SUMMARY
Mitochondria are vital organelles that require sophisticated homeostatic mechanisms for maintenance. Intercellular transfer of damaged mitochondria is a recently identified strategy broadly used to improve cellular health and viability. Here, we investigate mitochondrial homeostasis in the vertebrate cone photoreceptor, the specialized neuron that initiates our daytime and color vision. We find a generalizable response to mitochondrial stress that leads to loss of cristae, displacement of damaged mitochondria from their normal cellular location, initiation of degradation, and transfer to Müller glia cells, a key non-neuronal support cell in the retina. Our findings show transmitophagy from cones to Müller glia as a response to mitochondrial damage. Intercellular transfer of damaged mitochondria represents an outsourcing mechanism that photoreceptors use to support their specialized function.
Graphical abstract
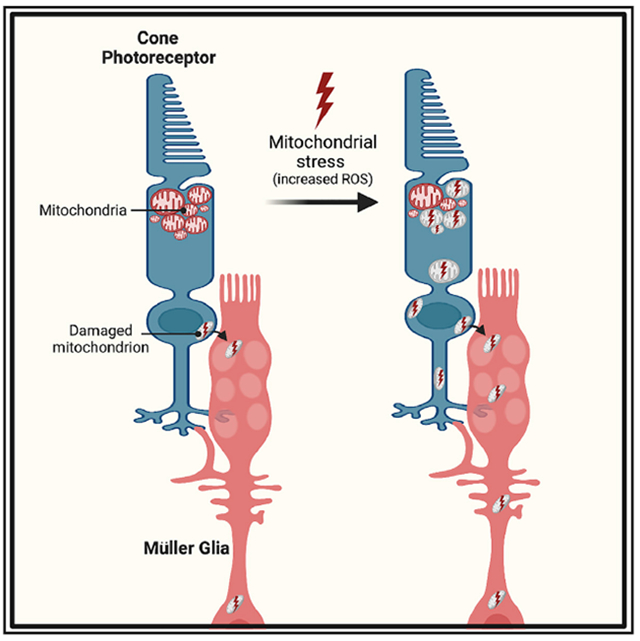
In brief
Hutto et al. show that mitochondrial stress in cone photoreceptors triggers selective movement of damaged mitochondria away from their normal location in the ellipsoid region of the cone. These damaged cone mitochondria can be exported to Müller glia cells, where they undergo degradation.
INTRODUCTION
Mitochondrial dysfunction is a pathological marker for neurodegenerative diseases. Mitophagy is a key strategy that cells use to maintain healthy mitochondria. One form of mitophagy involves the transfer of damaged mitochondria for transcellular degradation (transmitophagy). In neurons, transmitophagy was first reported between retinal ganglion cell axon mitochondria and adjacent astrocytes.1 Recent findings demonstrate the physiological role of this process in Alzheimer’s disease2 and in maintaining healthy dopaminergic neurons.3 Transmitophagy is also critical for nonneuronal cells. In adipocytes, the transfer of damaged mitochondrial fragments to macrophages mitigates the negative consequences of thermogenesis-induced reactive oxygen species (ROS).4
Photoreceptor (PR) cells are the postmitotic retinal neurons responsible for detecting light and initiating shape-detecting vision. Mitochondria are abundant in PRs presumably due to the significant energy requirements of these cells, particularly in the dark.5 Mitochondria are concentrated in the ellipsoid region of the cell and are thought to remain in this region.6 PRs reside in an oxidative environment near the choroidal blood supply. The high energy demands, oxidative stress, and long-lived presence of PRs necessitate robust strategies to remove and replenish damaged mitochondria.
Here, we developed three stress models to determine how mitochondrial homeostasis in cone PRs is affected by chronic stress. We exploited the zebrafish model due to the abundance of cones and live-imaging capabilities. In addition, recent work indicates that cone PRs are more sensitive to oxidative stress than rods.7 Cones also use more ATP than rods,8 likely making them more susceptible to mitochondrial dysfunction.
Our results show that cone mitochondria respond similarly to different stress paradigms. This includes migration away from the ellipsoid, acidification, and transcellular transfer to Müller glia (MGs). This phenomenon is also detected in unstressed cones but at lower levels. Our research has uncovered transmitophagy in cones as a response to mitochondrial damage and it adds to the repertoire of functions attributed to MGs. We propose that this response increases under stress and helps explain the robustness of the cone PR.
RESULTS
Mitochondria in cone PRs are mislocalized after stress
We examined mitochondrial mislocalization across different mitochondrial stress paradigms using cone-specific gnat2:mtm-Kate2 transgenic zebrafish to visualize cone mitochondria.
We induced stress by treating zebrafish larvae with chloramphenicol, an inhibitor of mitochondrial RNA translation9 (Figures S1A–S1C), or cold (16°C), which increases ROS in fish neural tissue10 (Figure S1D). Both stresses caused mislocalization of mitochondria away from the ellipsoid (Figures 1A and 1B).
Figure 1. Stress causes mislocalized mitochondria in cones.
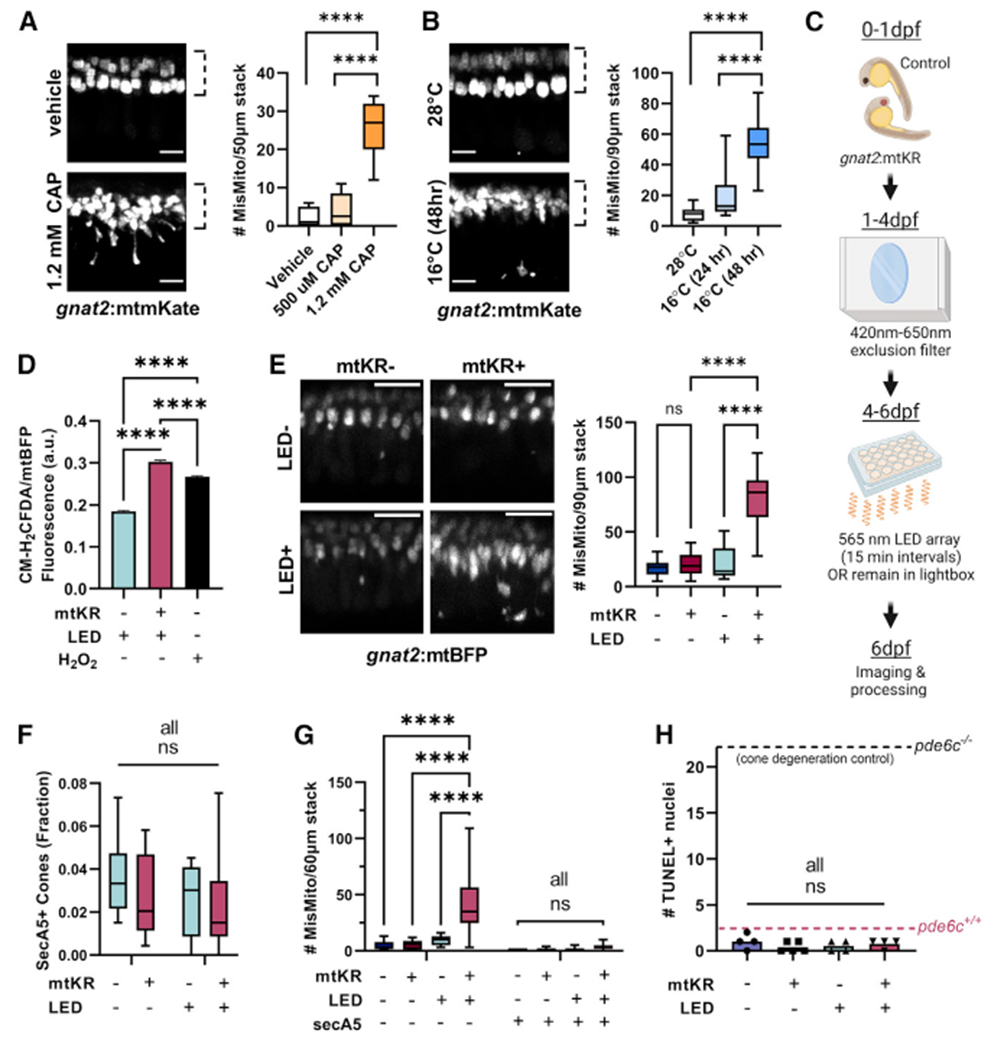
(A) Chloramphenicol (CAP) causes cone mitochondria (gnat2:mtmKate) to migrate away from the ellipsoid region (dotted line). Mislocalized mitochondria were quantified per fish. n = 11 fish for vehicle and 1.2 mM CAP conditions and n = 8 for 500 μM CAP. ****p < 0.0001 using one-way ANOVA. Scale bar: 5 μm.
