–118. doi: 10.1099/0022-1317-81-1-109. PubMed PMID: 10640548. [DOI] [PubMed] [Google Scholar]
- Donham K.J., VanDerMaaten M.J., Miller J.M., Kruse B.C., Rubino M.J. Seroepidemiologic studies on the possible relationships of human and bovine leukemia: brief communication. J. Natl. Cancer Inst. 1977;59(3):851–853. doi: 10.1093/jnci/59.3.851. [DOI] [PubMed] [Google Scholar]
- Faschinger A., Rouault F., Sollner J., Lukas A., Salmons B., Günzburg W.H., et al. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 2008;82(3):1360–1367. doi: 10.1128/jvi.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber B.K., Derse D., Athanassopoulos A., Campbell M., Pavlakis G.N. Cross-activation of the Rex proteins of HTLV-I and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol. 1989;1(3):318–328. PubMed Central PMCID: 2562124. [PubMed] [Google Scholar]
- Ferrer J.F. Vol. 12. Bulletin of the Pan American Health Organization [Internet]; 1978. pp. 304–309.https://www.scopus.com/inward/record.uri?eid=2-s2.0-0018217776&partnerID=40&md5=50500c1237a0986630fe696e64ff2bea (Advances in bovine leukemia). [Google Scholar]
- Ferrer J.F. Bovine leukosis: natural transmission and principles of control. J. Am. Vet. Med. Assoc. 1979;175(12):1281–1286. PubMed PMID: 231025. [PubMed] [Google Scholar]
- Flórez J.C.R., Peláez E.A.P., Marín N.T., López J.C.E., Corrales J.C.G. Prevalence of bovine leukosis virus in water buffaloes in West-central Colombia. Rev. Mex. Cienc. Pecu. 2021;12(2):419–436. doi: 10.22319/RMCP.V12I2.5459. [DOI] [Google Scholar]
- Gillet N.A., Willems L. Whole genome sequencing of 51 breast cancers reveals that tumors are devoid of bovine leukemia virus DNA. Retrovirology. 2016;13(1):75. doi: 10.1186/s12977-016-0308-3. PubMed Central PMCID: PMC5095936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet N., Florins A., Boxus M., Burteau C., Nigro A., Vandermeers F., et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology. 2007;4 doi: 10.1186/1742-4690-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanna M., Carlos U.J., María U.A., Gutiérrez M.F. Bovine leukemia virus gene segment detected in human breast tissue. Open J. Med. Microbiol. 2013;3(1):84–90. doi: 10.4236/ojmm.2013.31013. [DOI] [Google Scholar]
- Graves D.C., Ferrer J.F. In vitro transmission and propagation of the bovine leukemia virus in monolayer cell cultures. Cancer Res. 1976;36(11 Pt 1):4152–4159. PubMed PMID: 61801. [PubMed] [Google Scholar]
- Heeney J.L., Valli P.J., Jacobs R.M., Valli V.E. Evidence for bovine leukemia virus infection of peripheral blood monocytes and limited antigen expression in bovine lymphoid tissue. Lab. Invest. 1992;66(5):608–617. PubMed PMID: 1315405. [PubMed] [Google Scholar]
- Heneine W., Woods T.C., Sinha S.D., Khan A.S., Chapman L.E., Schonberger L.B., et al. Lack of evidence for infection with known human and animal retroviruses in patients with chronic fatigue syndrome. Clin. Infect. Dis. 1994;18 doi: 10.1093/clinids/18.Supplement_1.S121. S121-S5. [DOI] [PubMed] [Google Scholar]
- Hill A.B. The environment and disease: association or causation? Proc. R. Soc. Med. 1965;58(5):295–300. PubMed PMID: 14283879; PubMed Central PMCID: PMC1898525. [PMC free article] [PubMed] [Google Scholar]
- Indik S., Günzburg W.H., Kulich P., Salmons B., Rouault F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology. 2007;11(4:73) doi: 10.1186/1742-4690-4-73. PubMed Central PMCID: PMC2169256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi D., Buehring G.C. Are viruses associated with human breast cancer? scrutinizing the molecular evidence. Breast Cancer Res. Treat. 2012;135(1):1–15. doi: 10.1007/s10549-011-1921-4. Epub 20120125PubMed PMID: 22274134. [DOI] [PubMed] [Google Scholar]
- Kamihira S.S., TK K., Minami S., Uemura A., Akamatsu N., et al. Proviral status of HTLV-1 integrated into the host genomic DNA of adult T-cell leukemia cells. Clin. Lab. Haematol. 2005;27(4):235–241. doi: 10.1111/j.1365-2257.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- Khalilian M., Hosseini S.M., Madadgar O. Bovine leukemia virus detected in the breast tissue and blood of Iranian women. Microb. Pathog. 2019;135 doi: 10.1016/j.micpath.2019.103566. Epub 20190625PubMed PMID: 31252065. [DOI] [PubMed] [Google Scholar]
- Khalilian M., Hosseini S.M., Madadgar O. Bovine leukemia virus detected in the breast tissue and blood of Iranian women. Microb. Pathog. 2019;135 doi: 10.1016/j.micpath.2019.103566. [DOI] [PubMed] [Google Scholar]
- Khan Z., Abubakar M., Arshed M.J., Aslam R., Sattar S., Shah N.A., et al. Molecular investigation of possible relationships concerning bovine leukemia virus and breast cancer. Sci. Rep. 2022;12(1):1–8. doi: 10.1038/s41598-022-08181-5. PubMed Central PMCID: PMC8907172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson J.S., Glenn W.K. Multiple oncogenic viruses are present in human breast tissues before development of virus associated breast cancer. Infect. Agent Cancer. 2017;12(55) doi: 10.1186/s13027-017-0165-2. PubMed Central PMCID: PMC5644159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kim Y., Kang C.S., Cho D.H., Shin D.H., Yum Y.N., et al. Investigation of the bovine leukemia virus proviral DNA in human leukemias and lung cancers in Korea. J. Korean Med. Sci. 2005;20(4):603–606. doi: 10.3346/jkms.2005.20.4.603. PubMed Central PMCID: PMC2782155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendez P.A., Martinez Cuesta L., Nieto Farías M.V., Shen H.M., Dolcini G.L., Buehring G.C., et al. Bovine leukemia virus presence in breast tissue of Argentinian females and its association with cell proliferation and prognosis markers. Multidiscip. Cancer Investig. 2018;2(4):16–24. doi: 10.30699/acadpub.mci.4.16. [DOI] [Google Scholar]
- Mirsky M.L., Da Y., Lewin H.A. Detection of bovine leukemia virus proviral DNA in individual cells. PCR Methods Appl. 1993;2(4):333–340. doi: 10.1101/gr.2.4.333. PubMed PMID: 8391891. [DOI] [PubMed] [Google Scholar]
- Nogueira APMdS. Detecção do DNA do vírus da leucose bovina (VLB) em tecidos mamários humanos 2020. Available from: https://repositorio.ufpe.br/handle/123456789/40376.
