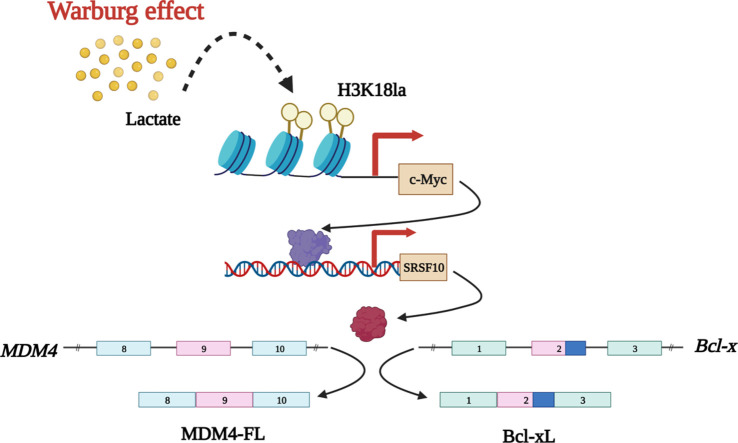
Highlights
-
•
Metabolic state of breast cancer cells affects the transcriptome.
-
•
High glycolytic rate promotes H3K18la-mediated c-Myc upregulation.
-
•
c-Myc modulates SRSF10 expression in response to intracellular lactate levels.
-
•
Attenuating glycolytic rate impedes c-Myc-SRSF10 axis.
Keywords: Epigenetics, Warburg effect, Histone modifications, Tumor metabolism, c-Myc, SRSF10
Abstract
Due to the enhanced glycolytic rate, cancer cells generate lactate copiously, subsequently promoting the lactylation of histones. While previous studies have explored the impact of histone lactylation in modulating gene expression, the precise role of this epigenetic modification in regulating oncogenes is largely unchartered. In this study, using breast cancer cell lines and their mutants exhibiting lactate-deficient metabolome, we have identified that an enhanced rate of aerobic glycolysis supports c-Myc expression via promoter-level histone lactylation. Interestingly, c-Myc further transcriptionally upregulates serine/arginine splicing factor 10 (SRSF10) to drive alternative splicing of MDM4 and Bcl-x in breast cancer cells. Moreover, our results reveal that restricting the activity of critical glycolytic enzymes affects the c-Myc-SRSF10 axis to subside the proliferation of breast cancer cells. Our findings provide novel insights into the mechanisms by which aerobic glycolysis influences alternative splicing processes that collectively contribute to breast tumorigenesis. Furthermore, we also envisage that chemotherapeutic interventions attenuating glycolytic rate can restrict breast cancer progression by impeding the c-Myc-SRSF10 axis.
Graphical abstract
Introduction
Cancer is a malady characterized by the uncontrolled proliferation of neoplastic cells beyond the realm of normal tissue development. During the evolution of this transformation, cells accumulate various genetic and epigenetic mutations to adapt cellular metabolism that supports the requirements of autonomous rapid growth. Therefore, altered metabolism is one of the critical hallmarks of cancer, as the metabolic requirements of cancer cells differ substantially from their normal counterparts. Interestingly, mounting evidence suggests that approximately every identified oncogene regulates the targets directly linked with metabolic adaptations [1], [2], [3]. Moreover, the aberrant profile of metabolites, in turn, supports the expression of oncogenes to initiate a vicious cycle that forms the cornerstone of carcinogenesis [4], [5], [6]. Therefore, a precise assessment of the interplay between metabolism and oncogenic factors is of paramount importance.
Seminal work by Otto Warburg in the 1950s laid the foundation for numerous studies initiated to decipher the altered tumor metabolism [7]. It is observed that many cancer types exhibit an elevated glycolytic rate which is associated with high lactate production [8,9]. Being identified as the most commonly diagnosed type of cancer, breast cancer accounted for 11.7% of the new cancer cases in 2020, affecting over 2.3 million people worldwide [10]. Moreover, it is demonstrated that higher grades of breast cancers are associated with enhanced lactate concentration, making studying the implications of lactate in breast cancer physiology indispensable [11]. Lactate, considered merely an unwanted by-product of aerobic glycolysis for decades, is increasingly being investigated for its function as an oncometabolite [12], [13], [14], [15]. Since the discovery of the ‘lactate shuttle theory’ in the mid 1980s, a plethora of emerging studies have verified that lactate acts as a signal to alter the metabolic landscape of neoplastic and non-neoplastic cells within the tumor microenvironment (TME) to promote cancer progression [16], [17], [18], [19], [20].
Interestingly, recent eminent work by Zhang and co-workers led to the striking discovery of a previously unreported histone modification, lactylation, derived from cellular lactate [21]. Histone lysine lactylation (Kla), observed to be enriched in a dose-dependent manner in response to exogenous and endogenous lactate, has been demonstrated to serve as an epigenetic modification that stimulates gene transcription in human and mouse cells. Although only a few reports have shown the cancer-promotive role exhibited by histone lactylation [14,[21], [22], [23], [24]], and to date, there is a lack of cancer cell line-derived epigenomic and transcriptomic data which would demonstrate the global distribution of histone lactylation marks and the genes regulated by this epigenetic modification. Therefore, the functional contributions of histone lactylation in cancer onset and progression strongly warrant further investigations. Undertaking such studies will aid in implementing new strategies to subside the expression of oncogenes responsive to cellular metabolic state. Considering the increasing efforts and novel strategies employed to target various subtypes of breast cancer [25], [26], [27], [28], [29], we explored the gene regulations mediated by histone lactylation by performing our experiments using cell lines that represent two of the commonly detected and widely studied types of breast cancer- luminal A (MCF7) and basal subtype (HCC1806).
By comparing the transcriptome of MCF7 (exhibiting lactate-rich metabolome) and its PKM2 knockout mutant cells (resembling a lactate-deficient metabolome), we identified that the oncogenic transcription factor c-Myc expression is sensitive to intracellular lactate. Experimental investigations uncovered that elevated lactate production resulted in histone H3 lysine 18 lactylation (H3K18la)-mediated c-Myc upregulation. Furthermore, by transcriptionally inducing serine/arginine-rich splicing factor 10 (SRSF10) expression, c-Myc influenced the alternative splicing outcomes of MDM4 and Bcl-x, thus acting as a switch connecting altered metabolism and alternative splicing reprogramming.
Materials and methods
Cell culture
Human breast cancer cell lines MCF7 and HCC1806 were procured from American Type Culture Collection (ATCC). MCF7 and HEK293T were cultured at 37 °C, 5% CO2 in DMEM (Invitrogen, 12800017). RPMI-1640 (Invitrogen, 23400021) was used to culture HCC1806. The culture media for all the cell lines was supplemented with 10% fetal bovine serum (FBS; Sigma, F7524), 100 units/ml of penicillin and streptomycin (Invitrogen, 15140122), and 2 mM/ l-glutamine (Invitrogen, 25030081). The Boris Binding Site mutant (BBS Mut) MCF7 and HCC1806 cell lines were generated as previously stated [30,31]. The media and cell culture conditions for CRISPR/Cas9 mutants were identical to the wild-type cells.
Molecular cloning
The PKM2 and PFKFB3 overexpression constructs were generated as previously mentioned [30,31]. Briefly, PKM2 was amplified using MCF7 cDNA as a template and cloned between NotI and SalI sites of the plasmid pCMV-3tag-1a. Similarly, PFKFB3 was amplified using MCF7 cDNA as a template and cloned between NotI and EcoRI sites of pCMV-3tag-1a plasmid. c-Myc was also amplified from MCF7 cDNA and cloned between BamHI and EcoRI sites in pCMV-3tag-1a. The sequence of the obtained construct was verified using Sanger sequencing. The details of the primers used are provided in Table S2.
Luciferase reporter assay
The c-Myc and SRSF10 promoter fragments were amplified using MCF7 genomic DNA as a template and individually subcloned between the KpnI and HindIII sites of the pGL3 basic vector (Promega, E1751). The sequence of the obtained constructs was verified using Sanger sequencing. The details of the primers used for generating luciferase constructs are enlisted in Table S2. Cells were seeded in a 24-well plate and allowed to attach overnight. The wells were co-transfected with different c-Myc or SRSF10 promoter constructs along with pRL-TK renilla luciferase plasmid. 24 h post-transfection, the cells were lysed, and luciferase activity was determined. The relative luciferase activity was calculated by dividing the firefly luciferase activity by the renilla luciferase activity.
