SP100 is an antiviral protein that restricts the productive stage of human papillomavirus (HPV) and multiple other viruses, and viruses in turn block SUMO-1-mediated stabilization of SP100 and promotes its degradation (Table S1). Interferon (IFN) signaling could still produce more SP100 through transcription to counteract viruses.1 Viruses also disable the transcriptional up-regulation of SP100 to achieve persistent infection in hosts. Chromosome-19 miRNA cluster (C19MC) miRNAs confer variable levels of resistance to different types of viral infections2 and here we use SP100 mRNA as our target for understanding the tumor context in which it is expressed or suppressed, and its relationship with C19MC-directed antiviral response miRNAs in human skin cutaneous melanoma (SKCM-TCGA). We show that, high SP100 mRNA expression reflects better survival in melanoma patients and that, the genomic landscape of the SP100 gene is subjected to copy number alteration in SP100Low melanomas with recurrent breakpoints in chromosome-2q between SP100 and SP110 gene loci and centromere. Besides, the C19MC miRNA-520G promotes SP100 mRNA expression and impedes melanin biosynthesis with down-regulated SLC45A2 and increased HTR2B mRNAs which are known indirect regulators of the tyrosine pool and melanin biosynthesis (Table S1).
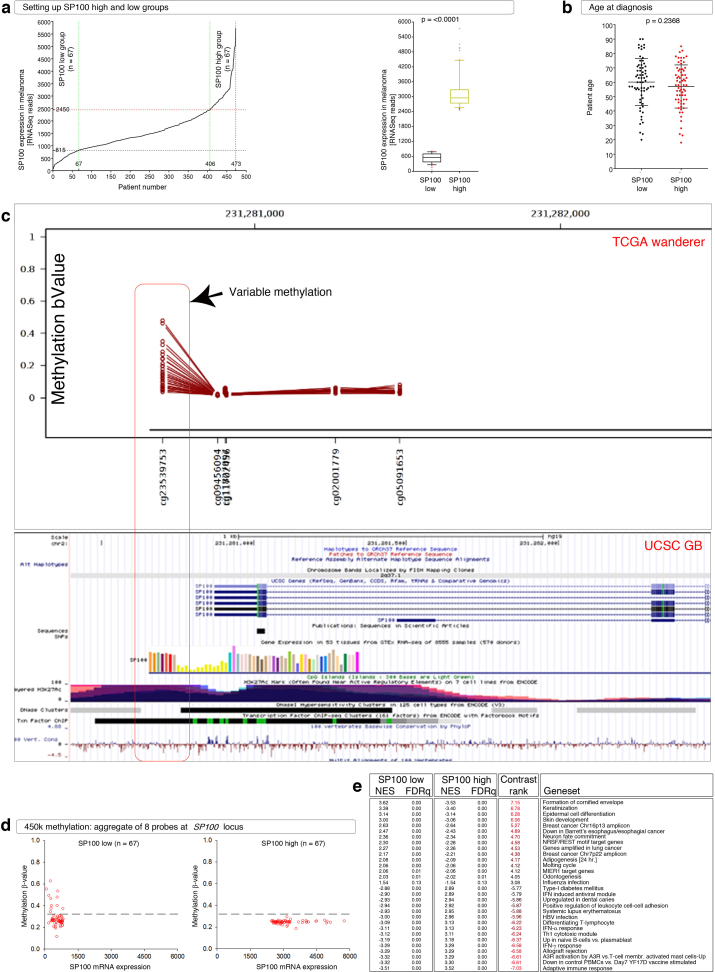
SP100 is known to be up-regulated with IFN targets, particularly in melanoma3 and viruses are sporadically detected in melanoma (Table S2). The patients with melanomas were classified into SP100High and SP100Low groups (n = 67 each), without statistically significant difference in patient ages at diagnosis between both groups and with balanced patient gender and significant difference in overall survival (Fig. 1A; Fig. S1A, B).
Figure 1.
