Abstract
Wnt signaling plays a major role in regulating cell proliferation and differentiation. The Wnt ligands are a family of 19 secreted glycoproteins that mediate their signaling effects via binding to Frizzled receptors and LRP5/6 coreceptors and transducing the signal either through β-catenin in the canonical pathway or through a series of other proteins in the noncanonical pathway. Many of the individual components of both canonical and noncanonical Wnt signaling have additional functions throughout the body, establishing the complex interplay between Wnt signaling and other signaling pathways. This crosstalk between Wnt signaling and other pathways gives Wnt signaling a vital role in many cellular and organ processes. Dysregulation of this system has been implicated in many diseases affecting a wide array of organ systems, including cancer and embryological defects, and can even cause embryonic lethality. The complexity of this system and its interacting proteins have made Wnt signaling a target for many therapeutic treatments. However, both stimulatory and inhibitory treatments come with potential risks that need to be addressed. This review synthesized much of the current knowledge on the Wnt signaling pathway, beginning with the history of Wnt signaling. It thoroughly described the different variants of Wnt signaling, including canonical, noncanonical Wnt/PCP, and the noncanonical Wnt/Ca2+ pathway. Further description involved each of its components and their involvement in other cellular processes. Finally, this review explained the various other pathways and processes that crosstalk with Wnt signaling.
Keywords: β-catenin, Canonical Wnt, Noncanonical Wnt, Signal transduction, Signaling crosstalk
Introduction
Wnt signaling is an important evolutionarily conserved pathway that regulates a diverse range of cellular activities. The importance of these pathways in cell proliferation, differentiation, and migration has led to extensive study of its various components, but there is still much to be learned and discovered about this pathway and its various interactions with other pathways. The extent of the research on this pathway has led to it becoming the subject of many evolving therapies. Many of its components have been implicated in the development of treatments for various conditions, such as cancer, neurodegenerative diseases, congenital disorders, and even diabetes and heart disease. Wnt signaling is truly a fundamental pathway to much of human development and health.
The discovery of the first Wnt gene dates back 40 years ago to 1982 during experiments intended to discover proto-oncogenes via activation by proviruses.1,2 These experiments led to the discovery of int1, which causes a tumor of mammary epithelial cells when activated.1 Eventually, it was discovered that mice and human int1 homologs shared 99% of their amino acid sequences, elucidating the high degree of conservation of the int1 proto-oncogene.3 Prior to this, Sharma and Chopra identified a gene that codes for the development of the wings in Drosophila melanogaster, naming the gene wingless (Wg).4 Additionally, molecular hybridization identified a similar int1 homologue in Drosophila.5 This gene was initially called Dint1, but isolation and sequencing of clones of the Dint1 gene determined that it was identical to the Wg gene.5, 6, 7 Later, it was decided that the int1 and Wg genes would be named together as Wnt1 to reduce confusion with the other int genes. Int2 was renamed to FGF3, int3 was renamed to Notch4, and int4 is now called Wnt3A.8 The Wnt ligands are now known to be a group composed of 19 glycoproteins that each bind receptors at the cell surface to trigger intracellular signaling cascades to modulate gene expression.9, 10, 11, 12, 13 Each of the Wnt ligands is a cysteine-rich protein that is 350–400 amino acids in length, with an N-terminal signal sequence targeting them for secretion (Table 1).14
Table 1.
List of Wnt genes and orthologs in humans and model organisms.
| Human |
Mouse |
Xenopus |
Zebrafish |
Drosophila |
||
|---|---|---|---|---|---|---|
| Wnt Gene | Chr | Ortholog | Chr | Ortholog | Ortholog | Ortholog |
| WNT1 | 12 | Wnt1 | 15 | Wnt1 | Wnt1 | Wg |
| WNT2 | 7 | Wnt2 | 6 | Wnt2a | Wnt2 | |
| WNT2B/13 | 1 | Wnt2b/13 | 3 | Wnt2b | Wnt2b | |
| WNT3 | 17 | Wnt3 | 11 | Wnt3 | Wnt3 | |
| WNT3A | 1 | Wnt3a | 11 | Wnt3a | ||
| WNT4 | 1 | Wnt4 | 4 | Wnt4 | Wnt4a & Wnt4b | |
| WNT5A | 3 | Wnt5a | 14 | Wnt5a | ||
| WNT5B | 12 | Wnt5b | 6 | Wnt5b | Wnt5b | |
| WNT6 | 2 | Wnt6 | 1 | Wnt6 | DWnt6 | |
| WNT7A | 3 | Wnt7a | 6 | Wnt7a | Wnt7 & Wnt7a | DWnt2 |
| WNT7B | 22 | Wnt7b | 15 | Wnt7b | ||
| WNT8A | 5 | Wnt8a | 18 | Wnt8a | Wnt8a | DWnt8/WntD |
| WNT8B | 10 | Wnt8b | 19 | Wnt8b | Wnt8b | |
| WNT9A | 1 | Wnt9a/14 | 11 | DWnt4 | ||
| WNT9B | Wnt9b/15 | 11 | ||||
| WNT10A | 2 | Wnt10a | 1 | Wnt10a | Wnt10a | DWnt10 |
| WNT10B/12 | 12 | Wnt10b | 15 | Wnt10b | Wnt10b | |
| WNT11 | 11 | Wnt11 | 7 | Wnt11 & Wnt11R | Wnt11 | |
| WNT14 | 1 | |||||
| WNT16 | 7 | Wnt16 | 6 | Wnt16 | ||
Wnt ligand family: canonical vs. noncanonical
Canonical and noncanonical Wnt ligands
Wnt signaling can be categorized into two pathways, the β-catenin-dependent pathway (canonical) and the β-catenin-independent pathway (noncanonical).15,16 The canonical pathway is important for inducing cell proliferation, differentiation, and maturation.16 It is also vital in producing proper body-axis specifications.17 The pathway is activated via the Wnt1 class ligands, which include Wnt2, Wnt3, Wnt3a, and Wnt8a.17 The canonical pathway is associated with the transport of β-catenin to the nucleus upon Wnt binding to the Frizzled (Fz or Fzd) receptor and the coreceptors LDL-receptor-related proteins 5 and 6 (LRP5 and LRP6)18,19 (Fig. 1). The Fz receptor contains a cysteine-rich domain (CRD) that is used to bind Wnt.20 In mammals, 10 Fz receptors have been identified.17,21
Figure 1.
The canonical Wnt signaling pathway. The left panel (A) demonstrates the activated Wnt signaling cascade, while the right side portrays the inhibited Wnt signaling cascade. Wnt binds to the Fz receptor and LRP5/6 co-receptor. This activates Dvl to cause the dissociation of Axin from the destruction complex, causing β-catenin to be stabilized and enter the nucleus. β-Catenin can then displace the inhibitory TLE/Groucho complexes, enabling TCF/LEF to transcribe the target genes. PP2A can also enhance Wnt signaling by dephosphorylating β-catenin, APC, and Axin. The result is the preservation of β-catenin by preventing ubiquitination and proteasomal breakdown. In the absence of Wnt signaling (B), the destruction complex breaks down β-catenin and inhibits gene transcription. Several other proteins also contribute to the inhibition of Wnt signaling. Dkk1 associates with Krm1 or Krm2 and LRP5/6, causing endocytosis of the LRP5/6 co-receptor. Wise/sclerostin binds to LRP5/6 to inhibit proper Wnt association with the coreceptor. xCer-L and WIF-1 both bind to Wnt ligands to inhibit signaling. IGFBP-4 functions as a competitive inhibitor of Wnt signaling by associating with LRP6 and Fz8, while sFRPs complex with Fz receptors to prevent Wnt ligand binding. The illustration was inspired by and created in BioRender.
If Wnt ligand binding does not occur, a destruction complex that is normally inhibited by Wnt removes the β-catenin. This destruction complex is composed of adenomatous polyposis coli (APC) protein, Axin, serine/threonine kinase glycogen synthase kinase 3 (GSK-3), casein kinase 1 (CK1), the E3-ubiquitin ligase β-TrCP, and protein phosphatase 2A (PP2A).22 While PP2A can impact Wnt signaling either positively or negatively in a cellular context-specific fashion,23 CK1 phosphorylates β-catenin at the Ser45 residue first, in a process called “priming”. This enables GSK-3 to phosphorylate the Ser33, Ser37, and Thr41 residues, which ultimately creates the binding site for the β-TrCP protein.22,24 The β-TrCP protein functions as an adaptor protein that complexes with Skp1/Cullin machinery to ubiquitinate β-catenin, enabling the destruction of β-catenin by the proteasome.22,24,25
The binding of Wnt to the Fz receptors and LRP 5/6 transports disheveled protein (Dvl) to the cell membrane, leading to phosphorylation of the cytoplasmic tails of LRP 5/6. The LRP 5/6 can then bind Axin, removing it from the destruction complex, thus causing the complex to disassemble and release the β-catenin.14,26,27 This results in the stabilization and accumulation of β-catenin in the cytoplasm, and then its translocation to the nucleus and binding with the transcription factors TCF/LEF (T-cell factor/Lymphoid enhancer factor) and thus gene expression.14,26,28 Without β-catenin, the TCF/LEF complex is joined with the transducing-like enhancer protein (TLE/Groucho), which recruits HDACs, leading to transcriptional repression. However, β-catenin binding to TCF/LEF displaces the TLE/Groucho complexes and leads to the recruitment of activators to modify the interacting proteins, such as CBP/p300, Pygo, BCL9, and BRG129 (Fig. 1).
In contrast with the canonical pathway, noncanonical Wnt signaling is β-catenin-independent, as previously mentioned, and involves the Wnt5a type ligands, which include Wnt 4, Wnt5a, Wnt5b, Wnt6, Wnt7a, and Wnt11 15,17. Additionally, the noncanonical pathway involves different functions in comparison to the canonical pathway, such as dictating cellular polarization and migration.17,30 Noncanonical Wnt signaling follows two distinct pathways, the Wnt/planar cell polarity (PCP) pathway and the Wnt/calcium (Ca2+) pathway.17,30
The Wnt/PCP pathway, like the canonical pathway, predominantly uses Fz receptors to bind Wnt (Fig. 2). These Fz receptors utilize several coreceptors, including protein tyrosine kinase 7 (PTK7),31 muscle-skeletal receptor tyrosine kinase (MUSK),32 tyrosine kinase-like orphan receptor (ROR1/ROR2),33 tyrosine kinase related receptor (RYK),34 syndecan,35,36 and glypican.37,38 However, the Wnt/PCP pathway also uses Celsr1 and Vangl2 receptors, although the ligand-receptor binding interaction is still relatively unknown39 (Fig. 2). The Fz receptors in this pathway bind Wnt ligands and phosphorylate Dvl, leading to the recruitment of Inversin (Invs).40 The polarity protein Par6 interacts with Dvl, and Smad ubiquitination regulatory factor (Smurf) is recruited by the phosphorylated Dvl and binds to Par6. Smurf then ubiquitinates Prickle, a protein that normally inhibits Wnt/PCP signaling, targeting it for proteasomal destruction.41 The breakdown of Prickle enables Dvl to associate with the Dvl-associated activator of morphogenesis (DAAM). This complex can then activate Ras homologue gene-family member A (RhoA), but not Rac1 or Cdc24.15,39 DAAM also activates Profilin.42 Rac1 activates JNK, which phosphorylates and activates c-Jun to go to the nucleus and initiate gene expression.43,44 JNK also activates CapZ-interacting protein (CapZIP) via phosphorylation.45 RhoA activates RHO-associated coiled-coil-containing protein kinase (ROCK) and diaphanous 1 (DIA1).46,47 ROCK activates the myosin II regulatory light chain (MRLC).48,49 CapZIP, MRLC, DIA1, and profilin all contribute to actin polymerization, which is vital to cell polarity and migration.39 The Celsr1 receptor appears to function similarly to the Fz receptor, where Wnt binding ultimately causes activation of Dvl to stimulate the same signaling cascade. On the other hand, the binding of Wnt on Vangl2 causes dissociation of a complex of Dvl, Prickle, and inturned (Intu), enabling Dvl to complex with Invs39 (Fig. 2).
Figure 2.
The noncanonical Wnt/PCP pathway. The binding of Wnt ligands leads to the phosphorylation of Dvl, which recruits Invs, Par6, and Smurf. Smurf ubiquitinates the inhibitory protein Prickle, targeting it for destruction. Dvl can then associate with DAAM, activating Rac1, profilin, and RhoA. Rac1 activates JNK, which phosphorylates c-Jun and CapZIP. c-Jun then goes to the nucleus to stimulate gene transcription. RhoA activates ROCK and DIA1, with the latter activating MRLC. CapZIP, MRLC, DIA1, and profilin all stimulate actin polymerization. Celsr1 stimulates Dvl due to Wnt binding like the Fz receptor. Wnt binding to the Vangl2 receptor causes dissociation of Prickle and Intu from Dvl, which can then bind to Invs. The illustration was inspired by and created in BioRender.
The Wnt/Ca2+ pathway is predominantly activated by the Wnt5a ligand and Fzd2 receptor50,51 (Fig. 3). The binding of the Wnt5a ligand to Fzd2 triggers G protein to activate phospholipase C (PLC).52,53 PLC then cleaves phosphatidylinositol-4,5-bisphosphate (PtdInsP2 or PIP2) into diacylglycerol (DAG) and inositol-1,4,5-trisphosphate (InsP3 or IP3). DAG, along with Ca2+, activates protein kinase C (PKC) to stimulate cell–division cycle 42 (Cdc42), which causes actin polymerization to contribute to cell polarization and migration.39 Meanwhile, IP3 binds to inositol-1,4,5-trisphosphate receptors (InsP3Rs) on the membrane of the ER, stimulating Ca2+ release through the Ca2+ channels and increasing cytoplasmic Ca2+ levels.39,54 Stromal interaction molecule 1/2 (STIM1/2) detects the decrease in Ca2+ in the endoplasmic reticulum (ER) and activates Orai family proteins (Orai1, Orai2, or Orai3) on the plasma membrane to mediate store-operated Ca2+ entry (SOCE).39,54 Sarcoplasmic/ER Ca2+ ATPases (SERCAs) then pump Ca2+ back into the ER.39,54 The increased cytosolic Ca2+ from IP3 binding to InsP3R not only stimulates PKC but also stimulates calcineurin and CAMKII. Calcineurin activates the nuclear factor of activated T cells (NFAT), which causes gene transcription.55,56 On the other hand, CAMKII stimulates TGF-β-activated protein kinase 1 (TAK1), which then activates Nemo-like kinase (NLK).57 NLK phosphorylates TCF, inhibiting the β-catenin/TCF complex and preventing gene transcription58 (Fig. 3).
Figure 3.
The noncanonical Wnt/Ca2+ pathway. Wnt binding to the Fz receptor leads to G protein-mediated activation of PLC. PLC cleaves PIP2 into IP3 and DAG. IP3 binds to IP3 receptors (InsP3R) on the ER membrane to stimulate Ca2+ release. STIM1/2 detects this decrease in ER Ca2+ levels and activates Orai proteins in the plasma membrane to bring more Ca2+ into the cell, where SERCA can pump Ca2+ back into the ER. DAG can then activate PKC in the presence of Ca2+ and PKC can stimulate Cdc42 to enhance actin polymerization. The elevated intracellular Ca2+ level also stimulates calcineurin and CAMKII. Calcineurin activates NFAT, causing gene transcription. CAMKII activates TAK1, which activates NLK, when then phosphorylates TCF, preventing β-catenin-mediated gene transcription. The illustration was inspired by and created in BioRender.
Biological conservation of Wnt signaling
The Wnt signaling pathway is highly conserved. Wnt was first identified in Drosophila as the Wg protein due to mutations in the gene causing lack of wing and haltere development.4 The Wg gene is also called Dint1, as it was shown to be homologous to Int1 found in mice. This was the first connection discovered between Wnt found in Drosophila and Wnt found in vertebrates.7 In an experiment that proved the Wg gene is homologous to Int1 found in mice, van Ooyen et al sequenced the human and mouse Int1 genes in blocks of 50 nucleotides3 (Table 1). Comparison between the mouse and human Int1 genes revealed conservation of the splice sites, TATA box, and polyadenylation signal.3 Both proteins were also 370 amino acids long, with the only differences being found in the hydrophobic N-terminus.3 Homology was also established in the non-coding sequences.3 The overall homology between mouse and human Int1 homologs was 99%, revealing the conservation of the protein across species.3
Studies of cnidarians and sponges have also revealed the conservation of the Wnt signaling pathway, as the pathway was shown to be involved in the control of axis polarity.59 Petersen and Reddien further clarified that Wnt signaling is broadly used in primary body axis development.59 The Porcupine gene (Porc) codes for a transmembrane protein in the ER, allowing the processing and distribution of Drosophila Wg in vitro.60 Porc is an acyltransferase that palmitoylates the Wnt protein in the ER for secretion.61 Porc is also important for localizing Drosophila Wnt3 in the embryonic CNS.60 Tanaka et al isolated mouse Porc (Mporc) and Xenopus (Xporc) and analyzed them alongside homologous human MG61, C. elegans Mom-1, and Drosophila Porc. It was discovered that the Porc homologs among the vertebrates were well conserved via comparison of amino acid sequences, while the Drosophila Porc had an additional hydrophilic N-terminal sequence. This study revealed the conservation of Porc across species.60 Furthermore, injection of Mporc RNA into Drosophila embryos with Drosophila Porc resulted in some rescue of the embryos, albeit to a reduced extent compared to Drosophila Porc RNA. This study revealed the conservation of Porc and Porc function across multiple species.60
Biological assays for canonical vs. noncanonical Wnt classification
Various biological assays are used to identify components of the canonical and noncanonical pathways. From a broader perspective, the secondary-axis formation (i.e., axis duplication) analysis is one of the prototypes of canonical Wnt assays and can be used to assay activators and inhibitors of the canonical pathway, due to its role in proper axis specification.62 Luciferase reporter assays revealed that PTK7, a part of the noncanonical pathway, inhibits canonical Wnt signaling by precipitating Wnt3a and Wnt8 in the canonical pathway.63 Luciferase reporter assays can also be used to detect TCF/LEF activation,64,65 which are proteins that are mainly involved in the canonical pathway.28 Luciferase reporter assay could be used to form a high throughput screen for Wnt/β-catenin signaling.65 Enzyme-linked immunosorbent assay (ELISA) is an assay that can be used to detect Dickkopf-1 (DKK-1), an inhibitor of the canonical pathway.66 Another type of analysis is the Western blot analysis, which can be used to detect the up-regulation of the canonical proteins β-catenin, Dvl, APC, and GSK-3 in the retina of mice.66 β-Catenin levels can also be detected using immunodetection assays.67
The Wnt inhibitory factor-1 (WIF-1), which is a member of the secreted Frizzled-related protein (sFRP) family, can inhibit both the canonical and noncanonical pathways by binding directly to Wnts, preventing them from binding to the Wnt receptor.68 Soft agar assay and Western blotting can be used to detect inhibition of osteosarcoma cell growth as a result of WIF-1 overexpression, providing insight into the regulation of both pathways.64
In the noncanonical Wnt pathway, Wnt5a is the most prominent ligand, using the ROR family of tyrosine kinases as receptors.69 A member of the kinesin family, Kif26b, is a downstream effector of the Wnt5a-ROR pathway, mediating cell migration during embryonic development.70 Hence, a Wnt5a-ROR-Kif26b (WRK) reporter assay was developed and could be used to measure the degree of Wnt5a-ROR signaling in real-time, utilizing a combination of flow cytometry, Western blot, and time-lapse microscopy.70 A similar test involves a GFP-Kif26b reporter cell line combined with flow cytometry to detect the levels of Wnt5a in the cells.71 The GFP signal enables quantitative analysis of the cells to detect Wnt5a expression, although the assay is sensitive to cell density.71
Controlled secretion of Wnt proteins
Wnt ligands are highly lipidated in the ER, limiting the range of diffusion to localize the ligand to its recipient cell.72 Specifically, the acyltransferase Porc palmitoylates the Wnt in the ER.61 The mom-1 gene codes for a similar acyltransferase in C. elegans.73 The Wnt ligands are secreted via a Wntless (Wls) transporter, which is a conserved transmembrane protein in the Golgi apparatus that was characterized in Wg-sending cells in Drosophila.74 Banziger et al transfected embryonic kidney cells (HEK-293T) with the Wnt3a expression vector and cultured them with siRNA to knock down the expression of the human WLS (hWLS) gene. Assaying the level of Wnt3a protein revealed that siRNA-treated hWLS (sihWLS) cells could not activate the Wnt pathway due to, at least in part, the lack of secretion of Wnt3a in the absence of the hWLS gene. This study established the importance of Wls for the secretion of Wnt ligands. Additionally, cell surface heparan sulfate proteoglycans (HSPGs) are also involved in Wnt signaling. Glypicans and syndecans compose the protein core of HSPGs and are covered in heparan sulfate chains.75 In Drosophila, a glypican called division abnormally delayed (dally) functions as a coreceptor for the Drosophila frizzled 2 (Dfz2) receptor.76 The dally gene specifically encodes the protein core of the HSPGs in Wg/Wnt signaling.76 Glypicans like dally also participate in transporting Wnt ligands toward target cells.74
Wnt receptors, co-receptors, and accessory proteins
Cognate receptors: the frizzled (Fz) proteins
Frizzled (Fz) is a seven-pass transmembrane receptor that binds Wnt ligands.77 The Fz genes generate Fz receptors, each of which ranges from 500 to 700 amino acids long, with a CRD on the N-terminus and a 40 to 100 amino acid-long hydrophilic linker region.78 The seven transmembrane domains are hydrophobic alpha-helices.78 Fz protein localizes in the plasma membrane, with the cysteine-rich N-terminus oriented extracellularly and the carboxy-terminus oriented intracellularly towards the cytoplasm.21,79,80 The intracellular carboxy-terminus is variable in length and not well conserved between the different Fz receptor types.78 Fz is also glycosylated79 and there are 10 Fz genes organized into four clusters.78 By amino acid sequence, Fzd1, Fzd2, and Fzd7 are 75% similar; Fzd5 and Fzd8 are 70% similar; Fzd4, Fzd9, and Fzd10 are 65% similar; and Fzd3 and Fzd6 are 50% similar.81 Comparison of amino acid sequences between clusters yields only 20%–40% similarity.78 The relative lack of sequence similarity suggests a lack of genomic conservation between the Fz receptors.
