Abstract
Reactive oxidative species (ROS) production-driven ferroptosis plays a role in acute kidney injury (AKI). However, its exact molecular mechanism is poorly understood. Scavenger receptor CD36 has important roles in oxidizing lipids, lipid accumulation, metabolic syndrome, and insulin resistance in chronic kidney disease, but its roles remain unexplored in AKI. The present study investigated the role and mechanism of CD36 in regulating proximal tubular cell ferroptosis and AKI. The expression of CD36 was found to be significantly up-regulated in AKI renal tissues and correlated with renal function, which might serve as an independent biomarker for AKI patients. Moreover, in adult mice subjected to AKI, deletion of CD36 (CD36−/−) induced tubular cell ROS accumulation, ferroptosis activation, and renal injury. Mechanistically, combining LC-MS/MS, co-IP, and ubiquitination analyses revealed that CD36 could specifically bind to ferroptosis suppressor protein 1 (FSP1) and regulate its ubiquitination at sites K16 and K24, leading to FSP1 degradation and progression of ferroptosis in AKI. The present results emphasize a novel mechanism of CD36 in cisplatin-induced AKI. The discovery of the special CD36 roles in promoting ferroptosis and AKI development by regulating the ubiquitination of FSP1 in proximal tubular cells may be potential therapeutic targets for AKI. Moreover, CD36 may play a key role in the progression of AKI. Therefore, targeting CD36 may provide a promising treatment option for AKI.
Keywords: Acute kidney injury, CD36, Ferroptosis, FSP1, Ubiquitin-dependent degradation
Introduction
Acute kidney injury (AKI) is a global disease with high morbidity and mortality in clinical patients.1 It is characterized by a sharp decline in renal function within 48 h2 accompanied by a decrease in glomerular filtration rate and an increase in serum creatinine (Scr) and blood urea nitrogen (BUN) levels.3 Cisplatin is one of the major causes of clinical AKI. Since the precise molecular mechanisms involved were not completely clear, very limited strategies for AKI prevention or therapy are available at present. Thus, it is urgent to find novel biomarkers and therapeutic targets to diagnose and treat AKI.
Ferroptosis is a type of regulated cell death that was identified in 2012. Unlike apoptosis and necrosis, ferroptosis results from the iron-dependent and fatal accumulation of lipid hydroperoxides. Overwhelming reactive oxidative species (ROS) production is considered to be an important driver of ferroptosis.4,5 ROS-driven ferroptosis has been widely reported in many diseases, such as neurological diseases, ischemia-reperfusion injury, and AKI.6 Recent studies have found that ferroptosis in proximal tubules is also one of the important pathological processes in AKI.7 However, the molecular mechanism of ROS-driven ferroptosis in AKI is still not fully understood.
CD36 is a B scavenger receptor and a transmembrane glycoprotein that is expressed on the surface of many cell types. In the kidney, CD36 can be expressed on the proximal tubular epithelium, podocytes, and mesangial cells.8, 9, 10 Interestingly, CD36 is distributed in the cell membrane and localizes to the endosomes, endoplasmic reticulum, and mitochondria.11,12 Moreover, it is a multi-ligand receptor protein capable of binding a variety of ligands, such as oxidized low-density lipoprotein (ox-LDL), long-chain fatty acids, and oxidized lipids.13 Current research on CD36 signaling in kidney injury has been on metabolic inflammation, energy reprogramming, apoptosis, and fibrosis, with the main focus on chronic kidney disease.14,15 To date, very few studies have assessed the relationship between CD36 and AKI. Jong et al have observed that human CD36 overexpression is a risk factor in proximal tubules in folic acid-induced AKI. Mechanistically, CD36 overexpression is associated with fibrotic factor-aggravated renal tubular epithelial cell damage.16 In addition, ROS and mitochondrial dysfunction in proximal renal tubular cells play a critical role in the pathophysiological mechanism of AKI.17 Studies have shown that excessive ROS production in mitochondria can damage mitochondrial proteins and aggravate the occurrence of ferroptosis.18 There is robust scientific evidence that CD36 regulates the production of ROS and mediates ox-LDL and lipid homeostasis, but little is known about the relationship between CD36 and ferroptosis, especially in AKI.
The present study showed that CD36 was up-regulated in renal tissues of AKI patients and mice and was closely correlated with the clinical renal function of AKI patients. In addition, the therapeutic potential of CD36 deficiency against oxidative stress and CD36 deficiency-induced reno–protection was found to be ferroptosis-dependent in CD36 knockout (KO) (CD36−/−) mice. The mechanism research demonstrated that CD36 could bind to ferroptosis suppressor protein 1 (FSP1), an established factor related to ferroptosis resistance, and regulate FSP1 ubiquitination at K16 and K24 sites, promoting tubular cell oxylipin accumulation and ferroptosis and leading to the progression of AKI. The present study findings provide novel insights into the underlying molecular mechanism of how CD36 contributes to the progression of AKI, revealing the possibility for CD36 to serve as an alternative diagnostic marker and therapeutic target for AKI.
Materials and methods
Ethics statement
The present study was approved and supervised by the Ethics Committee of Chongqing Medical University (The Second Affiliated Hospital of Chongqing Medical University). Informed consent and ethics approval were obtained prior to surgery (ethics approval number: 2021.606). All efforts were made to reduce the number of animals used and to minimize their suffering.
Animal models
WT male mice (background: C57/BL6J) were obtained from the Experimental Animal Center of Chongqing Medical University. Male total CD36 deletion (CD36−/−, C57BL/6J background; 6–8-week-old, 18–22 g) mice were provided by Maria Febbraio (Lerner Research Institute, Cleveland, OH). WT and CD36−/− mice were respectively randomized into control and treatment groups (n = 6 per group). The animals were kept at the Specific Pathogen-Free Laboratory Animal Center of Chongqing Medical University with a 12 h/12 h light and dark cycle and ∼60% humidity, at 25 °C, and with free access to drinking water and food. The treatment groups were administered with a single intraperitoneal injection of cisplatin (20 mg/kg in normal saline; Sigma-Aldrich, St. Louis, MO, USA). The WT groups were injected intraperitoneally with normal saline (20 mg/kg). All mice were sacrificed 72 h after cisplatin or normal saline injection. Mice were humanely sacrificed after the completion of the experiments, and blood and kidney samples were collected. Tissue samples were collected, fixed by immersion in 4% paraformaldehyde, and snap-frozen in liquid nitrogen for the subsequent experiments.
Human samples
Human kidney tissue and blood samples were collected in the experimental studies. The adjacent noncancerous and AKI tissues were respectively assigned to the normal (n = 6) and AKI (n = 6) groups. The patients from the Second Affiliated Hospital of Chongqing Medical University were evaluated using magnetic resonance imaging and pathological examinations. All patients provided written informed consent prior to surgery. All samples were removed aseptically and frozen in liquid nitrogen until further use. Scr levels, serum BUN levels, and glomerular filtration rate were measured by an automatic biochemical analyzer.
Renal function evaluation
BUN and Scr levels in mice were assessed to evaluate renal function. They were measured using a urea and creatinine assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's manual.
Kidney pathological evaluation
Kidney tissue (normal and AKI) samples from each group were fixed, paraffin-embedded, and cut into 4-μm thick sections. The sections were then stained with periodic acid-Schiff (PAS) and hematoxylin and eosin (HE). Three observers who were blinded to the treatment conditions scored the degree of morphological involvement in the pathological sections of AKI patient and mouse samples. The kidney pathology score was used to evaluate the severity of the renal injury. Each pathological section was scored using 10 randomly selected fields of view, with every field including 100 renal tubules. The scoring rules were as follows: i) brush border lost, ii) tubular dilation/flattening, iii) tubular cast formation, iv) vacuolization, and v) interstitial edema. The score was proportional to the severity of AKI.