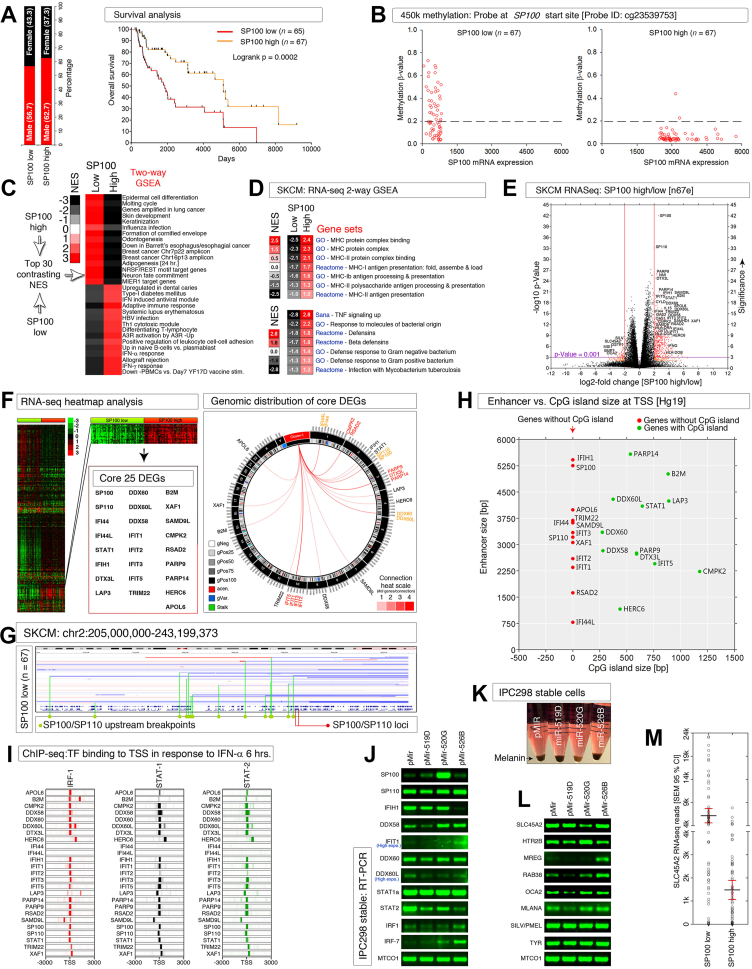
C19MC miRNA-520G restores SP100 mRNA expression and inhibits melanin production in melanoma. (A) SP100-based grouping of melanoma samples with balanced gender (left) but the KM curve showing significant difference between SP100High and SP100Low groups in overall survival. (B) 450k methylation of SP100 gene at the transcription start site probe (the only variable probe among 8; Fig. S1C) showing hypomethylation in SP100Low melanomas below 0.2 (β-value) equivalent to SP100High group. (C, D) Two-way GSEA analysis-based translation of SP100High and SP100Low melanoma coding transcriptomes showing top contrasting signatures enriched with least FDRq values (C) and antimicrobial antigen presentation machinery (D). (E) Volcano plot showing the differentially expressed genes between SP100High and SP100Low group melanomas. Red data points are having significance below the p-value of 0.001 and orange data points are having significance with a P-value between 0.05 and 0.001. [n = 67 each (n67e)]. (F) Identification of 25 core cluster-wide differentially expressed genes between SP100High and SP100Low group melanomas by RNA-seq heatmap analysis (left panel) and their genomic distribution shown by circos plot (right panel). (G) Copy number variation differences of SP100 and SP110 genes in SP100Low group melanomas (for comparison with the SP100High group, please see Fig. S2B). Copy number alterations are associated with clustered or focal upstream or downstream potential breakpoints (indicated by green lines from green dots). Red dots mark the locus/loci of indicated genes within the panel. (H) Presence or absence of CpG island and enhancer H3K27Ac mark at -3 and +3 kb flanking regions of transcription start site (TSS) of 25-Sp100 core co-expressed genes based on UCSC Hg19 annotation. (I) Heatmap showing the presence or absence of transcription factor binding in IFN-α treated K-652 cells [ChIP-seq for IRF-1, STAT-1, and STAT-2 at -3 + 3 kb flanking regions of transcription start site (TSS)] of 25-Sp100 core co-expressed genes based on UCSC Hg19 annotation. Scale: base pairs. Note that there was no binding at IFI44 and IFI44L genes. (J) RT-PCR analysis of selected Sp100-core co-expressed genes and their regulators in IPC298 melanoma cells stably overexpressed with C19MC miRNAs (miRNA expression verified by qRT-PCR). (K) Lower level of melanin accumulation in miR-520G stable IPC-298 cells (photograph). (L) RT-PCR analysis of melanin/DOPAmine pathway genes in IPC298 stable melanoma cells. (M) Aligned dot plot of SLC45A2 (tyrosine transporter) mRNA showing under-expression of it in SP100High compared to SP100Low group TCGA melanomas. CI, confidence interval.