The importance of Fz receptors in the Wnt/PCP pathway was characterized by Fz mutations in Drosophila that caused abnormal wing hair patterns and polarity disruptions, revealing the importance of Fz in the Wnt/PCP pathway.82,83 Loss of function and overexpression mutations in Fz altered the assembly location of F-actin in wing development, indicating that Fz is important in cytoskeletal development.79 The misorientation of hairs also revealed the importance of Fz in not only receiving the Wnt signal but also propagating it in a proximal-distal direction.84 Mutations in Fz receptors also cause the defective orientation of the ommatidia (individual units composing a compound eye).85
Mutations in the frizzled 4 (Fzd4) receptor causes familial exudative vitreoretinopathy (FEVR), a disease that generally results in the lack of retinal angiogenesis. This leads to a variety of symptoms, including retinal fibrosis, detachment, and dysplasia.86 Mutations in Fzd1 and Fzd2 in mice caused cleft palate and ventricular septal defects (VSD). These mutations also affected neural tube closure and inner ear development. Fzd7 mutations also caused VSDs but were more commonly associated with a kinked tail.87 Knockout of Fzd5 in mice led to embryonic lethality due to placental insufficiency.88 Loss of Fzd5 also caused retinal cell death and excessive mesenchymal cells in the vitreous cavity among other issues.89 Loss of Fzd8 alone causes no phenotypic change, although the loss of even a single Fzd8 allele can increase the severity and penetrance of the ocular effects from loss of Fzd5.90 Homozygous loss of Fzd9 has yielded B-cell developmental abnormalities, defective visuospatial learning, and bone mass reduction compounded by osteoblast dysfunction.21 Homozygous Fzd6 deletions cause randomized hair follicles, generating waves, whorls, and tufts.91 Homozygous deletion of Fzd3 in mice causes inappropriate development of peripheral and central axons, leading to the inability to detect thermal and mechanical stimuli from the feet.92 Concomitant deletions in Fzd3 and Fzd6 cause neural tube closure defects and misorientation of inner ear hair cells, indicating their similar functions.93
Co-receptors: LRP5 and LRP6 proteins
Low-density-lipoprotein receptor-related proteins 5/6 (LRP5/6) are single-pass transmembrane proteins that function as coreceptors for Fz receptors.19,94 The binding of the Wnt ligand triggers the dimerization of Fz and LRP5/6 94. This leads to the phosphorylation of the cytoplasmic tail of LRP5/6 at five conserved PPPSP (also called the PPPSPXS) motifs.95,96 Several protein kinases are involved in this phosphorylation, including GSK-3, which is part of the destruction complex that normally binds β-catenin in the absence of Wnt signaling.94,97 The PPPSP motifs bind Axin from the destruction complex, inhibiting β-catenin phosphorylation and subsequent ubiquitination that leads to proteasomal breakdown.95,96,98 LRP5 and LRP6 are homologous and expressed in embryogenesis.19 However, LRP6 is more important during embryogenesis, as evidenced by mice with homozygous deletion of Lrp6 demonstrating axial skeleton truncation, neural tube closure defects like spina bifida, as well as midbrain and hindbrain malformations.99 On the other hand, homozygous deletion of Lrp5 caused low bone mass due to decreased osteoblast activity, defective capillary cell apoptosis in the eye, defective clearance of chylomicrons, and impaired insulin secretion.100,101
Accessory Wnt binding proteins at the cell membrane
R-spondins
Roof plate specific-spondins (R-spondins) are four members of a larger family that contain thrombospondin type 1 repeats (TSR-1).102 The R prefix was given based on the first R-spondin due to its expression in the boundary of the roof plate and neuroepithelium in the dorsal neural tube.103 The R-spondins also contain an N-terminal signal peptide, two furin-like (FU1 and FU2) cysteine-rich domains near the N-terminus, and a C-terminal region with many positively charged amino acids.102,104 The R-spondins can bind to leucine-rich repeat-containing G-protein coupled receptors 4–6 (Lgr4-6).105 R-spondins bind Lgr4-6 via their FU2 domain and can bind ZNRF3 and RNF43 via their FU1 domain.106
ZNRF3 and RNF43
ZNRF3 and RNF43 are E3 ubiquitin ligases that target Wnt receptors for destruction to decrease Wnt signaling responses.107 ZNRF3 is a member of the ZNRF protein family, a family of E3 ubiquitin ligases that contain a zinc finger and a RING domain.108 R-spondins can bridge ZNRF3/RNF43 and LGR4/5/6, inhibiting ZNRF3/RNF43 activity via auto-ubiquitination and membrane clearance.106,109 In this manner, R-spondins act as Wnt agonists. RNF is a homolog of ZNRF43 that also contains a RING domain for its function as an E3 ubiquitin ligase.109
Derailed/Ryk
Derailed/Ryk (related to tyrosine kinase) is a member of the atypical tyrosine kinase family consisting of a Wnt inhibitory factor (WIF) domain extracellularly, an atypical kinase domain intracellularly, and a PDZ binding motif. Derailed is the Drosophila homolog, while Ryk is found in mammals.110 Interestingly, the tyrosine kinase domain is considered atypical due to sequence variations in the normally conserved tyrosine kinase residues. The domain lacks tyrosine kinase activity.111 The WIF domain of Ryk binds to Wnt1 and Wnt3a to activate TCF for the transcription of target genes.110 Ryk also forms a ternary complex with Wnt1 and Fz using its extracellular WIF domain, while the intracellular kinase domain binds to Dvl using its PDZ binding motif to activate TCF in response to Wnt3a stimulation.110 In Drosophila, Derailed was found to be important in learning and memory.112 Derailed/Ryk was also found to be important in axon guidance via interaction with Wnt5.113 The C. elegans homolog, lin-18, is important in determining vulval cell fate patterning.114
Receptor tyrosine kinase-like orphan receptors (RORs)
RORs are members of the receptor tyrosine kinase (RTK) family that are highly conserved and consist of two members, ROR1 and ROR2.115 ROR1 and ROR2 are single-pass transmembrane receptors with an intracellular tyrosine kinase domain and a proline-rich domain (PRD) that is flanked by two serine–threonine rich domains.116 The extracellular side of the ROR contains an immunoglobulin (Ig)-like domain, a CRD, and a Kringle domain (KRD). The CRD of ROR1 and ROR2 is like those on Fz receptors.117 ROR1 and ROR2 play crucial roles in embryonic development. Mice lacking ROR2 expression have shortened limbs and tails, facial abnormalities, and dwarfism among other issues.118 Mutations in ROR2 also cause Robinow syndrome and Brachydactyly type B in humans.119, 120, 121 Wnt5a binds ROR2, causing heterodimerization of ROR2 with Fz2 using its CRD. RORs can activate the noncanonical Wnt/JNK-PCP pathway and inhibit the canonical β-catenin/TCF pathway.122 However, the components of the Wnt/ROR pathways are still mostly unknown.122 ROR1 is expressed in B-lymphocyte precursors and can rarely cause precursor-B acute lymphoblastic leukemia (B-ALL).123 ROR1 and ROR2 in mice have been known to play an important role in the development of the nervous system.124
Gpr124 and Reck
Gpr124 is a G protein-coupled receptor (GPCR) and Reck is a glycosylphosphatidylinositol-anchored glycoprotein.125 Reck binds to Wnt7, creating the Reck/Wnt7 complex that binds to Gpr124. This complex then joins with the Fz receptor and LRP5/6 coreceptor to stimulate canonical Wnt/β-catenin signaling.125, 126, 127 The Gpr124 and Reck coactivators are vital in the development of the blood–brain barrier (BBB).128 Gpr124 knockout mice demonstrated microvascular hemorrhage and lethality in the embryo. Interestingly, Gpr124 deletion did not affect BBB integrity in adult mice.128
Intracellular mediators of Wnt signaling
The intracellular component of the Wnt signaling pathway is composed of several proteins for each pathway. This section will give a brief overview of each component of the canonical and two noncanonical Wnt pathways, looking further at the additional functions of the components outside of Wnt signaling. A description of the functions in Wnt signaling and the signaling cascade will be described in the canonical vs. noncanonical Wnt section. The inhibitors will also be discussed later. For the canonical pathway, the binding of Wnt to Fz and LRP5/6 mediates signal transduction to the nucleus via β-catenin (Fig. 1).
Intracellular mediators of the canonical pathway
β-Catenin was discovered to have two functions, one of them being its association with α- and γ-catenin to link Ca2+-dependent cell adhesion molecules (CAMs) to cytoskeletal structures.129 The term catenin was given to these three molecules due to its linkage of the CAM, E-cadherin, to cytoskeletal structures.129 The second function of β-catenin is its role in Wnt signaling, which was discovered through the analysis of its Drosophila homolog, Armadillo (Arm). Seminal screens for mutations that caused altered segmentation of Drosophila embryos revealed the signaling potential of β-catenin.130 Additionally, mutations in Wg in Drosophila caused a corresponding decrease in Arm, leading to altered segment polarity, further delineating the link between Wnt signaling and β-catenin.131 Further studies would later reveal that β-catenin mediates its effects via the TCF/LEF transcription factors, stimulating the transcription of target genes.132
β-Catenin is a 781 amino acid long protein with 12 Arm repeats, which forms a super-helical structure composed of multiple α-helices with a hydrophobic core.133 The superhelix contains a large, positively charged groove that allows β-catenin to interact with cadherins, TCF, and APC.133, 134, 135, 136, 137, 138, 139 β-Catenin then binds TCF and LEF to mediate gene transcription (Fig. 4). Interestingly, TCF is considered a transcriptional repressor, while LEF is considered a transcriptional activator.140 TCF/LEF binds to DNA via its HMG box domain, which recognizes a sequence called the Wnt/Wg response element (WRE) on the DNA.141,142 The HMG domain binds to the WRE in the minor groove of the DNA, causing the DNA to bend.142 In addition to the HMG domain, TCF/LEF also contains a nuclear localization signal (NLS) that makes nonspecific contacts with the phosphate backbone of the DNA, increasing TCF/LEF affinity for the DNA.142
Figure 4.
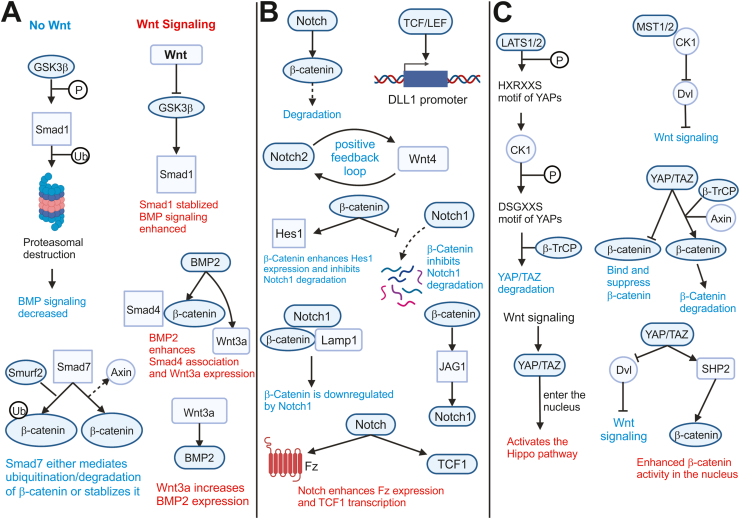
β-Catenin protein interactions. Some of these proteins were not discussed in this paper due to space constraints. Among those that have been discussed in this paper, there are notable inhibitors and activators. GSK-3β, CK1, APC, Axin, and β-TrCP are inhibitors of β-catenin as part of the destruction complex. PP2A has dual effects; it can dephosphorylate β-catenin to prevent ubiquitination and stabilize β-catenin, while it can also dephosphorylate GSK-3β, which can then inhibit β-catenin. YAP/TAZ is another inhibitor, as it can either bind to and suppress β-catenin without affecting its levels or associate with the destruction complex. Smad7 and Smurf2 can complex with β-catenin to ubiquitinate and degrade the protein. SUFU can export β-catenin from the nucleus. On the other hand, there are several activators of β-catenin, proteins that are activated by β-catenin, and proteins that assist with the functions of β-catenin. Smad3 is a chaperone protein that transports β-catenin into the nucleus. TCF/LEF are transcription factors that are activated by β-catenin. α- and γ-catenin join with β-catenin to link CAMs like E-cadherin to cytoskeletal structures, strengthening cell adhesion. It is noteworthy that some of the protein interactions are species-, tissue-, and/or context-dependent. The illustration was inspired by the Wnt homepage created and maintained by the Nusse Lab at Stanford University (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/protein_interactions) and reference 137.
Intracellular mediators of noncanonical Wnt/PCP pathway
Dvl proteins
There are two noncanonical Wnt signaling pathways, the Wnt/PCP pathway, and the Wnt/Ca2+ pathway. The Wnt/PCP pathway involves several additional receptors and co-receptors that will be discussed in later sections. The first intracellular mediator of Wnt/PCP signaling is Dvl. All Dvl proteins contain three conserved domains, a DIX domain at the N-terminus, a central PDZ domain, and a DEP domain at the carboxy-terminus.143 In between the DIX and PDZ domains is a “basic region” composed of conserved Ser and Thr residues. Between the PDZ and DEP domains is a “proline-rich region".143 The DIX domain primarily activates the canonical pathway, enabling dynamic polymerization of Dvl to form puncta, which can then interact with Axin to prevent it from mediating the destruction of β-catenin.144 Although further research needs to be conducted to delineate the exact process of this interaction between Dvl and Axin, it has been theorized that the DIX domain of Dvl interacts with the similar DIX domain on Axin to induce a conformational change in Axin or relocates Axin.144 The PDZ domain is involved in both the canonical and noncanonical pathways.145 It binds the Fz receptor at its C-terminal conserved Lys-Thr-X-X-X-TRP (KTXXXW) motif.146 This motif is required for the activation of the canonical pathway, although the molecular mechanisms are poorly understood.147 The DEP domain activates the noncanonical pathway by mediating the interaction between Dvl and DAAM1.143 The DEP domain also translocates the Dvl protein to the plasma membrane after Wnt stimulation.148
Dvl has an NLS located between the PDZ and DEP domain (aside from the proline-rich region), and an NES (nuclear export signal) located between the DEP and C-terminus.143 Increasing evidence indicates that nuclear Dvl protein is critical for β-catenin and TCF factors to form a complex.149 Dvl has been shown to interact with many other transcriptional factors including FOXK1/2, TAZ, and HIPK1, as well as gene promoters such as CYP19A1.149 Furthermore, Dvl1 was shown to interact with EZH2 while Dvl3 interacts with chromatin-modifying enzymes such as KMT2D in cancer cells. In addition, Dvl proteins are extensively modified post-translationally by phosphorylation, lysine acetylation, and methylation, as well as ubiquitination, although the functional roles of Dvl post-translational modifications remain to be fully investigated.149
Inversin (Inv) protein
Inversin (Inv) is a 1062 amino acid long protein with characteristic 15 successive ankyrin repeats. It was first identified in Inv mutant mice that had situs inversus (reversed left/right polarity), underdeveloped tubules, and cyst formation in the kidneys.150 This discovery linked Inv to Wnt signaling. Simons et al used glutathione S-transferase (GST) fusion protein containing the PDZ domain of Dvl to characterize the effect of Inv. The Inv directly interacted with the GST fusion protein. Inv also formed a protein complex with Dvl, indicating that it could inhibit the canonical pathway.151 Inv also participates in the noncanonical pathway, interacting with the planar cell polarity (PCP) pathway proteins Strabismus (Stbm) and Prickle (Pk) like the Drosophila PCP protein Diego.151 Inv mutant mice developed renal cysts due to unopposed canonical Wnt signaling, which causes overgrowth of cells without terminal differentiation of renal tubular epithelial cells.151
Par6 protein
Par6 serves as a polarity protein and a scaffolding protein with other molecules.152 The scaffolding function is beneficial in complexing with Dvl and Smurf in the noncanonical pathway.41 Its cell polarity function is mediated by the G-protein-activated phospholipase C-Beta (PLC-β) interacting with Par proteins like Par6 through multiple PDZ domains that can bind the extreme C-terminal S/TXL motifs of PLC-β.152 This activates the PLC-β to hydrolyze PIP2 into IP3 and DAG, two important secondary messengers. IP3 and DAG play important roles in regulating cell polarity and asymmetric cell division.152
Smurf1 and Smurf2 proteins
Smurfs (Smurf1 and Smurf2) are E3 ubiquitin ligases of the C2-WW-HECT family of proteins.153 They were first identified in the ubiquitination and degradation of R-Smads in the BMP pathway to antagonize TGF-β/BMP signaling.154 Smurf1 contains a phospholipid/Ca2+ binding domain on its N-terminus, two WW domains for binding to PPXY (also called PY) motifs on other proteins, and a HECT domain on the C-terminus for ubiquitination of target proteins.154 Smurf1 targets Smad1 and Smad5 using its WW domains, which bind to the PY motifs on Smad1 and Smad5, enabling their degradation.154 Smurf2 uses the same mechanism to degrade Smad1 and Smad2.155 Smurf1 can also be recruited by Par6 to target RhoA for degradation, establishing the proper cell polarity needed for cell movement.156 Smurf1-induced RhoA degradation in tight junctions leads to their dissolution and enables TGF-β dependent epithelial–mesenchymal transition.157 These results indicate the importance of Smurfs in noncanonical Wnt/PCP signaling.
