Summary
Circadian clocks are 24-hour endogenous oscillators in physiological and behavioral processes. Though recent transcriptomic studies have been successful in revealing the circadian rhythmicity in gene expression, the power calculation for omics circadian analysis have not been fully explored. In this paper, we develop a statistical method, namely CircaPower, to perform power calculation for circadian pattern detection. Our theoretical framework is determined by three key factors in circadian gene detection: sample size, intrinsic effect size and sampling design. Via simulations, we systematically investigate the impact of these key factors on circadian power calculation. We not only demonstrate that CircaPower is fast and accurate, but also show its underlying cosinor model is robust against variety of violations of model assumptions. In real applications, we demonstrate the performance of CircaPower using mouse pan-tissue data and human post-mortem brain data, and illustrate how to perform circadian power calculation using mouse skeleton muscle RNA-Seq pilot as case study. Our method CircaPower has been implemented in an R package, which is made publicly available on GitHub (https://github.com/circaPower/circaPower).
Keywords: circadian analysis, cosinor model, experimental design, power calculation
1 |. INTRODUCTION
Circadian rhythms are endogenous ~24 hour oscillations of behavior, physiology, and homeostasis in adaption to the diurnal cycle caused by the earth’s daily rotation. The circadian clock is found in virtually all cells throughout the body and controls oscillations in a wide variety of physiological processes, including sleep-wake cycles, body temperature, and melatonin secretion1,2,3,4. From the literature, the mechanism that drives circadian rhythms is a transcriptionial-translational feedback loop encoded by a set of core clock genes 5, including CLOCK, BMAL1 as the transcriptional activators; and period family (PER1, PER2, PER3) and cryptochrome family (CRY1, CRY2) as the major inhibitors. In addition to core clock genes, genome-wide transcriptomic studies have revealed additional circadian genes in post-mortem brain6,7, skeletal muscle8, liver9, and blood10. Transcriptomic circadian analyses in human11, mouse12, and baboon13 have shown that the circadian pattern in gene expression could be tissue-specific. Beyond transcriptomic data, circadian rhythmicity was also discovered in other types of omics data including DNA methylation14, ChIP-Seq (chromatin immunoprecipitation assays with sequencing)15, proteomics16, and metabolomics17. From epidemiology and animal studies, the disruption in clock and circadian gene expression was found to be linked to diseases including type 2 diabetes18, cancer19,20, sleep10, major depression disorder21, aging6, schizophrenia7, and Alzheimer’s disease22.
As the circadian omics studies have become increasingly popular over the years (Figure. 1), the experimental design of such circadian omics studies has come into focus23,24, where the design refers to the distribution of the collected Zeitgeber time (ZT; standardized diurnal time with ZT0/ZT24 for the beginning of day and ZT12 for the beginning of night). In this paper, we consider two types of sampling design: passive and active sampling design. In passive design, investigators have no control of the collected ZT. Such a passive design is commonly seen in studies with human tissues that are difficult to obtain (e.g., post-mortem brain tissues6,7,21) and the irregular sampling distribution should be considered in power calculation. In contrast, investigators have full control of the sample collection time in an active sampling design. Such an active design is commonly seen in animal studies 12 or human blood studies 10. In the literature, 6 time points (every 4 hours) per cycle across one or multiple full cycles have been widely adopted in many actively designed studies25,26,27. Hughes et al.28 recommended evenly sampling at least 12 time points per cycle (i.e., every 2 hours) across 2 full cycles. For the ease of discussion, we refer to this type of design as the “evenly-spaced sampling design”. Though these empirical practices and guidelines were presented and well-received, limited quantitative benchmarks are available. To address this, Ness-Cohn et al.29 developed a user-friendly website, TimeTrial, which allows researchers to explore the effects of experimental design on cycling detection. Although multiple circadian detection methods are allowed, the results are benchmarked through simulation using classification error rate and area under the curve of a ROC curve. However, a statistical method that enables exact power calculation is still lacking. Previous studies have reported the lack of overlapping circadian genes because of smaller number of samples9,30, indicating statistical power, i.e., the probability of successfully detecting the underlying circadian pattern, is not fully considered/justified. Thus, an analytical method that allows exact power calculation under different experimental designs is urgently needed. In addition, all these prior works on circadian study design were discussed within the scope of active design, where investigators have control of sample collection time. In the scope for passive design, there are no guidelines in the literature. It is unclear whether and how the irregular ZT distribution will impact the circadian power calculation.
Figure 1.
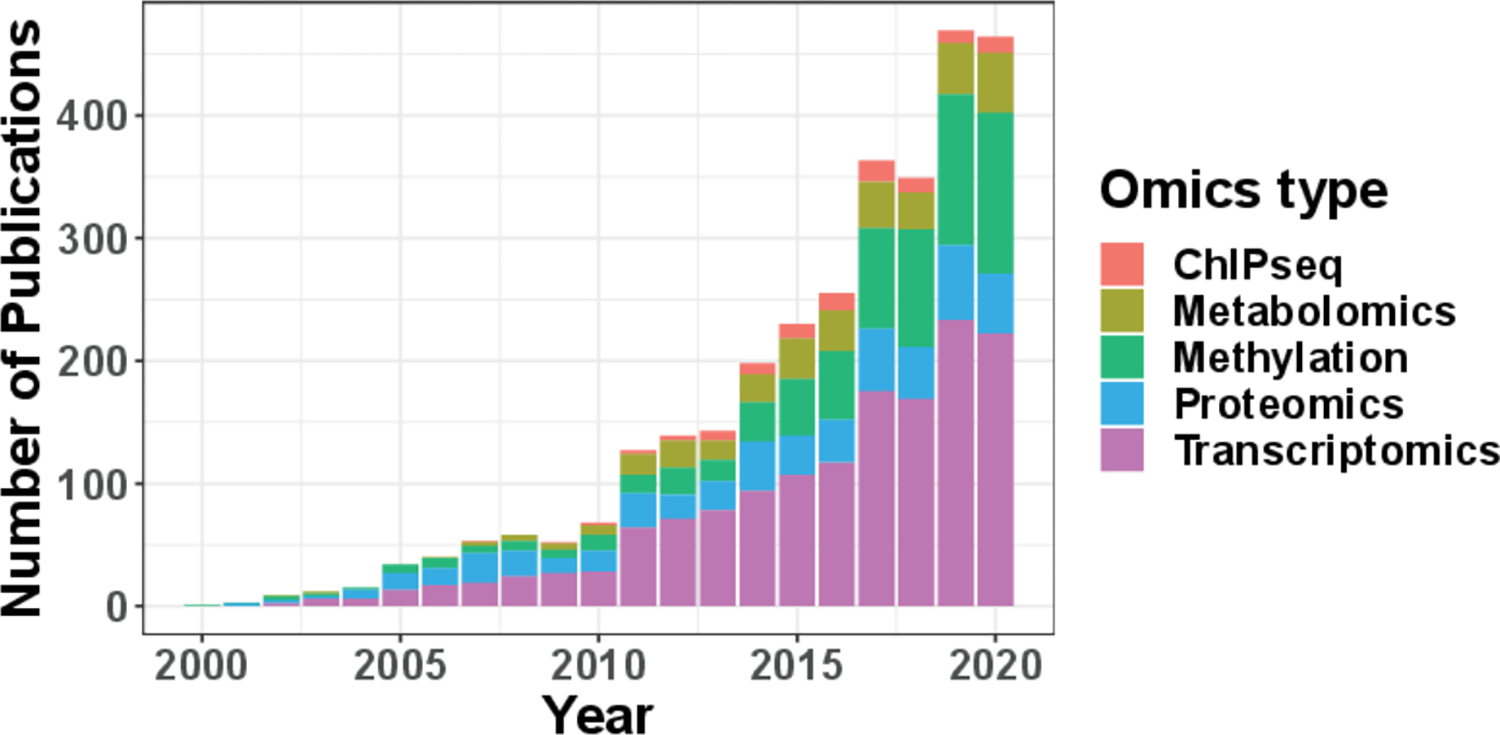
Annual number of publications on PubMed that contain the keywords “circadian/clock” and one of the following omics type: “ChIPseq”, “Metabolomics”, “Methylation”, “Proteomics”, “Transcriptomics”.