Immunohistochemistry (IHC) analysis
Renal tissues were fixed, blocked, sliced, and incubated with primary antibody against CD36 (NB400-144, Novus, USA) overnight at 4 °C. After the incubation, the slices were washed thrice with phosphate-buffered saline (PBS) for 2 min each. Then, the reacted slices were incubated with the corresponding secondary antibody at room temperature for 20 min and washed with PBS thrice for 2 min each. After PBS washing and slight drying, the sections were treated with the newly-prepared DAB solution for 30 s. For semi-quantitative analyses of the IHC-processed kidney specimens, images were collected and analyzed using the open-source software Image Pro Plus (Media Cybernetics, USA).
Immunofluorescence staining for ROS
Frozen renal tissues were cryo-sectioned at a thickness of 8 μm and then incubated with Cy3 (1:500) for 30 min at 37 °C in the dark. After washing three times with PBS (pH 7.4) for 5 min each time, the cell nuclei were counterstained with DAPI and incubated for 10 min at 37 °C and kept in the dark. The cells were viewed using confocal laser-scanning microscopy (Nikon Eclipse C1, Japan).
Immunofluorescence studies
Cells were fixed in 4% paraformaldehyde for 15 min after treatment and washed with PBS buffer twice for 5 min each. They were then blocked with 1% bovine serum albumin (BSA) solution at room temperature for 1 h and washed with PBS buffer twice for 5 min each. The cells were then incubated with each of the primary antibodies (anti-CD36, catalog #NB400-144, Novus, USA; anti-Aifm2, catalog #sc-377120, Santa Cruz, CA) overnight at 4 °C and washed. Next, the cells were incubated with a secondary antibody for 1 h, and the nuclei were subsequently counterstained with DAPI for 5 min. Finally, they were observed under a microscope equipped for epi-illumination with 5 × , 20 × , and 50 × objectives (Nikon Eclipse E800). The procedure for kidney tissue immunofluorescence staining was the same as that used for the IHC staining. The samples were incubated with primary antibodies (anti-NGAL, ab216462, Abcam; anti-Laminin, ab133645, Abcam) overnight at 4 °C and a secondary antibody for 1 h at 22 °C. The images were collected and analyzed with a UV microscope equipped with epi-illumination.
Cell culture and treatments
TCMK-1, a tubular epithelial mouse cell line, was maintained in Dulbecco's Modified Eagle Medium Nutrient Mixture F-12 supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. All cells were maintained in a 37 °C incubator with 5% CO2. The TCMK-1 cell treatment groups were then administered with 20 μM cisplatin (Sigma-Aldrich, catalog #: P4394) for 24 h. FSP1 inhibitor (iFSP1) was purchased from MCE (HY-136057, MCE, China). The cells were incubated with iFSP1 (3 μM) for 24 h. In addition, CD36-overexpressing TCMK-1 cells generated in our laboratory were also used for parallel studies. Knockdown studies included the use of CD36 siRNA (Shanghai GeneBio, China; 50 nM) in cells transfected with Lipofectamine 2000 reagent (Invitrogen, catalog #: 11668-019) according to the manufacturer's instructions. Supplementary materials provide more information on the viral sequences (Table S1).
Transmission electron microscopy (TEM)
Mouse renal tissues were collected 72 h after cisplatin or normal saline injection. Tissues were fixed with an electron microscope fixing solution. The renal cortices were cut into small pieces (1 mm3) and fixed with an electron microscope fixing solution. The samples were then dehydrated and embedded. Ultra-thin sections were stained and observed with a transmission electron microscope (JEM-1400 plus, Japan Electron Optics Laboratory Co., Ltd.; magnification, 10,000 × and 30,000 × ). Each group contained at least six images.
Real-time polymerase chain reaction (PCR)
Total RNA samples were extracted from kidney tissue using a High-Purity RNA Rapid Extraction Kit (Accurate Biotechnology, China). RNA samples were reverse-transcribed into cDNA using reverse transcription kits (Takara, Japan). Then, cDNA samples were reacted with 1× Fast SYBR Green Master Mix for quantitative PCR (Takara, Japan). All of the genes were normalized using β-actin in the same sample. Relative expression levels were calculated according to the standard 2−ΔΔCt method. The applied primer sequences are listed in Table S1.
Western blot studies
Renal tissues and TCMK-1 cells were lysed with RIPA buffer (Beyotime, catalog #: P0013B, China) containing 1% PMSF (Beyotime, catalog #ST506, China) for 30 min. The samples were centrifuged for 15 min at 4 °C and 12,000 g to remove insoluble residues. Protein concentrations were quantified using a BCA protein assay (Solarbio, catalog #PC0020, China). Total protein samples were separated using 12.5% SDS-PAGE. After electrophoresis, the proteins were transferred to a PVDF membrane (MerckMillipore, Darmstadt, Germany, catalog #ISEQ00010) and blocked with 5% nonfat dry milk for 1–2 h. They were then incubated overnight with primary antibodies against CD36 (1:1000, ab133625, Abcam), ACSL4 (1:10,000, ab155282, Abcam), GPX4 (1:1000, ab125066, Abcam), Xct (1:1000, ab175186, Abcam), NGAL (1:1000, ab216462, Abcam), FSP1 (1:1000, 20886-1-AP, ProteinTech, China), and β-actin (1:2000, GB15001, Servicebio, China) at 4 °C. The membranes were then washed again thrice with TBST solution and incubated with secondary antibodies. Finally, the blots were treated with an ECL reagent (catalog #: 17046, ZENBIO, China).
Detection of ROS in TCMK-1 cells
The dihydroethidium (DHE) assay for detecting ROS was used for the detection of intracellular ROS levels in TCMK-1 cells. After treatment with or without 20 μM cisplatin for 24 h, cells were stained with 5 μM DHE and incubated at 37 °C for 20 min in the dark. The stained cells were washed with PBS three times, collected, and evaluated using flow cytometry (Beckman Coulter, CA) at the excitation and emission wavelengths of 288 and 535 nm, respectively. The relative fluorescence intensity was taken as the average of values from three repeated experiments.
Co-immunoprecipitation (Co-IP) and mass spectrometry
The total protein samples from TCMK-1 cells were harvested 24 h after incubation with cisplatin and lysed in RIPA and PMSF (RIPA: PMSF = 100: 1). The lysates were centrifuged for 15 min at 4 °C. The collected supernatants were incubated with indicated antibodies on a shaker overnight at 4 °C. The antibodies included CD36 (1:50, ab133625, Abcam), FSP1 (1:50, 20886-1-AP, ProteinTech, China), and IgG (1:50, CST). IgG was used as a negative control. After the first incubation, the IP complexes were incubated with Sepharose beads (Beyotime, China) for 2 h at 4 °C on a rotator, followed by centrifugation. The beads-antibody-antigen complex was washed thrice with lysis buffer, followed by centrifugation. The supernatant was then collected. Finally, the complex was mixed with an equal volume of 1× SDS-PAGE sample loading buffer (Beyotime, China), boiled for 5 min at 95 °C, and analyzed using western blotting. The samples were used for tandem mass spectrometry analysis by Jinkairui Biotechnology company (PTM.BIO Lab, Wuhan, China).
