Comprehensive characterization of mast cells in lung adenocarcinoma elucidates their heterogeneity and identifies interplay between mast cells and CCR2+ T cells that is associated with a favorable prognosis.
Abstract
Mast cells constitute indispensable immunoregulatory sentinel cells in the tumor microenvironment. A better understanding of the regulation and functions of mast cells in lung adenocarcinoma (LUAD) could uncover therapeutic approaches to reprogram the immunosuppressive tumor microenvironment. Here, we performed flow cytometry and single-cell RNA sequencing (scRNA-seq) of patient LUAD samples to comprehensively characterize LUAD-infiltrating mast cells. Mast cells exhibited functional heterogeneity and were enriched in LUAD with ground-glass opacity features (gLUAD). The mast cells in gLUAD exhibited proinflammatory and chemotactic properties while those in radiologically solid LUAD (sLUAD) were associated with tumor angiogenesis. Mast cells were an important source of CCL2 and correlated with the recruitment of CCR2+ CTL, a specific subcluster of preexhausted T cells with tissue-resident memory phenotype and enhanced cytotoxicity. Increased infiltration of mast cells and CCR2+ CTLs and their colocalization showed a strong association with favorable prognosis after surgery but were not associated with improved survival after chemotherapy. Collectively, these findings reveal a key role of mast cells in LUAD and their potential cross-talk with CTLs, suggesting that targeting mast cells may be an immunotherapeutic strategy for LUAD.
Significance:
Comprehensive characterization of mast cells in lung adenocarcinoma elucidates their heterogeneity and identifies interplay between mast cells and CCR2+ T cells that is associated with a favorable prognosis.
Introduction
Lung cancer progression is the leading cause of cancer-related mortality, and lung adenocarcinoma (LUAD) is the most common histologic subtype of lung cancer (1). The progression of LUAD is thought to follow multistep progress: pure ground-glass opacity (pGGO) progresses to mixed ground-glass opacity (mGGO: the emergence of a radiologically solid component), which eventually progresses to solid LUAD (sLUAD; refs. 2, 3). We have elucidated the genomic and transcriptional dynamics associated with this progression in early-stage LUAD (4) and found that GGO featured LUAD (gLUAD) is a unique clinical LUAD subtype (5) that is characterized by a relatively indolent nature and favorable prognosis (6–8). However, the heterogeneity of the tumor microenvironments between gLUAD and sLUAD remains largely unexplored (9, 10).
The tumor microenvironment is a complex ecosystem (11, 12) in which tumor-infiltrating myeloid cells constitute a key component of the immune cell population (13, 14), including mast cells (MC; ref. 15). For a long time, the culprit role of MCs in allergic diseases, and the concomitant lack of knowledge about their functions in the tumor microenvironment, prevented MCs from becoming a focus of research in the field of tumor immunology (16, 17), although robust accumulation of MCs has been observed in many cancer types (18–20). This situation has changed dramatically over the past decade, as many functions of MCs have begun to be revealed, including functions in tumor angiogenesis and microenvironment remodeling (21, 22). However, the underlying mechanism by which MCs function in tumor microenvironment is poorly understood (23), and no research has focused on the heterogeneity and function of MCs in the LUAD tumor microenvironment.
Here, by combining high-dimensional flow cytometry (FCM), single-cell RNA sequencing (scRNA-seq), multi-immunofluorescence (mIF), and functional experiments, we comprehensively elucidated the heterogeneity, phenotypes, functions, spatial distributions, and metabolic activities of LUAD infiltrating MCs. MCs in gLUAD (gMC) exhibited proinflammatory and chemotactic properties, while sLUAD-infiltrating MCs (sMC) were associated with angiogenesis. Our results confirmed that tumor progression decreased the recruitment of MCs. Our results also revealed the prognostic value of MCs, as well as the role of MCs in predicting the response to tyrosine kinase inhibitor (TKI) therapy but not chemotherapy. MCs were the primary cellular source of CCL2 in gLUAD, and increased MCs numbers correlated with an increase in the infiltration of highly cytotoxic CCR2+ CTLs via the CCL2–CCR2 axis.
Materials and Methods
Patient cohorts and cell lines
Tissue microarray (TMA) cohort was constructed using specimens from two independent cohorts of patients with LUAD (n = 162 and n = 163, respectively) at Fudan University Shanghai Cancer Center (Shanghai, China) from January 2011 to April 2013. All samples from patients with LUAD were prereviewed histologically by hematoxylin and eosin (H&E) staining, and representative areas with small round lymphocyte infiltrate were premarked in the paraffin blocks, away from necrotic and hemorrhagic materials. TMA was constructed on slides with sections 4 mm in thickness and 1.5 mm in diameter.
The flow cytometry cohorts included blood, tumor, and paired normal lung tissues, which were obtained from Fudan University Shanghai Cancer Center (Shanghai, China) from September 2020 to December 2021. We used the flow cytometry cohorts to profile the immune landscape of LUAD (FCM profiling cohort) and study the phenotype and function of CCR2+ CTLs (FCM CCR2 cohort). The detailed information of two flow cytometry cohorts is provided in Supplementary Table. S1.
The data of TKI cohort was downloaded (NCBI BioProject #PRJNA591860; ref. 24) to study the correlation between MC infiltration and TKI treatment response. The detailed information is provided in the Supplementary Table S1 (TKI cohort sheet).
GFP-A549 cells were a gift from Wu Liunan (Nanjing Medical University, Nanjing, China; on October 23, 2021). In accordance with the Declaration of Helsinki, human LUAD specimens in this study were collected after obtaining written informed consent and were approved by the ethics committee and Institutional Review Boards (IRB) of Fudan University Shanghai Cancer Center (IRB20082239).
Flow cytometry and cell sorting
Tumor and paired normal lung tissues were dissected, mechanically minced, treated with RPMI1640 supplemented with collagenase IV (1 mg/mL; Gibco) and DNase I (100 μg/mL, Sigma) for 1 hour at 37°C and filtered to single-cell suspension (70 μm, Corning). Fresh peripheral blood samples were enriched by Lymphoprep density gradient centrifugation, washed, and resuspended in PBS to gain peripheral mononuclear cells. Live/dead staining was performed using the Zombie Yellow Fixable Viability Kit (BioLegend). For extracellular staining, antibodies were incubated with cells for 20 minutes in FACS buffer. For intracellular staining, cells were fixed with the FOXP3/Transcription Factor Staining Buffer Set (eBiosciences) and then stained with intracellular antibodies, as per the manufacturer's instructions. For the evaluation of in vitro cytokine production, isolated tumor-infiltrated CTLs plated with PMA 50 ng/mL plus ionomycin 1 mg/mL plus Brefeldin A 3.0 mg/mL in 96-well plates and then stained as described above. Data were collected using a LSR Fortessa (BD Biosciences) and analysis was performed using FlowJo soft (v.10.5.3, TreeStar). A complete list of antibodies can be found in Supplementary Data (Supplementary List S1).
For the sorting of CTLs, CCR2+CTLs, and CCR2-CTLs, single-cell suspension from tissues or peripheral blood were stained with PE-CCR2 (K036C2, BioLegend), PECy5-CD3 (UCHT1, BioLegend), FITC-CD4 (SK3, BioLegend), APCFire750-CD8a (HIT8a, BioLegend), APC-TCRVa7.2 (3C10, BioLegend) for 20 minutes at 4°C and sorted on a BD FACSAria II (BD Biosciences) into CD3+CD8+, CCR2+CD8+ and CCR2-CD8+ T cells. The gating strategy of flow sorting is shown in the Supplementary Fig. S1A. The purity of sorted CTLs is shown in the Supplementary Fig. S1B.
For the sorting of MCs (25), single-cell suspension from tissues was first enriched by magnetic CD117 bead separation according to the manufacturer's instructions (Miltenyi Biotec). To achieve final purity, the enriched cells were stained with specific antibodies (CD45, CD117, FceRIa, CD3/CD19/CD56/CD66b) and 4′,6-diamidino-2-phenylindole (DAPI). Live CD45+Lin-CD117+FceRIa+ MCs were sorted with BD FACSAria II (BD Biosciences). The flow plots (before magnetic bead enrichment; MACS, after MACS, after flow sorting) are shown in the Supplementary Fig. S1C.
Multiplex IF staining and analysis
Briefly, lung sections or TMA sections were deparaffinized and rehydrated successively, followed by microwave antigen retrieval and incubation in 3% H2O2 to block endogenous peroxidase activity. After preincubation with 10% normal goat serum (NGS), sections were serially stained with primary antibodies and secondary antibodies. A complete list of antibodies can be found in Supplementary Data (Supplementary List S1). The signal for antibody complexes was visualized by their corresponding Opal Fluorophore Reagents (PerkinElmer). The negative and positive controls for CCR2, CCL2, TNFα, and VEGFA are shown in the Supplementary Fig. S1D.
Slides were scanned using the PerkinElmer Vectra3 platform at 20× resolution and analyzed by inform (v2.3, PerkinElmer using machine learning algorithm with a visual cutoff followed by single-cell–based mean pixel fluorescence intensity to achieve accuracy and calculate. For the quantitative analysis, cells that were higher than the threshold of both CCL2 and tryptase were considered CCL2+ MCs and cells that were higher than the threshold of tryptase but lower than the threshold of CCL2 were considered CCL2− MCs. The quantification of positively stained cells and spatial distance analysis were processed by R script. For the spatial distance analysis, we calculated the number of nearest neighbor cells (within the circular area with a radius of 20 μm) on the assigned coordinates of each cell and by R script using the spatstat package.