To fill in these research gaps, we propose a model-based approach to accurately calculate the circadian power (namely CircaPower), based on the cosinor model31,32. It assumes the expression level of a gene is a sinusoidal function of the circadian time 31. The biological rationale for using a cosinor model is that the circadian rhythm is amenable to adapt the cycles in the environment5, including the day-light cycle, the tides, the phases of the moon, the seasons, etc33. Since the day-light cycle is the leading environmental factor that governs circadian rhythms, a cosinor wave model is widely used to mimic the cosinor cycle of the day-light intensity and many previous literatures6,7,34 have used this model to identify biological meaningful findings. In the literature, there are several other algorithms developed for circadian rhythm detection. Lomb-Scargle periodograms 35 and COSOPT36 are more complicated parametric models. By assuming mixture of multiple cosinor curves with distinct periods, these methods facilitate the detection of oscillating transcripts with irregular shape. ARSER37, RAIN38 and JTK CYCLE39 are non-parametric methods, which are free of modeling assumption and more powerful to capture irregular curve shapes. Both parametric and non-parametric algorithms were widely applied in transcriptomic studies, and comparisons of these algorithms have been conducted in several review studies28,24,40,41. Although the complex methods have advantages to detect irregular curves beyond cosinor models, the power calculation using these methods are not always feasible since the effect size and data variability are not explicitly defined in these models. In addition, concerns about the accuracy of the statistical inference for these complex methods have been raised41. To be specific, the p-values generated by many of these methods may not be correct (i.e., do not follow a uniform distribution (0, 1) under the null), implying a potential inflated or deflated type I error rate. Therefore, we propose the power calculation framework based on the cosinor model, because of its flexibility in deriving closed-form test statistics and accurate statistical inference. We acknowledge that other complex parameters models and non-parametric approaches are also popular with their own unique merit, and exploring circadian power calculation using these complex methods is one of our future directions.
To the best of our knowledge, this is the first theoretical methodology developed for circadian power calculation in omics data. The unique contribution of this paper includes: (i) identifying factors related to the statistical power of circadian rhythmicity detection, including sample size, intrinsic effect size and sampling design; (ii) developing CircaPower, an analytical solution based on a closed-form formula, for fast and accurate circadian power calculation; (iii) demonstrating via simulations that the evenly-spaced sampling design is superior because of its phase-invariant property, which is also corroborated by theoretical proofs; (iv) illustrating how to calculate statistical power and to design a circadian experiment with pilot data via a case study; (v) collecting, calculating, and summarizing the intrinsic effect sizes of existing human and animal studies, which serves as a useful reference resource when no pilot data is available; and (vi) providing an open-source R package.
The superior performance of our method is demonstrated in comprehensive simulation studies, as well as multiple transcriptomic applications in human and mouse. We demonstrate the performance of CircaPower using continuous gene expression data throughout this manuscript, but our method is also applicable in single biomarker data or other types of continuous omics data, including but not restricted to ChIP-Seq, DNA methylation, proteomics, and metabolomics.
2 |. METHODS
The CircaPower framework assumes the relationship of the expression level (continuous data type) of a gene and the Zeitgeber time (ZT) fits a sinusoidal wave curve, and is based on the statistics of a cosinor model31. Below we introduce the model notations, the construction of the statistics, the null and alternative distribution of the statistics, the closed-form formula for circadian power calculation, and factors affecting the power calculation of circadian rhythmicity detection.
2.1 |. Notations and basic model
As illustrated in Figure. 2a denote as a cosine function of (i.e., ; as the ZT; as the MESOR (Midline Estimating Statistic Of Rhythm, a rhythm-adjusted mean); as the amplitude. is the frequency of the sinusoidal wave, where . Without loss of generality, we set period hours to mimic the diurnal period. is the phase shift of the sinusoidal wave curve. Whenever there is no ambiguity, we will omit the unit “hours” in period, phase, and other related quantities. Due to the periodicity of a sinusoidal wave, are not identifiable when . Therefore, we will restrict . Under the current cosine formula, also denotes the peak time .
Figure 2.
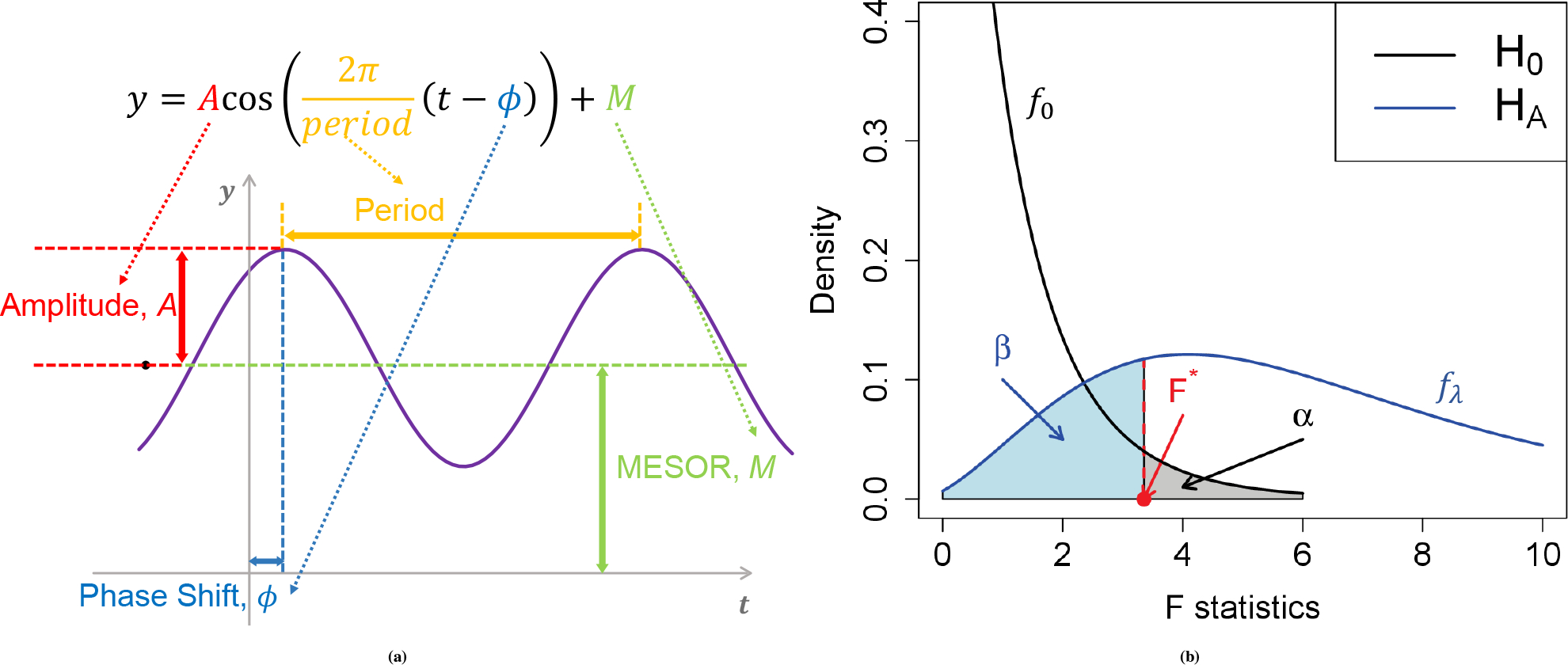
Basic sinusoidal model and the power/type I error discriminatory curve. (a) shows a sinusoidal wave curve underlying circadian rhythmicity power calculation framework. (b) shows the relationship between power and type I control in detecting circadian rhythmicity. The black curve represents the density function of the statistics under the null distribution (no circadian pattern) whose expectation is ; the blue curve represents the density function of the statistics under the alternative distribution whose expectation is . The difference in expectation between and is . The red dashed line represents the decision boundary (i.e., ) such that the type I error rate is controlled at (shaded gray). The corresponding type II error is the area with lightblue color and the detection power is .
