Abstract
Purpose:
Germline pathogenic variants in CHEK2 confer moderately elevated breast cancer risk (odds ratio, OR ∼ 2.5), qualifying carriers for enhanced breast cancer screening. Besides pathogenic variants, dozens of missense CHEK2 variants of uncertain significance (VUS) have been identified, hampering the clinical utility of germline genetic testing (GGT).
Experimental Design:
We collected 460 CHEK2 missense VUS identified by the ENIGMA consortium in 15 countries. Their functional characterization was performed using CHEK2-complementation assays quantifying KAP1 phosphorylation and CHK2 autophosphorylation in human RPE1–CHEK2-knockout cells. Concordant results in both functional assays were used to categorize CHEK2 VUS from 12 ENIGMA case–control datasets, including 73,048 female patients with breast cancer and 88,658 ethnicity-matched controls.
Results:
A total of 430/460 VUS were successfully analyzed, of which 340 (79.1%) were concordant in both functional assays and categorized as functionally impaired (N = 102), functionally intermediate (N = 12), or functionally wild-type (WT)–like (N = 226). We then examined their association with breast cancer risk in the case–control analysis. The OR and 95% CI (confidence intervals) for carriers of functionally impaired, intermediate, and WT-like variants were 2.83 (95% CI, 2.35–3.41), 1.57 (95% CI, 1.41–1.75), and 1.19 (95% CI, 1.08–1.31), respectively. The meta-analysis of population-specific datasets showed similar results.
Conclusions:
We determined the functional consequences for the majority of CHEK2 missense VUS found in patients with breast cancer (3,660/4,436; 82.5%). Carriers of functionally impaired missense variants accounted for 0.5% of patients with breast cancer and were associated with a moderate risk similar to that of truncating CHEK2 variants. In contrast, 2.2% of all patients with breast cancer carried functionally wild-type/intermediate missense variants with no clinically relevant breast cancer risk in heterozygous carriers.
Translational Relevance.
Protein-truncating germline variants in the CHEK2 gene confer a moderate breast cancer risk; however, the association of missense variants with breast cancer risk remains unknown. Thus, the majority of missense variants are clinically inconclusive variants of uncertain significance (VUS). We analyzed 430 CHEK2 missense VUS identified during a routine germline genetic testing of patients with cancer in 15 countries. Using two parallel functional assays, the functional analysis concordantly categorized 340/430 variants identified in 3,660 (82.5%) out of 4,436 carriers found in 73,048 female patients with breast cancer and 88,658 controls from 10 countries. Subsequent case–control analysis showed that only carriers of functionally impaired missense variants (0.5% of all patients with breast cancer) were associated with a clinically significant, moderate breast cancer risk (OR, 2.83; 95% confidence interval, 2.35–3.41), comparable with the risk for the protein-truncating variants. Our results will allow a conclusive interpretation for the majority of CHEK2 germline missense variants identified in patients with breast cancer from different ethnicities worldwide.
Introduction
The checkpoint kinase 2 gene (CHEK2) codes for nuclear serine/threonine protein kinase CHK2 phosphorylating numerous intracellular proteins in response to DNA damage and several other stress signals (1). Since its discovery in 1999, recurrent germline CHEK2 variants have been associated with breast cancer predisposition (2–4). In addition, subsequent studies indicated increased risk of other malignancies, including prostate, kidney, thyroid, and colon cancers (5, 6).
The prevalence of germline CHEK2 variants differs among patients with breast cancer worldwide (6). Two recent large studies demonstrated that CHEK2 is the second most frequently altered breast cancer predisposition gene among patients of European descent, surpassed by BRCA2 and followed by BRCA1 in frequency of germline pathogenic variants (7, 8). However, to high breast cancer risk associated with alterations in BRCA1 or BRCA2, both studies confirmed association of germline pathogenic CHEK2 variants with moderate breast cancer risk with odds ratio (OR) 2.5 and a cumulative lifetime breast cancer risk of 25%–30%.
Because the increased breast cancer risk, CHEK2 analysis has become a routine component of germline gene panels for identification of individuals at risk (9). Although c.1100delC or other CHEK2 variants leading to protein truncations or aberrant pre-mRNA splicing are considered clearly pathogenic, the vast majority of CHEK2 missense variants are clinically inconclusive (variants of uncertain significance, VUS). The presence of VUS impairs the clinical utility of diagnostic panel analysis and negatively influences patients’ perception of germline genetic testing (GGT; refs. 10–12). Moreover, the risk associated with germline missense variants in breast/ovarian cancer predisposition genes may differ from that in truncating variants (13–15).
To increase our understanding of the clinical relevance of CHEK2 VUS, we collected missense variants reported by members of the international ENIGMA (Evidence-based Network for the Interpretation of Germline Mutant Alleles) consortium (16) and analyzed the variants by functional assays in a human cell line model (17). We then collected 12 case–control datasets from the ENIGMA consortium members, including 161,706 patients with breast cancer and population-matched controls from 10 countries, and examined the breast cancer risk for carriers of functionally stratified CHEK2 missense variants.
Materials and Methods
Collection of CHEK2 missense variants
The ENIGMA consortium members were requested to report all CHEK2 missense variants (annotated using the NCBI reference sequence NM_007194.4) identified in patients with any cancer type that were analyzed by a routine GGT approved by local ethical committees. The variants were obtained between June 5, 2019 and August 24, 2022 and are listed in Supplementary Table S1.
Datasets of patients with breast cancer and controls
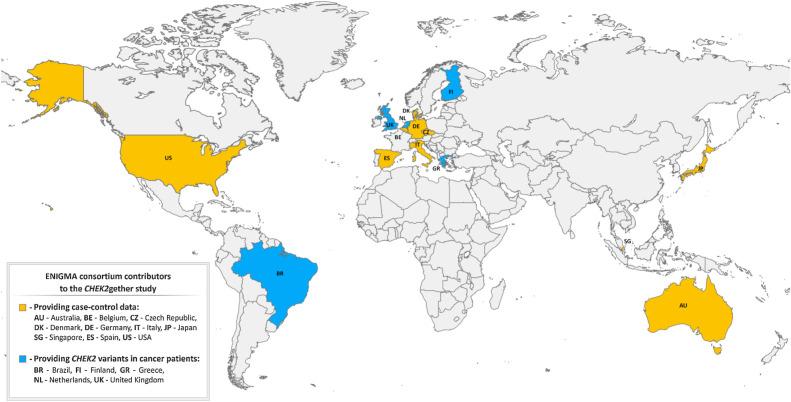
To estimate the breast cancer risk associated with CHEK2 missense VUS, we collected 12 case–control datasets from 10 countries (Fig. 1) that included 73,048 breast cancer cases and 88,658 population-matched controls. The description of case–control datasets, including enrollment, ethical statements, ethnicity, and CHEK2 analysis is provided in the Supplementary Table S2. Germline genetic analyses were conducted in accordance with the Declaration of Helsinki. Informed written consent for the participation in the studies, approved by institutional review boards, was obtained from each subject.
Figure 1.
Geographical origin of analyzed CHEK2 missense variants.
Cell lines
Human non-transformed hTERT-RPE1 cells (ATCC CRL-4000, RRID: CVCL_4388) and their derivatives RPE1-CHEK2-KO cells with knockout (KO) of the endogenous CHEK2 gene (17) were grown in DMEM media supplemented with 6% FBS (Gibco), penicillin (100 U/ml) and streptomycin (100 mg/mL). Cell lines were authenticated by using STR profiling and were regularly checked for the absence of mycoplasma infection using MycoAlert Plus reagent (Lonza; Cat# 75860–358). Where indicated, cells were exposed to ionizing radiation (IR; dose 5 Gy) generated by X-RAD 225XL (Precision).
Plasmids
Coding sequence of the wild-type human CHEK2 tagged at the N-terminus by FLAG sequence was cloned in frame into XhoI/EcoRI sites of pEGFP-C1 plasmid (Clontech; RRID: Addgene_165830). Subsequently, a panel of individual CHEK2 mutants was generated by gene synthesis and was verified by a Sanger sequencing (Synbio Technologies).
Antibodies
The following antibodies were used: Phospho-S473-KAP1 (BioLegend Cat# 654102, RRID: AB_2561782), phospho-S516-CHK2 (Cell Signaling Technology Cat# 2669, RRID: AB_330146), CHK2 (Abcam Cat# ab109413, RRID: AB_10863751), KAP1 (GeneTex Cat# GTX102227, RRID: AB_2037323), PCNA (Santa Cruz Biotechnology Cat# sc-56, RRID:AB_628110), goat anti-mouse (Thermo Fisher Scientific Cat# A-11004, RRID: AB_2534072), and goat anti-rabbit Alexa568 (Thermo Fisher Scientific Cat# A-11011, RRID: AB_143157).