(B) Cold (16°C) causes cone mitochondria to migrate from the cone ellipsoid (dotted line). Mislocalized mitochondria were quantified per fish. n = 10 28°C, n = 6 16°C (24 h), and n = 20 16°C (48 h) fish. ****p < 0.0001 using one-way ANOVA. Scale bar: 5 μm.
(C) Schematic for gnat2:mtKillerRed (mtKR) experiments. Embryos were put in a chamber reducing 420–650 nm light to prevent KillerRed activation while maintaining light/dark cycles. At 4 dpf, fish were either kept in the chamber or put on a plate above an LED array of 565 nm light. After 48 h, fish were processed/imaged.
(D) CM-H2DCFDA fluorescence in cone mitochondria (gnat2:mtBFP) shows increased mitochondrial ROS upon gnat2:mtKR activation. Fluorescence was normalized to mtBFP to correct for depth in the eye. ****p < 0.0001 with one-way ANOVA. mtKR−: 26,035 mitochondrial clusters from 8 eyes, mtKR+: 24,144 clusters from 13 eyes, and H2O2: 40,632 clusters from 15 eyes.
(E) Only mtKR fish activated by the LED array (mtKR+, LED+, see protocol in C) have increased cone mitochondrial mislocalization. Cone mitochondria labeled with gnat2:mtBFP. Scale bar: 10 μm. n = 13 fish for mtKR−, LED− and mtKR+, LED+; n = 16 for mtKR+, LED−; and n = 14 for mtKR−, LED+. ****p<0.0001 with one-way ANOVA.
(F) Quantification of secreted Annexin5+ (secA5; apoptotic) cones, identified by the presence of gnat2:mtBFP. Neither LED treatment nor mKR activation is associated with cone cell death. Total cones in a 60 μm stack were estimated by quantification of ellipsoid mitochondrial clusters across n = 5 fish. All comparisons are not significant (ns) with two-way ANOVA. mtKR−, LED−: n = 6; mtKR−, LED+: n = 9; mtKR+, LED−: n = 8; mtKR+, LED+: n = 19.
(G) Increases in mislocalized mitochondria (gnat2:mtBFP) upon mtKR activation were only observed in non-apoptotic cells (secA5−). Few mislocalized mitochondria were found in apoptotic cells. Same n as (F). ****p < 0.0001 with a two-way ANOVA.
(H) TUNEL+nuclei across a 10 μm slice from 6 dpf eyes. No difference detected regardless of LED exposure or the presence of mtKR. Positive control is the pde6c−/− cone degeneration model (black dotted line) and negative control is pde6c+/+ wild-type (WT) sibling (magenta dotted line). n = 4 for all but mtKR+, LED− (n = 5). All comparisons are ns with one-way ANOVA.
Since chloramphenicol and cold affect whole larvae, we developed a method that induces stress only in cone PRs. We generated a transgenic zebrafish strain that expresses mitochondrial-targeted Killer Red (KR) only in cone PRs (gnat2:mtKR). KR produces superoxide radicals when exposed to 540–590 nm light.11 We paired this transgene with gnat2:mtBFP fish to visualize cone mitochondria. To prevent KR activation during early development, we minimized exposure to 420–650 nm light up to 4 days postfertilization (dpf) before then exposing larvae to 565 nm LED pulses from 4 to 6 dpf (Figure 1C). Our activation strategy increased ROS production in cones (Figure 1D) and caused significant mislocalization of mitochondria in larvae with activated mtKR (mtKR+, LED+) but not in larval cones with inactivated mtKR (mtKR+, LED−) or in larval cones treated with LED exposure but not expressing mtKR (mtKR−, LED+) (Figure 1E).
To ensure that the mitochondrial mislocalization we observed is not caused by cell death, we used both Tg(TBP-GAL4;UAS:-secA5-YFP) fish12 and TUNEL staining. Secreted Annexin5 in the secA5-YFP fish accumulates on the outer surface of the plasma membrane of early apoptotic cells. Neither mtKR expression or LED treatment significantly increased the number of secreted Annexin5 positive (secA5+) cone PRs (identified by gnat2:mtBFP expression; Figures 1F and S1E). There were very few mislocalized mitochondria in secA5+ cells; the increase in mislocalized mitochondria of activated mtKR fish occurred in secA5− cells (Figure 1G). Similarly, TUNEL staining did not increase (Figures 1H and S1F). In contrast, we observed increased cell death and more mislocalized mitochondria in dying cells after chloramphenicol or cold treatment (see Figures S1G–S1L). Because of these findings, we conducted our studies primarily with gnat2:mtKR fish, and we used the mtKR−, LED+ fish as the control for mtKR+, LED+ fish.
Mislocalized cone mitochondria can be found both inside and outside of cones
mtKR LED activation led to substantial visible morphological changes to mitochondria (Figure 2A). Many cone mitochondria appeared translucent due to reduced cristae and possible swelling (Figure 2A, right panel). Abnormal mitochondria resided both within and outside of the ellipsoid region, unlike healthy electron-dense mitochondria, which we only observed within the ellipsoid region (Figure 2B). In controls, there were few unhealthy mitochondria outside of the ellipsoid. All mitochondria outside the ellipsoid appeared damaged, as in response to mitochondrial Ca2+ stress.13
Figure 2. Mislocalized mitochondria are abnormal and reside in and outside of cone photoreceptors.
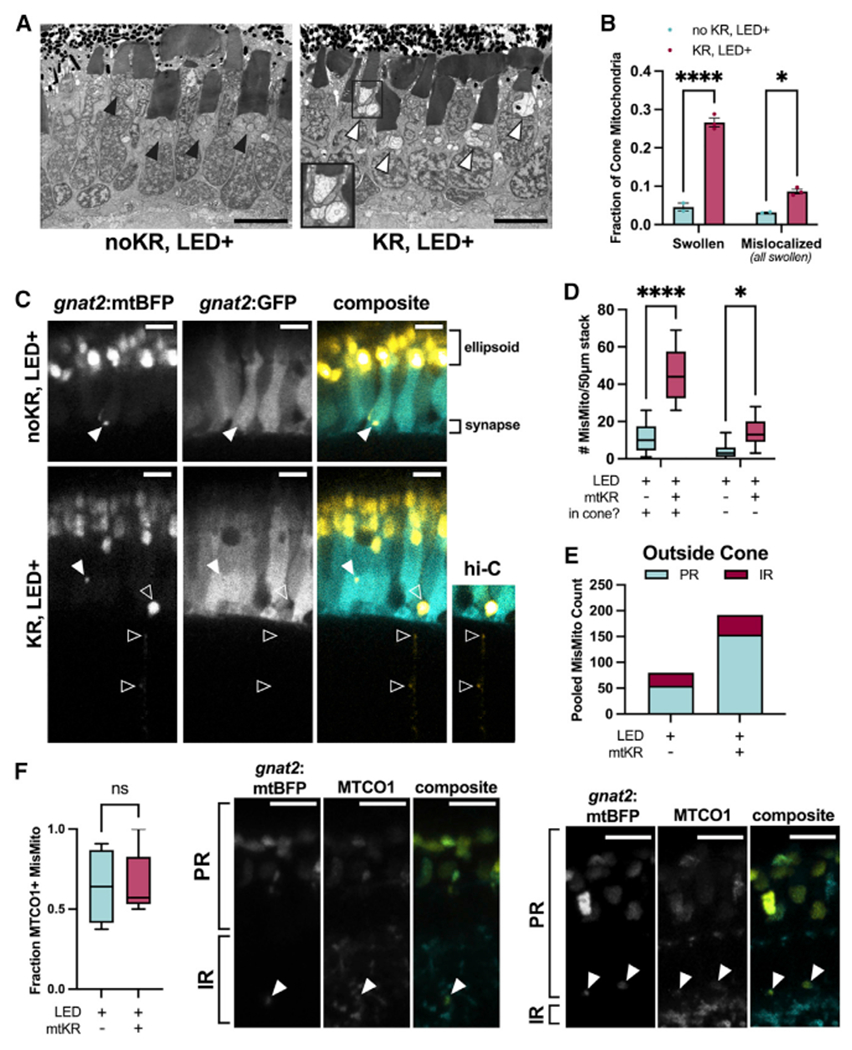
(A) Transmission electron micrographs taken at the optic nerve. Cones with activated mKR have many mitochondria with disturbed, swollen morphology both in and outside of the ellipsoid clusters; an inset (black box) shows a cone ellipsoid containing multiple swollen mitochondria. Black arrowhead: ellipsoid cluster with typical mitochondria morphology, white arrowhead: ellipsoid cluster with swollen mitochondria. Scale bar: 5 μm.
(B) Quantification of individual cone mitochondria from slices in (A). A greater fraction of mitochondria are swollen and mislocalized in mKR+ cones. All mislocalized mitochondria had swollen morphology. n = 2 kR− fish (641 and 759 total mitochondria) and n = 3 kR+ fish (520, 613, and 566 total mitochondria). *p < 0.05 and ****p < 0.0001 using a two-way ANOVA.
