Abstract
CRISPR/Cas9 is an effective gene editing tool with broad applications for the prevention or treatment of numerous diseases. It depends on CRISPR (clustered regularly interspaced short palindromic repeats) as a bacterial immune system and plays as a gene editing tool. Due to the higher specificity and efficiency of CRISPR/Cas9 compared to other editing approaches, it has been broadly investigated to treat numerous hereditary and acquired illnesses, including cancers, hemolytic diseases, immunodeficiency disorders, cardiovascular diseases, visual maladies, neurodegenerative conditions, and a few X-linked disorders. CRISPR/Cas9 system has been used to treat cancers through a variety of approaches, with stable gene editing techniques. Here, the applications and clinical trials of CRISPR/Cas9 in various illnesses are described. Due to its high precision and efficiency, CRISPR/Cas9 strategies may treat gene-related illnesses by deleting, inserting, modifying, or blocking the expression of specific genes. The most challenging barrier to the in vivo use of CRISPR/Cas9 like off-target effects will be discussed. The use of transfection vehicles for CRISPR/Cas9, including viral vectors (such as an Adeno-associated virus (AAV)), and the development of non-viral vectors is also considered.
Keywords: Clinical trials, CRISPR/Cas9, Gene therapy, Non-viral vectors, Viral vectors
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) was discovered in Escherichia coli and described as a 1664 nucleotide sequence by Ishino and colleagues in 1987.1 Before its functional application, two critical studies on genes adjacent to the CRISPR locus2 were done. The discovery of foreign viral DNA in CRISPR spacers3 directly proposed the role of CRISPR as an innate immune system in bacteria to fight viruses.4 This hypothesis was confirmed by experiments in Barango's laboratory, and the function of the CRISPR sequences was finally determined.5 Due to its excellent ability to target and cut specific DNA sequences, CRISPR was widely introduced as a gene editing tool.6 Compared to previous gene editing tools like zinc finger nucleases (ZFNs) and transcription-activating effector nucleases (TALENs), CRISPR-based gene-editing tools perform extremely better and are easier to handle.7 Due to the development of this technique, Emmanuel Charpentier and Jennifer Doudna received the Nobel Prize in 2020.8,9
CRISPR-Cas systems are classified into two main classes and six sub-types. CRISPR-Cas9 as a member of type II is the simplest and most widely used in genetic research due to the presence of a single protein with endonuclease activity (Cas9). In this system, the Cas9 protein can target the desired sequence using an engineered guide RNA (gRNA) which makes a smooth double-stranded cut in three nucleotides sequence upstream at the target site which is called Protospacer-Adjacent Motif (PAM). PAM is located immediately downstream of the crRNA binding site of gRNA in the incomplete strand and is important in target recognition by Cas9. The PAM sequence (5′-NGG) is not present in CRISPR arrays and is only located on the foreign invading genome; this is the reason why Cas9 cannot cut the bacterial genome.10,11
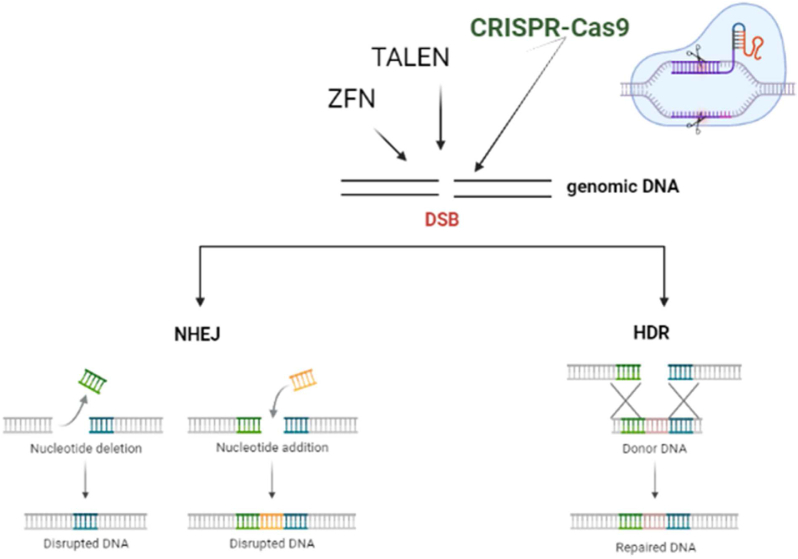
As shown in Figure 1, intracellularly double-stranded breaks (DSBs) can be repaired either by non-homologous end joining (NHEJ),12 a pathway that shows irregular break ends, or through Homology Directed Repair (HDR),13 a pathway that requires homologous patterns, which is carried out in gene repair, addition, or replacement.14 The wide potential of CRISPR lies in the directed action of Cas9 through an engineered sgRNA or crRNA/tracer RNA, allowing the researcher to target a wide range of genomic sequences.15
Figure 1.
ZFNs, TALENs, and CRISPRs lead to DSBs in the genome and are repaired mainly by two pathways: (i) non-homologous end joining (NHEJ): error-prone; (ii) homology-based repair (HDR).
This technique has the potential to treat a large number of genetic diseases like cancers.16,17 In this article, the current status of CRISPR applications in clinical trials and delivery methods will be reviewed. Finally, several challenges and opportunities facing researchers in this field will be elaborated.
Efforts to improve CRISPR/Cas9 technology
After the rapid development and remarkable success of this technique, researchers have tried tore-engineer and expand CRISPR/Cas9. CRISPR interference (CRISPRi), protein knockdown Cas9,18 Cas9 nickase (nCas9), and dead Cas9 (dCas9) are some of these examples. Cas9 nickase (nCas9) has mutations in its nuclease domain which helps to create the cut in a single-stranded DNA.19 A year after the creation of dead Cas9 (dCas9), the endonuclease-free Cas9 was designed to increase the quality of expression, particularly for CRISPRa.20 Subsequently, more precise and reliable CRISPR/Cas9-based gene editing strategies without introducing DSBs, such as base editing21,22 and primary editing23 were developed. The base modification techniques, Cytidine Counting Base Editor (CBE) and Adenine Base Editor (ABE) typically consist of nCas9/dCas9 evolved cytidine deaminase/adenine deaminase. CBE promotes the conversion of CG to TA while ABE promotes the conversion of AT to GC, and the current established nNme2-CBE framework seems to have higher efficiency and flexibility.24
Ultimately, gene editing tools are still limited in creating specific base conversions. Therefore, some editing tools have been developed using changes in the transcriptions of dCas9. In this regard, gRNA primers were used to realize precise modification with less off-target effect.23 With changes in CRISPR/Cas9 gene editing tools that are efficient and safe, they have the potential to enter clinical trials. A CRISPR-related transposase that not only cuts the DNA but also performs high-quality editing has been developed.25 In terms of in vivo gene therapy, various CRISPR frameworks such as light- and chemical-activated Cas9 have been explored.24,25 Also, CRISPR modifiers with smaller sizes have been investigated recently.26
Ex vivo genome editing
Ex vivo genome editing is a useful approach in which the genome of specific cells is altered in vitro, and then the modified cells are transplanted into the patient to induce a regenerative effect. This approach differs from in vivo genome modification approaches, where CRISPR/Cas9 or other genome-editing components are specifically delivered into the cell environment and perform their beneficial effect in situ.27,28 Compared to the in vivo techniques, the ex vivo modification method requires more steps (such as cell isolation, expansion, transfection, fixation, and transplantation) and may be more suitable for focusing on a specific organ rather than the whole organism.29 Notably, it avoids the enormous challenges of in vivo transfection that have been widely discussed.30,31 In addition, the ex vivo approach may have certain safety advantages, especially its low off-target rate. In vivo approaches should look after the unintended off-targets, either in the context of unintended transfer to non-target cell lines or in the form of unintended mutations in the genome. The ex vivo approach circumvents this issue by precisely altering the target cell, providing an opportunity to screen the effective alteration. In this context, the ex vivo applications of CRISPR/Cas9 for better genome editing and its methods and related references are summarized in Table 1.
Table 1.