Immunofluorescence microscopy
RPE1–CHEK2-KO cells seeded on a glass bottom 96-well plate (Eppendorf) were transfected with an empty EGFP plasmid, wild-type pEGFP–CHEK2 or mutant pEGFP–CHEK2 using X-tremeGENE HP DNA Transfection Reagent (Roche). Cells were fixed with 4% paraformaldehyde 24 hours after transfection, permeabilized by 0.2% Triton X-100 in PBS for 7 minutes and blocked with 3% BSA in PBS at room temperature. Fixed cells were incubated with the KAP1-pS473 antibody or CHK2-pS516 antibody for 2 hours at room temperature, three times washed with PBS and incubated with the secondary antibodies and DAPI (Supplementary Methods). After washing with PBS, samples were mounted by Vectashield H-1000 and imaged using a ScanR microscope (Olympus) equipped with an ORCA-285 camera and 40×/1.3 NA objective. Mean intensities of the nuclear KAP1-pS473 (pKAP1) or CHK2-pS516 (pCHK2) signals were analyzed in GFP-positive cells using ScanR analysis software (Olympus).
Functional categorization of CHEK2 missense variants
For both assays, only pEGFP–CHEK2-transfected cells were selected. To avoid potential bias due to expression of the studied CHK2 isoforms at supraphysiological levels, only those cells expressing low levels of GFP, ranging within the linear pKAP1 to GFP signal for wt-plasmid transfected cells in each plate, were analyzed for the pKAP1 assay. In this analysis window, the enzymatic activity of each CHK2 variant was determined as an average value of the pKAP1/GFP ratio from >300 individual cells normalized to the wild-type CHK2. This step allowed to merge the outputs of individual plates from the screen.
For pCHK2 autophosphorylation assay, cells in the upper quartile of GFP signal for each variant were excluded, and only cells with GFP intensities lower than 200 A.U. were analyzed. Enzymatic activity of CHK2 variants was counted from >150 individual cells normalized to the wild-type CHK2 for each variant and determined as a “b” coefficient in a linear regression equation (y = a + bx; x corresponds to GFP, y corresponds to pCHK2 signals). All analyses were performed in RStudio version 4.2.1 (RRID: SCR_000432).
All relative enzymatic activity values higher than the lowest wild-type replica were considered functionally wild-type (WT)–like. All variants with enzymatic activity values lower than the highest catalytically dead control (the in-frame exon 7 deletion resulting in in frame deletion of 18 amino acids in kinase domain of CHK2, and EGFP, respectively) were considered functionally impaired. Variants scoring between the lowest wild-type and the highest impaired were categorized as “intermediate” (Supplementary Methods).
Intracellular localization of CHK2 isoforms
The localization of all expressed CHK2 isoforms was assessed as the nuclear-to-cytoplasm ratio, calculated as the mean nuclear GFP signal intensity divided by the mean GFP signal intensity in a circular region outside the nucleus. Average of the nuclear-to-cytoplasm ratios from >300 individual cells were calculated for each analyzed variant (Supplementary Table S1). A set of 12 protein-truncating CHEK2 variants lacking the nuclear localization signal (amino acids 515–522) served as controls with impaired nuclear localization.
Statistical analyses
Variants for each “functional group” were combined to create a new variable used for the statistical testing in the form of a burden test. Variants were tested using the two-sided Fisher's exact test with P < 0.05 considered statistically significant. The meta-analysis of subgroups of populations was performed in R-studio 4.2.0 (RRID:SCR_000432 and RRID:SCR_001905) in library meta. The statistically significant P value (P < 0.05) and I2 > 75% indicated a heterogeneous sample. The analysis results were visualized using funnel and forest plots.
Data availability
All frequency data of CHEK2 VUS provided by the collaborating centers, kinase and localization assay data used to generate the figures' graphs and plots are provided in Supplementary Tables S1–S3. Raw data from high-content microscopy experiments are available upon request from the corresponding author.
Results
Identification of CHEK2 missense variants
We and others have previously demonstrated that quantification of CHK2 kinase activity in human cells allows reliable scoring of the functional consequences of germline CHEK2 missense variants (17, 18). The ensuing case–control analyses examining cancer risk in carriers of the functionally scoring variants showed promising results consistent with the functional categorization (17, 18). However, both studies were limited by the small number of CHEK2 VUS analyzed or their selective ascertainment. Therefore, we aimed to perform a comprehensive analysis of unselected missense variants identified in routine GGT of cancer susceptibility. To this end, we collected 460 unique CHEK2 germline missense VUS (Supplementary Table S1) identified in oncology patients by 20 members of the ENIGMA consortium from 15 countries (Fig. 1).
Validation of cell-based assays for detection of CHK2 activity
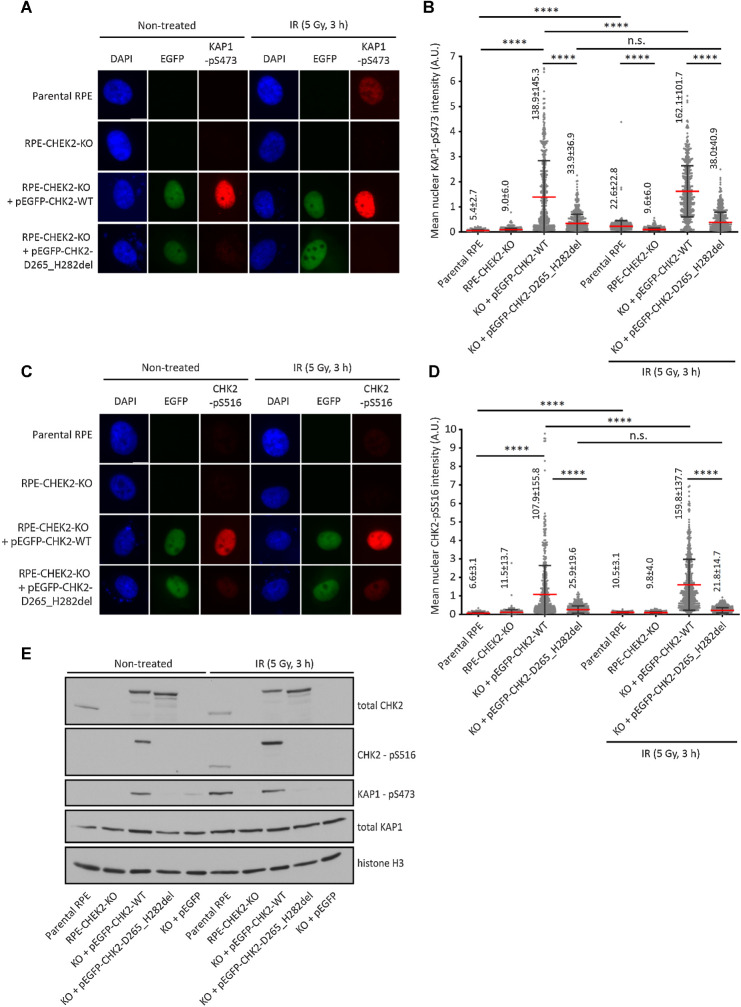
To improve functional characterization of the identified CHEK2 VUS, we established two complementary functional assays for quantification of the catalytic activity of CHK2 in human, diploid, non-transformed RPE1 cells. We used previously described RPE1–CHEK2-KO cells with inactivated CHEK2 and transiently transfected them with plasmids coding for EGFP-tagged CHEK2 variants (17). Using high-content microscopy, we quantified the phosphorylation of the KRAB-associated protein 1 (KAP1) at S473, which is an established substrate of CHK2 (refs. 17, 19; Fig. 2A and B). We observed a low level of KAP1-pS473 in parental RPE1 cells, and the signal significantly increased after exposure of cells to IR, which is an activator of the ATM/CHK2 pathway (Fig. 2B). In contrast, we observed only background KAP1-pS473 signal in RPE1–CHEK2-KO cells and it was not responsive to IR (Fig. 2B). Upon transfection of the wild-type EGFP-CHK2, we observed a high nuclear KAP1-pS473 signal that further increased after exposure of cells to IR (Fig. 2B). Importantly, transfection of the catalytically dead EGFP-CHK2 p.D265_H282del variant did not increase the KAP1-pS473 signal, confirming specificity of the assay (Fig. 2B). Using the same approach, we determined the level of CHK2 autophosphorylation at S516 (refs. 20–23; Fig. 2C and D). The CHK2-pS516 signal of was low at endogenous levels of CHK2; nevertheless, a clearly detectable signal was observed in cells transfected with the wild-type EGFP-CHK2 and it was further increased upon exposure of cells to IR (Fig. 2D). In contrast, CHK2-pS516 signal was significantly lower in cells transfected with the catalytically dead CHK2, and it did not respond to IR (Fig. 2D). Similar results were obtained using detection of KAP1-pS473 and CHK2-pS516 by immunoblotting (Fig. 2E). We conclude that the nuclear KAP1-pS473 and CHK2-pS516 signal corresponds to the activity of EGFP-CHK2 transfected to RPE1–CHEK2-KO cells. Although the transfected EGFP-CHK2 was expressed at slightly higher levels than endogenous CHK2, phosphorylation of the substrates did not reach saturation as further increase of the signal was observed upon exposure to IR. We also noted that transfection of EGFP-CHK2 yielded a strong and reproducible KAP1-pS473 and CHK2-pS516 signal in basal conditions, and therefore we performed the functional screening of CHK2 VUS without exposing cells to IR.
