Abstract
Purpose:
Cancer vaccines represent a novel treatment modality with a complementary mode of action addressing a crucial bottleneck for checkpoint inhibitor (CPI) efficacy. CPIs are expected to release brakes in T-cell responses elicited by vaccination, leading to more robust immune responses. Increased antitumor T-cell responses may confer increased antitumor activity in patients with less immunogenic tumors, a subgroup expected to achieve reduced benefit from CPIs alone. In this trial, a telomerase-based vaccine was combined with pembrolizumab to assess the safety and clinical activity in patients with melanoma.
Patients and Methods:
Thirty treatment-naïve patients with advanced melanoma were enrolled. Patients received intradermal injections of UV1 with adjuvant GM-CSF at two dose levels, and pembrolizumab according to the label. Blood samples were assessed for vaccine-induced T-cell responses, and tumor tissues were collected for translational analyses. The primary endpoint was safety, with secondary objectives including progression-free survival (PFS), overall survival (OS), and objective response rate (ORR).
Results:
The combination was considered safe and well-tolerated. Grade 3 adverse events were observed in 20% of patients, with no grade 4 or 5 adverse events reported. Vaccination-related adverse events were mostly mild injection site reactions. The median PFS was 18.9 months, and the 1- and 2-year OS rates were 86.7% and 73.3%, respectively. The ORR was 56.7%, with 33.3% achieving complete responses. Vaccine-induced immune responses were observed in evaluable patients, and inflammatory changes were detected in posttreatment biopsies.
Conclusions:
Encouraging safety and preliminary efficacy were observed. Randomized phase II trials are currently ongoing.
Translational Relevance.
While checkpoint inhibitors (e.g., pembrolizumab) confer substantial outcome improvements for patients with melanoma, further advancements are needed, especially for patients with poorly immunogenic tumors. Herein, we report a phase I clinical trial investigating a therapeutic cancer vaccine, UV1, in combination with pembrolizumab in patients with advanced melanoma. Therapeutic vaccination can potentially strengthen the antitumor T-cell response, aiming to extend immunotherapy efficacy to patients deprived of robust spontaneously formed T-cell responses. Therefore, we aimed to assess whether the combination therapy led to clinical responses in patients without inflamed tumor phenotypes typically associated with poorer response to checkpoint inhibition.
Introduction
Prospects for patients with advanced melanoma have improved considerably with the introduction of checkpoint inhibitors (CPI). Through their inhibition of immune checkpoints that suppress T-cell expansion and activation, CPIs, such as anti–PD-1 and anti–CTLA-4, have demonstrated the robust antitumor potential of patients’ immune systems (1). Despite historically meager outcomes for patients with advanced melanoma, the overall survival (OS) is significantly increased in patients treated with anti–PD-1 alone or in combination with anti–CTLA-4, with a median OS of 36.9 and 72.1 months, respectively (2). The objective response rates (ORR) were also superior with combined CPI therapy at 58% versus 45% with anti–PD-1 monotherapy. Although the efficacy of combination therapy is encouraging, it also represents a considerable increase in toxicity, with 59% versus 21% of patients experiencing treatment-related grade ≥ 3 adverse events for combination and monotherapy, respectively (3). Novel CPI combinations may offer increased efficacy while maintaining a moderate toxicity profile, such as anti–LAG-3 and anti–PD-1 (4). Nevertheless, most patients still do not achieve durable clinical benefits from CPIs, combinations, or monotherapies, and agents with a complementary mode of action that increases the clinical efficacy while maintaining tolerable toxicity profiles are urgently needed.
The mode of action of CPIs relies on the reactivation and expansion of spontaneous, tumor-induced T-cell responses (5). Hence, the overall immunogenicity of tumors can predict clinical benefit, as measured by the tumor mutational burden (TMB; ref. 6), neoantigen clonality (7), PD-L1 expression (8), and IFNγ gene signature (9). Patients with poorly immunogenic tumors are expected to have fewer spontaneously primed immune responses, thereby reducing the benefit of checkpoint inhibition alone. Therefore, emerging combination strategies are largely aimed at augmenting de novo T cell priming to extend immunotherapy efficacy to more patients.
Therapeutic cancer vaccines (TCV) represent a treatment modality that is well-positioned to stimulate antitumor T-cell responses to compensate for low tumor immunogenicity. By mounting de novo T-cell responses targeting antigens derived from either tumor-specific mutations or aberrantly expressed proteins, TCVs can potentially contribute to solving a central challenge of improving CPI efficacy (10). Telomerase is an enzyme expressed by cancer cells that enables replicative immortality, a hallmark of cancer (11). Telomerase expression is present in 85% to 90% of all tumor types and can be considered an almost pan-cancer antigen for immunotherapy (12–14). Recent evidence suggests that telomerase-specific immune responses are clinically meaningful because their spontaneous occurrence is associated with prolonged progression-free survival (PFS), OS, and response to CPI in melanoma (15). While these spontaneous T-cell responses are only observed in a subset of patients, vaccination against telomerase reverse transcriptase (hTERT) has led to immune responses in 91% of patients in a phase I clinical trial of melanoma (16). The vaccine, named UV1, comprises three synthetic long peptides covering a 54 amino acid sequence derived from the active site of hTERT. Vaccination with UV1 has been shown to induce high-frequency and durable T-cell responses across three completed phase I trials (17). Building on the scientific rationale for combining a TCV with CPIs, anti-hTERT immune responses occurred more frequently and rapidly when vaccination was combined with the anti–CTLA-4 CPI ipilimumab (17). Across the completed phase I trials covering 52 patients, 6 patients experienced hypersensitivity reactions. These reactions had occurred after a minimum of nine vaccinations with UV1 and the adjuvant GM-CSF (75 μg).
In this phase I clinical trial, UV1–103, patients with advanced melanoma received a combination of the UV1 vaccine with adjuvant GM-CSF and the anti–PD-1 monoclonal antibody pembrolizumab. The current study enrolled patients in two cohorts based on the adjuvant dose level (37.5 and 75 μg) to assess a potential contribution from the adjuvant to the overall safety profile. Both cohorts received up to eight UV1 vaccinations. A vaccine peptide dose of 300 μg was selected on the basis of robust immune responses and a favorable safety profile from the completed trials. The primary objective was to assess the safety of the treatment combination, with secondary objectives including parameters of clinical efficacy and immunologic responses.
Patients and Methods
Patients
Patients aged ≥18 years with a histologically confirmed diagnosis of unresectable stage III (B/C) or IV melanoma, according to the American Joint Committee on Cancer (AJCC) version 8, were eligible for enrollment. Eligible patients had not received systemic therapy, except BRAF or MEK inhibitors. Evaluable tumors according to the RECIST version 1.1, and available tumor tissue for sampling were both required. Other eligibility criteria included an Eastern Cooperative Oncology Group (ECOG) performance status score of 0 or 1 and adequate renal, hepatic, and hematologic functions. The key exclusion criteria were uveal or ocular melanoma, immunodeficiency, a history of autoimmune diseases, and active infection requiring systemic therapy. Patients with brain metastases that required treatment other than surgery or radiation therapy were excluded from the study. All patients provided written informed consent prior to enrollment.