Site-directed mutagenesis
The site-directed mutant construct of the SRSF10 promoter was generated using oligonucleotides harboring desired mutations in the c-Myc binding site located from −160 bp to −168 bp relative to the transcription start site (TSS). The wild-type SRSF10 promoter construct was used as a template to generate the mutant construct. The sequence of the site-directed mutagenesis (SDM) construct was verified by Sanger sequencing. The primers used for obtaining the SDM construct are enlisted in Table S2. A similar protocol, as mentioned for the luciferase assay, was followed for transfecting and obtaining readings of the SDM construct.
Immunoblotting
The cells were lysed in urea lysis buffer (8 M urea, 2 M thiourea, 2% CHAPS, 1% DTT) supplemented with 1 × protease inhibitor cocktail (PIC; leupeptin 10–100 M, pepstatin 1 M, EDTA 1–10 mM, AEBSF 1 mM) at 4 °C for 30 min and centrifuged for 2 h at maximum speed (16,900 g). The supernatant was separated, quantified, and an equal concentration of protein samples was loaded. After separation, proteins were electro-transferred on an active PVDF membrane. Following the transfer, the blots were incubated overnight at 4 °C with suggested dilutions of primary antibodies, followed by 1 h incubation with secondary antibody. The Odyssey membrane Scanning equipment was used to scan the blots. The bands were quantified using GelQuant software (version 1.8.2). The details of the antibodies used are provided in Table S1.
Quantitative RT-PCR
Total RNA was extracted according to the manufacturer's instructions using TRIZol reagent (Ambion, 15596018). The concentration was measured using an Eppendorf BioSpectrometer, and 2 µg of total RNA was reverse transcribed using the Invitrogen SuperScript® III First-Strand Synthesis System (18080–051). Amplifications were carried out in duplicates using the GO taq QPCR master mix (Promega, A6002) and the Roche light cycler 480 II according to the manufacturer's instructions. Primers were designed using IDT PrimerQuest tool (https://www.idtdna.com) and are listed in Table S3. Using the formula 2^(Ct_control-Ct_target), the average cycle thresholds from three independent biological replicates were computed and normalised to the housekeeping control gene RPS16. Student's t-test was performed to compare gene/exon expression levels between two groups. P< 0.05 was considered statistically significant.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as previously mentioned [31]. Approximately 10 million cells were crosslinked, scaped in PBS, lysed, and sonicated. 25 μg of sheared chromatin was immunoprecipitated with an antibody of interest following overnight incubation at 4 °C. The immunoprecipitated protein-DNA complexes and 5% input were purified to eliminate proteins and the eluted DNA was analysed by qRT-PCR using GO taq QPCR master mix (Promega, A6002) in triplicate using primers specific for c-Myc or SRSF10 promoter. Each experiment was performed at least thrice, and normalizations were performed using the formula 2^ (Ct input – Ct immunoprecipitation). The obtained values were normalized to relative rabbit IgG and control IP values. The primers were designed using IDT PrimerQuest tool (https://www.idtdna.com) and are listed in Table S3. Significance between the two groups was calculated using Student's t-test, with a value of <0.05 considered statistically significant.
RNA interference
3 × 105 cells were seeded per well of a six-well culture plate and allowed to attach for 24 h. The lentivirus containing small hairpin RNA (shRNA) (Sigma, Mission Human Genome shRNA Library) against the target gene was inoculated in the presence of 8 μg/ml polybrene (Sigma, H9268) containing media. Cells were selected for 72 h using 1 μg/ml puromycin (Sigma, P9620) and subsequently used for various experiments. The sequence of shRNAs used in this study is listed in Table S4.
Lactate assay
Following treatment, an equal number of WT BBS and BBS Mut MCF7 and HCC1806 cells were lysed using ice-cold assay buffer provided in the lactate assay kit (Sigma, MAK064–1). Lactate quantification was carried out using the deproteinized lysates according to the manufacturer's instructions. The readings were taken at room temperature with a microplate reader set to 450 nm optical density.
Extracellular flux assays
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined using Seahorse XF HS mini analyzer. 5 × 103 cells per well were seeded and allowed to attach overnight in an 8-well Seahorse XFp mini cell culture plate. Cells were then washed and incubated with XF assay medium supplemented with 1 mM pyruvate (Sigma, S8636), 2 mM l-glutamine (Invitrogen, 25030081), 10 mM glucose (Gibco, A24940–01) and incubated for 1 h at 37 °C in CO2-free incubator. OCR and ECAR estimation was performed as per the manufacturer's instructions. OCR was assessed in response to oligomycin (1.5 μM), FCCP (0.5 μM), and rotenone/antimycin A (0.5 μM), and ECAR was assessed in response to rotenone/antimycin A (0.5 μM), and 2-deoxy-d-glucose (2-DG; 50 mM). Finally, the readings were normalized to the respective protein concentrations.
Clonogenic assay
5 × 103 cells were seeded in each well of a 6-well cell culture plate and were cultured for 10 days. The media was replaced every 72 h. The cells were washed thrice with 1X PBS and then fixed using 4% formaldehyde for 15 min at RT. The cells were then stained with 0.05% crystal violet solution prepared in 10% ethanol. The cells were then gently washed thrice with 1X PBS, and the plates were air-dried for 15 min and imaged.
Wound healing assay
3 × 105 cells were seeded in a 6-well plate. A 200 µl sterile pipette tip was used to create a wound upon reaching confluency. The plate was then washed twice with 1X PBS to remove debris. The images of the same region were captured at 0, 24, and 48 h with an inverted microscope.
Generation of spheroid cultures
The spheroids were generated as previously described [31]. Briefly, 50μL of Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning, 356230) was spread in a well of 96-well cell culture plate and allowed to solidify at 37 °C for 1 h. 7 × 103 cells were resuspended in 100μL of cell-culture media supplemented with 1 μg/mL hydrocortisone (Sigma, H0888) and 5 μg/mL insulin (Sigma, I1882) and added over the solidified matrix. Media containing 10% GFR matrigel was then subsequently overlayed as the topmost layer and incubated at 37 °C in the presence of 5% CO2. The media containing hormones was replaced every 72 h, and images were captured using Thermo Scientific EVOS FL Auto 2 imaging system.
Breast cancer sample collection
Tumor and adjacent normal tissue pairs were collected from patients undergoing surgery for breast cancer at Bansal Hospital, Bhopal, India. For the study, approval was granted by the Institute Ethics Committee of Indian Institute of Science Education and Research Bhopal. Informed consent was obtained from all the patients. The tissue samples were snap frozen immediately after surgery and stored at −80 °C until use. Clinical characteristics of patients used in the study are provided in Table S7.
Human transcriptome array 2.0 data analysis
The CEL files (GSE190401) were analyzed using Transcriptome Array Console 4.0 (Invitrogen, version 4.0.2.15) using the gene+exon–SST-RMA method of summarization. Genes with thresholds of absolute fold-change >2, P<0.05, and false discovery rates (FDR) <0.05 were selected as differentially expressed genes (DEGs). The volcano plot for DEGs and violin plot for differentially expressed epigenetic factors and transcription factors were generated using GraphPad Prism 8 software. A heat map for differentially expressed epigenetic factors and transcription factors was generated using the online tool Morpheus (https://software.broadinstitute.org/morpheus), and the over-representation analysis (ORA) for the GO biological processes was generated using ShinyGo 0.76.2 [32]. FDR <0.05 was considered significant while obtaining enriched GO terms.
Survival data analysis
Recurrence-free survival of c-Myc was analyzed in cohort GSE9195 using the online tool Kaplan–Meier Plotter (www.kmplot.com). The best possible cut-off was used as the upper and lower quartile. Samples were divided into high- and low-expression groups and compared for recurrence-free survival.
ChIP-seq data analyses
The available ChIP-seq data for H3K18la (GSE115354) and c-Myc (GSM2501566) were downloaded from Gene Expression Omnibus database. The raw fastQ files were downloaded and trimmed by Trimmomatic (v0.39) with default parameters. STAR (v2.7.3a) aligner was used to uniquely align the reads to GRCh38. Peak calling was done using MACS2 v2.1.2.