Clinical trials involving ex vivo or in vivo CRISPR-based genome editing.
| Status | NCT Number | Diseases | Target gene | Intervention/treatment | Clinical phase | Delivery method |
|---|---|---|---|---|---|---|
| Completed | NCT04191148 | Urinary tract infections | E. coli genome | LBP-EC01 | Phase I | crPhage cocktail - in vivo |
| Unknown | NCT03728322 | Thalassemia genetic diseases | Hemoglobin subunit beta (HBB) | No | Phase I | ex vivo |
| Active, not recruiting | NCT03655678 | Thalassemia genetic diseases | BAF chromatin remodeling complex subunit 11 A (BCL11A) | CTX001 | Phase II/III | Electroporation - ex vivo |
| Active not recruiting | NCT04205435 | Beta-thalassemia | Hemoglobin subunit beta (HBB) | β-globin restored autologous HSC | Phase I/II | Electroporation - ex vivo |
| Enrolling by invitation | NCT05143307 | HIV | Undisclosed | EBT-101 | Phase I | AAV9 - in vivo |
| Concerned | 2018-001320-19 | Sickle cell disease, hematological diseases, hemoglobinopathies | HBB | CTX001 | Phase I/II | Electroporation - ex vivo |
| Active | NCT03745287 | Sickle cell disease, hematological diseases, hemoglobinopathies | BAF chromatin remodeling complex subunit 11 A (BCL11A) | CTX001 | Phase II/III | Electroporation - ex vivo |
| Active | ChiCTR2100052858 | Transfusion dependent beta-thalassemia | Undisclosed | RM-001 | Phase I | Undisclosed - ex vivo |
| Active | NCT04037566 | Leukemia lymphocytic acute in relapse, acute lymphocytic leukemia (ALL) refractory lymphoma, B-cell, CD19 positive, ALL | CD19 molecule, HPK1 hematopoietic progenitor kinase 1 (also known as MAP4K1) | Cyclophosphamide/Fludarabine | Phase I | Lentivirus (LV) and electroporation - ex vivo |
| Active | NCT04560790 | Herpes simplex virus refractory keratitis | UL8/UL29 | BD111 adult single group dose | Phase I/II | mRNA transfection - in vivo |
| Active | NCT03872479 | Blindness, Leber congenital amaurosis | Centrosomal protein 290 (CEP290) | EDIT-101 | Phase I/II | Adeno-associated virus (AAV5) - in vivo |
| Active | NCT05210530 | Type 1 diabetes | – | VCTX210A unit | Phase I | Ex vivo |
| Active | NCT05120830 | Hereditary angioedema | Kallikrein B1 (KLKB1) | Biological NTLA-2002 | Phase I/II | Lipid nanoparticles - in vivo |
| Active | NCT04601051 | Hereditary transthyretin amyloidosis | Transthyretin (TTR) | NTLA-2001 | Phase I | Lipid nanoparticles - in vivo |
Clinical trials involving ex vivo CRISPR-based genome editing
The primary clinical trial on CRISPR-based ex vivo genome editing endeavored to treat human immunodeficiency virus 1 (HIV-1) infection.32 Disruption of CCR5, which encodes a critical receptor for the viral compartment, was initiated by nucleofection of CCR5-centered ribonucleoprotein complexes in patient-derived hematopoietic stem cells and progenitor cells (HSPCs). A 27-year-old man with HIV-1 disease and severe lymphoblastic leukemia was treated as such which brought about effective and long-term engraftment of CRISPR-edited HSPCs. The efficacy of CCR5 disruption increased from 5.2% to 8.3% in bone marrow cells over 19 months which did not reach the desired goal (ClinicalTrials.gov, NCT03164135). Recently, a clinical trial attempting to treat serious single-gene diseases with CRISPR-based genome editing systems with promising results has been reported.33 Sickle cell disease and beta-thalassemia refer to a group of acquired hemoglobinopathies caused by changes in the gene of the beta-hemoglobin subunit (HBB) that resulted in misfolded beta-globin proteins. In this treatment approach, rather than the pathogenic HBB gene, BCL11A gene enhancer which is a translational variant to inhibit gamma-globin has been considered to re-establish fetal hemoglobin production, and compensate for the altered HBB gene. Patient-derived HSPCs were modified with CRISPR-Cas9 with a sgRNA targeting the BCL11A enhancer to create gene-edited HSPCs called CTX001. One subject with transfusion-dependent beta-thalassemia and one subject with sickle cell disease were examined with CTX001 after myeloablation. High altered allele frequencies and fetal hemoglobin levels were maintained, and both patients were cured of disease-related transfusion. However, actual adverse effects were observed in both patients like neutropenic pneumonia, hepatic vein-obstructive infection with sinus obstruction, sepsis with neutropenia, kidney stones, and gastric agony. This ongoing test is almost consistent with preliminary findings from additional trials (ClinicalTrials.gov, NCT03655678, and NCT03745287).
Furthermore, CRISPR-based ex vivo genome editing is additionally relevant to the treatment of malignant cancers.34, 35, 36 The basic concept is to promote common antitumor responses of cytotoxic T cells by depleting the modulatory genes of some checkpoints via CRISPR-Cas9. Two clinical trials using this technique were recently published with promising results. In one of them, scientists attempted to treat various advanced and incurable cancers including multiple myeloma and liposarcoma by CRISPR-Cas9 genome editing.34
In their experiment, T cells were isolated from cancerous patients and engineered with CRISPR-Cas9 to suppress the endogenous T cell receptor (TCR) and immune-modulated cell transit protein 1 (PD-1). Specifically, deletion of the TCR α chain (TRAC), TCR β chain (TRBC), and PDCD1 genes were initiated by electroporation of ribonucleoprotein complexes in patient-derived T cells. The modified T cells, called “NYCE” (CRISPR 3X NY-ESO-1-transduced cells) were then injected into patients intravenously. A total of 3 patients were treated with stable engraftment of edited T cells. The frequencies of target gene mutations in peripheral blood mononuclear cells were around 5%–10%, without significant off-target changes or actual adverse effects. Tumor recurrence was limited to the early stages of treatment, and all tumors inevitably progressed at the end of the trial (ClinicalTrials.gov, NCT03399448). In another comparative trial, non-small cell lung cancer was tried to be cured with CRISPR-engineered patient-derived T cells targeting the PD-1 gene.35
PD-1 gene-associated exon disruption was induced by electroporation of Cas9-and sgRNA-encoding plasmids into patient-derived T cells. A total of 12 treated patients demonstrated the development of modified T cells, even though the average gene modification rate was very low (5.8%). No significant changes were seen outside the target or in real contrast situations. Clinically, two patients had stable disease at 8 weeks, all patients had an infection, and 11 patients had a progression of infection (ClinicalTrials.gov, NCT02793856).
In vivo genome editing
Initially reported with CRISPR-based in vivo gene editing, this innovation was linked to transthyretin amyloidosis, or ATTR amyloidosis, which resulted from the accumulation of misfolded transthyretin (TTR) protein in different organs and tissues.37
ATTR amyloidosis is a monogenic infection and almost all TTR proteins form amyloid fibrils in different organs and are an ideal target for CRISPR-based in vivo gene editing. NTLA-2001, a liver-trophic lipid nanoparticle framework containing Cas9 mRNA and sgRNA targeting human TTR gene was offered to decrease circulating TTR amyloid forms in humans. Six patients with TTR mutations and tactile polyneuropathy were treated with a single injection of NTLA-2001, which reduced serum TTR levels up to 52% in the low-injected dose group and 87% in the high-injected dose group after 4 weeks without significant side effects. To confirm the efficacy and long-term safety of the treatment, all patients were followed up (ClinicalTrials.gov, NCT04601051).
Although detailed information is not available, numerous research groups have tried to utilize CRISPR-based in vivo gene editing to treat Leber's innate amaurosis 10 as a monogenic infection disease (ClinicalTrials.gov, NCT03872479).
Clinical trials utilizing ex vivo and in vivo CRISPR-based gene editing is summarized in Table 1. Moreover, a broad investigation of animal models for advanced gene editing applications is reviewed in the following section.
Animal models in CRISPR applications
A lot of knowledge regarding CRISPR-based genome editing approaches is acquired through the implication of animal models. Some examples are referred to here. Leber's congenital amaurosis (LCA, OMIM #204000) comprises a group of early-onset retinal dystrophies of childhood, each subtype results from changes of distinctly different genes. LCA sort 10 (LCA10, OMIM #611755) is caused by a mutation in the gene of CEP290. CRISPR-based genome editing has been used in mouse LCA10 models. Wild-type CEP290 expression was successfully re-established by subretinal infusion of a single AAV encoding both SaCas9 and sgRNA.38
LCA2 (LCA2, OMIM #204100) is done by RPE65 transfection. The rd12 mouse genetically edited with LCA2 was under subretinal infusion with two AAVs encoding SpCas9, sgRNA, and donor DNA. The as-described mouse was effectively treated with subretinal infusion of adenine base editors utilizing RNPs,39 intein-mediated part AAV vectors,40 and prime editors utilizing trans-splicing part AAV vectors.41 Retinitis pigmentosa (OMIM #268000), which alludes to a heterogeneous gathering of acquired visual infections that result in dynamic retinal degeneration, comprises 92 diverse phenotypes and is caused by changes in over 200 genes. AAV-mediated gene transfection to treat retinitis pigmentosa is one of the approved treatments in gene therapy history.42
Fundamental studies on CRISPR-based genome editing began in 2016. Since then, numerous CRISPR-based approaches (each focusing on different genes like Nrl Mertk, Pde6b, Rho, and RPGR),43, 44, 45, 46, 47, 48, 49 have been done in animal models. In particular, the Nrl gene was terminated via NHEJ and inhibited by an approach called CRISPR blockades, both were mediated by AAV vectors. The Mertk gene was edited by a novel strategy called homology-independent targeted integration that focuses on integration. The Pde6b gene was modified via homology repair, and the Rho gene via NHEJ. These two later genes were activated by in vivo electroporation using Cas9-encoding plasmids. Hereditary tyrosinemia type 1 (HT1, OMIM #276700), a fatal inherited disorder caused by mutations in fumaryl acetoacetate hydrolase (FAH), results in the accumulation of deleterious metabolites leading to severe liver damage. CRISPR-based genome editing using human HT1 models in mice was initiated in 2014 which modulates pathogenic mutations.50 Gene-edited hepatocytes showed a growth advantage over other mutated hepatocytes which can repopulate the liver in a truly regenerative manner. After this work, Cas9 modifications (NmeCas9,51 St1Cas952), base editors,53,54 and prime editors41 have effectively protected the deadly HT1 phenotype in grown-up mouse models.