Trial design
This trial was an open-label, multicenter phase I study (NCT03538314). The primary endpoint was the safety and tolerability of UV1 vaccine in combination with pembrolizumab. Secondary endpoints included assessment of immune responses towards UV1 peptides, PFS, ORR, best overall response rate (BOR), duration of response (DOR), and OS. Biological samples were collected for exploratory analysis of biomarkers related to the efficacy of UV1 and pembrolizumab. The study was approved by competent regulatory authorities and institutional review boards, and conducted in accordance with the Declaration of Helsinki.
Treatment
The UV1 vaccine (Ultimovacs ASA, Oslo, Norway) consists of three peptides, one 30-mer (p719-20) and two 15-mers (p725 and p728) in equimolar amounts, and is produced as a sterile aqueous solution, lyophilized, and stored at 2°C to 8°C. The peptides were reconstituted in water before intradermal administration. The vaccine adjuvant GM-CSF (sargramostim), in the form of preservative-free powder (lyophilized Leukine, Partner Therapeutics, Lexington, MA), was administered intradermally at the same injection site (lower abdomen) 10 to 15 minutes prior to UV1 administration.
All patients received intradermal injections of adjuvant GM-CSF and 300 μg UV1, hereafter referred to as the UV1 vaccination. Two different doses of GM-CSF were investigated, 37.5 μg (Cohort 1, n = 20) and 75 μg (Cohort 2, n = 10). The first three UV1 vaccinations were administered on week 1 (days 1, 3, and 5). From week 2, UV1 vaccination was followed by an intravenous infusion of 200 mg pembrolizumab every 3 weeks for up to eight UV1 vaccinations (Supplementary Fig. S1). The injected volumes were 100 μL and 150 μL for UV1 and GM-CSF, respectively. The infusion was administered at least 1 hour after UV1 vaccination. Pembrolizumab treatment was continued after completion of the UV1 vaccination according to the FDA label and as determined by the investigator.
Assessments
Clinical assessments
Treatment-emergent adverse events (TEAE) were defined as adverse events that occurred after the first UV1 vaccination and up to 3 months after the last vaccination. Adverse events were graded according to the NCI Common Terminology Criteria for Adverse Events version 4.03. Safety was evaluated by physical examination and the assessment of blood samples and vital signs. Objective tumor responses were assessed according to immune-related response criteria (iRECIST). CT was conducted at baseline, at the end of UV1 vaccination (week 14), and 3 and 9 months after the last UV1 vaccination (Supplementary Fig. S1). Thereafter, the patients were followed-up for survival and subsequent new anticancer treatment (after pembrolizumab) for up to 5 years after the first UV1 vaccination, every 3 months for the second year, and twice a year thereafter. During the second year, CT performed as part of the patient's standard of care were requested.
Survival was defined as the time from the initiation of the UV1 vaccination to death from any cause. iPFS was defined as the time from the initiation of UV1 vaccination to objective tumor progression according to iRECIST or death, whichever occurred first. ORR was defined as the proportion of patients who had a complete response (iCR) or partial response (iPR) as the best response, whereas DOR was defined as the time from the first documented response (iCR or iPR) to radiologic progression according to iRECIST or death. The data cutoff date was October 5, 2022.
Immunologic assessment
Proliferation assay:
The UV1-specific immune response was measured in peripheral blood mononuclear cells (PBMC). PBMCs were prepared from whole blood samples (BD Vacutainer CPT Cell Preparation tubes) and collected at baseline, 4 times during the UV1 vaccination period, and 4 times during the follow-up period (30 days and 3, 6, and 9 months after the last UV1 vaccination). At the start of the study (July 2018), the centrifuged CPT vacutainers were shipped to a U.S. central laboratory for isolation and cryopreservation. The procedure was modified in April 2020, after which the samples were isolated and cryopreserved at each site before being shipped to the central laboratory for storage. All samples were sent to Norway for immune response analysis. The UV1-specific T-cell response was measured by a proliferation assay (3H-Thymidine incorporation), as previously described (17). Briefly, thawed PBMCs were stimulated with UV1 peptides (peptide 725; hTERT 691–705 (RTFVLRVRAQDPPPE), peptide 719–20; hTERT 660–689 (ALFSVLNYERARRPGLLGASVLGLDDIHRA), peptide 728; hTERT 651–665 (AERLTSRVKALFSVL; Corden Pharma, Switzerland) at a concentration of 10 μmol/L for each peptide. On day 3, IL2 (20 U/mL) and IL7 (5 ng/mL) were added. After 10 to 14 days, the T cells were restimulated with peptide-loaded autologous irradiated antigen-presenting cells, and proliferation was determined by 3H-Thymidine incorporation assay, with all conditions tested in triplicates. The stimulation index (SI), which is the ratio of the mean counts in the wells with T cells stimulated with or without UV1 peptides, was calculated. If the SI at any time point was ≥3 for any of the three UV1 peptides or a mixture, the patient was considered an immune responder. Patients with a UV1-specific T-cell response at baseline (SI ≥ 3) required doubling of the SI in at least one postvaccination sample or an increase in the number of peptides recognized to be defined as vaccine responders. Staphylococcus aureus enterotoxin C3 (SEC3) was used as a positive control to determine immunocompetence. IR-evaluable samples had SEC3 SI ≥ 10.
Tumor tissue specimens
Tumor biopsies, one formalin-fixed, paraffin-embedded (FFPE) sample, and two snap-frozen samples were collected at baseline and the end of UV1 treatment (Supplementary Fig. S1). DNA and RNA were extracted from the snap-frozen biopsies (Novogene, Cambridge, United Kingdom). Whole-exome sequencing (WES) was performed on tumor- and whole blood–derived DNA using 400 ng as starting material. Briefly, the genomic DNA was fragmented and prepared as libraries containing duel-indexed sequencing barcodes. The pre-capture libraries were enriched using SureSelect Human All Exon V6 capture baits, according to the manufacturer's protocol (Agilent Technologies). Post-capture libraries were sequenced using a NovaSeq 6000 instrument (Illumina). The RNA samples were processed using the TruSeq Stranded Total RNA Library Prep kit (Illumina) using 500 ng as starting material. RNA sequencing was performed using the NovaSeq 6000 instrument (Illumina).
WES data were processed using a custom-developed bioinformatics pipeline (18) that aligns sequencing data to the human reference genome (GRCh38) to conduct variant calling and annotation (Ensembl 95). TMB was computed according to the best practices outlined in (19). Immunogenic neoantigens were predicted using an artificial intelligence platform, NEC Immune Profiler (NEC OncoImmunity, Oslo, Norway). Using both WES and RNA sequencing data, the software computed the likelihood of each somatic mutation giving rise to immunogenic neoantigens by predicting the key determinants for antigen presentation and binding to HLA alleles in individual patients. The computed antigen presentation score (AP) threshold was set at 0.6. Differentially expressed gene (DEG) analysis was performed on kallisto abundance files imported by tximport and analyzed using DEseq2 (v1.38.1) and R (v4.2.2). The pairing of the samples (obtained from the same patient) was included in the design formula. To improve the fold change estimation, shrinkage was performed using the Apeglm method. DEGs were determined at the following thresholds: log2FC > 0.5; Padj. < 0.05. Gene set enrichment analysis (GSEA) was performed using Webgestalt [WebGestalt: WEB-based GEne SeT AnaLysis Toolkit (RRID:SCR_006786)] and the Gene Ontology biological processes database.