- Olaya-Galán N.N. Virus de la leucosis bovina (VLB) y evidencias de su potencial zoonótico 2021. Available from: doi: 10.48713/10336_33690. [DOI]
- Olaya-Galan N.N., Salas-Cárdenas S.P., Rodriguez-Sarmiento J.L., Ibañez-Pinilla M., Monroy R., Corredor-Figueroa A.P., et al. Risk factor for breast cancer development under exposure to bovine leukemia virus in Colombian women: a case-control study. PLoS One. 2021;16(9 September) doi: 10.1371/journal.pone.0257492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A., Jaworski J.P., Douville R.N. Regulation of expression and latency in BLV and HTLV. Viruses. 2020;12(10) doi: 10.3390/v12101079. Epub 20200925PubMed PMID: 32992917; PubMed Central PMCID: PMC7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.A., Jaing C.J., Pierce Campbell C., Magliocco A., Xiong Y., Magliocco G., et al. Molecular evidence of viral DNA in non-small cell lung cancer and non-neoplastic lung. Br. J. Cancer. 2016;115(4):497–504. doi: 10.1038/bjc.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewick N., Durkin K., Artesi M., Marçais A., Hahaut V., Griebel P., et al. Cis-perturbation of cancer drivers by the HTLV-1/BLV proviruses is an early determinant of leukemogenesis. Nat. Commun. 2017;8:15264. doi: 10.1038/ncomms15264. Epub 20170523PubMed PMID: 28534499; PubMed Central PMCID: PMC5457497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J., Casey J.W., Boyd A.L., Gonda M.A., Cockerell G.L. Isolation of bovine leukemia virus infected endothelial cells from cattle with persistent lymphocytosis. Lab. Invest. 1991;65(2):192–202. PubMed PMID: 1652665. [PubMed] [Google Scholar]
- Schwingel D., Andreolla A.P., Erpen L.M.S., Frandoloso R., Kreutz L.C. Bovine leukemia virus DNA associated with breast cancer in women from South Brazil. Sci. Rep. 2019;9(1):2949. doi: 10.1038/s41598-019-39834-7. Epub 20190227PubMed PMID: 30814631; PubMed Central PMCID: PMC6393560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.P., Dock N.L., Ehrlich G.D., Sninsky J.J., Brothers C., Gillsdorf J., et al. Evaluation of HIV type 1 western blot-indeterminate blood donors for the presence of human or bovine retroviruses. AIDS Res. Hum. Retrovir. 1995;11(3):409–414. doi: 10.1089/aid.1995.11.409. PubMed PMID: 7786586. [DOI] [PubMed] [Google Scholar]
- Trujillo Piñeros M.A. Presencia de anticuerpos contra el virus de la leucosis bovina (VLB) en mujeres colombianas 2019. Available from: http://hdl.handle.net/10554/46182.
- Uribe A., Gutiérrez M., Ochoa-Cruz A. Vol. 11. Universitas Scientiarum [Internet]; 2006. pp. 31–40.https://revistas.javeriana.edu.co/index.php/scientarium/article/view/4968 (Estudio Del Potencial Zoonótico Del Virus De La Leucosis Bovina y Su Presencia En Casos De Cáncer De Seno). 01/01[pp.]. Available from: [Google Scholar]
- Velandia Álvarez S. Virus de la leucosis bovina: evaluación de la presencia del provirus en sangre humana y análisis in silico de los receptores celulares involucrados en su interacción 2021. Available from: doi: 10.11144/Javeriana.10554.59062. [DOI]
- Willems L., Gegonne A., Chen G., Burny A., Kettmann R., Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987;6(11):3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. PubMed PMID: 2828028; PubMed Central PMCID: PMC553795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Heremans H., Chen G., Portetelle D., Billiau A., Burny A., et al. Cooperation between bovine leukaemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 1990;9(5):1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. PubMed PMID: 2158445; PubMed Central PMCID: PMC551852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Avila D.E., Zapata-Benavides P., Cedillo-Rosales S., Avalos-Ramírez R., Zarate-Ramos J.J., Riojas-Valdés V., et al. Serological detection of bovine leukemia virus in slaughterhouse workers from San Nicols de los Garza, Nuevo Len, Mxico. Afr. J. Microbiol. Res. 2013;7(24):3042–3048. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.