(C) Imaging of Tg(gnat2:mtBFP, gnat2:GFP) fish reveals that mislocalized cone mitochondria reside inside cones (filled arrowheads) and partially or completely outside of cones (unfilled arrowheads). Hi-C, high-contrast to enhance visibility of mitochondria. Scale: 5 μm.
(D) Quantification of mislocalized mitochondria by colocalization of cone GFP; cone− includes mitochondria that appear partially or completely out of cones (see C). mtKR increases cone mitochondria number both inside and outside cones. *p < 0.05 and ****p < 0.0001 using a two-way ANOVA. n = 13 fish each condition.
(E) Pooled cone-mislocalized mitochondria totals across all fish binned by retinal region. IR, inner retina; PR, photoreceptor layer.
(F) Immunohistochemistry images of mislocalized mitochondria (filled arrows) colocalized with MTCO1 in PR and inner retina IR. Fraction of mislocalized cone mitochondria colocalized with mitochondrial protein, MTCO1. mKiller Red−: 74 mislocalized mitochondria from 4 eyes mtKillerRed+: 104 mislocalized mitochondria from 5 eyes. Comparisons are ns with unpaired t test. Scale: 5 μm.
We predicted that mislocalized mitochondria would be found primarily within cone PRs. To our surprise, some were partially or completely outside cone PRs (Figures 2C and 2D). These were present with and without mtKR, but mtKR increased the number of both intra- and extracellular mislocalized mitochondria (Figure 2D). Mislocalized cone mitochondria were present even outside the PR layer (Figures 2C and 2E). The identity of mislocalized fluorescence was further confirmed as mitochondria by colocalization with the mitochondrial protein cytochrome c oxidase 1 (MTCO1) (Figure 2F).14
MGs contain mislocalized cone mitochondria
These findings prompted us to ask if cone mitochondria were present within MGs. MG cells are resident glia that extend radially across all cellular layers of the retina and have many essential roles.15 MGs closely associate with PRs; their outer limiting membrane (OLM), which is formed by tight junctions between MGs, forms extensive contacts with PR inner segments.
After mtKR activation, mitochondria from cones were within MG processes (visualized using Tg(GFAP:GFP) fish) in the PR layer and in the inner retina (IR) closer to the MG cell body (Figures 3A and 3B; Video 1). The total number of mislocalized mitochondria both inside and outside of MGs increased compared with controls (Figure 3C), although the fraction of cone mitochondria within MGs was not significantly different between cones with mtKR vs. cones without it (Figure S2A). In vivo imaging captured migration of a cone mitochondrion into an MG (Figure 3D).
Figure 3. Mislocalized cone mitochondria are found in Müller glia (MG) cells.
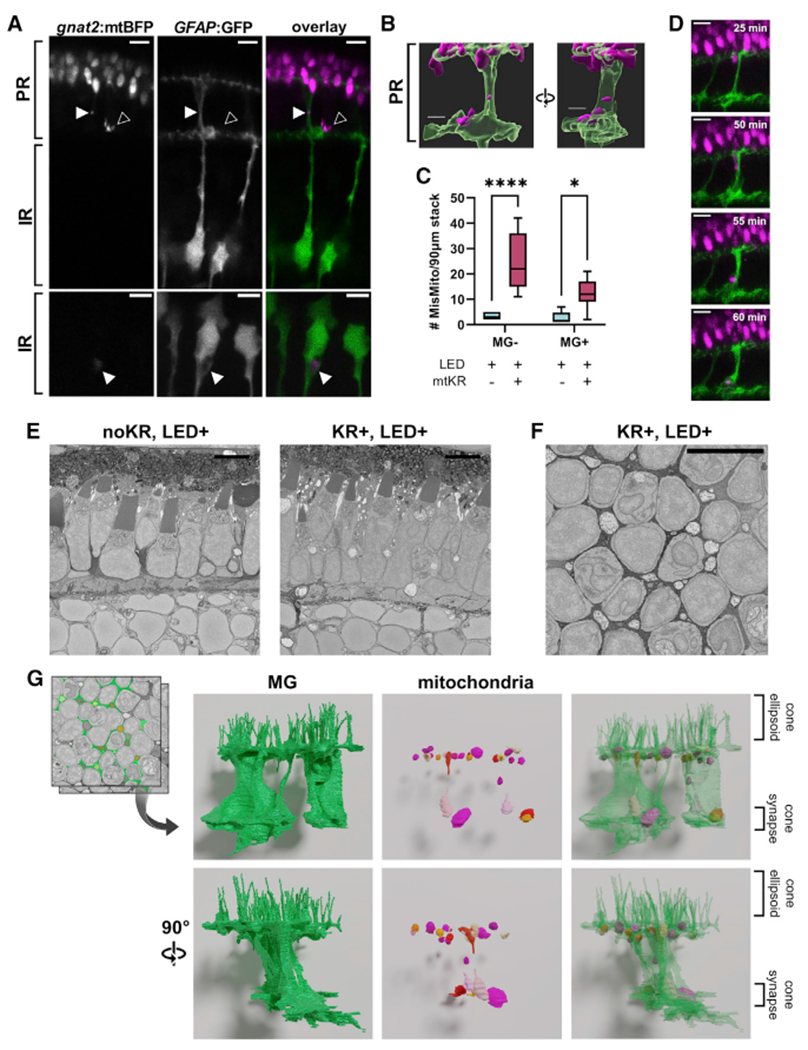
PR, photoreceptor layer; IR, inner retina; OLM, outer limiting membrane.
(A) In vivo imaging offish expressing gnat2:mtBFP and GFAP:GFP shows mislocalized mitochondria both inside (filled arrow) and outside (unfilled arrow) MGs. Both images were obtained from mtKR+, LED+ fish. Scale: 5 μm.
(B) 3D reconstruction of mislocalized mitochondrion encapsulated by an MG process from the first panel of (A). Scale: 3 μm.
(C) Quantification of mislocalized cone mitochondria classified by colocalization with MGs. Activation of mtKR increases both MG+ and MG− mislocalized mitochondria. *p < 0.05 and ****p < 0.0001 using a two-way ANOVA. n = 6 noKR fish, n = 7 kR+ fish.
(D) Images from a 4 h, 5 min interval in vivo timelapse of a Tg(gnat2:mtBFP, GFAP:GFP,gnat2:mtKR) fish post-LED treatment. Scale: 5 μm.
(E) Longitudinal micrographs from SBFEM for mtKR− and mtKR+ fish (both LED treated). The mtKR fish contains several swollen, electron-lucent mitochondria between cone cell bodies near the photoreceptor nuclear layer and synapse. Scale: 5 μm.
(F) Transverse section from SBFEM stack of a mtKR+ fish at the OLM where abundant swollen/lucent mitochondria can be observed between cone cells in MG. Scale: 5 μm.
(G) 3D reconstruction of MG cytosol (green) and the swollen mitochondria (multicolor) observed both at the OLM further down MG processes.
These findings indicate that many cone mitochondria that leave cones are taken up by MGs. The increase in the number of cone mitochondria in MGs due to ROS production in cone mitochondria suggests that we have revealed a transfer process that normally occurs under basal conditions and that increases in response to stress.
As further support for a normal transfer process that is stimulated by stress, we used a new mtKR activation strategy with a 24 h LED treatment followed by shielding from activating wavelengths. Transfer of mitochondria to MGs increased during the first 24 h to approximately 40% before stabilizing (Figure S2B). We asked whether microglia, which can phagocytize apoptotic PRs and debris, also take up mislocalized cone mitochondria. In both control and mtKR-activated fish containing the microglial transgene Tg(mpeg1:GFP)gl22,16 there were very few cone mitochondria detected in microglia (Figure S2C), consistent with the low levels of cell death associated with mtKR activation.
Longitudinal sections from serial block-face (SBF) scanning electron microscopy (EM) stacks of activated mtKR retinas revealed several mislocalized mitochondria between cones that resemble the damaged morphology of mitochondria within cones (Figure 3E). A top-down stack collected from an activated mtKR fish shows an abundance of damaged mitochondria between cones at the position of the OLM (Figure 3F). 3D reconstruction confirmed that these cone mitochondria are indeed within MGs and found near the OLM as well as near the PR synaptic layer (Figure 3G; Videos S2 and S3). We also captured what appeared as extrusion of damaged mitochondria (Figure S2D). Reconstruction suggests entry into a neighboring MG cell (Figure S2E; Videos S4 and S5). Additional mitochondrial extrusion events from other cones are documented by single EM images (Figures S2F and S2G).
Cone mitochondria in Müller cells are targets of mitophagy
We then used cone-specific expression of the lysosomal marker LAMP1-GFP (gnat2:LAMP1-GFP) to determine if autophagy machinery is recruited to mislocalized mitochondria in cones (Figure 4A). While some mislocalized mitochondria colocalized with cone LAMP1 in both control and mtKR fish, ROS stress triggered a more robust increase in LAMP1− mislocalized mitochondria (Figure 4B). Accordingly, a smaller fraction of mislocalized mitochondria were LAMP1+ in mtKR fish (Figure 4C).