FFPE biopsies were assessed using the PD-L1 IHC 22C3 pharmDx antibody for Autostainer Link 48 (Agilent, catalog no. SK006, RRID:AB_2889976). Biopsies were considered evaluable if at least 100 viable tumor cells were present. PD-L1 was considered for partial or complete cell membrane staining of tumor cells, which was perceived as distinct from cytoplasmic staining. Samples were PD-L1–positive with a tumor proportion score of ≥1%. A blinded experienced pathologist performed the sample quality and positivity evaluation.
FFPE biopsies were used for multiplex immunofluorescence staining as previously described (20). Four-micrometer sections were stained using a custom 8-color assay (Akoya Biosciences, Marlborough, M) and a fully automated Leica Bond RXm system (Leica Biosystems, Buffalo Grove, IL). The slides were deparaffinized, rehydrated, and rinsed with distilled H2O. The custom multiplex immunofluorescence panel consisted of antibodies against CD4 (Abcam, catalog no. ab133616, RRID:AB_2750883), CD8 (Thermo Fisher Scientific, catalog no. MA5–13473, RRID:AB_11000353), FoxP3 (Cell Signaling Technology, catalog no. 98377, RRID:AB_2747370), TERT (Abcam, catalog no. ab32020, RRID:AB_778296), GRZB (Agilent, catalog no. M7235, RRID:AB_2114697), Ki-67 (Agilent, catalog no. M7240, RRID:AB_2142367), and the Sox10/S100 cocktail (Akoya, EP268–1/ 4C4.9). Staining was performed using an amplification HRP-polymer system and Opal fluorophore dyes (Akoya, OP-000003). Cell nuclei were visualized by staining with 4′,6-diamidino-2-phenylindole (Spectral DAPI, Akoya). The slides were mounted with Prolong Diamond Antifade Mountant (Thermo Fisher, Waltham, MA) and imaged at ×20 magnification (0.5 μm/pixel) using the Vectra Polaris Automated Quantitative Pathology Imaging System (Akoya Biosciences, Marlborough, MA). A pathologist manually reviewed and curated each image to exclude artifacts and stain defects. The detection level was set to 0.1 counts/mm2.
Statistics
No formal statistical hypothesis testing was planned, and all statistical analyses were performed retrospectively. The sample size (n) represents the number of patients or samples analyzed. Survival analyses were performed using the Kaplan–Meier method. All statistical analyses were performed using GraphPad Prism, version 9.2.0. (RRID:SCR_002798). Statistical significance was set at P < 0.05.
Data availability
Relevant data are provided in the article or Supplementary Materials. Additional data are available on request. The raw sequencing data are available via the European Genome-Phenome Archive (ega-archive.org) with the accession number EGAS00001007210.
Results
Patients and treatment
Between July 2018 and August 2020, 30 patients with advanced cutaneous melanoma were recruited from three U.S. sites. The median age was 70.5 years (range, 30–87), and 70% of the patients were male. The majority (63%) had an ECOG performance status score of 0. Eleven (37%) patients had stage III B or C disease and the remaining 19 (63%) had stage IV disease. At baseline, nine patients (31%) had elevated lactate dehydrogenase (LDH) levels. Ten patients (37%) had a BRAF V600 mutation, and 8 patients (36%) had PD-L1–positive (≥1%) tumors. TMB was low [0–5 mutations per megabase (Mb)] in 8 patients (47%; Table 1). All patients were treatment-naïve for unresectable disease.
Table 1.
Patient baseline characteristics.
| Characteristic | Cohort 1 37.5 μg GM-CSF N = 20 | Cohort 2 75 μg GM-CSF N = 10 | Combined N = 30 |
|---|---|---|---|
| Median age (range) - years | 69.5 (30–81) | 73.5 (50–87) | 70.5 (30–87) |
| Male sex - no. (%) | 13 (65) | 8 (80) | 21 (70) |
| ECOG performance status - no. (%) | |||
| 0 | 14 (70) | 5 (50) | 19 (63) |
| 1 | 6 (30) | 5 (50) | 11 (37) |
| Elevated LDH – no. (%)a | 7 (37) | 2 (20) | 9 (31) |
| Stage – no. (%) | |||
| IIIB | 1 (5) | 1 (10) | 2 (7) |
| IIIC | 7 (35) | 2 (20) | 9 (30) |
| IV | 12 (60) | 7 (70) | 19 (63) |
| M1a | 3 (15) | 2 (20) | 5 (17) |
| M1b | 3 (15) | 2 (20) | 5 (17) |
| M1c | 5 (25) | 3 (30) | 8 (27) |
| M1d | 1 (5) | 0 | 1 (3) |
| Liver metastasis - no. (%) | 3 (15) | 1 (10) | 4 (13) |
| BRAF V600E status – no. (%)b | |||
| Mutated | 6 (35) | 4 (40) | 10 (37) |
| PD-L1 status – no. (%)c | |||
| Positive (≥ 1%) | 7 (53) | 1 (11) | 8 (36) |
| TMB - no. (%)d | |||
| High (≥ 20 mutations/Mb) | 1 (9) | 2 (33) | 3 (18) |
| Intermediate (6–19 mut/Mb) | 4 (36) | 2 (33) | 6 (35) |
| Low (1–5 mutations/Mb) | 6 (55) | 2 (33) | 8 (47) |
aOne patient did not have baseline LDH registered; the denominator for Cohort 1 is 19, combined 29.
bThree patients had missing BRAF status; the denominator for Cohort 1 is 17, combined 27.
cEight Patients had either no available or non-evaluable samples for PD-L1 testing; the denominator for Cohort 1 is 13, combined 22.
dThirteen patients had either no available or non-evaluable samples for TMB testing; the denominator for Cohort 1 is 11, combined 17.
The patients were enrolled in two cohorts based on the adjuvant dose received. The first 20 patients (Cohort 1) received 37.5 μg GM-CSF, while the remaining 10 (Cohort 2) received 75 μg GM-CSF. Baseline characteristics were balanced between the two cohorts (Table 1). Most patients completed all eight UV1 vaccinations according to the protocol (80%). The reason for treatment discontinuation was disease progression in 5 patients and an adverse event (chorioretinitis) in 1 patient.
Safety
TEAEs were observed in almost all patients (93%) and 70% experienced treatment-related TEAE. The most common treatment-related TEAEs were fatigue (33%), injection site reaction (20%), hypothyroidism (20%), colitis (17%), and diarrhea (17%; Table 2). Grade 3 treatment-related TEAEs were observed in 20% of the patients, and no grade 4 or 5 adverse events were reported. Only one grade 3 adverse event was considered to be possibly related to UV1 or GM-CSF (arthritis). The safety profiles were similar between the two cohorts.
Table 2.