For a given sample , is the total number of samples), denote by the expression value of a gene and the observed ZT. We assume the following sinusoidal wave function:
| (1) |
where is the error term for sample ; we assume ‘s are identically and independently distributed (i.i.d.) from , where is the noise level. To benchmark the goodness of sinusoidal wave fitting, we define the coefficient of determination
where , , , , with , , and being the fitted value for , , and in Equation 1 under least squares loss. ranges from 0 to 1, with 1 indicating perfect sinusoidal wave fitting, and 0 indicating no fitting at all. Denote , . Equivalently, we could re-write Equation 1 as
| (2) |
where , and , which turns into a linear regression problem.
According to the linear model theory, the statistics for the circadian model in Equation 1 can be derived as
| (3) |
where is number of independent samples, is number of parameters (i.e., , and in Equation 1.
2.2 |. Power calculation
2.2.1 |. Analytical power calculation
Under the null hypothesis of no circadian fitting (i.e., in Equation 1), from the linear model theory, , where denotes a regular distribution with degrees of freedom and (see Appendix A.1 for details). Under the alternative hypothesis (i.e., or in Equation 11), , where denotes a non-central F distribution with with noncentrality parameter (see Appendix A.2 for details). Figure. 2b shows the relationship between the null and alternative distributions of the statistics for the circadian model. By assuming the type I error rate at the rejection boundary is , the relationship between and the power is
where represents cumulative density function of evaluated at , and represents cumulative density function of evaluated at (see Appendix A for details).
As shown in the caption of Figure. 2b the non-centrality parameter controls the degree of separation of the null distribution and the alternative distribution . The larger the is, the more likely the alternative distribution will be away from the null distribution, and the higher power a gene will achieve. We thus define as the total effect size for the circadian power calculation. By inspecting the total effect size , this non-centrality parameter can be decomposed into three parts: (i) sample size , (ii) intrinsic effect size (closely relate to the goodness of fit statistics ), and (iii) sampling design effect . We then discuss the impact of each of these components on the circadian rhythmicity power calculation:
Sample size: As expected, given fixed and , a larger sample size will result in a larger total effect size , and achieve a higher statistical power.
Intrinsic effect size: Intuitively, a larger circadian amplitude with smaller residual variability will lead to a better sinusoidal curve fitting (i.e. larger ). Our formula suggests that circadian fitting parameters and work together as an intrinsic effect size and has a quadratic effect on the total effect size .
Sampling design effect: The sampling design effect is more complicated, because it involves both observed and the unknown parameter phase shift . It should be noted that the values of can either be unique, meaning that each sample has a distinct circadian time, or replicated, indicating that several replicates are taken at each time point. In general, given an arbitrary circadian sampling design, we need to estimate before performing power calculation. Fortunately, the power calculation for the evenly-spaced sampling design is independent of the phase value (i.e., phase-invariant). For example, Hughes et al.28 recommended a collection of 12 time points (every 2 hours) per cycle across 2 full cycles, which belongs to the evenly-spaced sampling design. Such active design is commonly seen in animal studies or human blood studies, where researchers can control the exact time to sacrifice the animal or to collect blood. The following theorem (phase-invariant property) shows that the sampling design effect is a constant under the one-period one-sample evenly-spaced design, i.e., are evenly spread within one period with only one sample per time point.
Theorem 1
(Phase-invariant property - one-period one-sample). Assuming there is a total of ZT points within a circadian period , which are ordered such that for all . If , and is evenly-spaced over the period (i.e., for all , is a fixed time interval, , then regardless of the value for , we have
The proof is given in Appendix B It can immediately be extended to the following corollary.
Corollary 1
(Phase-invariant property - multi-period multi-sample). For multi-period (two or more cycles) multi-sample evenly-spaced design, the sampling design effect is phase-invariant.
This is because the multi-period multi-sample evenly-spaced design just replicates the one-period one-sample evenly-spaced design, therefore the average of them remains to be 1/2.
Remark 1.
The even spread/space design invariance property is a mathematical derivation resulting from the equation , which holds true regardless of the phase. The intuition behind this is based on the cosinor function’s curvatures, which dictate that the sample’s contribution to power is more significant when is at a peak or trough, resulting in , and less significant when is at MESOR, resulting in . If the samples are evenly distributed across the period, their effects will be even out when averaged over, and the final sampling design effect .
2.2.2 |. Assumptions underlying the circadian modeling framework
The proposed circadian modeling framework has two underlying assumptions: (i) the relationship between the expression level of a gene and the ZT follows a sinusoidal wave curve; (ii) the error terms of each sample on top of the sinusoidal wave curve follows independent and identical Gaussian distribution. We discuss the implication of sinusoidal assumption on sampling design in Section 3.2 and demonstrate that the statistics is robust against various types of violation of model assumptions in Appendix C and Figure. C1.
3 |. SIMULATIONS
Throughout simulation and real application, we control type I error for circadian power calculations to account for potential multiple comparisons.
3.1 |. Comparison between CircaPower and Monte-Carlo simulation
Without the proposed analytical method CircaPower, a conventional method for circadian detection power calculation is by Monte-Carlo simulation (MC), which assumes known , , , and , .
We compare CircaPower with the MC algorithm described in Appendix D. For both methods, the ZT points are simulated from one-period one-sample evenly-spaced design for the ease of discussion, which enjoys the phase-invariant property (, Theorem 1). Since phase shift and MESOR have no impact on circadian detection power calculation in this case, we fix and . We evaluate their power derived at a grid of and . Note that for CircaPower, we only need the underlying parameters , , and to perform power calculation, which does not rely on the simulated dataset. We simulate the data for the purpose of evaluating the MC algorithm.