Figure 2.
Validation of KAP1-pS473 and CHK2-pS516 antibodies. A, Parental RPE, RPE1–CHEK2-KO cells or RPE1–CHEK2-KO cells transfected with the wild-type or mutant pEGFP–CHEK2 were left untreated or were exposed to ionizing radiation (5 Gy, 3 hours). After fixation, cells were probed with KAP1-pS473 antibody. Representative images are shown. B, Quantification of A. The mean nuclear intensity of the KAP1-pS473 signal is plotted. Each dot represents one cell; more than 300 cells were analyzed. Red line, error bars and numbers indicate mean ± SDs. Statistical significance was evaluated by the Mann–Whitney test (****, P < 0.0001). A representative experiment is shown from two independent replicates. C, Cells were grown and treated as in A and were probed with CHK2-pS516 antibody. Representative images are shown. D, Quantification of C. The mean nuclear intensity of the CHK2-pS516 signal is plotted. Each dot represents one cell; more than 300 cells were analyzed. Red line, error bars and numbers indicate mean ± SDs. Statistical significance was evaluated by the Mann–Whitney test (****, P < 0.0001). A representative experiment is shown from two independent replicates. E, Cells were grown and treated as in A. Whole-cell lysates were analyzed by immunoblotting with indicated antibodies.
Functional assessment of CHEK2 missense variants
Kinase activity analyses quantified phosphorylation of KAP1 (KAP1 assay) and autophosphorylation of CHK2 (CHK2 assay) using a high-content immunofluorescence microscopy that also allowed assessment of the intracellular localization of analyzed variants. Both assays were successfully performed for 430/460 (93.5%) variants (Supplementary Table S1). The remaining 30 variants were excluded from the analysis because the variants affected the first/last two coding nucleotides in an exon, and thus we cannot exclude that their functional consequence may result in aberrant splicing rather than in an amino acid change (16 variants) or due to poor expression (12 variants) or poor growth (2 variants) in RPE1 cells.
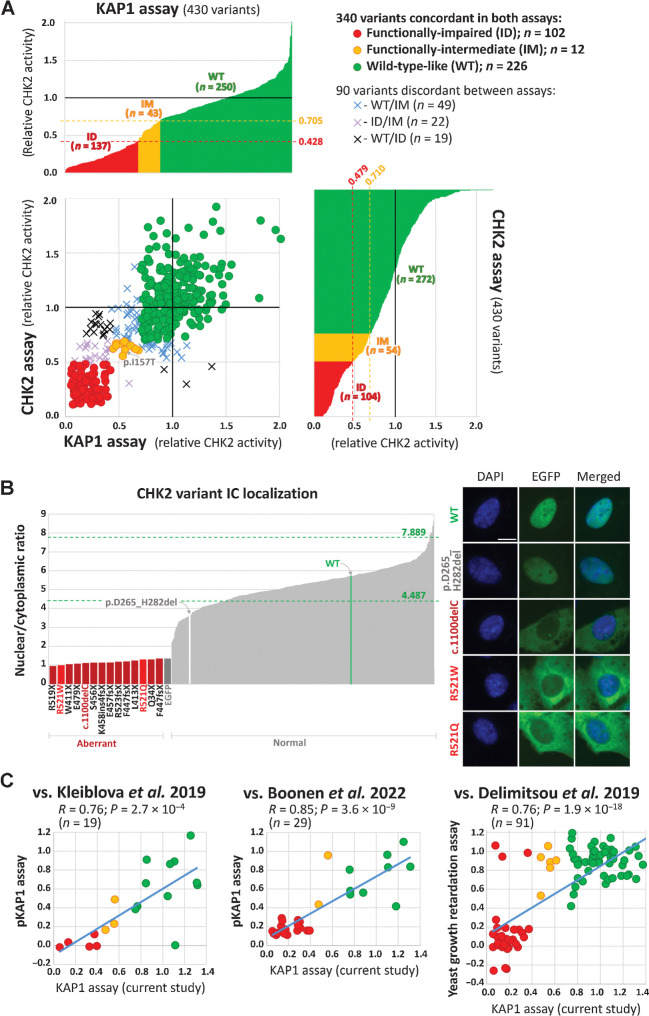
The functional categorization in KAP1 and CHK2 assays were in accord for 340/430 (79.1%) successfully analyzed variants (Fig. 3A). Two thirds (226/340; 66.5%) of concordant variants scored functionally wild–type-like, whereas 12/340 (3.5%) and 102/340 (30.0%) scored functionally intermediate and functionally impaired, respectively. Among the 102 functionally impaired variants, we also included two variants, p.R521W and p.R521Q (scored WT/intermediate and intermediate/WT in kinase assays, respectively), which were the only analyzed missense variants with severely impaired nuclear localization (Fig. 3B). It is noteworthy that the most common c.470C>T (p.I157T) variant scored intermediate in both kinase assays that is consistent with KAP1 assay performed previously (17) and with its low (clinically unimportant) but statistically significant association with breast cancer risk (OR∼1.5) documented in heterozygous p.I157T carriers (24). Discordant categorization between KAP1 and CHK2 assays was observed for 90/430 (20.9%) variants with the WT/intermediate (49 variants) being the most common. A visualization of 430 successfully analyzed CHEK2 missense variants is provided in Fig. 4, particular values for kinase assays and the nuclear-to-cytoplasmic ratio describing an intracellular localization are shown in Supplementary Table S1.
Figure 3.
Kinase KAP1 and CHK2 assays (A). The bar graphs show results of kinase assays for 430 CHEK2 missense variants. In both assays, variants with normalized relative CHK2 activity (mean WT-activity = 1) exceeding that of the weakest signal of WT replicas (not shown) were categorized functionally WT-like, variants with normalized signal intensity lower than the strongest signal for any of kinase-dead/empty EGFP vector controls (in-frame exon 7 deletion–p.D265_H282del; not shown) were categorized as functionally impaired. Variants with normalized CHK2 activities between these ranges were categorized functionally intermediate (0.428–0.705 and 0.479–0.710 for KAP1 and CHK2 assay, respectively; indicated by red and yellow dashed lines). Scatterplot combines results from both assays showing 340 concordant (circles) and 90 discordant (crosses) variants. The nuclear-to-cytoplasmic ratio (B) bar graph (left) displays all missense variants and a set of protein-truncating CHEK2 variants (dark red bars at left, zoomed part of the graph). The missense variants, p.R521W and p.R521Q, with an aberrant localization are highlighted as bright-red bars; the arrows denote WT (green bar) and catalytically-dead in-frame p.D265_H282del variant (white bar). The highest and lowest mean nuclear/cytoplasmic ratio values from all WT replicates are indicated by green dashed lines. Of all missense variants analyzed by ScanR microscopy, only codon 521 alterations revealed aberrant intracellular localization with intense cytoplasmic positivity (right), reminiscent of mislocalization of the c.1100delC (p.T367fsX; size bar, 10 μm) variant. In comparison, the in-frame deletion p.D265_H282del revealed normal intranuclear accumulation, similar to WT. C, Scatter plots depicting correlations between assays performed in this study and previous analyses of CHEK2 VUS. Studies of Kleiblova et al. (17) and Boonen et al. (18) used phosphorylation of KAP1 as a functional readout whereas the study of Delimitsou et al. (25) used a yeast growth retardation assay. The dots are colored according to the results of the KAP1 assay in this study (red, impaired; yellow, intermediate; green, wild-type–like). Blue line represents linear regression, R, correlation coefficient; P, P value. The scatter plot does not show the p.Arg512Trp variant classified by Boonen et al. as intermediate with impaired nuclear localization in our localization assay.