Twenty-one patients were censored because these tissues in the TMA cores fell off in whole or part, and only 304 patients were included in the final analysis. The detailed information is provided in the Supplementary Table S1 (TMA cohort sheet). Among the 304 TMA cohort, only patients who received adjuvant chemotherapy were enrolled in the chemotherapy cohort (Supplementary Table S1, chemotherapy cohort). In addition, because the gene mutations were not detected in all the tissues, the number of EGFR cohort, KRAS cohort, and p53 cohort are 303, 299, and 238.
scRNA-seq and analysis
We obtained scRNA-seq data from two published studies. For one study (10), one GGO scRNA-seq data was censored because of failure of successful download. For the other study (26), we excluded two lung squamous cell carcinomas (SCC). A total of 28 scRNA-seq data [6 normal lung tissues (nLung), 15 GGO, and 5 solid LUAD] were enrolled into the final analysis. To validate our key results, four nLung and five pathologically diagnosed solid LUAD samples were collected at our department. Primary tumor tissue samples were transported in ice-cold tissue storage solution (Miltenyi) immediately after surgical resection and the single-cell suspensions were prepared as described above. Single-cell suspensions were suspended with ice-cold ACK lysis buffer (Thermo Fisher Scientific), filtered with a 40-μm nylon mesh, and then were enriched for CD117+ cells by magnetic bead-separation according to the manufacturer's instructions (Miltenyi Biotec). Because of the limitation of GGO volume and cell counts after digestion, five GGO samples were not enriched by CD117+ microbeads. All the samples were sequenced by 10× Chromium single-cell platform (10× Genomics). The patients’ information was available in the Supplementary Table. S1F (scRNA-seq cohort).
Raw data were processed using Cell Ranger V3 and then aligned to human reference genome (GRCh38). Further quality control was performed on the basis of the total UMI count (>2,000), the number of detected genes (200–5,000) and proportion of mitochondrial gene count per cell (<10%), and proportion of mRNA coding protein (>80%). After normalizing the gene expression matrices by NormalizeData function in Seurat package, we processed single-cell data for dimension reduction and unsupervised clustering to project cells into two-dimensional space by Uniform Manifold Approximation and Projection (UMAP), and the FindAllMarkers function in Seurat was used to classify and annotate on the basis of expression of canonical cell type markers.
To identify differentially expressed genes between two MCs clusters, we used Wilcox. Test in R to evaluate the significance of each gene and set Padj = 0.05 as the cut-off value. We next performed the pathway enrichment analysis of those differentially expressed genes by using clusterProfiler V3.19.0 [enricher function, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets, or MCs activation gene sets]. To delineate the metabolic landscape of MCs, we performed scMetabolism designed to easily quantify single-cell metabolic activity (https://github.com/wu-yc/scMetabolism).
To infer the differentiation trajectory of MCs, we used monocle (V2.14.0) to predict the pseudotime of each MCs (method = “DDRTress”, ordering_genes = marker genes). We used CellPhoneDB to infer cell–cell interaction between MCs and lymphocytes. We selected the significantly differential crosstalk ligand–receptor pairs and used Circlize (V0.4.10) to visualize the results.
RNA-seq and The Cancer Genome Atlas data
The bulk RNA-seq data presented in this study was from our previously published studies (4, 27). Briefly, the study cohort contains RNA-seq data from tumor and matched normal tissues of 197 patients with LUAD. Of them, 150 samples with RNA integrity number (RIN) ≥ 5.0 were included in the final analysis, including 16 adenocarcinomas in situ (AIS), 52 minimally invasive adenocarcinomas (MIA) and 82 invasive adenocarcinomas (LUAD). For the study of the transcriptional correlation between MCs (TPSAB1) and other immune cells, data from all 150 tumor samples and paired adjacent normal tissues were analyzed. For survival analysis, only 82 patients with LUAD were included because all AIS/MIA patients had 100% recurrence-free survival after complete surgery. Raw data can be found at the European Genomephenome Archive (EGA) with the accession number EGAS00001004006 (https://ega-archive.org/studies/EGAS00001004006). Besides, we also fetched bulk RNA-seq data of patients with LUAD from the Gene Expression Omnibus (GEO) under accession number GSE111907 for independent validation.
MC stimulation and cytokine detection
MCs sorted from GGO or solid LUAD were resuspended in cell culture medium (StemPro-34 culture media, GIBCO) supplemented with glutamine (2 mmol/L), penicillin (100 U/mL), streptomycin (100 μg/mL), and human SCF (100 ng/mL, STEMCELL Technologies) at a concentration of 1×106 cells/mL and aliquoted into two groups (stimulated group and unstimulated group) in the 48-well plates. The stimulated group was ready for FcεRI-mediated stimulation by IgE sensitization (100 ng/mL, 12 hours, MCE). Cells were washed twice (300 × g, 10 minutes, room temperature) to remove excess of IgE cell and sensitized overnight with 100 ng/mL human biotinylated IgE (G7–26, BD Biosciences; refs. 28, 29). As a control, IgE and subsequent biotinylated IgE were not added in unstimulated group. After incubation, the supernatants were pipetted into 1.5 mL microcentrifuge tubes and centrifuged (14,000 × g, 10 minutes). The cell-free supernatants from stimulated and unstimulated groups were both collected and stored at −80°C until assayed. The cytokine or chemokine contents were measured by Luminex according to the manufacturer's instructions. Forty-five cytokines were detected at the same time and data were provided in the Supplementary Table S2, including BDNF, EGF, Eotaxin, FGF-basic, GM-CSF, GROα, HGF, IFNγ, IFNα, IL1RA, IL1β, IL1α, IL2, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12 p70, IL13, IL15, IL17A, IL18, IL21, IL22, IL23, IL27, IL31, IP10, LIF, MCP1, MIP1α, MIP 1β, NGFβ, PDGF BB, PLGF, RANTES, SCF, SDF1α, TNFα, TNF β, VEGFA, VEGFD.
Transwell migration assay
Migration assays were performed with Transwell cell culture inserts with a 5-μm pore size (Corning 3421). Fresh LUAD tissues were minced into 2 mm3 pieces in 1.5 mL tube, centrifuged at 10,000 × g for 10 minutes to gain the supernatant liquid, which were then filtered through a sterile 0.22-μm syringe filter to harvest the tumor supernatants (TS). Cell culture medium (Blank group), TS, unstimulated MCs (Usti-MC group), and stimulated MCs (Sti-MC group) according to previous description were added to the bottom compartments of the Transwell migration chamber (600 μL). CTLs from peripheral blood (1 × 105/100 μL) were stimulated with T-Activator CD3/CD28 Dynabeads (11131D, Gibco by Thermo Fisher Scientific) and then were added to the top compartments. After 4-hour incubation, cells in the bottom well were collected, stained with anti-CD3, anti-CD4, and anti-CD8 antibodies and analyzed by flow cytometry. The migration rate was calculated by the total number of CTLs dividing by the number of migrated CTLs in the bottom well.
In vitro migration blocking assay
To identify the potential role of the CCL2/CCR2 axis in mediating the migration of CTLs, sorted MCs were preincubated with or without the CCL2 antagonist (Carlumab, CNTO 888, 30 ug/mL, MCE; ref. 30) or CCR2 antagonist (INCB3344, 15 nmol/L, Selleck Chemicals; ref. 31) for 30 minutes after adequate stimulation as mentioned above. Then, they were seeded in the bottom compartments (600 μL). CCR2+ or CCR2− CTLs from peripheral blood were aliquoted to 96-well U-bottom plates (1 × 105/100 μL per well), stimulated with T-Activator CD3/CD28 Dynabeads (2 μL per well, 11131D, Gibco by Thermo Fisher Scientific) and were then added to the top compartments (5 μm, pore size). After 4-hour incubation, cells in the bottom compartments were collected, stained with anti-CD3, anti-CD4, and anti-CD8 antibodies, and counted by flow cytometry analysis. The migration rate was calculated by the total number of CTLs dividing by the number of migrated CTLs in the bottom well.
Cytotoxicity assay
CCR2+ or CCR2− CTLs from LUAD were aliquoted to 96-well U-bottom plates (1 × 105/100 μL per well) and stimulated with T-Activator CD3/CD28 Dynabeads (11131D, Gibco by Thermo Fisher Scientific) for 24 hours (32). GFP-A549 cells were seeded into 96-well plates (1×105/well) in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and cocultured with no CTLs, CCR2− CTLs, or CCR2+ CTLs at ratios of 1:1. After incubation for two days, cells were harvested and tumor cell line survival and apoptosis were evaluated by Annexin V (APC) and propidium iodide (PI) according to manufacturer's instructions.
Statistical analysis
Statistical analysis was performed with R (v3.6.2), SPSS (v22, IBM), and Prism 9.0 software. The statistical tools, methods, and threshold for each analysis are explicitly described with the results or detailed in the figure legends or Materials and Methods.
Data and code availability
The scRNA-seq data analyzed in this study is available at Sequence Read Archive (SRA) database under accession number SRP438272 as an NCBI BioProject #PRJNA973717. Public scRNA-seq data are accessible at the GSA of the BIG Data Center, Beijing Institute of Genomics (http://bigd.big.ac.cn/gsa-human) under accession number HRA000154 and the NCBI GEO depository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127465) under the accession number GEO: GSE127465. TKI cohort data are available as an NCBI BioProject (#PRJNA591860). RNA-seq data from our center are available at the European Genomephenome Archive (EGA) with the accession number EGAS00001004006 (https://ega-archive.org/studies/EGAS00001004006) and external RNA-seq data were available from the GEO under accession number GSE111907. Data and code used for analysis are available as a Code Ocean capsule (https://codeocean.com/capsule/6874338/tree/v1). All other raw data are available upon request from the corresponding author.