Figure. D2a shows that the power calculated from CircaPower is almost identical to the MC algorithm, corroborating the correctness of the closed-form solution in CircaPower. In addition, the power increases with respect to (i) larger , (ii) larger , and (iii) smaller , which are consistent with our theoretical formula of the total effect size .
As discussed in Section 2.2.1 the two curve fitting parameters and work together as the intrinsic effect size and therefore can be reduced to one parameter in MC approach. We further validate this observation by co-varying and simultaneously (i.e., ) while keeping their ratio as a constant (i.e., ). Figure. D2b shows that the power trajectories remain the same, indicating the proposed intrinsic effect size is sufficient to capture the goodness-of-fit of the model for the power calculation.
In terms of computing time, to generate all the results in Figure. D2a, it takes 1.84 seconds for the CircaPower using 1 CPU thread on a regular PC (8th Gen Intel Core i5–8250U Quad-Core processor, 1.60 GHz), while it requires 8 hours for the MC algorithm using the same computing resource. With parallel computing, the computing time reduces to 0.13 seconds for CircaPower using 40 CPU threads on a Linux server (Intel Xeon Gold 6130, 2.10GHz), while it still needs 24 minutes for the MC algorithm.
3.2 |. Impact of sampling design on CircaPower
Since the sample collection scheme for active design and passive design are quite different, we will discuss them separately. For active designs, we vary the intrinsic effect and . For passive designs, we vary the intrinsic effect and . The maximal sample size we use for the active design is smaller than that of the passive design, because the estimated intrinsic effect is usually higher in animal studies compared with human studies (see Table 1). Due to the fact that not all designs have the phase-invariant property, we also vary the phase shift .
Table 1.
Intrinsic effect sizes for public available transcriptomic circadian data, including 3 passively designed human post-mortem brain studies and 14 actively designed mouse studies from 20 types of tissues. These data are processed using the cosinor method31. Two types intrinsic effect sizes are used: (i) median of the 7 core circadian genes; (ii) minimum of the top 100 significant circadian genes. These intrinsic effect sizes can be used as a reference resource when investigators need to perform power calculation without any pilot data.
| Organism | Study | Data Availability | Tissue | Sample Size | Median r of the 7 core circadian genes | Minimum r of the top 100 circadian genes |
|---|---|---|---|---|---|---|
|
| ||||||
| Homo sapiens | Chen6 | GSE71620 | Pre-frontal cortex (BA11) | 147 | 0.91 | 0.46 |
| Pre-frontal cortex (BA47) | 147 | 0.77 | 0.44 | |||
|
| ||||||
| Ketchesin34 | GSE160521* | Striatum (NAc) | 59 | 0.71 | 1.06 | |
| Striatum (caudate) | 59 | 1.04 | 0.82 | |||
| Striatum (putamen) | 59 | 0.83 | 1.02 | |||
|
| ||||||
| Seney7 | Common Mind Consortium* | Pre-frontal cortex | 104 | 0.79 | 0.55 | |
|
| ||||||
| Mus musculus | Aguilar-Arnal42 | GSE49638 | Fibroblast | 18 | 2.38 | 2.94 |
|
| ||||||
| Bray43 | GSE10045 | Atrium | 32 | 3.43 | 1.46 | |
| Ventricle | 32 | 0.96 | 1.09 | |||
|
| ||||||
| Cho44 | GSE34018 | Liver | 12 | 2.84 | 4.11 | |
|
| ||||||
| Gerstner45 | GSE78215 | Cerebral cortex | 34 | 2.46 | 2.18 | |
|
| ||||||
| Hoogerwerf46 | GSE10644 | Colon | 18 | 1.86 | 1.56 | |
|
| ||||||
| Hughes9 | GSE11922 | Fibroblast | 48 | 1.27 | 1.02 | |
| GSE11923 | Liver | 48 | 2.00 | 2.77 | ||
|
| ||||||
| Mari47 | GSE52333 | Liver | 18 | 4.11 | 3.39 | |
|
| ||||||
| Masri48 | GSE73222 | Liver | 18 | 3.83 | 2.47 | |
|
| ||||||
| Masri49 | GSE57830 | Liver | 36 | 2.69 | 2.20 | |
|
| ||||||
| Na50 | GSE11516 | Liver | 36 | 3.65 | 3.69 | |
|
| ||||||
| Nikolaeva51 | GSE27366 | Kidney | 12 | 2.39 | 2.61 | |
|
| ||||||
| Paschos52 | GSE35026 | Adipose | 12 | 2.48 | 2.58 | |
|
| ||||||
| Solanas53 | GSE84580 | Satellite | 24 | 3.64 | 2.30 | |
| Epidermal | 20 | 5.03 | 2.62 | |||
|
| ||||||
| Zhang12 | GSE54650 | Adrenal gland | 24 | 5.17 | 2.27 | |
| Aorta | 24 | 5.55 | 2.29 | |||
| Brainstem | 24 | 3.90 | 2.07 | |||
| Brown fat | 24 | 5.05 | 2.72 | |||
| Cerebellum | 24 | 3.52 | 2.01 | |||
| Heart | 24 | 4.47 | 2.82 | |||
| Hypothalamus | 24 | 2.67 | 1.74 | |||
| Kidney | 24 | 6.33 | 3.65 | |||
| Liver | 24 | 3.51 | 3.67 | |||
| Lung | 24 | 5.78 | 3.47 | |||
| Muscle | 24 | 3.58 | 2.23 | |||
| White fat | 24 | 5.35 | 2.28 | |||
|
| ||||||
| GSE54651* | Adrenal gland | 8 | 5.29 | 5.40 | ||
| Aorta | 8 | 3.97 | 5.23 | |||
| Brainstem | 8 | 2.21 | 4.16 | |||
| Brown fat | 8 | 4.15 | 5.94 | |||
| Cerebellum | 8 | 4.06 | 4.86 | |||
| Heart | 8 | 4.73 | 6.23 | |||
| Hypothalamus | 8 | 2.19 | 4.19 | |||
| Kidney | 8 | 5.11 | 6.44 | |||
| Liver | 8 | 4.40 | 6.16 | |||
| Lung | 8 | 4.57 | 5.41 | |||
| Muscle | 8 | 5.26 | 5.24 | |||
| White fat | 8 | 3.71 | 3.87 | |||
denotes RNA-Seq data and others are microarray data.
For a typical active design, researchers usually need to control the number of (i) ZT points per cycle; (ii) replicates at each time point within a cycle; and (iii) cycles. Because of the periodicity property of the sinusoidal curve in the cosinor model, (ii) and (iii) are statistically equivalent. Therefore, for the ease of discussion, we summarize the following two key parameters for an active design: (i) number of ZT points per cycle ; (ii) total number of samples . The number of replicated samples (at the same ZT across all cycles) could be calculated as .