Prickle (Pk) protein
Prickle (Pk) is also involved in the Wnt/PCP pathway. Pk is considered a type 1 polarity gene along with Dvl and Fz because it affects the body surface and is believed to directly establish tissue polarity. On the other hand, type 2 and 3 tissue polarity genes affect specific body regions and are believed to interpret the polarity that the type 1 genes have established.158 All Pk proteins contain three LIM motifs and a conserved domain called Prickle Espinas Testin (PET).158 The LIM motifs are cysteine-rich domains with two zinc fingers that are joined by an amino acid spacer.159 The LIM domains bind target proteins and enable protein–protein interactions.158 The PET domain is monomeric and works with the LIM motifs to target Dvl to the cell membrane to facilitate its function in the Wnt/PCP pathway.160 Pk also has a prenylation motif on its C-terminus that is important but not required for its localization to the plasma membrane.161
Dvl-associated activator of morphogenesis (DAAM)
DAAM is a conserved actin nucleator that is a member of the formin family.162 DAAM members contain a GTPase binding domain (GBD), a diaphanous inhibitory domain (DID), an N-terminal dimerization domain (DD), a coiled-coil (CC), an FH1 domain, an FH2 domain, and a diaphanous autoregulatory domain (DAD). DAAM is highly expressed in tissues like the CNS, somites, dermomyotomes, and the heart and is important in organ symmetry. Specifically, it causes left-right (LR) symmetry due to its modulation of the myosin 1D (Myo1D) function. Drosophila Myo1D induces dextral twisting, which is important for orienting the native LR organs in larvae. The absence of Myo1D leads to situs inversus.163 DAAM nucleates F-actin, promoting the assembly of an F-actin network that enables Myo1D to induce chirality.162 Interference of DAAM with RNAi suppressed the expected 180-degree dextral rotation of the larval body, reducing it to just 90°. This experiment revealed the importance of DAAM in inducing proper chirality in Drosophila larvae.162 The FH2 domain is particularly important in actin nucleation and polymerization, while the FH1 domain interacts with profilin-actin during actin filament elongation.42
Profilin protein
Profilin is a protein associated with non-muscle actin (β- and γ-actin) and is involved in the control of actin polymerization. It was first isolated from calf spleens as a small protein that accompanied an actin-containing complex.164 When complexed with actin, profilin is called profilactin.165 Profilin contains a core of seven-stranded β-pleated sheets with α-helical N- and C-termini on one side, and two shorter α-helices on the other. The N- and C-termini form the poly-l-proline (PLP) binding surface, which enables profilin to bind actin and factors that participate in actin nucleation and elongation.166 Profilin inhibits spontaneous actin nucleation and polymerization by sequestering G-actin. However, it can also promote actin filament elongation using its PLP binding domain to interact with other proteins such as formins, Ena/vasodilator-stimulated phosphoprotein (VASP), Arp2/3-dependent Wiskott–Aldrich syndrome protein (WASP), and WASP family verprolin-homologous protein (WAVE) family.166,167 Profilin also facilitates the exchange of ADP for ATP on actin monomers, further enhancing polymerization.166,167 Interaction with formins allows profilin to associate with microtubules and increases the rate of depolymerization.168 With profilin as a mediator, DAAM1 can mediate cytoskeletal reorganization and cell movement for important processes such as gastrulation.169
Rac1 and RhoA proteins
Rac1 and RhoA are members of the Rho family of small GTPases.170 Both proteins are involved in a wide variety of cell processes, such as motility, proliferation, migration, and polarity via their regulation of actin polymerization.171 Both have conserved GDP/GTP binding domains called the G domain and a C-terminal region that contains a CAAX motif.172 The binding of GTP to Rac1 causes two regions (amino acids 25–40 and 60–76) called switch I and II to undergo a conformational change to interact with specific downstream effectors in the signaling cascade.171 Rac1 and RhoA can also undergo post-translational modification to prenylate the C-terminal CAAX motif, enabling membrane interaction.171 This association with the membrane is what enables Rac1 and RhoA to stimulate downstream signaling cascades to modulate cellular functions.170 The exchange of GDP for GTP is mediated by Guanine Nucleotide Exchange Factors (GEFs) and GTPase activating proteins (GAPs), triggering the active state of Rac1 and RhoA to interact with downstream proteins.171,173 Guanosine dissociation inhibitors (GDI) can conceal the C-terminal isoprenyl motif in a hydrophobic pocket, sequestering Rac1 in the cytoplasm and preventing downstream activation.173,174 GDI also prevents GDP/GTP exchange to keep both in their inactive states.171,174 RhoA and Rac1 are spatially separated, with Rac1 active towards the leading edge of the cell, while RhoA is active towards the lagging edge.170 The two are also temporally segregated, with RhoA activity peaking before Rac1 in a coordinated cycle of protrusion and retraction. Rac1 and RhoA can antagonize each other mutually to coordinate this effect.175 Dysregulation of Rac1 and RhoA have been linked to cancer, as well as cognitive and cardiovascular diseases.172
Rho-associated coiled-coil kinase (ROCK)
ROCK (also called RhoA/Rho kinase) is a Ser/Thr kinase that is stimulated by Rho.46,47,176,177 ROCK comes in two isoforms, ROCK1 and ROCK2. ROCK1 is 1354 amino acids long, while ROCK2 is 1388 amino acids long.178 They share 64% of their primary amino acid sequences, 92% homology in the kinase domains, and only 55% homology in the coiled-coil domains.178 The N-terminal region contains the ROCK kinase domains.178 The C-terminal region contains the coiled-coil domain and a pleckstrin homology (PH) domain; this region binds to the catalytic kinase domain to inhibit its activity. The coiled-coil domain contains a Rho-binding region that enables GTP-bound RhoA to disrupt the binding of the C-terminus and the kinase domain.179 The C-terminal region can also be cleaved by caspases during apoptosis to activate ROCK.180,181 Activated ROCK can associate with mammalian DIA (mDia) to stimulate actin cytoskeletal reorganization.177,182 ROCK proteins are generally expressed in many tissues and phosphorylate target proteins on R/KXXS/T or R/KXS/T amino acid motifs.178 ROCK proteins are involved in forming stress fibers composed of bundles of F-actin and myosin II.183 Focal adhesion complexes bind these fibers to the inner plasma membrane.184 The activation of ROCK by caspases gives ROCK a crucial role in forming membrane blebs during apoptosis.180,181 ROCK also plays a role in embryonic development, inducing cell migration, differentiation, and axis formation through its expression in the cardiac mesoderm, lateral plate mesoderm, and neural plate.178
c-Jun N-terminal kinase (JNK)
JNK is a member of the three mitogen-activated protein kinase (MAPK) pathways that control cell proliferation, migration, and differentiation.185 The MAPK pathways are all activated by a series of phosphorylation reactions. JNK is activated by the MAP2K enzymes MKK4 and MKK7.186 Scaffold proteins just as JNK interacting protein 1 (JIP1) facilitate rapid activation of the JNK pathway.187 The JNK pathway can be inactivated by dual-specificity phosphatases (DUSPs).188 The JNK family contains three genes that can be spliced into 10 isoforms, namely, JNK1, JNK2, and JNK3. JNK1 and JNK2 are expressed in a variety of tissues, while JNK3 is expressed in the brain, heart, and testis.185 The protein products of these three genes are about 400 amino acids long, with a canonical Ser/Thr kinase domain.189 JNK is best known for inducing apoptosis via stimulating the mitochondrial release of cytochrome C to activate caspase and trigger apoptosis.190 However, the effects of JNK are context-specific, as JNK can phosphorylate anti-apoptotic Bcl-2 to promote apoptosis, but it can also phosphorylate pro-apoptotic BAD protein to prevent apoptosis.185 The dysregulation of JNK is linked to multiple diseases, such as neurodegenerative disorders, cancer, and autoimmune diseases. JNK3 is being investigated as a potential target for the treatment of CNS disorders.191
C-Jun is the major substrate for JNK.185 c-Jun is a subunit of the transcription factor, activator protein 1 (AP-1).192 Anti-c-Jun antibodies caused partial G0 arrest in the cell cycle,193 while overexpression produced a greater transition into the S, G2, and M phases.194 Indeed, c-Jun is crucial in the regulation of the G1/S phase transition.195 c-Jun also plays an important role in both inducing and inhibiting apoptosis.196 Endogenously, c-Jun inhibits the expression of apoptosis-inducing genes and maintains cell survival, particularly p53 196,197. However, JNK signaling can cause c-Jun to induce apoptosis via survival factor removal.198 JNK phosphorylates the Ser63 and Ser73 residues of the c-Jun activation domain, causing AP-1 transcriptional activity to increase.199 This can cause the induction of apoptosis, although AP-1 is also involved in the inhibition of apoptosis as well depending on the tissue and developmental stage.196 As one might assume, c-Jun, along with JNK has been implicated in many cancers.185,197
CapZ-interacting protein (CapZIP)
CapZIP is a protein detected in muscle extracts that interacts with the F-actin capping protein CapZ. Capping proteins normally inhibit actin polymerization, preserving the actin monomer pool.200 In humans, CapZIP is phosphorylated at Ser-179 and Ser-244 by mitogen-activated protein kinase-activated protein kinase 2 (MAPKAP-K2).45 CapZIP serves as a substrate for stress-activated protein kinases (SAPKs), which can also phosphorylate it at several sites (Ser-68, Ser-83, Ser-108, and Ser-216).45 Cell exposure to various stressful events causes the activation of several MAPKs, including SAPKs, JNK1, and JNK2. When under stress, SAPK3 and SAPK4 will phosphorylate CapZIP, triggering the dissociation of CapZIP from CapZ and enabling CapZ to modify actin filaments.45 JNK1 can also phosphorylate CapZIP, although slower and less extensively.45 Northern blot analysis reveals that CapZIP is expressed highly in skeletal muscle but to a lesser extent in cardiac muscle. In other organs such as the brain, lung, and liver, it is hardly detectable at all.45 Multiple-tissue expression arrays revealed that CapZIP is expressed in immune organs like the thymus, spleen, lymph nodes, and bone marrow.45 CapZIP is also expressed in immune cells and has been isolated from B cells, as well as leukemia and lymphoma cell lines.45 CapZIP also regulates ciliogenesis via its upstream regulator Dvl. Dvl binds to ERK7 and CapZIP, functioning as a scaffold for a MAPK family member, ERK7 (also called MAPK15), to phosphorylate CapZIP, enabling ciliogenesis.201
Myosin II regulatory light chain (MRLC)
MRLC is a regulatory component of myosin II. Myosin light chain kinase (MLCK) phosphorylates the Ser19 residue of MRLC, and sometimes can even phosphorylate the Thr18 residue.202 This enhances the activity of the actin-activated Mg2+ ATPase in myosin II, increasing the assembly and stability of myosin II filaments. The dephosphorylation of MRLC increases myosin II activity and stability more so than monophosphorylation.203,204 However, both forms of MRLC are required for organizing stress fibers during interphase and forming the contractile ring in cell division.202 Dephosphorylation is also important for the disassembly of previous myosin filaments to form new ones.205 ROCK1 can phosphorylate MRLC to facilitate the generation of traction for cell motility.206 ROCK proteins also dephosphorylate MRLC,205 enabling the colocalization of MRLC with actin filaments in mitotic and interphase cells.202 Diphosphorylated MRLC also induces the formation of thick actin bundles containing myosin II, while unphosphorylated MRLC inhibits actin bundle formation.202 These findings emphasize the importance of MRLC in actin organization.
Diaphanous 1 (DIA1)
DIA1 (also called mDia in mammals) is a member of the diaphanous-related formins (DRFs) that are Rho-GTPase binding proteins.46 DIA1 contains a novel formin homology (FH) 2 domain that protects the barbed end of the actin filament from capping proteins and enables rapid assembly of actin subunits.207 DIA1 also has an FH1 domain that binds profilin-bound G-actin to bring it closer to the barbed end for elongation of the actin filament.207,208 DIA1 contains a Rho-GTPase binding domain in the N-terminal region and a Diaphanous-autoregulatory domain (DAD) in the C-terminal region. The N-terminal regulatory region is usually bound to DAD, causing autoinhibition. However, the binding of RhoA relieves this inhibition.209,210 DIA1 is involved in a variety of processes, such as mechanotransduction, cell polarity, migration, and even exocrine vesicle secretion.47 DIA1 mutations have been associated with deafness, cancer, and mental retardation.47 Mutations in formins like DIA1 have also been implicated in cancer metastasis due to the loosening of cellular adhesion. Expression levels of DIA1 have correlated with the stage and metastasis of cancer cells.210
Intracellular mediators of the Wnt/Ca2+ pathway
Phospholipase C (PLC)
PLC is activated by G protein due to Wnt ligand binding in the Wnt/Ca2+ pathway.52,53 The PLC family contains 13 different members with various functions and different structures. Although these members usually have little amino acid sequence homology, they have conserved EF-hand domains, PH domains, and C2 domains.211 They also all have catalytic X and Y domains.211 The PH domain is located towards the N-terminus and mediates the recruitment of PLC to the plasma membrane via binding to PIP2.212 The EF-hand motifs are part of the catalytic core of PLC along with X, Y, and C2 213. The EF-hand motif undergoes a conformational change upon binding of Ca2+ to PLC, revealing binding sites for other ligands.214 The X and Y domains form a triosephosphate isomerase (TIM) barrel-like structure composed of alternating α- and β-pleated sheets.213 The X domain contains all the catalytic residues, while the Y domain is important in modulating the preference of PLC for PIP2, as well as two other ligands PIP and PI.215 The C2 domains are formed from an eight-stranded β-pleated sandwich.213 Upon Ca2+ binding, C2 domains can mediate PLC binding to phospholipids to mediate signal transduction and membrane trafficking.216 Hokin et al used P32 to detect how phospholipid levels changed during enzyme secretion in pancreas slices with the addition of acetylcholine or carbamylcholine. They discovered that phospholipid activity increased five-to nine-fold.217
Unbeknownst to them at the time, the enzyme responsible for the increased phospholipid level was PLC.214 PLC cleaves PIP2 into IP3, which can then induce the intracellular release of Ca2+.218 PLC can be activated by a wide variety of receptors, such as B-cell receptors, T-cell receptors, Fc receptors, tyrosine kinase receptors, and G-protein coupled receptors. Due to the variety of receptors, this means that PLC can be activated by a wide variety of ligands, such as neurotransmitters, hormones, and histamine.214 Aside from IP3, the other product of PIP2 cleavage is DAG, which serves as a secondary messenger to activate Ca2+-dependent protein kinase C (PKC) to phosphorylate numerous downstream effectors and activate a wide array of cellular functions, such as cell polarization, proliferation, as well as learning and memory.219,220 Through Ca2+ release, PLC can regulate cell proliferation, differentiation, motility, gene expression, and other functions.214
Protein kinase C (PKC)
PKC is a family of protein kinases involved in a wide variety of diseases, such as diabetes, cancer, and heart disease.221 There have been 518 protein kinase genes identified.222 PKC, in particular, phosphorylates Ser and Thr residues.221 PKC is activated by DAG, but can also be activated by phorbol esters, which are tumor promoters that mimic the action of DAG.221,223 Specifically, DAG and phorbol esters bind to the C1 domain on PKC, which contains a cysteine-rich sequence that resembles a DNA-binding zinc finger domain.224 PKC contains a regulatory region in the N-terminal half and a catalytic region in the C-terminal half. The C1 and C2 domains are located in the regulatory region and bind to the catalytic region, inhibiting its activity.221 The catalytic kinase region contains C3 and C4 domains.221 An important aspect of PKC is that, upon activation, it translocates to various cellular locations, joining with specific anchoring proteins at each site of action.225 With phorbol esters that irreversibly activate PKC isoenzymes in a nonselective manner, PKC was discovered to regulate many cellular functions.221 Some of these include cell proliferation, cell death, regulation of ion channels and receptors, cell-to-cell contact, and increasing gene transcription.221 However, this early work using phorbol esters does not reflect the effects of DAG due to the irreversibility of phorbol ester binding. The lack of selectivity of phorbol esters also means that they cannot identify the function of each PKC isoenzyme.221 Isoenzyme-specific inhibitors are still under development and undergoing trials, but this endeavor has proven to be difficult.221,223
Cdc42 protein
Cdc42 is a member of the Rho family of small GTPases with roles in actin cytoskeleton regulation, cell motility, cell polarity, and cell cycle progression among other functions.226 Cdc42 was characterized in Saccharomyces cerevisiae, a species of yeast, with G25K being its mammalian and human homolog.227,228 Cdc42 has a P-loop, two switch regions (switch I and switch II), a polybasic region at the C-terminus, and a CAAX box for posttranslational geranylgeranylation.226 Cdc42 has been found in distinct pools in the Golgi apparatus, ER, and plasma membrane.229,230 The Golgi pool functions in three ways; it serves as a reservoir of Cdc42, functions independently from the plasma membrane pool to control protein transport from the Golgi apparatus, and coordinates with the pool to dictate cell polarity.231 Cdc42 modulates the Golgi-to-ER transport via actin regulation.232 The plasma membrane pool serves to regulate cell polarity via actin cytoskeletal rearrangement.233 At the ER, Cdc42 is necessary for tubule fission during ER remodeling.234 The activation of Cdc42 is regulated by GEFs, but unlike other Rho GTPases, Cdc42 is a hydrolase that can hydrolyze the GTP to GDP in the presence of GAP, even though GAP normally exchanges GDP for GTP.235,236 GDI inhibits Cdc42 like other Rho GTPases, via preventing GDP/GTP exchange.174 However, GDI also functions as a chaperone, delivering Cdc42 to its proper location and preventing its degradation.230 Cdc42 is currently under investigation as a therapeutic target for the treatment of cancer, although there are few Cdc42 mutations and no driver mutations that have been linked to cancer.226
Ca2+/calmodulin (CAM)-dependent kinase II (CAMKII)
CAMKII is a Ser/Thr protein kinase237 that was first characterized as a Ca2+-dependent regulator by Schulman and Greengard in 1978.238 There are over 80 known CAMKs.222 CAMKII is encoded by four different genes (α, β, γ, δ) in eukaryotes.239 CAMKII monomers form 12 subunit holoenzymes, with a C-terminal association domain that brings the N-terminal catalytic kinase domains together to fold into two rings of six subunits each.237,239 There is a regulatory segment that follows the kinase domain and is joined to a linker region that then connects to the association domain.237,240 The regulatory domain forms an α-helix to block the catalytic domain of each subunit.237 Additionally, the T286 phosphorylation site is sequestered in a hydrophobic groove, preventing autophosphorylation that would up-regulate CAMKII activity. Upon Ca2+/CAM binding to the regulatory domain, the regulatory segment is removed, enabling progressive autophosphorylation of the T286 site and increasing CAMKII activity.237 Normally, in the autoinhibited (compact) state, CAM cannot bind237,240,241; it is only when the compact state is in equilibrium with the non-autoinhibited (extended) state that CAM can bind the regulatory domain.237
Between the four CAMKII genes found in humans, the enzymes produced share 95% of their amino acid sequences in the kinase domains and 80% homology in the hub domains, with the linker region being the primary variable component.240 When two adjacent subunits are activated by Ca2+/CAM, they can phosphorylate one another on Thr286, causing increased Ca2+-independent activity.240
CAMKII has a role in adaptive contractive response during aerobic exercise.242 It is also involved in glucose production, cell cycle progression, and vascular smooth muscle function.243 Continuous activation of CAMKII can cause cardiac myocyte apoptosis, heart failure, and cardiac arrhythmia.244 CAMKII has also been identified as being able to spread the inflammatory response caused by damage to heart muscle.243 The progression of cardiomyopathy and even Chagas disease caused by Trypanosoma cruzi is mediated by CAMKII signaling.243,245
TGF-β activated kinase 1 (TAK1)
TAK1 is a member of the MAPK kinase kinase (MAPKKK) family. It is a Ser/Thr kinase that was originally discovered to be a mediator of BMP and TGF-β signaling.246,247 TAK1 can be activated by many cytokines, such as TNF-α, TGF-β, TLRs, and IL-1 248,249. TAK1 activation causes the phosphorylation of TAK1, which leads to the activation of NF-kB, JNK, ERK, p38, and MAPKs.248,249 TAK1 is also involved in T- and B-cell signaling,248 as well as angiogenesis during embryonic development.249 Due to its role in immune and inflammatory processes, TAK1 has been implicated in multiple cancers, such as lymphoma and neuroblastoma, as well as colon, ovarian, and pancreatic cancers.250 In addition, a blockade of TAK1 leads to p53 up-regulation, indicating the importance of TAK1 in controlling cellular stresses.251 Activation of TAK1 requires three proteins, TAK1-binding protein 1 (TAB1), TAB2, and TAB3. TAB1 serves as an adaptor protein located on the N-terminal kinase domain of TAK1, while TAB2 and TAB3 will only bind the C-terminal TAK-binding domain after stimulation.249 TAB1 overproduction leads to increased TAK1 activity, but TAB1 deficiency has minor downstream effects.249 TAB2 and TAB3 are not required early on in TAK1 activation but are required for sustained TAK1 activation.249 In mice, TAK1 has been demonstrated to be a regulator of TNF signaling in the skin and modulates skin inflammation. It was found that a lack of TAK1 caused keratinocyte death due to absent NF-kB and JNK-mediated cell survival signaling.252 Recently, a selective TAK1 inhibitor called Takinib has been developed; it is activated by ATP and competitively inhibits TAK1 by binding to its ATP-binding pocket.253 Another TAK1 inhibitor called piperidylmethyloxychalcone (PMOC) also functions in the same manner.254
Nemo-like kinase (NLK)
NLK is a conserved Ser/Thr MAPK.255 It is the mammalian homolog of the Drosophila nemo gene, which was identified in Drosophila as important for the rotation of photoreceptor clusters in eye morphogenesis.256 NLK has a longer N-terminal region that is rich in histidine, proline, alanine, and glutamine.257 NLK can phosphorylate TCF4 to prevent the β-catenin/TCF complex from binding to DNA and initiating gene transcription. TAK1 can also stimulate NLK to phosphorylate TCF4, so NLK functions as a downstream effector of TAK1 in the repression of Wnt/β-catenin signaling.58 Although TAK1 and NLK can repress canonical Wnt signaling, CAMKII from the noncanonical Wnt/Ca2+ pathway activates TAK1 in order to do so, indicating crosstalk between the two pathways.57 Furthermore, TAB2 can serve as a scaffold for TAK1 and NLK, enabling cooperative interaction for the inhibition of canonical Wnt signaling.258 NLK can also associate with NLK-associated RING finger protein (NARF), which is an E3 ubiquitin ligase whose action is up-regulated by NLK kinase activity.259 NARF can ubiquitinate TCF/LEF for degradation by the proteasome.259 NLK is involved in other pathways as well, such as Notch signaling and STAT protein signaling.259 Due to the function of NLK in Wnt signaling and other pathways, it is crucial in regulating cell proliferation, apoptosis, migration, and other functions.259 For instance, NLK was found to inhibit the growth and migration of non-small cell lung cancer by restoring the expression of E-cadherin, which normally suppresses migration and invasion.260 However, analysis of colorectal cancer (CRC) cells found that NLK activity was increased, and only half of the CRC cells were apoptotic compared to non-CRC cells, indicating that NLK may have an anti-apoptotic function.261 NLK has also been identified as a pathological effector in mouse hearts, leading to progression toward heart failure and other cardiac conditions.262 This makes NLK a potential therapeutic target for various diseases.
Calcineurin (CaN)
CaN is a conserved Ser/Thr phosphatase that is activated by increased intracellular Ca2+ 263. It was first discovered by Wang and Desai in 1976 as a protein that counteracted the activation of bovine brain cyclic nucleotide phosphodiesterase.264 CaN is a heterodimer consisting of calcineurin A (the catalytic subunit) and calcineurin B (the regulatory subunit).265 The catalytic domain of calcineurin A is towards the N-terminal end, while the C-terminus contains three regulatory domains, the calcineurin B binding domain, the calmodulin-binding domain, and the autoinhibitory domain that binds to the active site when Ca2+/calmodulin is absent.265 Calcineurin B contains four Ca2+ binding EF-hand motifs.265 The mechanism of calcineurin B activation is still unknown.266 It is also structurally homologous to CAM.263 CaN along with CAM binds to Ca2+, then CAM binds CaN. This complex is what forms the active phosphatase.263 Although there are multiple CAM-modulated kinases, CaN is the only phosphatase that is directly activated by Ca2+ 263. CaN is best known for regulating the transcription of IL-2, dephosphorylating the transcription factor NF-ATp in response to an increase in intracellular Ca2+ via T cell receptor activation, and stimulating NF-ATp. CaN also controls cellular Ca2+ sequestration and cytokinesis.266 The immunosuppressive drugs FK506 and cyclosporin A can both target CaN and were key to identifying its functions.266 CaN is also important in the regulation of neurotransmitter release in neuromuscular junctions (NMJs).267 Hypertrophic and dilated cardiomyopathy have both been linked to CaN overactivation.268
Antagonists and inhibitory regulators of Wnt signaling
Extracellular antagonists of Wnt signaling
Secreted frizzled-related proteins (sFRPs)
The largest family of secreted Wnt inhibitors are the sFRPs, which are structurally like the CRD ligand-binding domain of the Fz receptors.269 The first sFRP discovered was the Frizzled motif associated with bone development (Frzb).270 The sFRPs are about 295–346 amino acids long, with a CRD at the N-terminus.269 These CRDs share 30%–50% sequence similarity with the CRDs of Fz receptors, with 10 cysteine residues linked by disulfide bridges. The C-terminus has a netrin-related motif (NTR) that functions as a heparin-binding domain.271 Studies of Xenopus embryos revealed that Frzb binds to Wnt1 and Wnt8, sequestering them away from the Fz and LRP5/6 receptor complex.272, 273, 274 sFRPs can also form a nonfunctional complex with Fz receptors to prevent Wnt binding and signaling.275,276 Both the CRD and NTR domains are important in Wnt inhibition.277 The CRD domain binds to Wnt,273 while the NTR domain mimics the function of the entire sFRP1 to bind to Wnt ligands and inhibit Wnt signaling.278
Wnt-inhibitory factor 1 (WIF-1)
WIF-1 was first identified as an expressed sequence tag in the human retina279 found in fish, amphibians, and mammals.279 It contains a 150 amino acid long N-terminal signal sequence called the WIF domain, five epidermal growth factor (EGF)-like repeats, and a 45 amino acid hydrophilic domain at the carboxy-terminus.279 The complete WIF-1 protein is 379 amino acids long.279 Analysis of Xenopus embryo assays found that WIF-1 binds Wnt8, while in Drosophila clone-8 cells, WIF-1 binds Wg, although not as strongly as with Wnt8.279 WIF-1 inhibits the binding of Wnt8 to Dfz2, blocking Wnt signaling.279
Dickkopf (Dkk) proteins
The Dickkopf (Dkk) proteins contain four members in vertebrates (Dkk1-4). Dkk1 was the first protein of its family to be discovered through experiments determining its importance in embryonic head formation and as a Wnt antagonist.280 The Dkk proteins are glycoproteins composed of 255–350 amino acids.281 Interestingly, although the Dkks all have a signal sequence and two conserved CRDs, there is little sequence similarity between them otherwise.280 Dkks primarily modulate the canonical Wnt signaling pathway via binding to the LRP5/6 co-receptor. Dkk1 associates with one of two single-pass transmembrane proteins called Kremen 1 or 2 (Krm1 or Krm2) while binding to LRP5/6. This interaction allows the endocytosis of the LRP5/6 receptor.19,281 Analysis of Dkk1 in cardiogenesis suggests that Dkk1 can activate JNK, indicating that Dkk1 may be involved in the noncanonical Wnt/PCP pathway as well.282
Wise and sclerostin (SOST) proteins
Wise and SOST are both members of a subfamily of cysteine knot proteins as they both contain cysteine knot motifs.283 Both Wise and SOST are also BMP antagonists.283,284 Wise protein is composed of 206 amino acids with 38% sequence homology with sclerostin.284 Wise is expressed in a wide variety of tissues, including branchial arches, rat endometrium, developing testes, and more.285 The sclerostin polypeptide is about 190 amino acids long with a cysteine knot formed from its flexible N- and C-terminal regions.286 Sclerostin is expressed in osteocytes to regulate bone formation. Deletions in the SOST gene that codes for sclerostin can result in van Buchem disease, which is characterized by a high bone mass due to loss of inhibition of bone formation.286 Reporter assays demonstrate that Wise can block Wnt1, Wnt3a, and Wnt10b.285 Wise also binds to the LRP6 coreceptor, preventing the binding of Wnt8.284 Additionally, Wise can function intracellularly to prevent LRP6 trafficking to the cell surface.287 Wise can also bind LRP4 to inhibit Wnt signaling.288 Sclerostin binds LRP5/6, specifically binding to LRP5 at the YWTD-EGF repeat domains, inhibiting Wnt signaling.289,290
Cerberus
Cerberus is an abundant organizer-specific gene that was isolated from Xenopus and can induce ectopic head formation.291 It also plays an important role in cardiogenesis during vertebrate embryonic development.292 Cerberus, like Wise and SOST, is in the cysteine knot superfamily.293,294 Long-form Cerberus (xCer-L) binds to Wnt8 and could inhibit Wnt signaling, but short-form Cerberus (xCer-S) cannot do the same.294 Further study is required to better understand the effects of Cerberus on the Wnt pathway.
