Abstract
Cartilage development is controlled by the highly synergistic proliferation and differentiation of growth plate chondrocytes, in which the Indian hedgehog (IHH) and parathyroid hormone-related protein-parathyroid hormone-1 receptor (PTHrP-PTH1R) feedback loop is crucial. The inositol-requiring enzyme 1α/X-box-binding protein-1 spliced (IRE1α/XBP1s) branch of the unfolded protein response (UPR) is essential for normal cartilage development. However, the precise role of ER stress effector IRE1α, encoded by endoplasmic reticulum to nucleus signaling 1 (ERN1), in skeletal development remains unknown. Herein, we reported that loss of IRE1α accelerates chondrocyte hypertrophy and promotes endochondral bone growth. ERN1 acts as a negative regulator of chondrocyte proliferation and differentiation in postnatal growth plates. Its deficiency interrupted PTHrP/PTH1R and IHH homeostasis leading to impaired chondrocyte hypertrophy and differentiation. XBP1s, produced by p-IRE1α-mediated splicing, binds and up-regulates PTH1R and IHH, which coordinate cartilage development. Meanwhile, ER stress cannot be activated normally in ERN1-deficient chondrocytes. In conclusion, ERN1 deficiency accelerates chondrocyte hypertrophy and cartilage mineralization by impairing the homeostasis of the IHH and PTHrP/PTH1R feedback loop and ER stress. ERN1 may have a potential role as a new target for cartilage growth and maturation.
Keywords: Cartilage development, ER stress, ERN1, IHH, PTHrP/PTH1R
Introduction
Cartilage formation is a dynamic cellular process involving different types of cartilage, such as hyaline cartilage, fibrous cartilage, and elastic cartilage.1, 2, 3 Endoplasmic reticulum stress (ER stress) is a protective mechanism involved in the entire process of cartilage differentiation.4 The unfolded protein response (UPR), which is initiated to promote cell survival, directly participates in the degradation and refolding of abnormal proteins to protect cells by maintaining the stability of the environment in the ER.5,6 IRE1α is a transmembrane protein located on the ER membrane and serves as a type I transmembrane serine/threonine protein kinase and a site-specific endoribonuclease, which splices XBP1u to form XBP1s.7, 8, 9, 10 Our previous research showed that the IRE1α/XBP1s signaling pathway participates in the process of cartilage formation and bone growth.11,12 Furthermore, the expression profile of IRE1α and the phosphorylation IRE1α were increased in the articular cartilage of osteoarthritis patients compared with that in the normal.13 However, the mechanism underlying IRE1α regulation of cartilage maturation remains unknown.
It is well established that the Indian hedgehog (IHH) and parathyroid hormone-related protein-parathyroid hormone-1 receptor (PTHrP-PTH1R) negative feedback loop is a pivotal signaling pathway that regulates cartilage development. IHH can be synthesized and secreted by anterior hypertrophic chondrocytes and regulates chondrocyte differentiation. IHH promotes the transformation of chondrocytes from proliferation to hypertrophy. Furthermore, elevated IHH levels promote the secretion of PTHrP by the perichondrium. PTHrP can bind to its receptor PTH1R and then inhibits the hypertrophy and maturation of chondrocytes, thereby forming a negative feedback loop that dynamically regulates cartilage development.14, 15, 16 The role of PTHrP signaling depends on the location and amount of PTH1R, a G protein-coupled receptor highly expressed in the pre-hypertrophic zone. A decrease in PTHrP signaling is directly reflected by a decrease in PTH1R expression, indicating stage terminal differentiation.17,18
In the current study, the role of IRE1α in cartilage morphological changes, proliferation and differentiation of chondrocytes, and endochondral ossification were investigated in vitro and in vivo. Furthermore, we explored the imbalance of the PTHrP/PTH1R and IHH feedback loop, caused by ERN1 deficiency, in cartilage development. Finally, we elucidated the mechanism that ERN1 deficiency affects PTH1R and IHH leading to abnormal chondrocyte hypertrophy and cartilage maturation.
Methods
Cells
The human chondrocyte C28/I2 cells were provided by Professor Chuanju Liu (New York University). The chondrogenic cell line ATDC5 was provided by Professor Lin Chen (Daping Hospital, Army Medical University, Chongqing). C28/I2 cells19 were cultured in complete DMEM medium (Gibco, California, USA), C3H10T1/2 cells were cultured in complete MEM-α medium (Gibco, California, USA), and ATDC5 cells were cultured in complete DMEM/F12 medium (Gibco, California, USA), in a 5% CO2 incubator at 37 °C. The complete medium was supplemented with 10% fetal bovine serum (FBS) (BI, C04001050X10) and 10 U/mL penicillin-streptomycin (Beyotime, C0222). Trypsinase (0.5%) was used to trypsinize the cells at a density of 70%–80%.
Micromass culture
The micromass culture was performed as described previously.20 Briefly, the ATDC521 or C3H10T1/2 cells were trypsinized and then resuspended in DMEM/F12 or MEM-α medium with 10% FBS at a concentration of 106 cells per mL, and six drops of 100 μL of cells were placed in a 60-mm tissue culture dish (Becton Dickinson). After 2-h incubation at 37 °C, 1 mL of medium containing 10% FBS and BMP2 protein (300 ng/mL) was added. The media was replaced approximately every 2–3 days.
siRNA transfection and adenovirus infection
A mixture of 5 μL/mL PEI (Sigma-Aldrich, Cat No. 408727) reagent and 100 nM ERN1 small interfering RNA (siRNA) was evenly added to the complete medium (siERN1 group) when the C28/I2 and ATDC5 cell density reached 50%. The same amount of scrambled siRNA was used to set up a negative control group (NC group). siRNA was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The siRNA sequences are listed in Table S2.
Adenovirus IRE1α siRNA and adenovirus encoding IRE1α were constructed, respectively, using methods described previously.11,12 All constructs were verified by nucleic acid sequencing and subsequent analysis was performed using BLAST software at http://www.ncbi.nlm.nih.gov/BLAST/. AdPTH1R was a gift from Prof. Quan Yuan (West China Hospital of Stomatology, Sichuan University, Chengdu, China).
Real-time polymerase chain reaction (PCR) and quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted from tissues or cells. PCR was performed according to the manufacturer's instructions (Vazyme, P112-01). The Mouse Direct PCR kit (Bimake, B40013) was used to detect the genotype of mouse tissue samples using PCR. Quantitative real-time PCR was performed using a CFX-connect fast real-time PCR system (Bio-Rad, USA). The results were analyzed by using the Ct (threshold cycle) method, and the relative expression levels of the genes were normalized to those of GAPDH, which served as an internal control, using the comparative 2-△△Ct method. The sample mixture was composed of 5 μL SYBR (Vazyme, Q711-03), 1 μL primers, 1 μL cDNA, and 3 μL ddH2O. The touchdown cycling conditions were as follows: 1) 95 °C for 3 min; 2) 95 °C for 10 s; 3) 58 °C for 30 s; 4) 39 cycles of steps 2 and 3; 5) 65 °C for 5 s; 6) 95 °C for 5 s. The primers used in PCR and qPCR are listed in Table S3.