Overview of treatment-related TEAEs.
| Cohort 1 37.5 μg GM-CSF N = 20 | Cohort 2 75 μg GM-CSF N = 10 | Combined N = 30 | ||||
|---|---|---|---|---|---|---|
| Number of patients (percent) | ||||||
| Adverse event | Any grade | Grade 3 | Any grade | Grade 3 | Any grade | Grade 3 |
| Related to treatmenta | ||||||
| Any | 14 (70.0) | 5 (25.0) | 7 (70.0) | 1 (10.0) | 21 (70.0) | 6 (20.0) |
| Occurring in more than 1 patient or grade ≥ 3 | ||||||
| Fatigue | 7 (35.0) | 0 | 3 (30.0) | 0 | 10 (33.3) | 0 |
| Injection site reaction | 3 (15.0) | 0 | 3 (30.0) | 0 | 6 (20.0) | 0 |
| Hypothyroidism | 5 (25.0) | 0 | 1 (10.0) | 0 | 6 (20.0) | 0 |
| Colitis | 4 (20.0) | 1 (5.0) | 1 (10.0) | 1 (10.0) | 5 (16.7) | 2 (6.7) |
| Diarrhea | 2 (10.0) | 0 | 3 (30.0) | 0 | 5 (16.7) | 0 |
| Pruritus | 2 (10.0) | 0 | 2 (20.0) | 0 | 4 (13.3) | 0 |
| Hyperthyroidism | 3 (15.0) | 1 (5.0) | 1 (10.0) | 0 | 4 (13.3) | 1 (3.3) |
| Rash | 2 (10.0) | 0 | 1 (10.0) | 0 | 3 (10.0) | 0 |
| Arthritis | 2 (10.0) | 2 (10.0) | 0 | 0 | 2 (6.7) | 2 (6.7) |
| Dyspnea | 2 (10.0) | 0 | 0 | 0 | 2 (6.7) | 0 |
| Chorioretinitis | 1 (5.0) | 1 (5.0) | 0 | 0 | 1 (3.3) | 1 (3.3) |
| Diabetes mellitus | 1 (5.0) | 1 (5.0) | 0 | 0 | 1 (3.3) | 1 (3.3) |
Note: TEAEs were defined as all adverse events that occurred after the first dose of study medication up to 3 months ± 2 weeks after the last vaccine dose. Patients with events in more than one category were counted once within each category.
aThe investigators determined the relatedness of adverse events to study drugs.
Efficacy
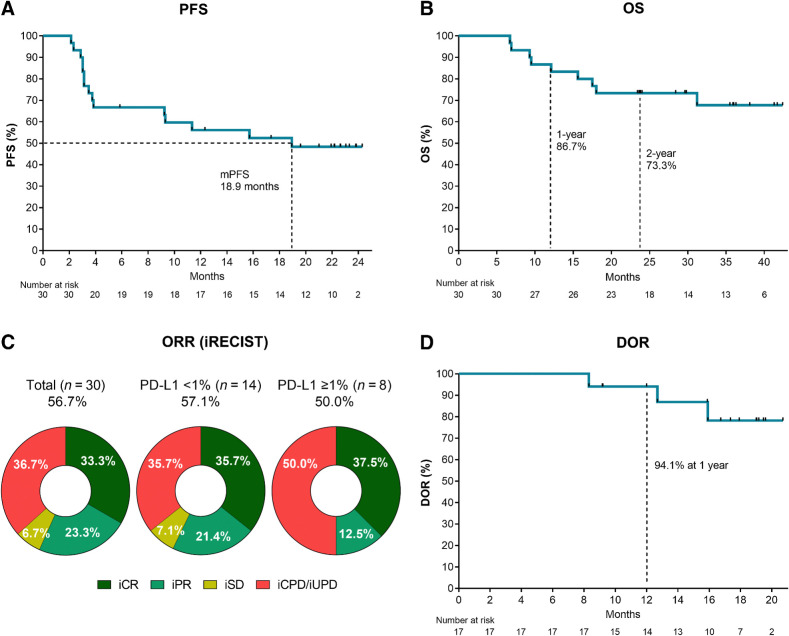
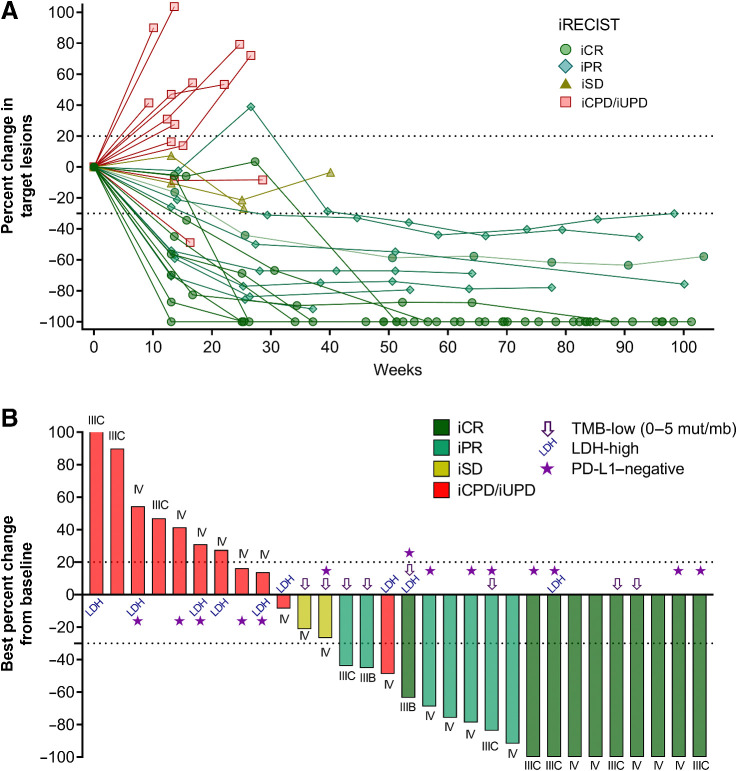
With a median follow-up time of 29.0 months (range, 6.7–42.4), the median iPFS was 18.9 months (95% CI, 3.5–no estimate), and the median OS was not reached (Figs. 1A and B). The 1- and 2-year OS rates were 86.7% and 73.3%, respectively. The ORR per iRECIST was 56.7% (95% CI, 37.4–74.5%). Complete responses were observed in 10 patients (33.3%) and partial responses in 7 patients (23.3%). Two patients achieved stable disease as the best overall response (iBOR; 6.7%) and the remaining patients experienced progressive disease (36.7%; Fig. 1C). iPFS, OS, and ORR were similar between the two cohorts (Supplementary Fig. S2 and Supplementary Table S1). Most objective responses were durable and persisted for up to 2 years (Figs. 1D and 2A). The median DOR was not reached, and only 3 of the 17 patients with a clinical response had progressed at the time of reporting (Fig. 1D). Two patients with iPR progressed after 8.3 and 12.7 months in response, respectively, and 1 patient achieving an iCR progressed after 15.9 months in response. Objective responses were observed across AJCC stages, in patients with LDH above the upper limit of normal, PD-L1–negative, and TMB-low tumors (Figs. 1C and 2B). The ORR in PD-L1–negative patients (n = 14) was 57.1%, with a CR rate of 35.7% (Fig. 1C).