Results
MCs are enriched in gLUAD and associated with a favorable prognosis
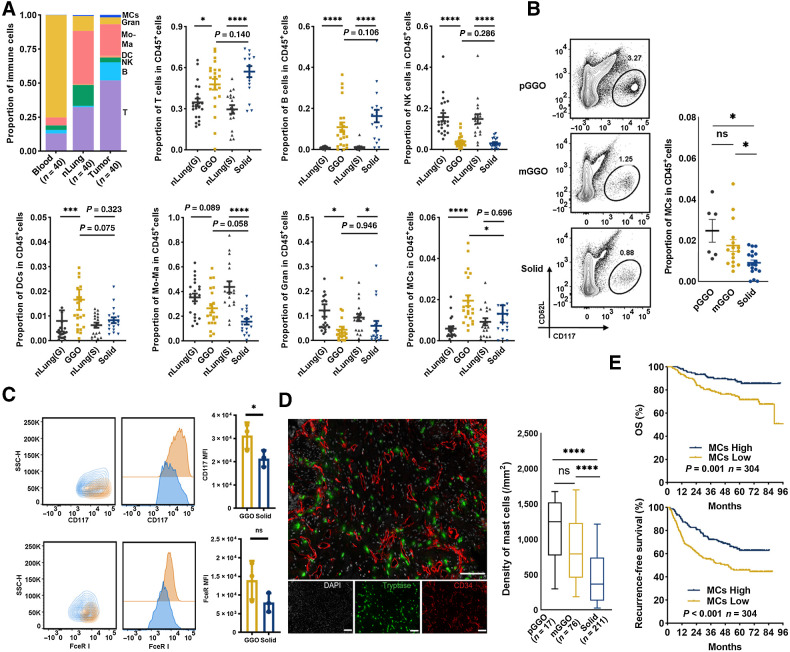
To characterize the immune cell landscape in LUAD, we employed high-dimensional FCM to study a prospective cohort of patients with LUAD undergoing surgical resection for curative intent [120 samples: 40 paired peripheral blood, nLungs, and LUAD samples; Supplementary Table S1], and we identified seven major immune subtypes (Fig. 1A): T cells, B cells, natural killer (NK) cells, dendritic cells (DC), monocytes-macrophages (Mo-Ma), granulocytes (Gran), and MCs. The gating strategy is shown in Supplementary Fig. S2A. Compared with paired nLung samples, LUAD samples were infiltrated with higher numbers of T cells, B cells, and MCs, while the infiltration of NK cells was decreased (Fig. 1A). gLUAD samples were infiltrated with a higher percentage of MCs than sLUAD, which was not observed for other immune cells (Fig. 1A). We next focused on MCs as their role in LUAD has not been studied (33).
Figure 1.
The accumulation and prognostic significance of LUAD-infiltrating MCs. A, Flow identification of LUAD immune cell composition (gating strategy shown in Supplementary Fig. S1A) and quantification as a proportion of CD45+ cells (n = 40 for peripheral blood, nLung, and LUAD). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; Mann–Whitney. B, Representative flow plots showing identification of MCs by FCM (left, additional replicates and full gating in Supplementary Fig. S1C), proportion of pGGO-, mGGO-, or sLUAD-infiltrating MCs in CD45+ cells (right). *, P < 0.05; Mann–Whitney. C, Representative CD117 and FcεRI expression of gMCs (SSClowCD117hiFcεRIhi; yellow) and sMCs (SSChiCD117lowFcεRIlow; blue). D, Two-plex staining panel showing the distribution of MCs (left) and MCs quantification as density (cell counts per mm2; right). ****, P < 0.0001; two-tailed unpaired t test. E, The prognostic value of MCs in surgically resected LUAD by mIF. P values were determined by log-rank test, and patients were stratified by median values. ns, nonsignificant.
LUAD is thought to follow a multistep progression, from pGGO to mGGO, and eventually to sLUAD (Supplementary Fig. S2B), thus, we evaluated the dynamics of MCs infiltration during this process. The infiltration of MCs, defined as CD45+Lin-CD117+FcεR I+ CD62low (Supplementary Fig. S2C), decreased during this process, with pGGO samples containing the highest proportion of MCs and sLUAD containing the lowest proportion of MCs (Fig. 1B). Interestingly, gMCs exhibited higher fluorescence intensity of CD117, and sMCs had higher side-angle light scatter (SSC) than gMCs (Fig. 1C). Furthermore, gMCs had higher fluorescence intensity of HLA-DR and CD80 while lower fluorescence intensity CD86 and PD-L1 than sMCs (Supplementary Fig. S3A), suggesting heterogeneity between gMCs and sMCs.
We then experimentally validated the presence and the increase in MCs fraction in gLUAD by IHC staining on a large LUAD cohort. Consistent with the FCM results, LUAD progression (pGGO-mGGO-solid) was associated with decreased accumulation of MCs (Fig. 1D). Consistent with previous evidence (34), most MCs were located proximal to blood vessels (Fig. 1D). Besides, we found that there was an inverse correlation between MC density and tumor size (R = −0.35, P < 0.0001; Supplementary Fig. S3B and S3C).
In light of the less “aggressive” nature and favorable prognosis of gLUAD, we then investigated the prognostic value of MCs in surgically resected LUAD. A higher density of MCs infiltration was correlated with prolonged overall survival (OS) and recurrence-free survival in LUAD cohort with 304 patients by IF staining (Fig. 1E). Furthermore, we validated their prognostic role in another RNA-seq cohort (n = 82) from our department and external The Cancer Genome Atlas cohort (n = 404). The level of MCs marker (TBSAB1: encodes MCs-specific tryptase) was correlated with favorable survival in two individual cohorts (Supplementary Fig. S3D and S3E). In summary, enhanced MCs abundance was observed in gLUAD and associated with favorable survival in LUAD.
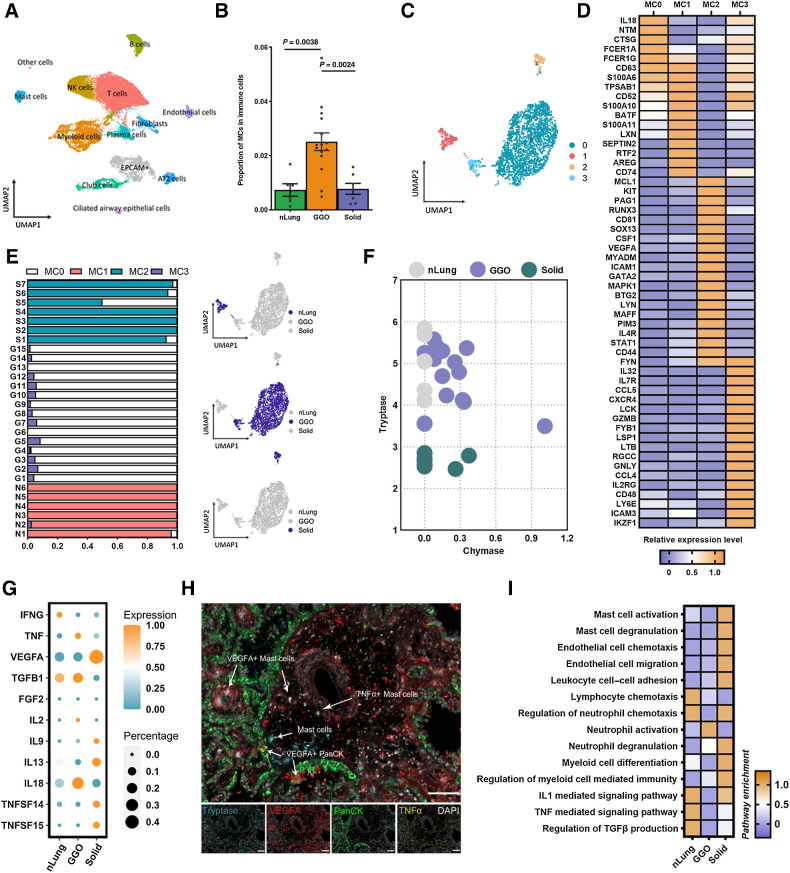
scRNA-seq reveals heterogeneous MCs in LUAD
Next, to delineate the heterogeneity of LUAD infiltrating MCs, we obtained scRNA-seq data from 28 samples (6 nLung, 15 gLUAD, and 7 sLUAD; refs. 10, 26), focusing on MCs (Fig. 2A; Supplementary Table S3), after strict quality control and filtration. Consistent with results from FCM and mIF, gLUAD showed a preference for the accumulation of MCs (Fig. 2B). Integrating the SingleR, manual marker-based annotation, and unbiased clustering of gene expression, we identified four distinct MCs clusters (Fig. 2C), designated as MC0 to MC3. All MCs clusters strongly expressed the MC-specific proteases tryptase (TPSAB1 and TPSB2) and CPA3, while the expression of chymase was observed only in a fraction of MC0 and MC3 (Supplementary Fig. S4A).
Figure 2.
Transcriptional heterogeneity of LUAD-infiltrating MCs. A, UMAP depiction of clustered cell populations from nLung (n = 6), GGO (gLUAD; n = 15), and solid (n = 7) samples. Each dot corresponds to a single cell, and the dots are colored according to cell type. B, Quantification of identified MC population as a proportion of the immune cell population. C, UMAP projection of four identified MC clusters derived from nLung, gLUAD, and sLUAD. D, Log-fold change (logFC) in the expression of genes that are significantly enriched in each identified MC cluster. E, Identification of tissue source (nLung, gLUAD, and sLUAD) and proportion of each MC cluster. F, MC-specific canonical protease expression in MCs derived from nLung, gLUAD, and sLUAD samples. G, Mean and percent expression of the cytokines in MCs from nLung, gLUAD, and sLUAD samples. H, Representative example of a LUAD section stained by mIF with anti-TPSAB1 (blue), anti-PanCK (green), anti-TNF (yellow), and anti-VEGFA (red) antibodies. Arrows, specific cell types. I, Enrichment (z scores) of differentially expressed genes in MCs from nLung, gLUAD, and sLUAD samples by GO or KEGG analysis.