Western blot
For Western blot analysis, RIPA buffer containing protease and phosphatase inhibitors were used to prepare whole-cell lysates. Protein lysates (30 μg) were first separated using 10% SDS-PAGE and then, transferred onto polyvinylidene difluoride (PVDF) membranes (0.45 μm, Millipore), which were subsequently blocked using 5% BSA at 37 °C for 2 h. After incubation with the primary antibodies overnight at 4 °C, 0.1% TBST was used to wash all membranes three times, 5 min each time. This was followed by HRP-anti-mouse IgG (Abcam, ab6789) or HRP-anti-rabbit IgG (Abcam, ab6721) incubation at 37 °C for 1 h, and the proteins were detected using chemiluminescence. The grey value of each membrane was quantitatively evaluated by Image Processing and Analysis in Java (Image J) software. The antibodies used in Western blot are listed in Table S4.
Transgenic mice
ERN1flox/flox mice were purchased from Shanghai Biomodel Organism Science & Technology Development Company Ltd., and Col2Cre mice were a kind gift from Prof. Lin Chen (Daping Hospital, Army Medical University; Chongqing, China). All mouse strains were placed on a full C57BL/6 J background.
Primary cultures of murine chondrocytes and explant culture
Articular cartilage of WT mice, ERN1 CKO mice or ERN1 control mice at age of 2 weeks was digested in type II collagenase (Worthing, LS004174) at a final concentration of 1 mg/mL at 37°C overnight under aseptic conditions, and then centrifuged to collect and extract primary chondrocytes. Explants from cartilage tissues of the knee joints of 8 WT mice at age of 2 weeks were removed under aseptic conditions. Primary chondrocytes and explants were cultured in complete DMEM/F12 (Gibco) medium with 10% fetal bovine serum (FBS) and 10 U/mL penicillin-streptomycin in a 5% CO2 incubator at 37 °C.
EdU and TUNEL
C28/I2 cells were inoculated onto coverslips in a 24-well plate with EdU fluid (Beyotime, C0078S) at 37 °C for 2 h. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.3% Triton X-100 at room temperature for 10 min. A mixture of click reaction buffer, CuSO4, azide 594, and click additive solution was added to the coverslips at room temperature for 30 min. Then, the cells on coverslips were stained with Hoechst33342 (Beyotime, C1026) for 5 min.
C28/I2 cells were treated with 4% paraformaldehyde for 30 min and 3% Triton X-100 at room temperature for 5 min. Tunel fluid (Beyotime, C1090) was placed in each well at 37 °C for 1 h. After washing with PBS, a confocal laser scanning microscope (Thermo Fisher Scientific, USA) was used to detect immunofluorescence staining.
Luciferase reporter assay and CHIP
The PTH1R or IHH luciferase reporter plasmid and XBP1s or negative control plasmid were transfected into 293T cells by using PEI. The activity of firefly and renilla luciferases was quantified using a dual luciferase reporter gene detection system (Promega Corp., Madison, WI, USA). To standardize the difference in transfection, firefly luciferase activity was normalized to that of renilla luciferase.
C28/I2 cells were submerged in DMEM containing 1% formaldehyde and incubated at 37 °C for 10 min. Fixation was stopped by the addition of 0.125 M glycine. The cell lysate was harvested and sheared with 12 pulses of 3% amplitude. Each pulse consisted of a 2.5-s sonication followed by a 7.5-s rest on ice to prevent heat build-up. The sample was incubated with 2 μg of anti-XBP1s (BioLegend, Cat. No. 647502) antibody or IgG (Cat. No. 2729) overnight at 4 °C on a vertical mixer. The enriched DNA fragments were harvested by using methods according to the manufacturer's instructions for the ChIP assay kit (Beyotime, P2078). The IHH (−747 to −553) and PTH1R (−1338 to −1137) promoter regions were amplified. The amount of immunoprecipitated DNA in each sample was represented as a percentage of the total amount of input chromatin, which was equivalent to 100%. The primers used in ChIP are listed in Table S1.
ALP staining
C3H10T1/2 cells were inoculated in a 6-well plate, and 4% paraformaldehyde was used to fix cells for 3 min. ALP mixture (Beyotime, P0321S) was put in each well for 15 min. After washing with PBS, the cells were stained again with the Nuclear Fast Red Solution. The positive cells were observed by microscope.
IHC staining, IF staining, and histological staining
Paraffin sections of the mouse joint were de-waxed and rehydrated. The sections were then incubated with a primary antibody at 4 °C overnight for IHC and IF. The immunohistochemistry kit (ZSGB-BIO, PV-9001) was used to analyze protein expression following a standardized protocol. The sections were incubated with DAB and hematoxylin for IHC staining, or specific secondary antibodies and DAPI for IF staining. The hematoxylin-eosin (HE) and Safranin–Fast Green stainings were performed according to the manufacturer's instructions for HE Staining Kit (Solarbio, G1120) and Modified Safranine O-Fast Green FCF Cartilage Stain Kit (Solarbio, G1371).
Intra-articular injection in mice
Three-week-old WT mice were used to construct the intra-articular injection model. The mice were randomly divided into the following three groups: i) the left knee joints received an injection of 10 μL PBS (NC group) and the right knee joints received an injection of 10 μL 4μ8C (MCE) at a concentration of 5 mg/mL (4μ8C-treated group), ii) the left knee joints received an injection of 5 μL 4μ8C at a concentration of 5 mg/mL and 5 μL PBS (4μ8C-treated group) and the right knee joints received an injection of 5 μL 4μ8C at a concentration of 5 mg/mL and 5 μL GDC-0449 (MCE) at a concentration of 2 mg/mL (4μ8C + GDC-0449-treated group), iii) the left knee joints received an injection of 10 μL PBS (NC group) and the right knee joints received an injection of 10 μL AdERN1 adenovirus (AdERN1 group).22, 23, 24, 25, 26 The injection was administered once weekly for four consecutive weeks. Samples were collected when mice were 8 weeks old.
Statistical analysis
Data were analyzed using the Statistical Package for the Social Sciences version 16.0. Experimental data were shown as the mean ± standard deviation (SD). All quantitative data were presented as the means and 95% confidence intervals (CIs). The difference between two groups in terms of intra-articular injection and explants was quantitated using the paired t-test, and the difference between two groups in the other experiments was quantitated using the independent-samples t-test.
Results
The morphological characteristics of skeletal development in ERN1-deficient mice
According to our previous study,13 we detected and found that the protein level of IRE1α in human degenerated cartilage was increased compared to that in the normal cartilage (P = 0.012) (Fig. S1). Then after the ERN1flox/+Col2Cre, ERN1flox/floxCol2Cre (ERN1 CKO), and ERN1flox/flox mice were identified correctly (Fig. S2–4), we compared the phenotypes of ERN1 CKO mice with those of ERN1flox/flox littermates (control mice) to explore the role of ERN1 in postnatal skeletal growth. The measure of body length (including tail length) (P = 0.007) (Fig. 1A, B), as well as X-ray examination results, showed that the average body length of ERN1 CKO mice increased by 6.1%, 5.1%, and 4.3% at 4 (P = 0.004), 8 (P = 0.012), and 12 (P = 0.015) weeks after birth (Fig. 1C–F). Meanwhile, the average weight of ERN1 CKO mice was greater than that of control mice at different weeks (P = 0.045) (Fig. 1G). The length of the femur was also increased by 8.9%, 9.2%, and 9% in mutant mice as compared to that in control mice at 4 (P = 0.022), 8 (P = 0.026), and 12 (P = 0.015) weeks after birth (Fig. 1H, I). This is consistent with the results of Alcian blue and Alizarin red full mouse skeleton staining, which showed that the body length, vertebrae, ribs, radius, femurs, tibia, and digital bone of ERN1 CKO mice were longer than that of control mice (Fig. 1J, K).