Figure 1.
Efficacy read-out. Kaplan–Meier plots of (A) progression-free survival (iPFS) and (B) OS in all patients (N = 30). C, Donut plot showing the ORRs in the total population (N = 30) and subgroups according to tumor biopsy PD-L1 positivity. D, Kaplan–Meier plot showing the DOR (n = 17).
Figure 2.
Spider and waterfall plots. A, Spider plot showing the percent change in tumor size from baseline. One line represents 1 patient, color- and symbol-coded by best overall response according to iRECIST. B, Waterfall plot depicting the maximum percent change in tumor size from baseline. One bar represents one patient, color-coded according to the best overall responses according to iRECIST. Symbols indicate patients with baseline LDH-high, TMB-low, and PD-L1–negative tumor biopsies.
Objective responses across different biomarker populations
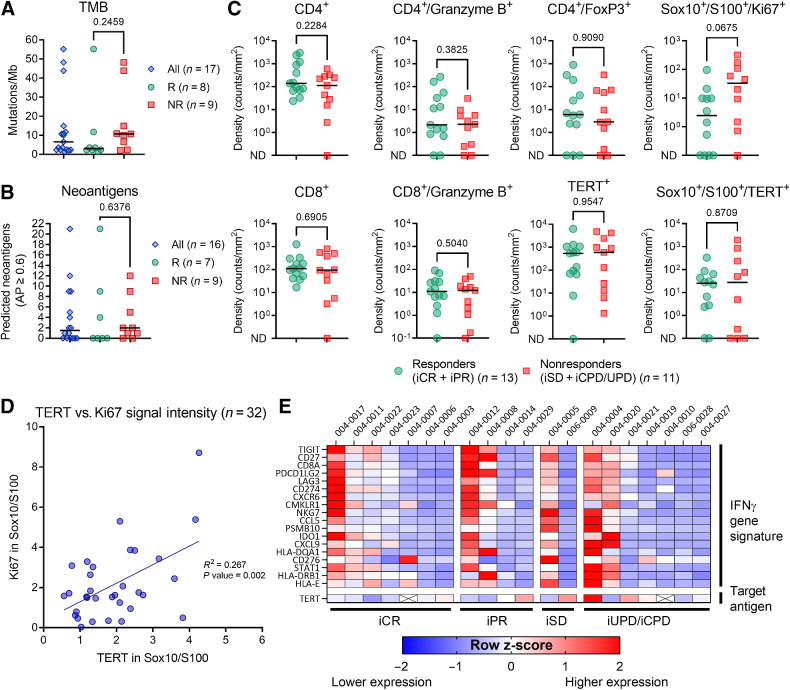
Baseline tumor biopsies were assessed for the levels of various biomarkers associated with CPI efficacy. The baseline median TMB was 6.6 mutations/Mb (range, 1.3–55.2), and the median number of predicted immunogenic neoantigens was 1.5 (range, 0–21; Fig. 3A and B). Clinical responders did not exhibit higher baseline TMB or neoantigen scores than nonresponders (Mann–Whitney test, median TMB 3.0 vs. 10.7; P value = 0.246; median number of neoantigens 0.0 vs. 2.0; P value = 0.638; Fig. 3A and B). To further investigate the baseline immunologic status of tumor biopsies, custom multiplex immunofluorescence staining was performed to evaluate the presence of CD4, CD8, Granzyme B, FoxP3, hTERT, Ki67, and the melanoma marker Sox10/S100. The results showed an equal distribution of most marker-defined cell subset densities in patients who achieved either a response or no response to the combination therapy (Fig. 3C). hTERT protein expression was documented in all except one baseline biopsy, and hTERT and melanoma marker (Sox10/S100) double-positive cells were detected in all except five biopsies (77.3%). Ki67 expression in melanoma cells was higher in nonresponders than in responders, although the difference was not statistically significant (Mann–Whitney, median density 2.5 vs. 33.0; P value = 0.068). A significant correlation was observed between the signal intensities of hTERT and Ki67 in melanoma cells (linear regression, R2 = 0.267; P value = 0.002; Fig. 3D).
Figure 3.
Baseline tumor characteristics in clinical responders and nonresponders. A, TMB and (B) predicted neoantigen in all patients with available baseline biopsy. C, Baseline multiplex immunofluorescence staining of T cells (CD4+ and CD8+), regulatory CD4 T cells (CD4+/FoxP3+), granzyme B+, hTERT+, Ki67+, and melanoma cells (Sox10+/S100+). Two samples with no Sox10/S100 stained cells were excluded from the analysis of co-staining melanoma cells and hTERT and Ki67. D, Among all available biopsies (both baseline and week 14), there was a significant correlation between signal intensity of Ki67 and TERT within melanoma cells (simple linear regression, R2 = 0.267; P = value 0.002). E, Baseline levels of the 18-gene IFNγ gene signature expression according to iRECIST category. Heat map shows the calculated z-score, representing the relative expression levels across patient samples. Unless otherwise described, all reported P values represent results from the unpaired nonparametric test (Mann–Whitney). ND, not detected. R, clinical responder. NR, clinical nonresponders.
The expression levels of the IFNγ transcriptional expression signature (9), were analyzed. Patients who achieved iCR or iPR were not enriched for the IFNγ gene signature at baseline (Fig. 3E). RNA expression of TERT was detected in all but two biopsies (patients 004–0007 and 004–0010), and the baseline hTERT expression level was evenly distributed between the different response categories (Fig. 3E). A relatively high density of hTERT and melanoma marker double-positive cells (235 counts/mm2) was observed in one of the two biopsies without detectable TERT RNA, whereas the other lacked Sox10/S100 stained cells.
Increase in anti-hTERT T-cell responses, tumor inflammatory markers, and TILs
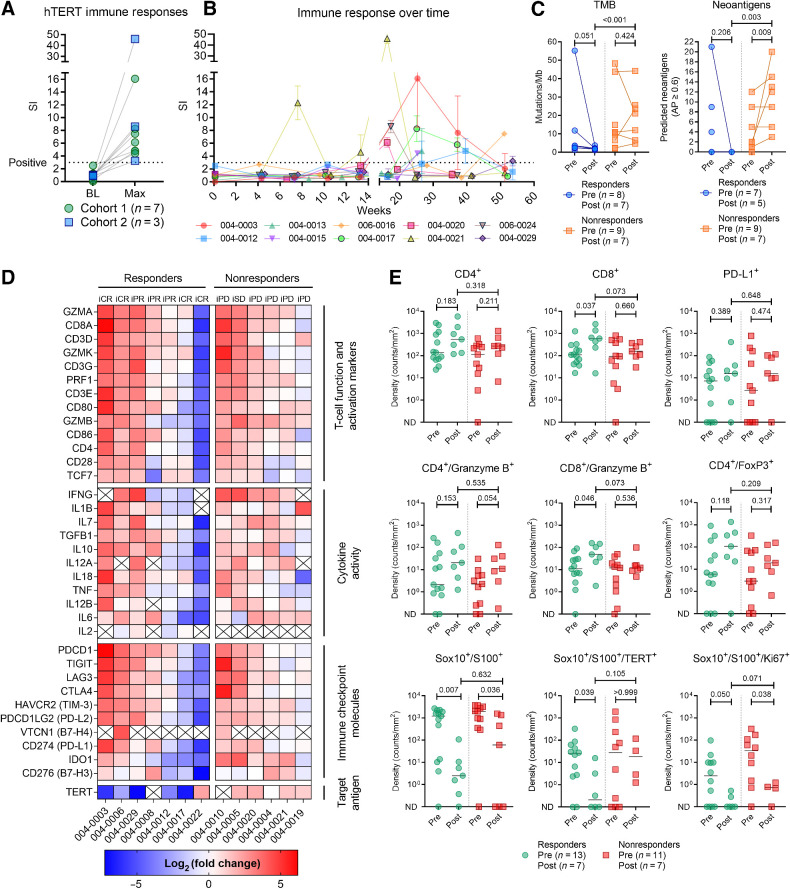
Owing to the initially poor quality of PBMC samples, logistical changes were implemented during the recruitment and follow-up periods to improve the yield and viability. These changes allowed the detection of anti-hTERT immune responses in 10 patients, albeit at later time points than anticipated on the basis of previous experience (Fig. 4A and B) (17). The median highest SI in immune responders was 6.8 (range, 3.3–46.0), and the median time to first immune response was 25.3 weeks (range, 7.6–53.6).