Figure 4. Cone mitochondria degradation occurs in MGs.
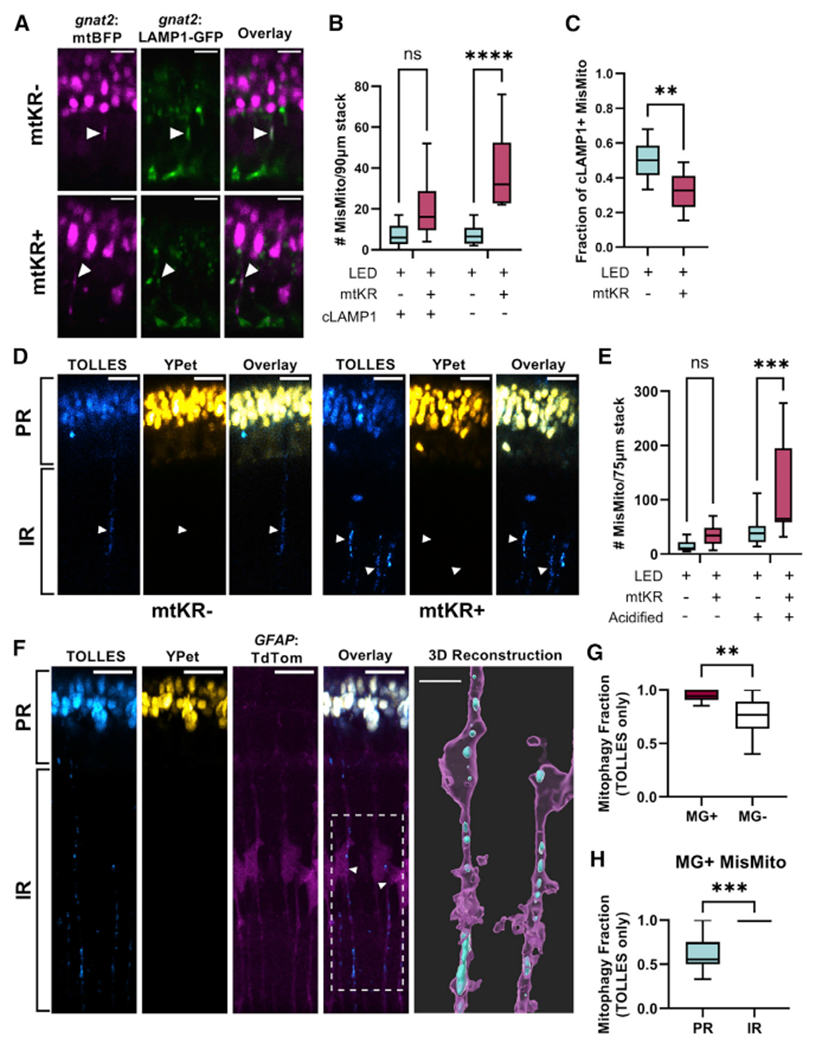
(A) In vivo imaging of Tg(gnat2:mtBFP, gnat2:-LAMP1-GFP) fish; mislocalized mitochondria colocalized with cone lysosomes (filled arrows) in mtKR− and mtKR+ fish. Scale: 5 μm.
(B) Quantification of (A) shows a marked increase in mislocalized mitochondria unassociated with cone lysosomes upon ROS stress. cLAMP1, cone LAMP1. n = ****p < 0.0001 using a two-way ANOVA. n = 10 fish each condition.
(C) Fraction of cLAMP1+ mislocalized mitochondria from data in (A) and (B). While mtKR have more mislocalized mitochondria, a lower fraction is associated with cone lysosomes compared with controls. n = 10 mtKR+ fish, n = 9 mtKR− fish (1 excluded from fraction due to <5 total mislocalized mitochondria). **p < 0.01 using Welch’s t test.
(D) In vivo imaging of Tg(gnat2:mtSRAI) with and without mtKR (LED treated). PR, photoreceptor layer; IR, inner retina. Mitochondria that have lost YPet fluorescence (acid and protease sensitive) are in acidified compartments. Arrowheads indicate “trails” of acidified cone mitochondria in the IR. Scale: 10 μm.
(E) Quantification of acidified mislocalized mitochondria from dataset in (D). 3D volumes were generated in Imaris, manually binned as mislocalized/ellipsoidal, and identified as acidified if mean fluorescence TOLLES > Ypet. noKR n = 12; KR+ n = 9 fish. ***p < 0.001 with two-way ANOVA.
(F) In vivo imaging of normal, unstressed Tg(gnat2:mtSRAI, GFAP:TdTomato) fish (no mtKR, no LED). Trails of TOLLES+ mislocalized cone mitochondria colocalize with MG processes in the IR. Scale: 10 μm. Imaris was used to generate a 3D reconstruction (scale: 5 μm) of two of these MGs; their cell bodies are noted by an arrowhead and the ROI by a dotted line in the overlay fluorescent image (magenta: MG, cyan: TOLLES+ MisMito).
(G) Mitophagy fraction (acidified/total) of mislocalized mitochondria inside and outside MGs in unstressed fish. Mitochondria were manually binned by presence of YPet (see STAR Methods). n = 11 fish. **p < 0.01 with Welch’s t test.
(H) Mitophagy fraction (acidified/total) of cone mitochondria in MG, binned by location in the retina. Mitochondria observed in inner retinal portions of MG were acidified. n = 11 fish. ***p < 0.001 with Welch’s t test.
To investigate mitophagy further, we used an in vivo mitophagy marker: mt-SRAI.17 mt-SRAI is a fluorescence resonance energy transfer (FRET) sensor with TOLLES and YPet. The ratio of TOLLES:YPet fluorescence is low in healthy mitochondria and high in acidified/degraded mitochondria. Using gnat2:mtSRAI fish, we did find mitochondria undergoing degradation within the PR layer and IR. (Figure 4D). Activated mtKR fish had significantly more acidified mislocalized mitochondria than controls (Figure 4E). A much higher fraction of mislocalized mitochondria were targeted for degradation than ellipsoid mitochondria in both control and mtKR fish, though an increase in the fraction of degraded mitochondria was noted in the ellipsoid of mtKR fish (Figure S3A). Thus, cone mitochondrial stress increases mislocalization and degradation of mitochondria. This finding combined with the lack of robust cone-derived LAMP1 recruitment to mislocalized mitochondria suggests that cells other than cones may be responsible for disposing of cone mitochondria.
mtKR is not compatible with additional red fluorophore tags, so we used unstressed fish to assess the localization of cone mitochondria with gnat2:mtSRAI. Overall, mislocalized cone mitochondria outside of cone PRs (gnat2:TdTomato) were much more likely to be acidified than those inside cones, and they were often present in long trails consistent with radially oriented MGs (Figures S3B and S3C). To verify this, we used gnat2:mtSRAI and GFAP:TdTomato fish. Trails of acidified cone mitochondrial remnants spanned the IR along MG processes (Figure 4F; Video S6). Correlative light and electron microscopy further confirmed that the cone-derived mitochondrial material in these long trails resided within cells in the center of the inner nuclear layer (Figure S3F). Cone mitochondria in MGs were overwhelmingly acidified compared with their mislocalized counterparts not in MGs (Figure 4G). Furthermore, of the cone mitochondria inside MGs, those in processes in the PR layer were far less likely to be acidified compared with those in the IR (Figure 4H; 100% acidified in the IR of all fish). These results indicate that while mitophagy of mislocalized mitochondria can occur within cones, it is more extensive in MGs. As initial confirmation of this hypothesis, we detected MG-derived acidified and unacidified phagosomes using a fish with the tandem marker for autophagosomes, GFAP:GFP-mCh-LC3 fish (Figures S3D and S3E). We found that after cold stress, a much greater fraction of the mislocalized cone mitochondria in MGs were associated with acidified phagosomes than with unacidified phagosomes.
DISCUSSION
The mtKR transgenic model
KR was a particularly useful tool that we used to examine the effects of oxidative stress on cone PR mitochondria. KR has been used to destroy cells selectively,11,18 but it also has been used to study physiologic responses to oxidation.19,20 We targeted KR specifically to cone PR mitochondria, and we constructed an optogenetic stimulus array for chronic activation of KR. Our EM analyses of these fish indicated that PRs appeared normal despite obvious morphological defects in cone mitochondria. Cristae were reduced, and mitochondria appeared large and translucent but not fragmented. We confirmed that LED-treated KR fish had elevated mitochondrial ROS compared with control fish, but this did not increase cone cell death. This model is a valuable tool for studying cone responses to elevated ROS beyond the larval stage examined here and in the context of retinal diseases. Although we detected significant increases in mitochondrial stress with KR and LED compared with LED alone, one potential caveat that requires further investigation is how LED treatment could influence circadian activity. Shorter LED treatments than used in this study could alleviate this concern.