IGFBP-4
IGFBP-4 is a member of the insulin-like growth-factor-binding proteins (IGFBPs) that bind and modulate insulin-like growth factors (IGFs). IGFBP-4 is important in cardiomyogenesis in vitro.295 IGFBP-4 interacts with LRP6 and Fz8, functioning as a competitive inhibitor of Wnt3a binding.295 In this manner, IGFBP-4 inhibits the canonical Wnt pathway.295,296 Furthermore, there are six IGFBP members. Although IGFBP-4 is the most powerful Wnt inhibitor, IGFBP-1, -2, and -6 also have Wnt inhibitor activity, albeit modestly. On the other hand, IGFBP-3 and -5 have no Wnt inhibition activity.295 Although some cancer cell lines in vitro saw a decrease in cell proliferation when treated with IGFBP-4, decreased levels of the protein increased the risk of breast cancer, and overexpression of IGFBP-4 caused prostate cancer growth in vivo.295
Intracellular negative regulators of Wnt signaling
Adenomatous polyposis coli (APC)
APC is a tumor suppressor gene that is heavily associated with colorectal cancer (CRC).297 The APC protein is 2843 amino acids long,297 with a central region spanning about 1000 amino acids containing motifs that bind β-catenin or Axin.22 In humans, APC contains four 15-mers and seven 20-mers. The 20-mer repeats are phosphorylated by GSK-3β and CK1, enhancing the affinity of APC for β-catenin.22 Interspersed with the 20-mer repeats are three Ser-Ala-Met-Pro (SAMP) repeats that bind to Axin.298,299 Full-length APC or APC with SAMP repeats was found to protect β-catenin from dephosphorylation by PP2A, ensuring the destruction of β-catenin.299 The N-terminus of APC has a dimeric coiled-coil domain, with a heptad-repeat region that is crucial for the dimerization of APC.300 APC also contains an armadillo repeat domain (APC-arm) that enables APC to regulate the actin cytoskeleton and microtubules during cell polarization and migration.301,302 The C-terminal region of APC binds several proteins, such as microtubulin, indicating its role in microtubule assembly.303 APC mutation is best known for causing familial adenomatous polyposis (FAP), a disease characterized by a family history of colorectal polyps and cancer. FAP is inherited in an autosomal dominant manner via a germline mutation.304
Axin proteins
Axin is a scaffold protein that was initially discovered as a protein product of the mouse Fused (Fu) gene, inhibiting Wnt signaling and regulating embryonic axis formation.305 There are two Axin homologs in eukaryotic organisms, Axin1 and Axin2, which are often collectively referred to as Axin. Axin1 is vital for embryonic viability and is widely expressed, while Axin2 is limited in its distribution to certain tissues.306,307 Axin1 was identified in mice as a locust causing kinky tail phenotypes, while Axin2 was identified due to its interactions with GSK-3β and β-catenin, as well as its homology to Axin1. The N-terminus contains the RGS (regulation of G-protein signaling) domain, which binds to APC.298 It is important to note that this region is homologous to regulators of G-protein signaling, but does not actually regulate any known G-protein.298 The RGS domain binds the third SAMP repeat and is highly conserved among Axins but is not conserved in other RGS proteins.298 Axin contains a C-terminal DIX domain that enables it to form homodimers with other Axin proteins, as well as heterodimers with Dvl.308,309 This interaction via the DIX domains is how Dvl can inhibit Axin and enable transduction of Wnt/β-catenin signaling.144 In between the RGS and DIX domains are regions that allow Axin to bind to β-catenin, GSK-3β, and CK1, forming a complex that will target β-catenin for degradation.307 Axin is important in many developmental processes, including anterior–posterior axis formation, organogenesis, neuronal proliferation and differentiation, synapse formation, and several others.307 Both Axin1 and Axin2 can function as the scaffold of the β-catenin destruction complex. They share the RGS and DIX domains, as well as the Tankyrase binding domain that maintains the Axin protein's stability.306 Interestingly, Axin2 is a target of β-catenin mediated gene transcription, but not Axin1. Due to this fact, Axin2, in particular, has been of interest in cancers caused by aberrant Wnt signaling activation, which leads to high levels of Axin2 expression.306 Axins also interact with many other pathways. Its interactions with p53 are important in stimulating transcription in p53-dependent target genes, shedding light on the importance of Axin as a tumor suppressor.310 Axin1 and Axin2 both inhibit Wnt signaling and stimulate TGF-β signaling. However, TGF-β signaling can inhibit Axin1 and Axin2 expression, leading to enhanced Wnt signaling that increases chondrocyte maturation.311
Glycogen synthase kinase-3 (GSK-3) proteins
GSK-3 is a conserved Ser/Thr kinase that was first identified in rabbit skeletal muscle.312,313 There are two forms of GSK-3 in mammals, GSK-3α and GSK-3β, which are 98% homologous in the internal kinase domain, albeit with different N-terminal regions314 and C-terminal regions.315 Defects in GSK-3α in mice caused impaired locomotion and coordination, as well as psychiatric disorders.316 GSK-3β defects, on the other hand, are embryonically lethal,317,318 indicating that GSK-3β may be more important than GSK-3α. GSK-3 inhibits glycogen synthase via phosphorylation, thereby inhibiting glycogen synthesis.319 GSK-3 is inactivated by insulin, so GSK-3 dysregulation has been implicated in type II diabetes.318 Since both Wnt and insulin signaling pathways involve GSK-3β, this suggests that there is potential crosstalk between them. AKT phosphorylates and inhibits GSK-3 320 and Wnt causes GSK-3 dislocation from the destruction complex.321 GSK-3β is rich in the brain and is involved in neurogenesis, neurotransmission, and regulation of synaptic plasticity.322 However, it has also been implicated in the formation of neurofibrillary tangles in Alzheimer's disease.322 GSK-3β can be inhibited by lithium, which is used as a mood stabilizer for treating bipolar disorder.323 GSK-3β also enhances apoptosis via up-regulation of p53.324 GSK-3 can also modulate inflammation downstream of TLR signaling; activation of GSK-3 leads to the production of the pro-inflammatory cytokines IL-6, IL-1B, and IFNy, while inhibition of GSK-3 leads to the production of anti-inflammatory IL-10.325
Casein kinase 1 (CK1)
Casein kinase 1 (CK1) is a large family of conserved Ser/Thr kinase found in eukaryotes, with seven isoforms in humans (α, γ1, γ2, γ3, δ, ε, and α-like).326 CK1 is involved in a diverse range of cellular processes, such as vesicular trafficking, DNA repair, cell proliferation, and apoptosis.327 The kinase domain is conserved, but the N- and C-terminal domains vary in length among the CK1 family members.326 The N-terminal domain of CK1 is its catalytic domain, while the C-terminal domain has been associated with an inhibitory function, as CK1 with a truncated C-terminus led to an increase in catalytic domain activity.328,329
CK1 is generally considered constitutively active.330 The reason is unknown, but phosphatases have been inferred to be involved in this activity.329,330 Phosphorylation of CK1 either by itself or by other kinases inhibits its catalytic activity.330 Autophosphorylation on the C-terminus results in a phosphopeptide that binds to the catalytic domain.330 CK1 isoforms are regulated by scaffold proteins, which can sequester CK1 at several subcellular locations or bind CK1 allosterically to promote or inhibit CK1 activation.330 Although CK1 is also involved in the Hedgehog, NF-kB, and Hippo signaling pathways,326,327,330 Wnt signaling is perhaps its best-characterized process.330 CK1 phosphorylates β-catenin on Ser45, priming it for subsequent phosphorylation on Thr41, Ser37, and Ser33 by GSK-3.331 β-TrCP can then ubiquitinate β-catenin for proteasomal destruction.22,24
CK1 also has a role in modulating p53 due to its inhibitory interacting proteins, mouse double minute homologue 2 and 4 (MDM2 and MDM4), being substrates of CK1. Phosphorylation of these substrates by CK1-δ and CK1-ε leads to p53 activation.332 However, other studies have demonstrated that the knockdown of CK1-α can also activate p53.333 This suggests that different isoforms could have opposite effects on p53 signaling. CK1 was also the first kinase discovered to regulate the circadian rhythm, where the positive regulatory complex CLOCK-BMAL activates the transcription of PER orthologs PER1-3, and cryptochrome proteins CRY1-2. The PER and CRY proteins then inhibit the CLOCK-BMAL complex.330 This completes the circadian cycle, but then CK1 can phosphorylate PER proteins for degradation, starting the cycle again.330 Additionally, CK1 is involved in mitosis, regulating spindle positioning upon being delivered to spindles by the CK1-specific binding protein FAM83D.334 CK1 has been prominently linked to neurodegeneration and cancer.330
β-Transducin repeat-containing proteins (β-TrCP)
β-TrCP is the substrate recognition subunit for the Skp1/Cullin1/F-box protein (SCF) E3 ubiquitin ligases.335 β-TrCP (also called FWD1 in mammals) is part of the Fbw (F-box/WD40 repeat containing) protein family, whose members share an N-terminal F-box motif and seven C-terminal WD40 repeats. Other F-box proteins include Fbl (F-box/leucine-rich repeats) and Fbx (F-box/unknown motifs).335 These motifs are highly conserved, and humans have two isoforms, β-TrCP1 and β-TrCP2.335 β-TrCP functions as an intracellular receptor to bind phosphorylated β-catenin at the phosphorylated sites, but this interaction is more efficient in the presence of Axin.336 In addition, Axin does not bind well with β-catenin without β-TrCP, suggesting that the three form a ternary complex that requires all three members for optimal stability.336 The F-box motif is used to bind β-TrCP to Skp1, which can then ubiquitinate β-catenin for degradation.337 However, the F-box domain is not required to bind β-catenin, as the binding to β-catenin and Axin is mediated by the WD40 repeat domains.336
SCF uses β-TrCP to determine substrate specificity.336 SCF ubiquitinates substrates like β-catenin at DSGXXS destruction motifs, with both Ser residues needing to be phosphorylated for SCF-mediated ubiquitination.335 β-TrCP/SCF is also involved in the NF-kB signaling pathway. NF-kB is usually retained in the cytoplasm by a family of inhibitory molecules called IkBs. These can then be phosphorylated by IkB kinase (IKK) and subsequently ubiquitinated by SCF for degradation, releasing NF-kB to translocate to the nucleus.338 β-TrCP has been implicated in cell division due to β-TrCP knockout mice having impaired progression through mitosis, with spermatocytes accumulating at metaphase I.339 This effect is attributed to the buildup of early mitotic inhibitor 1 (Emi1), which inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C).
Normally β-TrCP/SCF would degrade Emi1 to allow progression through mitosis, but this does not happen without β-TrCP recognizing the DSGXXS destruction motif.340 Due to the oncogenic risks of NF-kB in inducing gene products controlling signaling proliferation and suppressing apoptosis,341 β-TrCP/SCF is linked to oncogenesis and anticancer therapy resistance.335 β-TrCP/SCF is also under investigation for targeting disease-causing proteins for degradation.335
Protein phosphatase 2A (PP2A)
PP2A is a member of the phosphoprotein phosphatase (PPP) family and is involved in cell cycle regulation.342 It is a Ser/Thr phosphatase.343 PP2A is a heterotrimeric enzyme consisting of A, B, and C subunits. A is the scaffolding subunit, B is the regulatory subunit, and C is the catalytic subunit.343 The core of PP2A is composed of A and C subunits, which both have α and β isoforms that are homologous to each other, although the α isoform is the more commonly expressed variant.343 The A subunit contains 15 huntingtin-elongation-A subunit-TOR (HEAT) tandem repeats, with each HEAT repeat consisting of a pair of antiparallel helices, which assemble to form a curved structure.344 These helices form a hydrophobic inner ridge that facilitates B and C binding. They also stack with a hinge region between HEATs 12 and 13 to give the A subunit flexibility.343 The catalytic activity of the C subunit is thought to be mediated by two Mn2+ ions,343 coordinated by six conserved residues (two aspartate, one asparagine, and three histidine residues) along with a catalytic water molecule.342 Modifications of the C-terminal tail of the C subunit are needed for B subunit binding, although the B subunit has different classes and each requires its own unique modification.345
There are four classes of B subunits, namely, the B55 family (B), the PR72 family (B′), the B56 family (B″), and the Striatin family (B‴). They all vary widely in structure, allowing for diverse substrate specificity for a wide range of cellular functions.343 Furthermore, although A and C subunits are expressed ubiquitously, B subunit expression levels vary significantly across different cells and tissues.343 PP2A has dual roles in Wnt signaling. It can dephosphorylate β-catenin, APC, and Axin, enhancing Wnt signaling.342 However, it also inhibits Wnt signaling by dephosphorylating the Ser9 residue on GSK-3β after being recruited by heat shock cognate 40 (HSC40), activating GSK-3β.346
PP2A can also activate GSK-3β indirectly by inhibiting protein kinase B (AKT), which initially phosphorylates GSK-3β to inhibit it.320,346 Due to the different PP2A holoenzymes that exist, PP2A has a wide variety of functions. For instance, PP2A-B55 is crucial to the prevention of entry into mitosis and must be inactivated for cell division to begin.342 However, PP2A holoenzymes are also involved in the disassembly and reassembly of the Golgi apparatus mediated by phosphorylation and dephosphorylation during mitosis. Specifically, dephosphorylation of the Golgi matrix protein GM130 by PP2A induces Golgi reassembly at the end of mitosis.347 Mutations in PP2A have been associated with defects in brain development and are causally linked to intellectual disability and neurodevelopmental disorders.348 Abnormalities in PP2A have also been linked to Alzheimer's disease.349 It is also a factor in cancer onset, such as the progression of COPD in lung cancer when inactivated.350
Groucho
Groucho is a transcriptional corepressor that was first identified in 1968 by a mutation in Drosophila that caused clumps of extra bristles to form above the eyes. Transducin-like enhancer of split (TLE) protein is the human homolog.351 TLE/Groucho is involved in a variety of processes, such as eye development, segmentation, and sex determination.351 TLE/Groucho is characterized by a conserved glutamine-rich (Q) domain on the N-terminus, and a conserved WD-repeat domain on the C-terminus, with a less conserved central region dividing the two.351 In the nucleus, TCF/LEF are bound to TLE/Groucho via the N-terminal glutamine-rich (Q) domain on TLE/Groucho.352 The Q domain has an α-helical coiled-coil motif that enables TLE/Groucho to tetramerize, which is required for transcriptional repression.352 The Q domain binds to the HMG domain of TCF/LEF.352 The C-terminal WD-repeat domains function to interact with transcription factors and are also important in chromatin condensation.353 TLE/Groucho also contains a central glycine/proline (GP)-rich domain that recruits histone deacetylases (HDACs) to repress transcription.351,354
TAK1 and NLK
As mentioned previously, from the noncanonical pathway, TAK1 and NLK can inhibit canonical Wnt signaling via TAK1 activation of NLK to phosphorylate TCF4.58 NARF can also inhibit canonical Wnt signaling by ubiquitinating TCF/LEF for proteasomal degradation.259 Additionally, the coexpression of the noncanonical Wnt ligand, Wnt5a, in hepatocellular carcinoma (HCC) cells, resulted in a three-fold decrease in TCF activity. However, the mechanism that caused this is unknown.355
Crosstalk between Wnt and major signaling pathways
Crosstalk with bone morphogenetic protein (BMP) signaling
BMPs are members of the transforming growth factor-beta (TGF-β) family of cytokines.356 In TGF-β/BMP signaling, the binding of a ligand to type I and II receptors brings them together, where receptor II phosphorylates the receptor I kinase domain, propagating the signal via phosphorylation of the receptor-regulated Smad (R-Smad) signal transducing proteins.357 Of the five R-smads (Smad1, -2, -3, -5, and -8), Smad1, Smad5, and Smad8 are activated by BMP signaling, while Smad4 is the co-mediator Smad (Co-Smad).357 The R-Smads form complexes with Smad4 to translocate into the nucleus and regulate gene transcription.357,358 BMPs are vital in embryogenesis and homeostasis and are known to induce bone formation.11,356,359, 360, 361, 362, 363
BMP receptors determine the strength of BMP signaling through the C-terminal phosphorylation on Smad1, which contains four GSK-3 phosphorylation sites. GSK-3 can phosphorylate Smad1, leading to its ubiquitination and destruction, which reduces the strength of BMP signaling. Wnt signaling can also inhibit GSK-3β, stabilizing Smad1 and enabling BMP signaling.356 Smad7 is an inhibitory Smad, primarily down-regulating R-Smad activation via the recruitment of the E3 ubiquitin ligases: Smurf1 and Smurf2. Smurf1 and Smurf2 target Smads and the TGF-β receptors for degradation.357 Smad7 can complex with β-catenin and Smurf2, which causes ubiquitination and subsequent degradation of β-catenin364 (Fig. 5).
Figure 5.
Crosstalk between Wnt signaling and other major signaling pathways. The (A) left, (B) middle, and (C) right panels show the Wnt/BMP crosstalk, Wnt/Notch crosstalk, and the Wnt/Hippo crosstalk, respectively. Note that not all crosstalk interactions between each of these pathways are represented in the image due to space constraints. In Wnt/BMP crosstalk, the absence of Wnt signaling allows GSK-3β to phosphorylate Smad1, which is ubiquitinated and degraded. The activation of Wnt signaling inhibits GSK-3β, stabilizing Smad1 and enhancing BMP signaling. BMP2 can up-regulate Wnt3a and enhance β-catenin binding with Smad4. Wnt3a can also up-regulate BMP2 expression. Smad7 can either recruit Smurf2 to ubiquitinate β-catenin for degradation or cause Axin to dissociate from β-catenin, stabilizing it. For Wnt/Notch crosstalk, Notch can complex with β-catenin and promote its degradation. TCF/LEF can regulate the expression of the DLL1 Notch receptor ligand. Notch2 and Wnt4 are involved in a positive feedback loop. β-Catenin can enhance Hes1 transcription and inhibit the degradation of Notch1. β-Catenin can also up-regulate transcription of the JAG1 Notch ligand. Notch1 can complex with β-catenin and Lamp1 to promote lysosomal degradation of β-catenin. Notch can increase both Fz receptor expression and transcription of TCF1. Finally, in Wnt/Hippo crosstalk, there are many interactions as well. LATS1/2 can phosphorylate the HXRXXS motifs of YAP to signal to CK1 to phosphorylate the DSGXXS destruction motifs, leading to the recruitment of β-TrCP-mediated degradation of YAP. MST1/2 can sequester CK1, preventing phosphorylation of Dvl and inhibiting Wnt signaling. YAP/TAZ can bind to and suppress β-catenin while preserving its stability, although other reports indicate that YAP/TAZ is associated with Axin as part of the destruction complex. YAP/TAZ can also inhibit Dvl, reducing Wnt signaling. However, YAP can also interact with SHP2 to enhance β-catenin activity in the nucleus. Wnt signaling can cause YAP/TAZ to transcribe Hippo pathway target genes. The illustration was inspired by and created in BioRender.