Statistical analysis
GraphPad Prism 8 was used for all statistical analyses. Unless otherwise specified, all data are presented as mean ±SD and were analysed using Student's t-test. The statistical methods for each analysis are provided in the figure legends or the materials and methods sections. P-values less than 0.05 were deemed significant. ns = not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Results
Oncometabolite lactate influences breast cancer transcriptome
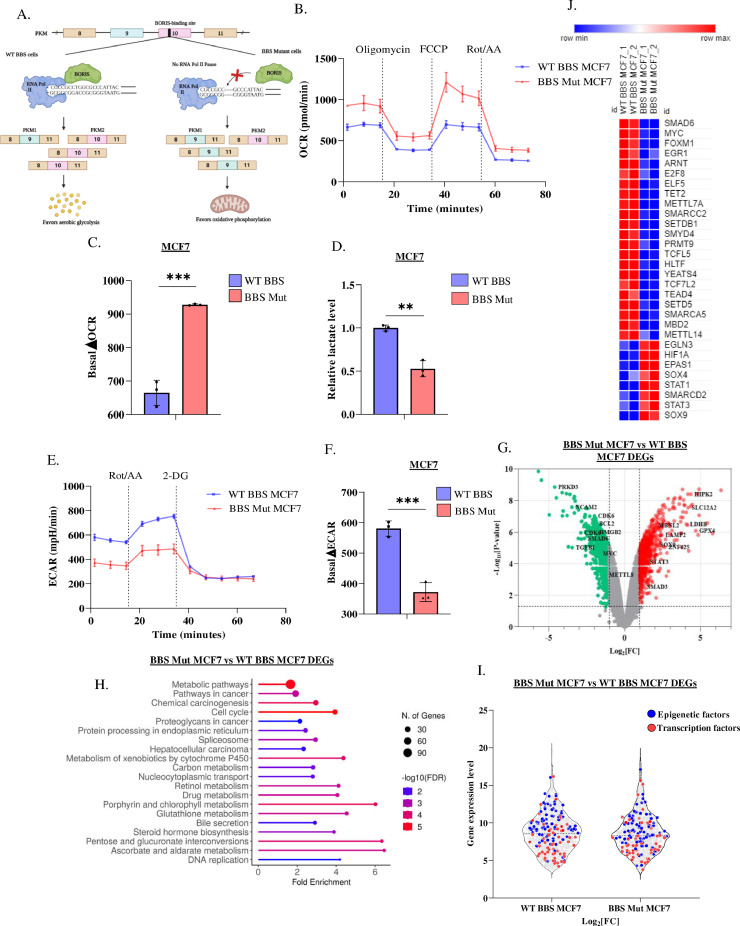
The rate of glycolysis- the principal producer of lactate, is essentially determined by the activity of its rate-limiting enzyme pyruvate kinase (PKM). Known to undergo alternative splicing, the M2 isoform of PKM (PKM2; overexpressed in various cancers) favours aerobic glycolysis. Hence, enhanced expression of PKM2 is strongly associated with higher lactate production [33]. Therefore, to identify genes that are regulated in a lactate-dependent manner, we employed our previously reported model system generated in the breast cancer cells lines, where we knocked out the expression of PKM2 by mutating the BORIS-binding site (BBS) present in the exon 10 of PKM to establish stable lactate-deficient cell lines [30,31] (Fig. 1A).
Fig. 1.
Cancer-associated metabolic affects transcriptome. (A) Schematic representation of CRISPR/Cas9 strategy employed to generate PKM2 knockout breast cancer cell lines. (B) Real-time OCR in WT BBS and BBS Mut MCF7 cell lines. (C) Quantification of ΔOCR in the WT BBS and BBS Mut MCF7 cell lines. (D) Intracellular lactate production in WT BBS and BBS Mut MCF7 cell lines estimated by lactate assay. (E) Real-time ECAR analysis in WT BBS and BBS Mut MCF7 cell lines. (F) Quantification of ΔECAR in the WT BBS and BBS Mut MCF7 cell lines. (G) Volcano plot depicting DEGs in BBS Mut vs. WT BBS MCF7. (H) The top 20 enriched terms for biological processes for the DEGs obtained from the microarray data. (I) Violin plot depicting the differentially expressed epigenetic factors (blue points) and transcription factors (red points) in BBS Mut MCF7 cells. (J) Heat map representation of top differentially expressed transcription and epigenetic factors in the BBS Mut MCF7 cells. Data are represented as mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns= not significant.
To verify if PKM2 knockout (BBS Mut) cell lines of MCF7 and HCC1806 exhibited elevated oxidative phosphorylation (OXPHOS), we measured the oxygen consumption rate (OCR) using a Seahorse XFp analyzer. Our results demonstrated that the BBS Mut cells clearly exhibited higher OCR (Fig. 1B, S1.A). Moreover, the significant basal OCR difference (ΔOCR) denoted that due to a complete lack of PKM2 expression, BBS Mut cell lines displayed enhanced reliance on OXPHOS (Fig. 1C, S1.B). Notably, upon performing lactate assay, we observed that the WT BBS cell lines had substantially higher intracellular lactate, which resulted in an enhanced extracellular acidification rate (ECAR) compared to their respective BBS Mut cell lines (Fig. 1D, 1E, S1.C, S1.D). Furthermore, the basal ECAR difference (ΔECAR) confirmed that the WT BBS cells exhibit lactate-rich metabolome (Fig. 1F, S1.E). Our data demonstrate that knocking out PKM2 expression significantly decreases lactate production, thereby making this model system ideal for studying the global transcriptomic alterations possibly induced by oncometabolite lactate.
Interestingly, the analysis of Human Transcriptome Array 2.0 (HTA 2.0) data (GSE190401) of the WT BBS and BBS Mut MCF7 cells revealed that a total of 1962 genes (coding and non-coding) were differentially expressed in BBS Mut MCF7 cells from which, 1146 and 816 genes were downregulated and upregulated, respectively (Fig. S1.F). The significant events (P < 0.05) corresponding to the coding genes with fold change > 2 (highlighted with red points) and < −2 (highlighted with green points) are marked in Fig. 1G. Moreover, GO terms for biological processes enrichment analysis of differentially expressed genes (DEGs) revealed that the transcription of genes related to processes such as metabolic and cancer-related pathways, cell cycle, and spliceosome was extensively affected, underscoring the probable contributions of metabolic lactate in regulating the expression of a plethora of genes involved in diverse cancer-associated pathways (Fig. 1H). As epigenetic alterations and differential transcription are the two fundamental processes that predominantly define cellular transcriptome, we further analysed the DEGs to investigate if the metabolic landscape modulated the expression of epigenetic and transcription factors. Interestingly, 64 transcription factors and 76 epigenetic factors were identified to be differentially expressed in BBS Mut MCF7 cells (Fig. 1I, Tables S5 and S6).
In conclusion, our microarray data suggest an intriguing possibility that intracellular lactate levels can affect the expression of a plethora of genes in breast cancer cells. Moreover, our data demonstrate that by regulating the expression of epigenetic and transcription factors, oncometabolite lactate can exert wide-reaching effects that can promote breast tumorigenesis.