High-throughput RNA sequencing and informatics analysis
Total RNA from AKI and normal mice was isolated using a TRIzol Reagent (Invitrogen, China) according to the manufacturer's instructions. RNA quality, purity, and quantity were assessed by Shanghai NovelBio Bio-Pharm Technology Co., Ltd. For quality control, clean reads were obtained from the raw reads by removing the adaptor sequences, reads with >5% of ambiguous bases (noted as N), and low-quality reads containing more than 20% of bases. High-throughput RNA sequencing was performed on an Illumina HiSeq X Ten Sequencing System. The differentially expressed genes (DEGs) were filtered using the significant threshold value (P-value), fold change (FC), and false discovery rate (FDR). A total of 2154 DEGs were identified between the control and AKI groups (Table S2). Then, pathway analysis was used to identify the significant pathway for the differential genes according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Fisher's exact test was utilized to select the significant pathway. The significance threshold was defined by the P-value and FDR under the following criteria: FC ≥ 1.5 and FDR <0.05. Gene expression data are available in the Gene Expression Omnibus database with accession no. GSE 212678.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software, Inc.). Data were represented as mean ± standard error of the mean (SEM). The data were analyzed using a student's t-test followed by one-way ANOVA when two groups were compared. Tukey's post hoc test was used to compare differences between multiple groups. A P-value <0.05 indicated a statistically significant difference (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Results
CD36 is highly expressed in AKI and is correlated with patient renal function
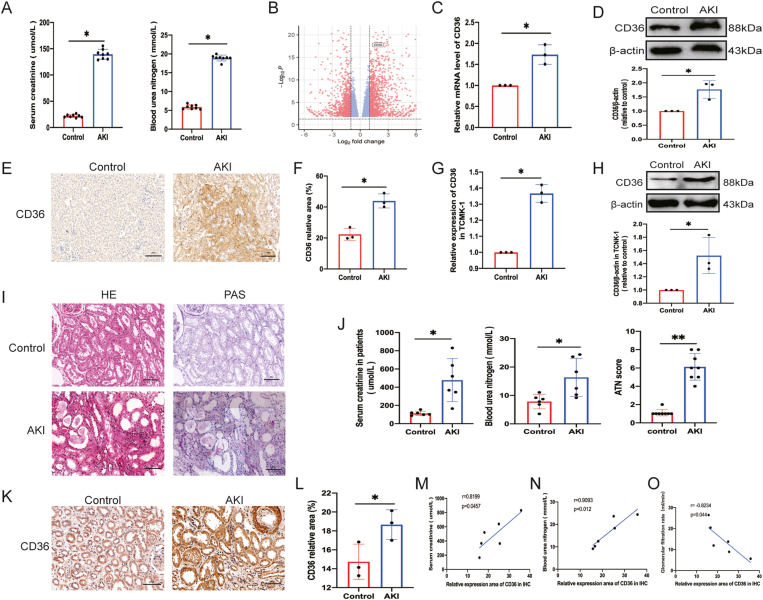
To investigate the expression pattern of CD36 in AKI, cis-AKI, and control mice were used in the present study. Compared to the controls, the AKI mice exhibited elevated Scr (about four-fold higher) and BUN (three-fold higher) levels after cisplatin injection (Fig. 1A). To gain insight into the AKI-associated mRNA, the transcriptome expression profiles were analyzed in renal tissues of AKI and control mice using RNA-seq. The results showed that 1183 mRNAs were up-regulated, while 971 mRNAs were down-regulated with cut-off criteria of FC ≥ 1.5 and FDR <0.05 (Fig. 1B and Table S2). KEGG pathway analysis was performed with these differentially expressed mRNAs to reveal the significantly enriched signaling pathways (Fig. S1). Among these differentially expressed mRNAs, CD36 was significantly increased in AKI mice. Previous work has shown that CD36 regulates the production of ROS and mediates ox-LDL and lipid homeostasis, which are related to AKI.18,42 Hence, CD36 was the focus of further studies. To demonstrate the expression of CD36 in AKI mice, qRT-PCR, western blotting, and immunohistochemistry staining were utilized. As expected, qRT-PCR (Fig. 1C), western blotting (Fig. 1D), and immunohistochemistry (Fig. 1E, F) results showed that CD36 up-regulation was about 1.5-fold higher in the AKI group. Moreover, CD36 expression was detected in TCMK-1 cells after cisplatin treatment to validate the above results. The qRT-PCR (Fig. 1G) and Western blot (Fig. 1H) results showed that CD36 expression was increased. In addition, sections derived from AKI patients' renal puncture samples (n = 6) and normal controls (n = 6) were used to detect the expression levels of CD36 in AKI patients (Fig. 1I, J). The immunohistochemistry results indicated that CD36 expression was increased by about two-fold in AKI samples (Fig. 1K, L). Furthermore, the relationship between the expression level of CD36 and the clinical characteristics of these AKI patients was analyzed. Pearson's correlation analysis showed that CD36 expression was significantly positively associated with Scr (Fig. 1M), BUN (Fig. 1N), and eGFR (Fig. 1O). Together, these findings suggested that CD36 was significantly up-regulated in AKI and might be a diagnostic marker for AKI patients.
Figure 1.
CD36 expression level was increased in AKI. (A) Cisplatin-induced AKI with elevated Scr and BUN levels (n = 8; ∗P < 0.05 vs. control group). (B) Transcriptome sequencing results indicated that CD36 gene was up-regulated. (C) CD36 mRNA level in mouse kidney (n = 3) quantified via qPCR. (D) Western blotting was used to detect CD36 expression in mouse kidney tissues (n = 3). (E) Representative images of immunohistochemical staining and quantitative statistics of relative CD36 area in mice (n = 3). (F) Quantitative statistical analysis of relative CD36 area from (E). (G) CD36 mRNA level in TCMK-1 cells (n = 3) quantified via qPCR. (H) Western blotting was used to detect CD36 expression in TCMK-1 cells (n = 3). All data are expressed as means ± standard deviation. ∗P < 0.05 and ∗∗P < 0.01 vs. control group. (I) Representative CD36 images of immunohistochemical and pathological sections (HE and PAS) in kidney tissue samples from clinical patients. Scale bars = 100 μm. (J) Renal function levels, including Scr, BUN, and GFR in clinical samples. (K) Representative immunohistochemical staining images of relative CD36 area in human AKI sample. (L) Quantitative statistical analysis of relative CD36 area from (K). (M–O) Pearson correlation analysis between CD36 expression and renal function.