Phenylketonuria (PKU, OMIM #261600) is an autosomal recessive liver illness caused by mutations within the phenylalanine hydroxylase gene, which may cause mental impediment due to the neurotoxicity of metabolites. In grown-up mouse models, intravenous infusion of AAVs encoding an intein-split cytosine base editor effectively reestablished blood phenylalanine levels and switched the PKU-associated hide color.55
Afterward, the ordinary homology-directed repair approach reduced the side effects with the assistance of chemical modifiers.56,57 Ornithine transcarbamylase (OTC) deficiency (OMIM #311250) is an X-linked metabolic clutter characterized by hyperammonemia and is caused by mutations within the OTC gene (OMIM ∗300,461). Employing a double AAV framework containing SaCas9-encoding sequences, sgRNA-encoding sequences, donor DNA, and OTC editions with homology-directed repair, lead to more survival in mouse models.58
Duchenne muscular dystrophy (DMD, OMIM #310200) is an acquired X-linked infection caused by changes within the dystrophin locus. The application of a CRISPR-based genome editing tool to regulate mutations and restore dystrophin expression in mouse zygotes was initiated in 2014.59 Following this initial work, many other studies investigated the exact properties of CRISPR-Cas9 restoration of dystrophin expression in rat,60, 61, 62 dog,63 and pig64 models of DMD. Furthermore, adenine base modification successfully reversed DMD pathology in mouse embryos and adult mouse models.65
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder in which the dynamic transmission of motor neurons is accompanied by loss of movement. Several causative features have been distinguished as early congenital ALS, and mutations in SOD1 (OMIM ∗ 147,450) are believed to be initiators for most cases of ALS type 1 (ALS1, OMIM #105400). Recently, intravenous infusion of AAV encoding SaCas9 and SOD1-targeting sgRNA appeared to delay disease appearance and enhance the applicability in ALS mouse models.66
Glycogen capacity illness Ia (GSD1A, OMIM #232200), too known as von Gierke illness, is caused by pathogenic mutations within the glucose-6-phosphatase alpha subunit (G6PC) gene. This mutation causes glycogen accumulation all over the body. Recently, the profoundly dominant G6PC p. R83C mutation was subjected to CRISPR-based genome editing in vivo in mouse models using two AAVs; one encoding SaCas9 and the other encoding sgRNA67 which knocked down the G6Pase gene.
Hutchinson–Gilford progeria disorder (HGPS, OMIM #176670) is caused by changes in the gene of layer A (LMNA). Recently, the LMNA c.1824C > T mutation was found in more than 90% of patients with HGPS. This was edited in transgenic mouse models using AAVs encoding conformations of the adenine base editor and resulted in more vascularization and life expectancy.68 This report demonstrated the potential of modern genome editors to specifically create point mutations for the treatment of inherited disorders.
CRISPR/Cas9 delivery platforms
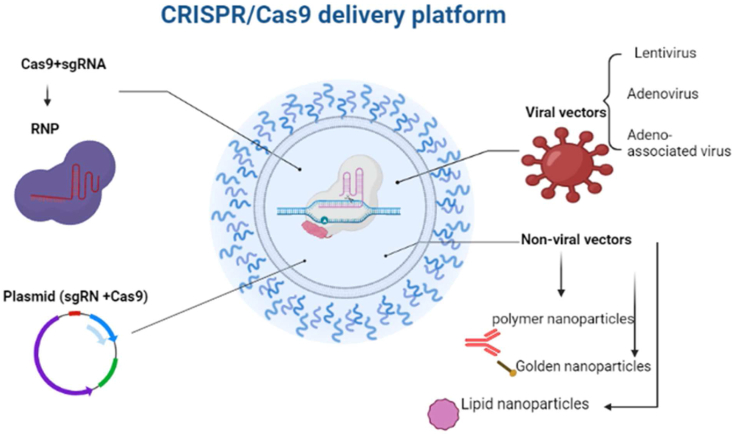
To fully exploit the editing potential of CRISPR/Cas9, they must be successfully delivered into target cells or tissues using appropriate vectors.69 Various transfer methods are shown in Figure 2. In the following section, the advantages and disadvantages of each transfer method will be reviewed.
Figure 2.
CRISPR/Cas9 delivery platforms showing viral and non-viral vectors, plasmid, and RNP.
Viral vectors
Recombinant viral vectors have been developed by exploiting the capacity of viruses to insert exogenous genetic material into cells in order to transfer beneficial properties into infected cells/tissues (Table 2).70 Among several viral vectors, adeno-associated viruses (AAV), lentivirus, and adenovirus play an important role in genome editing therapies and have been widely used in clinical models and trials. Although viral vectors do not cause severe human disease, they can induce immune responses, which may reduce transfection efficiency.71 Another problem with viral vectors is the capacity to permanently restore DNA in the host genome, which may lead to off-target effects and translocations.72
Table 2.
Viral vectors for CRISPR/Cas9 delivery system.
| Delivery | Packaging capacity | Advantages | Disadvantages |
|---|---|---|---|
| Lentivirus | Approximately 10 kb | High transduction efficiency; large cargo size; low immunogenicity; ability to transduce dividing and non-dividing cells in different tissues | Non-specific DNA integration causes cancer risk; complex packaging structure |
| Adenovirus | Approximately 8–10 kb | Efficient delivery; large cargo size | Inflammatory response |
| Adeno-associated virus | Approximately 4.7 kb | Multiple serotypes; low immunogenicity; ability to transduce dividing and non-dividing cells in different tissues | Pre-existing neutralizing antibodies; long-term expression of Cas9 causes off-target effects |
Adenovirus
Adenovirus is a double-stranded DNA virus with 80–100 nm dimensions. Its genome is about 34–43 kb in length and can store about 8 kb of exogenous DNA in itself.73 Due to its extraordinary capacity to carry large heritable cargos, the transfection ability of the adenovirus vector-mediated CRISPR/Cas9 module can be enhanced by providing additional targeting signals.74 Nowadays some adenoviral vectors can accumulate the target DNA up to 37 kb by reducing their primary genomic material.75 Adenovirus can infect both dividing and non-dividing cells, but an important issue is that its genome is not coordinated with cells, which reduces off-target effects and non-intended mutations.76 Regarding pathogenicity, the adenovirus vectors can stimulate the immune response,77 even though this reaction may promote the cytotoxicity effect on tumor cells, the neutralizing agent reaction induced by B cell action is not related to subsequent vector transfer.78 Therefore, reducing the immune response promoted by adenoviral vector will significantly increase the safety and effectiveness of the transfer by this vector. The use of poly (lactic acid/glycolic acid) copolymer for encapsulation of recombinant adenovirus vectors reduces the immunogenicity of adenoviruses and enhances near-neutralizing antibodies in vitro. This information will be used to improve more advanced viral vectors.79
Genetically engineered mouse models (GEMMs) of human cancer are imperative instruments to analyze the atomic components of tumorigenesis.80 Transferring CRISPR/Cas9 into cells by physical methods utilizing adenovirus vectors actuates particular chromosomal improvements to produce a mouse model of Eml4-Alk-driven lung cancer.81 This methodology extends how researchers develop human cancer models by rearranging complex and time-consuming hereditary genomic controls. Thus, adenoviral vectors have been utilized to improve gene editing techniques focusing on Pten in a mouse with nonalcoholic steatohepatitis (NASH). In this model mice infused with an adenoviral vector showed signs of hepatomegaly and NASH after 4 months. Indeed, the signs of typical adenoviral vector-related immunotoxicity in the liver appeared while adenoviral vectors intervened in effective Pten gene editing, giving a novel strategy to imitate human liver malady in mice.82 GEMMs produced by site-specific recombinase innovation are non-efficient and time-consuming, but adenoviral vector-mediated CRISPR/Cas9 gene editing can successfully deliver numerous subtypes of delicate tissue sarcoma in wild-type mice and GEMMs. Whole-exome sequencing validated that sarcomas produced utilizing CRISPR/Cas9 are comparative to those produced utilizing conventional recombinase innovation, demonstrating the system's potential to quickly create cancers with comparative genotypes and phenotypes as conventional techniques.83
Adeno-associated viruses (AAVs)
AAVs comprise an icosahedral protein capsid with a breadth of ∼26 nm and an ssDNA genome of ∼4.7 kb.84 AAV vectors have interesting properties, such as the need to be virulence (living in other cells in order to be replicated), long-term gene expression, and the capacity to live in dividing and non-dividing cells, so they are widely used for in vivo transfection systems.84,85 In the expansion of the AAV family, AAV vectors are characterized by serotype phenotyping and have variable tropism focusing on different organs.92,132 Even though AAVs are excellent delivery vehicles, they have some shortcomings in their use to deliver CRISPR/Cas9 in vivo. The ideal AAV transmission capacity is estimated to be 4.1–4.9 kb. Even though vectors are larger than their genome size, packaging efficiency is severely reduced.86 For example, the SpCas9 protein is ∼4.2 kb, and recombinant AAV must also contain critical regulatory components for gene expression, so AAVs cannot be used to provide extensive gene regulation.84