Transfer and degradation of mitochondria
A significant proportion of cone mitochondria not in PRs were in MGs. 3D reconstructions of confocal and SBF scanning EM images together with in vivo time-lapse imaging indicated that these mitochondria had been transferred to MGs. To dissect the fate of mislocalized mitochondria, we used transgenic strains with markers often used to assess stages of mitophagy: LAMP1, SRAI, and LC3.17,21,22 Acidification of mislocalized mitochondria, while occurring in cones, was especially pronounced outside of cones and in MGs. We also observed MG-derived LC3 accumulating on cone mitochondria that had been transferred to MGs. These results suggest that cone mitophagy operates near capacity and that, under stress, PRs dramatically increase the transfer of damaged mitochondria to MGs. This mechanism represents a significant process for PRs, since degradation occurred primarily outside of the ellipsoid region; in the ellipsoid, most mitochondria are unacidified (only 0.097% ± 0.035% acidified in control fish and 0.33% ± 0.082% acidified in mtKR+, by volume; see Figure S4A). These findings also align with previous work in mitoQC and mCh-GFP-LC3 mouse models showing enrichment of mitolysosomes and autophagosomes in the outer nuclear layer, away from the ellipsoid of PRs and where MG processes extend between PRs.23
Transmitophagy was first observed in retinal neurons and astrocytes.1 Axonal protrusions filled with damaged mitochondria from retinal ganglion cells (RGCs) were surrounded by astrocytes near the optic nerve head (ONH). The evidence for transfer of damaged mitochondria to astrocytes was the presence of RGC-derived acidified mitochondria, some of which were surrounded by Lamp1 inside of astrocytes at the ONH. These findings are similar to ours, although we require the development of more markers to fully compare the details of these two seemingly related processes. The use of astrocytes and not MGs reflects the specific association between astrocytes and RGC axons, whereas MGs ensheath the RGC cell body.24 Neuron-astrocyte transmitophagy also occurs in Parkinson’s and Alzheimer’s disease.2,3 Release of mitochondria from cells can elicit an inflammatory response.25 The lack of stress-induced microglia activation in our study suggests that a direct connection, either vesicular or tubular, may facilitate transport of mitochondria from cones to MGs. Our EM data thus far also support vesicular transfer based on appearance of apparent budding from cones to MGs (Figures S2D–S2G; Videos S4 and S5).
In other neuronal systems, not only has the transfer of damaged mitochondria for disposal and recycling been reported but the reverse mitochondrial transfer from glia back to neurons is associated with neuronal survival after injury both in vitro and in vivo.26 Transfer of mitochondria isolated from macrophages was also found to reduce neuropathic and inflammatory pain mediated through CD200 receptor signaling within the peripheral nervous system.27 In both studies, the purpose of the transfer is more than just to equilibrate ATP. Thus, transmitophagy represents a sub-topic within the broader field of mitochondrial transfer, and transfer of healthy mitochondria to rejuvenate cells is also important. Future studies in the mtKR model could assess whether any retinal glia supplement their mitochondria to cones.
Transmitophagy is a broadly used pathway not confined to the nervous system. For example, cardiomyocytes damaged by ischemia-reperfusion release large vesicles termed exophers that contain dysfunctional mitochondria. In the absence of macrophages that clear these damaged extruded mitochondria, resident cardiomyocyte mitochondria produce less ATP, which compromises cardiac function.28 Similarly, when macrophages are depleted and not available to clear oxidatively damaged mitochondria released by brown adipocytes, cold-induced thermogenesis is impaired.4 These studies support the hypothesis that transmitophagy is protective and prevents intracellular accumulation of dysfunctional mitochondria.
PR specialization and outsourcing functions
PR neurons are optimized for light detection and signal transmission; they employ specialized biochemical mechanisms for vesicular transport both for the outer segments,29 where phototransduction occurs, and for synaptic transmission.30 PRs use their cellular resources to maintain precise and robust responses to light by outsourcing cellular functions. For example, they are metabolically coupled with the retinal pigment epithelium both for nutrient cycling31 and for regeneration of 11-cis retinal after light exposure.32 We propose that the transfer of damaged mitochondria is another outsourcing mechanism that helps PR survival by preserving resources and limiting damage to minimize disturbance to the essential functions of light detection and signaling.
Limitations of the study
Our findings stimulate many future exciting questions. We have not defined the signaling mechanism leading to specific removal of damaged mitochondria from the ellipsoid, the mechanisms of movement within the PR, how mitochondria are transferred to glia, or the details of the transcellular degradation process. Further, our strategies were optimal for experiments using larvae. Verifying our findings in adults needs to be a separate study. Understanding how PRs maintain healthy mitochondria may help to develop strategies to treat and prevent disease and broadly inform research in the relatively new field of intracellular mitochondrial transfer in biology.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Susan Brockerhoff (sbrocker@uw.edu).
Materials availability
All unique and stable reagents generated in this study will be made available on request, but we may require payment and/or a completed materials transfer agreement (per guidelines of the University of Washington), especially if there is potential for commercial application. Transgenic fish strains will be provided upon request if swimming copies are available. The authors will freely distribute all plasmids used to generate transgenic fish lines.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
Experiments with zebrafish were authorized by the University of Washington Institutional Animal Care and Use Committee. All fish used in this study were maintained in the University of Washington South Lake Union aquatics facility at 27.5°C on a 14/10 h light/dark cycle. Fish used for confocal imaging and TUNEL staining were maintained in the Roy−/− genetic background, while fish used for western blotting, immunohistochemistry, qPCR, and electron microscopy were maintained in the AB or Roy+/− genetic background (iridophore pigmentation present). The pde6cw59 fish strain is described.33 Animals used in this study were in the larval stages prior to sexual maturation.
METHOD DETAILS
Transgenic zebrafish generation
Six transgenic zebrafish lines were generated for this study: Tg(gnat2:mtmKate2), Tg(gnat2:mtBFP), Tg(gnat2:mtKillerRed), Tg(gnat2:LAMP1-GFP), Tg(gnat2:mtSRAI), and Tg(GFAP:GFP-mCh-LC3). Gateway-Tol2 assembly was used for generation of all constructs. For cone-specific expression, we used the zebrafish gnat2 promoter cloned into a 5′ entry vector.34 All targeting of fluorophores to cone mitochondria was accomplished using the mitochondrial F0-ATPase targeting sequence cloned into a pME vector (cloned from Su9-EGFP, a gift from David Chan, Addgene plasmid # 23214; RRID:Addgene_23214). To generate the Tg(gnat2:mtBFP) line, we used a 3′ entry vector containing mTagBFP from Nicholas Cole (Addgene plasmid # 75175; http://n2t.net/addgene:75175; RRID:Addgene_75175).42 For the Tg(gnat2:mtmKate2) line, we used a 3′ entry vector containing mKate2 from Anna Planas & Tomas Santalucia (Addgene plasmid # 48345; http://n2t.net/addgene:48345; RRID:Addgene_48345).43 For generation of Tg(gnat2:KillerRed), the KillerRed DNA was gifted to us by Pablo Peixoto; the original mitochondrial targeting sequence of the construct was not used and instead was replaced with the Su9 sequence.11 The construct was cloned into a destination vector with a sBFP2 heart marker (a gift from Cecilia Moens) for aid in transgenic identification without activation of KillerRed. For generation of Tg(gnat2:mtSRAI), the SRAI sensor DNA was obtained from RIKEN DNA Bank (#RDB18223) and cloned into a 3′ entry vector; the original mitochondrial targeting sequence of the construct was not used and instead was replaced with the Su9 sequence.17 We obtained the LAMP1 DNA from Alex V. Nechiporuk.39 GFP-mCh-LC3 DNA was generated as previously described.40 The zebrafish GFAP promoter, p5E-gfap, was a gift from Judith Eisen (Addgene plasmid # 82401; http://n2t.net/addgene:82401; RRID:Addgene_82401).42 All constructs were injected into embryos at the 1-cell stage with Tol2 transposase mRNA. Larvae mosaic for the transgene were raised to adulthood to identify founder carriers. Single F0 founders were used to generate F1 fish that were screened for a single insertion of the transgene; fish from the F2 generation and onward were used in this study.