CD36 deficiency prevents AKI development
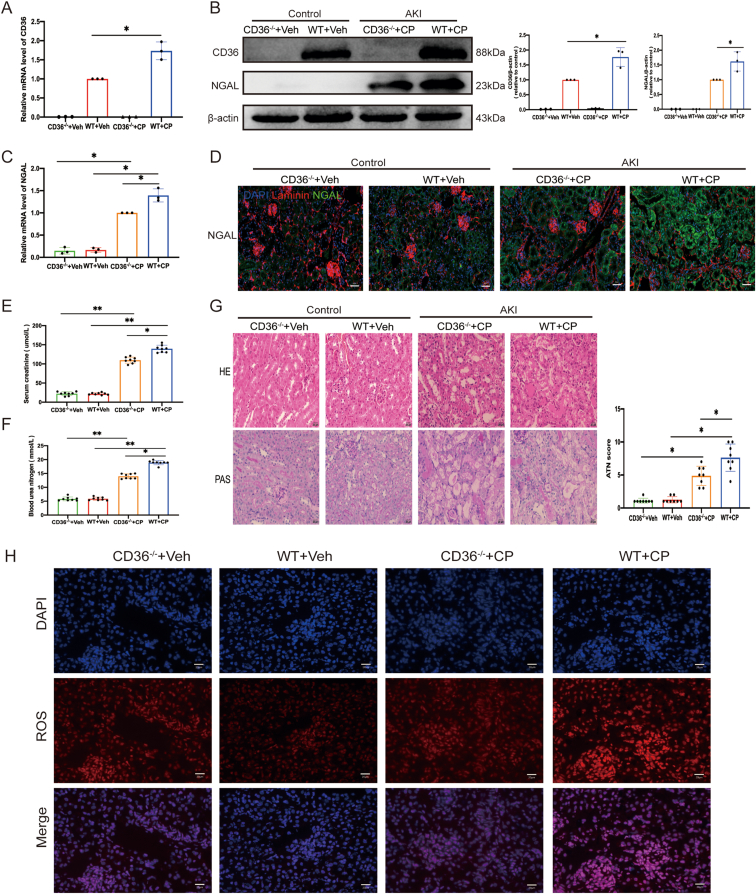
To determine the role of CD36 in AKI, CD36−/− mice were used to induce AKI with cisplatin (Fig. 2A, B). The progression of AKI after 72 h determined by measuring the level of AKI progression marker neutrophil gelatinase-associated lipocalin (NGAL),19, Scr level, BUN level, and pathological feature changes in renal tissues was significantly more relieved in the CD36−/− AKI group than in the AKI mice (Fig. 2B–G). The expression of AKI marker NGAL was increased about 1.3-fold in the AKI group and decreased in the CD36−/− AKI group, as shown by western blotting (Fig. 2B), qRT-PCR (Fig. 2C), and immunofluorescence (Fig. 2D) results. The Scr and BUN levels in the cisplatin injection groups of CD36−/− mice were significantly lower than those in the WT group with cisplatin injections (Fig. 2E, F). Furthermore, the pathological feature changes including tubular epithelium disruption, loss of staining border, and even cast formation in the tubular lumina in the CD36−/− mice were alleviated after cisplatin treatment compared to the WT AKI mice (Fig. 2G). Ferroptosis is promoted by excessive ROS accumulation in cells. Thus, ROS levels were determined in vivo. ROS levels in the kidneys of CD36−/− mice after injection with cisplatin were decreased compared to those in the WT mice with AKI (Fig. 2H).
Figure 2.
CD36 deficiency prevents AKI development. (A) Quantification of kidney CD36 mRNA levels by qPCR. (B) Western blotting was used to detect the expression of CD36 and NGAL in kidney tissues of four mouse groups (n = 3). (C) Quantification of kidney NGAL mRNA levels by qPCR. (D) Representative immunofluorescent NGAL-stained kidney sections. Scale bar = 20 μm. (E, F) Scr and BUN levels in four groups of mice (n = 8). (G) Representative photomicrographs of HE and PAS staining images in four mouse groups. All data are expressed as means ± standard deviation. ∗P < 0.05 and ∗∗P < 0.01 vs. control group. (H) ROS kidney levels were measured by Cy3 staining. Stained cells were viewed by confocal laser-scanning microscopy. Scale bar = 20 μm.
CD36 acts as an inducer of ferroptosis by promoting lipid peroxidation in AKI in vivo and in vitro
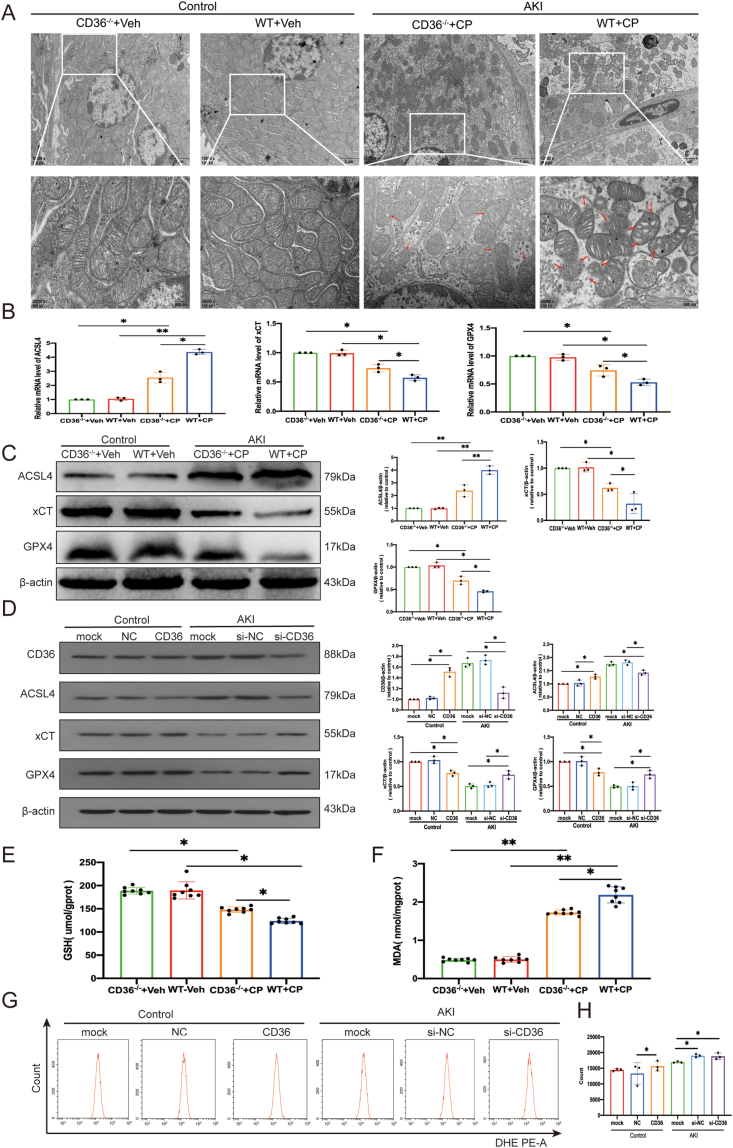
Similar to other cellular death processes, ferroptosis is associated with unique morphological changes. It mainly manifests as obvious mitochondrial shrinkage with increased membrane density and reduction in or vanishing of mitochondrial cristae, which is a different process from other modes of cell death.20 The relationship between ferroptosis and AKI was demonstrated in our previous study.21 Based on that data, CD36 may play a role in the ferroptosis of AKI. Therefore, the function of CD36 in ferroptosis was explored in vivo and in vitro. TEM results revealed that the mitochondria membranes were continuous and intact in the renal tissues of the normal group. The mitochondria were smaller in size and the outer membrane was ruptured in the renal tissues of AKI mice, showing typical ferroptosis changes. Compared to the AKI group, less mitochondrial damage was observed in the CD36−/− AKI mouse group (Fig. 3A). Moreover, the established ferroptosis markers ACSL4, xCT, and GPX4 were detected in all mouse groups. The qRT-PCR and western blotting results showed that the pro-ferroptosis protein ACSL4 level was increased, while levels of anti-ferroptosis proteins xCT and GPX4 were decreased in the AKI group compared to their levels in the control group. ACSL4 level was significantly lower, and xCT and GPX4 levels were higher in CD36−/− AKI mice than in WT AKI mice (P < 0.05) (Fig. 3B, C). Consistent with in vivo studies, the knockdown of CD36 expression by siRNA (siCD36) in TCMK-1 cells with the treatment of cisplatin increased the expression of ACSL4 and decreased the expression of xCT and GPX4. On the contrary, overexpression of CD36 with lentivirus in cisplatin-treated TCMK-1 cells showed the opposite results, with down-regulation of ACSL4 and overexpression of xCT and GPX4 (Fig. 3D). Collectively, these findings demonstrate that CD36 is a positive regulator of ferroptosis in AKI.
