Abstract
Human herpesvirus 8/Kaposi sarcoma-associated virus (HHV-8/KSHV) contains, in addition to genes required for viral replication, a unique set of nonstructural genes which may be part of viral mimicry and contribute to viral replication and pathogenesis in vivo. Among these, HHV-8 encodes four open reading frames (ORFs) that showed homology to the transcription factors of the interferon regulatory factor (IRF) family. The ORF K9, viral IRF 1 (vIRF-1), has been cloned, and it was shown that, when overexpressed, it down modulates the interferon-mediated transcriptional activation of the interferon-stimulated gene 15 (ISG 15) promoter, and the role of vIRF-1 in viral mimicry was implied. However, the molecular mechanism of this effect has not been clarified. Here, we extend this observation and show that vIRF-1 also downregulates the transcriptional activity of IFNA gene promoter in infected cells by interfering with the transactivating activity of cellular IRFs, including IRF-1 and IRF-3. We further show that ectopic expression of vIRF-1 in NIH 3T3 cells confers resistance to tumor necrosis factor alpha-induced apoptosis. While vIRF-1 is unable to bind DNA with the same specificity as cellular IRFs, we demonstrate by in vitro binding assay that it can associate with the family of cellular IRFs, such as IRF-1 and the interferon consensus sequence binding protein. vIRF-1 interaction domain was localized between amino acids (aa) 152 and 243. While no binding between the full-size IRF-3 and vIRF-1 could be detected by the same assay, we show that vIRF-1 also targets the carboxy-terminal region (aa 1623 to 2414) of the transcriptional coactivator p300 which could also bind IRF-3 and IRF-1. These results demonstrate that vIRF-1 can modulate the transcription of the IFNA genes by direct heterodimerization with members of the IRF family, as well as by competitive binding with cellular transcription factors to the carboxy-terminal region of p300.
The human herpesvirus 8/Kaposi Sarcoma-associated herpesvirus (HHV-8/KSHV) may be the causal factor in Kaposi sarcoma, AIDS-associated body cavity-based lymphoma or pleural effusion lymphoma, and multicentric Castelman’s disease. HHV-8 is a gammaherpesvirus, such as Epstein-Barr virus (EBV) (4) and herpesvirus saimiri, which are oncogenic (1). The analyses of HHV-8 genomic sequences (44) showed that this virus contains, in addition to genes required for viral replication, a unique set of nonstructural genes which may be part of viral mimicry and essential for viral replication and pathogenicity in vivo.
Two of the HHV-8 analogues of cellular IRFs, open reading frame (ORF) K9-encoded viral interferon (IFN) regulatory factor (vIRF; named vIRF-1 in this study) and vIRF-2 (11), have been cloned. The expression of vIRF-1 can be induced by tetradecanoyl phorbol acetate (TPA) treatment in BCBL-1 cells (30); the presence of vIRF-1 antisense RNA reduced the expression of several HHV-8 lytic genes, including interleukin-6 (IL-6) in TPA-treated BCBL-1 cells (26). In that study it was suggested that vIRF-1 plays an important role in the regulation of HHV-8 replication cycle. Several groups (15, 26, 38, 57) have shown that vIRF-1 can function as a repressor on promoters containing IFN-sensitive response element and that NIH 3T3 cells constitutively overexpressing vIRF-1 gained the ability to grow in soft agar and to form tumors in nude mice. These data indicate that vIRF-1, like IRF-2, behaves as an oncogene. However, the molecular mechanism by which vIRF-1 downmodulates IFN-stimulated activation of IFN-stimulated gene (ISG) promoters or confers to NIH 3T3 cells the ability to grow in soft agar and in nude mice has not been clarified. We have recently cloned and characterized a second HHV-8 encoded vIRF, vIRF-2, that encodes a short protein of 163 amino acids (aa) (11). vIRF-2 is a DNA binding protein with a specificity distinct from that of cellular IRF, since it binds to oligonucleotide corresponding to the NF-κB site. In a transient-transfection assay, vIRF-2 inhibits the virus-mediated induction of promoters of IFN genes as well as RelA-stimulated activity of the human immunodeficiency virus long terminal repeat. Thus, the properties and, consequently, the biological functions of vIRF-1 and vIRF-2 seem to be distinct.
Transcriptional factors of the IRF family have been shown to play an essential role in the regulated expression of IFN and ISGs (34). All the cellular IRF proteins identified show homology in their 5′ DNA binding domain (DBD), which is characterized by five highly conserved tryptophan (W) repeats; three of these repeats contact DNA recognizing the GAAA sequence (14). The carboxy-terminal halves of these proteins are diverse. IRF-1 was originally identified by its ability to bind the GAAAGT sequence present in multiple copies in the promoter of IFNB gene, and it was proposed that IRF-1 serves as a positive activator of IFN genes in virus-infected cells, while a closely related homologue, IRF-2, acts as a repressor (17). However, homozygous deletion of the IRF-1 gene has not affected the virus-mediated activation of IFNA and IFNB genes expression (29, 42), whereas it downmodulated the expression of IL-12 and the antiviral effects of IFNs (43). Another member of this family, p48, interacts with phosphorylated STAT1 and STAT2 transcription factors forming ISGF3 complex (6, 7) in IFN-treated cells. This complex binds to the IFN-stimulated response element (ISRE) in the promoter of ISGs and activates their transcription. Homozygous deletion of p48 in mice abolishes sensitivity of these mice to the antiviral effect of IFNs and impairs the induction of IFNA and IFNB genes in a cell-type-specific manner (19, 22). Several other members of the IRF family have been identified. The IFN consensus sequence binding protein (ICSBP) is expressed exclusively in cells of immune system, including monocytes, B cells, and T cells (9). It complexes with IRF-1 and thus represses the transactivation mediated by IRF-1 (33, 48, 53). In mice, homologous deletion of ICSBP results in the deregulation of hematopoiesis and lymphopoiesis (20) and the defective expression of IL-12 (16). IRF-3 is expressed constitutively in most of the tissues and cell types (3) and strongly cooperates with virus in stimulation of IFN gene expression. In infected cells, IRF-3 is phosphorylated and transported into the nucleus where it binds the transcriptional coactivator p300/CBP (27, 55). It appears, therefore, that IRF-3 plays a critical role in virus-mediated signaling (21, 32, 46, 47). Another IRF that was shown recently to play a critical role in the induction of IFNA genes is IRF-7 (2, 28, 45, 51). IRF-7 also strongly cooperates with the virus-mediated induction of IFN genes, particularly IFNA genes, and was shown also to play a role in the regulated expression of viral QP promoter of the EBV-encoded EBNA-1 gene (36, 56).