Elevated c-Myc expression is maintained by promoter-level histone lactylation
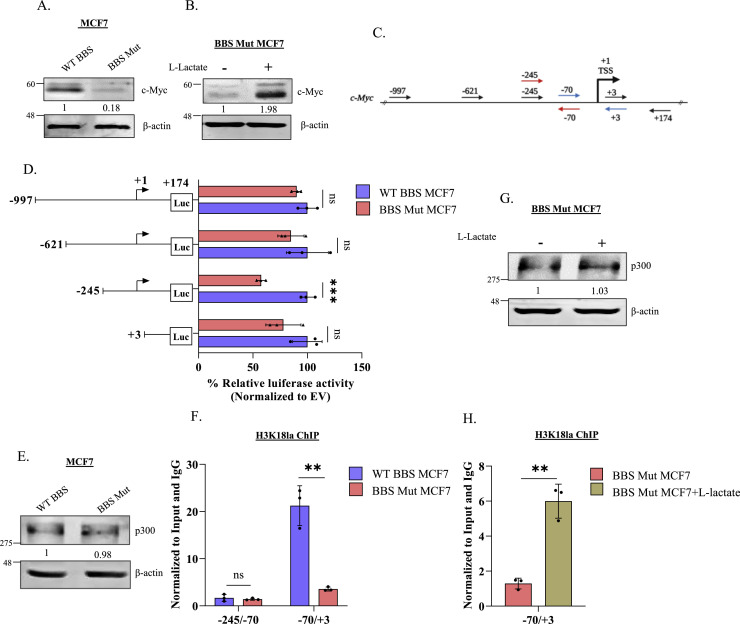
Interestingly, the oncogenic transcription factor c-Myc was one of the most downregulated genes in BBS Mut MCF7 cells (Fig. 1J). Found to be overexpressed in 46% of the primary breast tumors, and 30–50% of high-grade tumors, c-Myc acts as a signaling hub in regulating multiple cellular processes that drive breast cancer progression [34], [35], [36], [37], [38], [39], [40]. To investigate the intriguing possibility that c-Myc expression is responsive to metabolic lactate, we performed qRT-PCR and immunoblotting analysis and observed that c-Myc was significantly downregulated in the BBS Mut cell lines (Fig. S2.A, 2A, S2.B). Next, as a rescue experiment, we subjected the BBS Mut cells to 15 mM l-lactate treatment for 24 h and interestingly observed heightened induction of c-Myc which confirmed that intracellular lactate drives c-Myc expression (Fig. 2B, S2.C).
Fig. 2.
c-Myc undergoes promoter-level histone lactylation. Immunoblot analysis of c-Myc in (A) WT BBS and BBS Mut MCF7 cells and (B) on subjecting BBS Mut MCF7 cells to 15 mM l-lactate treatment for 24 h. (C) Schematic representation of c-Myc promoter. The primers used to generate deletional constructs for luciferase assay are represented using black arrows. The primers used for ChIP qRT-PCR have been marked using red and blue arrows. (D) Luciferase assay performed using the deletional constructs of the c-Myc promoter depicting a dampened activity of −245 to +174 fragment in the BBS Mut MCF7 cells. (E) Immunoblot analysis of p300 in WT BBS and BBS Mut MCF7 cells. (F) H3K18la ChIP assay performed in WT BBS and BBS Mut MCF7 cells. (G) Immunoblot analysis of p300 upon subjecting BBS Mut MCF7 cells to 15 mM l-lactate treatment for 24 h. (H) H3K18la ChIP assay performed in BBS Mut MCF7 cells subjected to 15 mM l-lactate treatment for 12 h. Data are represented as mean± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns= not significant.
Recently, it was demonstrated that metabolic lactate can influence the histone lactylation status to regulate gene expression [21]. Therefore, we analysed the chromatin immunoprecipitation sequencing (ChIP-seq) data available for histone H3 lysine 18 lactylation (H3K18la; GSE115354) and observed extensive enrichment of this epigenetic modification on the c-Myc promoter (Fig. S2.D). Next, we generated multiple deletional constructs of the c-Myc promoter for identifying the active segment primarily responsible for maintaining high c-Myc expression. Therefore, using luciferase assay, we estimated the activity of the approximately 1Kb sequence upstream of the transcriptional start site (TSS) and 174 bp sequence downstream of TSS of the c-Myc promoter (Fig. 2C). As depicted in Fig. 2C, a series of four deletional fragments were cloned in pGL3 basic vector: −997 to +174, −621 to +174, −245 to +174 and +3 to +174. Luciferase assay outcome demonstrated that the region between −245 to +3 exhibited decreased activity in the BBS Mut cell lines to suggest the probable involvement of this region in maintaining low c-Myc expression in the BBS Mut cells (Fig. 2D, S2.E).
Previous reports have identified acetyltransferase p300 as a potential histone lysine lactylation ‘writer’ enzyme [14,41]; therefore, we performed an immunoblotting analysis of p300. No alterations in the p300 expression were detected across all the WT BBS and BBS Mut cell lines (Fig. 2E, S2.F). Further, we performed H3K18la ChIP assay to check the promoter-level histone lactylation status of the −245 to +3 region using the primers sets indicated with red and blue arrows in Fig. 2C. Notably, the promoter region between −70 to +3 exhibited drastically decreased H3K18la marks in the BBS Mut MCF7 cells (Fig. 2F). Furthermore, upon subjecting the BBS Mut MCF7 cells l-lactate treatment, we observed a significant enrichment in H3K18la status of −70 to +3 fragment without any alterations in p300 expression (Fig. 2G, S2.G, 2H).
Collectively, our results demonstrate that the state of high intracellular lactate, as observed during elevated glycolytic rate, leads to H3K18la enrichment in −70 to +3 promoter region to upregulate c-Myc expression in breast cancer cells.
SRSF10 is a c-Myc target gene
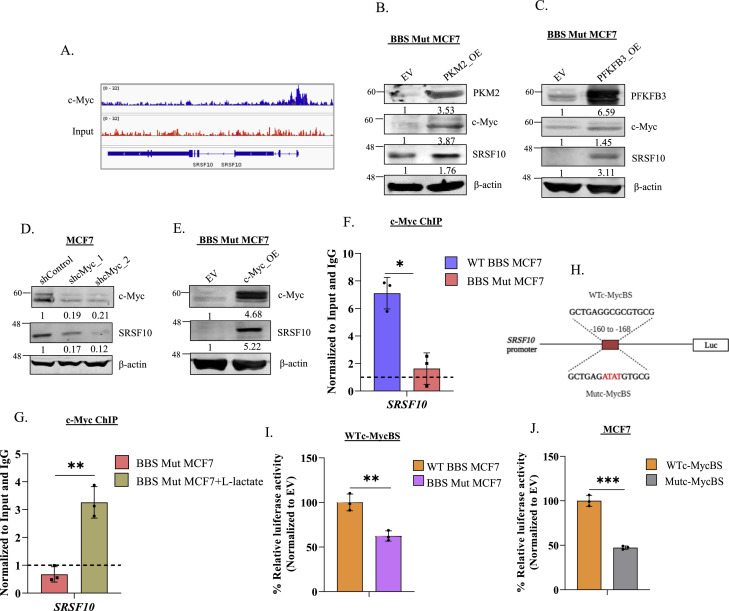
Next, we sought to elucidate the significance of high c-Myc expression in cancers exhibiting elevated aerobic glycolysis. Mounting evidence suggests that c-Myc transcriptionally regulates the expression of numerous splicing factors [42], [43], [44], [45]. Moreover, our microarray results indicated that the transcription of spliceosome-related genes was affected in the lactate-deficient BBS Mut MCF7 cells (Fig. 1H). Therefore, we hypothesized that in response to lactate, c-Myc might induce the expression of splicing factors to function as a vital link between metabolism and alternative splicing. To investigate this, we analyzed c-Myc ChIP-seq data (GSM2501566) and observed its enrichment on the promoter of several SRSF members, with prominent occupancy on the SRSF10 promoter (Fig. 3A, Table S8). Subsequently, we investigated SRSF10, as the immunoblotting screen of SRSFs confirmed that only SRSF10 was consistently downregulated in both the BBS Mut cell lines compared to its expression in the corresponding WT BBS cell lines, thus resembling the expression pattern of c-Myc (Fig. S3.A, S3.B).
Fig. 3.
c-Myc regulates SRSF10 expression. (A) c-Myc ChIP-seq analysis demonstrating its enrichment on SRSF10 promoter. Immunoblot analysis of c-Myc and SRSF10 upon overexpressing (B) PKM2 and (C) PFKFB3 in BBS Mut cell lines. (D) Immunoblot analysis demonstrating the effect of c-Myc knockdown on SRSF10 expression in WT BBS MCF7 cells. (E) Immunoblot assay performed upon overexpressing c-Myc in BBS Mut MCF7 cells. c-Myc ChIP assay performed in (F) WT BBS and BBS Mut MCF7 cells and (G) upon subjecting the BBS Mut MCF7 cells to 15 mM l-lactate treatment for 12 h. (H) Schematic representation of SRSF10 promoter luciferase construct depicting the wild-type c-Myc binding site (WTc-MycBS) and mutated c-Myc binding site (Mutc-MycBS). Luciferase assay of (I) WTc-MycBS SRSF10 construct performed in WT BBS and BBS Mut MCF7 cells and (J) WTc-MycBS and Mutc-MycBS SRSF10 constructs performed in WT BBS MCF7 cells. Data are represented as mean± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns= not significant.