Figure 3.
CD36 deficiency was resistant to ferroptosis. (A) Representative transmission electron microscopy images. Mitochondrial outer membrane rupture or mitochondrial crest injury are indicated by red arrows. (B) Quantification of ferroptosis biomarker ACSL4, xCT, and GPX4 mRNA levels by qPCR (n = 3). (C) Western blotting was used to detect the expression of ferroptosis biomarkers ACSL4, xCT, and GPX4 in kidney tissues of four mouse groups (n = 3). Data are expressed as means ± standard deviation (SD). ∗P < 0.05 and ∗∗P < 0.01 vs. control group. (D) After CD36 overexpression or silencing and treatment with normal saline and cisplatin, western blotting was used to detect the expression of CD36 and ferroptosis biomarkers ACSL4, xCT, and GPX4 in TCMK-1 cells; quantitative statistical analysis for each protein (n = 3) was also performed. (E, F) MDA and GSH levels in mice (n = 8). (G) Change in ROS generation in TCMK-1 cells. ROS levels were determined by flow cytometry. (H) Mean ROS fluorescence intensity was expressed as mean ± SD of three independent measurements. ∗P < 0.05 vs. the indicated sample (n = 3).
Increased levels of oxidative stress may induce ferroptosis by targeting membrane lipids. Thus, lipid peroxidation by ROS was analyzed by measuring Malondialdehyde (MDA) content, which is an end-product of oxidized lipids.22 The data showed that MDA levels were significantly increased in AKI mice, while MDA values in CD36−/− AKI mice were about 0.6-fold of those in the WT AKI mice (Fig. 3E). Moreover, GSH content was examined because reduced GSH level is a major intracellular antioxidant.23 The level of GSH in WT AKI mice was about 0.8-fold the level in CD36−/− AKI mice (Fig. 3F). In addition, previous studies have shown that ferroptosis is induced by lipid peroxidation, which is ROS-mediated lipid damage. Therefore, ROS levels in each group were measured, showing that the mean fluorescence intensity increased after treatment with cisplatin for 24 h (Fig. 3G, H). After treatment with cisplatin, the siCD36 group showed a significant decrease in ROS levels compared to the si-NC group (P < 0.01).
CD36 directly binds to FSP1 and promotes its degradation via ubiquitination
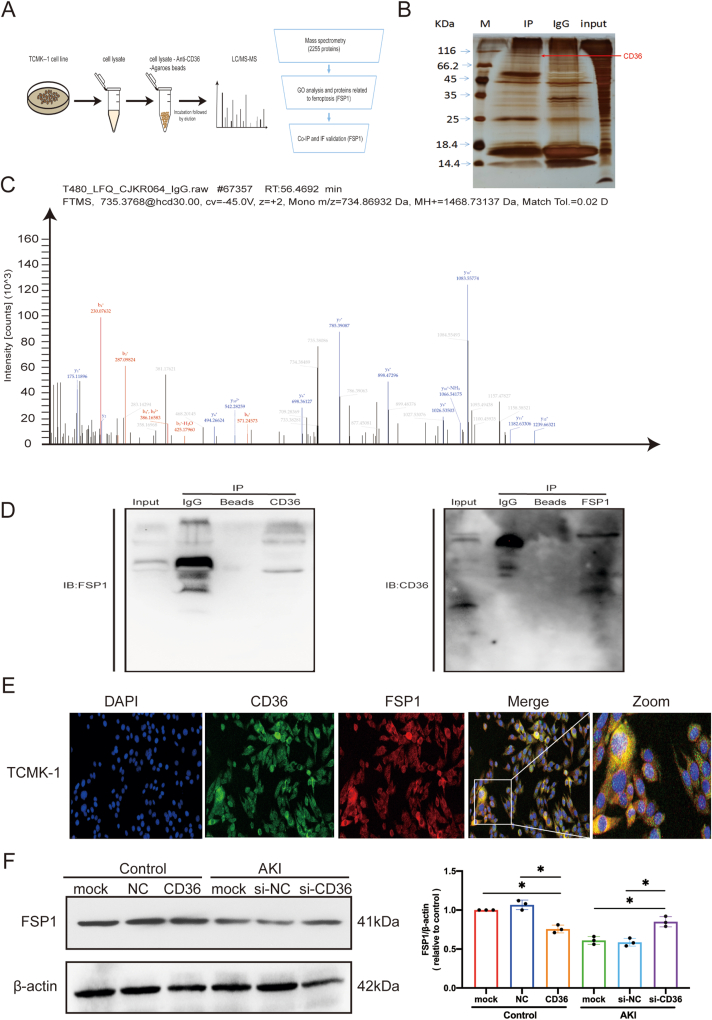
The above findings urged us to explore how CD36 affects ferroptosis in AKI. Therefore, LC-MS/MS analysis was performed on the co-immunoprecipitated CD36 complexes in TCMK-1 cells (Fig. 4A, B). With the cut-off criteria for unique peptides of ≥1 and confidence level of ≥95%, the analysis identified 2255 unique proteins that were co-immunoprecipitated with anti-CD36. All of the identified proteins are listed in Table S3. Procedural filtering showed one potential CD36-interacting protein FSP1, which is a ferroptosis resistance protein (Fig. 4C). To further validate the result of mass spectrometry analysis, co-IP assays were conducted and confirmed the interaction between CD36 and FSP1 (Fig. 4D). The co-localization assay revealed that CD36 and FSP1 were co-located in the cytoplasm of TCMK-1 cells, providing evidence for their interaction (Fig. 4E). Moreover, ectopic up-regulation of CD36 led to the degradation of FSP1 in TCMK-1 cells, while down-regulation of CD36 resulted in the increased enrichment of FSP1 in TCMK-1 cells treated with cisplatin (Fig. 4F).
Figure 4.
Interacted CD36 proteins were screened using LC-MS/MS. (A) Mass spectrometry process flowchart. (B) Silver staining map of the co-IP assay with CD36 antibody in TCMK-1 cells. The first lane represents marker, the second lane represents co-IP using CD36 antibody, the third lane is IgG antibody, and the fourth lane is IP products of CD36 in TCMK-1 cells. (C) MS spectra for FSP1 fragment identified by LC-MS/MS. (D) Total protein samples from TCMK-1 cells were extracted and interacting CD36 proteins and FSP1 were detected via co-IP. Western blotting using anti-FSP1 and anti-CD36 antibodies showed that CD36 interacts with FSP1. (E) Representative immunofluorescence staining images of CD36 (green) and FSP1 (red) in TCMK-1 cells. (F) FSP1 expression after transfection with CD36 small interfering RNA (siRNA).