Thorough examination of the transcriptional signature and differential gene expression results (Fig. 2D; Supplementary Table S4) indicated that MC0 cells were characterized by high expression of IL18, high-affinity IgE receptors (FCERIA, FCER1G), and the MCs protease-encoding transcript CTSG, suggesting a proinflammatory phenotype. The MC1 cluster closely resembled quiescent MCs (35), a cluster of MCs with a TPSAB1hiAREGhi phenotype; the AREG gene encodes a protein that maintains epithelial homeostasis. MC2 was characterized by high expression of VEGFA (Fig. 2D), which is a key gene that is associated with MC-derived tumor angiogenesis. In particular, MC2 was KIThiFCER1Alo phenotype, which is associated with MC proliferation in active eosinophilic esophagitis (35). In addition, the MC2 cluster was enriched for IL4R, which encodes the receptor for the Th2 cytokine IL4, in addition to RUXN3, GATA2, and STAT1 (Fig. 2D), although the functional relevance is unknown. It is noteworthy that MC3 represented a cluster of chemotactic MCs, marked as higher expression of chemokines (CCL4, CCL5) and adhesion molecules (CD48, ICAM3). We then validated the existence of four MC transcriptional states in our own scRNA-seq data. We identified five subclusters of MCs and named them MC-New1 to MC-New5 (MC-N1 to MC-N5, Supplementary Fig. S4B). MC-N1 was main from GGO and MC-N3 was main from nLung. We then performed cluster analysis combining previous four MC clusters and new five MC clusters. We found that MC-N5 and chemotactic MC3 were clustered together, which were characterized by high expression of CCL2, CCL4, and CCL5 (Supplementary Fig. S4C). MC-N1, which was close to MC0, highly expressed IL18, high-affinity IgE receptors (FCERIA and FCER1G), resembling previously identified proinflammatory MCs. MC-N2 was TPSAB1hiAREGhi, which may resemble quiescent MCs like MC1. MC-N3 and MC2 were characterized by high expression of VEGFA (Supplementary Fig. S4C), which was associated with tumor angiogenesis.
To evaluate the underlying origin and cellular differentiation of the MC clusters, we performed Monocle2 pseudotime analysis, which revealed a ternary branched structure and confirmed the direct stratification of the differentiation trajectory of the MC subclusters (Supplementary Fig. S4D): MC0 and MC1 represented the trunk and quiescent end, MC2 represented the end state of angiogenic MCs, and MC3 represented the end state of chemotactic status. These results indicated that LUAD-infiltrating MCs were profoundly affected by their local microenvironment, which endowed them with distinct developmental trajectories and phenotypes; this finding was consistent with the development and maturation of MCs occurring locally (36). Collectively, we identified four heterogeneous MCs clusters with distinct transcriptional characteristics and developmental trajectories: proinflammatory MC0, quiescent MC1, angiogenic MC2, and chemotactic MC3.
gMCs and sMCs exhibit distinct phenotypes and cytokine secretion patterns
The maturation and function are largely shaped by the local microenvironment, so we first examined the tissue source of the four identified MCs clusters. Interestingly, MC clusters exhibited remarkable tissue specificity, with quiescent MC1 mainly found in nLung samples, gLUAD samples containing proinflammatory MC0 and chemotactic MC3, and angiogenic MC2 mainly found in sLUAD samples (Fig. 2E). This finding inspired us to further examine whether distinct tissue-derived MCs belonged to different MCs subtypes: the MCT subset (mucosal MCs, expressing tryptase alone) and the MCTC subset (connective tissue MCs, expressing both tryptase and chymase; ref. 37). As expected, MCs from nLung samples belonged exclusively to the MCT subset (Fig. 2F), which was consistent with the commonly assumed lung-resident MCs phenotype. However, it was interesting to note that gMCs predominantly belonged to the MCTC subset, while most sMCs were classified into the MCT subset (Fig. 2F).
The spectrum of activation receptor expression, mediator release, and cytokine secretion, which is key to determining MCs function, is shown in Fig. 2G and Supplementary Fig. S4E and S4F. gMCs secreted more TNF and IL18 while less VEGFA and IL13 than sMCs (Fig. 2G), while no significant difference was observed in the secretion of other cytokines (Supplementary Fig. S4F). TNF derived from MCs and IL18 have been reported to enhance antitumor immunity through the regulation of DC functionality and T-cell priming by TNF (38) and through the stimulation of innate lymphocytes and antigen-experienced, but not naïve T cells by IL18 (39). Conversely, MC-derived VEGFA plays a critical role in tumor angiogenesis (34, 40), and the immunosuppressive role of IL13 is reported to involve skewing the antitumor response towards a Th2 response, thus suppressing Th1 immune responses (41). These results revealed the contrasting role of gMCs and sMCs. A higher ratio of TNF+/VEGFA+ MCs has recently been reported to define an antitumor MCs phenotype and correlates with a better prognosis in nasopharyngeal cancer (33). As expected, a higher ratio of TNF+/VEGFA+ was observed in gMCs than sMCs (Supplementary Fig. S4G), indicating the antitumor role of gMCs. IHC staining confirmed the existence of VEGFA+ MCs and TNF+ MCs (Fig. 2H). Besides, we found that MCs were located both in the tumor foci and perivascular tumor stroma (Fig. 2H). We then validated the secretion of TNFα and VEGFA by MCs using cytokine secretion detection. sMCs expressed more VEGFA than gMCs and less TNFα (Supplementary Fig. S4H, although the statistical difference was not significant for TNFα). Together, these findings suggest that gMCs and sMCs belonged to different MCs subsets that exhibited huge differences in receptor expression, mediator secretion, and cytokine secretion. In particular, gMCs were proinflammatory and chemotactic phenotype and sMCs were associated with tumor angiogenesis.
gMCs and sMCs exhibit distinct functions and metabolic activities
To better understand the functional differences of MCs imprinted by the microenvironment of distinct LUAD types, we used pathway enrichment analysis (Supplementary Table S5). This revealed that sMCs exhibited upregulation of genes involved in MC activation and degranulation (Fig. 2I), which was consistent with our in situ observations (Fig. 1D). In addition, we observed that sMCs were associated with endothelial cell chemotaxis and migration, neutrophil chemotaxis and degranulation, and regulation of TGFβ production, while gMCs were involved in lymphocyte chemotaxis (Fig. 2I). These results indicated that gMCs and sMCs play different immunomodulatory roles in the lung cancer microenvironment. According to computed classic functional scores, sMCs harbored the highest scores of immunosuppression, antigen processing and presentation, inflammation, phagocytosis, interleukin signaling pathway, and complement, in contrast to gMCs and MCs in nLung (Supplementary Fig. S4I; Supplementary Table S5). This may, to some degree, indicate that sMCs actively participate in tumor-associated inflammation and immune suppression.
Furthermore, we investigated the transcriptional genes involved in MC metabolism between gMCs and sMCs by ssMetabolism (Supplementary Table S6; ref. 42). MCs from nLung, gLUAD, and sLUAD samples were distinguished by large differences in metabolic activity. gMCs exhibited higher activity of oxidative phosphorylation and the citrate cycle (TCA cycle), while sMCs displayed a strong metabolic preference for lipid and fatty acid and sulfur metabolism (Supplementary Fig. S4J). Collectively, these results revealed functional and metabolic differences between gMCs and sMCs.
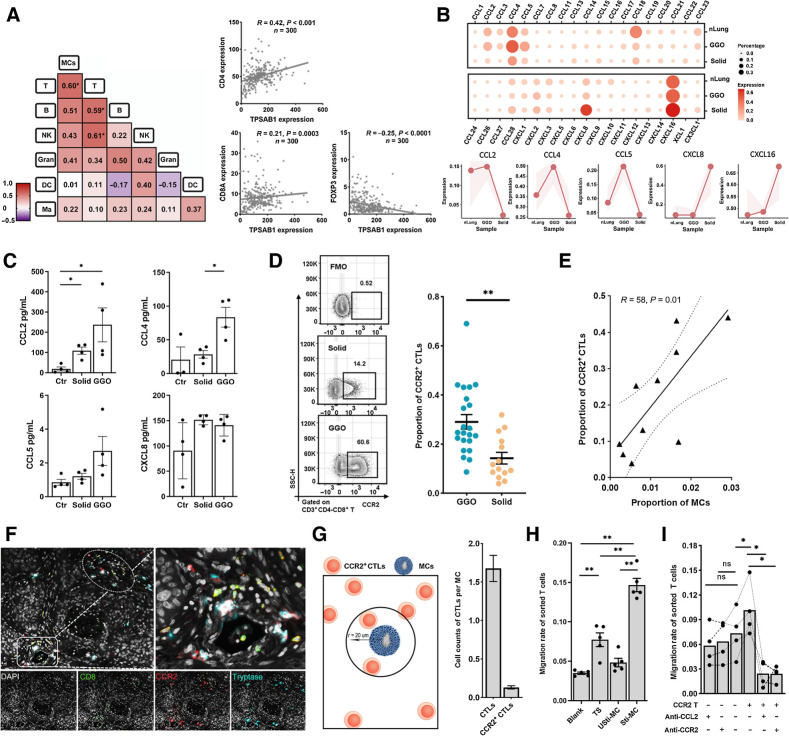
MCs are associated with the enrichment of CCR2+ CTLs in LUAD
As gMCs expressed a large number of chemokines and were associated with lymphocyte chemotaxis, we then explored whether they are potentially responsible for the recruitment of other immune cells in LUAD. We first assessed the quantitative correlation between tumor-infiltrating MCs and other immune cell subpopulations by FCM. T cells were the only subpopulations that correlated with MCs (Fig. 3A), indicating the relevance of their infiltrating levels. This observation was also validated by the positive correlation between the expression levels of MC markers (TPSAB1) and T-cell markers (CD4 and CD8A), while the negative correlation of the transcriptional levels of MC markers and Treg markers (FOXP3) by the analysis of bulk RNA-seq data from a cohort of 300 patients diagnosed with LUAD (Fig. 3A; Supplementary Table S7). These findings indicated the potential interaction of MCs with T cells in LUAD. We then performed the ligand-receptor–based MCs–lymphocytes crosstalk analysis and found that MCs interact most closely with the ZNF683+ effector T-cell subpopulation (Supplementary Fig. S5A), which are memory-like precursors that are defined by their single-cell transcriptional state (43). Interestingly, CCR2 was exclusively expressed in ZNF683+CD8+ T cells (Supplementary Fig. S5B).