As rescue experiments, we overexpressed two critical glycolytic enzymes to enhance lactate production in BBS Mut cells. Re-introduction of PKM2 and overexpression of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3)- a regulator of phosphofructokinase kinase-1 (PFK-1; which catalyzes the committed step of glycolysis) stimulated c-Myc and SRSF10 expression in BBS Mut cell lines (Fig. 3B, 3C, S3.C, S3.D). Next, c-Myc knockdown significantly reduced SRSF10 levels in WT BBS cell lines (Fig. 3D, S3.E). Moreover, enforced c-Myc expression increased the endogenous SRSF10 expression in BBS Mut cell lines (Fig. 3E, S3.F), and p300 knockdown also hampered c-Myc and SRSF10 expression to confirm the H3K18la-driven regulation of c-Myc-SRSF10 axis (Fig. S3.G).
Subsequently, to identify c-Myc binding sites, we analysed the SRSF10 promoter using the JASPAR database, which predicted the existence of two highly conserved overlapping c-Myc binding sites located between −160 to −168 bp [46]. c-Myc ChIP assay revealed its remarkable enrichment on SRSF10 promoter in the WT BBS MCF7 cells compared to the BBS Mut MCF7 cells (Fig. 3F). Moreover, as l-lactate treatment stimulates c-Myc expression (Fig. 2B, S2.C), we performed c-Myc ChIP assay upon treating the BBS Mut MCF7 cells with 15 mM l-lactate. Results demonstrated an augmented c-Myc occupancy at the SRSF10 promoter in the l-lactate treated cells (Fig. 3G). Further, a 113 bp fragment spanning the wild-type c-Myc binding site (WTc-MycBS) present in the SRSF10 promoter was cloned upstream of a firefly luciferase (FLuc) coding sequence in pGL3 basic vector (Fig. 3H). The FLuc activity of WTc-MycBS was significantly decreased in the BBS Mut cell lines (Fig. 3I, S3.H). Next, using site-directed mutagenesis PCR, we generated a construct of harboring mutations in the c-Myc binding site (Mutc-MycBS) (Fig. 3H). When transfected to WT BBS cells, the Mutc-MycBS construct exhibited substantially diminished FLuc activity compared to the WTc-MycBS construct to indicate c-Myc regulates SRSF10 expression in breast cancer cells (Fig. 3J, S3.I).
c-Myc-SRSF10 axis drives alternative splicing of MDM4 and Bcl-x
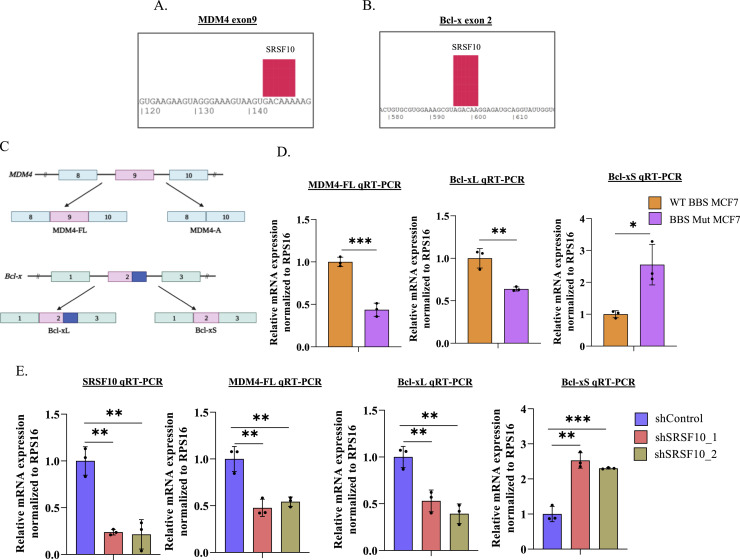
To understand the functional contributions of the c-Myc-SRSF10 axis, we investigated the alternative splicing events regulated by SRSF10. Previous studies have identified BCL2-like 1 (Bcl-x) and Murine Double Minute 4 (MDM4) as targets of SRSF10 [47], [48], [49], [50]. Using the SpliceAid database, we identified SRSF10 binding sites on the exons 9 and 2 of MDM4 and Bcl-x pre-mRNA, respectively (Fig. 4A, 4B) [51], and therefore, examined the mRNA abundance of MDM4-FL isoform and the two Bcl-x isoforms (Bcl-xL and Bcl-xS) (Fig. 4C). We observed high mRNA abundance of MDM4-FL and Bcl-xL in WT BBS cells (Fig. 4D, S4). Furthermore, BBS Mut cell lines exhibited decreased MDM4-FL, Bcl-xL, and increased Bcl-xS isoform abundance (Fig. 4D, S4). Notably, SRSF10 knockdown in WT BBS MCF7 cells led to decreased MDM4-FL and Bcl-xL isoforms with a concomitant enhancement in Bcl-xS isoform at mRNA level (Fig. 4E). Collectively, our results demonstrate that high SRSF10 expression supported by elevated glycolytic rate causes inclusion of MDM4 exon 9 and favors the selection of proximal 5′ splice site of Bcl-x.
Fig 4.
c-Myc-SRSF10 axis regulates alternative splicing of MDM4 and Bcl-x. (A) Predicted SRSF10 binding site on exon 9 of MDM4. (B) Predicted SRSF10 binding site on exon 2 of Bcl-x. (C) Schematic representation of MDM4 and Bcl-x splicing. (D) RPS16 normalized qRT-PCR analysis of MDM4-FL, Bcl-xL, and Bcl-xS in WT BBS and BBS Mut MCF7 cells. (E) RPS16 normalized qRT-PCR analysis of SRSF10, MDM4-FL, Bcl-xL, and Bcl-xS upon performing SRSF10 knockdown in WT BBS MCF7 cells. Data are represented as mean± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns= not significant.
Targeting aerobic glycolysis restricts the c-Myc-SRSF10 axis to impede the proliferation of breast cancer cells
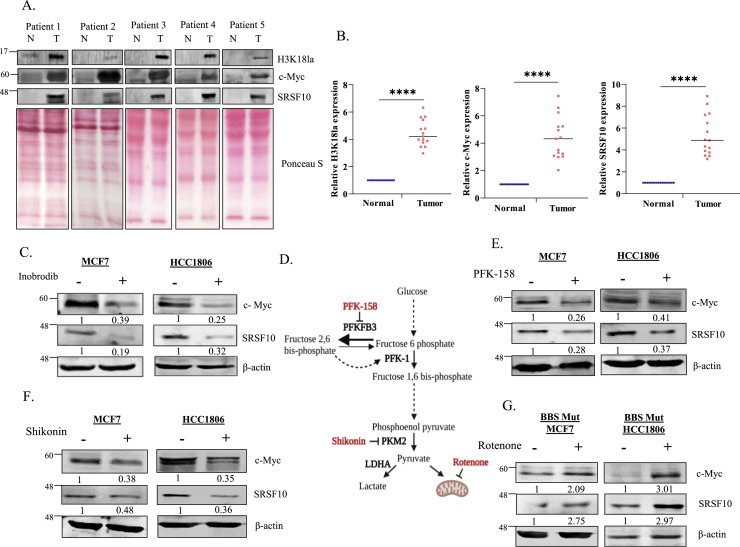
To investigate the clinical relevance of this study, we examined the protein abundance of H3K18la, c-Myc, and SRSF10 in 15 breast tumor samples. Immunoblot analysis demonstrated that H3K18la abundancy, as well as c-Myc and SRSF10 expression, was significantly elevated in the tumor tissues (Fig. 5A, 5B), highlighting the vitality of targeting the c-Myc-SRSF10 axis for therapeutic interventions of cancers exhibiting high glycolytic dependency. Upon treating the WT BBS cells with inobrodib (a p300 inhibitor currently under clinical trial for the treatment of various cancer types [52], [53], [54], [55]), we observed a substantial reduction in c-Myc and SRSF10 expression, underscoring the vital role of H3K18la-dependent epigenetic regulation of c-Myc (Fig. 5C). A plethora of research has highlighted targeting cancer metabolism as a desirable approach to restrict cancer progression [56]. Therefore, we treated the cell lines under study with a panel of metabolic inhibitors (Fig. 5D). PFK-158 and shikonin, which are potent inhibitors of PFKFB3 and PKM2, respectively, are known to attenuate glycolytic rate to restrict lactate production [57,58]. PFK-158 and shikonin treatment were individually observed to downregulate c-Myc and SRSF10 expression (Fig. 5E, 5F). Demonstrated to enhance the glycolytic rate and lactate pool, rotenone treatment enhanced c-Myc and SRSF10 expression in BBS Mut cell lines (Fig. 5G) [59].