Figure 1.
Integral morphology assessment of skeletal phenotypes in ERN1 conditional knockout mice. (A, B) Body length of ERN1 CKO and control mice at 2 weeks. ∗P < 0.05, n = 6. (C) Body length of mice at 4 weeks, 8 weeks, and 12 weeks by X-ray. (D–F) Quantitation of body length at 4 weeks, 8 weeks, and 12 weeks. ∗P < 0.05, ∗∗P < 0.005, n = 6. (G) Body weight of ERN1 CKO and control mice at 1 week and 3 weeks. P > 0.05, ∗P < 0.05, n = 5. (H) Femur length of ERN1 CKO and control mice at 4 weeks. (I) Quantification of femur length at 4 weeks, 8weeks, and 12 weeks. ∗P < 0.05, n = 6. (J, K) Skeletons staining by Alcian blue and Alizarin red at 2 weeks.
Loss of ERN1 in mouse chondrocytes promotes chondrocyte hypertrophy
HE and Safranin O-Fast Green staining results showed that the proportion of the hypertrophic zone to the total zone (HZ/TZ) of the cartilage growth plate of ERN1 CKO mice at 1 (P = 0.002), 4 (P = 0.001), and 8 (P = 0.010) weeks after birth was increased compared to that of control mice (Fig. 2A, B), whereas no change was observed in the proportion of the proliferation zone to the total zone (PZ/TZ) of the cartilage growth plate at 1, 4, and 8 weeks between the two mouse groups (Fig. S5A). Both of the length of TZ and the HZ of ERN1 CKO mice at 4 (P < 0.001, P < 0.001, respectively) and 8 (P = 0.019, P < 0.001, respectively) weeks as well as the proportion of the HZ were also longer than those of control mice (Fig. 2C, D). Although ERN1 did not affect cartilage development at the embryonic stage in mice (Fig. S5B), the HZ of the growth plate of the ERN1 CKO mice exhibited an obvious enhancement after birth compared to that of control mice.
Figure 2.
Histological assessment of the cartilage growth plate in ERN1 conditional knockout mice and ERN1 deficiency in mouse chondrocytes accelerates chondrocyte hypertrophy. (A) HE staining of Tibia sections at 1 week, 4 weeks, and 8 weeks. (B) Quantification of hypertrophic zone (HZ)/growth plate length (TZ) ratio at 1 week, 4 weeks, and 8 weeks. ∗P < 0.05, ∗∗P < 0.005, n = 5. (C) SafrainO-Fast Green staining of Tibia sections at 4 weeks and 8 weeks. (D) Quantification of TZ and HZ of Tibia sections at 4 weeks and 8 weeks. ∗P < 0.05, ∗∗∗P < 0.001, n = 5. (E) The mRNA levels of ERN1, COL2A1, Aggrecan, COL10A1, and MMP13 were detected by qPCR in primary chondrocytes isolated from ERN1 CKO and control mice. ∗P < 0.05, ∗∗P < 0.005, n = 3. (F) The protein levels of IRE1α, COL10A1, and MMP13 in rib tissues at 2-week-old mice were detected by Western blot. (G) Quantification of IRE1α, COL10A1, and MMP13. ∗P < 0.05, ∗∗∗P < 0.001, n = 3. (H–K) IHC staining and statistical analysis of MMP13, COL10A1, Aggrecan, and COL2A1 at 4-week-old mice. ∗P < 0.05, ∗∗P < 0.005, n = 5. Scale bar = 200 μm.
Next, qPCR results showed that the levels of collagen type II alpha 1 chain (COL2A1) (P = 0.048) and Aggrecan (P = 0.042) were decreased, while those of COL10A1 (P = 0.043) and matrix metallopeptidase 13 (MMP13) (P = 0.021) were increased in ERN1-deficient primary chondrocytes (Fig. 2E). ERN1 knockdown in C28/I2 cells showed increased expression of COL10A1 (P = 0.006) and MMP13 (P = 0.017) and decreased expression of COL2A1 (P = 0.003) (Fig. S6). Western blot results also showed up-regulated protein levels of COL10A1 (P = 0.006), and MMP13 (P = 0.007) in the cartilage tissues of ERN1 CKO mice at 2 weeks compared to control mice (Fig. 2F, G). The levels of MMP13 (P = 0.005) and COL10A1 (P = 0.037) were up-regulated (Fig. 2H, I; Fig. S7A), while those of COL2A1 (P = 0.042) and Aggrecan (P = 0.003) were down-regulated in ERN1 CKO mice compared to those of control mice by IHC (Fig. 2J, K; Fig. S7B). The expression of transcription factor SOX9 was also decreased in ERN1 CKO mice (Fig. S7C). High-density micromass culture of ATDC5 cells test was used to simulate the process of chondrocyte differentiation and the result showed inhibited expression of IHH (P = 0.005), MMP13 (P = 0.004), and COL10A1 (P = 0.004) after ERN1 was overexpressed during bone morphogenetic protein 2 (BMP2)-induced chondrocyte differentiation (Fig. S8A–C).
ERN1 deficiency in chondrocytes promotes chondrocyte proliferation and inhibits apoptosis in growth plates
Knockdown of ERN1 in C28/I2 chondrocytes up-regulated the mRNA levels of proliferation-related genes, including proliferating cell nuclear antigen (PCNA) (P = 0.019) and CyclinB1 (P = 0.041) (Fig. 3A–C), whereas ERN1 overexpression using adenovirus infection in vitro down-regulated the levels of PCNA (P = 0.025) and CyclinB1 (P = 0.001) (Fig. 3D–F). In addition, the p-IRE1α inhibitor 4μ8C27,28 promoted the expression of PCNA (P = 0.007), CyclinB1 (P = 0.018), and CyclinD (P < 0.001) (Fig. 3G). The 5-Ethynyl-2′-deoxyuridine (EdU) assay showed cell proliferation ability of the 4μ8C-treated group was higher than the control group (P = 0.016) (Fig. 3H, I). Immunofluorescence (IF) staining results showed the up-align Ki67 expression in the growth plate of ERN1-deficient mice (Fig. 3J). Furthermore, the TUNEL assay exhibited the number of apoptotic cells in the 4μ8C-treated group was lower than that in the control group (P = 0.019) (Fig. 3K, L). IHC represented the expression of p-ERK, a classical signal molecule related to cell proliferation,29 which was higher in the growth plate of ERN1-deficient mice (Fig. 3M). Western blot also confirmed the result of IHC (P = 0.007) (Fig. 3N, O). In addition, we found that compared to the control, the mRNA levels of SNORC (C2ORF82) (P = 0.004) and CLEC3A (P < 0.001) were down-regulated whereas the mRNA level of CHI3L1 (P = 0.002) was up-regulated in ERN1 CKO group, indicating the functional imbalance of effector chondrocytes and regulatory chondrocytes30 in ERN1 CKO chondrocytes (Fig. S9). These data demonstrate that ERN1 negatively regulates the proliferation and differentiation of chondrocytes in postnatal growth plates.
Figure 3.