The Tg(gnat2:GFP), Tg(GFAP:TdTomato) and Tg(gnat2:TdTomato) zebrafish lines are described.34,35,36 Tg(GFAP:GFP) and Tg(mpeg1:GFP)gl22 fish lines are from the Zebrafish International Resource Center (from the Raymond and Leischke labs, respectively).16,37 The Tg(TBP-GAL4;UAS:secA5-YFP) line was generated by Randy Peterson’s lab and gifted to our facility by Diana Mitchell.12,38
Western blotting
Protein isolation and western blotting were performed using standard procedures44 with the following details. 6 larval bodies obtained at 6 dpf were pooled in each independent sample for protein isolation. 40 μg of protein from each of these pooled samples was loaded per well. For primary antibodies, MTCO1 (Abcam, ab14705, RRID:AB_2084810) and SDHB (Abcam, ab14714, RRID:AB_301432) were both used at a 1:1000 dilution. The secondary antibody IRDye 680RD donkey anti-mouse IgG (H + L) (LI-COR Biosciences, 925–32212, RRID: AB_2716622) was used at 1:5000 dilution.
DNA isolation and qPCR
Total DNA was isolated from pooled larval samples (10 larvae each, 6 dpf) using a DNeasy Blood & Tissue kit (Qiagen Cat#69504). An Applied Biosystems 7500 Fast Real-Time PCR System in conjunction with iTaq Universal SYBR Green Supermix (Bio-Rad, 1,725,120) was used for qPCR measurements according to the manufacturer’s instructions. Relative quantification of mitochondrial DNA content was performed by normalizing CT values of mitochondrially-encoded NADH dehydrogenase 1 (mt-ND1) to nuclear-encoded DNA polymerase subunit gamma-1 (polg1) within each sample using previously characterized primers.28 Sample measurements were conducted in three technical replicates for each biological sample, and 5 biological replicates per condition were assayed.
Immunohistochemistry and TUNEL staining
Zebrafish larvae were collected at 6 dpf and fixed in 4% paraformaldehyde. Larvae were cryoprotected using sucrose gradients and embedded in OCT. 10μm sections were cut and warmed for 2 h. Sections were stained using a standard protocol.13 (Antibodies: [1:1000] MTCO1 antibody (abcam 14,705) and [1:500] Tag-FP antibody (FluoTag-X2 anti-TagFP Alexa Fluor 647 N0502-AF647-L)).
TUNEL assay was performed using EMD Millipore ApopTag Fluorescein In Situ Apoptosis Detection Kit (S71100) with the following modifications: hydrated sections with IHC PBS (pH 7.4) for 5 min, permeabilized with 20 μg/mL Proteinase K for 5 min, 5 min IHC PBS (pH 7.4) wash, 5 min incubation with equilibration buffer, 2 h TdT enzyme solution incubation, 2 washes with Stop/Wash buffer followed by IHC PBS (pH 7.4) wash, and 1 h Anti-Digoxigenin Conjugate incubation. Nuclei were stained using Hoechst 33,342, trihydrochloride trihydrate (10 μM; Life Technologies, H3570). Images collected on Leica SP8 (RRID: SCR_018169).
Stress treatments
For chloramphenicol treatments (CAP, Thermo Scientific, Cat#AC227920250), larvae were incubated in either 500 μM or 1.2mM CAP from 3 dpf to 6dpf with ethanol vehicle (0.1% v/v), using previously reported effective concentrations as a ref.45 Media was refreshed with treatment each day. For cold stress treatments, zebrafish were placed in a covered benchtop incubator set to 16°C (Benchmark Scientific, Cat# H2200-HC) fitted with a 4100K lightbulb for a 14/10 h light/dark cycle. Fish were kept in the incubator starting at 4 dpf (48 h treatment) or 5 dpf (24 h treatment). Confocal imaging was conducted at ambient temperature.
For cone-specific ROS generation in Tg(gnat2:mtKillerRed) fish, embryos were kept in a chamber outfitted with a UV-blue bandpass optical filter (Newport, Cat# FSR-BG3) to ensure fish were kept on the standard light/dark cycle but KillerRed would not be activated. At 4dpf, fish were sorted using blue heart fluorescence to identify Tg(gnat2:mtKillerRed) fish and either kept in the chamber or set up on the stimulating LED array. For stimulation of KillerRed, we constructed the optogenetic stimulus array with 565nm LED Stars (LED Supply, CREEXPE2-COL-X, PC Amber), an aluminum heatsink (Uxcell,a14111400ux0256), an LED driver (LED Supply, 0A011-D-V-xxx), and a power supply (Meanwell, LRS-200-24). LEDs were attached to the heatsink using heat-resistant glue in an array to fit under the 12 wells of a 48-well plate. The heatsink was fitted with 3D-printed housing to suspend the plate over the array. Two small fans were placed on opposite sides of the heat sink to disperse warm air. A mechanical outlet timer was used to set the intervals of LED stimulation (15 min on, 15 min off). Fresh media was replaced every 24 h and fish were collected for analysis at 6 dpf.
ROS measurements
ROS were detected using the dye CM-H2DCFDA (Thermo Fischer, Cat#C6827). Larval eyes were enucleated at 6 dpf and incubated in an oxygenated, supplemented Ringer’s media previously described that optimizes retinal tissue viability ex vivo.46 Eyes from each experimental group were pooled and incubated in the same media containing 50 μM CM-H2DCFDA for 30 min while covered on a gentle rocker. For H2O2 positive controls, this incubation media was supplemented with 10 mM H2O2. After incubation, eyes were washed three times with supplemented Ringer’s media and embedded with the lens facing downward in 0.5% low melting point agarose for confocal imaging. Embedded eyes were covered with the supplemented Ringer’s media throughout imaging.
Transmission and serial block-face scanning electron microscopy
Zebrafish larvae were fixed and embedded in blocks of resin at 6 dpf for TEM or SBEM imaging.13 Samples were imaged using a JEOL JEM-1230 transmission electron microscope or Zeiss Sigma VP scanning electron microscope. The step size for the SEM was 50nm and the thin sections imaged on the TEM were 80 nm. Z-stacks containing Müller glia and mitochondria were aligned and manually traced using the TrakEM2 plugin in ImageJ (RRID:SCR_008954)47 (for using TrakEM2 in general)48,49; (for aligning). 3-D reconstructions and animations were created using Blender (RRID:SCR_008606).
Confocal imaging of live larvae
For confocal imaging experiments, larvae were maintained in embryo media containing 0.003% 1-phenyl 2-thiourea (PTU, Sigma-Aldrich P7629) starting at 24 h post-fertilization. At 6dpf zebrafish larvae were anesthetized using 0.02% (w/v) Tricaine then transferred to 0.5% low melting point agarose containing embryo media with 0.003% PTU and 0.02% (w/v) Tricaine (Sigma-Aldrich, E10521). Larvae were positioned in agarose in a Petri dish mounted to a slide, then covered with embryo media and 0.02% (w/v) tricaine to prevent drying out. Imaging was performed with a Leica SP8 (RRID: SCR_018169). For all experiments other than Figure 4F, detectors were set to standard mode for quantitative imaging. For Figure 4F, due to the dim fluorescence of both SRAI in the inner retina and Tg(GFAP:TdTom) at the 6 dpf stage, detectors were changed to BrightR collection for a larger dynamic range. For all experiments, eye volume and the fluorophore intensity determined how much useful imaging depth was available, which was identical in controls for that experiment.
Image analysis and processing
All analyses of confocal images were conducted on images that were not processed with Leica’s Lightning algorithm. Lightning processing for presentation of images was only used for the dim signals in Figure 4F. Image analysis was conducted using ImageJ + Fiji software (SCR_002285) and researchers were blinded to sample identity using the “Blind Analysis Tools” plugin developed by Astha Jaiswal and Holger Lorenz.41 When identifying mislocalized mitochondria, researchers were blinded to other image channels. Colocalization analyses of mislocalized mitochondria were performed qualitatively on blinded samples after mitochondria identification and subsequent unblinding of the additional channels. For co-localization with microglia, mislocalized cone mitochondria were identified blinded to sample identity and blinded to other fluorescence channels, then mitochondria were assessed for colocalization with microglia while remaining blinded to sample identity. Imaris 9.9 (RRID:SCR_007370) was used for 3D surface reconstructions of fluorescence confocal data. For SRAI quantification, which required comparing the mean fluorescence of TOLLES and YPet for objects in 3D space, Imaris 9.9 was used to generate 3D surfaces of mitochondria in an image with the same surface algorithm applied to all samples. The total mitochondrial surface was divided into ellipsoid and mislocalized mitochondria manually by using the 3D reconstruction in tandem with a 3D slicer of original images for reference. Mitochondrial volumes with a mean fluorescence signal in the TOLLES channel higher than the YPet channel were designated as acidified. For the SRAI experiment in Figure 4F, imaging in BrightR mode prevented quantification of the channels in this manner and instead mitochondria were binned as positive or negative for YPet fluorescence.