Additionally, Smad7 can interact with Axin, and this Smad7-Axin complex can cause GSK-3β and β-catenin to dissociate from Axin while also inhibiting the recruitment of Smurf2 to β-catenin. The overall effect is to stabilize β-catenin, which complexes with E-cadherin to strengthen cell-to-cell adhesion.365 BMP receptor type 1a (BMPr1a) induces the expression of SOST to limit cancellous bone accrual by inhibiting Wnt signaling. However, BMPr1a-deficient osteocytes experience a decreased expression of SOST, indicating the role of BMPr1a in modulating bone growth through Wnt signaling.358 Loss of BMPr1a in mouse rib bones also caused decreased levels of Dkk1, indicating Dkk1 serves as another downstream target of BMPr1a.366 BMP9 induces the formation of ectopic bone via the recruitment of Runx2 and β-catenin to the osteocalcin promoter. Knockdown of β-catenin prevented osteogenic differentiation of MSCs via BMP9, establishing the importance of Wnt signaling in mediating BMP9 activity.367
Wnt3a and BMP9 both induce alkaline phosphatase (ALP) activity in MSCs while enhancing one another's ALP induction, suggesting that they act synergistically to induce bone formation.367 Between the two, Wnt3a induces ALP activity to a stronger degree and earlier than BMP9, demonstrating the crosstalk between the two pathways in osteogenic differentiation.367 Furthermore, Wnt3a and BMP9 both up-regulate CTGF/CCN2, a gene involved in osteogenic differentiation.363
BMP2 stimulates LRP5 expression and inhibits β-TrCP expression. This leads to an increase in β-catenin levels in osteoblasts and promotes osteogenic differentiation.368 BMP2 also increased the expression of the canonical Wnt ligands, Wnt1, Wnt3a, and Wnt4.368 The BMP2-mediated up-regulation of Wnt3a and enhancement of the interaction of Smad4 with β-catenin reveal the importance of BMP2 in the modulation of Wnt signaling.369 Interestingly, Wnt3a signaling or overexpression of β-catenin/TCF4 can increase transcription of BMP2, adding further complexity to the interplay of BMP and Wnt signaling.370 BMP2 and Wnt3a exhibit a cooperative effect to increase the transcription of osteogenic genes (Id1, Dlx5, Msx2, Osx, and Runx2).371
BMP2 can also antagonize Wnt signaling by promoting Smad1 binding to Dvl1, preventing the activation of β-catenin.372 BMP2, along with BMP4, can also induce Wnt8 expression in Xenopus mesoderm.373 Perhaps unsurprisingly, mutations in BMP genes can lead to several bone disorders, such as skeletal developmental defects and inappropriate ossification.374 BMP signaling has also been a therapeutic target in cardiovascular diseases such as atherosclerosis, pulmonary arterial hypertension, and anemia of chronic disease.375
Crosstalk with TGF-β signaling
TGF-β signaling coordinates with Wnt signaling in development and homeostasis. The presence of both signaling pathways is necessary for the initial inductive activity of BMPs.376 TGF-β and Wnt signaling regulate each other's ligand production, and the Smad/β-catenin/LEF complex regulates a group of shared target genes, further delineating the cooperativity of the two pathways.356 Eger et al conducted a study of polarized mouse mammary cells that underwent epithelial–mesenchymal transition (EMT) induction by an inducible c-fos estrogen receptor (FosER) oncoprotein. The inhibition of both TGF-β and Wnt signaling caused a reversal of cells that underwent EMT back to their polarized forms, while inhibition of only one pathway led to a partial rescue of the epithelial features. This indicates the cooperativity of TGF-β and Wnt signaling in the EMT.377 Furthermore, Nawshad et al demonstrated in palate medial-edge epithelial cells (MEE) that TGF-β3 signaling forms Smad2-P-Smad4-LEF1 transcription complexes that repress the E-cadherin gene during EMT.378 The mediation of the EMT via β-catenin and TGF-β is dependent on cyclic AMP-responsive element-binding protein (CREB)-binding protein (CBP). One target gene in EMT is α-smooth muscle actin (α-SMA), which requires β-catenin for the induction of transcription via TGF-β signaling. CBP promotes the interaction between Smad3 and β-catenin after TGF-β1 activation, leading to the transcription of α-SMA. This signaling mechanism has been implicated in pulmonary fibrosis and is being explored as a therapeutic target.379 TGF-β1 signaling also causes the translocation of β-catenin to the nucleus via Smad3 in MSCs. The stimulation of the TGF-β receptor causes phosphorylation of Smad3, disrupting its interactions with GSK-3β. Smad3 can then function as a chaperone to shuttle β-catenin into the nucleus.380
Crosstalk with Notch signaling
Notch signaling is a conserved cell–cell communication mechanism involved in determining cell fate and cell lineage.381 In mammals, there are four Notch receptors (Notch1–4) that are homologous to the single Notch receptor in Drosophila, and five Delta/Serrate/Lag-2 (DSL) ligands (DLL1, DLL3, DLL4, JAG1, and JAG2).382,383 Ligand binding leads to the opening of the negative regulatory domain (NRR). This enables the cleavage of the Notch intracellular domain (NICD) by the multiprotein γ-secretase complex. The NICD can then translocate to the nucleus and form a Notch transcriptional activation complex (NTC) with RBPJ and co-activators of the Mastermind-like (MAML) family. Without Notch, RBPJ interacts with transcriptional repressors, while the binding of Notch enables transcription.382,383
In Drosophila, Notch signaling promotes Wg expression at the wing margin, while Wg promotes Serrate and Delta expression in wing patterning. Notch has also been demonstrated to down-regulate Wnt signaling by complexing with β-catenin and promoting its degradation.384 Notch2 is involved in a positive feedback loop with Wnt4, up-regulating Wnt4 expression to promote differentiation of progenitor cells in the nephrons of mice.385 The Wnt pathway transcription factors LEF1/TCF regulate the expression of DLL1, establishing the importance of Wnt signaling in the regulation of Notch signaling.386 Additionally, Wnt/β-catenin signaling also regulates the transcription of the Notch target gene Hes1, which encodes a basic helix-loop-helix transcriptional repressor.387
β-Catenin can bind to Notch1 and the NICD, which enhances the transcription of Hes1 while also preventing ubiquitin-dependent degradation of Notch1 and NICD.388 On the other hand, Notch1 can down-regulate β-catenin by forming a complex with β-catenin and then colocalizing with the lysosomal protein Lamp1. Lysosomal activity can then decrease the levels of β-catenin.389 GSK-3β phosphorylates NICD, stabilizing Notch and positively regulating its signaling activity.390 CAMKII induced by Wnt5a signaling can phosphorylate the Notch signaling corepressor, silencing mediator or retinoic acid, and thyroid hormone receptor (SMRT). This causes SMRT to dissociate from RBPJ and get degraded by the proteasome. CAMKII also enhances binding between NICD and RBPJ. The overall effect of CAMKII is to increase Notch signaling.391 Dvl down-regulates Notch signaling by inhibiting the CSL (RBPJ) transcription factors that mediate Notch signaling392 (Fig. 5).
Notch-regulated ankyrin repeat protein (NRARP) is a negative regulator of Notch signaling but can also up-regulate Wnt signaling via stabilizing LEF1.393 In both colorectal and renal cancers, Wnt/β-catenin signaling up-regulates transcription of the Notch ligand Jagged1 (JAG1), leading to Notch activation.394,395 In hematopoietic progenitor cells (HPCs), Notch signaling up-regulates the expression of Fz receptors to enhance dendritic cell (DC) differentiation.396 Notch signaling also increases transcription of TCF1, increasing mediation of Wnt signaling to stimulate T-cell specification.397 Notch dysregulation can lead to kidney fibrosis and chronic kidney disease (CKD).381 Defective Notch signaling can also cause congenital heart disease but has become a potential therapeutic target for heart regeneration.398 Liver fibrosis, chronic liver disease, and osteosarcoma are also attributable to aberrant Notch signaling.399,400
Crosstalk with the Hippo/Yap pathway
The Hippo pathway plays an important role in regulating cell fate and tissue structure.401 The Hippo pathway in Drosophila contains four core components, namely, the NDR family protein kinase Warts (Wts), the WW domain-containing protein Salvador (Sav), the Ste20-like protein kinase Hippo (Hpo), and the adaptor protein Mob as a tumor suppressor (Mats). The mammalian orthologs for each of these are LATS1/2 kinase (Wts), SAV1 (Sav), MST1/2 (Hpo), and MOB1 (Mats). MST1/2 forms heterodimers with SAV1 for the phosphorylation of SAV1, MOB1, and LATS1/2. This enables LATS1/2 to phosphorylate yes-associated protein (YAP) and WW domain-containing transcription regulator protein 1 (TAZ). YAP/TAZ are orthologs of the transcriptional coactivator called Yorkie (Yki), which normally stimulates transcription. The phosphorylation of YAP/TAZ by LATS1/2 and the Hippo kinase cascade inhibits transcription.401 The Hippo pathway interacts with Wnt signaling in several ways.
Phosphorylation of the HXRXXS motif of Ser381 of YAP by LATS1/2 can provide a signal for CK1 to phosphorylate a phosphodegron, which is a DSGXXS destruction motif. This causes the recruitment of β-TrCP, which leads to YAP ubiquitination and eventual degradation, inhibiting YAP and preventing gene transcription.402 TAZ is also degraded via phosphorylation by CK1 in the phosphodegron and subsequent ubiquitination and degradation.403 CK1 can also be bound by MST1/2, causing sequestration of CK1 and preventing it from phosphorylating Dvl, leading to inhibition of Wnt signaling.404 An analysis of overexpression of YAP/TAZ in cells found that YAP/TAZ bound and suppressed β-catenin without affecting β-catenin levels, suggesting the preservation of β-catenin stability.405 However, other reports indicate that when Wnt signaling is inactivated, YAP/TAZ normally binds β-catenin as part of the destruction complex. This association of YAP/TAZ to the destruction complex (via Axin, specifically) recruits β-TrCP, degrading β-catenin.406 Further experimentation may be needed to determine whether YAP/TAZ affects the stability of β-catenin or not.
When Wnt signaling is activated, YAP/TAZ is released from this complex and moves to the nucleus to activate the Hippo pathway.406,407 YAP/TAZ can also bind to Dvl, suppressing its phosphorylation and downstream effects on Wnt signaling.405 YAP can also interact with Src homology 2 domain tyrosine phosphatase (SHP2), an amino acid phosphatase that enhances β-catenin activity in the nucleus. This enables YAP to dampen Wnt signaling strength.407 These findings indicate that YAP/TAZ can serve as mediators or antagonists of Wnt signaling, where activation of Wnt signaling enables YAP/TAZ to transcribe Hippo pathway target genes, while inactivation leads to the antagonism of β-catenin.405, 406, 407 The noncanonical Wnt5a/b and Wnt3a ligands can activate YAP/TAZ as mediators of noncanonical Wnt signaling.408 Dysregulation of the Hippo pathway causes resistance to apoptosis and even chemotherapeutic treatments, leading to tumorigenesis, metastasis, and cancer relapse.401 The inhibition of Wnt signaling by Hippo is also crucial in controlling heart size by limiting cardiomyocyte proliferation.409
Crosstalk with Hedgehog (Hh) signaling
The Hedgehog (Hh) pathway can be divided into canonical and noncanonical pathways. The canonical pathway is activated by the Shh ligand binding to the transmembrane protein Patched (Ptch1), which normally inhibits another transmembrane protein called Smo. The binding of Shh leads to Ptch1 degradation, causing Smo accumulation at the primary cilium (PC) which stimulates a signaling cascade. This leads to the Gli family of proteins translocating to the nucleus and transcribing genes, including Ptch1 for negative feedback and Gli1 for positive feedback.410 Noncanonical signaling is independent of Gli and can be further subdivided into two types, type I which modulates Ca2+ and the actin cytoskeleton via downstream actions of Smo, and type II which increases cell proliferation and survival and is Smo-independent.410
Hh signaling can recruit CK1 to phosphorylate Smo to increase its cell-surface accumulation, leading to stronger Hh signaling.411,412 CK1 can also stabilize Gli by phosphorylating Ser/Thr-rich motifs in the N- and C-terminal regions of Gli to prevent degradation by HIB.413 Additionally, CK1 can phosphorylate and activate the Fused (Fu) protein kinase, a kinase that phosphorylates Gli for activation.414 Further regulation of Hh signaling can be induced by GSK-3β, which can phosphorylate suppressor of fused (SUFU), which normally binds the Gli transcription factors and prevents them from entering the nucleus. Phosphorylation of SUFU by GSK-3β prevents SUFU from binding to the Gli transcription factors, presenting a positive regulatory role for GSK-3β in Hh signaling.415 One of the downstream genes that are transcribed by the Hh signaling cascade is sFRP1. Specifically, Gli1 and Gli2 are responsible for the transcription. In other words, the activation of Hh signaling inhibits Wnt signaling via the production of sFRP1, indicating a regulatory role of the Hh pathway for the Wnt pathway.416 Furthermore, SUFU can also decrease β-catenin levels via nuclear export and in turn down-regulate TCF-dependent transcription.417
Crosstalk with fibroblast growth factor (FGF) signaling
Fibroblast growth factors (FGFs) are a family of 22 polypeptide growth factors in seven subfamilies that are involved in diverse cellular processes. The binding of FGFs to FGF receptors (FGFRs) causes receptor dimerization that is promoted by heparan sulfate. This leads to autophosphorylation of FGFR that can activate the MAPK, phosphatidylinositol-3 kinase/Akt (PI3K/Akt), and phospholipase C-γ (PLC-γ) pathways.418 They also have interactions with the Wnt signaling pathway. Akt from the PI3K/Akt pathway phosphorylates Ser9 on GSK-3β, decreasing its activity. This reduces the ability of GSK-3β to phosphorylate SNAIL for degradation. SNAIL usually represses transcription of the E-cadherin gene, CDH1, which maintains cell adhesion. In this manner, FGF signaling can stimulate the EMT via decreasing E-cadherin levels by GSK-3β inhibition.419 Activation of the PI3K/Akt pathway by FGF also releases β-catenin from a complex composed of E-cadherin, β-catenin, and α-catenin, enabling β-catenin to translocate to the nucleus.419 FGF18 and FGF20 are target genes of the canonical Wnt pathway, indicating that Wnt signaling can mediate FGF signaling activation.419 In the development of the anterior heart field (AHF) in mice, Wnt/β-catenin signaling regulates the expression of FGF3, 10, 16, and 20 in AHF progenitor cells.420 Mesenchymal FGF signaling is vital in Wnt2a expression stabilizing β-catenin, and Wnt/β-catenin signaling sustains expression of FGFR1c/2c and responsiveness to FGF9, creating a positive feedback loop for mesenchymal proliferation in the lung.421
Crosstalk with parathyroid hormone (PTH) signaling
Parathyroid hormone (PTH) is a protein implicated in bone remodeling and acts through two G-protein coupled pathways, the Gs/cAMP/protein kinase A (PKA) pathway and the Gq/PLC/Ca2+/protein kinase C (PKC) pathway.422 The bone repair effects of PTH are partly mediated by Wnt signaling, as PTH-treated bones had increased numbers of β-catenin-expressing osteoblastic cells in the fracture callus. These fracture calluses also demonstrated increased levels of Wnt4, 5a, 5b, 10, and 11.423 PTH also enhances the transcriptional activity induced by β-catenin by promoting the expression of Smad3.424 Additionally, PTH appears to be able to stimulate Wnt/β-catenin signaling to prevent apoptosis, but this does not require PTH, suggesting the need for further studies to clarify this role.424 PTH can stimulate osteoclastogenesis in an opposing manner to Wnt signaling, which inhibits osteoclastogenesis. PTH and Wnt signaling modulates β-catenin in different manners to cause these opposite effects.425 The PTH receptor (PTH1R) can recruit Dvl and activate β-catenin signaling without Wnt or LRP5/6. This association of PTH1R and Dvl is what enables PTH-induced osteoclastogenesis.425
Crosstalk with other pathways
Wnt signaling can interact with multiple other pathways in addition to the BMP, Notch, and Hippo signaling pathways. Analysis of mouse hepatocytes found that high insulin levels can stimulate the Wnt/β-catenin pathway to promote lipogenic gene expression. Insulin activates stearoyl-CoA-desaturase 1 (SCD1), an enzyme involved in lipogenesis that supplies palmitoleate for Porc, enabling it to acylate Wnt ligands for secretion.426 Insulin can also enhance Wnt signaling by activating Akt to phosphorylate and inactivate GSK-3β, which leads to increased glycogen storage, glucose clearance, and insulin sensitivity. As such, GSK-3β inhibitors have become a therapeutic target in the treatment of type II diabetes.318
PP2A is another component of Wnt signaling that is involved in other pathways. For instance, it is involved in the mechanistic target of the rapamycin (mTOR) pathway, which is vital in cell growth and metabolism.342 PP2A can negatively regulate the mTOR signaling pathway by dephosphorylating and inactivating Akt, which is normally activated by mTOR. PP2A can also inhibit IRS1, which is upstream of Akt. However, mTOR can also inhibit PP2A, suggesting a complex interaction between Wnt signaling and mTOR signaling.342 PP2A also has a major role in both positively and negatively regulating the MAPK pathway. In terms of negative regulation, PP2A can bind the phosphotyrosine-binding (PTB) domain of Shc, an adaptor protein that acts as a signal transducer in the Ras/MAPK pathway. PP2A can inhibit MAPK activation, but this association of PP2A to Shc can be alleviated by growth factor stimulation or small-t antigen expression.427 For positive regulation, the PR130 regulatory subunit of PP2A can form a holoenzyme that complexes with SRC homology 2 domain-containing inositol polyphosphate phosphatase (SHIP2). This complex stabilizes the EGF receptors (EGFR) that are necessary for the ERK/MAPK signaling pathway.428
Conclusions and future directions
The study of Wnt signaling has progressively provided more insight into this complex pathway. The structures and many interactions of the Wnt signaling components have been established, providing a breadth of knowledge about the crosstalk capability of Wnt signaling with a heavy emphasis on its vast effects in different organs. The conservation of the Wnt ligands, Fz receptors, and coreceptors, has further driven home the fundamental importance of the Wnt signaling pathway. Additionally, the antagonists of Wnt signaling also have functions beyond just the Wnt pathway, adding further complexity to the interactions of Wnt signaling with other pathways.
However, many details are still needed to be clarified. For the 19 Wnt ligands, it is not completely clear how each ligand interacts with Fz receptors and LRP5/6 coreceptors. The specificity of the interaction between these receptors and the Wnt ligands needs to be further clarified. Further research continues to uncover more and more pathways with which Wnt signaling interacts, yet the number of pathways and proteins that crosstalk with Wnt signaling is still not fully determined. Furthermore, the mechanisms of many Wnt signaling interactions with other pathways are not fully elucidated. The functions of each component of the Wnt signaling pathway are diverse and may still not be completely known yet.
Due to the implication of dysregulated Wnt signaling in diseases such as cancer, therapeutics have been developed. However, like the Wnt signaling pathway and components, the therapeutics also require caution in their function and use. Both activators and inhibitors of the Wnt signaling pathway can come with their own risk of tumorigenesis or unintentional inhibition of important cellular functions, which can lead to organ dysfunction. Furthermore, if an activator or inhibitor is affected by treatment, it may be unknown what potential effects can occur if that activator or inhibitor is also involved in another pathway. Additional research is needed to further clarify the therapeutic effects of various treatment options, as well as ensure their safety.
Although the current analysis of Wnt signaling is not completely exhaustive, the broad amount of research on the Wnt signaling pathway has provided us with significant insight into the system and its many functions in living organisms over the last 40 years. Hopefully, in the future, more research will be performed to provide more knowledge on the Wnt signaling pathway, leading to further advancements in biology, medicine, and human health.
Conflict of interests
Tong-Chuan He is the Editor-in-Chief of Genes & Diseases. To minimize bias, he was excluded from all editorial decision-making related to the acceptance of this article for publication. The remaining authors declare no conflict of interests.
Funding
The reported work was supported in part by research grants from the National Institutes of Health (No. CA226303 to TCH and No. DE030480 to RRR) and the American Shoulder and Elbow Surgeons PJI Research Grant (LLS). JF was supported in part by research grants from the Natural Science Foundation of China (No. 82102696), the 2019 Science and Technology Research Plan Project of Chongqing Education Commission (China) (No. KJQN201900410), and the 2019 Funding for Postdoctoral Research (Chongqing Human Resources and Social Security Bureau No. 298). WW was supported by the Medical Scientist Training Program of the National Institutes of Health (No. T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (No. P30CA014599) and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health through Grant Number 5UL1TR002389. TCH was also supported by the Mabel Green Myers Research Endowment Fund and The University of Chicago Orthopaedics Alumni Fund. Funding sources were not involved in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication.
Acknowledgements
The authors apologize to the investigators whose work cannot be cited in the manuscript due to space constraints. We here acknowledge the investigators for their contribution to this work.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Tong-Chuan He, Email: tche@uchicago.edu.
Lewis L. Shi, Email: lshi@bsd.uchicago.edu.