Loss of ERN1 promotes chondrocytes proliferation in vivo and in vitro. (A–C) The mRNA levels of ERN1, CyclinB1, and PCNA were detected by qPCR in C28/I2 cells after knocking down ERN1 by transfecting siRNA for 48 h ∗P < 0.05, ∗∗P < 0.005, n = 6. (D–F) The mRNA levels of ERN1, CyclinB1, and PCNA were detected by qPCR in C28/I2 cells after overexpressing ERN1 by infecting AdERN1 for 48 h ∗P < 0.05, ∗∗P < 0.005, n = 6. (G) The mRNA levels of CyclinB1, CyclinD1, and PCNA were detected by qPCR in C28/I2 cells after inhibiting p-IRE1α with 25 μM 4μ8C for 48 h ∗P < 0.05, ∗∗∗P < 0.001, n = 3. (H) EdU of C28/I2 cells after treating with 10 μM 4μ8C for 48 h. (I) Statistical analysis of EdU. ∗P < 0.05, n = 3. (J) IF of Ki67 in 2-week-old mice. (K) TUNEL of C28/I2 cells after treating with 15 μM 4μ8C for 48 h. (L) Statistical analysis of TUNEL. ∗P < 0.05, n = 3. (M) IHC of p-ERK of tibia sections in 4-week-old mice. (N) The protein levels of p-ERK and ERK in rib tissues at 2-week-old mice were detected by Western blot. (O) Quantification of p-ERK/ERK. ∗P < 0.05, n = 3. Scale bar = 200 μm.
Loss of ERN1 in chondrocytes accelerates endochondral ossification
In ERN1-deficient mice, the expression of osteogenic genes dentin matrix acidic phosphoprotein (DMP) (P < 0.001), osteoprotegerin (OPG) (P = 0.001), and Runt-related transcription factor 2 (RUNX2) (P < 0.001) was up-regulated in the primary chondrocytes of ERN1-deficient mice (Fig. 4A). Results of IHC showed that RUNX2 was increased in ERN1 CKO mice (P = 0.026) (Fig. 4B). Furthermore, the results of in vitro experiments showed that ERN1 knockdown up-regulated the expression of DMP (P = 0.003), OPG (P = 0.040), and RUNX2 (P = 0.006) in C3H10T1/2 cells (Fig. 4C), whereas ERN1 overexpression down-regulated the expression of these genes (P = 0.002, P < 0.001, P < 0.001, respectively) (Fig. 4D). Alkaline phosphatase (ALP) staining was lower in the ERN1 overexpressing group than in the control group (Fig. 4E).
Figure 4.
ERN1 promotes the chondrocyte ossification and mineralization. (A) The mRNA levels of DMP, OPG, and RUNX2 were detected in primary chondrocytes isolated from the cartilage of ERN1 CKO and control mice by qPCR. ∗∗P < 0.005, ∗∗∗P < 0.001, n = 6. (B) The expression of RUNX2 of growth plate was detected by IHC staining in 4-week-old WT mice. ∗P < 0.05, n = 4. Scale bar = 200 μm. (C) The mRNA levels of DMP, OPG, and RUNX2 in C3H10T1/2 cells after inhibiting ERN1 were detected by qPCR. ∗P < 0.05, ∗∗P < 0.005, n = 6. (D) The mRNA levels of DMP, OPG, and RUNX2 in C3H10T1/2 cells infected by AdERN1 were detected by qPCR. ∗∗P < 0.005, ∗∗∗P < 0.001, n = 6. (E) The ALP staining of AdERN1-infected C3H10T1/2 cells. Scale bar = 200 μm.
The PTHrP/PTH1R and IHH feedback loop imbalance in the ERN1-deficient mice
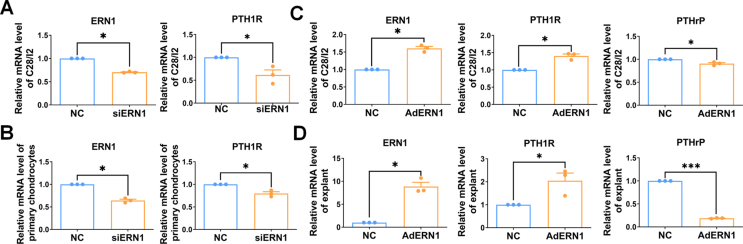
To clarify how ERN1 regulates cartilage development, we analyzed the mRNA profile of chondrogenesis-related genes in wild-type (WT) mice. The mRNA levels of ERN1, IHH, PTHrP, PTH1R and COL2A1 consistently peaked at 2 weeks, and those of ERN1, IHH, PTHrP, and PTH1R showed similar trends. However, the change trends of the mRNA levels of XBP1, COL2A1, and COL10A1 were not similar to those of ERN1, IHH, PTHrP, and PTH1R. The mRNA expression of XBP1 and COL2A1 increased after 1 week and declined after 2 weeks (Fig. 5A). The results of qPCR showed that the mRNA levels of IHH (P = 0.019) and PTHrP (P = 0.033) were increased, while PTH1R (P = 0.001) was decreased in the primary chondrocytes of ERN1 CKO mice (Fig. 5B). Western blot results also confirmed that the protein level of IHH (P = 0.020) was up-regulated whereas PTH1R (P = 0.041) was down-regulated in the cartilage of ERN1 CKO mice (Fig. 5C, D). The protein level of IHH (P = 0.012) was down-regulated while PTH1R (p = 0.019) was up-regulated in C28/I2 cells after overexpressing ERN1 (Fig. 5E, F). The qPCR results showed that the expressions of ERN1 (P = 0.007) and PTH1R (P = 0.025) were decreased following the knockdown of ERN1 in human C28/I2 cells and mouse primary chondrocytes (P = 0.006, P = 0.037, respectively) (Fig. S10A, B). In contrast, the levels of ERN1 and PTH1R were increased following ERN1 overexpression using adenovirus infection in C28/I2 cells (P = 0.008, P = 0.019, respectively) and explants (P = 0.013, P = 0.013, respectively), while PTHrP was decreased in the ERN1 overexpressing group (P = 0.048, P < 0.001, respectively) (Fig. S10C, D). Similarly, both the IF and IHC results showed that PTH1R expression was reduced and IHH expression was increased in the growth plate of ERN1-deficient mice (Fig. 5G–I). Western blot showed that the increased protein level of MMP13 after ERN1 knockdown in the overexpressed-PTH1R C28/I2 cells (Fig. 5J–L).
Figure 5.
Loss of ERN1 impaired the homeostasis of PTHrP/PTH1R and IHH feedback loop. (A) The qPCR analysis of ERN1, IHH, PTHrP, PTH1R, XBP1, COL2A1, and COL10A1 in primary chondrocytes isolated from the cartilage of C57BL/6 WT mice at 1 week, 2 weeks, and 4 weeks. (B) The qPCR analysis of IHH, PTHrP, and PTH1R in primary chondrocytes isolated from the cartilage of ERN1 CKO and WT mice. ∗P < 0.05, ∗∗P < 0.005, n = 5. (C) Western blot analysis of IRE1α, IHH, and PTH1R in rib tissues isolated from ERN1 CKO and control mice at 2 weeks, and (E) in C28/I2 cells infected with AdERN1. (D, F) Quantification of IRE1α, IHH, and PTH1R. ∗P < 0.05, ∗∗P < 0.005, n = 3. (G) IF and (I) IHC of PTH1R in the cartilage growth plate of tibia tissues isolated from ERN1 CKO and control mice at 4 weeks. (H) IF of IHH in the cartilage growth plate of tibia tissues in 2-week-old mice. (J–L) Western blot analysis of IRE1α and MMP13 in C28/I2 cells infected with AdPTH1R with or without siERN1. ∗P < 0.05, n = 3. Scale bar = 200 μm.