Both IRF-1 and IRF-2 also modulate cell growth. Overexpression of IRF-1 has a growth-inhibitory effect and induces apoptosis (23, 49). In contrast, overexpression of IRF-2 in NIH 3T3 cells confers oncogenic transformation and tumor formation in nude mice, implicating IRF-2 as a potential oncogene. IRF-1, however, can reverse the IRF-2-induced transformation, as well as suppress c-myc and c-fos-induced transformation (18). Fibroblasts from IRF-1−/− mice show resistance to UV- and drug-induced apoptosis (49, 50). Since the deletion of IRF-1 on chromosome 5q31.3 is frequently found in leukemia, it was suggested that IRF-1 might function as a tumor suppressor gene (8, 54).
The aim of the present study was to further characterize the functional role of vIRF-1 in the expression of the early inflammatory gene (IFNA), as well as to determine the molecular mechanisms by which vIRF-1 exerts some of its biological effects. We have shown that in infected cells, vIRF-1 specifically represses the transcriptional activity of IFNA gene promoters, while, in uninfected cells, overexpression of vIRF-1 confers resistance to tumor necrosis factor alpha (TNF-α)-induced apoptosis. By using in vitro pull-down assay, we have further demonstrated a specific interaction between vIRF-1 and cellular IRFs, including IRF-1, and demonstrated that vIRF-1 contains an interaction-associated domain (IAD) by which it associates with IRF-1. We have further demonstrated an in vitro interaction between vIRF-1 and the C′-terminal domain of p300. We therefore conclude that, by specific interaction with IRF-1, vIRF-1 may lessen both the antiviral and antiapoptotic functions of IRF-1 while, by its interaction with p300, vIRF-1 may compete for binding of both IRF-1 and IRF-3 and so decrease their transactivating potential.
MATERIALS AND METHODS
Cell culture and transfections.
NIH 3T3 cells were grown in Dulbecco modified Eagle medium (DMEM) and 10% fetal bovine serum (FBS). The BCBL-1 cells were grown in RPMI supplemented with 10% FBS. In the transfection assays, subconfluent NIH 3T3 cells (2 × 105 cells/35-mm plate) were cotransfected with 1 μg of reporter chloramphenicol acetyltransferase (CAT) plasmid and 1 μg of each indicated expression plasmid by using the calcium phosphate coprecipitation method (31) or Superfect (Qiagen). The total amount of DNA was adjusted to 3 μg with empty vector pcDNA3.1 (Invitrogen) for each experiment. When indicated, cells were infected with Newcastle disease virus (NDV) (multiplicity of infection of 5) at 24 h after transfection for 16 h. Protein extracts were prepared by the freeze-thaw method at 48 h after transfection, and CAT assays were done as described previously (40). The same amount of protein was used for each reaction. Thin-layer chromatography plates were quantified by using a PhosphorImager (Molecular Dynamics).
Viability assay.
Cells (1.5 × 105/ml) in DMEM containing 10% FBS, 1 μg of actinomycin D per ml, and increasing concentrations of TNF-α were seeded (100 μl) into 96-well panels and grown in CO2 at 37°C for 24 h. Viable cells were measured by the MTT assay (12).
Plasmids. (i) Cloning of vIRF-1.
The vIRF-1 DNA corresponding to K9 ORF of HHV-8 was amplified from DNA extracted from KS tumor specimen by 25 cycles of PCR. Primers 5′-AGTAAGCTTGCGGGACAATGGACCCAGGCC and 3′-TTGTCTAGATTATTGCATGGCATCCCATAA were based on the published HHV-8 sequences; the resulting 1,368-bp vIRF-1 fragment was inserted into the HindIII and XbaI sites of pcDNA3.1 vector (Invitrogen) to construct pcDNA/vIRF-1. The cDNA was amplified by use of Pfu polymerase (Stratagene) and then sequenced. The comparison of the sequenced analysis with that deposited in GenBank indicated that the cloned KS/vIRF-1 contained five point mutations, four of which resulted in the amino acid change. Therefore, we have amplified and cloned vIRF-1 also from the genomic library of BCBL-1 cells. The DNA sequence of BCBL1/vIRF-1 was identical to that deposited in GenBank. Both the KS/vIRF-1 and the BCBL1/vIRF-1 had identical functional properties as seen in the transient-transfection assay, and neither of them was able to bind DNA. For consistency with the published results (26), the data presented here were obtained with BCBL1/vIRF-1-containing plasmids.
(ii) Construction of truncated and fusion vIRF-1.
To generate GST/vIRF-1 fusion proteins, full-length cDNA was amplified by PCR with the primers 5′-GCCGGAATTCAATGGACCCAGGC and 3′-CATCTCGAGGCATGGCATCCCATAA and inserted in frame into EcoRI-XhoI sites of pGEXT4 vector (Pharmacia). The N′-terminal 489-bp fragment coding for N′ vIRF-1 (aa 1 to 158) was amplified by PCR with the primers 5′-ATAGGATCCATGGACCCAGGCCAAAGACC and 3′-AGACTCGAGGTGCCTTTAAACGAGGCGTC. The C′-terminal 908-bp fragment of vIRF-1 (aa 152 to 449) was amplified by PCR with the 5′ primer TCTGGATCCGACGCCTCGTTTAAAGGCAC. The 3′ primer was identical to that used for amplification of glutathione S-transferase (GST)–vIRF-1. The 273-bp fragment vIRF-1A (aa 152 to 243) was amplified with primers 5′-TCTGGATCCGACGCCTCGTTTAAAGGCAC and 3′-AGACTCGAGCTAACAAGATGGCACGGGCGTTAC. The 464-bp fragment vIRF-1B (aa 152 to 307) was amplified with the primers 5′-TCTGGATCCGACGCCTCGTTTAAAGGCAC and 3′-AGACTCGAGCTATTGGGTAGCCATACCTGGCC, and the 461-bp fragment vIRF-1C (aa 295 to 449) was amplified with the primer 5′-ATAGGATCCGCCATGGCAGTGGGGTCTCCGGGCCAG and a 3′ primer identical to that used for the amplification of GST–vIRF-1. All of these vIRF-1 fragments were cloned into BamHI-XhoI sites of pGEXT4 vector. Amplifications were done by using 30 cycles with Pfu polymerase from the BCBL1/vIRF-1 expression plasmid. The constructed proteins were expressed in bacteria and were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to determine the correct molecular weight.
(iii) Construction of truncated p300 proteins.
The expression plasmids coding for amino- and C′-terminal parts of human p300 were provided by G. Nabel (37). The 1,161-bp fragment p300A (aa 1243 to 1630) was amplified by PCR with the primers 5′-TCAGGATCCGCCATGTTTGTTGAATGTACAGAGTGCGGA and 3′-ACTCTCGAGCTATGCATCCCGACCATCCAT. The 776-bp fragment p300B (aa 1623 to 1882) was amplified with the primers 5′-TCAGGATCCCCGATGGATGGTCGGGATGCGTTT and 3′-ACTCTCGAGCTACATGCTATTGGGAGGGGTA. The 1,596-bp fragment p300C (aa 1882 to 2414) was amplified with the primers 5′-TCAGGATCCGCCATGCCACCCTACTTGCCCAG and 3′-ACTCTCGAGCTAGTGTATGTCTAGTGTACTCTGTGAGAGG. All of these p300 fragments were cloned into BamHI-XhoI sites of vector pcDNA3.1. Amplifications were done by use of 30 cycles with Pfu polymerase from the p300 expression plasmid, and constructed proteins were checked by in vitro translation to determine the correct molecular weight.