Figure 4.
Anti-hTERT immune responses and impact of therapy on tumor gene expression, infiltration of lymphocytes, and tumor markers. A, Vaccine-induced T-cell responses against hTERT were documented in 10 patients. Figure shows the difference between baseline sample and highest SI achieved. B, Immune response development over time. C, TMB and neoantigen load in pre- and posttreatment biopsies. D, Change in tumor expression of genes related to T-cell function and activation, cytokine activity, immune checkpoint molecules, and hTERT. Heat map reflects the relative change in expression (transcripts per million) from baseline to week 14. The log2(fold change) was uncalculable for cells marked with “X” (zero transcripts per million in either baseline or week 14 biopsy). E, Comparison of immunofluorescence staining of biopsies at baseline and week 14. In the analysis of Sox10/S100 co-stained with Ki67 and TERT, six biopsies without any Sox10/S100 stained cells were excluded. All reported P values represent results from the unpaired nonparametric test (Mann–Whitney). ND, not detected.
Biopsies were collected at week 14 (End of Treatment visit), and we investigated the relative changes in TMB and neoantigen load from baseline. TMB was reduced and there were no predicted neoantigens in the five posttreatment biopsies from clinical responders. Conversely, for nonresponders, there was an increase in the number of predicted neoantigens (median 2.0 vs. 12.0; P value = 0.009; Fig. 4C).
The expression of genes related to T-cell function and activation, cytokine activity, and immune checkpoint molecules was assessed using RNA sequencing. A relative increase in expression across these categories was observed in both responders and nonresponders, indicating an impact of the therapy in nearly all patients, despite the apparent lack of objective responses in the progressors (Fig. 4D). The relative increase in the expression of T cell related genes was further supported by a GSEA of DEGs posttreatment, although it was less evident in nonresponders (Supplementary Fig. S3A and S3B). Interestingly, the RNA expression of TERT decreased in clinical responders and had increased in nonresponders.
On the basis of multiplex immunofluorescence staining, a significant increase in the density of CD8+ T cells and CD8+/granzyme B+ co-stained cells were observed in clinical responders posttreatment (median density 110 vs. 600; P value = 0.037; and median density 11 vs. 49; P value = 0.046, respectively; Fig. 4E). While Sox10+/S100+ cells were reduced in both responders and nonresponders, hTERT+ and Sox+/S100+ double-positive cells were significantly reduced in responders only (median density 25 vs. 0.2; P value = 0.039; Fig. 4E).
Discussion
The efficacy of CPIs relies on spontaneous antitumor immunity acquired through the natural recognition of mutated or conserved tumor-specific antigens by the patient's T cells. While many patients display sufficient spontaneous antitumor T-cell responses for CPI clinical benefit, the high variability of such responses remains a bottleneck for broader efficacy. Hence, novel combination strategies aim to augment antitumor T-cell responses through approaches such as induced immunogenic cell death (e.g., chemotherapy and radiotherapy), soluble cytokines (e.g., IL2 and IFNα), or vaccination strategies (21). While there are many potential ways to influence the immune system for therapeutic purposes, vaccines are distinctively positioned to instruct the immune system to mount de novo responses specific to clinically relevant tumor antigens. In this context, hTERT represents a promising therapeutic target because of its expression in 85% to 90% of all cancers. By establishing an immune response towards a common denominator of cancer, the immune response may remain relevant over time and across heterogeneous tumors. Previous studies on UV1 have shown that the induction of a CD4+ Th1-polarized immune response correlates with prolonged OS (17). Isolated vaccine-specific T-cell clones have shown in vitro recognition and killing of melanoma cells in an HLA-dependent manner (22). Furthermore, spontaneous anti-hTERT Th1 responses were associated with an increased response to checkpoint inhibition in a prospective melanoma study (15), substantiating the potential clinical impact of these immune responses.
Here, we report a phase I clinical trial, UV1–103, investigating the combination of UV1 vaccination and pembrolizumab in 30 patients with advanced melanoma. The combination was considered safe and well tolerated, with adverse event types and frequencies mostly as expected for pembrolizumab monotherapy. While adverse events deemed related to UV1 vaccination were generally mild injection site reactions, numerically higher rates of colitis were observed compared to the KEYNOTE-006 trial investigating pembrolizumab monotherapy (23), and gastrointestinal adverse events should be a focus for future studies. The highest adjuvant dose (75 μg) was well tolerated, and there were no observations of hypersensitivity reactions at either dose level, indicating that the reduction in number of vaccinations from previous trials was sufficient to mitigate this risk.
The median iPFS was 18.9 months (95% CI, 3.5–no estimate), and 73% of the patients were still alive after 2 years. Objective responses, according to iRECIST, were observed in 57% of the patients (95% CI, 37.4–74.5%), with 33% experiencing complete responses. For reference, after five years of follow-up, the KEYNOTE-006 trial reported a median PFS of 8.4 months (95% CI, 6.6–11.3), 2-year OS rate of 55.2%, and ORR of 42% (95% CI, 38.1–46.5), with complete responses observed in 14% of patients (24). It should be noted that the KEYNOTE-006 trial is by no means a perfect historical reference study, especially because it recruited almost only patients with stage IV disease, whereas the current trial included a substantial fraction of patients with stage III disease (37%).