References
- 1.Nusse R., Varmus H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 2.Nusse R., Varmus H. Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J. 2012;31(12):2670–2684. doi: 10.1038/emboj.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Ooyen A., Kwee V., Nusse R. The nucleotide sequence of the human int-1 mammary oncogene; evolutionary conservation of coding and non-coding sequences. EMBO J. 1985;4(11):2905–2909. doi: 10.1002/j.1460-2075.1985.tb04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma R.P., Chopra V.L. Effect of the Wingless (wg1) mutation on the wing and haltere development in Drosophila melanogaster. Dev Biol. 1976;48(2):461–465. doi: 10.1016/0012-1606(76)90108-1. [DOI] [PubMed] [Google Scholar]
- 5.Uzvölgyi E., Kiss I., Pitt A., et al. Drosophila homolog of the murine Int-1 protooncogene. Proc Natl Acad Sci U S A. 1988;85(9):3034–3038. doi: 10.1073/pnas.85.9.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker N.E. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 1987;6(6):1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rijsewijk F., Schuermann M., Wagenaar E., et al. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987;50(4):649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 8.Nusse R., Brown A., Papkoff J., et al. A new nomenclature for int-1 and related genes: the Wnt gene family. Cell. 1991;64(2):231. doi: 10.1016/0092-8674(91)90633-a. [DOI] [PubMed] [Google Scholar]
- 9.He T.C., Sparks A.B., Rago C., et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 10.He T.C., Chan T.A., Vogelstein B., et al. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.H., Liu X., Wang J., et al. Wnt signaling in bone formation and its therapeutic potential for bone diseases. Ther Adv Musculoskelet Dis. 2013;5(1):13–31. doi: 10.1177/1759720X12466608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammed M.K., Shao C., Wang J., et al. Wnt/β-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016;3(1):11–40. doi: 10.1016/j.gendis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K., Wang X., Zhang H., et al. The evolving roles of canonical WNT signaling in stem cells and tumorigenesis: implications in targeted cancer therapies. Lab Invest. 2016;96(2):116–136. doi: 10.1038/labinvest.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayat R., Manzoor M., Hussain A. Wnt signaling pathway: a comprehensive review. Cell Biol Int. 2022;46(6):863–877. doi: 10.1002/cbin.11797. [DOI] [PubMed] [Google Scholar]
- 16.Fan J., Wei Q., Liao J., et al. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8(16):27105–27119. doi: 10.18632/oncotarget.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chae W.J., Bothwell A.L.M. Canonical and non-canonical Wnt signaling in immune cells. Trends Immunol. 2018;39(10):830–847. doi: 10.1016/j.it.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 19.He X., Semenov M., Tamai K., et al. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131(8):1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 20.Janda C.Y., Waghray D., Levin A.M., et al. Structural basis of Wnt recognition by frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Chang H., Rattner A., et al. Frizzled receptors in development and disease. Curr Top Dev Biol. 2016;117:113–139. doi: 10.1016/bs.ctdb.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamos J.L., Weis W.I. The β-catenin destruction complex. Cold Spring Harbor Perspect Biol. 2013;5(1):a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J.J., Williams C.S. Protein phosphatase 2A in the regulation of Wnt signaling, stem cells, and cancer. Genes. 2018;9(3):121. doi: 10.3390/genes9030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang S., Hua F., Hu Z.W. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue J., Chen Y., Wu Y., et al. Tumour suppressor TRIM33 targets nuclear β-catenin degradation. Nat Commun. 2015;6:6156. doi: 10.1038/ncomms7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan P., Bonewald L.F. The role of the Wnt/β-catenin signaling pathway in formation and maintenance of bone and teeth. Int J Biochem Cell Biol. 2016;77:23–29. doi: 10.1016/j.biocel.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva-García O., Valdez-Alarcón J.J., Baizabal-Aguirre V.M. Wnt/β-catenin signaling as a molecular target by pathogenic bacteria. Front Immunol. 2019;10:2135. doi: 10.3389/fimmu.2019.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doumpas N., Lampart F., Robinson M.D., et al. TCF/LEF dependent and independent transcriptional regulation of Wnt/β-catenin target genes. EMBO J. 2019;38(2) doi: 10.15252/embj.201798873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan T., Rindtorff N., Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Chen Z., Tang Y., et al. The involvement of noncanonical Wnt signaling in cancers. Biomed Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.110946. [DOI] [PubMed] [Google Scholar]
- 31.Berger H., Wodarz A., Borchers A. PTK7 faces the Wnt in development and disease. Front Cell Dev Biol. 2017;5:31. doi: 10.3389/fcell.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messéant J., Dobbertin A., Girard E., et al. MuSK frizzled-like domain is critical for mammalian neuromuscular junction formation and maintenance. J Neurosci. 2015;35(12):4926–4941. doi: 10.1523/JNEUROSCI.3381-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh M., Katoh M. Molecular genetics and targeted therapy of WNT-related human diseases (Review) Int J Mol Med. 2017;40(3):587–606. doi: 10.3892/ijmm.2017.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bovolenta P., Rodriguez J., Esteve P. Frizzled/RYK mediated signalling in axon guidance. Development. 2006;133(22):4399–4408. doi: 10.1242/dev.02592. [DOI] [PubMed] [Google Scholar]
- 35.Pataki C.A., Couchman J.R., Brábek J. Wnt signaling cascades and the roles of syndecan proteoglycans. J Histochem Cytochem. 2015;63(7):465–480. doi: 10.1369/0022155415586961. [DOI] [PubMed] [Google Scholar]
- 36.van Andel H., Kocemba K.A., Spaargaren M., et al. Aberrant Wnt signaling in multiple myeloma: molecular mechanisms and targeting options. Leukemia. 2019;33(5):1063–1075. doi: 10.1038/s41375-019-0404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolluri A., Ho M. The role of glypican-3 in regulating Wnt, YAP, and Hedgehog in liver cancer. Front Oncol. 2019;9:708. doi: 10.3389/fonc.2019.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Moraes G.F.A., Listik E., Justo G.Z., et al. The Glypican proteoglycans show intrinsic interactions with Wnt-3a in human prostate cancer cells that are not always associated with cascade activation. BMC Mol Cell Biol. 2021;22:26. doi: 10.1186/s12860-021-00361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X., Zhang M., Xu F., et al. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. 2020;19:165. doi: 10.1186/s12943-020-01276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenny A., Reynolds-Kenneally J., Das G., et al. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat Cell Biol. 2005;7(7):691–697. doi: 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- 41.Narimatsu M., Bose R., Pye M., et al. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009;137(2):295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 42.Barkó S., Bugyi B., Carlier M.F., et al. Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J Biol Chem. 2010;285(17):13154–13169. doi: 10.1074/jbc.M109.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coso O.A., Chiariello M., Yu J.C., et al. The small GTP-binding proteins Rac 1 and Cdc 42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81(7):1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Mlodzik M. Wnt-Frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt) Annu Rev Cell Dev Biol. 2015;31:623–646. doi: 10.1146/annurev-cellbio-100814-125315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyers C.E., McNeill H., Knebel A., et al. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem J. 2005;389(Pt 1):127–135. doi: 10.1042/BJ20050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitzing T.M., Sahadevan A.S., Brandt D.T., et al. Positive feedback between Dia 1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21(12):1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kühn S., Geyer M. Formins as effector proteins of Rho GTPases. Small GTPases. 2014;5(3) doi: 10.4161/sgtp.29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamao K., Ono T., Matsushita M., et al. ZIP kinase phosphorylated and activated by Rho kinase/ROCK contributes to cytokinesis in mammalian cultured cells. Exp Cell Res. 2020;386(1) doi: 10.1016/j.yexcr.2019.111707. [DOI] [PubMed] [Google Scholar]
- 49.Oyama H., Nukuda A., Ishihara S., et al. Soft surfaces promote astrocytic differentiation of mouse embryonic neural stem cells via dephosphorylation of MRLC in the absence of serum. Sci Rep. 2021;11 doi: 10.1038/s41598-021-99059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slusarski D.C., Corces V.G., Moon R.T. Interaction of Wnt and a frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997;390(6658):410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 51.De A. Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin. 2011;43(10):745–756. doi: 10.1093/abbs/gmr079. [DOI] [PubMed] [Google Scholar]
- 52.Kühl M., Sheldahl L.C., Park M., et al. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16(7):279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- 53.McQuate A., Latorre-Esteves E., Barria A. A Wnt/calcium signaling cascade regulates neuronal excitability and trafficking of NMDARs. Cell Rep. 2017;21(1):60–69. doi: 10.1016/j.celrep.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Hooper R., Zhang X., Webster M., et al. Novel protein kinase C-mediated control of Orai 1 function in invasive melanoma. Mol Cell Biol. 2015;35(16):2790–2798. doi: 10.1128/MCB.01500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H.Y., Malbon C.C. Wnt signaling, Ca2+, and cyclic GMP: visualizing Frizzled functions. Science. 2003;300(5625):1529–1530. doi: 10.1126/science.1085259. [DOI] [PubMed] [Google Scholar]
- 56.Heit J.J., Apelqvist A.A., Gu X., et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443(7109):345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 57.Ishitani T., Kishida S., Hyodo-Miura J., et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23(1):131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishitani T., Ninomiya-Tsuji J., Nagai S., et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399(6738):798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 59.Petersen C.P., Reddien P.W. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139(6):1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka K., Okabayashi K., Asashima M., et al. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. Eur J Biochem. 2000;267(13):4300–4311. doi: 10.1046/j.1432-1033.2000.01478.x. [DOI] [PubMed] [Google Scholar]
- 61.Valenta T., Degirmenci B., Moor A.E., et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 2016;15(5):911–918. doi: 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- 62.Moon R.T., Kohn A.D., De Ferrari G.V., et al. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 63.Peradziryi H., Kaplan N.A., Podleschny M., et al. PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling. EMBO J. 2011;30(18):3729–3740. doi: 10.1038/emboj.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubin E.M., Guo Y., Tu K., et al. Wnt inhibitory factor 1 decreases tumorigenesis and metastasis in osteosarcoma. Mol Cancer Therapeut. 2010;9(3):731–741. doi: 10.1158/1535-7163.MCT-09-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao W.N., Cheng C., Theriault K.M., et al. A high-throughput screen for Wnt/β-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J Biomol Screen. 2012;17(9):1252–1263. doi: 10.1177/1087057112456876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z., Xiu Y., Qiu F., et al. Canonical Wnt signaling drives Myopia development and can be pharmacologically modulated. Invest Ophthalmol Vis Sci. 2021;62(9):21. doi: 10.1167/iovs.62.9.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abu-Remaileh M., Gerson A., Farago M., et al. Oct-3/4 regulates stem cell identity and cell fate decisions by modulating Wnt/β-catenin signalling. EMBO J. 2010;29(19):3236–3248. doi: 10.1038/emboj.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawano Y., Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116(Pt 13):2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 69.Ho H.H., Susman M.W., Bikoff J.B., et al. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A. 2012;109(11):4044–4051. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Susman M.W., Karuna E.P., Kunz R.C., et al. Kinesin superfamily protein Kif26b links Wnt5a-Ror signaling to the control of cell and tissue behaviors in vertebrates. Elife. 2017;6 doi: 10.7554/eLife.26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karuna E.P., Susman M.W., Ho H.H. Quantitative live-cell reporter assay for noncanonical Wnt activity. Bio Protoc. 2018;8(6) doi: 10.21769/BioProtoc.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mills K.M., Szczerkowski J.L.A., Habib S.J. Wnt ligand presentation and reception: from the stem cell niche to tissue engineering. Open Biol. 2017;7(8) doi: 10.1098/rsob.170140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rocheleau C.E., Downs W.D., Lin R., et al. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90(4):707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 74.Bänziger C., Soldini D., Schütt C., et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125(3):509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 75.Kjellén L., Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 76.Lin X., Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature. 1999;400(6741):281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 77.Wodarz A., Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 78.Huang H.C., Klein P.S. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004;5(7):234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park W.J., Liu J., Adler P.N. The frizzled gene of Drosophila encodes a membrane protein with an odd number of transmembrane domains. Mech Dev. 1994;45(2):127–137. doi: 10.1016/0925-4773(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y., Macke J.P., Abella B.S., et al. A large family of putative transmembrane receptors homologous to the product of the Drosophila tissue polarity gene frizzled. J Biol Chem. 1996;271(8):4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 81.Fredriksson R., Lagerström M.C., Lundin L.G., et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 82.Vinson C.R., Adler P.N. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature. 1987;329(6139):549–551. doi: 10.1038/329549a0. [DOI] [PubMed] [Google Scholar]
- 83.Wong L.L., Adler P.N. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J Cell Biol. 1993;123(1):209–221. doi: 10.1083/jcb.123.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adler P.N. The genetic control of tissue polarity in Drosophila. Bioessays. 1992;14(11):735–741. doi: 10.1002/bies.950141103. [DOI] [PubMed] [Google Scholar]
- 85.Zheng L., Zhang J., Carthew R.W. Frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121(9):3045–3055. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]
- 86.Gilmour D.F. Familial exudative vitreoretinopathy and related retinopathies. Eye. 2015;29(1):1–14. doi: 10.1038/eye.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu H., Smallwood P.M., Wang Y., et al. Frizzled 1 and frizzled 2 genes function in palate, ventricular septum and neural tube closure: general implications for tissue fusion processes. Development. 2010;137(21):3707–3717. doi: 10.1242/dev.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishikawa T., Tamai Y., Zorn A.M., et al. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development. 2001;128(1):25–33. doi: 10.1242/dev.128.1.25. [DOI] [PubMed] [Google Scholar]
- 89.Liu C., Nathans J. An essential role for frizzled 5 in mammalian ocular development. Development. 2008;135(21):3567–3576. doi: 10.1242/dev.028076. [DOI] [PubMed] [Google Scholar]
- 90.Liu C., Bakeri H., Li T., et al. Regulation of retinal progenitor expansion by Frizzled receptors: implications for microphthalmia and retinal coloboma. Hum Mol Genet. 2012;21(8):1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y., Badea T., Nathans J. Order from disorder: self-organization in mammalian hair patterning. Proc Natl Acad Sci U S A. 2006;103(52):19800–19805. doi: 10.1073/pnas.0609712104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hua Z.L., Jeon S., Caterina M.J., et al. Frizzled 3 is required for the development of multiple axon tracts in the mouse central nervous system. Proc Natl Acad Sci U S A. 2014;111(29):E3005–E3014. doi: 10.1073/pnas.1406399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y., Guo N., Nathans J. The role of Frizzled 3 and Frizzled 6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J Neurosci. 2006;26(8):2147–2156. doi: 10.1523/JNEUROSCI.4698-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 95.Tamai K., Zeng X., Liu C., et al. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13(1):149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 96.Wu G., Huang H., Garcia Abreu J., et al. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stamos J.L., Chu M.L., Enos M.D., et al. Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6. Elife. 2014;3 doi: 10.7554/eLife.01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mao J., Wang J., Liu B., et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7(4):801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 99.Pinson K.I., Brennan J., Monkley S., et al. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407(6803):535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 100.Kato M., Patel M.S., Levasseur R., et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157(2):303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujino T., Asaba H., Kang M.J., et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci U S A. 2003;100(1):229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Lau W.B.M., Snel B., Clevers H.C. The R-spondin protein family. Genome Biol. 2012;13(3):242. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kamata T., Katsube K., Michikawa M., et al. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim Biophys Acta. 2004;1676(1):51–62. doi: 10.1016/j.bbaexp.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 104.Kazanskaya O., Glinka A., del Barco Barrantes I., et al. R-Spondin 2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7(4):525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 105.Carmon K.S., Gong X., Lin Q., et al. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tocci J.M., Felcher C.M., García Solá M.E., et al. R-spondin-mediated WNT signaling potentiation in mammary and breast cancer development. IUBMB Life. 2020;72(8):1546–1559. doi: 10.1002/iub.2278. [DOI] [PubMed] [Google Scholar]
- 107.Zebisch M., Xu Y., Krastev C., et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4:2787. doi: 10.1038/ncomms3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Araki T., Milbrandt J. ZNRF proteins constitute a family of presynaptic E3 ubiquitin ligases. J Neurosci. 2003;23(28):9385–9394. doi: 10.1523/JNEUROSCI.23-28-09385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hao H.X., Xie Y., Zhang Y., et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 110.Lu W., Yamamoto V., Ortega B., et al. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119(1):97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 111.Hovens C.M., Stacker S.A., Andres A.C., et al. RYK, a receptor tyrosine kinase-related molecule with unusual kinase domain motifs. Proc Natl Acad Sci U S A. 1992;89(24):11818–11822. doi: 10.1073/pnas.89.24.11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dura J.M., Preat T., Tully T. Identification of linotte, a new gene affecting learning and memory in Drosophila melanogaster. J Neurogenet. 2007;21(4):307–320. doi: 10.1080/01677060701693479. [DOI] [PubMed] [Google Scholar]
- 113.Yoshikawa S., McKinnon R.D., Kokel M., et al. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 2003;422(6932):583–588. doi: 10.1038/nature01522. [DOI] [PubMed] [Google Scholar]
- 114.Inoue T., Oz H.S., Wiland D., et al. C. elegans LIN-18 is a ryk ortholog and functions in parallel to LIN-17/frizzled in Wnt signaling. Cell. 2004;118(6):795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 115.Menck K., Heinrichs S., Baden C., et al. The WNT/ROR pathway in cancer: from signaling to therapeutic intervention. Cells. 2021;10(1):142. doi: 10.3390/cells10010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Masiakowski P., Carroll R.D. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992;267(36):26181–26190. [PubMed] [Google Scholar]
- 117.Saldanha J., Singh J., Mahadevan D. Identification of a Frizzled-like cysteine rich domain in the extracellular region of developmental receptor tyrosine kinases. Protein Sci. 1998;7(7):1632–1635. doi: 10.1002/pro.5560070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Matsuda T., Nomi M., Ikeya M., et al. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech Dev. 2001;105(1–2):153–156. doi: 10.1016/s0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 119.Oldridge M., Fortuna A.M., Maringa M., et al. Dominant mutations in ROR2, encoding an orphan receptor tyrosine kinase, cause brachydactyly type B. Nat Genet. 2000;24(3):275–278. doi: 10.1038/73495. [DOI] [PubMed] [Google Scholar]
- 120.Afzal A.R., Rajab A., Fenske C.D., et al. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat Genet. 2000;25(4):419–422. doi: 10.1038/78107. [DOI] [PubMed] [Google Scholar]
- 121.van Bokhoven H., Celli J., Kayserili H., et al. Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat Genet. 2000;25(4):423–426. doi: 10.1038/78113. [DOI] [PubMed] [Google Scholar]
- 122.Petrova I.M., Malessy M.J., Verhaagen J., et al. Wnt signaling through the Ror receptor in the nervous system. Mol Neurobiol. 2014;49(1):303–315. doi: 10.1007/s12035-013-8520-9. [DOI] [PubMed] [Google Scholar]
- 123.Broome H.E., Rassenti L.Z., Wang H.Y., et al. ROR1 is expressed on hematogones (non-neoplastic human B-lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk Res. 2011;35(10):1390–1394. doi: 10.1016/j.leukres.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oishi I., Takeuchi S., Hashimoto R., et al. Spatio-temporally regulated expression of receptor tyrosine kinases, mRor1, mRor2, during mouse development: implications in development and function of the nervous system. Gene Cell. 1999;4(1):41–56. doi: 10.1046/j.1365-2443.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 125.Eubelen M., Bostaille N., Cabochette P., et al. A molecular mechanism for Wnt ligand-specific signaling. Science. 2018;361(6403) doi: 10.1126/science.aat1178. [DOI] [PubMed] [Google Scholar]
- 126.Posokhova E., Shukla A., Seaman S., et al. GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. 2015;10(2):123–130. doi: 10.1016/j.celrep.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vallon M., Yuki K., Nguyen T.D., et al. A RECK-WNT7 receptor-ligand interaction enables isoform-specific regulation of Wnt bioavailability. Cell Rep. 2018;25(2):339–349.e9. doi: 10.1016/j.celrep.2018.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chang J., Mancuso M.R., Maier C., et al. Gpr124 is essential for blood-brain barrier integrity in central nervous system disease. Nat Med. 2017;23(4):450–460. doi: 10.1038/nm.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8(6):1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wieschaus E., Nüsslein-Volhard C., Jürgens G. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster: III. Zygotic loci on the X-chromosome and fourth chromosome. Wilehm Roux Arch Dev Biol. 1984;193(5):296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 131.Riggleman B., Schedl P., Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 1990;63(3):549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- 132.Huber O., Korn R., McLaughlin J., et al. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59(1):3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 133.Huber A.H., Nelson W.J., Weis W.I. Three-dimensional structure of the Armadillo repeat region of β-catenin. Cell. 1997;90(5):871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 134.Graham T.A., Weaver C., Mao F., et al. Crystal structure of a β-catenin/Tcf complex. Cell. 2000;103(6):885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 135.Zhurinsky J., Shtutman M., Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000;113(Pt 18):3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
- 136.Eklof Spink K., Fridman S.G., Weis W.I. Molecular mechanisms of beta-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-beta-catenin complex. EMBO J. 2001;20(22):6203–6212. doi: 10.1093/emboj/20.22.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huber A.H., Weis W.I. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105(3):391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 138.Xing Y., Clements W.K., Kimelman D., et al. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17(22):2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ha N.C., Tonozuka T., Stamos J.L., et al. Mechanism of phosphorylation-dependent binding of APC to β-catenin and its role in β-catenin degradation. Mol Cell. 2004;15(4):511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 140.Hrckulak D., Kolar M., Strnad H., et al. TCF/LEF transcription factors: an update from the Internet resources. Cancers. 2016;8(7):70. doi: 10.3390/cancers8070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van de Wetering M., Cavallo R., Dooijes D., et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88(6):789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 142.Atcha F.A., Syed A., Wu B., et al. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27(23):8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sharma M., Castro-Piedras I., Simmons G.E., Jr., et al. Dishevelled: a masterful conductor of complex Wnt signals. Cell Signal. 2018;47:52–64. doi: 10.1016/j.cellsig.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schwarz-Romond T., Fiedler M., Shibata N., et al. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14(6):484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 145.Moon R.T., Shah K. Developmental biology: signalling polarity. Nature. 2002;417(6886):239–240. doi: 10.1038/417239a. [DOI] [PubMed] [Google Scholar]