(iv) Expression and reporter plasmids.
CMV/IRF-1 and CMV/IRF-2 expression plasmids were obtained from T. Taniguchi (University of Tokyo, Tokyo, Japan). The CMV/IRF-3 plasmid was as described previously (3). pSG5/ICSBP plasmid was obtained from K. Ozato (9). The IFNA4CAT reporter plasmid was described previously (41). In order to express deletion mutants of vIRF-1 in mammalian cells, the corresponding vIRF-1 fragments were subcloned from pGEX4T vector into the BamHI-XhoI sites of pCMV-TAG expression plasmid (Stratagene).
Preparation of GST fusion proteins.
Escherichia coli DH5α cells (200 ml at an optical density at 600 nm of 0.6) harboring the recombinant expression vectors were induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) at 37°C for 2.5 h. Cells were then washed with phosphate-buffered saline (PBS), resuspended in 10 ml of sonication buffer (PBS supplemented with 1% Triton X-100 and 1 mM phenymethylsulfonyl fluoride), and incubated with 10 mg of lysozyme (Sigma) per ml on ice for 15 min. The cell suspension was sonicated, and the lysate was cleared by centrifugation (12,000 × g for 10 min at 4°C). The supernatants were mixed with glutathione-agarose beads (200 μl of a 1:1 slurry in PBS) (Pharmacia) at 4°C for 1 h, and the beads were washed three times with ice-cold sonication buffer. The purity and quantity of fusion proteins were examined by Tricine–SDS–10% PAGE followed by Coomassie blue staining.
GST pull-down assay.
The 35S-labeled proteins were synthesized in vitro by using the coupled TNT T7 transcription-translation system (Promega) according to the manufacturer’s instructions. A total of 1 μg of nonlinearized expression plasmid was used in each reaction, followed by incubation (90 min, 30°C) in the presence of 4 μl of Translabel (DuPont) amino acid mixture. GST fusion proteins (0.5 μg) bound to glutathione-agarose beads were incubated with 10-μl reaction mixture aliquots of 35S-labeled proteins in 250 μl of binding buffer (10 mM Tris-Cl, pH 7.6; 100 mM NaCl; 0.1 mM EDTA, pH 8.0; 1 mM dithiothreitol [DTT]; 5 mM MgCl2; 0.05% Nonidet P-40; 8% glycerol; mammalian protease inhibitor cocktail [Sigma]) at 4°C for 90 min. After three washes (10 min at room temperature) with binding buffer, the proteins bound to the beads were solubilized in sample lysis buffer and resolved on Tricine–SDS–10% PAGE. When indicated, 1 μg of rabbit polyclonal antibody against the C-terminal peptide of human IRF-1 (Santa Cruz) was added to the binding reaction. The gel was dried and exposed to a PhosphorImager screen.
Electrophoretic mobility shift analysis.
DNA binding reactions were performed at room temperature for 30 min in 20 μl of binding buffer (12 mM Tris-HCl, pH 8.0; 2 mM MgCl2; 12 mM EDTA, pH 8.0; 40 mM NaCl; 6 mM DTT; 7% glycerol) containing 1 ng of GST–IRF-1 protein, the indicated amount of His6–vIRF-1 protein, 1 μg of poly(dI-dC), 1 μg of BSA, and 105 cpm of the indicated 32P-labeled double-stranded probe. Protein-DNA complexes were then resolved in a nondenaturing (50 mM Tris, 380 mM glycine, 2 mM EDTA) 7% polyacrylamide gel, dried, and exposed to a PhosphorImager screen. The sequence of PRD-I probe used was 5′-GAGAAGTGAAAGTGGGAACCCTCTCCTT.
RESULTS
vIRF-1 can inhibit IRF-1- or IRF-3-stimulated transcriptional activity of IFN-α4 promoter in infected cells.
Results from other laboratories (15, 26, 57), as well as our own previous data (38), showed that in a transient-expression assay vIRF-1 repressed the IFN-stimulated transcriptional activity of the ISRE-containing promoters of ISGs.
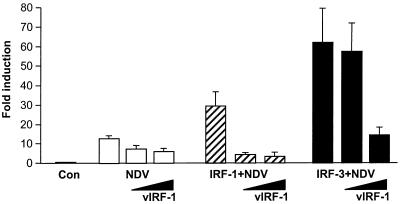
To determine whether the vIRF-1 repression is limited to the IFN-mediated stimulation of ISGs or whether it can also modulate the virus-stimulated expression of early inflammatory genes, such as type I IFN genes, we analyzed the effect of vIRF-1 on the virus-mediated stimulation of IFNA4 reporter plasmid in a transient-expression assay (41). This plasmid contains IFNA4 gene promoter region, including the virus-inducible elements with an IRF-E site but no NF-κB site. Constitutive expression of the IFNA4 CAT plasmid in NIH 3T3 cells was very low, but the transcriptional activity of the IFNA promoter was significantly stimulated (15-fold) upon infection with NDV (Fig. 1) (3). Cotransfection with vIRF-1-expressing plasmid inhibited the CAT expression by about two- to threefold. We have previously shown that, in a transient-transfection assay, virus-mediated stimulation of the IFNA4 gene promoter could be enhanced by cotransfection with IRF-1 (3), IRF-3 (21), or IRF-7 (2). While cotransfection with IRF-1 has increased virus-mediated activation of IFNA4 promoter threefold, IRF-3-enhanced virus stimulated the activity of IFNA4 promoter by sixfold (Fig. 1). Cotransfection with vIRF-1 effectively inhibited the IRF-1-mediated stimulation of the IFNA gene promoter in infected cells, indicating that vIRF-1 interfered with the transactivation potential of IRF-1. Similarly, synergism between virus- and IRF-3-mediated activation was also inhibited in cells overexpressing vIRF-1; however, the inhibitory effect by vIRF-1 could be demonstrated only at higher levels of vIRF-1 (Fig. 1). These data indicate that vIRF-1 interferes with the IRF-1- and IRF-3-mediated transcriptional activation of IFNA4 promoter in infected cells.
FIG. 1.
vIRF-1 inhibited synergistic activation of IFNA4 promoter by NDV and IRF-1 or IRF-3. The IFNA4 CAT reporter plasmid (1 μg) was cotransfected into NIH 3T3 cells with 1 μg of either pcDNA vector (Con), IRF-1, or IRF-3 expression plasmids, together with increasing amount of vIRF-1 expression plasmid (1 and 4 μg). Cells were infected with NDV 24 h after transfection for 16 h and analyzed for CAT activity. Error bars show standard errors for triplicate experiments.