Correlative light and electron microscopy (CLEM)
Zebrafish larvae expressing both Tg(gnat2:mtKillerRed) and Tg(gnat2:mtSRAI) were treated with PTU, raised on the LED array described above starting at 4dpf, and collected at 6 dpf. Whole larvae were incubated 30 min at room temperature (RT) on a rotator in fixative (2% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer), and washed with PBS. Following 2 h cryoprotection in 30% sucrose, larvae were embedded in OCT and frozen at −20°C. 60 μm cryosections were collected into PBS, washed with 2X saline sodium citrate (SSC) buffer, then incubated with RNase A (1:200 in 2X SSC; New England Biolabs, T3018-2) for 20 min at 37°C. Sections were washed 3 times in excess 2X SSC, then counterstained with propidium iodide (PI; 1 μg/mL; Life Technologies, P3566) for 5 min at RT.
For fluorescence imaging, individual sections were sandwiched between a slide and coverslip in a drop of 500 ng/mL PI in 2X SSC. TOLLES, YPet, and PI image stacks were collected with a Leica SP8 confocal using a 63X oil objective; XY resolution was 90 nm/pixel and Z resolution was 300 nm. After confocal imaging, sections were released into PBS and processed as described for SBEM imaging.14 SBEM images were collected using a VolumeScope SEM (Apreo, Thermo Fisher Scientific) at a voxel size of 8 × 8 × 50 nm.3 Alignment of confocal and SBEM stacks was performed in ImageJ with the BigWarp plugin50 using nuclei to assign landmarks.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
Numerical results in text are reported as mean ± standard error of the mean unless otherwise stated. Statistical tests were performed using Graphpad Prism v 9.4 software. For statistical analysis, replicates (n) were always defined as biological replicates, not technical replicates.
Information on what constitutes n (e.g., fish, mitochondria, cells) is listed in the figure legend of each experiment. For all box-and-whisker plots, the whiskers show the minimum and maximum values while the box boundaries represent the 25th and the 75th quartile with a line at the median. For one-way ANOVA tests, Tukey’s multiple comparisons test was used. For two-way ANOVA tests, šídák’s multiple comparisons test was used.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| MTCO1 | Abcam, ab14705 | RRID:AB_2084810 |
| SDHB | Abcam, ab14714 | RRID:AB_301432 |
| IRDye 680RD donkey anti-mouse IgG (H + L) | LI-COR Biosciences, 925–32212 | RRID: AB_2716622) |
| FluoTag-X2 anti-TagFP Alexa Fluor 647 | NanoTag Biotechnologies | N0502-AF647-L |
| Chemicals, peptides, and recombinant proteins | ||
| Chloramphenicol | Thermo Scientific | Cat#AC227920250 |
| dye CM-H2DCFDA | Thermo Fischer | Cat#C6827 |
| RNase A | New England Biolabs | Cat#T3018-2 |
| iTaq™ Universal SYBR® Green Supermix | Bio-Rad | Cat#1725120 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue kit | Qiagen | Cat#69504 |
| Millipore ApopTag Fluorescein In Situ Apoptosis Detection Kit | EMD Millipore | Cat#S71100 |
| Experimental models: Organisms/strains | ||
| Tg(gnat2:mtSRAI)w268 | This study | N/A |
| Tg(gnat2:mtBFP)w269 | This study | N/A |
| Tg(gnat2:mtKillerRed)w270 | This study | N/A |
| Tg(gnat2:mtmKate2)w271 | This study | N/A |
| Tg(gnat2:LAMP1-GFP)w272 | This study | N/A |
| Tg(GFAP:GFP-mCh-LC3)w273 | This study | N/A |
| pde6c w59 | Stearns et al.,33 | http://zfin.org/ZDB-ALT-080206-1 |
| Tg(gnat2:GFP)w206 | Kennedy et al.,34 | http://zfin.org/ZDB-ALT-181217-7 |
| Tg(GFAP:TdTomato) | Shin et al.,35 | N/A |
| Tg(gnat2:TdTomato) | Sloan et al.,36 | N/A |
| Tg(GFAP:GFP) | Bernardos e al.,37 | http://zfin.org/action/feature/view/ZDB-ALT-060623-4 |
| Tg(mpeg1:GFP)gl22 | Ellett et al.,16 | http://zfin.org/ZDB-ALT-120117-1 |
| Tg(TBP-GAL4;UAS:secA5-YFP) | van Ham et al.,12 Blume et al.,38 | N/A |
| Recombinant DNA | ||
| Su9-EGFP | Addgene plasmid # 23214 | RRID:Addgene_23214 |
| mTagBFP | Addgene plasmid # 75175 | RRID:Addgene_75175 |
| mKate2 | Addgene plasmid # 48345 | RRID:Addgene_48345 |
| SRAI | RIKEN DNA Bank | #RDB18223 |
| KillerRed | Bulina et al.,11 | N/A |
| p5E-gfap | Addgene plasmid # 82401 | RRID:Addgene_82401 |
| LAMP1 | Drerup et al.,39 | N/A |
| GFP-mCh-LC3 | George et al.,40 | N/A |
| Software and algorithms | ||
| Imaris 9.9 | Oxford Instruments | RRID:SCR_007370 |
| ImageJ | Schindelin et al.,41 | RRID:SCR_002285 |
| Blender | www.blender.org | RRID:SCR_008606 |
Highlights.
Mitochondrial stress triggers movement of cone mitochondria
Damaged cone mitochondria can be exported to Müller glia cells
Degradation of cone mitochondria occurs in Müller glia
ACKNOWLEDGMENTS
We thank Stanley Kim for managing our fish facility and Sharm Knecht for assisting with the SBF scanning EM (SBFSEM) required for the correlative light and electron microscopy (CLEM) analysis. Funding for this project was provided by P30EY001730 (UW Vision Core; Neitz), R01EY026020 (S.E.B.), F31EY033983 (K.M.R.), and T32EY007031 (R.A.H.). The graphical abstract was created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112115.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Davis C.h.O., Kim KY, Bushong EA, Mills EA, Boassa D, Shih T, Kinebuchi M, Phan S, Zhou Y, Bihlmeyer NA, et al. (2014). Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. USA 111, 9633–9638. 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lampinen R, Belaya I, Saveleva L, Liddell JR, Rait D, Huuskonen MT, Giniatullina R, Sorvari A, Soppela L, Mikhailov N, et al. (2022). Neuron-astrocyte transmitophagy is altered in Alzheimer’s disease. Neurobiol. Dis 170, 105753. 10.1016/j.nbd.2022.105753. [DOI] [PubMed] [Google Scholar]
- 3.Morales I, Sanchez A, Puertas-Avendaño R, Rodriguez-Sabate C, Perez-Barreto A, and Rodriguez M (2020). Neuroglial transmitophagy and Parkinson’s disease. Glia 68, 2277–2299. 10.1002/glia.23839. [DOI] [PubMed] [Google Scholar]
- 4.Rosina M, Ceci V, Turchi R, Chuan L, Borcherding N, Sciarretta F, Sánchez-Díaz M, Tortolici F, Karlinsey K, Chiurchiù V, et al. (2022). Ejection of damaged mitochondria and their removal by macrophages ensure efficient thermogenesis in brown adipose tissue. Cell Metabol. 34, 533–548.e12. 10.1016/j.cmet.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okawa H, Sampath AP, Laughlin SB, and Fain GL (2008). ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol 18, 1917–1921. 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoang QV, Linsenmeier RA, Chung CK, and Curcio CA (2002). Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis. Neurosci 19, 395–407. 10.1017/s0952523802194028. [DOI] [PubMed] [Google Scholar]
- 7.Azuma K, Koumura T, Iwamoto R, Matsuoka M, Terauchi R, Yasuda S, Shiraya T, Watanabe S, Aihara M, Imai H, and Ueta T (2022). Mitochondrial glutathione peroxidase 4 is indispensable for photoreceptor development and survival in mice. J. Biol. Chem 298, 101824. 10.1016/j.jbc.2022.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingram NT, Fain GL, and Sampath AP (2020). Elevated energy requirement of cone photoreceptors. Proc. Natl. Acad. Sci. USA 117, 19599–19603. 10.1073/pnas.2001776117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shichino Y, and Iwasaki S (2022). Compounds for selective translational inhibition. Curr. Opin. Chem. Biol 69, 102158. 10.1016/j.cbpa.2022.102158. [DOI] [PubMed] [Google Scholar]
- 10.Tseng YC, Chen RD, Lucassen M, Schmidt MM, Dringen R, Abele D, and Hwang PP (2011). Exploring uncoupling proteins and antioxidant mechanisms under acute cold exposure in brains of fish. PLoS One 6, e18180. 10.1371/journal.pone.0018180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, and Lukyanov KA (2006). A genetically encoded photosensitizer. Nat. Biotechnol 24, 95–99. 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 12.van Ham TJ, Mapes J, Kokel D, and Peterson RT (2010). Live imaging of apoptotic cells in zebrafish. Faseb. J 24, 4336–4342. 10.1096/fj.10-161018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutto RA, Bisbach CM, Abbas F, Brock DC, Cleghorn WM, Parker ED, Bauer BH, Ge W, Vinberg F, Hurley JB, and Brockerhoff SE (2020). Increasing Ca(2+) in photoreceptor mitochondria alters metabolites, accelerates photoresponse recovery, and reveals adaptations to mitochondrial stress. Cell Death Differ. 27, 1067–1085. 10.1038/s41418-019-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giarmarco MM, Brock DC, Robbings BM, Cleghorn WM, Tsantilas KA, Kuch KC, Ge W, Rutter KM, Parker ED, Hurley JB, and Brockerhoff SE (2020). Daily mitochondrial dynamics in cone photoreceptors. Proc. Natl. Acad. Sci. USA 117, 28816–28827. 10.1073/pnas.2007827117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reichenbach A, and Robison SR (1995). The Involvement on Muller Cells in Outer Retina (Springer Science & Business Media). [Google Scholar]