Fig. 5.
Glycolytic inhibitors attenuate the c-Myc-SRSF10 axis. (A) Immunoblotting analysis showing H3K18la, c-Myc, and SRSF10 expression levels in 5 representative breast cancer patient samples. (B) Quantification of H3K18la, c-Myc, and SRSF10 levels in 15 breast cancer patient-derived normal and tumor samples. (C) Immunoblot analysis showing the effect of inobrodib (p300 inhibitor) treatment on c-Myc and SRSF10 expression in WT BBS cell lines. (D) Schematic representation of various metabolic inhibitors and their targets used in the study. Immunoblot analysis depicting the effect of (E) PFK-158 and (F) Shikonin treatment on c-Myc and SRSF10 expression in WT BBS cell lines. (G) Immunoblot analysis depicting the effect of rotenone on c-Myc and SRSF10 expression in BBS Mut cell lines. Data are represented as mean± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns= not significant.
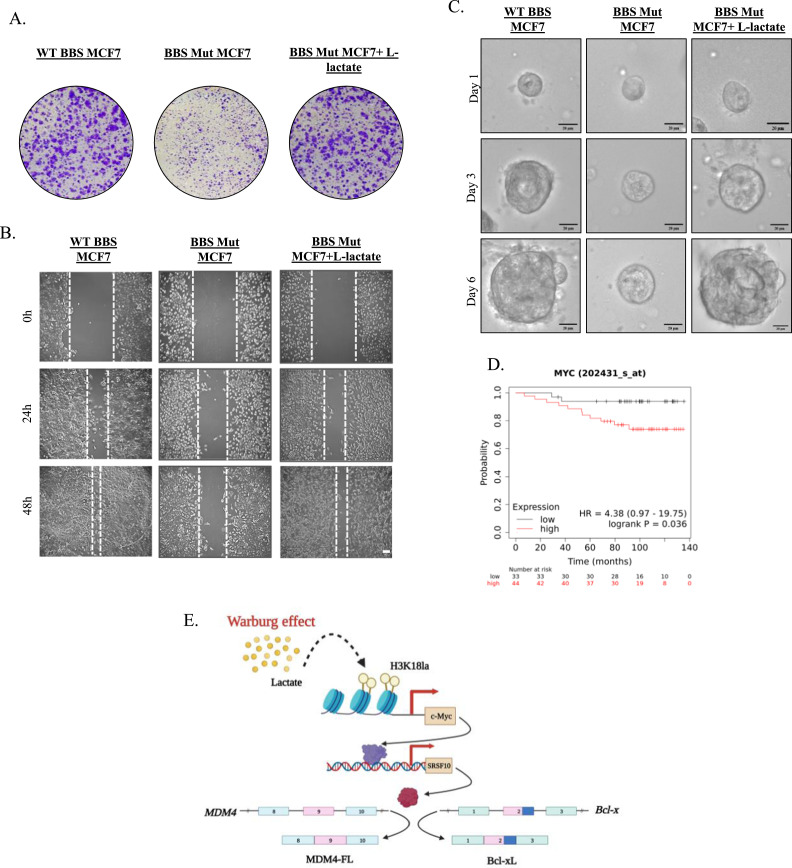
Furthermore, to evaluate if lactate-mediated upregulation of the c-Myc-SRSF10 axis promoted breast carcinogenesis, we performed clonogenic assay in WT BBS, BBS Mut, and BBS Mut cells cultured in media supplemented with l-lactate. Restricting lactate production by knocking out PKM2 led to a substantial reduction in cell proliferation, while this phenotype was reverted upon providing l-lactate externally (Fig. 6A, S5.A). Additionally, wound healing assay demonstrated that BBS Mut cells exhibited remarkably reduced proliferative and migratory capacity, which was rescued upon subjecting to l-lactate treatment (Fig. 6B, S5.B). To mimic the physiological conditions in an in-vitro model, we monitored the growth of the spheroids derived from the WT BBS, BBS Mut, and BBS Mut cells treated with l-lactate. As evident from the results, the BBS Mut cells formed remarkably smaller spheroids than their WT BBS counterparts. Moreover, supplementing l-lactate drastically enhanced the proliferation of the BBS Mut cells (Fig. 6C, S5.C). Furthermore, Kaplan-Meier survival analysis with The Cancer Genome Atlas (TCGA) dataset (GSE9195) showed that high c-Myc expression is associated with unfavorable patient outcomes (Fig. 6D), thus, underscoring the vitality of targeting c-Myc.
Fig. 6.
c-Myc-SRSF10 axis promotes proliferation of breast cancer cells. (A) Colony formation assay performed in WT BBS, BBS Mut, and BBS Mut MCF7 cells subjected to 5 mM l-lactate treatment for 10 days. (B) Cell migration analysed via wound healing assay in WT BBS, BBS Mut, and BBS Mut MCF7 cells subjected to 5 mM l-lactate treatment for 48 h. (C) Spheroid formation assay performed in WT BBS, BBS Mut, and BBS Mut MCF7 cells subjected to 5 mM l-lactate treatment for 6 days. (D) Kaplan–Meier curve showing significant association (p-value 0.036) of Recurrence Free survival with c-Myc expression in Breast Cancer TCGA dataset (GSE9195). (E) Graphical illustration of the H3K18la-driven c-Myc-SRSF10 axis.
Altogether, our data demonstrate that therapeutical interventions attenuating aerobic glycolysis can restrict the adverse repercussions of the c-Myc-SRSF10 axis.
Discussion
As metabolism is central to all life processes, any metabolic abnormality distorts the delicate balance between various cellular pathways [60]. As per the findings of Otto Warburg, the arterial glucose uptake is about 47–70% and 2–18% in cancer cells and normal cells, respectively; moreover, the cancer cells converted 66% of the uptaken glucose to lactate [61]. Considering the vast difference in the lactate-producing capabilities, it is imperative to understand the role of lactate beyond its metabolic function. The recently proposed ‘lactagenesis hypothesis’ suggests that lactate not only functions as fuel that supports the expansion of cancer biomass but also critically exhibits signaling properties, thus presenting itself as explanation and purpose of the Warburg effect in carcinogenesis [62]. Consequently, there has been a growing interest in abrogating the trafficking and exchange of lactate between the cancer cells and the other neoplastic and non-neoplastic cells within the TME to restrict cancer progression [16,17,[63], [64], [65], [66]].
As activation of c-Myc has been widely reported, the c-Myc-driven pathways are substantially elevated in aggressive and higher grades of breast cancer. Therefore, it is imperative to identify the poorly investigated epigenetic mechanisms enabling the maintenance of high c-Myc expression. Although San-Millan and co-workers previously showed that endogenous and exogenous lactate exposure can alter the expression of oncogenic transcription factors and cell division and proliferation-associated genes in breast cancer cells, the underlying mechanism was not investigated [67].
Recent years have witnessed an exponential rise in studies targeted at identifying the genes regulated by histone lactylation. Notably, emerging reports strongly suggest that histone lactylation, like histone acetylation, promotes gene activation [14,21,68,69]. Interestingly, the gene expression signatures associated with histone lactylation differ distinctly from those of histone acetylation [70]. Therefore, exploring the dynamic alterations in the global distribution pattern of histone lactylation in normal and neoplastic tissues can provide vital information regarding the cancer-associated histone lactylation signatures that may assist in uncovering novel therapeautic strategies to suppress the expression of oncogenes. Moreover, consistent with previous reports, our data shows that H3K18la is elevated in the tumor tissues, underscoring the vitality of decoding the mechanisms involved in regulating this histone modification [69]. Therefore, further understandings regarding the nature of histone lactylation-mediated alteration of transcriptome may provide fascinating insights into its potential applicability as a biomarker for predicting the stage and advancement of cancer.