We denote the active design scheme with points per cycle as FixTime and the one-period one-sample evenly-spaced design (i.e., FixTime-n) as the EvenSpace. For , Figure. 3a shows that (i) the power curves are the same regardless of phi, confirming the phase-invariant property; (ii) the power trajectories for different are also identical, which implies that under evenly-spaced sampling design with , the detection power only depends on the total number of samples but not the . Note that these arguments are purely based on the statistical power given sinusoidal wave assumption. In the perspective of curve fitting, smaller number of time points may not necessarily guarantee the goodness-of-fit for a sinusoidal curve, resulting in potentially false positive findings. To explore the impact of number of on the goodness-of-fit, we then simulate expression data from the sinusoidal model and perform non-parametric curve fitting (see Appendix E and Figure. E3. E4 for details). Considering both circadian power calculation and smooth curve fitting, our results suggest (i.e., every 4 hours for 24h period design) to be the minimum number of ZT points to fully capture the circadian rhythmicity pattern, which is commonly adopted in the literature.
Figure 3.
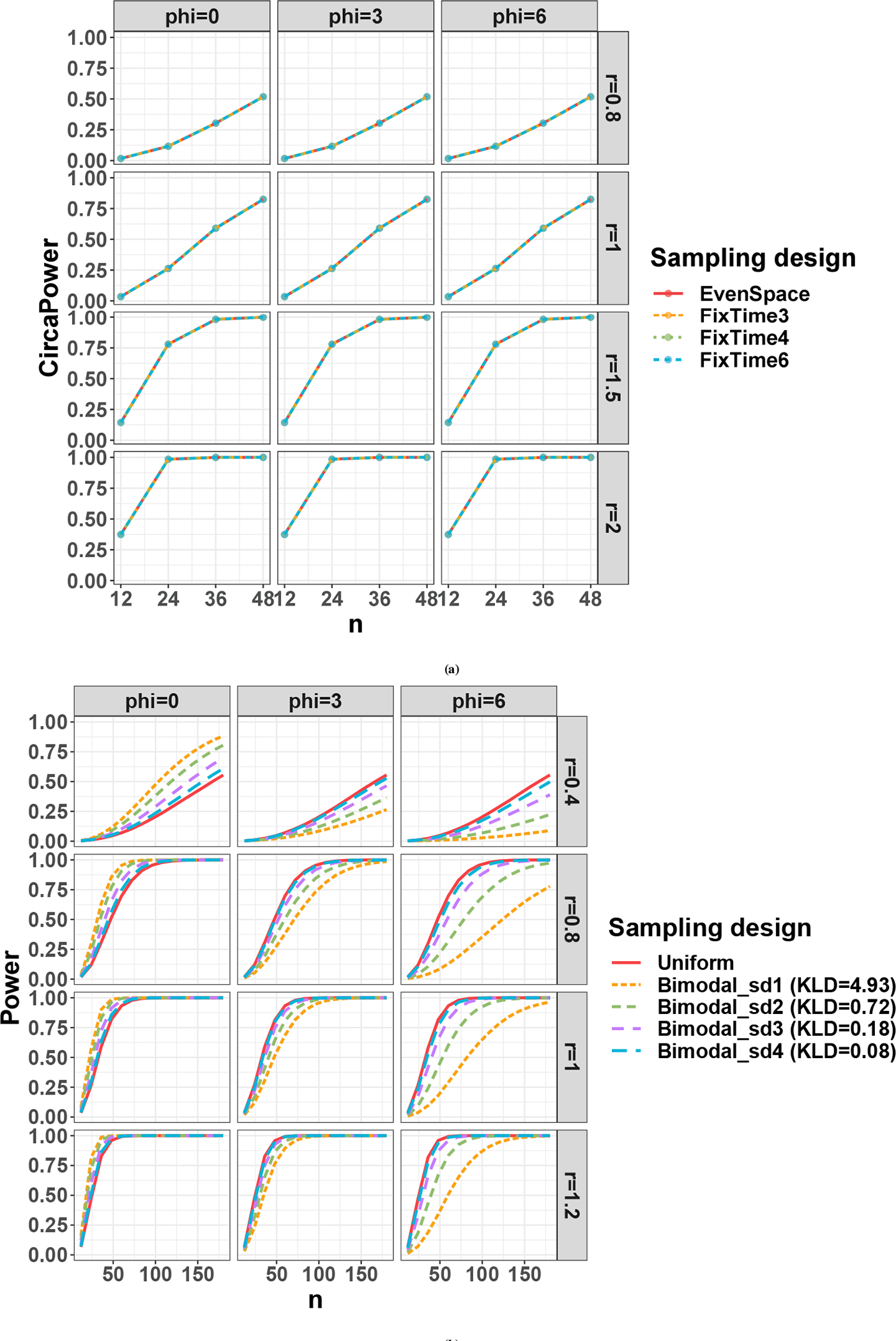
Sampling design effect on circadian power calculation. (a) shows the sampling design effect for active design; (b) shows the sampling design effect for passive design.
For passive designs, the collection of the ZT cannot be controlled. We therefore simulate from (i) uniform distribution (uniform design): ; and (ii) bimodal Gaussian distributions (bimodal designs): ; . We allow and calculate their corresponding Kullback-Leibler divergence (KLD) against the uniform distribution as a relative measurement to benchmark their divergence from the uniform. Figure. 3b shows that for the uniform design, the power trajectory is close to phase-invariant. This is expected since the uniform distribution is a random realization of the evenly-spaced sampling design, and the impact of phase on the individual will average out. The bimodal designs show phase-dependent circadian power trajectories and the impact of phase influence increases as the distribution deviates more from uniform (i.e., larger KLD). Specifically, the power loss of bimodal designs when is significant when KLD is 0.72 or greater (green and yellow curve) while negligible when KLD is only 0.18 or smaller (purple and blue curve). In fact, since the phase shift impacts the sampling design through , it achieves higher power if the mode of the ZT distribution occurs at the underlying peak/trough time. In real omics applications, circadian genes usually have different phase shift values over the day. If the collected ZT distribution is far away from the uniform distribution, the detection power of each circadian gene would be affected differently across the genome as a result of its unique phase shift.
4 |. REAL APPLICATIONS
4.1 |. CircaPower for human studies with passive design
We investigate the power trajectories of human studies using three human post-mortem brain transcriptomic studies (Chen6, Seney7 and Ketchesin34) with different time of death distributions. Detailed descriptions of each dataset can be found in the original papers. Briefly, Chen6 and Seney7 performed gene expression circadian analysis using microarray and RNA-seq respectively using pre-frontal cortex tissues; and Ketchesin34 performed RNA-seq gene expression circadian analysis with participants using dorsal and ventral striatum tissues.
To estimate the intrinsic effect sizes from the three brain studies, we apply the cosinor method31 to identify genes with rhythmic patterns and obtain estimates for their amplitude and noise level . We estimate the intrinsic effect sizes using the 7 core circadian genes, or the top 100 significant rhythmic genes (ranked by p-values from the cosinor method). The 7 core circadian genes include Arntl, Dbp, Nrld1, Nrld2, Perl, Per2, and Per3, which showed persistent circadian pattern across 12 mouse tissues12. The Homo sapiens section of Table 1 shows the estimated intrinsic effect sizes for: (i) median of the 7 core circadian genes; (ii) minimum of the 100 most significant circadian genes. The estimated intrinsic effect sizes for these three human studies range between 0.44 and 1.06 (see Table 1).