Figure 3.
MCs correlate with the infiltration of CCR2+ CTLs. A, Left, heat map showing the Pearson correlation of the immune cell population by FCM. Number representing the Pearson correlation index. Right, the transcriptional correlation of the transcriptional levels of MC marker (TPSAB1) and T-cell markers (CD4, CD8A, and FOXP3). B, The expression of chemokines in MCs from nLung, gLUAD, and sLUAD samples. The shaded areas represent the upper quantile and lower quantile. C, The secretion of CCL2, CCL4, CCL5, and CXCL8 by MCs detected by Luminex. Ctr, unstimulated MCs; solid, stimulated MCs from solid LUAD; GGO, stimulated MCs from GGO. D, Representative flow plots showing CCR2+ CTLs gating (left) and proportion of CCR2+ CTLs among tumor-infiltrating CTLs (right). FMO, fluorescence minus one. n = 36 (Mann–Whitney). E, Scatterplot showing the Pearson correlation of the proportion of CCR2+ CTLs (divided by the total CTL number) and MCs (divided by the total CD45+ cells) by FCM analysis (n = 10). F, Three-plex staining panel showing the spatial distribution of CCR2+ CTLs and MCs. G, Spatial analysis of the relationship between MCs and CTLs. Left, depiction of methodology for spatial analyses performed. The number of CTLs close to per MCs. H, The migration rate of CTLs (the ratio of migrated CTLs to total CTLs) in the transwell assay (n = 6, paired t test). Blank group, medium only; TS group, tumor supernatants; Usti-MC group, nonstimulated MCs; and Sti-MC group, stimulated MCs. I, The migration rate of CTLs (the ratio of migrated CTLs to total CTLs) in the migration blocking assay (n = 4, paired t test). CCR2 T−, CCR2− CTLs; CCR2 T+, CCR2+ CTLs. *, P < 0.05; **, P < 0.01; ns, nonsignificant.
As chemokines control immune cell trafficking into tumor, we then examined the profile of chemokine expression in LUAD-infiltrating MCs. Both gMCs and sMCs expressed a plethora of chemokines, especially CCL4, CCL18, and CXCL16 (Fig. 3B). Interestingly, gMCs ranked highest in the expression of the proinflammatory chemokines CCL2, CCL4, and CCL5 (Fig. 3B). Conversely, sMCs secreted more CXCL8 and CXCL16 (Fig. 3B). CXCL8 was reported to be responsible for the recruitment of immunosuppressive neutrophils and MDSCs by binding to CXCR1 and CXCR2 (44, 45) and promoting tumor angiogenesis by interacting with VEGFA. However, sLUAD samples were not found to be infiltrated with a higher proportion of neutrophils (Supplementary Fig. S5C), and the abundance of infiltration was not relevant (Fig. 3A), although previous evidence suggested that MCs may regulate neutrophil recruitment (46). We then validated these chemokines by Luminex detection (Fig. 3C). We confirmed that gMCs secreted more CCL2, CCL4, CCL5 (no statistical difference for CCL5) than sMCs. However, no statistically significant difference was found in the secretion of CXCL8.
The CCL2 was secreted by different cellular types; however, we unexpectedly found that MCs surpassed the other cell subtypes in the secretion of CCL2 and were the main cellular source of CCL2 in the gLUAD microenvironment (Supplementary Fig. S5D). The expression of CCL2 in MCs was confirmed by mIF staining (Supplementary Fig. S5E). As expected, more CCL2+ MCs were observed in gLUAD samples than in sLUAD samples from TMA cohort (Supplementary Fig. S5E). We then confirmed the expression of CCR2, which is the main receptor of CCL2, in LUAD-infiltrating CTLs by FCM and found that gLUAD samples were infiltrated with a higher proportion of CCR2+ CTLs than sLUAD samples (Fig. 3D). In addition, the increased density of infiltrating CCR2+ CTLs was also validated by IF analysis (Supplementary Fig. S5F). Furthermore, we identified a positive correlation between the proportion of infiltrating CCR2+ CTLs and MCs by FCM (Fig. 3E). The recruitment of CCR2+ CTLs by MCs was further visualized by the observation that CCR2+ CTLs were present in close proximity to MCs around vessels (Fig. 3F), which was consistent with previous observations of the colocation between MCs and T cells (47). Furthermore, we performed spatial analysis and found that 1.68 ± 1.75 CTLs were located in close proximity to per MC (within 20 μm) and 0.13 ± 0.21 CCR2+ CTLs per MC (Fig. 3G). However, no significant difference was found between GGO and solid LUAD (Supplementary Fig. S5G). Finally, we aimed to validate the chemotactic activity of MCs on CTLs by transwell assay. The migration rate of CTLs in the Sti-MC group was higher than the blank group (medium only), TS group, and Usti-MC group (Fig. 3H), suggesting that MCs might contribute to the recruitment of CTLs. Furthermore, to test whether the CCL2/CCR2 axis mediated the recruitment of CTLs by MCs, we used CCL2 antagonist (CNTO 888) and CCR2 antagonist (INCB3344) in the migration blocking assay. The migration rate of CCR2+ CTLs was higher than CCR2− CTLs by stimulated MCs. Both CCL2 and CCR2 antagonists decreased the migration of CCR2+ CTLs, while CCR2− CTLs were not affected (Fig. 3I).
Together, our results revealed a different chemokine expression pattern between gMCs and sMCs, and the quantitative and spatial relationship between MCs and T cells. In vitro transwell and migration blocking assays supported the notation that MCs might be involved in the recruitment of CCR2+ CTLs through the CCL2–CCR2 axis.
Differences in CCR2 expression cause differences in the recruitment of CTLs between gLUAD and sLUAD
To investigate whether there were other chemokine receptors, in addition to CCR2, that differentially mediated the recruitment of CTLs into gLUAD and sLUAD, we generated a comprehensive chemokine receptor profile of LUAD-infiltrating CTLs (Supplementary Fig. S5H). As expected, the expression of CCR5, CXCR3, and CXCR6 was markedly enriched in LUAD (both gLUAD and sLUAD) samples compared with paired peripheral blood and nLung samples (Supplementary Fig. S5H), which was consistent with previous evidence suggesting their critical roles in CTL tumor trafficking (44, 48). Although CCR10+ CTLs tended to accumulate in sLUAD samples, we observed that CCR2 was the only chemokine receptor that was specifically expressed on CTLs in gLUAD samples (Supplementary Fig. S5I). Interestingly, we observed that the gLUAD microenvironment increased recruitment of CCR2+ CTLs compared with the paired nLung microenvironment, while the sLUAD microenvironment resulted in decreased infiltration of CCR2+ CTLs (Supplementary Fig. S5J). This revealed that the differential recruitment of CTLs into gLUAD and sLUAD samples is mainly caused by differences in CCR2 expression, which to a certain degree, may be attributed to CCL2-secreting MCs that reside in gLUAD tissues.
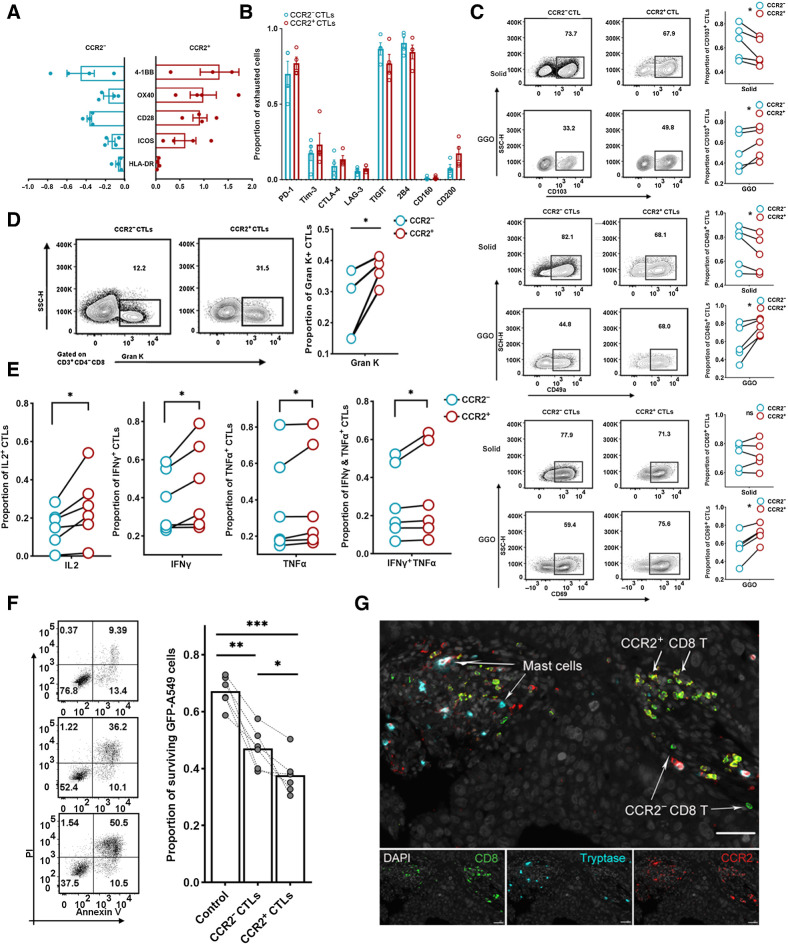
CCR2+ CTLs display enhanced cytotoxic potential
CTLs are the key to effectively eliminating cancer cells, so we speculated that the antitumor role of gMCs may involve the recruitment of CCR2+ CTLs. We next revealed the previously unreported characteristics of CCR2+ CLTs in LUAD, focusing on their phenotype, function, and spatial distribution.