To investigate whether ERN1 is crucial in cartilage differentiation in vivo, we constructed a mouse model using intra-articular AdERN1 injection and found that the length of PZ (P = 0.027) and PZ/TZ (P = 0.018) were increased, HZ/TZ (P = 0.018) was decreased in the AdERN1 group compared to those of the control group (Fig. S11). Results of explant culture with AdERN1 also showed that COL10A1 (P = 0.017) and MMP13 (P = 0.011) were decreased after ERN1 overexpression (Fig. S12).
ERN1 regulates chondrocyte hypertrophy by controlling the IHH and PTH1R feedback loop through XBP1s
ERN1 is a key regulator of ER stress. After treating the primary cartilage chondrocytes of ERN1-deficient mice with 10 μg/mL tunicamycin (TM), a basic ER stress inducer,31 we found that certain ER stress-associated molecules, including XBP1s (P = 0.002), nuclear factor-erythroid factor 2-related factor 2 (Nrf2) (P = 0.004), protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) (P < 0.001), and p-PERK (P = 0.002), were down-regulated, suggesting that TM could not activate ER stress normally in the chondrocytes of ERN1 deficient mice (Fig. 6A; Fig. S13). Meanwhile, the mRNA level of ERN1 (P < 0.001) was up-regulated, while IHH (P = 0.029) was down-regulated in C28/I2 cells after treatment with TM. Inhibiting ERN1 reversed the effect of TM on ERN1 (P = 0.001) and IHH (P = 0.009) (Fig. 6B). We treated ATDC5 cells with 1% insulin-transferrin-selenium (ITS) to simulate the process of chondrocyte differentiation and found that siERN1 and 4μ8C could inhibit the expression of ERN1 (P = 0.001, P = 0.002, respectively), PTH1R (P = 0.007, P = 0.007, respectively), XBP1u (P < 0.001, P = 0.002, respectively), XBP1s (P = 0.037, P = 0.048, respectively), and COL2A1 (P < 0.001, P < 0.001) (Fig. 6E). In addition, XBP1u can be spliced to produce the transcription factor XBP1s after activating the phosphorylation of IRE1α during ER stress. XBP1s can enter the nucleus to regulate the transcription and expression of related genes. PCR also revealed that ERN1 knockdown decreased the mRNA expression levels of XBP1s (P = 0.043) (Fig. 6C, D). Furthermore, overexpression of XBP1s up-regulates the transcription of pGL3-IHH (P = 0.044) or pGL3-PTH1R (P = 0.045) (Fig. 6F, G, J, K), immunoprecipitation (ChIP) confirmed that XBP1s could bind to the promoter regions of IHH (−747 to −553) (P = 0.048) and PTH1R (−1338 to −1137) (P = 0.032) (Fig. 6H, L). The qPCR showed that PTH1R (P < 0.001) and IHH (P = 0.002) were increased following XBPIs overexpression using adenovirus infection in C3H10T1/2 cells (Fig. 6I, M).
Figure 6.
IRE1α phosphorylation activity is required for PTH1R and IHH transcription. (A) The protein levels of XBP1s, Nrf2, PERK, and p-PERK were detected by Western blot in primary chondrocytes isolated from ERN1 CKO and control mice. (B) The mRNA levels of ERN1 and IHH were detected in C28/I2 cells after adding with 10 ug/mL Tunicamycin (TM) for 6 h ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.001, n = 3. (C, D) The expression of XBP1s was quantified by RT-PCR. ∗P < 0.05, n = 3. (E) The mRNA levels of ERN1, XBP1u, XBP1s PTH1R, and COL2A1 were detected by qPCR in ATDC5 cells after treating with 1% ITS for 5 days with or without 25 μM 4μ8C for 48 h ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.001, n = 3. (F, J) Prediction results of XBP1s binding sites in PTH1R or IHH promoter region from Human TFDB Database (http://bioinfo.life.hust.edu.cn/HumanTFDB/). (G) Luciferase reporter assays of PTH1R and (K) IHH in 293T cells, ∗P < 0.05, n = 3. (H) CHIP of PTH1R (−1338 to −1137) and (L) IHH (−747 to −553) in 293T cells. ∗P < 0.05, n = 3. (I) Expression of PTH1R and (M) IHH were detected by qPCR in C3H10T1/2 cells. ∗∗P < 0.005, ∗∗∗P < 0.001, n = 3.
PTH1R (P = 0.002) and IHH (P = 0.041) were inhibited at the mRNA level in explants or primary chondrocytes by 4μ8C, an inhibitor of IRE1α phosphorylation (Fig. 7A, B). After intra-articular injection with 4μ8C, the results showed that the PZ length (P = 0.026) and PZ/TZ (P = 0.019) in the growth plate of the 4μ8C-treated group decreased compared to those of the control group (Fig. 7C, D). This is consistent with the IHC results that the expression of PTH1R (P = 0.027) was decreased, whereas that of MMP13 (P = 0.039) was increased in the 4μ8C-treated group compared to those of the control group (Fig. 7E–H). We then determined the effect of 4μ8C combined with abaloparatide, a basic PTH1R activator,32,33 on cartilage. We observed that the expression of PTH1R (P = 0.015) and COL2A1 (P < 0.001) was increased, while that of MMP13 (P = 0.019) was inhibited by 4μ8C combined with abaloparatide in cartilage explants compared with that of the 4μ8C-treated group (Fig. 7I), suggesting that abaloparatide could rescue, at least partly, the expression of COL2A1 and MMP13 in the 4μ8C-treated cartilage tissues. Meanwhile, the intra-articular injection with PTH1R adenovirus promoted the proportion of proliferation zone (PZ/TZ, P = 0.032) and reduced the proportion of hypertrophic zone (HZ/TZ, P = 0.032) in growth plate in ERN1-deficient mice (Fig. 7J, K; Fig. S14). We next detected the effect of 4μ8C combined with vismodegib (GDC-0449), a typical Hedgehog inhibitor,34,35 on cartilage growth plates. The mRNA level of IHH (P = 0.003) was inhibited by GDC-0449 in primary chondrocytes (Fig. 7L). The results of intra-articular injection also showed that the TZ length (P = 0.049) and HZ length (P = 0.048) were increased after the combined treatment of 4μ8C with GDC-0449 (Fig. 7M, N).
Figure 7.
ERN1 regulates chondrocyte proliferation and hypertrophy through IHH and PTH1R. (A) The mRNA levels of PTH1R in explants and (B)IHH in primary chondrocytes of C57BL/6 WT mice were detected by qPCR after treating with 25 μM 4μ8C for 2 weeks (explants) or 48 h (primary chondrocytes). ∗P < 0.05, ∗∗P < 0.005, n = 5–6. (C) HE and Safrain O-Fast Green staining of tibia tissues in 8-week-old mice after intra-articular injection of 4μ8C. (D) Quantification of PZ and PZ/TZ ratio. ∗P < 0.05, n = 5. (E–H) PTH1R and MMP13 of tibia tissues in 8-week-old WT mice were detected by IHC, ∗P < 0.05, n = 5. (I) The qPCR analysis of PTH1R, COL2A1, and MMP13 in explants of C57BL/6 WT mice after treating by 50 μM 4μ8C with or without 1 μM Abaloparatide for 2 weeks. ∗P < 0.05, ∗∗∗P < 0.001, n = 5. (J) HE staining and SafrainO-Fast Green staining of tibia tissue in 8-week-old ERN1 CKO mice after intra-articular injection of AdPTH1R adenovirus. (K) Quantification of PZ/TZ ratio and HZ/TZ ratio. ∗P < 0.05, n = 4. (L) The qPCR analysis of IHH in primary chondrocytes of C57BL/6 WT mice after treating by 25 μM 4μ8C with or without 10 μM GDC-0449 for 48 h, ∗∗P < 0.005, n = 6. (M) HE and SafrainO-Fast Green staining of tibia tissues in 8-week-old mice after intra-articular injection of 4μ8C with or without GDC-0449. (N) Quantification of TZ and HZ. ∗P < 0.05, n = 5. Scale bar = 200 μm.