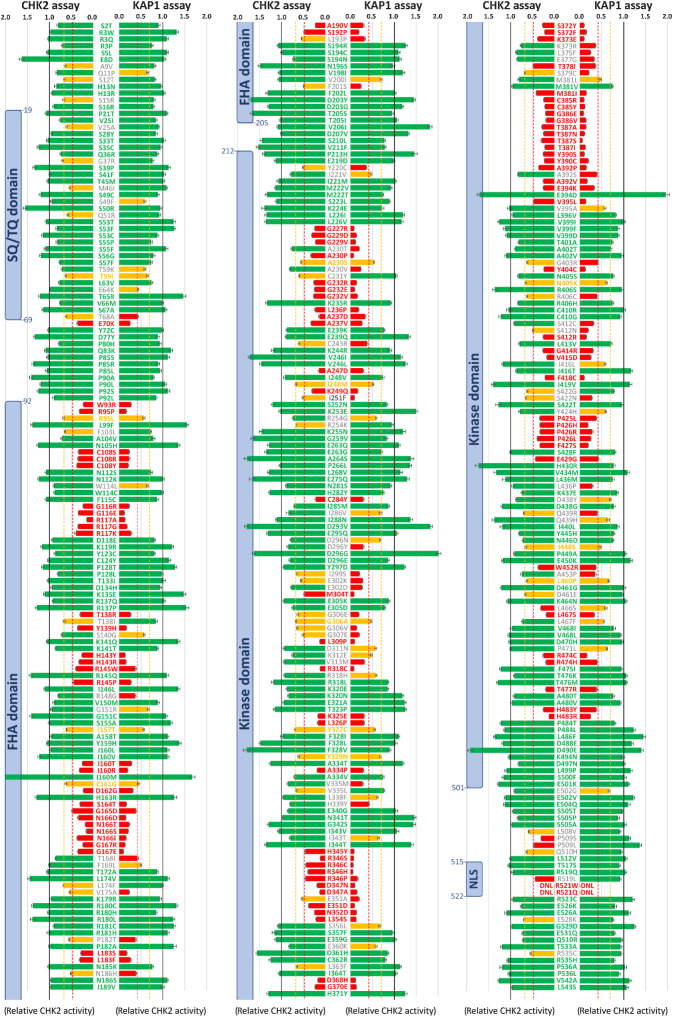
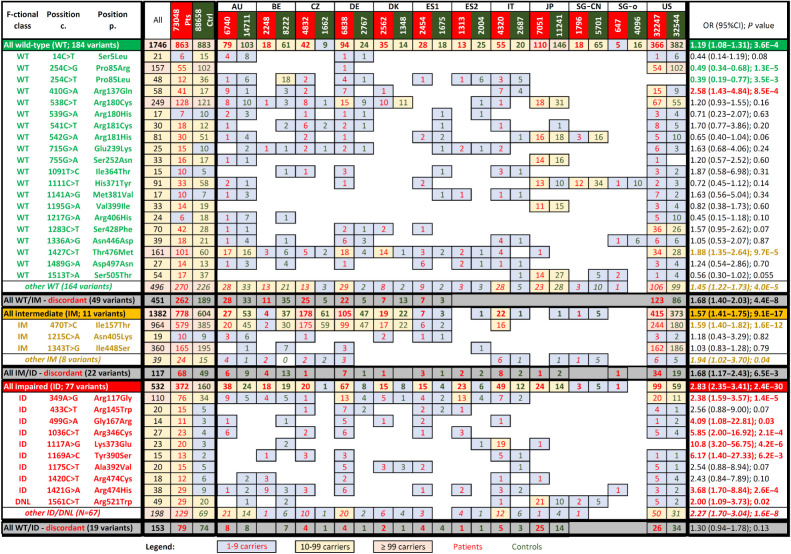
Figure 4.
Results of KAP1 and CHK2 kinase assays for 430 successfully analyzed missense CHEK2 variants (shown as an average relative CHK2 kinase activity). Bars are colored as functionally WT-like (green), intermediate (IM; yellow), and impaired (ID; red), respectively, with thresholds for IM variants (0.428 and 0.479) and ID variants (0.705 and 0.710) for KAP1 and CHK2 assays, respectively (dashed lines). Error bars represent standard errors of mean. Color/gray letters for protein variants indicate concordant/discordant functional assays result, respectively. Blue boxes denote conserved CHK2 domains. DNL, variants that do not localize into the nucleus.
From 340 missense VUS concordant in kinase assays, 19 and 30 overlapped with variants analyzed previously using KAP1 assay by Kleiblova and colleagues (17) and Boonen and colleagues (18), respectively, and 91 overlapped with VUS analyzed by Delimitsou and colleagues (25) using a yeast growth retardation assay. A significant correlation between the KAP1 assay and the results of all three previous studies (Fig. 3C) demonstrates reliability of the functional assessment. Comparison of functional data for 340 variants concordantly categorized in our assays that were analyzed in aforementioned studies is provided in the Supplementary Table S1.
Functionally categorized CHEK2 missense variants and female breast cancer risk
To explore how variants categorized by our functional assay results associated with female breast cancer risk, we used 12 case–control datasets (Supplementary Table S2) provided by ENIGMA consortium members from 10 countries (Fig. 1). The majority of the 161,706 individuals (patients with breast cancer and unaffected controls) were of European descent (n = 117,877; 73.0%). Individuals of Asian (n = 33,535; 20.7%), African-American (n = 8,942; 5.5%), and other races/ethnicities (n = 799; 0.8%) were less frequent. Ascertainment for patient subgroups were heterogeneous, including family/hospital based (40,801; 55.9%) and unselected (32,247; 44.1%) female patients with breast cancer.
Among all 161,706 individuals, there were 4,436 carriers (2.7%) of 377 unique CHEK2 missense VUS analyzed in functional assays. The majority (3,660/4,436; 82.5%) of VUS carriers were individuals carrying some of 272 variants that were categorized as concordant in both functional assays (Supplementary Table S1). Remaining 776 individuals were excluded, including 721 (16.3%) carriers of 78 variants discordantly categorized by kinase assays, 31 (0.7%) carriers of 13 variants suspected to interfere with pre-mRNA splicing, and 24 (0.5%) carriers of 14 variants that failed in the functional analysis. The baseline frequency of all 4,436 CHEK2 VUS carriers was significantly higher in patients over controls [3.4% vs. 2.3%; OR, 1.52; 95% confidence interval (CI), 1.43–1.61] setting a low but significant background breast cancer risk. Increased proportion of variant carriers in cases over controls was maintained also for 3,660 carriers of concordantly categorized variants (2.8% vs. 1.9%; OR, 1.50; 95% CI, 1.40–1.60) with similar background risk (Supplementary Tables S1 and S3).
First, we analyzed association with the breast cancer risks for the functionally characterized categories. In the burden analysis, the statistically significant OR gradually increased from 1.19 (95% CI, 1.08–1.31) to 1.57 (95% CI, 1.41–1.75), and 2.83 (95% CI, 2.35–3.41) for variants functionally characterized as WT-like, intermediate and impaired, respectively (Fig. 5). The non-overlapping 95% CIs indicate the reliable functional characterization discriminating between the risk categories.
Figure 5.
Presence of analyzed CHEK2 missense variants categorized according to the functional assays in patients with breast cancer (BC pts; red numbers) and matched controls (dark green numbers). The association with breast cancer risk (odds ratio; OR) were calculated for prevalent variants having ≥10 carriers among patients or controls, respectively. Colors of the numbers in the last column highlight significant association with moderate-or-higher risk (red; OR > 2), low risk (OR < 2), protective variants (green) or variants without significant impact on breast cancer risk (black). Gray rows display variants that were discordant in the kinase assays. DNL, variants that do not localize into the nucleus.
The results of the subsequent case–control analysis of individual prevalent variants (identified in ≥10 carriers in patients or controls; Fig. 5) showed that the association with breast cancer differed from the functional categorization for two WT-like variants, reaching a significant moderate risk for p.R137Q (OR, 2.58; 95% CI, 1.43–4.84) and a modestly elevated risk for p.T476M (OR, 1.88; 95% CI, 1.35–2.64), respectively. Other WT-like variants were not associated with significantly increased risk or exerted a protective effect (p.P85R and p.P85L). The risk of functionally intermediate variants was predominantly influenced by p.I157T, as this variant accounted for 964/1382 (69.8%) carriers in the intermediate category. None of other individual variants from the functionally intermediate group were significantly associated with breast cancer risk. Among variants categorized as functionally impaired, 7/10 variants were individually significantly associated with breast cancer and the proportion of carriers among cases outnumbered those among controls for the remaining three variants.
In addition, we performed the burden case–control analysis for variants with discordant results in the kinase assays (Fig. 5) and also for variants affecting border exon sites, which are suspected to affect splicing. The latter category (not shown in the Fig. 5) included 20 carriers in patients and 11 in controls and was associated with increased breast cancer risk (OR, 2.21; 95% CI, 1.06–4.61; P = 0.03).