Since ubiquitination plays a crucial role in protein degradation,24 LC/MS-MS was used to identify the FSP1 ubiquitination sites. The results demonstrated that K16 and K24 were the major FSP1 ubiquitination sites (Fig. 5A). It was thus hypothesized that FSP1 ubiquitination may be related to the relationship between CD36 and FSP1 in AKI. To ascertain the effect of AKI on FSP1 stability, cycloheximide (CHX) chase analysis and half-life test were performed in TCMK-1 cells with or without cisplatin treatment. The CHX chase analysis demonstrated that cisplatin-induced AKI led to a substantial decrease in FSP1 protein stability (Fig. 5B). In addition, the half-life test results showed that the FSP1 protein in the control group was significantly more stable (T1/2 = 6.25 h) than that in the cisplatin group (T1/2 = 3.45 h; Fig. 5C). Then, the effect of CD36 on FSP1 ubiquitination was determined in TCMK1 cells. FSP1 polyubiquitination, as demonstrated by expressing FSP1 with HA-tagged ubiquitin (HA-Ub), was dramatically attenuated by CD36 (Fig. 5D). In contrast, FSP1 ubiquitination was diminished by the depletion of CD36. In addition, the FSP1 protein level in TCMK1 cells was robustly enhanced by treatment with proteasome inhibitor MG132 (Fig. 5E). The mRNA levels of FSP1 remained stable with the addition of MG132 (Fig. 5F), indicating that endogenous FSP1 was also subject to significant proteasomal degradation. Moreover, based on the detection of CD36 and Ub in total lysates of cells, it was also demonstrated that CD36 promoted FSP1 ubiquitination degradation. This effect was abolished by MG132 treatment (Fig. 5G). Likewise, the amount of pulled down ubiquitinated FSP1 after FSP1 IP was lower in the precipitates of CD36 plus MG132 samples than in the vector plus MG132 samples (Fig. 5H). Overall, these results are consistent with the idea that CD36 promotes FSP1 polyubiquitination by physically interacting with FSP1 in AKI.
Figure 5.
Ubiquitination of FSP1 in vitro. (A) Positions of identified ubiquitination sites in TCMK-1 cells determined using mass spectrometry. Sequences marked with light red indicate ubiquitination positions. (B) TCMK-1 cells were treated with cycloheximide (CHX) at different time points. Cell lysates were harvested over 10 h and analyzed on SDS/PAGE followed by western blotting. (C) FSP1 protein from (B) half-life. (D) Plasmids encoding FSP1 and HA-ubiquitin (Ub) were co-transfected into TCMK-1 cells. Then, cell lysates were immunoprecipitated with anti-FSP1 or anti-HA and Western blot analysis was performed for FSP1 and HA showing ubiquitinated species of FSP1. (E) FSP1 protein level with different concentrations of MG132 was analyzed. (F) TCMK-1 cells were treated with MG132 at different concentrations. FSP1 mRNA levels were analyzed using real-time PCR. Results are shown as mean ± standard deviation from three independent experiments. Significance levels were estimated using ANOVA. ∗∗P < 0.01. P < 0.05 was considered statistically significant. ns, no significant difference. (G) Total lysates were analyzed for FSP1 and ubiquitinated proteins by immunoblotting using anti-FSP1 and anti-Ub antibodies. The decrease in FSP1 protein observed upon CD36 overexpression was abolished by MG132, while the ubiquitination of FSP1 was increased. (H) IP with FSP1 antibody and western blotting with anti-Ub show that ubiquitination of FSP1 was higher in CD36-overexpressing plus MG132-treated cells vs. control untreated cells as well as empty vector-transfected plus MG132-treated cells. IgG was used as a negative control.
CD36 regulates ferroptosis via FSP1 in AKI
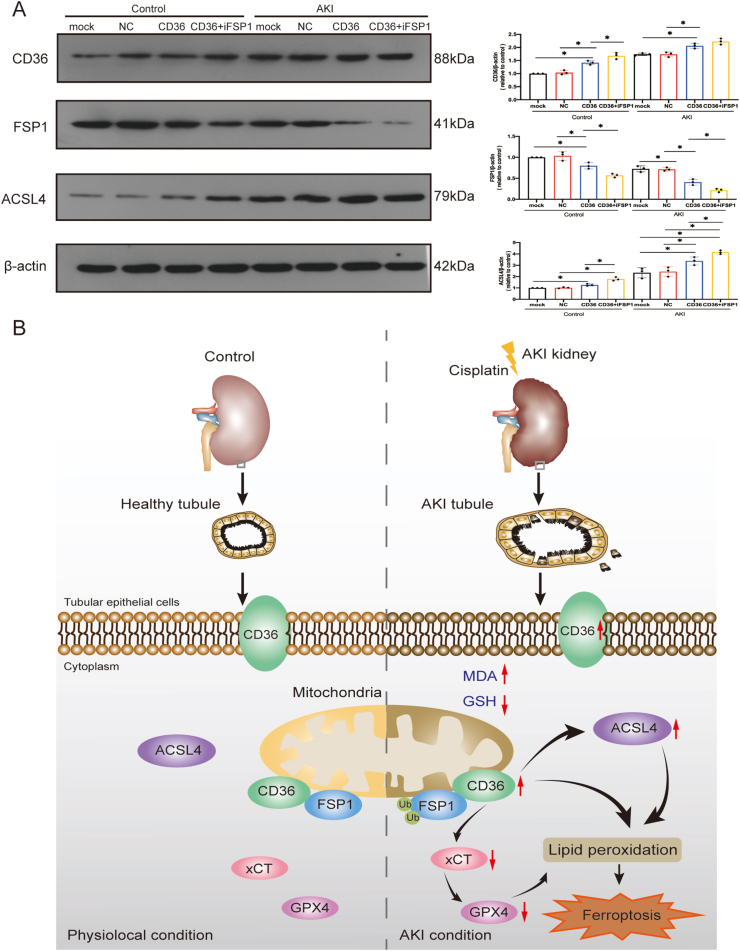
To examine the functional relationship between CD36 and FSP1 in AKI, iFSP1 was used in TCMK-1 cells when CD36 was overexpressed. The results showed that the expression of ferroptosis marker ACSL4 was increased in CD36-overexpressing TCMK-1 cells upon the treatment of cisplatin. Interestingly, the increasing effect was enhanced when CD36 overexpression was combined with the FSP1 inhibitor (Fig. 6A). Collectively, these results demonstrate that the ferroptosis-promoting effect induced by overexpression of CD36 can be regulated by iFSP1 in TCMK-1 cells treated with cisplatin. Mechanistically, the interaction between CD36 and ferroptosis-associated FSP1 was a key molecular event that triggered the activation of ferroptosis (Fig. 6B).
Figure 6.
CD36 regulates ferroptosis via FSP1 in AKI. (A) After CD36 overexpression or iFSP1 treatment, western blotting was used to detect the expression of CD36, FSP1, and ACSL4 in TCMK-1 cells, and quantitative statistical analysis for each protein (n = 3) was also performed. Data are expressed as means ± standard deviation. ∗P < 0.05 and ∗∗P < 0.01 vs. control group. (B) Mechanism diagram. Ferroptosis was inactivated in cells after AKI, resulting in CD36 overexpression, which promoted AKI after the ubiquitination of FSP1.
Discussion
The present study uncovered a novel AKI-related molecule CD36 that might contribute to proximal tubular cell ferroptosis. Of note, the expression of CD36 was substantially elevated in the renal tissues of AKI patients. Thus, high levels of CD36 might be an independent correlated factor for the kidney function of AKI patients. Moreover, CD36−/− AKI mouse and TCMK1 cell models further confirmed that lipid peroxidation and ferroptosis depended on the AKI-promoting effect of CD36 in vivo and in vitro. These findings revealed the role of CD36 in the progression of AKI, highlighting the possibility of CD36 being a promising biomarker for AKI.