Discussion
Cartilage is the bone precursor for most parts of the skeleton and participates in the entire process of bone growth and development from fetal life to maturity. The development of abnormal cartilage leads to a variety of diseases.36, 37, 38 During the process of cartilage formation and differentiation, IHH and PTHrP-PTH1R feedback loop play a vital role as paracrine molecules secreted by chondrocytes in different region. ERN1 is a key regulatory molecule in ER stress.39 Herein, we found that ERN1 CKO mice presented increased growth compared to WT mice. The longitudinal growth of bone mainly depends on the proliferation of chondrocytes in the growth plate, the transformation of proliferative chondrocytes into hypertrophic chondrocytes, and the regulation of the synthesis and degradation of extracellular matrix by chondrocytes.40,41 Therefore, the structure of the cartilage growth plate was examined. In the cartilage growth plate of ERN1 CKO mice, the proportion of chondrocytes in the HZ was larger than that in the control group. Meanwhile, the expression of the hypertrophic cartilage marker genes COL10A1 and MMP13 was higher than that of the control group (Figure 1, Figure 2). Using in vitro experiments to explore the relationship between ERN1 and chondrocyte proliferation and differentiation, we observed that the expression of Cyclin B, Cyclin D, PCNA and osteogenesis-related genes was up-regulated in siERN1-treated C28/I2 cells compared to that of control cells. Overexpression of ERN1 down-regulated the levels of proliferation and osteogenesis markers. It was also observed that the inhibition of p-IRE1α by 4μ8C up-regulated the expression of proliferation-related genes (Fig. 3). These results showed that the ability to proliferate chondrocytes to synthesize the extracellular matrix was decreased, although the proliferative ability of chondrocytes was increased after ERN1 was defected. Moreover, the transformation of proliferating chondrocytes into hypertrophic chondrocytes was accelerated during the development process, and the expression of COL10A1 and RUNX2 in hypertrophic chondrocytes was promoted. Therefore, the ability of proliferating cells to synthesize the extracellular matrix was impaired, although ERN1 deficiency promoted the proliferation of chondrocytes, and a large number of proliferating chondrocytes were transformed into hypertrophic chondrocytes, leading to excessive chondrocyte hypertrophy. The final manifestation was that the proportion of the proliferation area in the cartilage growth plate of ERN1 CKO mice was decreased, while that of the hypertrophic area was increased. The latest single-cell RNA-seq analysis research reported that there are three novel populations, named as effector chondrocytes, regulatory chondrocytes and homeostatic chondrocytes in human cartilage.30 The chondrocyte heterogeneity is closely associated with their functions. Furthermore, in ERN1-depleted chondrocytes, we found that the function homeostasis of effector chondrocytes and regulatory chondrocytes was disrupted. ECs and RegCs were enriched in different metabolisms, signaling pathways, and modulated cellular homeostasis and growth plate.
The IHH and PTHrP-PTH1R feedback loops synergistically regulate chondrocyte proliferation, differentiation, and endochondral bone growth. IHH and PTHrP, as paracrine molecules secreted by chondrocytes in different regions, exert their effects through binding to their respective receptors.14,15 Some studies showed that lack of IHH in the mouse could inhibit the proliferation of chondrocytes to promote the abnormal maturation of chondrocytes. IHH-deficient down-regulates the expression of hypertrophy-related markers, and inhibits chondrocyte differentiation and mineralization.42,43 In addition, some studies exhibited that PTH1R suppression leads to the abnormal development of cartilage due to abnormal chondrocyte hypertrophy.44 In our study, the results showed that lack of ERN1 promotes chondrocytes differentiation and endochondral ossification (Fig. 4). The expression of IHH was up-regulated, while that of PTH1R was down-regulated in the primary chondrocytes and cartilage tissues of ERN1-deficient mice. As an exocrine protein, PTHrP is regulated by the IHH signaling pathway. It has been confirmed that up-regulated IHH could promote the expression of PTHrP.15 We also observed that the expression of PTHrP was also up-regulated in ERN1 CKO mice. In summary, ERN1 knockdown could alter the expression profile of PTH1R, IHH, and PTHrP, and then disrupt the homeostasis of the PTHrP-IHH feedback loop (Fig. 5). It is demonstrated that the loss of ERN1 leads to an enhanced proportion of hypertrophic chondrocytes in the cartilage growth plate of ERN1 CKO mice.
After the initial clarification of the relationship between ERN1 and IHH, in which PTHrP-PTH1R represented a negative feedback loop, we explored the specific mechanism underlying the regulation of IHH and PTHrP-PTH1R by ERN1. It has been confirmed that ERN1 can produce the transcription factor XBP1s, which is bound to the promoter region (−1338 to −1137) of PTH1R and up-regulates its expression. The production of XBP1s was reduced in the absence of ERN1, which affected the expression of PTH1R (Fig. 6). It has been reported that the IRE1α-XBP1s signal axis regulates the expression of PTH1R in mouse embryonic fibroblasts (MEFs) after loss of ERN1.45 In addition, we observed that ERN1 knockout reduced the expression of PTH1R, mainly because ERN1 deficiency leads to loss of XBP1s, which in turn results in defective up-regulation of PTH1R by XBP1s. On the other side, ERN1 knockout promoted the expression of IHH in chondrocytes, while that XBP1s up-regulated the transcription of IHH. These results suggested that besides the IRE1α-XBP1s signal axis, another signaling pathway may participate in the dynamic regulation of IHH in chondrocytes.
The intra-articular injection of 4μ8C induced an increase in the proportion of hypertrophic chondrocytes, while reducing the proportion of proliferating chondrocytes in the cartilage growth plate, and injection with PTH1R adenovirus can reverse the abnormal transition from the proliferation to the hypertrophy in the growth plate in ERN1-deficient mice (Fig. 7). It is suggested that the IRE1α-XBP1s signal axis participates in regulating the expression of PTH1R, influencing the transformation of chondrocytes from proliferative to hypertrophic. In addition, it has been confirmed that the lack of IHH could inhibit the proliferation of chondrocytes to promote the abnormal maturation of chondrocytes.42,43 The results of intra-articular injection showed that the combination of 4μ8C with the IHH inhibitor GDC-0449 could further promote chondrocyte hypertrophy. As the core pathway of ER stress, IRE1α-XBP1s participates in the UPR when ER stress is activated.8 In the ERN1 null chondrocytes, ER stress could not be activated normally, and caused the abnormality of hypertrophy.
As summarized in the model of Figure 8, knocking out ERN1 specifically in chondrocytes could accelerate the transformation of proliferative into hypertrophic chondrocytes in the cartilage growth plate of mice. ERN1 negatively controls proliferation, hypertrophy, and chondrocyte differentiation in postnatal growth plates. ERN1 deficiency accelerates cartilage hypertrophy and mineralization by impairing the homeostasis of the IHH-PTHrP feedback loop.