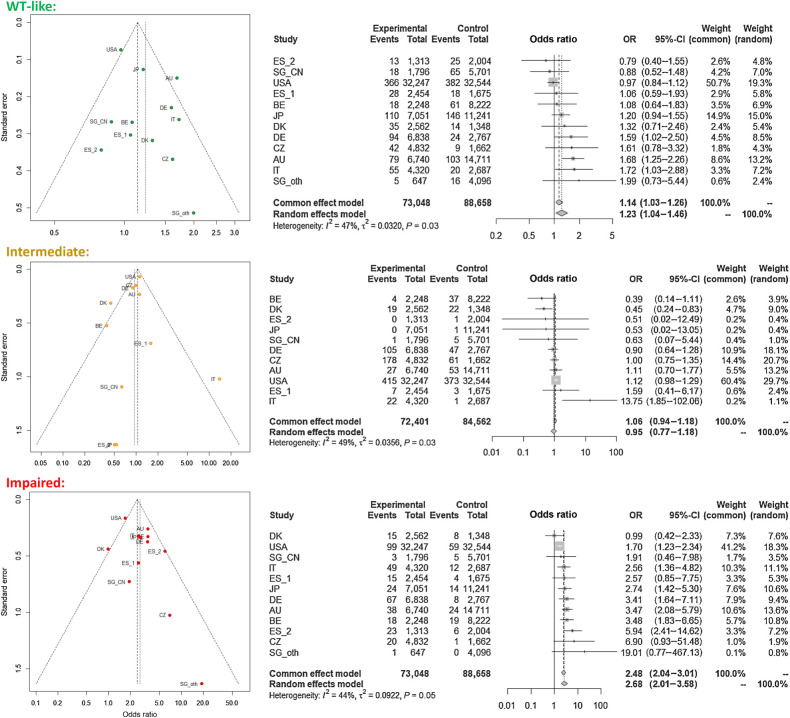
Meta-analyses of functionally categorized population datasets
We performed the meta-analysis to assess the magnitude of the risk in the groups of CHEK2 VUS carriers categorized according to the results of our functional assays (Fig. 6). The P values of <0.05 indicated substantial variability for the datasets in VUS functionally characterized as WT-like and intermediate. The variability was lower for the functionally impaired category retaining a significant moderate risk (OR, 2.68; 95% CI, 2.01–3.58 for random effect model) based on significantly increased risks in 7/12 individual population datasets. The risk for WT-like variants was marginally statistically significant but close to one and thus clinically irrelevant. The non-significant risk of the functionally intermediate group was affected by variable prevalence of the p.I157T variant, which was enriched in control populations from Belgium and Denmark but rare in Italian controls. The results of meta-analysis revealed large variability in case–control datasets but confirmed a low breast cancer risk in carriers of WT-like and intermediate variants, and a clinically considerable and significant breast cancer risk in carriers of variants that were categorized as functionally impaired.
Figure 6.
Funnel plot (left) and forest plots (right) for individual datasets of breast cancer cases and controls from 12 datasets (10 countries) stratified according to the functional categorization.
Discussion
GGT of the CHEK2 gene yields appreciable frequencies of pathogenic or likely pathogenic (P/LP) germline variants in breast cancer or other patients with cancer (6, 26). On the other hand, genetic testing has also revealed a large number of missense variants that are mostly classified as VUS, hindering its clinical utility. This is illustrated by review of recent (January, 2023) ClinVar database data, registering 593 unique frameshift/nonsense/splice-site CHEK2 alterations of which 566 (95.4%) received conclusive classification [564 P/LP and 2 benign/likely-benign (B/LB)]. In contrast, only 15/1,497 (1.0%) CHEK2 missense variants in ClinVar had non-conflicting conclusive classification (including 6 P/LP and 9 B/LB). Analysis of gnomAD data indicated that the overall frequencies of stop-gain/frameshift/splice-site (1.65%) and missense (1.87%) CHEK2 germline variants were comparable.
A large, international, case–control study of the Breast Cancer Association Consortium (BCAC; ref. 7) identified rare CHEK2 missense variants (i.e., variants with a population frequency of <0.001) in 2.0% patients with breast cancer and 1.4% controls, indicating slightly increased association with breast cancer for missense variants as a group (OR, 1.43; 95% CI, 1.31–1.57). Frequency of CHEK2 missense variants in our study was 3.4% in patients with breast cancer and 2.2% in controls (OR, 1.52; 95% CI, 1.43–1.61; Supplementary Table S3). When applying the rare variant definition from the BCAC study (i.e., excluding carriers of p.I157T, p.I448S, and p.R180C), missense variant frequency reduced to 2.2% in cases and 1.5% in controls and the association with breast cancer was comparable with that reported by BCAC (OR, 1.51; 95% CI, 1.40–1.62 for variants with frequency <0.001). This association indicates that in addition to functionally neutral variants, functionally impaired missense variants increasing breast cancer risk are enriched among patients with breast cancer. The clinical need urges for clear discrimination between the pathogenic and non-pathogenic variants as both may modify clinical management in their carriers (and relatives in case of pathogenic alterations).
The ACMG guidelines provide a generally adopted framework to standardize variant interpretation in clinical settings (27). However, additional credible methods for variant interpretation are required to effectively harness data from GGT. Validated functional assays have been considered as one of the most powerful tools to aid variant interpretation (28, 29). Our study is the largest and the most comprehensive functional analysis of real-world germline CHEK2 missense VUS found in patients with various cancer diagnoses and in controls. Corroborated by the case–control analysis of variant groups stratified according to the results of our functional assays, it provides several meaningful insights:
First, the CHEK2 functional analysis, based on a combination of KAP1/CHK2 assays with a high-content microscopy controlling the intracellular targeting of analyzed variant in human non-cancer cells, scored concordantly for 340 variants. This more than doubled the number of 160 CHEK2 missense VUS categorized so-far (reviewed recently in ref. 14) using various approaches. In comparison with our previous functional study (17) describing the KAP1-based CHEK2 analysis, we implemented improved calculations of relative KAP1-phosphorylation intensities normalized to CHK2 expression, added CHK2 autophosphorylation and localization assays in human cells and abandoned synthetic in vitro phosphorylation assays. Our current results showed a high correlation with the results from our previous KAP1 assay (17), the KAP1 assay by Boonen and colleagues (18) in mouse embryonic stem cells and also with yeast-based assay by Delimitsou and colleagues (25). To the yeast survival assays, our assays provide the assessment of CHK2 kinase activity in physiologic intracellular environment, including natural CHK2 substrates, inhibitors, and interactors.
Second, the burden case–control analysis allowed us to determine association of the functionally categorized missense variants with breast cancer. A clinically significant moderate association with breast cancer risk was observed for carriers of functionally impaired variant for both simplistic burden analysis (OR, 2.83; Fig. 5) and meta-analysis of individual population-specific datasets (OR, 2.68; Fig. 6). The burden case–control analysis of the most common truncating variant c.1100delC performed in our datasets revealed a similar association with breast cancer risk (OR, 2.73; 95% CI, 2.29–3.26; Supplementary Table S1). Comparable associations were noted in previous meta-analyses of c.1100delC in unselected breast cancer populations (OR, 2.75; 95% CI, 2.25–3.36 and OR, 2.89; 95% CI, 2.63–3.16; refs. 30, 31). Thus, we demonstrate that the breast cancer risk associated with functionally impaired CHEK2 missense variants is comparable with the risk associated with P/LP truncating variants. In addition, the study conferred that carriers of functionally impaired missense variants account for 0.5% patients with breast cancer overall. This was substantially lower than 1.3% predicted by Dorling and colleagues (32) in BCAC data analyzed by in silico prediction tools. However, the proportion of carriers varied among national datasets in our study, being highest in European patients (0.9%) and less frequent in patients from the USA (0.3%) and patients from Japan and Singapore (0.2%). On the other hand, 2.2% of all patients with breast cancer in this study were carriers of missense variants categorized as WT-like or intermediate with clinically irrelevant breast cancer risk.
Third, the case–control analyses of individual variants largely corresponded to their functional categorization. The association of p.R117G (the most common variant from the functionally impaired class and the sixth most frequent variant in our dataset) with breast cancer risk (OR, 2.38; 95% CI, 1.58–3.68) was similar to the risk of the entire functionally impaired subgroup. A comparable risk (OR, 2.26; 95% CI, 1.29–3.95) for p.R117G was reported by Southey and colleagues (33) in the Collaborative Oncological Gene-environment Study and recently by Dorling and colleagues (ref. 32; OR, 2.69; 95% CI, 1.46–4.94) analyzing the BCAC data. Interestingly, the p.I157T has been categorized as functionally intermediate with modestly elevated association with breast cancer risk (OR, 1.59; 95% CI, 1.40–1.82), which was comparable with previously published p.I157T meta-analyses with OR ranging between 1.28 and 1.48 in unselected patients with breast cancer (6). Of note, case–control analyses and the functional characterization were discrepant for several variants. The category of functionally WT-like variants included mostly variants with OR∼1.0, but two variants, p.R137Q and p.T476M (fully functional in our kinase assays), showed elevated associations with breast cancer risk in case–control analyses (OR = 2.58 and OR = 1.88, respectively). Although both variants were frequent (the tenth and the fourth most common in our study, respectively), they were virtually absent in Asian populations, and present with large variability in populations of European ancestry (Fig. 5). They have been subjected to functional analyses previously. Although p.R137Q was described as neutral by Sodha and colleagues (34) and Bell and colleagues (35), the functional data for p.T476M were rather discrepant (14). Recent case–control analysis of large cohort of patients with cancer, including 250 carriers of p.T476M, revealed only a modest association with the breast cancer risk (OR, 1.35; 95% CI, 1.03–1.77; P = 0.03) and no risk of other analyzed cancers (36). In the functionally intermediate subgroup, we included the p.I448S variant, which was considered intermediate in the KAP1 assay in Boonen and colleagues (18) and also the yeast assay in Delimitsou and colleagues (25). Prevalent in the US data, the variant had OR∼1 in the case–control analysis and it is classified a B/LB in ClinVar. In the functionally impaired subgroup, 7/10 variants showed concordantly significantly increased OR>2 in case–control analysis. These partially conflicting results indicate that larger case–control studies will be required to refine the risks for individual rare VUS but for population-specific variants, case–control data from founder populations will be of particular importance. In addition, extended functional assays would help to determine the boundary between WT-like and intermediate categories more precisely; however, it should be emphasized that both subgroups have low or negligible clinical impact, at least for carriers of heterozygous variants. Individual-level case–control analyses and/or additional functional/splicing assays will remain important to inform classification of functionally intermediate/WT-like variants because some individual variants may impact CHK2 function via mechanisms not surveyed by our assays. However, taken together, the functional analysis will facilitate clinical classification of germline CHEK2 missense VUS, in particular as evidence toward pathogenicity for variants concordantly categorized as functionally impaired.