When using AAVs for transfection, SpCas9 and sgRNA must be encoded on different vectors.58,87 Another important issue of AAV is the presence of pre-existing neutralizing antibodies against AAV in patients with previous AAV infection, which significantly reduces the transfection efficiency.88 In many cases, the combination of capsid regulation and genome regulation to provide an optimal AAV serotype vector can reduce the avidity of neutralizing antibodies, subsequently reducing resistant reactions and providing stepwise transfection efficiency.89 Furthermore, long-term expression of the AAV transgene may also be an opportunity, as the continuous expression of the Cas9 core may cause significant off-target effects.90 There are problems in the mass production of AAVs. Even though there are still several challenges to overcome, scientists have tried to explore AAV-mediated CRISPR transfection systems. The AAV dual-carrier framework effectively targets a single gene or multiple genes in the mouse brain and characterizes the effects of genome regulation on neurons,91 suggesting that AAV-mediated genome editing could be useful for brain diseases. As distinct AAV serotypes have broad tissue tropism, AAV-mediated genome editing can also be used to generate existing cancer models.92 Platt et al delivered a single AAV vector into the lungs of mice models to interfere with p53, Lkb1, and KrasG12D genes leading to adenocarcinoma. Furthermore, the use of AAV to deliver sgRNA to Cas9 knockout mice can be used for high-throughput in vivo editing to generate mouse models of the desired cancer.93
Lentivirus
Lentivirus is a subset of the retrovirus family, and its genome contains a single-stranded RNA of 7–12 kb.94 Lentiviral vectors achieve successful cell transfection in various cell types (dividing and non-dividing cells) with a reduction of required culture time for cell transfection. Compared to adenovirus or AAV vectors, lentivirus appears to have less cytotoxicity and immunogenicity and fewer adverse effects on transduced cells.95 Furthermore, lentiviruses are promising as in vivo transfection cargo due to their relative ease of use. Regularly, the lentivirus integrates its genome into the host genome, which enhances the time of transgene expression. Continuous expression of Cas9 increases the chance of off-target effects and prevents application in high-precision genome edition.96 Selective and integrase-deficient lentiviral vectors produced by integrase modification can significantly reduce the risk of unintended mutations.97 In preclinical studies, it seems that lentiviral delivery of Cas9 and direct RNA targeting of the KRAS gene have inhibited the proliferation of cancer cells.98 CRISPR/Cas9 viral delivery targeting BCR-ABL essentially inhibits cell growth, myelogenous leukemia cells, and tumorigenesis. Thus, therapies based on editing ABL may offer a potential technique for patients with imatinib-resistant myeloid leukemia.99 Lentiviruses have been approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA).100
Non-viral vectors
Safety issues remain a major bottleneck for the lack of widespread clinical use of viral vectors,101 safe reactivity,71 and stable expression.102 As an alternative, non-viral vectors for cancer therapy have been investigated due to their immunogenicity, high biocompatibility, amazing delivery capability, and large-scale production.103,104 Nanotechnology-based drug delivery frameworks develop broad applications of CRISPR/Cas9 therapy, advance safety, and provide a practical approach to overcoming the challenges of viral vectors (Table 3).
Table 3.
Nanotechnology-based delivery systems for CRISPR/Cas9.
| Delivery system | Cargo options | Advantages | Disadvantages |
|---|---|---|---|
| Polymer nanoparticles | RNP plasmid DNA; RNP complex; Cas9 mRNA; sgRNA; donor DNA | High biocompatibility; low immunogenicity; ability to reduce off-target effects; mass production; low cost | Toxicity; limited delivery efficiency |
| Golden nanoparticles | RNP plasmid DNA; RNP complex; Cas9 mRNA; sgRNA; donor DNA | High biocompatibility; low immunogenicity; ability to reduce off-target effects; mass production; low cost | Limited delivery efficiency |
| Lipid nanoparticles | RNP plasmid DNA; RNP complex; Cas9 mRNA; sgRNA; donor DNA | High biocompatibility; low immunogenicity; ability to reduce off-target effects; mass production; low cost | Degradation in vivo |
Lipid nanoparticles (LNPs)
LNPs are amphiphilic vehicles composed of various hydrophobic and hydrophilic moities, such as cationic or ionized lipids; neutral lipids such as phospholipids or cholesterol, and polyethylene glycol-lipids.105 LNPs were developed as carriers for the delivery of an array of particles into cells, particularly in nuclear transports. Since nucleic acids are relatively unstable outside the cell and carry many anions, they cannot easily cross the cell membrane. However, the ionic and polar head in cationic liposomes allows for easy delivery of nucleic acids into cells. Compared with conventional drug delivery, LNPs have interesting features including monitoring the cargo, targeted delivery systems, and reduced toxicity. These features have made LNPs attractive for the delivery of anticancer drugs.106 Pre-clinical trials found that LNPs can effectively transfer siRNA or mRNA107,108; therefore, LNPs seem to be a safe and suitable transfer method.109
Within the past years, numerous preclinical studies about CRISPR/Cas9 delivery using LNPs have been performed. Two fundamental strategies are utilized for LNP delivery of CRISPR/Cas9 components like transferring the Cas9/sgRNA plasmid and transferring the Cas9/sgRNA RNP complex. Cas9 mRNA and sgRNA can be effectively stacked on LNPs and precisely transported to the liver of mice while intervening in the mouse model for transthyretin (Ttr).110 Despite several advances, routine prediction, and planning of LNP delivery to target tissues other than the liver for precise gene modification remains a challenge. Organ-specific targeting allows nanoparticles to provide gene-editing frameworks for specific organs, which is expected to encourage the development of gene-regulating therapies.111
Polymeric nanoparticles
Polymeric materials have long blood circulation, as well as high bioavailability, amazing biocompatibility, and degradability, and thus they are considered powerful delivery tools.112 However, conventional strategies for sgRNA-Cas9 deliveries are RNPs which are in danger by proteases in cells. The protein core and the permeable polymer shell form a modern nanocapsule that can be programmed to be aberrantly degraded or stable at different pH values. Capsule degradation breaks down the outer shell and allows the core protein to enter the cell to perform its normal functions. Albeit its disadvantages, this strategy leads a set of proteins into cells and opens a new way for sgRNA delivery, Cas9-RNP, and cancer therapy.113 Furthermore, in 2019 Chen et al synthesized a glutathione cleavable covalent cross-linked polymer coating around the Cas9-RNP complex to create a nanocapsule. This nanocapsule performed the gene edition in vitro without any cytotoxicity evidence. Topical organization of the nanocapsules in mice produced effective gene edition capabilities.114 In further studies, Cas9-RNP was efficiently transfected into HEK293-T cells and colon cancer cells and induced genome modification. Crucially, nanoarray targeting of mutated KRAS in cancer cells can successfully suppress tumor growth and metastasis in tumor-bearing mouse models.115 Guo et al have knocked down the breast cancer oncogene lipocalin 2 (LCN2) in human TNBC cells through CRISPR-polymeric nanoparticles. LCN2 generally restricted the migration of mesenchymal phenotype of human TNBC cells and reduced their invasiveness.116 In another study, Zhang et al combined nanotechnology and genomic design to disrupt cyclin-dependent kinase 5 (Cdk 5) and reduce PD-L1 expression in tumor cells. They significantly inhibited murine melanoma development and TNBC lung metastasis.117 It seems that polymeric nanoparticles have great prospects and wide potential in CRISPR genome editing as a modern precision medicine for cancer treatment.
Gold nanoparticles (GNPs)
GNPs are another alternative carrier for transferring CRISPR/Cas9. GNPs can be combined with distinctive components such as nucleic acids, lipids, or polymers. They have relative biocompatibility and enter into various sorts of cells.118,119 Attaching different components of nucleic acids and glycoproteins on the surface of the GNPs makes beneficial applications.120 The pharmacokinetics of GNPs can be controlled by changing their size, shape, charge, and surface area.121,122 GNPs/Cas9/sgRNA complex can achieve ∼30% transfection efficiency, which provides a modern strategy for genomic research.123 While HDR-based therapies treat most inherited diseases, developing delivery vehicles that can activate HDR in the body has been challenging. A transfer vehicle composed of DNA-conjugated GNPs and cationic endosomal escape polymers can transfer Cas9-RNP to essential cells and stem cells. This complex, called CRISPR-Gold has actuated HDR in MDX mouse myoblasts with negligible off-target effects.124 Since safety is the main barrier to the entry of GNPs into the human body, it is important to investigate its interaction inside the cell. The uptake of GNPs by resistant mast cells stimulates the production of pro-inflammatory cytokines, suggesting that GNPs have an immunostimulatory effect.125 In many cells, the interaction of GNPs with different receptors on the surface of cells and different types of endocytosis depends on the regulatory changes caused by GNPs.126,127 In addition, due to the specific biophysical properties of metal particles, the charge and electrostatic field on the surface of the particles also fundamentally affect safety issues. Further research is needed to fully characterize the components involved in the interaction of GNPs with the immune system.