CCR2+ CTLs were highly activated, as characterized by the higher expression of costimulating molecules, such as 4–1BB, OX40, CD28, ICOS, and HLA-DR on these cells than on CCR2− CTLs (Fig. 4A). However, no significant difference in the expression of coinhibitory receptors other than TIGIT was observed between CCR2+ CTLs and CCR2− CTLs (Fig. 4B), suggesting no significant discrepancy in exhaustion status. We found that CCR2+ CTLs in the sLUAD samples expressed fewer tissue-resident memory (TRM)-associated markers, including CD103, CD49a, and CD69 than paired CCR2− CTLs, while gLUAD-infiltrating CCR2+ CTLs expressed more TRM markers (Fig. 4C). These results indicated a great effect of the tumor-specific microenvironment on CCR2+ CTLs—gLUAD microenvironment increased their tissue residency, and the sLUAD microenvironment decreased their abundance in the tissue. CCR2+ CTLs expressed a higher proportion of other chemokine receptors than CCR2− CTLs (Supplementary Fig. S6A), suggesting their potentially enhanced migration capability to the tumor microenvironment. Moreover, there was no significant difference in the expression of proliferation (Ki-67; Supplementary Fig. S6B) and apoptosis-associated markers (Bax and Bcl2; Supplementary Fig. S6C). The expression of another 28 membrane receptors and 14 T-cell–associated transcription factors were also evaluated (Supplementary Fig. S6D and S6E). Of note, CCR2+ CTLs expressed higher levels of CD26, CD127, CD9, and CD161 than CCR2− CTLs, which was consistent with previous evidence that CD26hi T cells expressed high levels of CCR2 with enhanced migration and antitumor capabilities (49).
Figure 4.
Characterization of LUAD-infiltrating CCR2+ CTLs. A, Bar plot showing the expression of costimulatory molecules. The x-axis value represents the FC of the expression in costimulation molecules on CCR2−CTLs or CCR2+ CTLs compared with bulk CTLs (n = 4). B, Box plot showing the expression of coinhibitory molecules (n = 4). C, Frequency of TRM+ (CD103, top; CD49a, middle; CD69, bottom) CCR2− CTLs or CCR2+ CTLs. Representative flow plots (left) and summary (right) of four independent experiments (paired t test). D, Frequency of granzyme K expressing CCR2− CTLs or CCR2+ CTLs (n = 4; paired t test). E, Frequency of cytotoxic molecule–secreted CCR2− CTLs or CCR2+ CTLs (n = 6; paired t test). F, Left, representative flow plots showing the apoptosis of cocultured GFP-A549 cell line. Right, proportion of surviving target cells (n = 4; Wilcoxon test). G, Representative example of LUAD section stained by mIF with anti-CCR2 (red), CD8 (green), and tryptase (blue) antibodies. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
On the basis of these phenotypic differences, we hypothesized that there were functional differences between CCR2+ and CCR2− CTLs. We found that a larger fraction of CCR2+ CTLs expressed granzyme K (GZMK) than CCR2− CTLs (Fig. 4D), suggestive of a preexhausted status in a previous study (50), while no differences were found in perforin, granzyme A, granzyme B, and granulysin (Supplementary Fig. S6F). After in vitro stimulation, CCR2+ CTLs produced a higher amount of IL2, IFNγ, TNFα, TNFα and IFNγ (Fig. 4E). Given the differences in functionality and phenotype, we reasoned that CCR2+ CTLs would have a superior ability to control tumors. Flow-sorted CCR2+ CTLs were more cytotoxic against GFP-A549 tumor-cell targets according to direct cytotoxicity assays after 48-hour coculture (Fig. 4F), indicating that they killed tumor cells more efficiently than CCR2− CTLs.
To explore the spatial distribution pattern of CCR2+CTLs, we performed mIF and found that CCR2+CTLs usually gathered within lymphocyte aggregate structure in LUAD (Fig. 4G), contrasting with the commonly assumed scattered distribution of CTLs in tumor microenvironment. Whether this lymphocyte aggregate structure was tertiary lymphoid structure and the mechanism of aggregate CCR2+ CTLs remained to be solved. Collectively, CCR2+ CTLs displayed a highly activated, nonexhausted, tissue-resident memory phenotype, with enhanced capability to kill tumor cells and usually gathered to form lymphocyte aggregate structure.
Less aggressive LUAD is enriched for CCL2+ MCs–CCR2+ CTLs
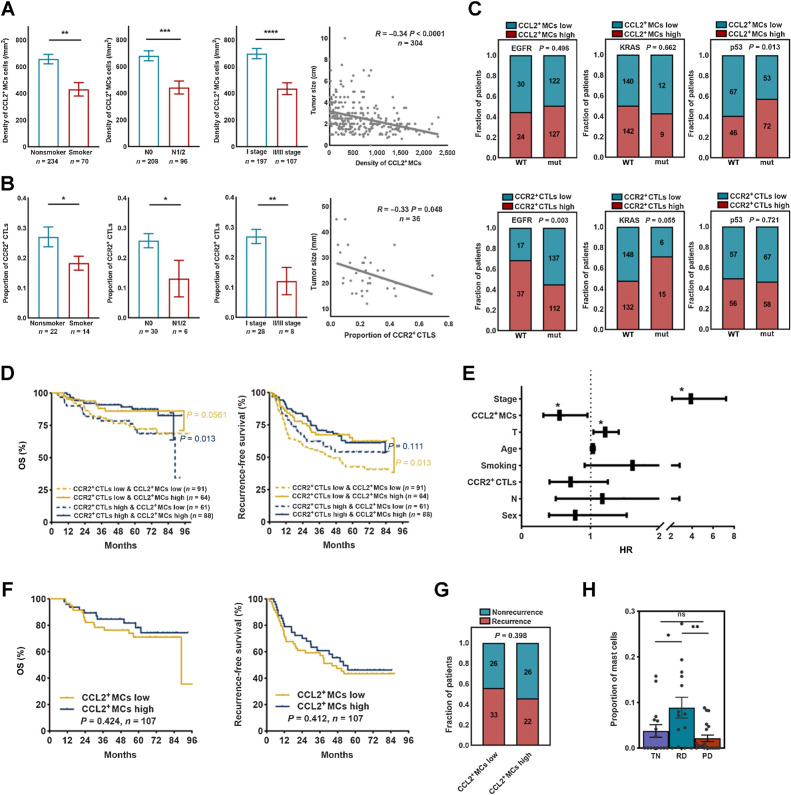
We then explored whether any particular clinical correlates were associated with the heightened presence of CCL2+ MCs–CCR2+ CTLs axis. A higher density of CCL2+ MCs and proportion of CCR2+ CTLs were observed in patients who were nonsmokers, who had smaller or lymph node–negative tumors, and who were at a lower pathologic stage (Fig. 5A and B), while no significant relationship was observed between CCL2+ MC density or CCR2+ CTL proportion and sex or age (Supplementary Fig. S7A). These results were limited to early-stage LUAD, so we next explored their value in metastatic LUAD. By analyzing scRNA-seq data of a metastatic LUAD TKI cohort (24), we found that MCs scarcely infiltrated extrapulmonary metastatic lesions in contrast with primary lesions (Supplementary Fig. S7B; Supplementary Table S1), suggesting that MCs may play an important role in controlling tumor progression and metastasis.
Figure 5.
MCs correlated with therapeutic response to TKI therapy but not chemotherapy. A, Clinic relevance of CCL2+ MCs (Mann–Whitney). B, Clinic relevance of CCR2+ CTLs (two-tailed unpaired t test). C, Oncogene mutation relevance of CCL2+ MCs or CCR2+ CTLs (χ2 test). D, The prognostic value of CCL2+ MCs–CCR2+ CTLs in surgically resected LUAD. P values were determined by the log-rank test. E, Nomogram showing independent prognostic factors for survival in surgically resected LUAD by Cox proportional hazard analyses (*, independent prognostic factors). F, The prognostic value of CCL2+ MCs in patients with LUAD treated with chemotherapy. P values were determined by the log-rank test. G, The correlation of risk of recurrence after chemotherapy and CCL2+ MCs. The P value was determined by the χ2 test. H, MC quantification as a proportion of immune cells (Mann–Whitney; n = 15 for the TN group; n = 14 for the RD group; and n = 20 for the PD group). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Identifiable driver oncogene mutations are a key determinant of tumor progression and therapeutic targets in LUAD; thus, we explored whether mutation background affected the infiltration of MCs and CTLs. CCR2+ CTLs accumulated in KRAS-mutated and EGFR wild-type (WT) tumors, while CCL2+ MCs were enriched in tumors with TP53 WT (Fig. 5C), suggesting a relationship between the tumor mutation background and the MC–CTL infiltration.