Interpretation of efficacy from a phase I trial should be done with caution, and the small sample size and non-randomized design were the main limitations of this study. Indeed, several agents that have shown initial promise in early-phase melanoma trials have later failed in the randomized setting (25, 26). Considering the encouraging read-out, the study population was assessed to determine whether it was biased toward those expected to benefit from pembrolizumab. Baseline biopsies were evaluated for several biomarkers associated with clinical outcomes of checkpoint inhibition. Remarkably, patients achieving clinical responses to the combination did not exhibit higher PD-L1 expression, higher TMB, a large volume of predicted immunogenic neoantigens, enrichment of the IFNγ gene signature, or higher tumor-infiltrating lymphocytes, all of which are measures of immunologically active tumors and have shown some degree of predictive value for CPI efficacy in melanoma in previous studies (5, 9, 27–29). Indeed, for patients with PD-L1–negative tumors, the KEYNOTE-006 trial showed an ORR of only 24% (95% CI, 16.4%–33.7%) with 5.8% achieving CR (30). In the UV1–103 study, the combination treatment yielded an ORR of 57% in this subgroup of patients, with 35.7% reaching iCR. The median TMB in the present study population was 6.6 mutations/Mb (range, 1.3–55.2). While higher TMB is associated with improved clinical response to pembrolizumab in melanoma (28), clinical responders in the UV1–103 trial exhibited lower TMB than nonresponders (3.0 vs. 10.7 mutations/Mb; P value = 0.246). Furthermore, four patients who achieved an objective response had no predicted high-quality neoantigens at the baseline. These data provide further support for vaccination against non-mutated tumor-associated antigens, mounting antitumor immune responses also in patients with a low TMB, where other vaccination approaches, such as personalized neoantigen vaccines, are less relevant. The lack of response in some high-TMB patients underscores the multifactorial nature of immunotherapy response, where an immunosuppressive tumor microenvironment or a disruption in HLA-antigen presentation may have contributed to progression (31, 32). Although melanoma is generally considered a more inflamed tumor type, the observations that seemingly non-inflamed tumors were responsive to the combination therapy indicate mobilization of novel antitumor immune responses that can lead to tangible tumor cell killing.
Poor PBMC sample quality and a limited number of evaluable patients restricted correlative analyses of immune responses, other biomarkers, and clinical outcomes. While results from a phase I melanoma trial with UV1 in combination with ipilimumab showed a median time to the first immune response of 4 weeks (17), immune responses in the current study were first documented in samples from later time points. This late immune response detection is likely due to the laboratory and shipment adjustments implemented during the study, which subsequently increased the quality of the samples. The immune responses may thus have occurred before and contributed to the earlier occurring clinical responses. All except one baseline biopsy exhibited hTERT expression based on RNA sequencing or immunofluorescence staining. Five biopsies did not display double-positive cells for hTERT and the melanoma marker Sox10/S100. This was unexpected, as previous studies have shown ubiquitous and homogeneous hTERT expression patterns in melanoma, with 95% of cases exhibiting more than 50% hTERT-positive melanoma cells (n = 85; ref. 33). As hTERT expression is linked to several tumorigenic features, including epithelial to mesenchymal transition, cancer cell stemness, and metastasis (34–40), and is associated with poor outcomes (41), these hTERT-negative melanoma cells may represent bystander cancer clones that contribute less to tumor growth. The significant correlation between Ki67 and hTERT in melanoma cells observed in our study supports this hypothesis, as Ki67 is a proliferation marker also associated with poorer melanoma prognosis (42).
The results of the UV1–103 trial offer optimism for therapeutic cancer vaccination to enhance the clinical efficacy of checkpoint inhibition without aggravating toxicity. These data support further investigation of the UV1 vaccine in combination with checkpoint inhibition. Randomized phase II clinical trials with UV1 are currently underway, implementing the eight-dose vaccination regime and the 75 μg adjuvant dose level. The trials evaluate UV1 in combination with various CPIs across multiple indications; non-small cell lung cancer (NCT05344209), mesothelioma (NCT04300244; ref. 43), ovarian cancer (NCT04742075), head and neck cancer (NCT05075122), and melanoma (NCT04382664; ref. 44).
Supplementary Material
Figure S1
Figure S2
Figure S3
Table S1
Table S2
Acknowledgments
We thank the patients and their families for participating in the study, Else Marit Inderberg and her lab at Oslo University Hospital for performing the immunomonitoring, and Wenche Rasch (Ultimovacs ASA) for study management. The trial was sponsored by Ultimovacs ASA, Oslo, Norway. EBE received grants from The Research Council of Norway (grant no. 298864).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
E.B. Ellingsen reports personal fees from Ultimovacs ASA and grants from Research Council of Norway during the conduct of the study. A. Mezheyeuski reports other support from Ultimovacs ASA during the conduct of the study. A. Gromadka reports employment with NEC OncoImmunity, an AI-based vaccine research company. T. Clancy reports employment with NEC OncoImmunity, an AI-based vaccine research company. Y. Zakharia reports grants and other support from Ultimovacs during the conduct of the study; Advisory Board member relationships with Bristol-Myers Squibb, Ultimovacs, Amgen, Novartis, Eisai, Exelixis, Castle Biosciences, AstraZeneca, Array, Bayer, Pfizer, Clovis, EMD Serono, and Myovant; grant/research support from Institution clinical trial support from NewLink Genetics, Pfizer, Exelixis, and Eisai; DSMC relationships with Janssen Research and Development; and Consultant honorarium from Pfizer and Novartis. No disclosures were reported by the other authors.
Authors' Contributions
E.B. Ellingsen: Conceptualization, formal analysis, methodology, writing–original draft. S. O'Day: Investigation. A. Mezheyeuski: Data curation, formal analysis, methodology, writing–review and editing. A. Gromadka: Software, formal analysis, writing–review and editing. T. Clancy: Formal analysis, supervision, methodology, writing–review and editing. T.S. Kristedja: Investigation. M. Milhem: Investigation. Y. Zakharia: Conceptualization, supervision, investigation, writing–review and editing.