Challenges and future opportunities
In the future, the use of base editors and core editors in human experiments will have a significant impact on health issues, because there are many inherited diseases caused by point mutations that can be cured by modern types of genome editors with extraordinary levels of minimal undesired off-targets. Some researchers are trying to expand the potential of base and core editors. A significant increase in the number of clinical trials using the CRISPR technique for genome editing has been seen in recent years. In this part, some challenges related to the CRISPR technique will be discussed. Due to the speed of research, a solution to these problems and complications will be also provided.
Challenges of CRISPR/Cas9
Delivery challenge
Even though CRISPR/Cas9 may be a developed gene editing innovation that has been utilized broadly, it keeps up numerous issues due to the off-target effects, efficiency, and safety challenges. In terms of CRISPR-based gene therapy, the challenges of the in vivo transfection methods are highlighted.27 A complete transfection strategy for CRISPR/cas9 should have high transfection efficiency, an exceptional focus on capacity, and ease of mass production. However, the current methods are still far from reaching these goals.58 Physical methods of CRISPR/Cas9 transfer are commonly used in vitro but are uncommon and non-efficient in vivo. Different methods like electroporation, microinjection, ultrasound-driven nanomotors,128 microfluidic/nanofluidic approaches,129 and nanocarriers130 have been widely used. For using CRISPR/Cas9 in human stem cell gene editing, electroporation is still the main choice in several studies.131 However, several physical approaches can deliver CRISPR/Cas9 in vivo. For example, hydrodynamic injection (HDI) was described as a novel approach for CRISPR delivery,50 but its application is limited because this strategy may cause damage during transfection.
Apart from the physical techniques, scientists are delivering CRISPR/Cas9 using diverse carriers. Along with the types of carriers, the CRISPR/Cas9 transfection strategy may be divided into two types: viral transfection methods and non-viral techniques. Due to the greater transfection efficiency, viral vectors are more common for CRISPR/Cas9 transfection. Adeno-associated virus (AAV) is the most common vector for gene therapy in vivo and in vitro due to its wide serotype, low immunogenicity, and toxicity,132 but its small carrying capacity (about 4.5–5 kb) must be improved. Compared to AAV, lentivirus (LV) and adenovirus (AdV) have the better, higher, and stronger capacity and transfer the additional genetic components. Therefore, one of the most important points about AdV is its capacity to transfect a wider range of cells than LV and AAV. In some cases, larger LV and AdV sizes may induce humoral responses and indeed cellular immunity.123 Other viral vectors have been recently developed and have distinct characteristics. For example, EBV vectors can be more stable in cells,133 and Sendai virus vectors are competent to infect a wider variety of cells,134 while baculovirus vectors have a higher transfect capacity.135 However, these vectors are currently used in ambient conditions, but they can still be used in vivo after further optimization.
Concerning non-viral techniques, liposomes are utilized frequently since they have been merchandised broadly. In other examples, some researchers attempted to utilize gold nanoparticles (AuNPs) as the vectors for CRISPR/Cas9 transfer. Other non-viral vectors like Lipofectamine RNAiMAX,136 PolyJet™ In Vitro DNA Transfection Reagent,137 and X-tremeGENE HP DNA Transfection Reagent137 are commercial and appropriate for in vitro or ex vivo tests. In terms of in vivo transfection, thousands of researchers are focusing on finding and synthesizing non-viral vectors with high efficiency and low cytotoxicity. These techniques include conventional nano-sized arrays (such as self-assembled micelles138 and polyethylene glycol-modified cationic lipid nanoparticles139 for CRISPR/Cas9 plasmid delivery), DNA nanostructures,140 and phosphorus nanosheets.141 For CRISPR/Cas9 transfection, receptor-mediated transfection methods (e.g., folate receptor-targeted liposomes of CRISPR/Cas9 plasmids142), cell-penetrating peptides (CPPs)-mediated transfection methods, and KASIM transfection methods (e.g., a hybrid of K9 protein)143 are used. Wang developed cationic α-helical polypeptide-based PEGylated nanoparticles to deliver CRISPR/Cas9 plasmids and sgRNA.144 Also, several delivery methods (e.g., R8-dGR did peptide-modified cationic liposome for delivery of CRISPR/Cas9 plasmids and sgRNA145 and near-infrared transformation-activated framework for CRISPR/Cas9 delivery146) were developed. In a recent study, a four-component formula [DOTAP (1,2-dioleoyl-3-trimethylammoniumpropane)/DOPE/cholesterol/Chol-PEG (cholesterol-polyethylene glycol)] liposome with the highest rate of transfection of Cas9/sgRNA resulted in 39% gene-editing efficiency to knockout GFP reporter.109
Still, several inconveniences within the field of non-viral procedures, like transfer obstructions and endosomal escape exist.123
In comparison to different vectors, the shapes of cargos moreover play an important role in CRISPR/Cas9 transfer. Cas9 is transferred within the shape of DNA or mRNA with sgRNA and layout sequence together. In the case of gene editing productivity, Yin and his colleagues transferred Cas9 mRNA by lipid nanoparticles whereas transferring sgRNA and making layout arrangement by AAV separately.147 In addition, Cas9 proteins can also be easily transfected into cells by hybridized strategies. This bypasses the risk of genome integrity and reduces the off-target effect due to the short half-life of the Cas9 protein, which is considered a safer approach for gene therapy.147 Consequently, the smaller the carrying capacity of transfection vectors is, less immune responses will occur; while there is a need for greater capacity to carry more CRISPR. However, multifunctional chimeric peptides due to their size flexibility, biocompatibility, and biodegradability have shown potential abilities in gene delivery that may be promising for CRISPR-based gene editing.148,149
Off-target effect
The off-target effect is one of the limitations of the CRISPR/Cas9 technique and is considered a critical risk in vivo gene therapy. Although several computer programs have optimized the design of sgRNA, its specificity cannot be fully guaranteed. In addition to sgRNA design, Cas9 protein also plays a vital role. As an example, the design of Cas9, such as protein transfer, may substantially reduce the off-target effect. The optimized Cas9 protein as well as using the SpCas9-HF1150 and eSpCas9151 are two approaches to reduce the off-target effect. Notably, the off-target effect remains unresolved in vivo and is strongly related to the transfer technique.
PAM limitation
As it was reviewed, the PAM sequence is fundamental for CRISPR/Cas9 and is largely dependent on Cas9 specificity. PAM sequence limits the design of sgRNA and reduces the adaptability of CRISPR/Cas9 with other systems. Even though an expanding number of CRISPR sorts are found, more PAM sequences have been discovered. The obligatory PAM addition still influences the design of sgRNA in a few circumstances. Subsequently, how to create a designable PAM is critical to broadening the application of CRISPR/Cas9.
Immune response
Even though there are not numerous reports about the extreme immune responses caused by Cas9, the antibodies against Cas9 have been broadly recognized in human bodies,152 which recommends the potential hazard of aggravation against CRISPR/Cas9-based gene therapies. Scientists pay more attention to the immunogenicity caused by vectors, particularly viral vectors, since the human body may have been exposed to them before. Collectively, the immune responses caused by CRISPR/Cas9 gene-editing framework are one of the major hazard components within the improvement of CRISPR-based gene therapy in vivo.
Multiple gene-editing
CRISPR/Cas9 is a proficient gene-editing system that changes one gene locus with a sgRNA at a single time. Subsequently, numerous gene-editing techniques which use CRISPR/Cas9 are dependent on different sgRNAs, which decreases the editing efficiency and increases the transfection problem. Later CRISPR/Cas12a has overcome this challenge, but other issues are still unsolved, including the cell inactivation and cell cycle capture after different gene-editing. In the future, convincing different gene-editing techniques will significantly enhance the gene therapy of polygenic maladies and cancers.
Future perspectives
CRISPR/Cas9 as a microbial intrinsic immunity framework has been developed as a robust gene-editing technology. Due to its precision and efficiency, CRISPR/Cas9 techniques can provide an incredible chance to treat a few gene-related diseases by deletion, insertion, regulation, and blocking different genes. Cas9-mediated gene therapy has been utilized to treat different non-cancerous maladies. Monogenetic diseases and X-linked illnesses caused by gene transfection are the most studied diseases by this technique which have even entered clinical trials. In this regard, the probability of both CVDs and NDDs has become lower by using CRISPR/Cas9. At the same time, the treatment of visual illnesses by Cas9 has developed into clinical stages.