Finally, the meta-analyses of functionally characterized categories indicated that although some case–control datasets were heterogeneous, meta-analyses findings corresponded to those from the burden analysis.
We are aware of several limitations of this study. Although we selected the cells with low CHK2 expression for the functional analysis of VUS, we cannot completely exclude that the assay could be affected by the expression of the studied CHK2 isoforms at supraphysiological levels; however, this concern is unlikely to affect the categorization of the functionally impaired variants. The carriers were identified by different genotyping approaches and variant reporting differed among contributing centers of ENIGMA members. The patient populations included hospital-based, high-risk, and unselected patients with breast cancer in individual datasets. This ascertainment bias could slightly overestimate the breast cancer risks for variant carriers. Because no other clinicopathological data were available, we could not analyze the associations of CHEK2 variants with clinicopathological variables.
In conclusion, we functionally categorized a majority of germline CHEK2 missense variants commonly occurring in patients with cancer and controls in various populations worldwide. The case–control analysis revealed that the breast cancer risk of functionally impaired germline variants is comparable with that of truncating CHEK2 P/LP variants and suggested that the clinical management of (breast) cancer prevention in both groups of carriers should be similar (37). Moreover, the comprehensive functional classification will foster development of clinical interpretation guidelines for germline CHEK2 variants and will allow exploration of the association of germline functionally impaired missense variants with other cancer types. The data may also serve as a predictive information for tailored anticancer therapy using PARP or immune checkpoint inhibitors in carriers of germline functionally impaired variants (38–40).
Supplementary Material
Detail description of the functional categorization of analyzed CHEK2 missense variants.
List of all analyzed CHEK2 variants with results of KAP1/CHK2 kinase and localization assays and the results from recent previously published functional analyses of the CHEK2 VUS.
Characteristics of 12 case-control datasets from the ENIGMA consortium partners.
Frequencies of all reported germline CHEK2 variant carriers and carriers of variants concordantly categorized by functional our kinase assays in breast cancer patients and controls in 12 analyzed population datasets.
Acknowledgments
This work was supported by the grant projects of the Czech Ministry of Health NV19–03–00279, DRO-VFN-64165; Charles University projects COOPERATIO and SVV260516; Ministry of Education, Youths and Sports of the Czech Republic grant EXCELES no. LX22NPO05102 funded by EU. L. Stolarova was partially supported by Czech Academy of Science (PPPLZ project L200522201). The CARRIERS study was supported by NIH grants R01 CA192393, R35 CA253187 and a specialized program of research excellence (SPORE) in breast cancer (P50 CA116201). Contract grant sponsor A. Vega: supported by Spanish Instituto de Salud Carlos III (ISCIII) funding, an initiative of the Spanish Ministry of Economy and Innovation partially supported by European Regional Development FEDER Funds (PI22/00589, PI19/01424; INT20/00071), the Autonomous Government of Galicia (Consolidation and structuring program: IN607B), by the Fundación Mutua Madrileña (call 2018), and by the AECC (PRYES211091VEGA). A.B. Spurdle was supported by an NHMRC Investigator Fellowship (APP177524).
The Biobank Japan, in Addition to Those Named in the Author List: Koichi Matsuda, Yoichiro Kamatani, Takayuki Morisaki, Akiko Nagai, Kaori Muto, Yoshinori Murakami, Yoichi Furukawa, Yuji Yamanashi, Yusuke Nakamura, (Institute of Medical Science, The University ofTokyo, Tokyo); Taisei Mushiroda, Yukihide Momozawa, Toshihiro Tanaka, Yozo Ohnishi, Michiaki Kubo (RIKEN Center for Integrative Medical Sciences, Yokohama); Shinichi Higashiue, Shuzo Kobayashi (Tokushukai Hospitals, Tokyo); Shiro Minami, Hiroki Yamaguhci (Nippon Medical School, Tokyo); Hajime Arai, Ken Yamaji, Yasushi Okazaki (Juntendo University, Tokyo); Satoshi Asai, Yasuo Takahashi (Nihon University, Tokyo); Tomoaki Fujioka, Wataru Obara (Iwate Medical University Iwate); Seijiro Mori, Shigeo Murayama (Tokyo Metropolitan Institute of Gerontology, Tokyo); Satoshi Nagayama, Yoshio Miki (The Cancer Institute Hospital of JFCR, Tokyo); Akihide Masumoto, Akira Yamada (Aso Iizuka Hospital, Fukuoka); Yasuko Nishizawa, Masahiko Higashiyama (Osaka International Center Institute, Osaka) Hiromu Kutsumi (Shiga University of Medical Science, Shiga); Yukihiro Koretsune (National Hospital Organization, Osaka National Hospital, Osaka); Takashi Yoshiyama (Fukujuji Hospital, Tokyo).
The CZECANCA consortium, in Addition to Those Named in the Author List: Marianna Borecka, Marta Cerna, Milena Hovhannisyan, Sandra Jelinkova, Petr Nehasil (Institute of Medical Biochemistry and Laboratory Diagnostics, First Faculty of Medicine and General University Hospital in Prague, Prague); Lenka Foretova, Eva Machackova (Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute, Brno); Vera Krutilkova, Spiros Tavandzis (AGEL Laboratories, Novy Jicin); Leona Cerna, Stepan Chvojka, Monika Koudova, Alena Puchmajerova (GENNET, Prague); Ondrej Havranek, Jan Novotny, Kamila Vesela, Michal Vocka (Institute of Biology and Medical Genetics, First Faculty of Medicine and General University Hospital in Prague, Prague); Lucie Hruskova, Renata Michalovska, Denisa Schwetzova, Zdenka Vlckova (GHC Genetics, Prague); Monika Cerna, Marketa Hejnalova, Nikol Jedlickova, Ivan Subrt, Tomas Zavoral (Institute of Medical Genetics, University Hospital Pilsen, Pilsen); Marcela Kosarova, Gabriela Vacinova (PRONATAL, Prague); Maria Janikova, Romana Kratochvilova, Vaclava Curtisova, Radek Vrtel (Department of Medical Genetics, University Hospital Olomouc, Faculty of Medicine and Dentistry, Palacky University, Olomouc); Ondrej Scheinost, Petra Duskova (Hospital Ceske Budejovice, Ceske Budejovice); Viktor Stranecky (Department of Paediatrics and Inherited Metabolic Disorders, First Faculty of Medicine and General University Hospital in Prague, Prague).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Contributor Information
Biobank Japan:
Koichi Matsuda, Yoichiro Kamatani, Takayuki Morisaki, Akiko Nagai, Kaori Muto, Yoshinori Murakami, Yoichi Furukawa, Yuji Yamanashi, Yusuke Nakamura, Taisei Mushiroda, Yukihide Momozawa, Toshihiro Tanaka, Yozo Ohnishi, Michiaki Kubo, Shinichi Higashiue, Shuzo Kobayashi, Shiro Minami, Hiroki Yamaguhci, Hajime Arai, Ken Yamaji, Yasushi Okazaki, Satoshi Asai, Yasuo Takahashi, Tomoaki Fujioka, Wataru Obara, Seijiro Mori, Shigeo Murayama, Satoshi Nagayama, Yoshio Miki, Akihide Masumoto, Akira Yamada, Yasuko Nishizawa, Masahiko Higashiyama, Hiromu Kutsumi, Yukihiro Koretsune, and Takashi Yoshiyama
Consortium CZECANCA:
Marianna Borecka, Marta Cerna, Milena Hovhannisyan, Sandra Jelinkova, Petr Nehasil, Lenka Foretova, Eva Machackova, Vera Krutilkova, Spiros Tavandzis, Leona Cerna, Stepan Chvojka, Monika Koudova, Alena Puchmajerova, Ondrej Havranek, Jan Novotny, Kamila Vesela, Michal Vocka, Lucie Hruskova, Renata Michalovska, Denisa Schwetzova, Zdenka Vlckova, Monika Cerna, Marketa Hejnalova, Nikol Jedlickova, Ivan Subrt, Tomas Zavoral, Marcela Kosarova, Gabriela Vacinova, Maria Janikova, Romana Kratochvilova, Vaclava Curtisova, Radek Vrtel, Ondrej Scheinost, Petra Duskova, and Viktor Stranecky
Authors' Disclosures