CD36 is a multifunctional glycoprotein and a member of the class B scavenger receptor family that can transport long-chain fatty acids.25 Numerous studies have indicated that CD36 mediates inflammation and oxidative stress26 and may serve as a significant therapeutic target in metabolic diseases.14 In addition, CD36 regulates proliferation, metastasis, and angiogenesis in multiple tumor cells.27 Currently, studies on CD36 in kidney diseases are relatively limited, mainly involving chronic kidney diseases and diabetic nephropathy.28 Jong et al have observed that human CD36 overexpression is a risk factor in proximal tubules in folic acid-induced AKI.16 However, the biological role and underlying mechanisms of CD36 in AKI remain largely unclear. Therefore, the accurate function and mechanism of CD36 in AKI need to be elucidated. In the present study, CD36 was shown to be up-regulated in the renal tissues of AKI mice using RNA-seq. The expression trend was consistent in the renal tissues of AKI mice and TCMK-1 cells treated with cisplatin. Moreover, the up-regulation of CD36 in renal tissues was positively correlated with kidney function indexes Scr and BUN in AKI patients. Therefore, these results suggest that CD36 might be an important factor in AKI.
Previous studies have shown that ferroptosis is essential in AKI induced by ischemia/reperfusion and cisplatin.21,29 Thus, discovering more ferroptosis genes may shed new light on the regulation of AKI. In the present study, AKI was induced in CD36 KO mice upon treatment with cisplatin. Less mitochondrial damage was observed in renal tissues in the CD36−/− AKI mice group using TEM. In addition, the level of pro-ferroptosis marker ACSL4 was significantly lower, while levels of anti-ferroptosis markers xCT and GPX4 were higher in CD36−/− AKI mice compared to the WT AKI mice. Furthermore, the data showed that MDA levels were significantly lower in CD36−/− AKI mice than in WT AKI mice. The GSH levels were significantly higher in CD36−/− AKI mice than in WT AKI mice. All of these data demonstrate that CD36 might be responsible for lipid peroxidation and ferroptosis in AKI.
Further exploration of the mechanism underlying the effect of CD36 on ferroptosis in AKI using LC-MS/MS analysis uncovered a ferroptosis-related CD36-interacting protein FSP1 that was previously not reported. Consistently, co-IP and co-localization assays validated the interaction between CD36 and FSP1. FSP1, also known as apoptosis-inducing factor mitochondria-associated 2, was initially described as a pro-apoptotic gene.30 It is mainly localized in the mitochondrial outer membrane and is distributed in the cytoplasm.31 FSP1 is a glutathione-independent ferroptosis suppressor that confers protection against ferroptosis elicited by GPX4 deletion. A previous study has indicated that the suppression of ferroptosis by FSP1 is mediated by coenzyme Q10, also known as ubiquinone.32 It has also revealed that ferroptosis suppression by FSP1 was mediated by ubiquinone, which traps lipid peroxyl radicals that affect lipid peroxidation. The FSP1 ubiquinone pathway exists as a stand-alone parallel system.32,33 Although the present study did not investigate the relationship between GPX4 and the FSP1 system, the interaction between FSP1 and CD36 was confirmed. Moreover, the present data showed that ectopic up- or down-regulation of CD36 led to degradation or increased enrichment of FSP1 in TCMK-1 cells. Combined with the results from a previous study, this verified that CD36 could inhibit the expression of FSP1 by binding with FSP1.
Ubiquitin is an evolutionarily conserved protein that post-translationally marks proteins for degradation and is found in all tissues of eukaryotic organisms. Ubiquitination degradation is crucial for a plethora of physiological processes, including cell survival and differentiation and death.24,34 The ubiquitin molecule is conjugated to target proteins and triggers the next multistep enzymatic cascade.35 Ubiquitination requires the sequential actions of three enzymes. In the first step of this enzymatic cascade, the ubiquitin-activating enzyme UBA1 (E1) adenylates and then binds to the residues of the ubiquitin molecule relying on the energy supply from ATP. Subsequently, the E1 enzyme then transfers a ubiquitin molecule to the ubiquitin-conjugating enzyme E2. Finally, the E2 enzyme transfers the ubiquitin to the target protein with the help of the ubiquitin ligase E3, resulting in the ubiquitination of the target proteins.36 Currently, there is growing evidence that ubiquitin modification is involved in ferroptosis.37, 38, 39 Previous studies have found that some key molecular participants of ferroptosis, such as GPX4, SLC7A11, and Nrf2, can be regulated by E3 and de-ubiquitylating enzymes in cancer or ischemia/reperfusion disease.39,40 In addition, some studies have also described ubiquitination involved in ischemia reperfusion-induced AKI.41,42 Regrettably, very few studies have focused on the ubiquitination of ferroptosis in AKI. The present study is the first to demonstrate FSP1 ubiquitination. It also determined the protein levels of FSP1 after treatment with different concentrations of MG132 (a proteasome inhibitor). The results showed that the FSP1 mRNA levels remained stable with the addition of MG132, indicating that MG132 prevented the degradation of FSP1. Then, the half-life of FSP1 was examined upon the treatment of CHX, a blocker of protein synthesis and eukaryotic translation.43,44 These results indicated that FSP1 protein was ubiquitinated, with ubiquitination sites located at K16 and K24. In addition, CD36 promotion of FSP1 ubiquitination degradation was analyzed using immunoblotting. Notably, unlike the typical ubiquitination site, these results provided evidence for ubiquitination occurring at K16 and K24 of the lysine residues. Some research has also presented evidence for ubiquitination occurring at K24 and K16, which have been reported as ubiquitination sites in virus infection and the immune system.45,46 Thus, it can be speculated that post-translational modifications occurring on different critical lysine residues may provide a possible explanation for the functional heterogeneity among ubiquitination proteins. Moreover, different disease models demonstrated different residues, implying a unique protein signature imparted by the disease state.
In conclusion, CD36 may play a key role in the progression of AKI. The interaction between CD36 and ferroptosis-associated FSP1 was a key molecular event that triggered the activation of ferroptosis. Understanding the molecular mechanism and signaling pathways of ferroptosis may provide new insights and treatment methods for AKI. The present results emphasize a novel CD36 mechanism in the AKI process. Targeting CD36 may provide a promising treatment option for AKI.
Author contributions
X.L., X.R., and Z.Z. designed this research study. Y.M., L.H., and F.T. performed the experiments. Q.C. and L.H. analyzed the data. C.W., X.Z., and P.Y. participated in the sample collection. Z.Z. and L.H. wrote and revised the paper. X.L. provided financial support. All authors reviewed and approved the manuscript.
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81873604), the Medical Scientific Research Project of the Chongqing Health Commission (China) (No. 2022GDRC005), and Chongqing Science and Technology Agency (China) (CSTB2022NSCQ-MSX0984).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Figure S1 Gene set enrichment analysis of differentially expressed genes in AKI mice.