CRISPR/Cas9 in the first attempt was used for cancer therapy. Combined with computational simulation methods, Cas9-based target screening gives an advanced approach to monitoring the cancer stages. Deleting oncogenes or turning on the tumor-silencing genes alone or in combination with each other, are the major approaches to treating cancers and viral disorders. Besides, a few investigations illustrated a few gene regulations, epigenetic factors, and micro-environmental genes which play crucial roles in cancer therapy. In some studies, CAR-T cells have been generated by the Cas9 system and reached successfully to clinical trial phases. Thus, CRISPR/Cas9 has shown up as a vigorous gene-editing tool. In some cases, a few issues keep unsolved like off-target effects, transfer challenges, PAM confinement, and immunogenicity. During gene therapy, the off-target effect and editing effectiveness are two of the most concerning issues since the off-target may cause unintended editions in ordinary genes and then lead to some illnesses. Also, the editing efficiency specifically influences the final impact. The transfer of CRISPR/Cas9 is crucial for CRISPR-based treatments. The transfection efficiency determines the proficiency of Cas9-mediated gene therapy to a significant degree. The targetability and transfer time are two factors related to the off-target effect.
As discussed before, expendable plans (such as transferring the Cas9 protein) and all-in-one plans (such as transferring the Cas9 plasmids and sgRNA at the same time) are two ways to reduce the off-target effect. Furthermore, re-engineered or optimized Cas9 proteins decrease the off-target effect as well. Compared with conventional CRISPR/Cas9-based gene editing tools, base edition and primary edition tools do not make DSBs when altering genes. This phenomenon will lead to fewer off-targets. Also, their edition efficiency will be improved using optimizing chemicals or prime editing guide RNA (pegRNA), recommending the awesome potential for clinical application. sgRNA design is still fundamental since it plays a key role in gene targeting. For optimizing sgRNA design, some rules and computer programs calculating some factors like the distance from the PAM sequence may be effective on sgRNA design. As studied, most clinical trials study gene editing in patient-derived cells ex vivo, such as the treatment of SCDs. This strategy reduces the hazard of off-target effect and transfer challenges but is not reasonable for all illnesses. The clinical application of CRISPR/Cas9 is still at an early stage, and off-target effects and transfer mechanisms remain two important challenges of Cas9 -based editing tools.
Compared with monogenetic illnesses, cancer therapy by Cas9-based systems is more challenging due to numerous gene mutations in cancerous cells, even though it is possible to apply numerous gene editing by CRISPR/Cas9. CRISPR/Cas9-mediated numerous gene editing technology is not broadly used in clinical applications. It is thought that it has a few potential issues, such as extreme off-target effects and the deletion of enormous DNA fragments.153 In this manner, novel approaches in multiple gene editing were developed to overcome these challenges. But for CRISPR/Cas9, other CRISPR frameworks, including Cas12a, Cas3 (with Cascade), Cas13, dCas9, and nCas9 are under attention.154
In addition, compared to Cas9, Cas12a has a site for the period II CRISPR framework. But Cas12a creates an amazing cut in comparison to Cas9, which could be an incredible advantage when joining DNA groupings. Within the Cas3 framework, the Cascade complex ties and recognizes the target DNA arrangement at the Cas3 recognition site in order to produce a single-strand DNA sequence. Based on the PAM sequence, Cas3 is more adaptable to target DNA sequences than Cas9. Different from Cas9, Cas12a, and Cas3, Cas13 is an RNA-guided that can alter single-strand RNA proficiently. Both dCas9 and nCas9 lose the nuclease action but keep up the capacity to target DNA groupings, so a part of re-engineered CRISPR/Cas9 devices, such as CRISPRi, CRIPARa, gene editing tools, preliminary editing techniques, etc., are based on dCas9 or nCas9. In conclusion, CRISPR/Cas9 is an efficient gene-editing tool but not an idealized therapy approach. CRISPR needs a better safety level. Cell treatment by Cas9 appears to be easier to design whereas it has a few inconveniences during in vivo gene therapy approaches. It is noteworthy that only hundreds of diseases can be treated by cell therapy, while a broader range of hereditary maladies can be cured by gene therapy tools like Cas9 which is one of the major viewpoints to be more developed in the future. Effectively and safely gene editing by CRISPR/Cas9 in vivo will be done with proper transfection systems within the following decade.
Conflict of interests
The authors declare no conflict of interests.
Funding
Financial support of this work was provided by Chongqing Medical University (China).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Tang X.D., Gao F., Liu M.J., et al. Methods for enhancing clustered regularly interspaced short palindromic repeats/Cas9-mediated homology-directed repair efficiency. Front Genet. 2019;10:551. doi: 10.3389/fgene.2019.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varble A., Meaden S., Barrangou R., et al. Recombination between phages and CRISPR-cas loci facilitates horizontal gene transfer in staphylococci. Nat Microbiol. 2019;4(6):956–963. doi: 10.1038/s41564-019-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto-Perez P., Bisanz J.E., Berry J.D., et al. CRISPR-cas system of a prevalent human gut bacterium reveals hyper-targeting against phages in a human virome catalog. Cell Host Microbe. 2019;26(3):325–335. doi: 10.1016/j.chom.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T., Tamman H., Coppieters 't Wallant K., et al. Direct activation of a bacterial innate immune system by a viral capsid protein. Nature. 2022;612(7938):132–140. doi: 10.1038/s41586-022-05444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Iranzo J., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalil A.M. The genome editing revolution: review. J Genet Eng Biotechnol. 2020;18(1):68. doi: 10.1186/s43141-020-00078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azimzadeh M., Mousazadeh M., Jahangiri-Manesh A., et al. CRISPR-powered microfluidics in diagnostics: a review of main applications. Chemosensors. 2021;10(1):3. [Google Scholar]
- 8.Thapliyal G., Bhandari M.S., Vemanna R.S., et al. Engineering traits through CRISPR/cas genome editing in woody species to improve forest diversity and yield. Crit Rev Biotechnol. 2022:1–20. doi: 10.1080/07388551.2022.2092714. [DOI] [PubMed] [Google Scholar]
- 9.Zittersteijn H.A., Harteveld C.L., Klaver-Flores S., et al. A small key for a heavy door: genetic therapies for the treatment of hemoglobinopathies. Front Genome Ed. 2021;2 doi: 10.3389/fgeed.2020.617780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez Rojo F., Nyman R.K.M., Johnson A.A.T., et al. CRISPR-Cas systems: ushering in the new genome editing era. Bioengineered. 2018;9(1):214–221. doi: 10.1080/21655979.2018.1470720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Schiel J.A., Maksimova E., et al. ErCas12a CRISPR-MAD7 for model generation in human cells, mice, and rats. CRISPR J. 2020;3(2):97–108. doi: 10.1089/crispr.2019.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., Alswillah T., Kapoor I., et al. USP14 is a deubiquitinase for Ku70 and critical determinant of non-homologous end joining repair in autophagy and PTEN-deficient cells. Nucleic Acids Res. 2020;48(2):736–747. doi: 10.1093/nar/gkz1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo N., Liu J.B., Li W., et al. The power and the promise of CRISPR/Cas9 genome editing for clinical application with gene therapy. J Adv Res. 2022;40:135–152. doi: 10.1016/j.jare.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A.K., Ramalingam S., Rao D.N., et al. Genome editing revolution in life sciences. Reson. 2021;26(7):971–998. [Google Scholar]
- 15.Hossain M.A. CRISPR-Cas9:a fascinating journey from bacterial immune system to human gene editing. Prog Mol Biol Transl Sci. 2021;178:63–83. doi: 10.1016/bs.pmbts.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M., Eshraghian E.A., Al Jammal O., et al. CRISPR technology: the engine that drives cancer therapy. Biomed Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.111007. [DOI] [PubMed] [Google Scholar]
- 17.Xing H., Meng L.H. CRISPR-cas9:a powerful tool towards precision medicine in cancer treatment. Acta Pharmacol Sin. 2020;41(5):583–587. doi: 10.1038/s41401-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi L., Larson M., Gilbert L., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Standage-Beier K., Zhang Q., Wang X. Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS Synth Biol. 2015;4(11):1217–1225. doi: 10.1021/acssynbio.5b00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanenbaum M., Gilbert L., Qi L., et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudelli N.M., Komor A.C., Rees H.A., et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komor A.C., Kim Y.B., Packer M.S., et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anzalone A.V., Randolph P.B., Davis J.R., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z., Chen S., Jia Y., et al. Efficient and high-fidelity base editor with expanded PAM compatibility for cytidine dinucleotide. Sci China Life Sci. 2021;64(8):1355–1367. doi: 10.1007/s11427-020-1775-2. [DOI] [PubMed] [Google Scholar]
- 25.Strecker J., Ladha A., Gardner Z., et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burstein D., Harrington L.B., Strutt S.C., et al. New CRISPR-cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mout R., Ray M., Lee Y.W., et al. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: progress and challenges. Bioconjugate Chem. 2017;28(4):880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho B.X., Loh S.J.H., Chan W.K., et al. In vivo genome editing as a therapeutic approach. Int J Mol Sci. 2018;19(9):2721. doi: 10.3390/ijms19092721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat Rev Genet. 2011;12(5):301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 30.Wang H.X., Li M., Lee C.M., et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev. 2017;117(15):9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 31.Glass Z., Lee M., Li Y., et al. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36(2):173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L., Wang J., Liu Y., et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med. 2019;381(13):1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 33.Frangoul H., Altshuler D., Cappellini M.D., et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med. 2021;384(3):252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 34.Stadtmauer E.A., Fraietta J.A., Davis M.M., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481) doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y., Xue J., Deng T., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26(5):732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 36.Jing Z., Zhang N., Ding L., et al. Safety and activity of programmed cell death-1 gene knockout engineered t cells in patients with previously treated advanced esophageal squamous cell carcinoma: an open-label, single-arm phase I study. J Clin Oncol. 2018;36(15_suppl):3054. [Google Scholar]