Expression of vIRF-1 in NIH 3T3 cells confers resistance to apoptosis.
IRF-1 was shown to inhibit cell growth and induce apoptosis. The fibroblast cells derived from mice with homozygous deletion of IRF-1 gene were resistant to drug- and UV-induced apoptosis (50). Since we have found that vIRF-1 interferes with the function of IRF-1 in infected cells, we wished to examine whether vIRF-1 could also interfere with the cellular function of IRF-1. The NIH 3T3 cell line that constitutively expressed transfected vIRF-1 (Fig. 2A) and showed a decreased sensitivity to the antiviral effect of mouse IFN-α and IFN-β (data not shown) was analyzed for its sensitivity to TNF-α-induced apoptosis (24). The NIH 3T3/vIRF-1 and the parental NIH 3T3 cells were treated with an increasing concentration of TNF-α in the presence of actinomycin D, and the cell viability was determined 24 h later by MTT assay (12). The results, shown in Fig. 2B, demonstrate that NIH 3T3/vIRF-1 cells were less susceptible to TNF-α-induced apoptosis than were the parental NIH 3T3 cells. At the TNF-α concentration of 4 pg/ml, no cell death could be detected in NIH 3T3/vIRF-1 cells, while more than 50% of the NIH 3T3 cells were killed. The same degree of difference in cell killing between the vIRF-1-expressing clone and the parental line was seen in cells treated with double-stranded RNA (data not shown).
FIG. 2.
NIH 3T3 cells expressing vIRF-1 showed resistance to TNF-α-mediated apoptosis. (A) Expression of vIRF-1 mRNA in the clone (3T3/vIRF-1) selected for studies of apoptosis (Northern blot). The NIH 3T3 cell line expressing vIRF-1 was generated as described in Materials and Methods. (B) Comparison of TNF-α-induced cell killing in control NIH 3T3 cells and NIH 3T3/vIRF-1 cells. Cells were treated with the indicated concentrations of mouse TNF-α and actinomycin D (1 μg/ml) for 24 h, and the cell viability was determined by MTT assay. The data represent an average (± the standard error of the mean) of two separate experiments done in triplicate.
These data further suggest that the expression of vIRF-1 interferes with the functions of IRF-1 even in the absence of viral infection.
vIRF-1 interacts with cellular IRF transcription factors in vitro.
It was previously reported by others that recombinant, full-length vIRF-1 was unable to bind ISRE elements and a positive regulatory domain (PRD-I) present in the IFNB promoter (15, 57). Since vIRF-1 contains an N′-terminal 85-aa peptide which is not present in any other cellular IRFs and which could interfere with binding of the putative DNA binding domain (DBD), we examined whether the removal of this peptide would restore DNA binding. However, the N′-terminal-truncated vIRF-1 expressed as a GST fusion protein (aa 85 to 449) was also unable to bind these probes (data not shown). These data indicated that the inhibition by vIRF-1 was not the result of competition with IRF-1 for the DNA binding as demonstrated for IRF-1 and IRF-2. It was shown that the activity of cellular IRFs could be modulated by their association either with other family members or other transcription factors (10, 39, 48). We therefore examined whether the observed inhibitory effect of vIRF-1 could be mediated by its interactions with cellular IRFs or other cellular transcription cofactors.
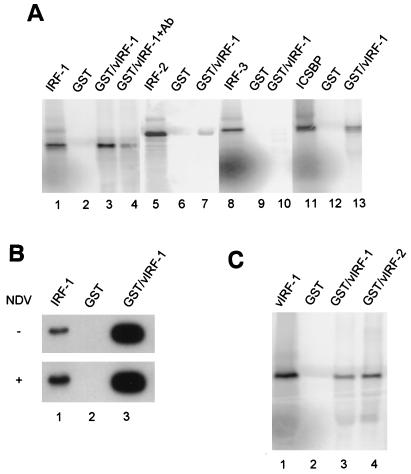
To analyze the interaction between vIRF-1 and the members of the IRF family of transcription factors, we used in vitro pull-down assay. GST–vIRF-1 fusion protein was immobilized on glutathione-agarose beads and incubated with the in vitro-synthesized 35S-labeled IRF proteins. The immobilized vIRF-1 bound strongly IRF-1 (Fig. 3A, lanes 1 to 3), and 21% of the input 35S-labeled IRF-1 was bound to immobilized vIRF-1. No binding was detected to control beads containing GST only. Furthermore, adding antibody that recognized a C′-terminal peptide of IRF-1 (lane 4) inhibited the binding of IRF-1 to GST–vIRF-1 by about fivefold. These data indicate that the observed binding of IRF-1 to vIRF-1 is specific. Only 0.2% of in vitro translated IRF-2 bound to the immobilized GST–vIRF-1, a level of binding which we considered to be insignificant (lane 7). We were also unable to detect any specific binding of in vitro-translated IRF-3 to GST–vIRF-1 (Fig. 3A, lanes 8 to 10). Although the binding of ICSBP to immobilized GST–vIRF-1 was lower than the binding of IRF-1, 6% of the input ICSBP was retained on GST–vIRF-1 beads (lanes 11 to 13). It should be mentioned at this point that Zimring et al. (57) failed to detect any association between in vitro-cotranslated vIRF-1 and IRF-1.
FIG. 3.
(A) Analysis of interactions between vIRF-1 and cellular IRF transcription factors by in vitro pull-down assay. Recombinant GST–vIRF-1 fusion protein was immobilized on glutathione-agarose beads and incubated with indicated 35S-labeled IRF proteins as described in Materials and Methods. In lanes 1, 5, 8, and 11, 20% of the respective in vitro-labeled input IRF was loaded onto the gel. Lanes 2, 6, 9, and 12 show binding to beads containing GST protein only. Lanes 3, 7, 10, and 13 show binding of IRF-1, IRF-2, IRF-3, and ICSBP, respectively. The binding in the presence of antibody (1 μg) against C-terminal peptide of IRF-1 is shown in lane 4. (B) Virus infection does not modulate IRF-1–GST–vIRF-1 interaction in cell lysates. NIH 3T3 cells were transfected with IRF-1 expression plasmid, and 24 h later cells were infected with NDV (multiplicity of infection of 5) for 16 h. Cell lysates from control (−) or infected cells (NDV) were then subjected to pull-down assay. GST–vIRF-1-bound proteins were resolved on SDS–10% PAGE and immunoblotted with anti-IRF-1 antibody (lane 3). Input (lane 1) represents 1% of total protein added into binding reaction. No detectable interaction was observed with control, GST-containing beads (lane 2). (C) Detection of vIRF-1 and vIRF-2 protein interactions. In vitro 35S-labeled vIRF-1 and vIRF-2 proteins were incubated with GST–vIRF-1 or GST bound to glutathione-agarose beads. Bound proteins were resolved by SDS–10% PAGE. Input vIRF-1 (lane 1) represents 20% of 35S-labeled vIRF-1 translation mixture added to beads.