References
- 1. Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. [DOI] [PubMed] [Google Scholar]
- 2. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, et al. Long-term outcomes with nivolumab plus ipilimumab or nivolumab alone versus ipilimumab in patients with advanced melanoma. J Clin Oncol 2022;40:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob J-J, Cowey CL, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2017;377:1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N Engl J Med 2022;386:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim JY, Kronbichler A, Eisenhut M, Hong SH, van der Vliet HJ, Kang J, et al. Tumor mutational burden and efficacy of immune checkpoint inhibitors: a systematic review and meta-analysis. Cancers (Basel) 2019;11:1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T-cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin Cancer Res 2014;20:5064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFNγ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin MJ, Svensson-Arvelund J, Lubitz GS, Marabelle A, Melero I, Brown BD, et al. Cancer vaccines: the next immunotherapy frontier. Nat Cancer 2022;3:911–26. [DOI] [PubMed] [Google Scholar]
- 11. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 12. Ellingsen EB, Mangsbo SM, Hovig E, Gaudernack G. Telomerase as a target for therapeutic cancer vaccines and considerations for optimizing their clinical potential. Front Immunol 2021;12:682492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shay JW, Bacchetti S. A survey of telomerase in human cancer. Eur J Cancer 1997;33:787–91. [DOI] [PubMed] [Google Scholar]
- 14. Kim N, Piatyszek M, Prowse K, Harley C, West M, Ho P, et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994;266:2011–5. [DOI] [PubMed] [Google Scholar]
- 15. Nardin C, Laheurte C, Puzenat E, Boullerot L, Ramseyer M, Marguier A, et al. Naturally occurring telomerase-specific CD4 T-cell immunity in melanoma. J Invest Dermatol 2022;142:435–44. [DOI] [PubMed] [Google Scholar]
- 16. Aamdal E, Inderberg EM, Ellingsen EB, Rasch W, Brunsvig PF, Aamdal S, et al. Combining a universal telomerase based cancer vaccine with ipilimumab in patients with metastatic melanoma: five-year follow up of a phase I/IIa trial. Front Immunol 2021;12:663865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellingsen EB, Aamdal E, Guren T, Lilleby W, Brunsvig PF, Mangsbo SM, et al. Durable and dynamic hTERT immune responses following vaccination with the long-peptide cancer vaccine UV1: long-term follow-up of three phase I clinical trials. J Immunother Cancer 2022;10:e004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anzar I, Sverchkova A, Stratford R, Clancy T. NeoMutate: an ensemble machine learning framework for the prediction of somatic mutations in cancer. BMC Med Genomics 2019;12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merino DM, McShane LM, Fabrizio D, Funari V, Chen S-J, White JR, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the friends of cancer research TMB harmonization project. J Immunother Cancer 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellingsen EB, Bounova G, Kerzeli I, Anzar I, Simnica D, Aamdal E, et al. Characterization of the T-cell receptor repertoire and melanoma tumor microenvironment upon combined treatment with ipilimumab and hTERT vaccination. J Transl Med 2022;20:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu S, Zhang T, Zheng L, Liu H, Song W, Liu D, et al. Combination strategies to maximize the benefits of cancer immunotherapy. J Hematol Oncol 2021;14:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inderberg-Suso EM, Trachsel S, Lislerud K, Rasmussen AM, Gaudernack G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. Oncoimmunology 2012;1:670–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus Ipilimumab in advanced melanoma. N Engl J Med 2015;372:2521–32. [DOI] [PubMed] [Google Scholar]
- 24. Robert C, Ribas A, Schachter J, Arance A, Grob J-J, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post hoc 5-year results from an open-label, multicenter, randomized, controlled, phase III study. Lancet Oncol 2019;20:1239–51. [DOI] [PubMed] [Google Scholar]
- 25. Long GV, Dummer R, Hamid O, Gajewski TF, Caglevic C, Dalle S, et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase III, randomized, double-blind study. Lancet Oncol 2019;20:1083–97. [DOI] [PubMed] [Google Scholar]
- 26. Diab A, Gogas HJ, Sandhu SK, Long GV, Ascierto PA, Larkin J, et al. 785O - PIVOT IO 001: first disclosure of efficacy and safety of bempegaldesleukin (BEMPEG) plus nivolumab (NIVO) vs NIVO monotherapy in advanced melanoma (MEL). Ann Oncol 2022; 33:S356–409. [Google Scholar]
- 27. Daud AI, Wolchok JD, Robert C, Hwu W-J, Weber JS, Ribas A, et al. Programmed death-ligand 1 expression and response to the anti–programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol 2016;34:4102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cristescu R, Aurora-Garg D, Albright A, Xu L, Liu XQ, Loboda A, et al. Tumor mutational burden predicts the efficacy of pembrolizumab monotherapy: a pan-tumor retrospective analysis of participants with advanced solid tumors. J Immunother Cancer 2022;10:e003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abbott CW, Boyle SM, Pyke RM, McDaniel LD, Levy E, Navarro FCP, et al. Prediction of immunotherapy response in melanoma through combined modeling of neoantigen burden and immune-related resistance mechanisms. Clin Cancer Res 2021;27:4265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carlino MS, Long GV, Schadendorf D, Robert C, Ribas A, Richtig E, et al. Outcomes by line of therapy and programmed death ligand 1 expression in patients with advanced melanoma treated with pembrolizumab or ipilimumab in KEYNOTE-006: a randomized clinical trial. Eur J Cancer 2018;101:236–43. [DOI] [PubMed] [Google Scholar]
- 31. Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 2018;6:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hazini A, Fisher K, Seymour L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J Immunother Cancer 2021;9:e002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bustos B DU, Torralba AS, Poveda PM, Simó GP, Farinos JS, Ros ML, et al. Telomerase expression in a series of melanocytic neoplasms. Actas Dermosifiliogr 2019;110:212–9. [DOI] [PubMed] [Google Scholar]
- 34. Im E, Yoon JB, Lee H-W, Chung KC. Human Telomerase Reverse Transcriptase (hTERT) positively regulates 26S proteasome activity. J Cell Physiol 2017;232:2083–93. [DOI] [PubMed] [Google Scholar]
- 35. Hu C, Ni Z, Li B-s, Yong X, Yang X, Zhang J-w, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut 2017;66:31–42. [DOI] [PubMed] [Google Scholar]
- 36. Saretzki G. Extra-telomeric functions of human telomerase: cancer, mitochondria and oxidative stress. Curr Pharm Des 2014;20:6386–403. [DOI] [PubMed] [Google Scholar]
- 37. Masutomi K, Possemato R, Wong JMY, Currier JL, Tothova Z, Manola JB, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci USA 2005;102:8222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z, Li Q, Li K, Chen L, Li W, Hou M, et al. Telomerase reverse transcriptase promotes epithelial–mesenchymal transition and stem cell–like traits in cancer cells. Oncogene 2013;32:4203–13. [DOI] [PubMed] [Google Scholar]
- 39. Zhang K, Guo Y, Wang X, Zhao H, Ji Z, Cheng C, et al. WNT/β-catenin directs self-renewal symmetric cell division of hTERThigh prostate cancer stem cells. Cancer Res 2017;77:2534–47. [DOI] [PubMed] [Google Scholar]
- 40. Hannen R, Bartsch JW. Essential roles of telomerase reverse transcriptase hTERT in cancer stemness and metastasis. FEBS Lett 2018;592:2023–31. [DOI] [PubMed] [Google Scholar]
- 41. Hugdahl E, Kalvenes MB, Mannelqvist M, Ladstein RG, Akslen LA. Prognostic impact and concordance of TERT promoter mutation and protein expression in matched primary and metastatic cutaneous melanoma. Br J Cancer 2018;118:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Q, Peng Z, Shen L, Shen L. Prognostic and clinicopathological value of Ki-67 in melanoma: a meta-analysis. Front Oncol 2021;11:737760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haakensen VD, Nowak AK, Ellingsen EB, Farooqi SJ, Bjaanæs MM, Horndalsveen H, et al. NIPU: a randomized, open-label, phase II study evaluating nivolumab and ipilimumab combined with UV1 vaccination as second line treatment in patients with malignant mesothelioma. J Transl Med 2021;19:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Day S, Bechter O, Lorigan P, Nyakas M. Abstract CT231: nivolumab and ipilimumab ± UV1 vaccine as first-line treatment in patients with malignant melanoma (INITIUM-trial). Cancer Res 2021;81:CT231–CT. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Table S1
Table S2
Data Availability Statement
Relevant data are provided in the article or Supplementary Materials. Additional data are available on request. The raw sequencing data are available via the European Genome-Phenome Archive (ega-archive.org) with the accession number EGAS00001007210.