- 37.Gillmore J.D., Gane E., Taubel J., et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385(6):493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 38.Maeder M.L., Stefanidakis M., Wilson C.J., et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25(2):229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 39.Jo D.H., Song D.W., Cho C.S., et al. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci Adv. 2019;5(10) doi: 10.1126/sciadv.aax1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang H.K., Jo D.H., Lee S.N., et al. High-purity production and precise editing of DNA base editing ribonucleoproteins. Sci Adv. 2021;7(35) doi: 10.1126/sciadv.abg2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang H., Jo D.H., Cho C.S., et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng. 2022;6(2):181–194. doi: 10.1038/s41551-021-00788-9. [DOI] [PubMed] [Google Scholar]
- 42.Smalley E. First AAV gene therapy poised for landmark approval. Nat Biotechnol. 2017;35(11):998–999. doi: 10.1038/nbt1117-998. [DOI] [PubMed] [Google Scholar]
- 43.Yu W., Mookherjee S., Chaitankar V., et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8 doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno A.M., Fu X., Zhu J., et al. In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol Ther. 2018;26(7):1818–1827. doi: 10.1016/j.ymthe.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goossens R., van den Boogaard M.L., Lemmers R.J.L.F., et al. Intronic SMCHD1 variants in FSHD: testing the potential for CRISPR-Cas9 genome editing. J Med Genet. 2019;56(12):828–837. doi: 10.1136/jmedgenet-2019-106402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vagni P., Perlini L.E., Chenais N.A.L., et al. Gene editing preserves visual functions in a mouse model of retinal degeneration. Front Neurosci. 2019;13:945. doi: 10.3389/fnins.2019.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latella M.C., di Salvo M.T., Cocchiarella F., et al. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids. 2016;5(11):e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakondi B., Lv W., Lu B., et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24(3):556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu S., Du J., Chen N., et al. In vivo CRISPR/Cas9-mediated genome editing mitigates photoreceptor degeneration in a mouse model of X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2020;61(4):31. doi: 10.1167/iovs.61.4.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H., Xue W., Chen S., et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibraheim R., Song C.Q., Mir A., et al. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 2018;19:137. doi: 10.1186/s13059-018-1515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agudelo D., Carter S., Velimirovic M., et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020;30(1):107–117. doi: 10.1101/gr.255414.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song C.Q., Jiang T., Richter M., et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng. 2020;4(1):125–130. doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Wang L., Huo Y., et al. Amelioration of an inherited metabolic liver disease through creation of a de novo start codon by cytidine base editing. Mol Ther. 2020;28(7):1673–1683. doi: 10.1016/j.ymthe.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villiger L., Grisch-Chan H.M., Lindsay H., et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24(10):1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 56.Richards D.Y., Winn S.R., Dudley S., et al. AAV-mediated CRISPR/Cas9 gene editing in murine phenylketonuria. Mol Ther Methods Clin Dev. 2020;17:234–245. doi: 10.1016/j.omtm.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin S., Ma L., Shao T., et al. Enhanced genome editing to ameliorate a genetic metabolic liver disease through co-delivery of adeno-associated virus receptor. Sci China Life Sci. 2022;65(4):718–730. doi: 10.1007/s11427-020-1744-6. [DOI] [PubMed] [Google Scholar]
- 58.Yang Y., Wang L., Bell P., et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long C., McAnally J.R., Shelton J.M., et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson C.E., Hakim C.H., Ousterout D.G., et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabebordbar M., Zhu K., Cheng J.K.W., et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bengtsson N.E., Hall J.K., Odom G.L., et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8 doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amoasii L., Hildyard J.C.W., Li H., et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362(6410):86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moretti A., Fonteyne L., Giesert F., et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. 2020;26(2):207–214. doi: 10.1038/s41591-019-0738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryu S.M., Koo T., Kim K., et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36(6):536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 66.Gaj T., Ojala D.S., Ekman F.K., et al. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci Adv. 2017;3(12):eaar3952. doi: 10.1126/sciadv.aar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnaoutova I., Zhang L., Chen H.D., et al. Correction of metabolic abnormalities in a mouse model of glycogen storage disease type Ia by CRISPR/Cas9-based gene editing. Mol Ther. 2021;29(4):1602–1610. doi: 10.1016/j.ymthe.2020.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koblan L.W., Erdos M.R., Wilson C., et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589(7843):608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilbie D., Walther J., Mastrobattista E. Delivery aspects of CRISPR/cas for in vivo genome editing. Acc Chem Res. 2019;52(6):1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kay M.A., Glorioso J.C., Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7(1):33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 71.Bessis N., GarciaCozar F.J., Boissier M.C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 72.Samulski R.J., Zhu X., Xiao X., et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10(12):3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan J., Kang D.D., Turnbull G., et al. Delivery of CRISPR-Cas9 system for screening and editing RNA binding proteins in cancer. Adv Drug Deliv Rev. 2022;180 doi: 10.1016/j.addr.2021.114042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maggio I., Zittersteijn H.A., Wang Q., et al. Integrating gene delivery and gene-editing technologies by adenoviral vector transfer of optimized CRISPR-Cas9 components. Gene Ther. 2020;27(5):209–225. doi: 10.1038/s41434-019-0119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palmer D., Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther. 2003;8(5):846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 76.Jager L., Ehrhardt A. Persistence of high-capacity adenoviral vectors as replication-defective monomeric genomes in vitro and in murine liver. Hum Gene Ther. 2009;20(8):883–896. doi: 10.1089/hum.2009.020. [DOI] [PubMed] [Google Scholar]
- 77.Byrnes A.P., Rusby J.E., Wood M.J.A., et al. Adenovirus gene transfer causes inflammation in the brain. Neuroscience. 1995;66(4):1015–1024. doi: 10.1016/0306-4522(95)00068-t. [DOI] [PubMed] [Google Scholar]
- 78.Yang Y., Greenough K., Wilson J.M. Transient immune blockade prevents formation of neutralizing antibody to recombinant adenovirus and allows repeated gene transfer to mouse liver. Gene Ther. 1996;3(5):412–420. [PubMed] [Google Scholar]
- 79.Beer S.J., Matthews C.B., Stein C.S., et al. Poly (lactic-glycolic) acid copolymer encapsulation of recombinant adenovirus reduces immunogenicity in vivo. Gene Ther. 1998;5(6):740–746. doi: 10.1038/sj.gt.3300647. [DOI] [PubMed] [Google Scholar]
- 80.Tuveson D.A., Jacks T. Technologically advanced cancer modeling in mice. Curr Opin Genet Dev. 2002;12(1):105–110. doi: 10.1016/s0959-437x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 81.Maddalo D., Manchado E., Concepcion C.P., et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D., Mou H., Li S., et al. Adenovirus-mediated somatic genome editing of pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther. 2015;26(7):432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J., Chen M., Whitley M.J., et al. Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma. Nat Commun. 2017;8 doi: 10.1038/ncomms15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18(5):358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo J., Luo Y., Sun J., et al. Adeno-associated virus-mediated cancer gene therapy: current status. Cancer Lett. 2015;356(2):347–356. doi: 10.1016/j.canlet.2014.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong J.Y., Fan P.D., Frizzell R.A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7(17):2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 87.Senís E., Fatouros C., Große S., et al. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9(11):1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 88.Sun J.Y., Anand-Jawa V., Chatterjee S., et al. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003;10(11):964–976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- 89.Huttner N.A., Girod A., Perabo L., et al. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 2003;10(26):2139–2147. doi: 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- 90.Vakulskas C.A., Behlke M.A. Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid Therapeut. 2019;29(4):167–174. doi: 10.1089/nat.2019.0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Swiech L., Heidenreich M., Banerjee A., et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winters I.P., Chiou S.H., Paulk N.K., et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun. 2017;8:2053. doi: 10.1038/s41467-017-01519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chow R.D., Guzman C.D., Wang G., et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20(10):1329–1341. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr Opin Biotechnol. 1998;9(5):457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 95.Kantor B., Bailey R.M., Wimberly K., et al. Methods for gene transfer to the central nervous system. Adv Genet. 2014;87:125–197. doi: 10.1016/B978-0-12-800149-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ling S., Yang S., Hu X., et al. Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat Biomed Eng. 2021;5(2):144–156. doi: 10.1038/s41551-020-00656-y. [DOI] [PubMed] [Google Scholar]