To further examine whether viral infection modifies the binding of IRF-1 to vIRF-1, we analyzed the binding of IRF-1 from cell lysates of transiently transfected NIH 3T3 cells with IRF-1 expressing vector and infected with NDV. As seen in Fig. 3B, a significant amount of IRF-1 was coprecipitated with GST–vIRF-1 beads from cell lysates of IRF-1-transfected NIH 3T3 cells (about 30% of input); however, the binding was not significantly different if the lysates were prepared from NDV-infected cells (Fig. 3B). Since viral infection stimulated the expression of endogenous IRF-1 gene, the IRF-1 signal in infected cells was higher than in uninfected cells. Virus stimulation therefore did not influence the interaction between IRF-1 and vIRF-1.
We further examined whether vIRF-1 was able to form homodimers or interacted with vIRF-2 by using the pull-down assay with 35S-labeled vIRF-1. As seen in Fig. 3C, vIRF-1 was retained both by GST–vIRF-1 and GST–vIRF-2 immobilized on agarose beads (lanes 3 and 4), while no vIRF-1 was pulled down by the control, GST-containing agarose beads (lane 2). These data indicate that, in vitro, vIRF-1 can form homodimers as well as interact with vIRF-2.
Identification of the vIRF-1 binding domain.
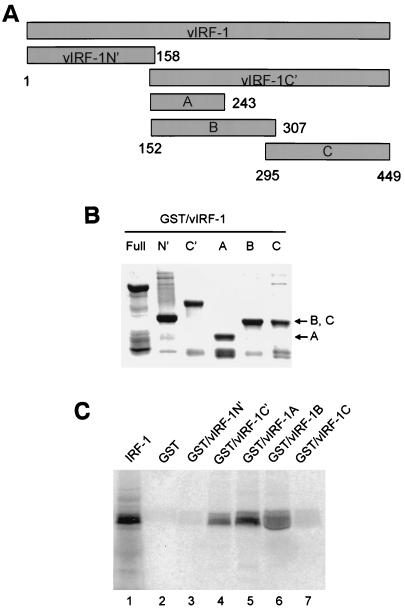
To determine which part of vIRF-1 protein interacted with IRF-1, we constructed GST fusion proteins with the N′-terminal (aa 1 to 158) and C′-terminal (aa 152 to 449) parts of vIRF-1. The C′-terminal part of vIRF-1 molecule was further divided to three peptides designated parts A (aa 152 to 243), B (aa 152 to 307), and C (aa 295 to 449) (Fig. 4A), which were also expressed as GST fusion proteins. The mobility and purity of these recombinant fusion peptides is shown in Fig. 4B. When used in the pull-down assay, GST–vIRF-1C′ was able to bind effectively in vitro-translated 35S-labeled IRF-1 (Fig. 4C, lane 4), while no binding of the labeled IRF-1 to GST–vIRF-1N′ was detected (lane 3), indicating that IRF-1 interacts specifically with the C′-terminal part of vIRF-1. To determine precisely the IAD of vIRF-1, we analyzed the binding of 35S-IRF-1 to the immobilized GST–vIRF-1A, -1B, and -1C fusion peptides. It can be seen (Fig. 4C, lanes 5 and 6) that IRF-1 binds to both immobilized GST–vIRF-1A and GST–vIRF-1B fusion peptides, while no interaction with GST–vIRF-1C peptide was observed (lane 7). Since the A and B peptides partially overlap, we conclude that the vIRF-1 IAD is located between aa 152 and 243.
FIG. 4.
Mapping of vIRF-1 interaction domain. (A) Positional scheme of vIRF-1 deletion mutants. Numbers correspond to amino acid boundaries of respective fragments. (B) Coomassie blue-stained gel illustrating the purity and size of the respective GST–vIRF-1 fusion fragments. (C) In vitro interaction between IRF-1 and different vIRF-1 fragments as detected by pull-down assay. 35S-labeled IRF-1 input (20%) and its binding to GST beads are shown in lanes 1 and 2, respectively. IRF-1 was pulled down by GST–vIRF-1C′ (lane 4) and not by GST–vIRF-1N′ (lane 3). Both GST–vIRF-1A (lane 5) and GST–vIRF-1B (lane 6) fragments actively bound IRF-1, while no IRF-1 binding was detected with GST–vIRF-1C fragment (lane 7).
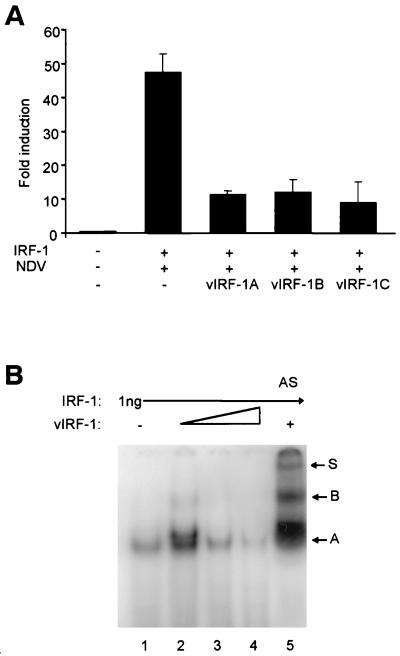
Using the transient-transfection assay, we assessed the ability of the C′-terminal peptides of vIRF-1 to inhibit transcriptional activity of the IFNA4 promoter. We cotransfected IFNA4 CAT reporter plasmid together with IRF-1 and vIRF-1A, -1B, and -1C (Fig. 4) expressing plasmids into NIH 3T3 cells, which were then infected with NDV. The results in Fig. 5A show that C′-terminal vIRF-1 peptides were able to downregulate the transcriptional activity of the IFNA4 promoter. All the C′-terminal peptides were better inhibitors than the full-length vIRF-1, indicating that, in the full-length protein, the IAD may be less accessible or actively regulated by other domains of the molecule. Interestingly, the C peptide of vIRF-1 that was not able to bind effectively to IRF-1 in the GST pull-down assay was also inhibitory. Since in our preliminary results the C peptide of vIRF-1 did not interact strongly with the p300 protein (data not shown), the observed inhibition of IFNA4 activity by the vIRF-1C peptide may be due to its interaction with other regulatory proteins. These data indicate that vIRF-1 peptides that are able to bind either IRF-1 or p300 can also downregulate their transactivating activities onto IFNA4 promoter in the transient-transfection assay.
FIG. 5.
(A) The C′-terminal peptides of vIRF-1 disrupt the specific activation of the IFNA4 promoter by NDV and IRF-1. The cotransfections were done as described in Fig. 1 by using 1 μg of each IRF-1, IFNA4 CAT, and the respective plasmid encoding the C′-terminal vIRF-1 peptides. The relative levels of vIRF-1 peptides in transfected cells were comparable (data not shown). Error bars show the standard errors for triplicate experiments. (B) vIRF-1 modulates the binding of IRF-1 to PRD-I oligonucleotide. Gel retardation assays were performed with purified recombinant GST–IRF-1 and His6–vIRF-1 proteins and 32P-labeled PRD-I probe. A total of 1 ng of GST–IRF-1 was preincubated alone (lane 1) or in the presence of 3, 5, or 7 ng of His6–vIRF-1 (lanes 2 to 4). The same reaction as in lane 2 was performed in the presence of 3 μl of vIRF-1 antiserum (AS, lane 5).