Figure 8.
Schematic diagram of the role of IRE1α/ERN1 in regulating IHH/PTH1R negative feedback loop in cartilage development. (A) A working model of IHH, PTHrP-PTH1R feedback loop during orchestrating cartilage development. (B)ERN1 knockout promoted transformation of chondrocytes from proliferation to hypertrophy by up-regulating IHH and inhibiting PTHrP-PTH1R. Transcription factor XBP1s, which is spliced from XBP1u by IRE1α, plays a role in ERN1 regulating PTHrP-IHH feedback loop.
Author contributions
All authors contributed to the writing of this manuscript. All authors approved the final manuscript prior to submission. F.J.G. and M.T.F. designed experiments. M.T.F., N.N.G., X.Y.L., D.Y.Y., Y.Y.Y., F.N.B., L.L., and R.J. carried out experiments. C.C. provided samples of osteoarthritis. X.L.L., F.T.L., H.B.Q., D.Y.Y., Y.Y.Y., N.N.G., M.T.F., and F.J.G. analyzed data. Q.Y.T. and Y.L.X. analyzed data and gave some good advice on the interpretation of data.
Prof. Guo FJ designed the manuscript and had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of interests
The authors declare that they have no conflict of interests.
Funding
Our experiments were supported by the National Natural Science Foundation of China (No. 81672209, 81871769, 82272550) and the Chongqing Science and Technology Bureau (China) (No. cstc2021jcyj-bshX0214).
Acknowledgements
The authors would like to thank Prof. Quan Yuan (State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, China) for providing adenovirus-PTH1R (AdPTH1R) for our study as a gift. The experimental procedures ensured the safety of practitioners in laboratory animal projects, conformed to human ethical standards and international practices, and were approved by the Animal Experimental Ethics Committee of Chongqing Medical University. All animal experiments conformed to the guidelines of Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.11.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Figure S1.
The protein level of IRE1α in human cartilage. (A) The expression of IRE1α in the human normal cartilage and the human degenerated cartilage were detected by Western blot. (B) Semiquantitative analysis results of Western blot. ∗P < 0.05, n = 3.
Figure S2.
Identification of ERN1 conditional knockout mice. (A) Identification of Flox genotypes in mice. (1) ERN1flox/+Col2Cre, (2) ERN1flox/floxCol2Cre, and (3) ERN1flox/flox. (B) Identification of Cre genotypes in ERN1flox/+Col2Cre, ERN1flox/floxCol2Cre, and ERN1flox/flox mice.
Figure S3.
Identification of FloxCre genotypes in different tissues of ERN1 CKO and control mice. M: DL2,000 DNA Marker; CKO: Col2-Cre, ERN1flox/flox; NC: ERN1flox/flox.
Figure S4.
Protein level of IRE1α in primary chondrocytes. Western blot of primary chondrocytes isolated from the cartilage of ERN1 CKO and control mice.
Figure S5.
Analysis on the growth plate of ERN1 CKO mice. (A) Quantification assay of proliferation zone (PZ)/growth plate length (TZ) ratio of ERN1 CKO and control mice at week 1, week 4, and week 8; (B) Quantification of growth plate at E15 (embryonic 15 days), E17 (embryonic 21 days), E19 (embryonic 15 days), E21 (embryonic 15 days). P > 0.05, n = 5.
Figure S6.
Knockdown or knockout of ERN1 promotes chondrocyte hypertrophy in C28/I2 cells. (A–C) The expression of COL2A1, COL10A1, and MMP13 were detected by qPCR in C28/I2 cells. ∗P < 0.05, ∗∗P < 0.005, n = 5-6.
Figure S7.
ERN1 knockout promotes the expression of MMP13 and COL10A1 and inhibits the expression of Aggrecan, COL2A1, and SOX9. (A–C) IHC and (D) negative control of IHC staining of growth plate in 4-week-old WT mice. Scale bar = 200 μm.
Figure S8.
ERN1 overexpression inhibits chondrocyte hypertrophy in ATDC5 cells. The expression of IHH (A), COL10A1 (B) and MMP13 (C) was detected by qPCR in AdERN1-infected ATDC5 cells for 1, 3, 5, 7 and 10 days. ∗P < 0.05, ∗∗P < 0.005, n = 3.
Figure S9.
ERN1 knockout impairs balance of effector chondrocytes and regulatory chondrocytes. (A–C) The mRNA levels of SNORC, CLEC3A, and CHI3L1 were detected by qPCR in primary chondrocytes isolated from the cartilage of ERN1 CKO and control mice. ∗∗P < 0.005, ∗∗∗P < 0.001, n = 3.
Figure S10.
ERN1 regulates the expression of PTHrP and PTH1R. (A) The qPCR analysis of ERN1 and PTH1R in C28/I2 cells after inhibiting ERN1. ∗P < 0.05, n = 3. (B) The qPCR analysis of ERN1 and PTH1R in primary chondrocytes of C57BL/6 WT mice after inhibiting ERN1. ∗P < 0.05, n = 3. (C) The qPCR analysis of ERN1, PTH1R, and PTHrP in C28/I2 cells after overexpressing ERN1. ∗P < 0.05, n = 3. (D) The qPCR analysis of ERN1, PTH1R, and PTHrP in explants of 2-week-old WT mice after overexpressing ERN1. ∗P < 0.05, ∗∗∗P < 0.001, n = 3.
Figure S11.
ERN1 overexpression affects the cartilage growth plate of WT mice. (A, B) HE staining of tibia tissue in 8-week-old WT mice after intra-articular injecting AdERN1 adenovirus, and quantification of PZ length, PZ/TZ ratio, and HZ/TZ ratio. ∗P < 0.05, n = 5.
Figure S12.
ERN1 overexpression inhibits the expression of MMP13 and COL10A1. (A) (Ⅰ, Ⅱ) HE staining of cartilage explants in 2-week-old WT mice; (Ⅲ, Ⅳ) the expression of COL10A1 was detected and observed by IHC; and (Ⅴ, Ⅵ) the enlarged images of Ⅲ and Ⅳ. (B) (Ⅰ, Ⅱ) Safrain O-Fast Green staining of cartilage explants in 2-week-old WT mice; (Ⅲ, Ⅳ) the expression of MMP13 was detected and observed by IHC; and (Ⅴ, Ⅵ) the enlarged images of Ⅲ and Ⅳ. (C) COL10A1 and MMP13 levels in cartilage explants were quantified by IHC. ∗P < 0.05, n = 3. Scale bar = 200 μm.
Figure S13.
Quantification of XBP1s, Nrf2, PERK and p-PERK in the chondrocytes of ERN1 deficient mice. ∗∗P < 0.005, ∗∗∗P < 0.001, n = 3.
Figure S14.
IF staining of tibia tissue in 8-week-old ERN1 CKO mice after intra-articular injection of AdPTH1R adenovirus. Scale bar = 200 μm.