- 97.Wanisch K., Yáñez-Muñoz R.J. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17(8):1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim W., Lee S., Kim H.S., et al. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 2018;28(3):374–382. doi: 10.1101/gr.223891.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S.H., Hsieh Y.Y., Tzeng H.E., et al. ABL genomic editing sufficiently abolishes oncogenesis of human chronic myeloid leukemia cells in vitro and in vivo. Cancers. 2020;12(6):1399. doi: 10.3390/cancers12061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.High K.A. Turning genes into medicines-what have we learned from gene therapy drug development in the past decade? Nat Commun. 2020;11:5821. doi: 10.1038/s41467-020-19507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baum C., Kustikova O., Modlich U., et al. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17(3):253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 102.Waehler R., Russell S.J., Curiel D.T. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8(8):573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pack D.W., Hoffman A.S., Pun S., et al. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 104.Pahle J., Walther W. Vectors and strategies for nonviral cancer gene therapy. Expet Opin Biol Ther. 2016;16(4):443–461. doi: 10.1517/14712598.2016.1134480. [DOI] [PubMed] [Google Scholar]
- 105.Witzigmann D., Kulkarni J.A., Leung J., et al. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Deliv Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sercombe L., Veerati T., Moheimani F., et al. Advances and challenges of liposome assisted drug delivery. Front Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mehnert W., Mäder K. Solid lipid nanoparticles: production, characterization and applications. Adv Drug Deliv Rev. 2001;47(2–3):165–196. doi: 10.1016/s0169-409x(01)00105-3. [DOI] [PubMed] [Google Scholar]
- 108.Pardi N., Hogan M.J., Pelc R.S., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hosseini E.S., Nikkhah M., Hosseinkhani S. Cholesterol-rich lipid-mediated nanoparticles boost of transfection efficiency, utilized for gene editing by CRISPR-Cas9. Int J Nanomed. 2019;14:4353–4366. doi: 10.2147/IJN.S199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Finn J.D., Smith A.R., Patel M.C., et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22(9):2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 111.Cheng Q., Wei T., Farbiak L., et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol. 2020;15(4):313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song R., Murphy M., Li C., et al. Current development of biodegradable polymeric materials for biomedical applications. Drug Des Dev Ther. 2018;12:3117–3145. doi: 10.2147/DDDT.S165440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan M., Du J., Gu Z., et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2010;5(1):48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 114.Chen G., Abdeen A.A., Wang Y., et al. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14(10):974–980. doi: 10.1038/s41565-019-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wan T., Chen Y., Pan Q., et al. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J Contr Release. 2020;322:236–247. doi: 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 116.Guo P., Yang J., Huang J., et al. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc Natl Acad Sci U S A. 2019;116(37):18295–18303. doi: 10.1073/pnas.1904697116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deng H., Tan S., Gao X., et al. Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm Sin B. 2020;10(2):358–373. doi: 10.1016/j.apsb.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gindy M.E., Prud'homme R.K. Multifunctional nanoparticles for imaging, delivery and targeting in cancer therapy. Expet Opin Drug Deliv. 2009;6(8):865–878. doi: 10.1517/17425240902932908. [DOI] [PubMed] [Google Scholar]
- 119.Jahangiri-Manesh A., Mousazadeh M., Taji S., et al. Gold nanorods for drug and gene delivery: an overview of recent advancements. Pharmaceutics. 2022;14(3):664. doi: 10.3390/pharmaceutics14030664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bowman M.C., Ballard T.E., Ackerson C.J., et al. Inhibition of HIV fusion with multivalent gold nanoparticles. J Am Chem Soc. 2008;130(22):6896–6897. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chompoosor A., Saha K., Ghosh P.S., et al. The role of surface functionality on acute cytotoxicity, ROS generation and DNA damage by cationic gold nanoparticles. Small. 2010;6(20):2246–2249. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahmad S., Zamry A.A., Tan H.T.T., et al. Targeting dendritic cells through gold nanoparticles: a review on the cellular uptake and subsequent immunological properties. Mol Immunol. 2017;91:123–133. doi: 10.1016/j.molimm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 123.Lino C.A., Harper J.C., Carney J.P., et al. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee K., Conboy M., Park H.M., et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yen H.J., Hsu S.H., Tsai C.L. Cytotoxicity and immunological response of gold and silver nanoparticles of different sizes. Small. 2009;5(13):1553–1561. doi: 10.1002/smll.200900126. [DOI] [PubMed] [Google Scholar]
- 126.Bastús N.G., Sánchez-Tilló E., Pujals S., et al. Homogeneous conjugation of peptides onto gold nanoparticles enhances macrophage response. ACS Nano. 2009;3(6):1335–1344. doi: 10.1021/nn8008273. [DOI] [PubMed] [Google Scholar]
- 127.Lee J.Y., Park W., Yi D.K. Immunostimulatory effects of gold nanorod and silica-coated gold nanorod on RAW 264.7 mouse macrophages. Toxicol Lett. 2012;209(1):51–57. doi: 10.1016/j.toxlet.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 128.Hansen-Bruhn M., de Ávila B.E.F., Beltrán-Gastélum M., et al. Active intracellular delivery of a Cas9/sgRNA complex using ultrasound-propelled nanomotors. Angew Chem Int Ed Engl. 2018;57(10):2657–2661. doi: 10.1002/anie.201713082. [DOI] [PubMed] [Google Scholar]
- 129.Hur J., Chung A.J. Microfluidic and nanofluidic intracellular delivery. Adv Sci. 2021;8(15) doi: 10.1002/advs.202004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sessions J.W., Skousen C.S., Price K.D., et al. CRISPR-Cas9 directed knock-out of a constitutively expressed gene using lance array nanoinjection. SpringerPlus. 2016;5(1):1521. doi: 10.1186/s40064-016-3037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song X., Liu C., Wang N., et al. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv Drug Deliv Rev. 2021;168:158–180. doi: 10.1016/j.addr.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 132.Samulski R.J., Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol. 2014;1(1):427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 133.Jiang S., Wang L.W., Walsh M.J., et al. CRISPR/Cas9-mediated genome editing in Epstein-Barr virus-transformed lymphoblastoid B-cell lines. CP Molecular Biology. 2018;121:31. 12.1–31.12.23. doi: 10.1002/cpmb.51. [DOI] [PubMed] [Google Scholar]
- 134.Park A., Hong P., Won S.T., et al. Sendai virus, an RNA virus with no risk of genomic integration, delivers CRISPR/Cas9 for efficient gene editing. Mol Ther Methods Clin Dev. 2016;3 doi: 10.1038/mtm.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mansouri M., Ehsaei Z., Taylor V., et al. Baculovirus-based genome editing in primary cells. Plasmid. 2017;90:5–9. doi: 10.1016/j.plasmid.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 136.Zuris J.A., Thompson D.B., Shu Y., et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu Z., Yu L., Zhu D., et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed Res Int. 2014;2014 doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lao Y.H., Li M., Gao M.A., et al. HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute. Adv Sci. 2018;5(7) doi: 10.1002/advs.201700540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang L., Wang P., Feng Q., et al. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017;9(10):e441. [Google Scholar]
- 140.Sun W., Ji W., Hall J.M., et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhou W., Cui H., Ying L., et al. Enhanced cytosolic delivery and release of CRISPR/Cas9 by black phosphorus nanosheets for genome editing. Angew Chem Int Ed Engl. 2018;57(32):10268–10272. doi: 10.1002/anie.201806941. [DOI] [PubMed] [Google Scholar]
- 142.He Z.Y., Zhang Y.G., Yang Y.H., et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum Gene Ther. 2018;29(2):223–233. doi: 10.1089/hum.2017.209. [DOI] [PubMed] [Google Scholar]
- 143.Kim S.M., Shin S.C., Kim E.E., et al. Simple in vivo gene editing via direct self-assembly of Cas9 ribonucleoprotein complexes for cancer treatment. ACS Nano. 2018;12(8):7750–7760. doi: 10.1021/acsnano.8b01670. [DOI] [PubMed] [Google Scholar]
- 144.Wang H.X., Song Z., Lao Y.H., et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc Natl Acad Sci U S A. 2018;115(19):4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li M., Xie H., Liu Y., et al. Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer. J Contr Release. 2019;304:204–215. doi: 10.1016/j.jconrel.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 146.Pan Y., Yang J., Luan X., et al. Near-infrared upconversion-activated CRISPR-Cas9 system: a remote-controlled gene editing platform. Sci Adv. 2019;5(4) doi: 10.1126/sciadv.aav7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yin H., Song C.Q., Dorkin J.R., et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cheraghi R., Alipour M., Nazari M., et al. Optimization of conditions for gene delivery system based on PEI. Nanomedicine J. 2017;4(1):8–16. [Google Scholar]
- 149.Alipour M., Hosseinkhani S. Design, preparation, and characterization of peptide-based nanocarrier for gene delivery. Methods Mol Biol. 2019;2000:59–69. doi: 10.1007/978-1-4939-9516-5_5. [DOI] [PubMed] [Google Scholar]
- 150.Kleinstiver B.P., Pattanayak V., Prew M.S., et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Slaymaker I.M., Gao L., Zetsche B., et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Charlesworth C.T., Deshpande P.S., Dever D.P., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li C., Brant E., Budak H., et al. CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ - Sci B. 2021;22(4):253–284. doi: 10.1631/jzus.B2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]