We examined whether the expression of SP100 mRNA in SP100High and SP100Low groups is different because of altered methylation. A substantial number of the SP100Low melanomas had hypomethylation (β value < 0.2) comparable to the SP100High group (Fig. 1B; Fig. S1C, D), indicating that more factors play a role in SP100 mRNA expression than methylation.
We examined the context of melanomas with SP100high and SP100low mRNA expression by two-way GSEA and found that the SP100Low melanomas exhibited up-regulation of gene signatures related to the ichthyosis phenotype (skin epidermis development, keratinization, cornified envelope, epidermal cell differentiation, and molting cycle) (Table S1) and enrichment in neuronal fate commitment and REST (Fig. 1C). Furthermore, the SP100Low melanomas exhibited a defective adaptive immune profile (down-regulated: adaptive immune response, allograft rejection, interferon-γ (IFN-γ), interferon-α (IFN-α), antiviral module, B cells, T cells, leukocyte adhesion, Th1 cytotoxic module, MHC-I, MHC-II, β-defensin, tumor necrosis factor-α (TNF-α), defense response to both Gram-positive and Gram-negative bacteria, including Mycobacterium tuberculosis) with a down-regulated type-I diabetes mellitus gene signature (Fig. 1C, D). Interestingly, SP100Low melanomas exhibited an influenza infection signature whereas SP100High melanomas exhibited a hepatitis-B virus signature (Fig. 1C). Notably, all these signatures are expressed with high contrast between SP100Low and SP100High groups (Fig. S1E).
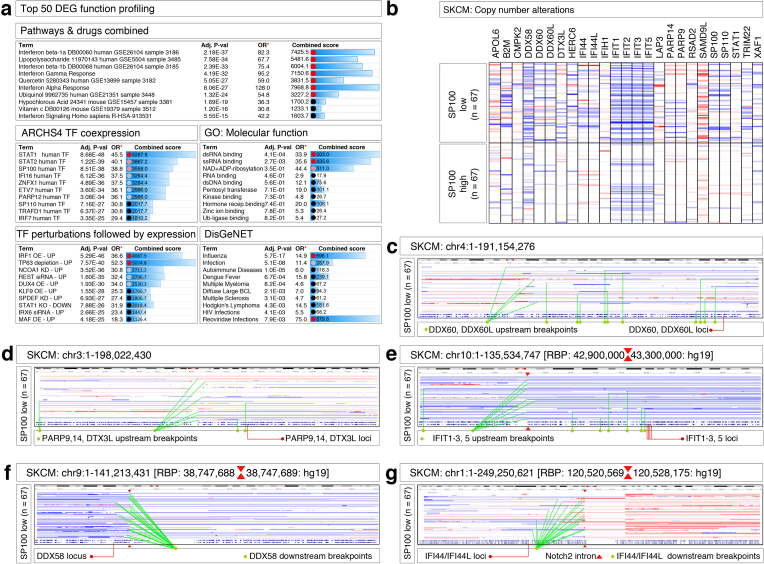
We next focused on the individual genes that are tightly co-expressed with SP100. Volcano plotting, heatmap, and EnrichR analyses indicated that a genome-wide distributed core 25 interferon-STAT-regulated genes (SP100, SP110, IFI44, IFI44L, STAT1, IFIH1, DTX3L, LAP3, DDX60, DDX60L, DDX58, IFIT1, IFIT2, IFIT3, IFIT5, TRIM22, B2M, XAF1, SAMD9L, CMPK2, RSAD2, PARP9, PARP14, HERC6, and APOL6) were significantly co-expressed with SP100 (Fig. 1E, F; Fig. S2A). The bacterial cell wall component lipopolysaccharide, dsRNA and ssRNA viruses, the food components, quercetin and vitamin-C, transcription factors STAT-1, STAT-2, IRF-1, REST, knockdown of p63 (TP63), could accompany the transcriptional up-regulation of the core 25 gene network in SP100High melanomas (Fig. S2A).