Multiple studies in the recent few years have identified the potent contribution of genes regulated in histone lactylation-dependent manner in affecting cancer outcome [14,22,24,69]; however, to date, there is a dearth of evidence investigating how does intracellular lactate modulate the expression of oncogenes within the cancer cells. For the first time, our study unveils the role of H3K18la-mediated regulation of oncogenes that promote breast cancer progression. We have revealed that oncometabolite lactate-driven epigenetic regulation of c-Myc influences SRSF10 expression in breast cancer cells. Notably, our investigations demonstrate that amongst various SRSF members, only SRSF10 exhibited an expression pattern similar to c-Myc in response to modulations of intracellular lactate to dictate the alternative splicing outcomes of MDM4 and Bcl-x. Furthermore, our results show that therapeautic interventions suppressing the rate of aerobic glycolysis can successfully inhibit the c-Myc-SRSF10 axis to serve as a potential strategy for impeding the advancement of glycolytic cancers. Although we have demonstrated the critical role of H3K18la in c-Myc regulation, future studies to define the differential distribution of H3K18la marks are necessary to better comprehend the impact of this histone modification in regulating global gene expression and its possible applicability as a biomarker for cancer diagnosis. Another limitation of this study is that lactyl-CoA, the lactate donor utilized for modifying the histone lysine residue, was not quantitatively measured. As the lactate produced by the elevated glycolytic rate has been previously positively correlated with histone lactylation levels [21], a quantitative estimation of the lactyl-CoA will perhaps provide a better understanding of the interplay between altered metabolism and epigenetic rewiring mediated by histone lactylation.
Conclusion
Our findings uniquely demonstrate the metabolic link bridging epigenetics and oncogene-induced alternative splicing to emphasize the complexity of tumor metabolism, which warrants further studies. Moreover, our data underscore the wide-reaching effects of altered metabolism that hold paramount importance in cancer pathogenesis.
CRediT authorship contribution statement
Madhura R. Pandkar: Investigation, Conceptualization, Methodology, Writing – original draft. Sommya Sinha: Investigation, Methodology, Formal analysis. Atul Samaiya: Resources. Sanjeev Shukla: Conceptualization, Methodology, Writing – original draft, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no competing interest with the contents of this article.
Acknowledgments
Acknowledgments
The graphical representations were generated using BioRender. The authors thank all the members of the Epigenetics and RNA Processing Lab, IISER Bhopal, for their critical inputs.
Funding
This work is supported by the Department of Biotechnology (DBT)/ Wellcome Trust India Alliance Fellowship Grant (IA/I/16/2/502719) and Science and Engineering Research Board (SERB) Grant (CRG/2021/004949, STR/2020/000093, IPA/2021/000148) to S. Shukla. M.R.P and S. Sinha were supported by the University Grants Commission Fellowship.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101758.
Appendix. Supplementary materials
References
- 1.Levine A.J., Puzio-Kuter A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science (80-.). Dec. 2010;330(6009):1340–1344. doi: 10.1126/SCIENCE.1193494/ASSET/0EE526FC-644A-4D18-8835-A4452F5CD9F8/ASSETS/GRAPHIC/330_1340_F2.JPEG. [DOI] [PubMed] [Google Scholar]
- 2.Locasale J.W., Cantley L.C., Heiden M.G.V. Cancer's insatiable appetite. Nat. Biotechnol. Oct. 2009;27(10):916–917. doi: 10.1038/nbt1009-916. 2009 2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagarajan A., Malvi P., Wajapeyee N. Oncogene-Directed Alterations in Cancer Cell Metabolism. Trends Cancer. Jul. 2016;2(7):365–377. doi: 10.1016/J.TRECAN.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Egervari G., Wang Y., Berger S.L., Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. Jun. 2018;19(9):563–578. doi: 10.1038/s41580-018-0029-7. 2018 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tummala K.S., et al. Hepatocellular Carcinomas Originate Predominantly from Hepatocytes and Benign Lesions from Hepatic Progenitor Cells. Cell Rep. Apr. 2017;19(3):584–600. doi: 10.1016/J.CELREP.2017.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W., et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. U. S. A. Jun. 2012;109(23):8983–8988. doi: 10.1073/PNAS.1203244109/SUPPL_FILE/PNAS.201203244SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/SCIENCE.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 8.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. Nov. 2004;4(11):891–899. doi: 10.1038/nrc1478. 2004 411. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz-López K.G., Castro-Muñoz L.J., Reyes-Hernández D.O., García-Carrancá A., Manzo-Merino J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. Nov. 2019;9 doi: 10.3389/FONC.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung H., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 11.Cheung S.M., et al. Lactate concentration in breast cancer using advanced magnetic resonance spectroscopy. Br. J. Cancer. May 2020;123(2):261–267. doi: 10.1038/s41416-020-0886-7. 2020 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhagat T.D., et al. Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. Elife. Oct. 2019;8 doi: 10.7554/ELIFE.50663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao L., et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2) Autophagy. Apr. 2018;14(4):671–684. doi: 10.1080/15548627.2017.1381804/SUPPL_FILE/KAUP_A_1381804_SM7949.PPTX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong J., et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell. May 2022;82(9):1660–1677. doi: 10.1016/J.MOLCEL.2022.02.033. e10. [DOI] [PubMed] [Google Scholar]
- 15.Brooks G.A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018;27(4):757–785. doi: 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Fiaschi T., et al. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. Oct. 2012;72(19):5130–5140. doi: 10.1158/0008-5472.CAN-12-1949/650617/AM/RECIPROCAL-METABOLIC-REPROGRAMMING-THROUGH-LACTATE. [DOI] [PubMed] [Google Scholar]
- 17.Sanità P., et al. Tumor-stroma metabolic relationship based on lactate shuttle can sustain prostate cancer progression. BMC Cancer. Mar. 2014;14(1):1–14. doi: 10.1186/1471-2407-14-154/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks G.A. Anaerobic threshold: review of the concept and directions for future research. Med. Sci. Sports Exerc. Feb. 1985;17(1):22–34. https://europepmc.org/article/med/3884959 Accessed: May 04, 2023. [Online]. Available. [PubMed] [Google Scholar]
- 19.Brooks G.A. Lactate shuttles in Nature. Biochem. Soc. Trans. Apr. 2002;30(2):258–264. doi: 10.1042/BST0300258. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y., et al. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget. 2017;8(34):57813–57825. doi: 10.18632/oncotarget.18175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang J., et al. Lactate Modulates Cellular Metabolism Through Histone Lactylation-Mediated Gene Expression in Non-Small Cell Lung Cancer. Front. Oncol. Jun. 2021;11:1235. doi: 10.3389/FONC.2021.647559/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y., et al. Numb/Parkin-directed mitochondrial fitness governs cancer cell fate via metabolic regulation of histone lactylation. Cell Rep. Feb. 2023;42(2) doi: 10.1016/J.CELREP.2023.112033. [DOI] [PubMed] [Google Scholar]
- 24.Pan L., et al. Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol. Res. Jul. 2022;181 doi: 10.1016/J.PHRS.2022.106270. [DOI] [PubMed] [Google Scholar]
- 25.Asleh K., Riaz N., Nielsen T.O. Heterogeneity of triple negative breast cancer: current advances in subtyping and treatment implications. J. Exp. Clin. Cancer Res. Sep. 2022;41(1):1–26. doi: 10.1186/S13046-022-02476-1. 2022 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzo A., et al. Ladiratuzumab vedotin for metastatic triple negative cancer: preliminary results, key challenges, and clinical potential. Expert Opin. Investig. Drugs. 2022;31(6):495–498. doi: 10.1080/13543784.2022.2042252. [DOI] [PubMed] [Google Scholar]
- 27.Santoni M., et al. Complete remissions following immunotherapy or immuno-oncology combinations in cancer patients: the MOUSEION-03 meta-analysis. Cancer Immunol. Immunother. Jun. 2023;72(6):1365–1379. doi: 10.1007/S00262-022-03349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzo A., Ricci A.D. Biomarkers for breast cancer immunotherapy: PD-L1, TILs, and beyond. Expert Opin. Investig. Drugs. 2022;31(6):549–555. doi: 10.1080/13543784.2022.2008354. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo A., et al. Immune-based combinations for metastatic triple negative breast cancer in clinical trials: current knowledge and therapeutic prospects. Expert Opin. Investig. Drugs. 2022;31(6):557–565. doi: 10.1080/13543784.2022.2009456. [DOI] [PubMed] [Google Scholar]