To demonstrate the power trajectories in real data, we vary intrinsic effect sizes , which roughly cover the estimated range of in the post-mortem brain studies. The ZT points are sampled 1000 times from the kernel density estimated from the observed time-of-death distributions in these three studies (see top panels of Figure. 4). Since investigators in these human post-mortem brain studies have no control of sample collection time (i.e., time of death) and can only accept passive sampling design, the detection power curves are not phase-invariant. We vary phase shift and use a confidence band to represent the range of power achieved across phase shifts (see bottom panels of Figure. 4). For each scenario (i.e., fixed , and ), the mean power among the 1000 times repetitions is reported. The power trajectory from an evenly-spaced design is also calculated as a comparison.
Figure 4.
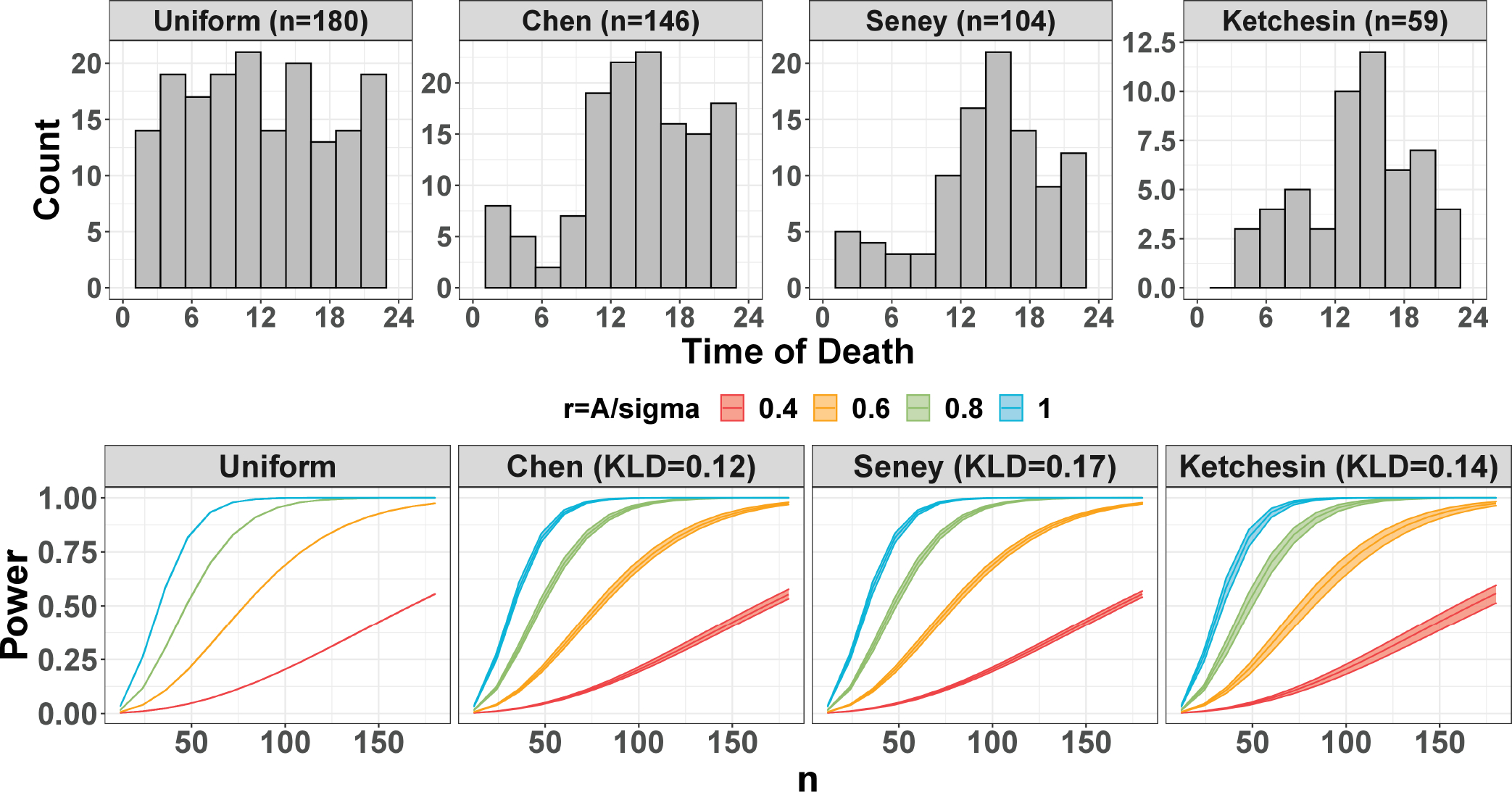
The circadian power calculation using publicly available human datasets. The top panel shows the time of death distribution for Chen, Seney, and Ketchesin. The bottom panel shows the mean power trajectories of different study designs over 1000 repetitions with different intrinsic effect sizes . The confidence bands represent the range of power achieved across phase values at for each scenario.
As expected, larger sample size and larger intrinsic effect size lead to a larger circadian power. In addition, the power trajectory is phase-invariant for the evenly-spaced sampling design with 0 band width, but not for the passive designs from the three human studies. As discussed in Section 3.2, the power will depend on the relationship between the mode of the ZT distribution and the underlying peak/trough time. To further demonstrate the impact of phase on the irregular ZT distribution, we fix , , while varying . As shown in Figure. E5. the power trajectories fluctuate across different when samples are drawn from irregular distributions in the three post-mortem studies, while the trajectory stays the same for evenly-spaced ZT. However, since the KLDs of the kernel densities estimated from the Chen, Seney and Ketchesin are relatively low (i.e., 0.12, 0.17, 0.14) compared with the bimodal designs in Section 3.2 the variation of power as a result of phase shift is small, with 3.8%, 4.4%, and 9.9% maximum drop, respectively.
4.2 |. CircaPower for animal studies with with active design
We next examine the power trajectories of actively designed mouse studies using 14 mouse gene expression circadian data42,43,44,45,46,9,47,48,49,50,51,52,53,12 from 20 types of tissues.Sample sizes of each study tissue are shown in Table 1. To estimate the intrinsic effect sizes of these tissues, we apply the cosinor method31 similarly to identify genes with rhythmic patterns and obtained estimates for their amplitude and noise level . The estimated intrinsic effect sizes for the median of the 7 core circadian genes and the minimum of the top 100 significant circadian genes are shown in the Mus musculus section of Table 1, ranging from 0.96 to 6.33, a much larger magnitude than previous human studies. This is reasonable since human studies are usually more heterogeneous in terms of genetics and environmental background. We thus fix the intrinsic effect sizes to be in our subsequent power calculation. Since these experiments employ an evenly-spaced active sampling design, the sampling design factor is a constant (i.e., , Corollary 1) regardless of the phase value.. As a result, we employ the one-sample one-period evenly-spaced design (see left panel of Figure. 5) for the purpose of power calculation. By further assuming the alpha levels to be , the power trajectories with respect to sample size is shown in Figure. 5 (right panel).
Figure 5.