Copy number analysis of this 25-core gene network revealed that 23 of these genes (except for APOL6 and SAMD9L) had increased copy number alterations (CNA loss) in the SP100Low group compared to the SP100High group (Fig. S2B). SP100 and SP110 genes at the middle of chromosome-2q were impacted by CNA point clusters between the centromere and SP100/SP110 loci (Fig. 1G). Likewise, DDX60 and DDX60L loci at the distal end of chromosome-4q were preceded by a cluster of CNA points up to the centromere in SP100Low group melanomas (Fig. S2C). In the case of PARP9, PARP14, and DTX3L loci on chromosome-3q or in the case of IFIT1, IFIT2, IFIT3, and IFIT5 loci on chromosome-10q multiple CNA points are clustered at the q arm close to the centromeres where the recurrent breakpoints (RBP) are mapped between the coordinates 42,900,000–43,300,000 (Hg19) on chromosome-10q (Fig. S2D, E). In the case of DDX58, the breakpoint is even precise where the DDX58 locus on chromosome-9p had numerous CNA points downstream but at a specific nucleotide [RBP: 38,747,688–38,747,689 (Hg19) on chromosome9] (Fig. S2F). In the case of IFI44 and IFI44L loci on chromosome-1p the recurrent CNA breakpoints mapped to Notch2 intronic region close to the centromere [RBP: 120,520,569–120,528,175 (Hg19)] (Fig. S2G). Therefore, the core 25 SP100 gene network expression is frequently impaired by copy number loss due to breakpoints that map close to centromeres.
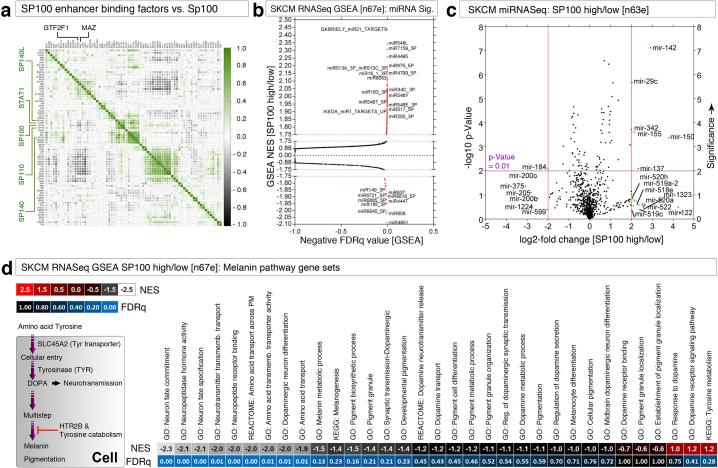
Notably, 13 out of 25-core genes lack CpG islands within the promoter/enhancer region which includes SP100 gene (Fig. 1G). A close examination of the correlative expression of gene products that bind to SP100 enhancer region across SP100High and SP100Low melanomas revealed that SP100 mRNA was correlatively expressed with STAT1 along with other SP100 family member mRNAs (Fig. S3A). We examined the promoter/enhancer binding ChIP-seq pattern of STAT-1, IRF-1, and STAT-2 under IFN-α treated conditions. All three transcription factors STAT-1, IRF-1, and STAT-2 bound to most of the 25-core gene transcription start sites (TSS) but excluded IFI44 and IFI44L gene TSSs (Fig. 1I).
Since C19MC miRNA-520G remodels IFN-γ-dependent transcription in the context of hepatocellular carcinoma4 and alters melanoma antigen expression,5 we examined the GSEA results for miRNA target gene expression. Many miRNA targets were up-regulated or down-regulated with a normalized enrichment score of ≥ 2 or ≤ 2 with the least FDRq values (Fig. S3B and Table S3). Intriguingly, a group of C19MC antiviral miRNAs (miR-520h, miR-519a-2, miR-518e, miR-1323, miR-520a, miR-522, miR-519c that share highly overlapping mature miRNA sequence with well-characterized miR-520G and miR-519D) are enriched greater than 2-fold in SP100High group compared to SP100Low group (Fig. S3C).