- 30.Singh S., et al. Intragenic DNA methylation and BORIS-mediated cancer-specific splicing contribute to the Warburg effect. Proc. Natl. Acad. Sci. U. S. A. 2017;114(43):11440–11445. doi: 10.1073/pnas.1708447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandkar M.R., et al. PKM2 dictates the poised chromatin state of PFKFB3 promoter to enhance breast cancer progression. NAR Cancer. Jun. 2023;5(3) doi: 10.1093/NARCAN/ZCAD032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge S.X., Jung D., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. Apr. 2020;36(8):2628–2629. doi: 10.1093/BIOINFORMATICS/BTZ931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillaumond F., et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110(10):3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chrzan P., Skokowski J., Karmolinski A., Pawelczyk T. Amplification of c-myc gene and overexpression of c-Myc protein in breast cancer and adjacent non-neoplastic tissue. Clin. Biochem. Oct. 2001;34(7):557–562. doi: 10.1016/S0009-9120(01)00260-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X., et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc. Natl. Acad. Sci. U. S. A. Feb. 2012;109(8):2790–2795. doi: 10.1073/PNAS.1100764108/SUPPL_FILE/PNAS.201100764SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh E., et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. Mar. 2004;6(4):308–318. doi: 10.1038/ncb1110. 2004 64. [DOI] [PubMed] [Google Scholar]
- 37.Blancato J., Singh B., Liu A., Liao D.J., Dickson R.B. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br. J. Cancer. Mar. 2004;90(8):1612–1619. doi: 10.1038/sj.bjc.6601703. 2004 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deming S.L., Nass S.J., Dickson R.B., Trock B.J. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br. J. Cancer. Dec. 2000;83(12):1688–1695. doi: 10.1054/bjoc.2000.1522. 2000 8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eilers M., Eisenman R.N. Myc's broad reach. Genes Dev. Oct. 2008;22(20):2755–2766. doi: 10.1101/GAD.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang C.V. MYC on the Path to Cancer. Cell. Mar. 2012;149(1):22–35. doi: 10.1016/J.CELL.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z., et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat. Metab. Jan. 2023;5(1):61–79. doi: 10.1038/s42255-022-00710-w. 2023 51. [DOI] [PubMed] [Google Scholar]
- 42.Zhao W., et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. Sep. 2019;27(3):919–933. doi: 10.1038/s41418-019-0423-5. 2019 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caggiano C., Pieraccioli M., Panzeri V., Sette C., Bielli P. c-MYC empowers transcription and productive splicing of the oncogenic splicing factor Sam68 in cancer. Nucleic Acids Res. Jul. 2019;47(12):6160–6171. doi: 10.1093/NAR/GKZ344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David C.J., Chen M., Assanah M., Canoll P., Manley J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010 doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbanski L., et al. MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. Nov. 2022;41(8) doi: 10.1016/J.CELREP.2022.111704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fornes O., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. Jan. 2020;48(D1):D87–D92. doi: 10.1093/NAR/GKZ1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X., et al. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. Apr. 2014;42(6):4019–4030. doi: 10.1093/NAR/GKT1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sohail M., et al. A novel class of inhibitors that target SRSF10 and promote p53-mediated cytotoxicity on human colorectal cancer cells. NAR cancer. Jun. 2021;3(2):1–22. doi: 10.1093/NARCAN/ZCAB019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav S., Pant D., Samaiya A., Kalra N., Gupta S., Shukla S. ERK1/2-EGR1-SRSF10 axis mediated alternative splicing plays a critical role in head and neck cancer. Front. Cell Dev. Biol. Sep. 2021;9:2468. doi: 10.3389/FCELL.2021.713661/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., et al. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. Jun. 2023;34(12):2198–2207. doi: 10.1128/MCB.01674-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piva F., Giulietti M., Nocchi L., Principato G. SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics. May 2009;25(9):1211–1213. doi: 10.1093/BIOINFORMATICS/BTP124. [DOI] [PubMed] [Google Scholar]
- 52.Nicosia L., et al. Potent Pre-Clinical and Early Phase Clinical Activity of EP300/CBP Bromodomain Inhibitor CCS1477 in Multiple Myeloma. Blood. Nov. 2022;140(Supplement 1):852–853. doi: 10.1182/BLOOD-2022-157535. [DOI] [Google Scholar]
- 53.Nicosia L., et al. Therapeutic Targeting of EP300/CBP By Bromodomain Inhibition in Acute Myeloid Leukemia. Blood. Nov. 2022;140(Supplement 1):8774–8775. doi: 10.1182/BLOOD-2022-166260. [DOI] [Google Scholar]
- 54.Welti J., et al. Targeting the p300/cbp axis in lethal prostate cancer. Cancer Discov. May 2021;11(5):1118–1137. doi: 10.1158/2159-8290.CD-20-0751/333557/AM/TARGETING-P300-CBP-AXIS-IN-LETHAL-PROSTATE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helminen L., Huttunen J., Aaltonen N., Niskanen E.A., Palvimo J.J., Paakinaho V. Chromatin Accessibility and Pioneer Factor FOXA1 Shape Glucocorticoid Receptor Action in Prostate Cancer. bioRxiv. Mar. 2023:2023. doi: 10.1101/2023.03.03.530941. 03.03.530941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stine Z.E., Schug Z.T., Salvino J.M., Dang C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. Dec. 2021;21(2):141–162. doi: 10.1038/s41573-021-00339-6. 2021 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Y., et al. Inhibition of PFKFB3 induces cell death and synergistically enhances chemosensitivity in endometrial cancer. Oncogene. Jan. 2021;40(8):1409–1424. doi: 10.1038/s41388-020-01621-4. 2021 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J., Xie J., Jiang Z., Wang B., Wang Y., Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene. 2011;30(42):4297–4306. doi: 10.1038/onc.2011.137. [DOI] [PubMed] [Google Scholar]
- 59.Hou W.L., et al. Inhibition of mitochondrial complex I improves glucose metabolism independently of AMPK activation. J. Cell. Mol. Med. Feb. 2018;22(2):1316. doi: 10.1111/JCMM.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandkar M.R., Dhamdhere S.G., Shukla S. Oxygen gradient and tumor heterogeneity: the chronicle of a toxic relationship. Biochim. Biophys. Acta - Rev. Cancer. 2021;1876(1) doi: 10.1016/j.bbcan.2021.188553. [DOI] [PubMed] [Google Scholar]
- 61.Warburg O., Wind F., Negelein E. THE METABOLISM OF TUMORS IN THE BODY. J. Gen. Physiol. Mar. 1927;8(6):519. doi: 10.1085/JGP.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.San-Millán I., Brooks G.A. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. Feb. 2017;38(2):119–133. doi: 10.1093/CARCIN/BGW127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonveaux P., et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 2008;118(12):3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morais-Santos F., et al. Targeting lactate transport suppresses in vivo breast tumour growth. Oncotarget. Aug. 2015;6(22):19177. doi: 10.18632/ONCOTARGET.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Draoui N., et al. Synthesis and pharmacological evaluation of carboxycoumarins as a new antitumor treatment targeting lactate transport in cancer cells. Bioorg. Med. Chem. Nov. 2013;21(22):7107–7117. doi: 10.1016/J.BMC.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Doherty J.R., Cleveland J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. Sep. 2013;123(9):3685. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.San-Millán I., Julian C.G., Matarazzo C., Martinez J., Brooks G.A. Is Lactate an Oncometabolite? Evidence Supporting a Role for Lactate in the Regulation of Transcriptional Activity of Cancer-Related Genes in MCF7 Breast Cancer Cells. Front. Oncol. Jan. 2020;9:1536. doi: 10.3389/FONC.2019.01536/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rho H., Terry A.R., Chronis C., Hay N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metab. Jul. 2023 doi: 10.1016/J.CMET.2023.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu J., et al. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. Dec. 2021;22(1):1–21. doi: 10.1186/S13059-021-02308-Z/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Izzo L.T., Wellen K.E. Histone lactylation links metabolism and gene regulation. Nat. Oct. 2019;574(7779):492–493. doi: 10.1038/d41586-019-03122-1. 2021 5747779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.