To test whether the formation of vIRF-1 and IRF-1 heterodimers affects the DNA binding activity of IRF-1, we subjected a preincubated mixture of purified recombinant GST–IRF-1 and His6–vIRF-1 proteins together with radioactively labeled DNA probe corresponding to the PRD-I binding site in the IFNB gene promoter to electrophoretic mobility shift analysis. As can be seen in Fig. 5B, incubation of 1 ng of GST–IRF-1 with PRD-I probe resulted in the formation of one distinct DNA-protein complex A (lane 1). When His6–vIRF-1 protein (3 ng) was added to binding reaction, the intensity of complex A increased and a new complex with a slower mobility was formed (lane 2). This new complex could represent a vIRF-1–IRF-1 heterodimer. However, when an increased amount of His6–vIRF-1 protein (5 and 7 ng) was preincubated with GST–IRF-1, the DNA binding activity of GST–IRF-1 diminished (lanes 3 and 4). To confirm the presence of His6–vIRF-1 protein in the GST–IRF-1 DNA complex, antiserum against vIRF-1 was added to the binding reaction. As seen in Fig. 5B, lane 5, in the presence of vIRF-1 antiserum, two new complexes with lower mobilities (B and S) were detected. These data indicate that at equal molar ratio, IRF-1–vIRF-1 heterodimer can bind PRD-I element, but an excess of vIRF-1 blocks binding of IRF-1 to DNA.
Interaction of vIRF-1 and IRF-1 with the transcription coactivator, p300.
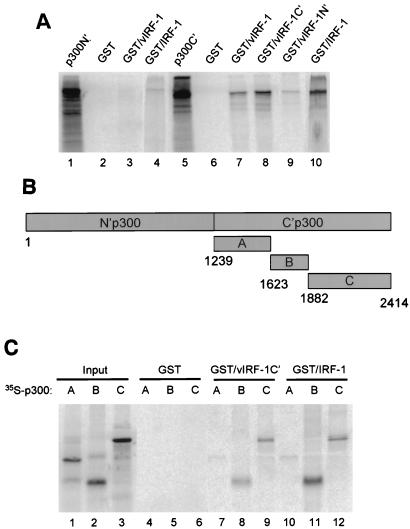
Interaction between transcription factors of the IRF family, IRF-3 and IRF-7, and transcription coactivator p300 has been shown to have a functional impact (2, 21, 27, 52, 55). Therefore, we tested the affinity of vIRF-1 for p300 by the pull-down assay. The N′- and C′-terminal parts of p300 (for details, see Material and Methods) were translated in vitro and subjected to pull-down assay on immobilized GST–vIRF-1 fusion proteins. The interaction between IRF-1 and p300 was analyzed by using the same assay. As shown in Fig. 6A, only C′-terminal part of p300 was able to bind to immobilized GST–vIRF-1 or GST–IRF-1 (lanes 7 and 10), while no significant interaction between the N′-terminal part of p300 and vIRF-1 or IRF-1 could be detected (lanes 3 and 4). We further found that the C′-terminal domain of vIRF-1 strongly binds to p300C′ (lane 8), whereas the binding of p300C′ to GST–vIRF-1N′ beads was very low and probably insignificant (lane 9). No binding of either the N′-terminal or the C′-terminal part of p300 to GST alone was detected (lanes 2 and 6). These data indicate that both vIRF-1 and IRF-1 bind in vitro to a C′-terminal half of p300 transcription coactivator. This region of p300 is targeted by a variety of transcription factors, including STAT-1, c-Fos, E1A, TFIIB, NF-κB, and kinase pp90 (13). To map precisely the region of p300 binding to vIRF-1 and IRF-1 proteins, we divided p300C′ molecule into three fragments: A (aa 1239 to 1622), B (aa 1623 to 1882), and C (aa 1882 to 2414) (Fig. 6B). The mobility and purity of in vitro-translated A, B, and C peptides on SDS gel is shown in Fig. 6C (lanes 1, 2, and 3). It can be seen (Fig. 6C) that in a pull-down assay only the B and C peptides were able to bind to the immobilized GST–vIRF-1C′ fusion protein (lanes 8 and 9). The B and C peptides also bound to the immobilized GST–IRF-1 fusion protein (lanes 11 and 12). No significant binding of A peptide to GST–vIRF-1C′ or GST–IRF-1 beads was observed (lanes 7 and 10). None of these peptides bound to GST beads only (lanes 4 to 6). These data indicate that both vIRF-1 and IRF-1 proteins bind to a C′-terminal region of p300 protein containing the C/H3 and Q-rich domains (aa 1623 to 2414) and may compete for the same binding site.
FIG. 6.
In vitro binding analysis of vIRF-1 and IRF-1 to p300. (A) Interaction of vIRF-1 and IRF-1 with the N′- and C′-terminal parts of p300. Binding of in vitro 35S-labeled N′-terminal half of p300 to immobilized GST–vIRF-1 and GST–IRF-1 is shown in lanes 3 and 4. The binding of the in vitro 35S-labeled C-terminal half of p300 to immobilized GST–vIRF-1 and GST–IRF-1 is shown in lanes 7 and 10. The binding of the C-terminal half of the p300 to N- and C-terminal parts of GST–vIRF-1 is shown in lanes 8 and 9, respectively. Lanes 2 and 6 show the binding to GST beads only, and lanes 1 and 5 show the input of the 35S-labeled N- or C-terminal part of p300 (20%). (B) Positional scheme of p300 deletion mutants. The numbers correspond to the amino acid boundaries of the respective fragments. (C) Mapping of p300 binding site for vIRF-1 and IRF-1 by in vitro pull-down assay. Recombinant GST–vIRF-1C′ and GST–IRF-1 fusion proteins were immobilized on glutathione-agarose beads and incubated with indicated 35S-labeled p300 peptides as shown in Fig. 6B. In lanes 1 to 3, 20% of the in vitro-labeled p300 fragments A, B, and C were analyzed on an SDS gel. Lanes 4 to 6 show the binding to beads containing GST protein only. Lanes 7 to 9 show the binding of the indicated p300 fragment to GST–vIRF-1C′ beads, and lanes 10 to 12 show the binding of the indicated p300 fragment to GST–IRF-1 beads.