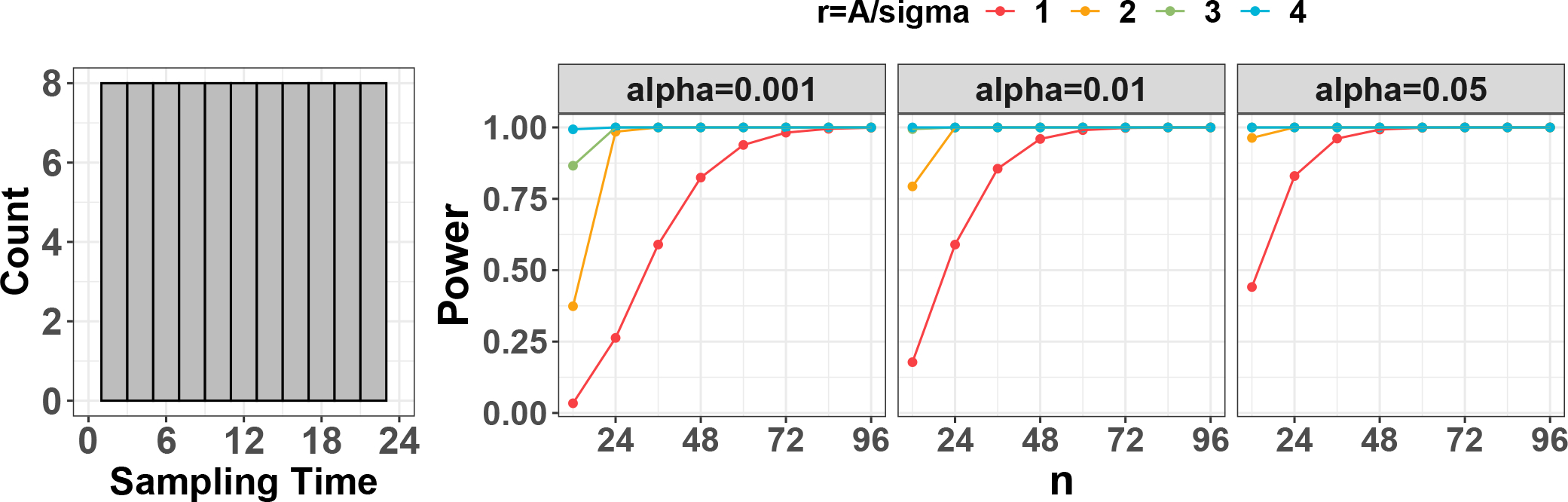
Circadian power calculation using publicly available mouse datasets. The left panel shows the distribution of one-sample one-period evenly-spaced design. The right panel shows the power trajectories for each of the type I error control assuming intrinsic effect sizes .
4.3 |. Case study: circadian power calculation using mouse pilot dataset
To demonstrate how to perform circadian power calculation using pilot dataset from scratch, we utilize a circadian gene expression data in mouse with skeletal muscle, which is part of the mouse pan-tissue gene expression circadian microarray data12. Detailed description of this dataset has been described previously8. Briefly, 24 mouse muscle samples were collected (every 2 hours) across 2 full cycles. With this pilot data, we perform genome-wide circadian rhythmicity detection using the cosinor method31. Under , we identify 716 significant genes showing circadian pattern. We similarly estimate the intrinsic effect sizes for: (i) median of the 7 core circadian genes; (ii) minimum of the top 100 significant circadian genes. The resulting intrinsic effect sizes are 3.58 and 2.23 respectively. By assuming different to be 0.05, 0.01, 0.001, the power curves with respect to sample size are shown in Figure. 6. We observe that can achieve 97.1% and 50.5% detection power for the two intrinsic effect sizes at .
Figure 6.
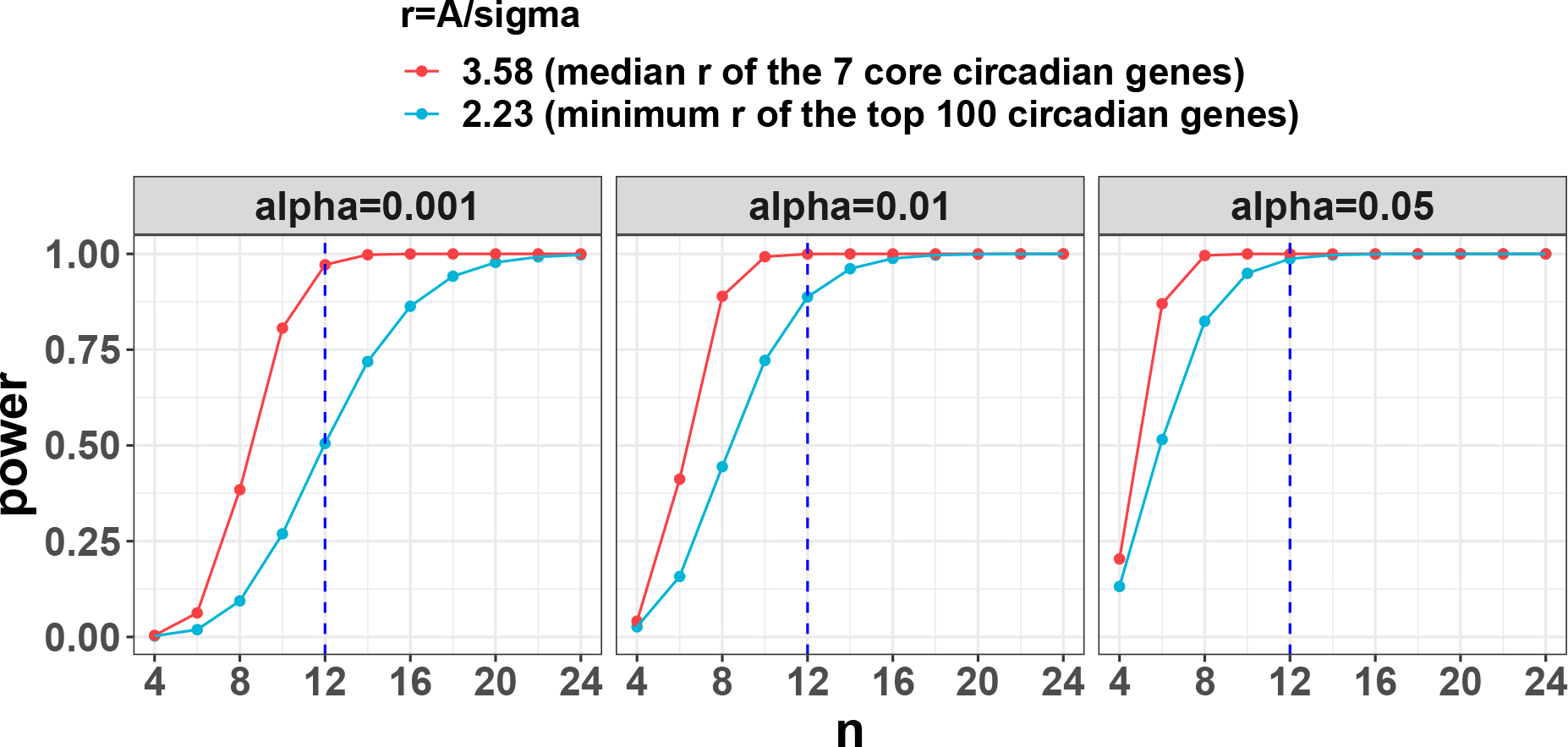
Case study: power trajectories using mouse muscle gene expression circadian data as the pilot data. The pilot data contains 24 mouse muscle samples collected every 2 hours across 2 full cycles. CircaPower is used to calculate the power. The intrinsic effect sizes were used as (i) median of the 7 core circadian genes; (ii) minimum of the top 100 significant circadian genes. By assuming different to be 0.05, 0.01, 0.001, the power trajectories with respect to sample size is shown in this figure. The blue dashed lines indicate the detection power when per cycle which is statistically equivalent to sampling every two hours across two full cycles.