- 146.Umbhauer M., Djiane A., Goisset C., et al. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signalling. EMBO J. 2000;19(18):4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wong H.C., Bourdelas A., Krauss A., et al. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell. 2003;12(5):1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pan W.J., Pang S.Z., Huang T., et al. Characterization of function of three domains in dishevelled-1:DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 2004;14(4):324–330. doi: 10.1038/sj.cr.7290232. [DOI] [PubMed] [Google Scholar]
- 149.Boligala G.P., Yang M.V., van Wunnik J.C., et al. Nuclear Dishevelled: an enigmatic role in governing cell fate and Wnt signaling. Biochim Biophys Acta Mol Cell Res. 2022;1869(10) doi: 10.1016/j.bbamcr.2022.119305. [DOI] [PubMed] [Google Scholar]
- 150.Mochizuki T., Saijoh Y., Tsuchiya K., et al. Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature. 1998;395(6698):177–181. doi: 10.1038/26006. [DOI] [PubMed] [Google Scholar]
- 151.Simons M., Gloy J., Ganner A., et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37(5):537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cai Y., Stafford L.J., Bryan B.A., et al. G-protein-activated phospholipase C-beta, new partners for cell polarity proteins Par 3 and Par 6. Oncogene. 2005;24(26):4293–4300. doi: 10.1038/sj.onc.1208593. [DOI] [PubMed] [Google Scholar]
- 153.Izzi L., Attisano L. Ubiquitin-dependent regulation of TGβ signaling in cancer. Neoplasia. 2006;8(8):677–688. doi: 10.1593/neo.06472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhu H., Kavsak P., Abdollah S., et al. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400(6745):687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 155.Lin X., Liang M., Feng X.H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad 2 in transforming growth factor-β signaling. J Biol Chem. 2000;275(47):36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 156.Sahai E., Garcia-Medina R., Pouysségur J., et al. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol. 2007;176(1):35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ozdamar B., Bose R., Barrios-Rodiles M., et al. Regulation of the polarity protein Par 6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307(5715):1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 158.Gubb D., Green C., Huen D., et al. The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev. 1999;13(17):2315–2327. doi: 10.1101/gad.13.17.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dawid I.B., Toyama R., Taira M. LIM domain proteins. C R Acad Sci III. 1995;318(3):295–306. [PubMed] [Google Scholar]
- 160.Sweede M., Ankem G., Chutvirasakul B., et al. Structural and membrane binding properties of the prickle PET domain. Biochemistry. 2008;47(51):13524–13536. doi: 10.1021/bi801037h. [DOI] [PubMed] [Google Scholar]
- 161.Veeman M.T., Slusarski D.C., Kaykas A., et al. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 162.Chougule A., Lapraz F., Földi I., et al. The Drosophila actin nucleator DAAM is essential for left-right asymmetry. PLoS Genet. 2020;16(4) doi: 10.1371/journal.pgen.1008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lebreton G., Géminard C., Lapraz F., et al. Molecular to organismal chirality is induced by the conserved myosin 1D. Science. 2018;362(6417):949–952. doi: 10.1126/science.aat8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carlsson L., Nyström L.E., Sundkvist I., et al. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115(3):465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 165.Tilney L.G. The polymerization of actin. III. Aggregates of nonfilamentous actin and its associated proteins: a storage form of actin. J Cell Biol. 1976;69(1):73–89. doi: 10.1083/jcb.69.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karlsson R., Dráber P. Profilin-A master coordinator of actin and microtubule organization in mammalian cells. J Cell Physiol. 2021;236(10):7256–7265. doi: 10.1002/jcp.30379. [DOI] [PubMed] [Google Scholar]
- 167.Pinto-Costa R., Sousa M.M. Profilin as a dual regulator of actin and microtubule dynamics. Cytoskeleton. 2020;77(3–4):76–83. doi: 10.1002/cm.21586. [DOI] [PubMed] [Google Scholar]
- 168.Nejedla M., Sadi S., Sulimenko V., et al. Profilin connects actin assembly with microtubule dynamics. Mol Biol Cell. 2016;27(15):2381–2393. doi: 10.1091/mbc.E15-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sato A., Khadka D.K., Liu W., et al. Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development. 2006;133(21):4219–4231. doi: 10.1242/dev.02590. [DOI] [PubMed] [Google Scholar]
- 170.Nguyen L.K., Kholodenko B.N., von Kriegsheim A. Rac 1 and RhoA: networks, loops and bistability. Small GTPases. 2018;9(4):316–321. doi: 10.1080/21541248.2016.1224399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kotelevets L., Chastre E. Rac 1 signaling: from intestinal homeostasis to colorectal cancer metastasis. Cancers. 2020;12(3):665. doi: 10.3390/cancers12030665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mosaddeghzadeh N., Ahmadian M.R. The RHO family GTPases: mechanisms of regulation and signaling. Cells. 2021;10(7):1831. doi: 10.3390/cells10071831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vigil D., Cherfils J., Rossman K.L., et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mehta P., Wavreille A.S., Justiniano S.E., et al. LyGDI, a novel SHIP-interacting protein, is a negative regulator of FcγR-mediated phagocytosis. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Machacek M., Hodgson L., Welch C., et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jacobs M., Hayakawa K., Swenson L., et al. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem. 2006;281(1):260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 177.Narumiya S., Thumkeo D. Rho signaling research: history, current status and future directions. FEBS Lett. 2018;592(11):1763–1776. doi: 10.1002/1873-3468.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Julian L., Olson M.F. Rho-associated coiled-coil containing kinases (ROCK):structure, regulation, and functions. Small GTPases. 2014;5 doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ishizaki T., Maekawa M., Fujisawa K., et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15(8):1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 180.Coleman M.L., Sahai E.A., Yeo M., et al. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3(4):339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 181.Sebbagh M., Renvoizé C., Hamelin J., et al. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3(4):346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 182.Leung T., Chen X.Q., Manser E., et al. The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol. 1996;16(10):5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Murakami T., Ishikawa H. Stress fibers in situ in proximal tubules of the rat kidney. Cell Struct Funct. 1991;16(3):231–240. doi: 10.1247/csf.16.231. [DOI] [PubMed] [Google Scholar]
- 184.Schiller H.B., Fässler R. Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 2013;14(6):509–519. doi: 10.1038/embor.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bode A.M., Dong Z. The functional contrariety of JNK. Mol Carcinog. 2007;46(8):591–598. doi: 10.1002/mc.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Grynberg K., Ma F.Y., Nikolic-Paterson D.J. The JNK signaling pathway in renal fibrosis. Front Physiol. 2017;8:829. doi: 10.3389/fphys.2017.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Avruch J. MAP kinase pathways: the first twenty years. Biochim Biophys Acta. 2007;1773(8):1150–1160. doi: 10.1016/j.bbamcr.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Patterson K.I., Brummer T., O'Brien P.M., et al. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418(3):475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 189.Zeke A., Misheva M., Reményi A., et al. JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol Mol Biol Rev. 2016;80(3):793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tournier C., Hess P., Yang D.D., et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288(5467):870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 191.Kumar A., Singh U.K., Kini S.G., et al. JNK pathway signaling: a novel and smarter therapeutic targets for various biological diseases. Future Med Chem. 2015;7(15):2065–2086. doi: 10.4155/fmc.15.132. [DOI] [PubMed] [Google Scholar]
- 192.Angel P., Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072(2–3):129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 193.Kovary K., Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11(9):4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pfarr C.M., Mechta F., Spyrou G., et al. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76(4):747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 195.Schreiber M., Kolbus A., Piu F., et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 197.Katiyar S., Casimiro M.C., Dettin L., et al. C-jun inhibits mammary apoptosis in vivo. Mol Biol Cell. 2010;21(23):4264–4274. doi: 10.1091/mbc.E10-08-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ham J., Eilers A., Whitfield J., et al. c-Jun and the transcriptional control of neuronal apoptosis. Biochem Pharmacol. 2000;60(8):1015–1021. doi: 10.1016/s0006-2952(00)00372-5. [DOI] [PubMed] [Google Scholar]
- 199.Kuan C.Y., Yang D.D., Roy D.R.S., et al. The Jnk 1 and Jnk 2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22(4):667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 200.Hernandez-Valladares M., Kim T., Kannan B., et al. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol. 2010;17(4):497–503. doi: 10.1038/nsmb.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Miyatake K., Kusakabe M., Takahashi C., et al. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat Commun. 2015;6:6666. doi: 10.1038/ncomms7666. [DOI] [PubMed] [Google Scholar]
- 202.Iwasaki T., Murata-Hori M., Ishitobi S., et al. Diphosphorylated MRLC is required for organization of stress fibers in interphase cells and the contractile ring in dividing cells. Cell Struct Funct. 2001;26(6):677–683. doi: 10.1247/csf.26.677. [DOI] [PubMed] [Google Scholar]
- 203.Ikebe M., Hartshorne D.J. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 1985;260(18):10027–10031. [PubMed] [Google Scholar]
- 204.Ikebe M., Koretz J., Hartshorne D.J. Effects of phosphorylation of light chain residues threonine 18 and serine 19 on the properties and conformation of smooth muscle myosin. J Biol Chem. 1988;263(13):6432–6437. [PubMed] [Google Scholar]
- 205.Watanabe T., Hosoya H., Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell. 2007;18(2):605–616. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Peng Y., Chen Z., Chen Y., et al. ROCK isoforms differentially modulate cancer cell motility by mechanosensing the substrate stiffness. Acta Biomater. 2019;88:86–101. doi: 10.1016/j.actbio.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 207.Goode B.L., Eck M.J. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 208.Paul A.S., Pollard T.D. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18(1):9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lammers M., Rose R., Scrima A., et al. The regulation of mDia1 by autoinhibition and its release by Rho∗GTP. EMBO J. 2005;24(23):4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu D., Fu X., Wang Y., et al. Protein diaphanous homolog 1 (Diaph1) promotes myofibroblastic activation of hepatic stellate cells by regulating Rab5a activity and TGFβ receptor endocytosis. FASEB J. 2020;34(6):7345–7359. doi: 10.1096/fj.201903033R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fukami K. Structure, regulation, and function of phospholipase C isozymes. J Biochem. 2002;131(3):293–299. doi: 10.1093/oxfordjournals.jbchem.a003102. [DOI] [PubMed] [Google Scholar]
- 212.Harlan J.E., Hajduk P.J., Yoon H.S., et al. Pleckstrin homology domains bind to phosphatidylinositol-4, 5-bisphosphate. Nature. 1994;371(6493):168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 213.Essen L.O., Perisic O., Cheung R., et al. Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature. 1996;380(6575):595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 214.Bill C.A., Vines C.M., Phospholipase C. Adv Exp Med Biol. 2020;1131:215–242. doi: 10.1007/978-3-030-12457-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ellis M.V., U S, Katan M. Mutations within a highly conserved sequence present in the X region of phosphoinositide-specific phospholipase C-delta 1. Biochem J. 1995;307(Pt 1):69–75. doi: 10.1042/bj3070069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Essen L.O., Perisic O., Lynch D.E., et al. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta 1. Biochemistry. 1997;36(10):2753–2762. doi: 10.1021/bi962466t. [DOI] [PubMed] [Google Scholar]
- 217.Hokin M.R., Hokin L.E. Enzyme secretion and the incorporation of P32 into phospholipides of pancreas slices. J Biol Chem. 1953;203(2):967–977. [PubMed] [Google Scholar]
- 218.Streb H., Irvine R.F., Berridge M.J., et al. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1, 4, 5-trisphosphate. Nature. 1983;306(5938):67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- 219.Sun M.K., Alkon D.L. Pharmacology of protein kinase C activators: cognition-enhancing and antidementic therapeutics. Pharmacol Ther. 2010;127(1):66–77. doi: 10.1016/j.pharmthera.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 220.Rosse C., Linch M., Kermorgant S., et al. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol. 2010;11(2):103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- 221.Mochly-Rosen D., Das K., Grimes K.V. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11(12):937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Manning G., Whyte D.B., Martinez R., et al. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 223.Cooke M., Magimaidas A., Casado-Medrano V., et al. Protein kinase C in cancer: the top five unanswered questions. Mol Carcinog. 2017;56(6):1531–1542. doi: 10.1002/mc.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ono Y., Fujii T., Igarashi K., et al. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86(13):4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mochly-Rosen D. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science. 1995;268(5208):247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- 226.Murphy N.P., Binti Ahmad Mokhtar A.M., Mott H.R., et al. Molecular subversion of Cdc 42 signalling in cancer. Biochem Soc Trans. 2021;49(3):1425–1442. doi: 10.1042/BST20200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Johnson D.I., Pringle J.R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111(1):143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Munemitsu S., Innis M.A., Clark R., et al. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol Cell Biol. 1990;10(11):5977–5982. doi: 10.1128/mcb.10.11.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Erickson J.W., Zhang C.J., Kahn R.A., et al. Mammalian Cdc 42 is a brefeldin A-sensitive component of the Golgi apparatus. J Biol Chem. 1996;271(43):26850–26854. doi: 10.1074/jbc.271.43.26850. [DOI] [PubMed] [Google Scholar]
- 230.Michaelson D., Silletti J., Murphy G., et al. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Farhan H., Hsu V.W. Cdc 42 and cellular polarity: emerging roles at the Golgi. Trends Cell Biol. 2016;26(4):241–248. doi: 10.1016/j.tcb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Luna A., Matas O.B., Martínez-Menárguez J.A., et al. Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell. 2002;13(3):866–879. doi: 10.1091/mbc.01-12-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Etienne-Manneville S. Cdc 42: the centre of polarity. J Cell Sci. 2004;117(Pt 8):1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 234.Lu M.S., Drubin D.G. Cdc 42 GTPase regulates ESCRTs in nuclear envelope sealing and ER remodeling. J Cell Biol. 2020;219(8) doi: 10.1083/jcb.201910119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hodge R.G., Ridley A.J. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17(8):496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 236.Salat-Canela C., Carmona M., Martín-García R., et al. Stress-dependent inhibition of polarized cell growth through unbalancing the GEF/GAP regulation of Cdc 42. Cell Rep. 2021;37(5) doi: 10.1016/j.celrep.2021.109951. [DOI] [PubMed] [Google Scholar]
- 237.Lisman J., Yasuda R., Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13(3):169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Schulman H., Greengard P. Ca2+-dependent protein phosphorylation system in membranes from various tissues, and its activation by calcium-dependent regulator. Proc Natl Acad Sci U S A. 1978;75(11):5432–5436. doi: 10.1073/pnas.75.11.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tombes R.M., Faison M.O., Turbeville J.M. Organization and evolution of multifunctional Ca2+/CaM-dependent protein kinase genes. Gene. 2003;322:17–31. doi: 10.1016/j.gene.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 240.Chao L.H., Stratton M.M., Lee I.H., et al. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin- dependent kinase II holoenzyme. Cell. 2011;146(5):732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bayer K.U., Schulman H. CaM kinase: still inspiring at 40. Neuron. 2019;103(3):380–394. doi: 10.1016/j.neuron.2019.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Burgos J.I., Yeves A.M., Barrena J.P., et al. Nitric oxide and CaMKII: critical steps in the cardiac contractile response to IGF-1 and swim training. J Mol Cell Cardiol. 2017;112:16–26. doi: 10.1016/j.yjmcc.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 243.Beckendorf J., van den Hoogenhof M.M.G., Backs J. Physiological and unappreciated roles of CaMKII in the heart. Basic Res Cardiol. 2018;113(4):29. doi: 10.1007/s00395-018-0688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Feng N., Anderson M.E. CaMKII is a nodal signal for multiple programmed cell death pathways in heart. J Mol Cell Cardiol. 2017;103:102–109. doi: 10.1016/j.yjmcc.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Weinreuter M., Kreusser M.M., Beckendorf J., et al. CaM Kinase II mediates maladaptive post-infarct remodeling and pro-inflammatory chemoattractant signaling but not acute myocardial ischemia/reperfusion injury. EMBO Mol Med. 2014;6(10):1231–1245. doi: 10.15252/emmm.201403848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Shibuya H., Iwata H., Masuyama N., et al. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17(4):1019–1028. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yamaguchi K., Shirakabe K., Shibuya H., et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270(5244):2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 248.Bosman M.C.J., Schepers H., Jaques J., et al. The TAK1-NF-κB axis as therapeutic target for AML. Blood. 2014;124(20):3130–3140. doi: 10.1182/blood-2014-04-569780. [DOI] [PubMed] [Google Scholar]
- 249.Xu Y.R., Lei C.Q. TAK1-TABs complex: a central signalosome in inflammatory responses. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.608976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tan L., Gurbani D., Weisberg E.L., et al. Structure-guided development of covalent TAK1 inhibitors. Bioorg Med Chem. 2017;25(3):838–846. doi: 10.1016/j.bmc.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zonneville J., Wong V., Limoge M., et al. TAK1 signaling regulates p53 through a mechanism involving ribosomal stress. Sci Rep. 2020;10:2517. doi: 10.1038/s41598-020-59340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Omori E., Matsumoto K., Sanjo H., et al. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J Biol Chem. 2006;281(28):19610–19617. doi: 10.1074/jbc.M603384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Totzke J., Gurbani D., Raphemot R., et al. Takinib, a selective TAK1 inhibitor, broadens the therapeutic efficacy of TNF-α inhibition for cancer and autoimmune disease. Cell Chem Biol. 2017;24(8):1029–1039.e7. doi: 10.1016/j.chembiol.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Park S.H., Kwak J.A., Jung S.H., et al. Piperidylmethyloxychalcone improves immune-mediated acute liver failure via inhibiting TAK1 activity. Exp Mol Med. 2017;49(11):e392. doi: 10.1038/emm.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Coulombe P., Meloche S. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim Biophys Acta. 2007;1773(8):1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 256.Choi K.W., Benzer S. Rotation of photoreceptor clusters in the developing drosophila eye requires the nemo gene. Cell. 1994;78(1):125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 257.Brott B.K., Pinsky B.A., Erikson R.L. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci U S A. 1998;95(3):963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Li M., Wang H., Huang T., et al. TAB2 scaffolds TAK1 and NLK in repressing canonical Wnt signaling. J Biol Chem. 2010;285(18):13397–13404. doi: 10.1074/jbc.M109.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yamada M., Ohnishi J., Ohkawara B., et al. NARF, an Nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF) J Biol Chem. 2006;281(30):20749–20760. doi: 10.1074/jbc.M602089200. [DOI] [PubMed] [Google Scholar]
- 260.Chen J., Lin Q., Ni T., et al. NLK interacts with 14-3-3ζ to restore the expression of E-cadherin. Oncol Rep. 2020;43(6):1845–1852. doi: 10.3892/or.2020.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chen X., Zhou Y., Wan Y., et al. The expression of NLK is functionally associated with colorectal cancers (CRC) J Cancer. 2021;12(23):7088–7100. doi: 10.7150/jca.62526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liu R., Khalil H., Lin S.J., et al. Nemo-like kinase (NLK) is a pathological signaling effector in the mouse heart. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Creamer T.P. Calcineurin. Cell Commun Signal. 2020;18:137. doi: 10.1186/s12964-020-00636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang J.H., Desai R. A brain protein and its effect on the CA2+-and protein modulator-activated cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1976;72(3):926–932. doi: 10.1016/s0006-291x(76)80220-3. [DOI] [PubMed] [Google Scholar]
- 265.Rusnak F., Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80(4):1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 266.Klee C.B., Ren H., Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273(22):13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 267.Tarasova E.O., Gaydukov A.E., Balezina O.P. Calcineurin and its role in synaptic transmission. Biochemistry Mosc. 2018;83(6):674–689. doi: 10.1134/S0006297918060056. [DOI] [PubMed] [Google Scholar]
- 268.Parra V., Rothermel B.A. Calcineurin signaling in the heart: the importance of time and place. J Mol Cell Cardiol. 2017;103:121–136. doi: 10.1016/j.yjmcc.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Cruciat C.M., Niehrs C. Secreted and transmembrane Wnt inhibitors and activators. Cold Spring Harbor Perspect Biol. 2013;5(3):a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hoang B., Moos M., Jr., Vukicevic S., et al. Primary structure and tissue distribution of FRZB, a novel protein related to Drosophila frizzled, suggest a role in skeletal morphogenesis. J Biol Chem. 1996;271(42):26131–26137. doi: 10.1074/jbc.271.42.26131. [DOI] [PubMed] [Google Scholar]
- 271.Chong J.M., Üren A., Rubin J.S., et al. Disulfide bond assignments of secreted frizzled-related protein-1 provide insights about Frizzled homology and netrin modules. J Biol Chem. 2002;277(7):5134–5144. doi: 10.1074/jbc.M108533200. [DOI] [PubMed] [Google Scholar]
- 272.Leyns L., Bouwmeester T., Kim S.H., et al. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997;88(6):747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lin K., Wang S., Julius M.A., et al. The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc Natl Acad Sci U S A. 1997;94(21):11196–11200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang S., Krinks M., Lin K., et al. Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88(6):757–766. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 275.Bafico A., Gazit A., Pramila T., et al. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem. 1999;274(23):16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 276.Bi Y., Huang J., He Y., et al. Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J Cell Biochem. 2009;108(1):295–303. doi: 10.1002/jcb.22254. [DOI] [PubMed] [Google Scholar]
- 277.Bhat R.A., Stauffer B., Komm B.S., et al. Structure-function analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J Cell Biochem. 2007;102(6):1519–1528. doi: 10.1002/jcb.21372. [DOI] [PubMed] [Google Scholar]
- 278.Lopez-Rios J., Esteve P., Ruiz J.M., et al. The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Dev. 2008;3:19. doi: 10.1186/1749-8104-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hsieh J.C., Kodjabachian L., Rebbert M.L., et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398(6726):431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 280.Glinka A., Wu W., Delius H., et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391(6665):357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 281.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25(57):7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 282.Pandur P., Läsche M., Eisenberg L.M., et al. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418(6898):636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 283.Ellies D.L., Viviano B., McCarthy J., et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21(11):1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 284.Itasaki N., Jones C.M., Mercurio S., et al. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130(18):4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 285.Lintern K.B., Guidato S., Rowe A., et al. Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem. 2009;284(34):23159–23168. doi: 10.1074/jbc.M109.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Paszty C., Turner C.H., Robinson M.K. Sclerostin: a gem from the genome leads to bone-building antibodies. J Bone Miner Res. 2010;25(9):1897–1904. doi: 10.1002/jbmr.161. [DOI] [PubMed] [Google Scholar]