Stable overexpression of C19MC miRNAs miR-519D, miR-520G, and miR-526B in the IPC-298 melanoma cell line in parallel to control pMIR vector revealed that the SP100 mRNA was tremendously up-regulated (independent of other IFN targets examined) by miR-520G but not by miR-519D or miR-526B (Fig. 1J). Interestingly, the cells stably overexpressing miR-520G accumulated less melanin pigment compared to control pMIR, miR-519D or miR-526B overexpressing stable cells, indicating that the miR-520G or its downstream targets are linked to inhibition of melanin biosynthesis or accumulation (Fig. 1K). Therefore, we examined the melanin pathway genes by RT-PCR and found that the tyrosine transporter SLC45A2 mRNA was strongly down-regulated by miR-520G overexpression which is accompanied by increased expression of HTR2B mRNA, a serotonin receptor gene (Fig. 1L).
We next examined the TCGA data for melanin pathway alterations between SP100High and SP100Low groups and found that the aminoacid-transport pathway (including tyrosine transport: a precursor of melanin biosynthesis) and various dopaminergic neuron signatures (DOPA/serotonin/5-HT, an intermediate product of tyrosine to melanin biosynthesis) are down-regulated in SP100High melanomas with robust NES and least FDRq values (Fig. S3D). Notably, SLC45A2 is under-expressed in SP100High than in the SP100Low group (Fig. 1M).
In summary, the induction of SP100 mRNA by C19MC miRNA-520G is linked to the inhibition of melanin biosynthesis, and repression of SP100 mRNA in patient melanomas reflects impeded ichthyosis signatures and antimicrobial immune response signatures.
Author contributions
G.G.J. and I.G. conceived the SP100 classification and profiling hypothesis to explore SP100 antiviral network. G.G.J. and A.S.B. conceived the SP100 link to antiviral C19MC. G.G.J. designed the study, performed bioinformatics analyses, generated the stable cell lines, generated & composited figures, and wrote the manuscript. A.S.B. and K.S.M.S. edited the manuscript. G.G.J. and I.G. tracked the 25-core gene network in the human genome. M.T.S. and N.M. performed PCRs. M.N. periodically tested the cells for mycoplasma. A.S.B. and E.R.F. contributed to the funding and supervision. All other authors contributed to the acquisition of reagents or helped with tissue culture. All authors read and agreed to the contents and submission of this manuscript. G.G.J., M.T.S., I.G., and N.M. share the first authorship.
Conflict of interests
A.B. has no conflict of interests related to this work but has advisory board relationships with Bayer and Deciphera. The remaining authors declare no conflict of interests.
Funding
Research reported in this publication was supported in part by the National Institutes of Health under award number K08CA255933.
Acknowledgements
The authors sincerely thank Dr. Sherman Weissman, Yale University, and Dr. Michael Snyder, Yale University & Stanford, USA & team members who contributed the IFN-α IRF-1/STAT-1/STAT-2 ChIP-seq data to ENCODE database. The authors sincerely thank the TCGA network for the SKCM data.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.02.047.
Contributor Information
Goodwin G. Jinesh, Email: goodwinjinesh@gmail.com.
Andrew S. Brohl, Email: Andrew.Brohl@moffitt.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
References
- 1.Grötzinger T., Sternsdorf T., Jensen K., et al. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur J Biochem. 1996;238(2):554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- 2.Delorme-Axford E., Donker R.B., Mouillet J.F., et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110(29):12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harel M., Ortenberg R., Varanasi S.K., et al. Proteomics of melanoma response to immunotherapy reveals mitochondrial dependence. Cell. 2019;179(1):236–250. doi: 10.1016/j.cell.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinesh G.G., Napoli M., Ackerman H.D., et al. Regulation of MYO18B mRNA by a network of C19MC miRNA-520G, IFN-γ, CEBPB, p53 and bFGF in hepatocellular carcinoma. Sci Rep. 2020;10(1):12371. doi: 10.1038/s41598-020-69179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinesh G.G., Napoli M., Smallin M.T., et al. Mutant p53s and chromosome 19 microRNA cluster overexpression regulate cancer testis antigen expression and cellular transformation in hepatocellular carcinoma. Sci Rep. 2021;11:12673. doi: 10.1038/s41598-021-91924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.