MCs are an independent factor predicting patient outcome
To evaluate the prognostic value of the MC–CTL axis in patients with LAUD, we first conducted a Kaplan–Meier analysis in a cohort with mIF data (n = 325). We found that a higher density of CCR2+ CTLs correlated with better recurrence-free survival (Supplementary Fig. S7C), while the correlation of CD8+ CTLs failed to reach statistical significance (Supplementary Fig. S7D). Next, combining the prognostic significance of CCR2+ CTLs and CCL2+ MCs, we found that the density of CCL2+ MCs could further stratify the survival of patients with both CCR2+ CTLs higher infiltration and low infiltration (Fig. 5D). These data highlight the prognostic value of CCL2+ MCs even in patients with low CCR2+ CTLs infiltration. Furthermore, Cox multivariate analyses validated the prognostic significance of CCL2+ MCs [HR, 0.551; 95% confidence interval (CI), 0.317–0.957; P = 0.034] independent of TNM stage and tumor size (Fig. 5E), suggesting that CCL2+ MCs were better than CCR2+ CTLs in the prediction of the oncologic outcomes of patients after surgical excision.
To assess the value of MC infiltration for predicting therapeutic response, we performed survival analysis of 107 patients with LUAD receiving adjuvant chemotherapy after surgery. Indeed, the density of CCL2+ MC infiltration had no impact on the stratification of either OS or recurrence-free survival (Fig. 5F). In addition, no significant differences in risk of recurrence were observed between LUAD with high CCL2+ MC infiltration and low CCL2+ MC infiltration (Fig. 5G). Furthermore, we aimed to investigate the predictive role of MCs in patients with LUAD who were treated with TKI therapy. Surprisingly, a notable accumulation of MCs was observed in patients at the residual disease (RD) state after treatment (Fig. 5H), which was not observed in patients at the progressive disease (PD) or before treatment (TN). This may, to some extent, highlight the critical role of the infiltration of MCs in TKI treatment efficacy. In summary, MC infiltration could independently predict a favorable prognosis and the recruitment of MCs correlated with response to TKI therapy, but not to adjuvant chemotherapy.
Discussion
In this study, we comprehensively characterized the transcriptional and functional heterogeneity of MCs as well as their dynamic changes during LUAD progression. Consistent with previous evidence (10), our study revealed that MCs were enriched in gLUAD and have undergone extensive reprogramming that endowed MCs with distinct immunoregulatory functions, in line with commonly assumed development of MCs occurring locally (36). LUAD-infiltrating MCs, which were associated with the recruitment of CCR2+ CTLs, correlated with favorable prognosis and therapeutic response to TKI treatment but not chemotherapy.
In our study, the infiltration of MCs correlated with prolonged survival and response to TKI treatment but not chemotherapy. The substantial heterogeneity and the presence of distinct MC subpopulations (16, 22) would account for the conflicting roles of MCs that have been reported across many cancer types (15, 51–53). However, to the best of our knowledge, no research has evaluated the heterogeneous subclusters of MCs in the tumor microenvironment. In addition to the quiescent MCs that were enriched in the nLung, we identified a gLUAD-enriched chemotactic MC subset and a sLUAD-enriched angiogenesis-associated MC subcluster. The stark transcriptional differences between gMCs and sMCs suggest both distinct functions and immunoregulatory roles that are imprinted by tissue-specific factors. The ratio of TNF+ versus VEGFA+ MCs suggested in nasopharyngeal carcinoma (33) would hardly be applicable in LUAD, in light of the scarce expression of TNF in LUAD-infiltrating MCs, although a higher ratio of TNF+/VEGFA+ MCs was observed in gMCs compared with sMCs.
The mastocytosis observed following effective TKI treatment could reflect a generalized allergic response promoted by TKI therapy. On the other hand, our data reveal exciting new roles for MCs as essential accessory cells in cancer treatment with TKI and highlight MCs as potential cellular targets for future cancer therapy. gMCs belong to MCTC subtype, while sLUAD decreases the recruitment of MCs, which are MCT subtype and tend to be activated and degranulated under the influence of sLUAD-related factors. It is thus reasonable to speculate that the increased recruitment of MCs and the prevention of their activation and degranulation would be a key MC target in the context of cancer therapy (54).
Previous studies have observed interactions between MCs and T cells, such as antigen presentation and T-cell priming (38, 55, 56), and the activation of MCs by Toll-like receptor 3 modulates CD8+ T-cell recruitment in a MC-deficient mouse model (57). However, evidence about the link between MCs and the recruitment of T cells in the tumor microenvironment is lacking. Here, on the basis of various lines of evidence in our study, we believe that MCs would correlate with the recruitment of CCR2+ CTLs. Further studies are needed to validate this process and reveal the underlying mechanism as well as confirm the crosstalk between MCs and CTLs. We reason that the CCL2+ MCs–CCR2+ CTLs axis would maintain long-term antitumor immunity against LUAD and could maintain gLUAD indolent nature from progression. Thus, our study may provide an innovative new paradigm involving a positive feedback loop between MCs and T cells that results in enhanced long-term antitumor immunity to LUAD.
CCR2 is a chemokine receptor that is known for its role in proinflammatory monocyte chemotaxis. However, the expression of CCR2 in CTLs remains elusive. We confirmed the expression of CCR2 in LUAD-infiltrating CTLs for the first time. Moreover, CCR2+ CTLs exhibited a highly activated and TRM phenotype and expressed high GZMK, indicating that these cells have a “precursor exhausted” memory-like phenotype (50). These observations may provide valuable insights for the manipulation of CCR2+ CTLs in immunotherapy, such as CAR T-cell therapy (58). Unexpectedly, CCR2+ CTLs usually formed lymphocyte aggregate structures, and CCR2 is believed to be a specific marker of peripheral helper T cells (TPH; ref. 59). Whether this lymphocyte aggregate as a tertiary lymphocyte structure (TLS; ref. 60) and the reason for CCR2+ CTLs aggregation need to be further explored.
In summary, we comprehensively profiled the heterogeneity of MCs clusters and highlighted their prognostic and predictive roles in LUAD. In particular, MCs appeared to correlate with the recruitment of CTLs into the tumor microenvironment. Our findings provide unique insights and a deeper understanding of the previously unrecognized role of MCs in the tumor microenvironment and raise the possibility of targeting MCs in the context of LUAD treatment.
Supplementary Material
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Acknowledgments
The authors thank Liunan Wu from Nanjing Medical University for providing GFP-A549 cell line, Dongning Rao, Yingcheng Wu, and Shuyi Ji from the Department of Liver Surgery, Zhongshan Hospital, Fudan University, for experimental and computational advice; Shan Jiang, Xiaoshan Liu, Yuesheng Guo, and Yamin Chen, members of the Zhang Laboratory for administrative and experimental support; GeGe Yuan from Nanjing Medical University for experimental and technical advice. This work was supported by the National Natural Science Foundation of China (81930073), the National Natural Science Foundation of China (82203037), the Shanghai Science and Technology Innovation Action Project (20JC1417200), the Cooperation Project of Conquering Major Diseases in Xuhui District (XHLHGG202101), and Shanghai Sailing Program (22YF1408900). This work was supported in part by National Natural Science Foundation of China (32070657) and National Key R&D Program of China (2019YFA0905900).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
F. Fan: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Gao: Conceptualization, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. Y. Zhao: Resources, data curation, software, formal analysis, supervision, funding acquisition, validation, visualization, methodology, writing–review and editing. J. Wang: Data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. L. Meng: Conceptualization, supervision, investigation, and methodology. J. Ma: Conceptualization, data curation, software, formal analysis, supervision, and methodology. T. Li: Software and methodology. H. Han: Supervision, investigation, methodology, writing–review and editing. J. Lai: Software, investigation, and methodology. Z. Gao: Supervision, investigation, and methodology. X. Li: Software, supervision, investigation, and methodology. R. Guo: Supervision and methodology. Z. Cao: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, and project administration. Y. Zhang: Conceptualization, supervision, investigation, methodology, project administration, writing–review and editing. X. Zhang: Conceptualization, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. H. Chen: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Yatabe Y, Borczuk AC, Powell CA. Do all lung adenocarcinomas follow a stepwise progression? Lung Cancer 2011;74:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y, Fu F, Chen H. Management of ground-glass opacities in the lung cancer spectrum. Ann Thorac Surg 2020;110:1796–804. [DOI] [PubMed] [Google Scholar]
- 4. Chen H, Carrot-Zhang J, Zhao Y, Hu H, Freeman SS, Yu S, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun 2019;10:5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ye T, Deng L, Wang S, Xiang J, Zhang Y, Hu H, et al. Lung adenocarcinomas manifesting as radiological part-solid nodules define a special clinical subtype. J Thorac Oncol 2019;14:617–27. [DOI] [PubMed] [Google Scholar]
- 6. Fu F, Zhang Y, Wen Z, Zheng D, Gao Z, Han H, et al. Distinct prognostic factors in patients with stage I non-small cell lung cancer with radiologic part-solid or solid lesions. J Thorac Oncol 2019;14:2133–42. [DOI] [PubMed] [Google Scholar]