- 287.Guidato S., Itasaki N. Wise retained in the endoplasmic reticulum inhibits Wnt signaling by reducing cell surface LRP6. Dev Biol. 2007;310(2):250–263. doi: 10.1016/j.ydbio.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 288.Ohazama A., Johnson E.B., Ota M.S., et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3(12) doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Li X., Zhang Y., Kang H., et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280(20):19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 290.Semënov M., Tamai K., He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280(29):26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 291.Bouwmeester T., Kim S., Sasai Y., et al. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382(6592):595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 292.Aykul S., Martinez-Hackert E. New ligand binding function of human Cerberus and role of proteolytic processing in regulating ligand-receptor interactions and antagonist activity. J Mol Biol. 2016;428(3):590–602. doi: 10.1016/j.jmb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Biben C., Stanley E., Fabri L., et al. Murine Cerberus homologue mCer-1:a candidate anterior patterning molecule. Dev Biol. 1998;194(2):135–151. doi: 10.1006/dbio.1997.8812. [DOI] [PubMed] [Google Scholar]
- 294.Piccolo S., Agius E., Leyns L., et al. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397(6721):707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhu W., Shiojima I., Ito Y., et al. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature. 2008;454(7202):345–349. doi: 10.1038/nature07027. [DOI] [PubMed] [Google Scholar]
- 296.Ueno K., Hirata H., Majid S., et al. IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int J Cancer. 2011;129(10):2360–2369. doi: 10.1002/ijc.25899. [DOI] [PubMed] [Google Scholar]
- 297.Neufeld K.L. Nuclear APC. Adv Exp Med Biol. 2009;656:13–29. doi: 10.1007/978-1-4419-1145-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Spink K.E., Polakis P., Weis W.I. Structural basis of the Axin-adenomatous polyposis coli interaction. EMBO J. 2000;19(10):2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Su Y., Fu C., Ishikawa S., et al. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell. 2008;32(5):652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 300.Day C.L., Alber T. Crystal structure of the amino-terminal coiled-coil domain of the APC tumor suppressor. J Mol Biol. 2000;301(1):147–156. doi: 10.1006/jmbi.2000.3895. [DOI] [PubMed] [Google Scholar]
- 301.Watanabe T., Wang S., Noritake J., et al. Interaction with IQGAP1 links APC to Rac 1, Cdc 42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7(6):871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 302.Jimbo T., Kawasaki Y., Koyama R., et al. Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol. 2002;4(4):323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- 303.Smits R., Kielman M.F., Breukel C., et al. Apc1638T: a mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev. 1999;13(10):1309–1321. doi: 10.1101/gad.13.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Half E., Bercovich D., Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis. 2009;4:22. doi: 10.1186/1750-1172-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zeng L., Fagotto F., Zhang T., et al. The mouse Fused locus encodes axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90(1):181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 306.Mazzoni S.M., Fearon E.R. AXIN1and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 2014;355(1):1–8. doi: 10.1016/j.canlet.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mallick A., Taylor S.K.B., Ranawade A., et al. Axin family of scaffolding proteins in development: lessons from C. elegans. J Dev Biol. 2019;7(4):20. doi: 10.3390/jdb7040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kishida M., Koyama S., Kishida S., et al. Axin prevents Wnt-3a-induced accumulation of beta-catenin. Oncogene. 1999;18(4):979–985. doi: 10.1038/sj.onc.1202388. [DOI] [PubMed] [Google Scholar]
- 309.Hsu W., Zeng L., Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274(6):3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 310.Rui Y., Xu Z., Lin S., et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 2004;23(23):4583–4594. doi: 10.1038/sj.emboj.7600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Dao D.Y., Yang X., Chen D., et al. Axin 1 and Axin 2 are regulated by TGF- and mediate cross-talk between TGF- and Wnt signaling pathways. Ann N Y Acad Sci. 2007;1116:82–99. doi: 10.1196/annals.1402.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Embi N., Rylatt D.B., Cohen P. Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur J Biochem. 1980;107(2):519–527. [PubMed] [Google Scholar]
- 313.Rylatt D.B., Aitken A., Bilham T., et al. Glycogen synthase from rabbit skeletal muscle. Amino acid sequence at the sites phosphorylated by glycogen synthase kinase-3, and extension of the N-terminal sequence containing the site phosphorylated by phosphorylase kinase. Eur J Biochem. 1980;107(2):529–537. [PubMed] [Google Scholar]
- 314.Lin J., Song T., Li C., et al. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta Mol Cell Res. 2020;1867(5) doi: 10.1016/j.bbamcr.2020.118659. [DOI] [PubMed] [Google Scholar]
- 315.Woodgett J.R. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9(8):2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Kaidanovich-Beilin O., Lipina T.V., Takao K., et al. Abnormalities in brain structure and behavior in GSK-3 alpha mutant mice. Mol Brain. 2009;2:35. doi: 10.1186/1756-6606-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hoeflich K.P., Luo J., Rubie E.A., et al. Requirement for glycogen synthase kinase-3 beta in cell survival and NF-kappaB activation. Nature. 2000;406(6791):86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 318.MacAulay K., Doble B.W., Patel S., et al. Glycogen synthase kinase 3 alpha-specific regulation of murine hepatic glycogen metabolism. Cell Metabol. 2007;6(4):329–337. doi: 10.1016/j.cmet.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 319.Roach P.J. Glycogen and its metabolism. Curr Mol Med. 2002;2(2):101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 320.Leung-Hagesteijn C., Mahendra A., Naruszewicz I., et al. Modulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1. EMBO J. 2001;20(9):2160–2170. doi: 10.1093/emboj/20.9.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Ding V.W., Chen R.H., McCormick F. Differential regulation of glycogen synthase kinase 3β by insulin and Wnt signaling. J Biol Chem. 2000;275(42):32475–32481. doi: 10.1074/jbc.M005342200. [DOI] [PubMed] [Google Scholar]
- 322.Beurel E., Grieco S.F., Jope R.S. Glycogen synthase kinase-3 (GSK3):regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Klein P.S., Melton D.A. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93(16):8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Watcharasit P., Bijur G.N., Song L., et al. Glycogen synthase kinase-3 beta (GSK3beta) binds to and promotes the actions of p53. J Biol Chem. 2003;278(49):48872–48879. doi: 10.1074/jbc.M305870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Souder D.C., Anderson R.M. An expanding GSK3 network: implications for aging research. GeroScience. 2019;41(4):369–382. doi: 10.1007/s11357-019-00085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Janovská P., Normant E., Miskin H., et al. Targeting casein kinase 1 (CK1) in hematological cancers. Int J Mol Sci. 2020;21(23):9026. doi: 10.3390/ijms21239026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Knippschild U., Krüger M., Richter J., et al. The CK1 family: contribution to cellular stress response and its role in carcinogenesis. Front Oncol. 2014;4:96. doi: 10.3389/fonc.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Carmel G., Leichus B., Cheng X., et al. Expression, purification, crystallization, and preliminary X-ray analysis of casein kinase-1 from Schizosaccharomyces pombe. J Biol Chem. 1994;269(10):7304–7309. [PubMed] [Google Scholar]
- 329.Gietzen K.F., Virshup D.M. Identification of inhibitory autophosphorylation sites in casein kinase I ε. J Biol Chem. 1999;274(45):32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 330.Fulcher L.J., Sapkota G.P. Functions and regulation of the serine/threonine protein kinase CK1 family: moving beyond promiscuity. Biochem J. 2020;477(23):4603–4621. doi: 10.1042/BCJ20200506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Marin O., Bustos V.H., Cesaro L., et al. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc Natl Acad Sci U S A. 2003;100(18):10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Dumaz N., Milne D.M., Meek D.W. Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett. 1999;463(3):312–316. doi: 10.1016/s0014-5793(99)01647-6. [DOI] [PubMed] [Google Scholar]
- 333.Elyada E., Pribluda A., Goldstein R.E., et al. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470(7334):409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 334.Fulcher L.J., He Z., Mei L., et al. FAM83D directs protein kinase CK1α to the mitotic spindle for proper spindle positioning. EMBO Rep. 2019;20(9) doi: 10.15252/embr.201847495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Fuchs S.Y., Spiegelman V.S., Kumar K.G. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23(11):2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 336.Kitagawa M., Hatakeyama S., Shirane M., et al. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18(9):2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Bai C., Sen P., Hofmann K., et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 338.Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 339.Guardavaccaro D., Kudo Y., Boulaire J., et al. Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev Cell. 2003;4(6):799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 340.Margottin-Goguet F., Hsu J.Y., Loktev A., et al. Prophase destruction of Emi 1 by the SCFβTrCP/slimb ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4(6):813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 341.Mayo M.W., Baldwin A.S. The transcription factor NF-kappaB: control of oncogenesis and cancer therapy resistance. Biochim Biophys Acta. 2000;1470(2):M55–M62. doi: 10.1016/s0304-419x(00)00002-0. [DOI] [PubMed] [Google Scholar]
- 342.Wlodarchak N., Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol. 2016;51(3):162–184. doi: 10.3109/10409238.2016.1143913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.O'Connor C.M., Perl A., Leonard D., et al. Therapeutic targeting of PP2A. Int J Biochem Cell Biol. 2018;96:182–193. doi: 10.1016/j.biocel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Groves M.R., Hanlon N., Turowski P., et al. The structure of the protein phosphatase 2A PR65/a subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96(1):99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 345.Longin S., Zwaenepoel K., Louis J.V., et al. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic subunit. J Biol Chem. 2007;282(37):26971–26980. doi: 10.1074/jbc.M704059200. [DOI] [PubMed] [Google Scholar]
- 346.Mitra A., Menezes M.E., Pannell L.K., et al. DNAJB6 chaperones PP2A mediated dephosphorylation of GSK3β to downregulate β-catenin transcription target, osteopontin. Oncogene. 2012;31(41):4472–4483. doi: 10.1038/onc.2011.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lowe M., Gonatas N.K., Warren G. The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J Cell Biol. 2000;149(2):341–356. doi: 10.1083/jcb.149.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Verbinnen I., Vaneynde P., Reynhout S., et al. Protein Phosphatase 2A (PP2A) mutations in brain function, development, and neurologic disease. Biochem Soc Trans. 2021;49(4):1567–1588. doi: 10.1042/BST20201313. [DOI] [PubMed] [Google Scholar]
- 349.Torrent L., Ferrer I. PP2A and alzheimer disease. Curr Alzheimer Res. 2012;9(2):248–256. doi: 10.2174/156720512799361682. [DOI] [PubMed] [Google Scholar]
- 350.Nader C.P., Cidem A., Verrills N.M., et al. Protein phosphatase 2A (PP2A):a key phosphatase in the progression of chronic obstructive pulmonary disease (COPD) to lung cancer. Respir Res. 2019;20:222. doi: 10.1186/s12931-019-1192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Chen G., Courey A.J. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249(1–2):1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 352.Daniels D.L., Weis W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 353.Chodaparambil J.V., Pate K.T., Hepler M.R.D., et al. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014;33(7):719–731. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Chen G., Fernandez J., Mische S., et al. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13(17):2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Yuzugullu H., Benhaj K., Ozturk N., et al. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer. 2009;8:90. doi: 10.1186/1476-4598-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Guo X., Wang X.F. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Shi Y., Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 358.He G., Shi Y., Lim J., et al. Differential involvement of Wnt signaling in Bmp regulation of cancellous versus periosteal bone growth. Bone Res. 2017;5 doi: 10.1038/boneres.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zhang F., Song J., Zhang H., et al. Wnt and BMP signaling crosstalk in regulating dental stem cells: implications in dental tissue engineering. Genes Dis. 2016;3(4):263–276. doi: 10.1016/j.gendis.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Luo W., Zhang L., Huang B., et al. BMP9-initiated osteogenic/odontogenic differentiation of mouse tooth germ mesenchymal cells (TGMCS) requires Wnt/β-catenin signalling activity. J Cell Mol Med. 2021;25(5):2666–2678. doi: 10.1111/jcmm.16293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Zhang L., Luo Q., Shu Y., et al. Transcriptomic landscape regulated by the 14 types of bone morphogenetic proteins (BMPs) in lineage commitment and differentiation of mesenchymal stem cells (MSCs) Genes Dis. 2019;6(3):258–275. doi: 10.1016/j.gendis.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zhang H., Wang J., Deng F., et al. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs) Biomaterials. 2015;39:145–154. doi: 10.1016/j.biomaterials.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Luo Q., Kang Q., Si W., et al. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279(53):55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 364.Millar S.E. Smad 7:licensed to kill β-catenin. Dev Cell. 2006;11(3):274–276. doi: 10.1016/j.devcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 365.Tang Y., Liu Z., Zhao L., et al. Smad 7 stabilizes beta-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J Biol Chem. 2008;283(35):23956–23963. doi: 10.1074/jbc.M800351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kamiya N., Kobayashi T., Mochida Y., et al. Wnt inhibitors Dkk 1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J Bone Miner Res. 2010;25(2):200–210. doi: 10.1359/jbmr.090806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Tang N., Song W.X., Luo J., et al. BMP-9-induced osteogenic differentiation of mesenchymal progenitors requires functional canonical Wnt/beta-catenin signalling. J Cell Mol Med. 2009;13(8B):2448–2464. doi: 10.1111/j.1582-4934.2008.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Zhang M., Yan Y., Lim Y.B., et al. BMP-2 modulates beta-catenin signaling through stimulation of Lrp5 expression and inhibition of beta-TrCP expression in osteoblasts. J Cell Biochem. 2009;108(4):896–905. doi: 10.1002/jcb.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Fischer L., Boland G., Tuan R.S. Wnt-3A enhances bone morphogenetic protein-2-mediated chondrogenesis of murine C3H10T1/2 mesenchymal cells. J Biol Chem. 2002;277(34):30870–30878. doi: 10.1074/jbc.M109330200. [DOI] [PubMed] [Google Scholar]
- 370.Zhang R., Oyajobi B.O., Harris S.E., et al. Wnt/β-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone. 2013;52(1):145–156. doi: 10.1016/j.bone.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Rodríguez-Carballo E., Ulsamer A., Susperregui A.R.G., et al. Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. J Bone Miner Res. 2011;26(4):718–729. doi: 10.1002/jbmr.260. [DOI] [PubMed] [Google Scholar]
- 372.Liu Z., Tang Y., Qiu T., et al. A dishevelled-1/Smad 1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J Biol Chem. 2006;281(25):17156–17163. doi: 10.1074/jbc.M513812200. [DOI] [PubMed] [Google Scholar]
- 373.Hoppler S., Moon R.T. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev. 1998;71(1–2):119–129. doi: 10.1016/s0925-4773(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 374.Wu M., Chen G., Li Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4 doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Morrell N.W., Bloch D.B., ten Dijke P., et al. Targeting BMP signalling in cardiovascular disease and anaemia. Nat Rev Cardiol. 2016;13(2):106–120. doi: 10.1038/nrcardio.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Luo K. Signaling cross talk between TGF-β/smad and other signaling pathways. Cold Spring Harbor Perspect Biol. 2017;9(1):a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Eger A., Stockinger A., Park J., et al. Beta-Catenin and TGFbeta signalling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene. 2004;23(15):2672–2680. doi: 10.1038/sj.onc.1207416. [DOI] [PubMed] [Google Scholar]
- 378.Nawshad A., Medici D., Liu C.C., et al. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad 2-Smad 4-LEF1 transcription complex. J Cell Sci. 2007;120(Pt 9):1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Zhou B., Liu Y., Kahn M., et al. Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP) J Biol Chem. 2012;287(10):7026–7038. doi: 10.1074/jbc.M111.276311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Jian H., Shen X., Liu I., et al. Smad 3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;20(6):666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Edeling M., Ragi G., Huang S., et al. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12(7):426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Wang H., Zang C., Liu X.S., et al. The role of Notch receptors in transcriptional regulation. J Cell Physiol. 2015;230(5):982–988. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Sprinzak D., Blacklow S.C. Biophysics of Notch signaling. Annu Rev Biophys. 2021;50:157–189. doi: 10.1146/annurev-biophys-101920-082204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Hayward P., Kalmar T., Arias A.M. Wnt/Notch signalling and information processing during development. Development. 2008;135(3):411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 385.Fujimura S., Jiang Q., Kobayashi C., et al. Notch 2 activation in the embryonic kidney depletes nephron progenitors. J Am Soc Nephrol. 2010;21(5):803–810. doi: 10.1681/ASN.2009040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Galceran J., Sustmann C., Hsu S.C., et al. LEF1-mediated regulation of Delta-like 1 links Wnt and Notch signaling in somitogenesis. Genes Dev. 2004;18(22):2718–2723. doi: 10.1101/gad.1249504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Borggrefe T., Lauth M., Zwijsen A., et al. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFβ/BMP and hypoxia pathways. Biochim Biophys Acta. 2016;1863(2):303–313. doi: 10.1016/j.bbamcr.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 388.Jin Y.H., Kim H., Ki H., et al. Beta-catenin modulates the level and transcriptional activity of Notch 1/NICD through its direct interaction. Biochim Biophys Acta. 2009;1793(2):290–299. doi: 10.1016/j.bbamcr.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 389.Kwon C., Cheng P., King I.N., et al. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13(10):1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Foltz D.R., Santiago M.C., Berechid B.E., et al. Glycogen synthase kinase-3 beta modulates Notch signaling and stability. Curr Biol. 2002;12(12):1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- 391.Ann E.J., Kim H.Y., Seo M.S., et al. Wnt5a controls Notch 1 signaling through CaMKII-mediated degradation of the SMRT corepressor protein. J Biol Chem. 2012;287(44):36814–36829. doi: 10.1074/jbc.M112.356048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Collu G.M., Hidalgo-Sastre A., Acar A., et al. Dishevelled limits Notch signalling through inhibition of CSL. Development. 2012;139(23):4405–4415. doi: 10.1242/dev.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Pinto I., Duque M., Gonçalves J., et al. NRARP displays either pro- or anti-tumoral roles in T-cell acute lymphoblastic leukemia depending on Notch and Wnt signaling. Oncogene. 2020;39(5):975–986. doi: 10.1038/s41388-019-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Rodilla V., Villanueva A., Obrador-Hevia A., et al. Jagged 1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106(15):6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Fendler A., Bauer D., Busch J., et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun. 2020;11:929. doi: 10.1038/s41467-020-14700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Zhou J., Cheng P., Youn J.I., et al. Notch and wingless signaling cooperate in regulation of dendritic cell differentiation. Immunity. 2009;30(6):845–859. doi: 10.1016/j.immuni.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Germar K., Dose M., Konstantinou T., et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108(50):20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.MacGrogan D., Münch J., de la Pompa J.L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol. 2018;15(11):685–704. doi: 10.1038/s41569-018-0100-2. [DOI] [PubMed] [Google Scholar]
- 399.Adams J.M., Jafar-Nejad H. The roles of Notch signaling in liver development and disease. Biomolecules. 2019;9(10):608. doi: 10.3390/biom9100608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Canalis E. Notch in skeletal physiology and disease. Osteoporos Int. 2018;29(12):2611–2621. doi: 10.1007/s00198-018-4694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Ma S., Meng Z., Chen R., et al. The hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 402.Zhao B., Li L., Tumaneng K., et al. A coordinated phosphorylation by lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Liu C.Y., Zha Z.Y., Zhou X., et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285(48):37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Xu F., Wang Y.L., Chang J.J., et al. Mammalian sterile 20-like kinase 1/2 inhibits the Wnt/β-catenin signalling pathway by directly binding casein kinase 1ε. Biochem J. 2014;458(1):159–169. doi: 10.1042/BJ20130986. [DOI] [PubMed] [Google Scholar]
- 405.Imajo M., Miyatake K., Iimura A., et al. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012;31(5):1109–1122. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Piccolo S., Dupont S., Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94(4):1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 407.Jiang L., Li J., Zhang C., et al. YAP-mediated crosstalk between the Wnt and Hippo signaling pathways (Review) Mol Med Rep. 2020;22(5):4101–4106. doi: 10.3892/mmr.2020.11529. [DOI] [PubMed] [Google Scholar]
- 408.Park H.W., Kim Y.C., Yu B., et al. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162(4):780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Heallen T., Zhang M., Wang J., et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Carballo G.B., Honorato J.R., de Lopes G.P.F., et al. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Jia J., Tong C., Wang B., et al. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432(7020):1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 412.Chen Y., Sasai N., Ma G., et al. Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9(6) doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Shi Q., Li S., Li S., et al. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc Natl Acad Sci U S A. 2014;111(52):E5651–E5660. doi: 10.1073/pnas.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zhou Q., Kalderon D. Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev Cell. 2011;20(6):802–814. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Takenaka K., Kise Y., Miki H. GSK3β positively regulates Hedgehog signaling through Sufu in mammalian cells. Biochem Biophys Res Commun. 2007;353(2):501–508. doi: 10.1016/j.bbrc.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 416.He J., Sheng T., Stelter A.A., et al. Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J Biol Chem. 2006;281(47):35598–35602. doi: 10.1074/jbc.C600200200. [DOI] [PubMed] [Google Scholar]
- 417.Meng X., Poon R., Zhang X., et al. Suppressor of fused negatively regulates β-catenin signaling. J Biol Chem. 2001;276(43):40113–40119. doi: 10.1074/jbc.M105317200. [DOI] [PubMed] [Google Scholar]
- 418.Ebeid M., Huh S.H. FGF signaling: diverse roles during cochlear development. BMB Rep. 2017;50(10):487–495. doi: 10.5483/BMBRep.2017.50.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Katoh M., Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5(9):1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 420.Cohen E.D., Wang Z., Lepore J.J., et al. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117(7):1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Volckaert T., De Langhe S.P. Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev Dynam. 2015;244(3):342–366. doi: 10.1002/dvdy.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Sutkeviciute I., Clark L.J., White A.D., et al. PTH/PTHrP receptor signaling, allostery, and structures. Trends Endocrinol Metab. 2019;30(11):860–874. doi: 10.1016/j.tem.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Kakar S., Einhorn T.A., Vora S., et al. Enhanced chondrogenesis and Wnt signaling in PTH-treated fractures. J Bone Miner Res. 2007;22(12):1903–1912. doi: 10.1359/jbmr.070724. [DOI] [PubMed] [Google Scholar]
- 424.Tobimatsu T., Kaji H., Sowa H., et al. Parathyroid hormone increases beta-catenin levels through Smad 3 in mouse osteoblastic cells. Endocrinology. 2006;147(5):2583–2590. doi: 10.1210/en.2005-1627. [DOI] [PubMed] [Google Scholar]
- 425.Romero G., Sneddon W.B., Yang Y., et al. Parathyroid hormone receptor directly interacts with dishevelled to regulate β-catenin signaling and osteoclastogenesis. J Biol Chem. 2010;285(19):14756–14763. doi: 10.1074/jbc.M110.102970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Cabrae R., Dubuquoy C., Caüzac M., et al. Insulin activates hepatic Wnt/β-catenin signaling through stearoyl-CoA desaturase 1 and porcupine. Sci Rep. 2020;10:5186. doi: 10.1038/s41598-020-61869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Ugi S., Imamura T., Ricketts W., et al. Protein phosphatase 2A forms a molecular complex with Shc and regulates Shc tyrosine phosphorylation and downstream mitogenic signaling. Mol Cell Biol. 2002;22(7):2375–2387. doi: 10.1128/MCB.22.7.2375-2387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Zwaenepoel K., Goris J., Erneux C., et al. Protein phosphatase 2A PR130/B''alpha 1 subunit binds to the SH2 domain-containing inositol polyphosphate 5-phosphatase 2 and prevents epidermal growth factor (EGF)-induced EGF receptor degradation sustaining EGF-mediated signaling. FASEB J. 2010;24(2):538–547. doi: 10.1096/fj.09-140228. [DOI] [PubMed] [Google Scholar]