References
- 1.Hoste E.A.J., Kellum J.A., Selby N.M., et al. Global epidemiology and outcomes of acute kidney injury. Nat Rev Nephrol. 2018;14(10):607–625. doi: 10.1038/s41581-018-0052-0. [DOI] [PubMed] [Google Scholar]
- 2.Martin R.K. Acute kidney injury: advances in definition, pathophysiology, and diagnosis. AACN Adv Crit Care. 2010;21(4):350–356. doi: 10.1097/NCI.0b013e3181f9574b. [DOI] [PubMed] [Google Scholar]
- 3.Basile D.P., Anderson M.D., Sutton T.A. Pathophysiology of acute kidney injury. Compr Physiol. 2012;2(2):1303–1353. doi: 10.1002/cphy.c110041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirschhorn T., Stockwell B.R. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–143. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Cao F., Yin H.L., et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z., Zhang H., Yi B., et al. VDR activation attenuate cisplatin induced AKI by inhibiting ferroptosis. Cell Death Dis. 2020;11(1):73. doi: 10.1038/s41419-020-2256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan X.Z., Varghese Z., Powis S.H., et al. Human mesangial cells express inducible macrophage scavenger receptor. Kidney Int. 1999;56(2):440–451. doi: 10.1046/j.1523-1755.1999.00587.x. [DOI] [PubMed] [Google Scholar]
- 9.Hua W., Huang H.Z., Tan L.T., et al. CD36 mediated fatty acid-induced podocyte apoptosis via oxidative stress. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Susztak K., Ciccone E., McCue P., et al. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med. 2005;2(2):e45. doi: 10.1371/journal.pmed.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonen A., Luiken J.J., Arumugam Y., et al. Acute regulation of fatty acid uptake involves the cellular redistribution of fatty acid translocase. J Biol Chem. 2000;275(19):14501–14508. doi: 10.1074/jbc.275.19.14501. [DOI] [PubMed] [Google Scholar]
- 12.Smith B.K., Jain S.S., Rimbaud S., et al. FAT/CD36 is located on the outer mitochondrial membrane, upstream of long-chain acyl-CoA synthetase, and regulates palmitate oxidation. Biochem J. 2011;437(1):125–134. doi: 10.1042/BJ20101861. [DOI] [PubMed] [Google Scholar]
- 13.Trites M.J., Febbraio M., Clugston R.D. Absence of CD36 alters systemic vitamin A homeostasis. Sci Rep. 2020;10 doi: 10.1038/s41598-020-77411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., Okamura D.M., Lu X., et al. CD36 in chronic kidney disease: novel insights and therapeutic opportunities. Nat Rev Nephrol. 2017;13(12):769–781. doi: 10.1038/nrneph.2017.126. [DOI] [PubMed] [Google Scholar]
- 15.Hou Y., Wu M., Wei J., et al. CD36 is involved in high glucose-induced epithelial to mesenchymal transition in renal tubular epithelial cells. Biochem Biophys Res Commun. 2015;468(1–2):281–286. doi: 10.1016/j.bbrc.2015.10.112. [DOI] [PubMed] [Google Scholar]
- 16.Jung J.H., Choi J.E., Song J.H., et al. Human CD36 overexpression in renal tubules accelerates the progression of renal diseases in a mouse model of folic acid-induced acute kidney injury. Kidney Res Clin Pract. 2018;37(1):30–40. doi: 10.23876/j.krcp.2018.37.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holditch S.J., Brown C.N., Lombardi A.M., et al. Recent advances in models, mechanisms, biomarkers, and interventions in cisplatin-induced acute kidney injury. Int J Mol Sci. 2019;20(12):3011. doi: 10.3390/ijms20123011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krabbendam I.E., Honrath B., Dilberger B., et al. SK channel-mediated metabolic escape to glycolysis inhibits ferroptosis and supports stress resistance in C. elegans. Cell Death Dis. 2020;11(4):263. doi: 10.1038/s41419-020-2458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buonafine M., Martinez-Martinez E., Jaisser F. More than a simple biomarker: the role of NGAL in cardiovascular and renal diseases. Clin Sci (Lond) 2018;132(9):909–923. doi: 10.1042/CS20171592. [DOI] [PubMed] [Google Scholar]
- 20.Stockwell B.R., Friedmann Angeli J.P., Bayir H., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L.L., Liao X.H., Sun H., et al. Augmenter of liver regeneration protects the kidney from ischaemia-reperfusion injury in ferroptosis. J Cell Mol Med. 2019;23(6):4153–4164. doi: 10.1111/jcmm.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye T., Shi H., Wang Y., et al. Contrasting changes caused by drought and submergence stresses in bermudagrass (Cynodon dactylon) Front Plant Sci. 2015;6:951. doi: 10.3389/fpls.2015.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Feng D., Wang Z., et al. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019;26(11):2284–2299. doi: 10.1038/s41418-019-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popovic D., Vucic D., Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Mao Y., Wang B., et al. Quercetin improving lipid metabolism by regulating lipid metabolism pathway of ileum mucosa in broilers. Oxid Med Cell Longev. 2020;2020 doi: 10.1155/2020/8686248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy D.J., Kuchibhotla S., Westfall K.M., et al. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc Res. 2011;89(3):604–613. doi: 10.1093/cvr/cvq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo X., Zheng E., Wei L., et al. The fatty acid receptor CD36 promotes HCC progression through activating Src/PI3K/AKT axis-dependent aerobic glycolysis. Cell Death Dis. 2021;12(4):328. doi: 10.1038/s41419-021-03596-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Y., Wang Q., Han B., et al. CD36 promotes NLRP3 inflammasome activation via the mtROS pathway in renal tubular epithelial cells of diabetic kidneys. Cell Death Dis. 2021;12(6):523. doi: 10.1038/s41419-021-03813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L., Yu P., Wang T.T., et al. Polydatin attenuates cisplatin-induced acute kidney injury by inhibiting ferroptosis. Oxid Med Cell Longev. 2022;2022 doi: 10.1155/2022/9947191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M., Xu L.G., Li X., et al. AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J Biol Chem. 2002;277(28):25617–25623. doi: 10.1074/jbc.M202285200. [DOI] [PubMed] [Google Scholar]
- 31.Ohiro Y., Garkavtsev I., Kobayashi S., et al. A novel p53-inducible apoptogenic gene, PRG3, encodes a homologue of the apoptosis-inducing factor (AIF) FEBS Lett. 2002;524(1–3):163–171. doi: 10.1016/s0014-5793(02)03049-1. [DOI] [PubMed] [Google Scholar]
- 32.Doll S., Freitas F.P., Shah R., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 33.Bersuker K., Hendricks J.M., Li Z., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaid S., Brandts C.H., Serve H., et al. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hershko A., Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 36.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 37.Harrigan J.A., Jacq X., Martin N.M., et al. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17(1):57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19(1):59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Wang Y., Li Z., et al. Regulation of ferroptosis pathway by ubiquitination. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.699304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma S., Sun L., Wu W., et al. USP22 protects against myocardial ischemia-reperfusion injury via the SIRT1-p53/SLC7A11-dependent inhibition of ferroptosis-induced cardiomyocyte death. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.551318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen L., Zhang Q., Tu S., et al. SIRT3 mediates mitofusin 2 ubiquitination and degradation to suppress ischemia reperfusion-induced acute kidney injury. Exp Cell Res. 2021;408(2) doi: 10.1016/j.yexcr.2021.112861. [DOI] [PubMed] [Google Scholar]
- 42.Meyer-Schwesinger C. The ubiquitin-proteasome system in kidney physiology and disease. Nat Rev Nephrol. 2019;15(7):393–411. doi: 10.1038/s41581-019-0148-1. [DOI] [PubMed] [Google Scholar]
- 43.Santos D.A., Shi L., Tu B.P., et al. Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 2019;47(10):4974–4985. doi: 10.1093/nar/gkz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu D., Osuka S., Zhang Z., et al. Bai1 suppresses medulloblastoma formation by protecting p53 from Mdm2-mediated degradation. Cancer Cell. 2018;33(6):1004–1016.e5. doi: 10.1016/j.ccell.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fantini D., Vascotto C., Marasco D., et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res. 2010;38(22):8239–8256. doi: 10.1093/nar/gkq691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chattopadhyay S., Kuzmanovic T., Zhang Y., et al. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity. 2016;44(5):1151–1161. doi: 10.1016/j.immuni.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.