- 7. Miyoshi T, Aokage K, Katsumata S, Tane K, Ishii G, Tsuboi M. Ground-glass opacity is a strong prognosticator for pathologic stage IA lung adenocarcinoma. Ann Thorac Surg 2019;108:249–55. [DOI] [PubMed] [Google Scholar]
- 8. Sun F, Xi J, Zhan C, Yang X, Wang L, Shi Y, et al. Ground glass opacities: Imaging, pathology, and gene mutations. J Thorac Cardiovasc Surg 2018;156:808–13. [DOI] [PubMed] [Google Scholar]
- 9. Lu T, Yang X, Shi Y, Zhao M, Bi G, Liang J, et al. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov 2020;6:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xing X, Yang F, Huang Q, Guo H, Li J, Qiu M, et al. Decoding the multicellular ecosystem of lung adenocarcinoma manifested as pulmonary subsolid nodules by single-cell RNA sequencing. Sci Adv 2021;7:eabd9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer 2019;19:9–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer 2016;16:447–62. [DOI] [PubMed] [Google Scholar]
- 14. Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20:662–80. [DOI] [PubMed] [Google Scholar]
- 15. Maciel TT, Moura IC, Hermine O. The role of mast cells in cancers. F1000Prime Rep 2015;7:09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. St John AL, Rathore APS, Ginhoux F. New perspectives on the origins and heterogeneity of mast cells. Nat Rev Immunol 2022;23:55–68. [DOI] [PubMed] [Google Scholar]
- 17. Galli SJ, Gaudenzio N, Tsai M. Mast cells in inflammation and disease: recent progress and ongoing concerns. Annu Rev Immunol 2020;38:49–77. [DOI] [PubMed] [Google Scholar]
- 18. Welsh TJ, Green RH, Richardson D, Waller DA, O'Byrne KJ, Bradding P. Macrophage and mast-cell invasion of tumor cell islets confers a marked survival advantage in non-small-cell lung cancer. J Clin Oncol 2005;23:8959–67. [DOI] [PubMed] [Google Scholar]
- 19. Marichal T, Tsai M, Galli SJ. Mast cells: potential positive and negative roles in tumor biology. Cancer Immunol Res 2013;1:269–79. [DOI] [PubMed] [Google Scholar]
- 20. Majorini MT, Colombo MP, Lecis D. Few but Efficient: The role of mast cells in breast cancer and other solid tumors. Cancer Res 2022;82:1439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Somasundaram R, Connelly T, Choi R, Choi H, Samarkina A, Li L, et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat Commun 2021;12:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cildir G, Yip KH, Pant H, Tergaonkar V, Lopez AF, Tumes DJ. Understanding mast cell heterogeneity at single cell resolution. Trends Immunol 2021;42:523–35. [DOI] [PubMed] [Google Scholar]
- 23. Komi DEA, Redegeld FA. Role of mast cells in shaping the tumor microenvironment. Clin Rev Allergy Immunol 2020;58:313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 2020;182:1232–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kulka M, Metcalfe DD. Isolation of tissue mast cells. Curr Protoc Immunol 2010;Chapter 7:Unit 7 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 2019;50:1317–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Y, Shang J, Gao J, Han H, Gao Z, Yan Y, et al. Increased tumor intrinsic growth potential and decreased immune function orchestrate the progression of lung adenocarcinoma. Front Immunol 2022;13:921761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Radinger M, Jensen BM, Swindle E, Gilfillan AM. Assay of mast cell mediators. Methods Mol Biol 2015;1220:307–23. [DOI] [PubMed] [Google Scholar]
- 29. Kuehn HS, Radinger M, Gilfillan AM. Measuring mast cell mediator release. Curr Protoc Immunol 2010;Chapter 7:Unit 7 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loberg RD, Ying C, Craig M, Yan L, Snyder LA, Pienta KJ. CCL2 as an important mediator of prostate cancer growth in vivo through the regulation of macrophage infiltration. Neoplasia 2007;9:556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang F, Feng W, Wang H, Wang L, Liu X, Wang R, et al. Monocyte-derived leukemia-associated macrophages facilitate extramedullary distribution of T-cell acute lymphoblastic leukemia cells. Cancer Res 2020;80:3677–91. [DOI] [PubMed] [Google Scholar]
- 32. Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 2017;214:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021;184:792–809. [DOI] [PubMed] [Google Scholar]
- 34. Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer 2008;8:618–31. [DOI] [PubMed] [Google Scholar]
- 35. Ben-Baruch Morgenstern N, Ballaban AY, Wen T, Shoda T, Caldwell JM, Kliewer K, et al. Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission. J Allergy Clin Immunol 2022;149:2062–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gurish MF, Austen KF. Developmental origin and functional specialization of mast cell subsets. Immunity 2012;37:25–33. [DOI] [PubMed] [Google Scholar]
- 37. Wernersson S, Pejler G. Mast cell secretory granules: armed for battle. Nat Rev Immunol 2014;14:478–94. [DOI] [PubMed] [Google Scholar]
- 38. Dudeck J, Ghouse SM, Lehmann CH, Hoppe A, Schubert N, Nedospasov SA, et al. Mast-cell-derived TNF amplifies CD8(+) dendritic cell functionality and CD8(+) T cell priming. Cell Rep 2015;13:399–411. [DOI] [PubMed] [Google Scholar]
- 39. Zhou T, Damsky W, Weizman OE, McGeary MK, Hartmann KP, Rosen CE, et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 2020;583:609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med 2013;273:114–27. [DOI] [PubMed] [Google Scholar]
- 41. Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol 2020;17:204–32. [DOI] [PubMed] [Google Scholar]
- 42. Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov 2022;12:134–53. [DOI] [PubMed] [Google Scholar]
- 43. Gueguen P, Metoikidou C, Dupic T, Lawand M, Goudot C, Baulande S, et al. Contribution of resident and circulating precursors to tumor-infiltrating CD8(+) T cell populations in lung cancer. Sci Immunol 2021;6:eabd5778. [DOI] [PubMed] [Google Scholar]
- 44. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Oliveira S, Reyes-Aldasoro CC, Candel S, Renshaw SA, Mulero V, Calado A. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J Immunol 2013;190:4349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dudeck J, Kotrba J, Immler R, Hoffmann A, Voss M, Alexaki VI, et al. Directional mast cell degranulation of tumor necrosis factor into blood vessels primes neutrophil extravasation. Immunity 2021;54:468–83. [DOI] [PubMed] [Google Scholar]
- 47. Galli SJ, Nakae S, Tsai M. Mast cells in the development of adaptive immune responses. Nat Immunol 2005;6:135–42. [DOI] [PubMed] [Google Scholar]
- 48. Di Pilato M, Kfuri-Rubens R, Pruessmann JN, Ozga AJ, Messemaker M, Cadilha BL, et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 2021;184:4512–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailey SR, Nelson MH, Majchrzak K, Bowers JS, Wyatt MM, Smith AS, et al. Human CD26(high) T cells elicit tumor immunity against multiple malignancies via enhanced migration and persistence. Nat Commun 2017;8:1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24:978–85. [DOI] [PubMed] [Google Scholar]
- 51. Nonomura N, Takayama H, Nishimura K, Oka D, Nakai Y, Shiba M, et al. Decreased number of mast cells infiltrating into needle biopsy specimens leads to a better prognosis of prostate cancer. Br J Cancer 2007;97:952–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pittoni P, Tripodo C, Piconese S, Mauri G, Parenza M, Rigoni A, et al. Mast cell targeting hampers prostate adenocarcinoma development but promotes the occurrence of highly malignant neuroendocrine cancers. Cancer Res 2011;71:5987–97. [DOI] [PubMed] [Google Scholar]
- 53. Strouch MJ, Cheon EC, Salabat MR, Krantz SB, Gounaris E, Melstrom LG, et al. Crosstalk between mast cells and pancreatic cancer cells contributes to pancreatic tumor progression. Clin Cancer Res 2010;16:2257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lichterman JN, Reddy SM. Mast cells: a new frontier for cancer immunotherapy. Cells 2021;10:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodriguez Cetina Biefer H, Heinbokel T, Uehara H, Camacho V, Minami K, Nian Y, et al. Mast cells regulate CD4(+) T-cell differentiation in the absence of antigen presentation. J Allergy Clin Immunol 2018;142:1894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stelekati E, Bahri R, D'Orlando O, Orinska Z, Mittrucker HW, Langenhaun R, et al. Mast cell-mediated antigen presentation regulates CD8+ T cell effector functions. Immunity 2009;31:665–76. [DOI] [PubMed] [Google Scholar]
- 57. Orinska Z, Bulanova E, Budagian V, Metz M, Maurer M, Bulfone-Paus S. TLR3-induced activation of mast cells modulates CD8+ T-cell recruitment. Blood 2005;106:978–87. [DOI] [PubMed] [Google Scholar]
- 58. Moon EK, Carpenito C, Sun J, Wang LC, Kapoor V, Predina J, et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin Cancer Res 2011;17:4719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshitomi H, Ueno H. Shared and distinct roles of T peripheral helper and T follicular helper cells in human diseases. Cell Mol Immunol 2021;18:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sautes-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer 2019;19:307–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Data Availability Statement
The scRNA-seq data analyzed in this study is available at Sequence Read Archive (SRA) database under accession number SRP438272 as an NCBI BioProject #PRJNA973717. Public scRNA-seq data are accessible at the GSA of the BIG Data Center, Beijing Institute of Genomics (http://bigd.big.ac.cn/gsa-human) under accession number HRA000154 and the NCBI GEO depository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127465) under the accession number GEO: GSE127465. TKI cohort data are available as an NCBI BioProject (#PRJNA591860). RNA-seq data from our center are available at the European Genomephenome Archive (EGA) with the accession number EGAS00001004006 (https://ega-archive.org/studies/EGAS00001004006) and external RNA-seq data were available from the GEO under accession number GSE111907. Data and code used for analysis are available as a Code Ocean capsule (https://codeocean.com/capsule/6874338/tree/v1). All other raw data are available upon request from the corresponding author.