DISCUSSION
HHV-8 is the first identified viral genome that contains ORFs homologous to the cellular transcriptional factors of the IRF family. While cellular IRFs are DNA binding proteins, neither the full-size vIRF-1 nor its 5′-end deletion mutant was able to bind oligonucleotides with specificities similar to those of the cellular IRFs. The vIRF-1 shows partial homology in its N-terminal region with a DBD of IRF-3 and IRF-4. However, only two of the five characteristic W repeats in cellular IRFs are preserved in vIRF-1. Nevertheless, it is unlikely that the amount of W repeat in the N terminus of vIRF-1 accounts entirely for the inability of this protein to bind DNA. This region of vIRF-1 is very similar to vIRF-2, which we have recently shown to be able to bind oligonucleotide corresponding to the NF-κB binding site (11). In contrast, vIRF-1 cannot bind to NF-κB probe (data not shown). However, it is possible that the C′-terminal part of the vIRF-1 may interact with and mask the binding domain of vIRF-1. The full-size mouse IRF-4 (PIP) is also unable to bind DNA because of the interaction between the N′-terminal and C′-terminal parts of the PIP protein (10). However, when this interaction is prevented either by deletion of the C-terminal part of the PIP protein or by its binding to another transcription factor, PU.1, the DNA binding capacity of PIP is restored (39). Experiments are in progress to examine whether 3′ deletion mutants of vIRF-1 regain the ability to bind DNA.
It was suggested previously that the expression of vIRF-1 may contribute to viral mimicry and enable HHV-8 to escape the antiviral effect of IFN since vIRF-1 was shown to downregulate IFN-stimulated transcriptional activity of various ISG promoters; however, the mechanism by which vIRF-1 exerts this effect was not clarified (15, 26, 38, 57). We have further extended this observation and shown in this study that vIRF-1 inhibited virus-mediated transcriptional activation of the IFNA4 promoter. Furthermore, vIRF-1 effectively downmodulated the synergistic interaction between virus and IRF-1 or IRF-3, indicating that vIRF-1 interferes with the transactivation ability of these two cellular IRFs. In the absence of demonstrable vIRF-1 DNA binding, we examined the possibility that vIRF-1 is blocking the transactivation activity of IRF-1 and IRF-3 by direct interaction with these proteins, as was demonstrated for ICSBP-mediated inhibition of IRF-1 activation (53). Our data show that GST–vIRF-1 interacts strongly with both in vitro-translated IRF-1 and IRF-1 present in the cellular extracts from infected and uninfected cells. However, in the same assay, no interaction between vIRF-1 and in vitro-translated IRF-3 was detected. There may be at least two reasons for this discrepancy. (i) IRF-3 is phosphorylated in infected cells (21, 27, 46, 55), and there is an indication of an interaction between the C′- and N′-terminal parts of the unphosphorylated IRF-3 peptide (unpublished results). Therefore, it is possible that IRF-3 has to be phosphorylated in the C′ terminus to uncover the interacting domain. (ii) It was shown that IRF-3 interacts with the C′-terminal part of the CBP/p300 transcriptional cofactor (27, 52, 55) and that this interaction is functional, since E1A inhibits IRF-3-mediated transactivation by targeting p300 (21). Our results show that vIRF-1 also interacts with C′-terminal part of p300, and thus the observed inhibition could be the result of competitive binding between IRF-3 and vIRF-1 to p300. Additional experiments are being performed to determine which of these mechanisms are operative.
The observation that vIRF-1 can inhibit virus-mediated activation of type I IFN gene promoter also in fibroblast cells lacking the IRF-1 gene (data not shown) further indicates that vIRF-1 inhibition is not limited to IRF-1-mediated activation (57). While vIRF-1 interacts strongly with IRF-1, it also binds p48 (data not shown) and ICSBP in vitro. Since the IFN-stimulated induction of ISGs is mediated by the ISGF3γ complex consisting of p48, STAT1, and STAT2 proteins, the interaction of vIRF-1 with p48 could result in the disruption of this complex; however, the formation of ISGF3γ complex in IFN-treated cells does not seem to be prevented in cells overexpressing vIRF-1 (37a). Studies with p48 and IRF-1 knockout mice revealed that the establishment of maximal antiviral state is impaired in cells that have deleted either of these genes and showed that both IRF-1 and p48 have essential and nonredundant functions in the antiviral response to IFNs (22). The interaction of vIRF-1 with ICSBP is also of interest since ICSBP interacts with IRF-1 and inhibits its transactivating ability in a cell-type-specific manner (48). Therefore, it is possible that binding of vIRF-1 to ICSBP could result in a potentiation of IRF-1 transcriptional activity.
The association of vIRF-1 with IRF-1 may alter not only the cellular responses to IFNs and viral infection but also a variety of cellular responses not related to viral infection. IRF-1 is a prototype transcription factor that can modulate the expression of a number of cellular genes, including those involved in the regulation of cell growth, susceptibility to transformation, and apoptosis. It was shown that the ectopic expression of IRF-1 inhibits cell growth, while overexpression of IRF-2 (35) and vIRF-1 (15) causes the oncogenic transformation in NIH 3T3 cells. It was also shown that the overexpression of IRF-1 could reverse the IRF-2-transformed NIH 3T3 cells to a normal phenotype (18). These data indicate that the balance between IRF-1 and its antagonists may significantly affect cells growth. Notably, we have shown in this study that the vIRF-1-overexpressing NIH 3T3 cells are resistant to TNF-α-induced apoptosis. As previously reported, similar observations were also made with cells that contained homozygous deletion of IRF-1 (22) or in which the expression of IRF-1 was impaired (25). It was shown that IRF-1 has an essential role in the regulation of the expression of p21waf cell cycle inhibitor. The promoter of p21waf contains three potential IRF-1 binding sites and can be activated by IRF-1. Basal expression of p21waf was found to be dramatically decreased both in IRF-1−/− mouse fibroblast (50), as well as in NIH 3T3 cells overexpressing vIRF-1 (15) that were tumorigenic. It remains to be determined whether the observed resistance of NIH 3T3/vIRF-1 cells to TNF-α-induced apoptosis could be entirely related to deregulation of p21waf or whether vIRF-1 by targeting CBP/p300 coactivator and displacing binding of IRF unrelated transcription factors downregulates the expression of proapoptotic genes as demonstrated recently in NIH 3T3 expressing catalytic variant of PKR (5).
In conclusion, our results suggest that, by inserting IRF-like ORF into viral genome, HHV-8 developed an effective mechanism of viral mimicry which allows the virus to both overcome the induction of type I IFN genes and to inhibit their functions. However, the expression of vIRF-1 can result in changes not directly related to viral mimicry. As shown in the present study, there are at least two mechanisms by which vIRF-1 modulates expression of cellular genes. First, the association of vIRF-1 with the cellular multifunctional factor IRF-1 alters its functional diversity in multiple cellular responses. Second, competitive binding of vIRF-1 with specific transcription factors to a discrete domain of the transcriptional coactivator p300 may also affect an expression of IRF-1-independent genes and result in a more general inhibition of gene transcription. Therefore, it will be important to identify target genes expression of which is modulated by vIRF-1.