- 16.Ellett F, Pase L, Hayman JW, Andrianopoulos A, and Lieschke GJ (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56. 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama H, Hama H, Nagasawa K, Kurokawa H, Sugiyama M, Ando R, Funata M, Yoshida N, Homma M, Nishimura T, et al. (2020). Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell 181, 1176–1187.e16. 10.1016/j.cell.2020.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Yano S, Tazawa H, Kishimoto H, Kagawa S, Fujiwara T, and Hoffman RM (2021). Real-time fluorescence image-guided oncolytic virotherapy for precise cancer treatment. Int. J. Mol. Sci 22, 879. 10.3390/ijms22020879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korzh V, Teh C, Kondrychyn I, Chudakov DM, and Lukyanov S (2011). Visualizing compound transgenic zebrafish in development: a tale of green fluorescent protein and KillerRed. Zebrafish 8, 23–29. 10.1089/zeb.2011.0689. [DOI] [PubMed] [Google Scholar]
- 20.Teh C, Chudakov DM, Poon KL, Mamedov IZ, Sek JY, Shidlovsky K, Lukyanov S, and Korzh V (2010). Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol 10, 110. 10.1186/1471-213X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuvel DJ, Li L, Krishnasamy Y, Gooz M, Takemoto K, Woster PM, Lemasters JJ, and Zhong Z (2022). Mitochondrial depolarization after acute ethanol treatment drives mitophagy in living mice. Autophagy 18, 2671–2685. 10.1080/15548627.2022.2046457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Indira D, Varadarajan SN, Subhasingh Lupitha S, Lekshmi A, Mathew KA, Chandrasekharan A, Rajappan Pillai P, Pulikkal Kadamberi I, Ramachandran I, Sekar H, et al. (2018). Strategies for imaging mitophagy in high-resolution and high-throughput. Eur. J. Cell Biol 97, 1–14. 10.1016/j.ejcb.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 23.McWilliams TG, Prescott AR, Villarejo-Zori B, Ball G, Boya P, and Ganley IG (2019). A comparative map of macroautophagy and mitophagy in the vertebrate eye. Autophagy 15, 1296–1308. 10.1080/15548627.2019.1580509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holländer H, Makarov F, Dreher Z, van Driel D, Chan-Ling TL, and Stone J (1991). Structure of the macroglia of the retina: sharing and division of labour between astrocytes and Muller cells. J. Comp. Neurol 313, 587–603. 10.1002/cne.903130405. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Gao Y, Liu J, Huang Y, Yin J, Feng Y, Shi L, Meloni BP, Zhang C, Zheng M, and Gao J (2021). Intercellular mitochondrial transfer as a means of tissue revitalization. Signal Transduct. Targeted Ther 6, 65. 10.1038/s41392-020-00440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayakawa K, Esposito E, Wang X, Terasaki Y, Liu Y, Xing C, Ji X, and Lo EH (2016). Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535, 551–555. 10.1038/nature18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Vlist M, Raoof R, Willemen HLDM, Prado J, Versteeg S, Martin Gil C, Vos M, Lokhorst RE, Pasterkamp RJ, Kojima T, et al. (2022). Macrophages transfer mitochondria to sensory neurons to resolve inflammatory pain. Neuron 110, 613–626.e9. 10.1016/j.neuron.2021.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Artuso L, Romano A, Verri T, Domenichini A, Argenton F, Santorelli FM, and Petruzzella V (2012). Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim. Biophys. Acta 1817, 1002–1011. 10.1016/j.bbabio.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Wensel TG, Potter VL, Moye A, Zhang Z, and Robichaux MA (2021). Structure and dynamics of photoreceptor sensory cilia. Pflügers Archiv 473, 1517–1537. 10.1007/s00424-021-02564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoreson WB (2021). Transmission at rod and cone ribbon synapses in the retina. Pflügers Archiv 473, 1469–1491. 10.1007/s00424-021-02548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley JB (2021). Retina metabolism and metabolism in the pigmented epithelium: a busy intersection. Annu. Rev. Vis. Sci 7, 665–692. 10.1146/annurev-vision-100419-115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palczewski K, and Kiser PD (2020). Shedding new light on the generation of the visual chromophore. Proc. Natl. Acad. Sci. USA 117, 19629–19638. 10.1073/pnas.2008211117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stearns G, Evangelista M, Fadool JM, and Brockerhoff SE (2007). A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J. Neurosci 27, 13866–13874. 10.1523/JNEUROSCI.3136-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy BN, Alvarez Y, Brockerhoff SE, Stearns GW, Sapetto-Rebow B, Taylor MR, and Hurley JB (2007). Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Invest. Ophthalmol. Vis. Sci 48, 522–529. 10.1167/iovs.06-0975. [DOI] [PubMed] [Google Scholar]
- 35.Shin J, Chen J, and Solnica-Krezel L (2014). Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 141, 3807–3818. 10.1242/dev.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloan JL, Achilly NP, Arnold ML, Catlett JL, Blake T, Bishop K, Jones M, Harper U, English MA, Anderson S, et al. (2020). The vitamin B12 processing enzyme, mmachc, is essential for zebrafish survival, growth and retinal morphology. Hum. Mol. Genet 29, 2109–2123. 10.1093/hmg/ddaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernardos RL, and Raymond PA (2006). GFAP transgenic zebrafish. Gene Expr. Patterns 6, 1007–1013. 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Blume ZI, Lambert JM, Lovel AG, and Mitchell DM (2020). Microglia in the developing retina couple phagocytosis with the progression of apoptosis via P2RY12 signaling. Dev. Dynam 249, 723–740. 10.1002/dvdy.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drerup CM, and Nechiporuk AV (2016). In vivo analysis of axonal transport in zebrafish. Methods Cell Biol. 131, 311–329. 10.1016/bs.mcb.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George AA, Hayden S, Stanton GR, and Brockerhoff SE (2016). Arf6 and the 5’phosphatase of Synaptojanin 1 regulate autophagy in cone photoreceptors. Inside Cell 1, 117–133. 10.1002/icl3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fowler DK, Stewart S, Seredick S, Eisen JS, Stankunas K, and Washbourne P (2016). A MultiSite gateway toolkit for rapid cloning of vertebrate expression constructs with diverse research applications. PLoS One 11, e0159277. 10.1371/journal.pone.0159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buj R, Iglesias N, Planas AM, and Santalucía T (2013). A plasmid toolkit for cloning chimeric cDNAs encoding customized fusion proteins into any Gateway destination expression vector. BMC Mol. Biol 14, 18. 10.1186/1471-2199-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bisbach CM, Hutto RA, Poria D, Cleghorn WM, Abbas F, Vinberg F, Kefalov VJ, Hurley JB, and Brockerhoff SE (2020). Mitochondrial Calcium Uniporter (MCU) deficiency reveals an alternate path for Ca(2+) uptake in photoreceptor mitochondria. Sci. Rep 10, 16041. 10.1038/s41598-020-72708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrnes J, Ganetzky R, Lightfoot R, Tzeng M, Nakamaru-Ogiso E, Seiler C, and Falk MJ (2018). Pharmacologic modeling of primary mitochondrial respiratory chain dysfunction in zebrafish. Neurochem. Int 117, 23–34. 10.1016/j.neuint.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giarmarco MM, Cleghorn WM, Hurley JB, and Brockerhoff SE (2018). Preparing fresh retinal slices from adult zebrafish for ex vivo imaging experiments. JoVE. 10.3791/56977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, and Douglas RJ (2012). TrakEM2 software for neural circuit reconstruction. PLoS One 7, e38011. 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saalfeld S, Fetter R, Cardona A, and Tomancak P (2012). Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9, 717–720. 10.1038/nmeth.2072. [DOI] [PubMed] [Google Scholar]
- 49.Saalfeld S, Cardona A, Hartenstein V, and Tomančák P (2010). As-rigid-as-possible mosaicking and serial section registration of large ssTEM datasets. Bioinformatics 26, i57–i63. 10.1093/bio-informatics/btq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogovic JA, Hanslovsky P, Wong A, and Saalfeld S (2016). Robust registration of calcium images by learned contrast synthesis. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 1123–1126. 10.1109/ISBI.2016.7493463. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