References
- 1.Zhou X., von der Mark K., Henry S., et al. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Crombrugghe B., Lefebvre V., Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13(6):721–728. doi: 10.1016/s0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 3.Eyre D.R., Muir H. The distribution of different molecular species of collagen in fibrous, elastic and hyaline cartilages of the pig. Biochem J. 1975;151(3):595–602. doi: 10.1042/bj1510595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron T.L., Bell K.M., Gresshoff I.L., et al. XBP1-independent UPR pathways suppress C/EBP-β mediated chondrocyte differentiation in ER-stress related skeletal disease. PLoS Genet. 2015;11(9) doi: 10.1371/journal.pgen.1005505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oakes S.A., Papa F.R. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calfon M., Zeng H., Urano F., et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida H., Oku M., Suzuki M., et al. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172(4):565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J.H., Li H., Yasumura D., et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 11.Guo F.J., Xiong Z., Han X., et al. XBP1S, a BMP2-inducible transcription factor, accelerates endochondral bone growth by activating GEP growth factor. J Cell Mol Med. 2014;18(6):1157–1171. doi: 10.1111/jcmm.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo F.J., Jiang R., Li X., et al. Regulation of chondrocyte differentiation by IRE1α depends on its enzymatic activity. Cell Signal. 2014;26(9):1998–2007. doi: 10.1016/j.cellsig.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Feng N., Liang L., Fan M., et al. Treating autoimmune inflammatory diseases with an siERN1-nanoprodrug that mediates macrophage polarization and blocks toll-like receptor signaling. ACS Nano. 2021;15(10):15874–15891. doi: 10.1021/acsnano.1c03726. [DOI] [PubMed] [Google Scholar]
- 14.Chijimatsu R., Saito T. Mechanisms of synovial joint and articular cartilage development. Cell Mol Life Sci. 2019;76(20):3939–3952. doi: 10.1007/s00018-019-03191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuhashi K., Ono W., Matsushita Y., et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563(7730):254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohba S. Hedgehog signaling in skeletal development: roles of Indian hedgehog and the mode of its action. Int J Mol Sci. 2020;21(18):6665. doi: 10.3390/ijms21186665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahon M.J., Donowitz M., Yun C.C., et al. Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature. 2002;417(6891):858–861. doi: 10.1038/nature00816. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L.H., Ma S., Sutkeviciute I., et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364(6436):148–153. doi: 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Shen S., Li Z., et al. MIR-140-5p affects chondrocyte proliferation, apoptosis, and inflammation by targeting HMGB1 in osteoarthritis. Inflamm Res. 2020;69(1):63–73. doi: 10.1007/s00011-019-01294-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Kong L., Carlson C.S., et al. Cbfa1-dependent expression of an interferon-inducible p204 protein is required for chondrocyte differentiation. Cell Death Differ. 2008;15(11):1760–1771. doi: 10.1038/cdd.2008.112. [DOI] [PubMed] [Google Scholar]
- 21.Yao Y., Wang Y. ATDC5: an excellent in vitro model cell line for skeletal development. J Cell Biochem. 2013;114(6):1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 22.Joronen K., Ala-aho R., Majuri M.L., et al. Adenovirus mediated intra-articular expression of collagenase-3 (MMP-13) induces inflammatory arthritis in mice. Ann Rheum Dis. 2004;63(6):656–664. doi: 10.1136/ard.2003.009720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi Z., Ghivizzani S.C., Lechman E., et al. Adverse effects of adenovirus-mediated gene transfer of human transforming growth factor beta 1 into rabbit knees. Arthritis Res Ther. 2003;5(3):R132–R139. doi: 10.1186/ar745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne K.A., Lee H.H., Haleem A.M., et al. Single intra-articular injection of adeno-associated virus results in stable and controllable in vivo transgene expression in normal rat knees. Osteoarthritis Cartilage. 2011;19(8):1058–1065. doi: 10.1016/j.joca.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans C.H., Ghivizzani S.C., Robbins P.D. Gene delivery to joints by intra-articular injection. Hum Gene Ther. 2018;29(1):2–14. doi: 10.1089/hum.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrat M.A., Ion G. Imaging IGF-I uptake in growth plate cartilage using in vivo multiphoton microscopy. J Appl Physiol. 2017;123(5):1101–1109. doi: 10.1152/japplphysiol.00645.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anspach L., Tsaryk R., Seidmann L., et al. Function and mutual interaction of BiP-, PERK-, and IRE1α-dependent signalling pathways in vascular tumours. J Pathol. 2020;251(2):123–134. doi: 10.1002/path.5423. [DOI] [PubMed] [Google Scholar]
- 28.Cross B.C., Bond P.J., Sadowski P.G., et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109(15):E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu N., Malemud C.J. Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated gene expression. Int J Mol Sci. 2019;20(15):3792. doi: 10.3390/ijms20153792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Q., Zheng Y., Zhang G., et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78:100–110. doi: 10.1136/annrheumdis-2017-212863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron T.L., Gresshoff I.L., Bell K.M., et al. Cartilage-specific ablation of XBP1 signaling in mouse results in a chondrodysplasia characterized by reduced chondrocyte proliferation and delayed cartilage maturation and mineralization. Osteoarthritis Cartilage. 2015;23(4):661–670. doi: 10.1016/j.joca.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Leder B.Z., O'Dea L.S., Zanchetta J.R., et al. Effects of abaloparatide, a human parathyroid hormone-related peptide analog, on bone mineral density in postmenopausal women with osteoporosis. J Clin Endocrinol Metab. 2015;100(2):697–706. doi: 10.1210/jc.2014-3718. [DOI] [PubMed] [Google Scholar]
- 33.Le Henaff C., Ricarte F., Finnie B., et al. Abaloparatide at the same dose has the same effects on bone as PTH (1-34) in mice. J Bone Miner Res. 2020;35(4):714–724. doi: 10.1002/jbmr.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan B., Zhang Z., Jin D., et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat Commun. 2016;7 doi: 10.1038/ncomms11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dlugosz A., Agrawal S., Kirkpatrick P. Vismodegib. Nat Rev Drug Discovery. 2012;11(6):437–438. doi: 10.1038/nrd3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decker R.S. Articular cartilage and joint development from embryogenesis to adulthood. Semin Cell Dev Biol. 2017;62:50–56. doi: 10.1016/j.semcdb.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan Y., Grodzinsky A.J. Cartilage diseases. Matrix Biol. 2018;71–72:51–69. doi: 10.1016/j.matbio.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michigami T. Regulatory mechanisms for the development of growth plate cartilage. Cell Mol Life Sci. 2013;70(22):4213–4221. doi: 10.1007/s00018-013-1346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han D., Lerner A.G., Vande Walle L., et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138(3):562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C.F., Samsa W.E., Zhou G., et al. Transcriptional control of chondrocyte specification and differentiation. Semin Cell Dev Biol. 2017;62:34–49. doi: 10.1016/j.semcdb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berendsen A.D., Olsen B.R. Bone development. Bone. 2015;80:14–18. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razzaque M.S., Soegiarto D.W., Chang D., et al. Conditional deletion of Indian hedgehog from collagen type 2alpha1-expressing cells results in abnormal endochondral bone formation. J Pathol. 2005;207(4):453–461. doi: 10.1002/path.1870. [DOI] [PubMed] [Google Scholar]
- 43.St-Jacques B., Hammerschmidt M., McMahon A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin T.J. Parathyroid hormone-related protein, its regulation of cartilage and bone development, and role in treating bone diseases. Physiol Rev. 2016;96(3):831–871. doi: 10.1152/physrev.00031.2015. [DOI] [PubMed] [Google Scholar]
- 45.Tohmonda T., Yoda M., Mizuochi H., et al. The IRE1α-XBP1 pathway positively regulates parathyroid hormone (PTH)/PTH-related peptide receptor expression and is involved in PTH-induced osteoclastogenesis. J Biol Chem. 2013;288(3):1691–1695. doi: 10.1074/jbc.C112.424606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.