ACKNOWLEDGMENTS
This study was supported by grants CA76946 (NCI) and AI19737 (NIAID) from the National Institutes of Health (P.M.P.).
We thank J. Nicholas for the λ phage library and the BCBL-1 cells and J. Hiscott, T. Taniguchi, and K. Ozato for the p65, IRF-1, IRF-2, and ICSBP expression plasmids, respectively, and G. Nabel for p300 plasmids. We also thank R. Pine for sharing his unpublished results.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Hones R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au W-C, Moore P A, LaFleur D W, Tomball B, Pitha P M. Characterization of intereferon regulatory factor-7, and its potential role in the transcriptional activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 3.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 5.Balachandran S, Kim C N, Yeh W C, Mak T W, Bhalla K, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluyssen H A, Muzaffar R, Vlieststra R J, van der Made A C, Leung S, Stark G R, Kerr I M, Trapman J, Levy D E. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc Natl Acad Sci USA. 1995;92:5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluyssen H A R, Durbin J E, Levy D E. ISGF3gamma p48, a specificity switch for IFN activated transcriptional factors. Cytokine Growth Factor Rev. 1996;7:11–17. doi: 10.1016/1359-6101(96)00005-6. [DOI] [PubMed] [Google Scholar]
- 8.Boultwood J, Fidler C, Lewis S, MacCarthy A, Sheridan H, Kelly S, Oscier D, Buckler V J, Wainscoat J S. Allelic loss of IRF1 in myelodysplasia and acute myeloid leukemia: retention of IRF1 on the 5q-chromosome in some patients with the 5q-syndrome. Blood. 1993;82:2611–2616. [PubMed] [Google Scholar]
- 9.Bovolenta C, Driggers P H, Marks M S, Medin J A, Politis A D, Vogel S N, Levy D E, Sakaguchi K, Appella E, Coligan J E, et al. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci USA. 1994;91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is import for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 11.Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 12.Cheung S C, Chattopadhyay S K, Hartley J W, Morse H C, Pitha P M. Aberrant expression of cytokine genes in peritoneal macrophages from mice infected with LP-BM5 MuLV, a murine model of AIDS. J Immunol. 1991;146:121–127. [PubMed] [Google Scholar]
- 13.Daniel P B, Walker W H, Habener J F. Cyclic AMP signaling and gene regulation. Annu Rev Nutr. 1998;18:353–383. doi: 10.1146/annurev.nutr.18.1.353. [DOI] [PubMed] [Google Scholar]
- 14.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 15.Gao S J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;85:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 16.Giese N A, Gabriele L, Doherty T M, Klinman D M, Tadesse-Heath L, Contursi C, Epstein S L, Morse H C., III Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J Exp Med. 1997;186:1535–1546. doi: 10.1084/jem.186.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 18.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 19.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Sok-PinTan R, Takasugi T, Kadokawa Y, Schindler C, Schreiber R D, Noguchi S, Taniguchi T. Regulation of IFN-α/β genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-α/β. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 20.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch K-P, Gabriele L, Waring J F, Bachmann M F, Zinkernagel R M, Morse H C I, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 21.Juang Y, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan R S, Takasugi T, Matsuyama T, Mak T W, Noguchi S, Taniguchi T. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type III interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 23.Kirchhoff S, Schaper F, Hauser H. Interferon regulatory factor 1 (IRF-1) mediates cell growth inhibition by transactivation of downstream target genes. Nucleic Acids Res. 1993;21:2881–2889. doi: 10.1093/nar/21.12.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig T M, Amakawa R, Kishihara K, Wakeham A, Potter J, Furlonger C L, Narendran A, Suzuki H, Ohashi P S, Paige C J, Taniguchi T, Mak T W. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 30.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 31.Mosca J D, Bednarik D P, Raj N B K, Rosen C A, Sodroski J G, Haseltine W A, Hayward G S, Pitha P M. Activation of human immunodeficiency virus by herpesvirus infection: identification of a region within the long terminal repeat that responds to a trans-acting factor encoded by herpes simplex virus 1. Proc Natl Acad Sci USA. 1987;84:7408–7412. doi: 10.1073/pnas.84.21.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson N, Marks M S, Driggers P H, Ozato K. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen H, Mustafa A, Hiscott J, Lin R. Transcription factor IRF-2 exerts its oncogenic phenotype through the DNA binding/transcription repression domain. Oncogene. 1995;11:537–544. [PubMed] [Google Scholar]
- 36.Nonkwelco C, Ruf I K, Sample J. The Epstein-Barr virus EBNA-1 promoter Qp requires an initiator-like element. J Virol. 1997;71:354–361. doi: 10.1128/jvi.71.1.354-361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 37a.Pine, R. Personal communication.
- 38.Pitha P M, Burysek L, Schafer S. Critical role of Kaposi sarcoma virus (HHV8)-encoded IRFs in virus replication and pathogenicity. J Interferon Cytokine Res. 1997;17(Suppl. 2):S37. [Google Scholar]
- 39.Pongubala J M, Atchison M L. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc Natl Acad Sci USA. 1997;94:127–132. doi: 10.1073/pnas.94.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj N B K, Au W-C, Pitha P M. Identification of a novel virus-responsive sequence in the promoter of murine interferon-α genes. J Biol Chem. 1991;266:11360–11365. [PubMed] [Google Scholar]
- 41.Raj N B K, Israeli R, Kellum M, Pitha P M. Upstream regulatory elements of murine α4 gene confer inducibility and cell type-restricted expression. J Biol Chem. 1989;264:11149–11157. [PubMed] [Google Scholar]
- 42.Reis L F, Ruffner H, Stark G, Aguetand M, Weissmann C. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruffner H, Reis L F, Naf D, Weissmann C. Induction of type I interferon genes and interferon-inducible genes in embryonal stem cells devoid of interferon regulatory factor 1. Proc Natl Acad Sci USA. 1993;90:11503–11507. doi: 10.1073/pnas.90.24.11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 46.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 47.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 48.Sharf R, Azriel A, Lejbkowicz F, Winograd S S, Ehrlich R, Levi B Z. Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J Biol Chem. 1995;270:13063–13069. doi: 10.1074/jbc.270.22.13063. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier M S, Aizawa S, Mak T W, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka N, Ishihara M, Lamphier M S, Nozawa H, Matsuyama T, Mak T W, Sizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumour suppression IRF-1 and p53 in response to DNA damage. Nature. 1996;382:555–568. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 51.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 52.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weisz A, Marx P, Sharf R, Appella E, Driggers P H, Ozato K, Levi B-Z. Human interferon consensus sequence binding protein a negative regulator of enhancer elements to interferon-inducible genes. J Biol Chem. 1992;267:25589–25596. [PubMed] [Google Scholar]
- 54.Willman C L, Sever C E, Pallavicini M G, Harada H, Tanaka N, Slovak M L, Yamamoto H, Harada K, Meeker T C, List A F, et al. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemia myelodysplasia. Science. 1993;259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- 55.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]