P. Zemankova reports grants from Charles University, Prague and Ministry of Health of the Czech Republic during the conduct of the study. M. Janatova reports grants from Ministry of Health of the Czech Republic and Charles University during the conduct of the study. J. Soukupova reports grants from Ministry of Health of the Czech Republic and Charles University during the conduct of the study. M. Barnard reports grants from Karin Grunebaum Cancer Research Foundation during the conduct of the study as well as personal fees from EpiExcellence LLC outside the submitted work. J. Brunet reports other support from GSK outside the submitted work. J.B. Chiang reports personal fees from AstraZeneca outside the submitted work. L. Cortesi reports personal fees from AstraZeneca, Pfizer, Novartis, Gilead, and Clovis and grants from MSD outside the submitted work. L. Devereux reports grants from NH&MRC outside the submitted work. C. Engel reports grants from German Cancer Aid and BMBF during the conduct of the study. D.G.R. Evans reports personal fees from AstraZeneca and Everythinggenetic Ltd. during the conduct of the study. J. Hauke reports grants from German Cancer Aid during the conduct of the study. C. Lázaro reports personal fees from AstraZeneca and Illumina outside the submitted work. N. Li reports grants from Cancer Council Victoria during the conduct of the study. S.L. Neuhausen reports grants from National Cancer Institute during the conduct of the study. H. Nevanlinna reports grants from Sigrid Juselius Foundation and Government Funding (VTR) of Helsinki University Hospital during the conduct of the study. J. Palmer reports grants from National Institutes of Health during the conduct of the study. M.E. Richardson reports personal fees from Ambry Genetics outside the submitted work. A.E. Toland reports grants from National Cancer Institute during the conduct of the study. A. Trentham-Dietz reports grants from National Cancer Institute during the conduct of the study. C.M. Vachon reports grants from GRAIL outside the submitted work. J.N. Weitzel reports grants from NIH during the conduct of the study as well as personal fees from Natera outside the submitted work. T. Zima reports grants from Charles University, Ministry of Education Youth and Sport CZ, and Ministry of Health CZ during the conduct of the study. J. Ngeow reports research funding from AstraZeneca. Y. Momozawa reports grants from Japan Agency for Medical Research and Development during the conduct of the study as well as personal fees from AstraZeneca, Sanofi, and ActMed outside the submitted work. F.J. Couch reports grants from NIH and BCRF, non-financial support from Ambry Genetics, and personal fees from AstraZeneca during the conduct of the study. L. Macurek reports grants from Czech Ministry of Health, Czech Academy of Science, and National Institute for Cancer Research during the conduct of the study. Z. Kleibl reports grants from Czech Ministry of Health, Charles University, and Czech Ministry of Education, Youths and Sports during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
L. Stolarova: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, writing–original draft, writing–review and editing. P. Kleiblova: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. P. Zemankova: Conceptualization, data curation, software, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. B. Stastna: Data curation, software, formal analysis, investigation, writing–review and editing. M. Janatova: Resources, data curation, software, formal analysis, investigation, writing–original draft, writing–review and editing. J. Soukupova: Resources, data curation, writing–original draft, writing–review and editing. M.I. Achatz: Data curation, software, formal analysis, investigation, writing–review and editing. C. Ambrosone: Data curation, writing–review and editing. P. Apostolou: Data curation, writing–review and editing. B.K. Arun: Data curation, writing–review and editing. P. Auer: Data curation, writing–review and editing. M. Barnard: Data curation, writing–review and editing. B. Bertelsen: Data curation, writing–review and editing. M.J. Blok: Data curation, writing–review and editing. N. Boddicker: Data curation, writing–review and editing. J. Brunet: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. E.S. Burnside: Data curation, writing–review and editing. M. Calvello: Data curation, writing–review and editing. I. Campbell: Data curation, writing–review and editing. S.H. Chan: Data curation, writing–review and editing. F. Chen: Data curation, writing–review and editing. J.B. Chiang: Data curation, writing–review and editing. A. Coppa: Data curation, writing–review and editing. L. Cortesi: Data curation, writing–review and editing. A. Crujeiras-González: Data curation, writing–review and editing. K. De Leeneer: Data curation, writing–review and editing. R. De Putter: Data curation, writing–review and editing. A. DePersia: Data curation, writing–review and editing. L. Devereux: Data curation, writing–review and editing. S. Domchek: Data curation, writing–review and editing. A. Efremidis: Data curation, writing–review and editing. C. Engel: Data curation, writing–review and editing. C. Ernst: Data curation, writing–review and editing. D.G.R. Evans: Data curation, writing–review and editing. L. Feliubadaló: Data curation, writing–review and editing. F. Fostira: Data curation, writing–review and editing. O. Fuentes-Ríos: Data curation, writing–review and editing. E.B. Gómez-García: Data curation, writing–original draft, writing–review and editing. S. González: Data curation, writing–original draft. C. Haiman: Data curation, writing–original draft. T.v.O. Hansen: Data curation, writing–original draft, writing–review and editing. J. Hauke: Data curation, writing–original draft, writing–review and editing. J. Hodge: Data curation, writing–review and editing. C. Hu: Data curation, writing–review and editing. H. Huang: Data curation, writing–review and editing. N.D.B. Ishak: Data curation, writing–review and editing. Y. Iwasaki: Data curation, writing–review and editing. I. Konstantopoulou: Resources, data curation, writing–original draft, writing–review and editing. P. Kraft: Resources, data curation, writing–original draft, writing–review and editing. J. Lacey: Data curation, writing–review and editing. C. Lázaro: Data curation, writing–review and editing. N. Li: Data curation, writing–review and editing. W.K. Lim: Data curation, writing–review and editing. S. Lindstrom: Data curation, writing–review and editing. A. Lori: Data curation, writing–review and editing. E. Martinez: Data curation, writing–review and editing. A. Martins: Data curation, writing–review and editing. K. Matsuda: Data curation, writing–review and editing. G. Matullo: Data curation, writing–review and editing. S. McInerny: Data curation, writing–review and editing. K. Michailidou: Data curation, writing–review and editing. M. Montagna: Data curation, writing–review and editing. A.N.A. Monteiro: Data curation, writing–review and editing. L. Mori: Data curation, writing–review and editing. K. Nathanson: Data curation, writing–review and editing. S.L. Neuhausen: Data curation, writing–review and editing. H. Nevanlinna: Data curation, writing–review and editing. J.E. Olson: Data curation, writing–review and editing. J. Palmer: Data curation, writing–review and editing. B. Pasini: Data curation, writing–review and editing. A. Patel: Data curation, writing–review and editing. M. Piane: Data curation, writing–review and editing. B. Poppe: Data curation, writing–review and editing. P. Radice: Data curation, writing–review and editing. A. Renieri: Data curation, writing–review and editing. N. Resta: Data curation, writing–review and editing. M.E. Richardson: Data curation, writing–review and editing. T. Rosseel: Data curation, writing–review and editing. K.J. Ruddy: Data curation, writing–review and editing. M. Santamariña: Data curation, writing–review and editing. E.S. Dos Santos: Data curation, writing–review and editing. L. Teras: Data curation, writing–review and editing. A.E. Toland: Data curation, writing–review and editing. A. Trentham-Dietz: Resources, data curation, writing–original draft, writing–review and editing. C.M. Vachon: Resources, data curation, writing–original draft, writing–review and editing. A.E. Volk: Data curation, writing–review and editing. N. Weber-Lassalle: Data curation, writing–review and editing. J.N. Weitzel: Data curation, writing–review and editing. L. Wiesmuller: Data curation, writing–review and editing. S. Winham: Data curation, writing–review and editing. S. Yadav: Data curation, writing–review and editing. D. Yannoukakos: Data curation, writing–review and editing. S. Yao: Data curation, writing–review and editing. V. Zampiga: Data curation, writing–review and editing. M. Zethoven: Data curation, writing–review and editing. Z.W. Zhang: Resources, data curation, writing–review and editing. T. Zima: Resources, data curation, supervision, writing–original draft, writing–review and editing. A.B. Spurdle: Resources, data curation, formal analysis, supervision, writing–original draft, writing–review and editing. A. Vega: Resources, data curation, formal analysis, writing–review and editing. M. Rossing: Resources, data curation, formal analysis, writing–review and editing. J. Del Valle: Resources, data curation, formal analysis, writing–review and editing. A. De Nicolo: Resources, data curation, formal analysis, supervision, writing–original draft, writing–review and editing. E. Hahnen: Resources, data curation, formal analysis, supervision, writing–original draft, writing–review and editing. K.B.M. Claes: Resources, data curation, formal analysis, writing–review and editing. J. Ngeow: Resources, data curation, formal analysis, writing–review and editing. Y. Momozawa: Resources, data curation, formal analysis, writing–review and editing. P.A. James: Resources, data curation, formal analysis, supervision, writing–review and editing. F.J. Couch: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. L. Macurek: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. Z. Kleibl: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol 2014;6:442–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 1999;286:2528–31. [DOI] [PubMed] [Google Scholar]
- 3. Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol 2008;26:542–8. [DOI] [PubMed] [Google Scholar]
- 4. Schmidt MK, Hogervorst F, van Hien R, Cornelissen S, Broeks A, Adank MA, et al. Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers. J Clin Oncol 2016;34:2750–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004;75:1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stolarova L, Kleiblova P, Janatova M, Soukupova J, Zemankova P, Macurek L, et al. CHEK2 germline variants in cancer predisposition: stalemate rather than checkmate. Cells 2020;9:2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breast Cancer Association C, Dorling L, Carvalho S, Allen J, Gonzalez-Neira A, Luccarini C, et al. Breast cancer risk genes - association analysis in more than 113,000 women. N Engl J Med 2021;384:428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med 2021;384:440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tung N, Domchek SM, Stadler Z, Nathanson KL, Couch F, Garber JE, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol 2016;13:581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clift K, Macklin S, Halverson C, McCormick JB, Abu Dabrh AM, Hines S. Patients' views on variants of uncertain significance across indications. J Community Genet 2020;11:139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scherr CL, Ramesh S, Getachew-Smith H, Kalke K, Ramsey K, Fischhoff B, et al. How patients deal with an ambiguous medical test: decision-making after genetic testing. Patient Educ Couns 2021;104:953–9. [DOI] [PubMed] [Google Scholar]
- 12. Monteiro AN, Bouwman P, Kousholt AN, Eccles DM, Millot GA, Masson JY, et al. Variants of uncertain clinical significance in hereditary breast and ovarian cancer genes: best practices in functional analysis for clinical annotation. J Med Genet 2020;57:509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Engel C, de la Hoya M, Peterlongo P, Yannoukakos D, Livraghi L, et al. Risks of breast and ovarian cancer for women harboring pathogenic missense variants in BRCA1 and BRCA2 compared with those harboring protein truncating variants. Genet Med 2022;24:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boonen R, Vreeswijk MPG, van Attikum H. CHEK2 variants: linking functional impact to cancer risk. Trends Cancer 2022;8:759–70. [DOI] [PubMed] [Google Scholar]
- 15. Young EL, Feng BJ, Stark AW, Damiola F, Durand G, Forey N, et al. Multigene testing of moderate-risk genes: be mindful of the missense. J Med Genet 2016;53:366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, Nathanson KL, et al. ENIGMA–evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 2012;33:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kleiblova P, Stolarova L, Krizova K, Lhota F, Hojny J, Zemankova P, et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int J Cancer 2019;145:1782–97. [DOI] [PubMed] [Google Scholar]
- 18. Boonen R, Wiegant WW, Celosse N, Vroling B, Heijl S, Kote-Jarai Z, et al. Functional analysis identifies damaging CHEK2 missense variants associated with increased cancer risk. Cancer Res 2022;82:615–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu C, Zhang S, Gao X, Gao X, Xu X, Lv Y, et al. Roles of kruppel-associated box (KRAB)-associated co-repressor KAP1 Ser-473 phosphorylation in DNA damage response. J Biol Chem 2012;287:18937–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu X, Chen J. Autophosphorylation of checkpoint kinase 2 at serine 516 is required for radiation-induced apoptosis. J Biol Chem 2003;278:36163–8. [DOI] [PubMed] [Google Scholar]
- 21. Olsen BB, Larsen MR, Boldyreff B, Niefind K, Issinger OG. Exploring the intramolecular phosphorylation sites in human Chk2. Mutat Res 2008;646:50–9. [DOI] [PubMed] [Google Scholar]
- 22. Gabant G, Lorphelin A, Nozerand N, Marchetti C, Bellanger L, Dedieu A, et al. Autophosphorylated residues involved in the regulation of human chk2 in vitro. J Mol Biol 2008;380:489–503. [DOI] [PubMed] [Google Scholar]
- 23. Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell 2009;35:818–29. [DOI] [PubMed] [Google Scholar]
- 24. Liu C, Wang Y, Wang QS, Wang YJ. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev 2012;13:1355–60. [DOI] [PubMed] [Google Scholar]
- 25. Delimitsou A, Fostira F, Kalfakakou D, Apostolou P, Konstantopoulou I, Kroupis C, et al. Functional characterization of CHEK2 variants in a Saccharomyces cerevisiae system. Hum Mutat 2019;40:631–48. [DOI] [PubMed] [Google Scholar]
- 26. Nielsen SM, Eccles DM, Romero IL, Al-Mulla F, Balmana J, Biancolella M, et al. Genetic testing and clinical management practices for variants in non-BRCA1/2 breast (and breast/ovarian) cancer susceptibility genes: an international survey by the Evidence-Based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) clinical working group. JCO Precis Oncol 2018;2:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 2019;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brnich SE, Rivera-Munoz EA, Berg JS. Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks. Hum Mutat 2018;39:1531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Y, Zhang F, Wang Y, Liu SC. CHEK2 1100delC variant and breast cancer risk in Caucasians: a meta-analysis based on 25 studies with 29,154 cases and 37,064 controls. Asian Pac J Cancer Prev 2012;13:3501–5. [DOI] [PubMed] [Google Scholar]
- 31. Liang M, Zhang Y, Sun C, Rizeq FK, Min M, Shi T, et al. Association between CHEK2*1100delC and breast cancer: a systematic review and meta-analysis. Mol Diagn Ther 2018;22:397–407. [DOI] [PubMed] [Google Scholar]
- 32. Dorling L, Carvalho S, Allen J, Parsons MT, Fortuno C, Gonzalez-Neira A, et al. Breast cancer risks associated with missense variants in breast cancer susceptibility genes. Genome Med 2022;14:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Southey MC, Goldgar DE, Winqvist R, Pylkas K, Couch F, Tischkowitz M, et al. PALB2, CHEK2, and ATM rare variants and cancer risk: data from COGS. J Med Genet 2016;53:800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sodha N, Mantoni TS, Tavtigian SV, Eeles R, Garrett MD. Rare germ line CHEK2 variants identified in breast cancer families encode proteins that show impaired activation. Cancer Res 2006;66:8966–70. [DOI] [PubMed] [Google Scholar]
- 35. Bell DW, Kim SH, Godwin AK, Schiripo TA, Harris PL, Haserlat SM, et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer 2007;121:2661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bychkovsky BL, Agaoglu NB, Horton C, Zhou J, Yussuf A, Hemyari P, et al. Differences in cancer phenotypes among frequent CHEK2 variants and implications for clinical care-checking CHEK2. JAMA Oncol 2022;8:1598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lowry KP, Geuzinge HA, Stout NK, Alagoz O, Hampton J, Kerlikowske K, et al. Breast cancer screening strategies for women with ATM, CHEK2, and PALB2 pathogenic variants: a comparative modeling analysis. JAMA Oncol 2022;8:587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kinget L, Bechter O, Punie K, Debruyne PR, Brems H, Clement P, et al. Multitumor case series of germline BRCA1, BRCA2 and CHEK2-mutated patients responding favorably on immune checkpoint inhibitors. Curr Oncol 2021;28:3227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iyevleva AG, Aleksakhina SN, Sokolenko AP, Baskina SV, Venina AR, Anisimova EI, et al. Somatic loss of the remaining allele occurs approximately in half of CHEK2-driven breast cancers and is accompanied by a border-line increase of chromosomal instability. Breast Cancer Res Treat 2022;192:283–91. [DOI] [PubMed] [Google Scholar]
- 40. Vittal A, Saha D, Samanta I, Kasi A. CHEK2 mutation in a patient with pancreatic adenocarcinoma-a rare case report. AME case reports 2021;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detail description of the functional categorization of analyzed CHEK2 missense variants.
List of all analyzed CHEK2 variants with results of KAP1/CHK2 kinase and localization assays and the results from recent previously published functional analyses of the CHEK2 VUS.
Characteristics of 12 case-control datasets from the ENIGMA consortium partners.
Frequencies of all reported germline CHEK2 variant carriers and carriers of variants concordantly categorized by functional our kinase assays in breast cancer patients and controls in 12 analyzed population datasets.
Data Availability Statement
All frequency data of CHEK2 VUS provided by the collaborating centers, kinase and localization assay data used to generate the figures' graphs and plots are provided in Supplementary Tables S1–S3. Raw data from high-content microscopy experiments are available upon request from the corresponding author.