5 |. DISCUSSION
In this paper, we propose an analytical framework, CircaPower, to calculate the statistical power for circadian gene detection. To the best of our knowledge, this is the first analytical method to perform circadian power analysis. In simulations, we not only demonstrate the CircaPower is fast and accurate, but also show that its underlying cosinor model is robust against violations of model assumptions. In real applications, we show the performance of the CircaPower in mouse studies and postmortem human studies. In addition, we obtain the estimated intrinsic effect sizes from publicly available human and mouse transcriptomic circadian data. These summarized intrinsic effect sizes can be used as a reference resource to facilitate investigators without pilot data to perform circadain power calculation. In case study, we also demonstrate circadian power calculation step-by-step given a pilot dataset.
Our method has several advantages. To begin with, the theoretical framework suggests that the power calculation is related to a total effect size, which can be decomposed into sample size, intrinsic effect size (representing goodness-of-fit of circadian curve), and sampling design factor. Moreover, the sampling design factor brings about the concept of active design and passive design when samples are collected. This is an important concept in circadian experiment design, since the ZT collection for human (passive design) and animal (active design) could be quite different. After that, we demonstrate the phase-invariant property of the evenly-spaced sampling design, which provides theoretical foundation for the design of many published circadian studies. In addition, the closed-form formula in CircaPower allows unique inverse calculation of sample size given desired power at fast computing speed compared with the conventional MC algorithm. In this paper, we also systematically examine the intrinsic effect sizes of published mouse or human gene expression circadian data, which could provide guidance for future researchers to design their transcriptomic circadian experiment when pilot data are not available. Although we present our work using transcriptomic data, CircaPower is applicable to other omics data, such as DNA methylation, ChIP-Seq proteomics, metabolomics, and clinical data (e.g., body temperature).
Our work has the following limitations and future work. Firstly, the CircaPower assumes a pre-fixed alpha level. We intentionally select a more stringent alpha (e.g., ) to account for multiple comparison when thousands of genes are tested simultaneously. Additional modeling is needed to extend for calculating genome-wide power calculation while controlling false discovery rate. Secondly, in addition to detecting genes with rhythmic pattern, another important research question is to identify differential circadian pattern32,54,55,56 (i.e., the circadian pattern is disrupted because of the treatment or condition), which will be another future direction. Thirdly, the Gaussian assumptions are widely assumed in biomedical research, and we have demonstrated that the cosinor model is robust against violations of Gaussian assumptions. If an investigator still worries about these assumptions, we would recommend data transformations (e.g. Box–Cox transformation) before applying our method. Our previous work32 has shown that the Box-Cox transformation can rescue the normality assumption for circadian rhythmicity detection using cosinor models. Similar justifications have been adopted in the literature. For example, though the student T test also assumes Gaussian assumptions, but it is still widely used in the literature, as long as there are methods to rescue the violation (i.e., data transformation). Lastly, as discussed in the introduction, both parametric and non-parametric models are popular and widely used in the literature. In the proposal, we only focus on the cosinor model for its simplicity and accurate statistical inference32. Further extending the current framework to a more flexible family of circadian pattern is of biological interests to the general circadian research field.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health [R21LM012752 and R01LM014142 to WZ and GT, R01MH111601 and P50DA046346 to WZ, GT and CM, R01HL153042 to KE and ZH, R01AR079220 to AL, KE and ZH, K01MH128763 to KK, NIH UL1TR001857 to MG].
APPENDIX
A. PROOF OF STATISTICS DISTRIBUTION UNDER THE ALTERNATIVE
A.1. Derivation of statistics distribution under the null
Lemma 1.
Under the null distribution in the linear model framework, , where denotes a regular F distribution with degrees of freedom and .
Proof.
The proof is given in57.
Therefore, under the null hypothesis that there is no circadian rhythmicity. i.e., in Equation 1, or equivalently, in Equation 2,
where 2 and are the degrees of freedom of the distribution, and denotes a regular distribution with non-centrality parameter 0
A.2. Derivation of statistics distribution under the alternative
Lemma 2.
If and is positive definite variance-covariance matrix for the dimensional multivariate normal distribution, then
where is the non-centrality parameter for the distribution with degree of freedom .
Proof.
The proof is given in57.
Lemma 3.
In a linear model framework, if the design matrix , the regression coefficient , there are hypotheses to be tested , where , represents the true parameters .
We have the following results:
where denotes a non-central F distribution with non-centrality parameter and degrees of freedom and .
Proof.
If we denote , and base on Lemma 2 we have:
where . In addition, since
therefore we could derive the following relationship58.
Theorem 2.
Under the sinusoidal model assumption (Equation 2), under the alternative hypothesis (there is a circadian fitting, i.e., or ), we have
where .
Proof.
Fitting the sinusoidal model Equation 2 into the linear model framework, the design matrix is
And
The regression coefficient is
The joint hypotheses are and , which is equivalent to the following:
By denoting , the hypothesis is equivalent to . According to Lemma 3 we have
B. PROOF OF THE PHASE-INVARIANT PROPERTY IN THE ONE-SAMPLE ONE-PERIOD EVENLY-SPACED DESIGN
Lemma 4.
For , , , define .
Proof.
See 59.
Corollary 2.
When , ;
With that being said, the evenly spaced design with time points will achieve the same design effect, regardless of phase
Proof.
Following Lemma 4, set , where , , then , .
Theorem 3
(Phase-invariant property - one-period one-sample). Assuming there is a total of ZT points within a circadian period , the ZT are ordered such that for all . If , and is evenly-spaced over the period (i.e., for all , is a fixed time interval, , then regardless of the value for ,
Proof.
C. ROBUSTNESS ANALYSIS OF STATISTICS
To examine the robustness of our method when the iid Gaussian assumption is violated, we investigate the type I error control of statistics in the following scenarios: (i) heavy tail error distribution (i.e., student T distribution); (ii) existence of outliers; (iii) non-independent Gaussian errors. For all these simulations, noisy genes are simulated with error term ‘s specified above. By declaring circadian rhythmicity at 5% nominal level, we will evaluate the actual type I error rate of the test from the cosinor model. Since CircaPower is built on the statistics for rhythmicity detection it will be benchmarked as robust if the actual type I error rate is close to the nominal level. The detailed simulateion setting is provided below:
Heavy tail error distribution. Instead of sampling the error term , and , we sample , where is the student T distribution with degree of freedom . In general, the smaller the is, the heavier tail the error distribution is. When , the error distribution becomes the Cauchy distribution, and when , the error distribution converges to standard Gaussian distribution (i.e., ). To evaluate the impact of heavy tail error distribution on CircaPower, we simulate a grid of . Figure. C1a shows that when there is no or mild violation of the Gaussian assumption (i.e., or ), the cosinor method achieves accurate type I error control (i.e., 5%). When there is moderate to severe violation of the Gaussian assumption (i.e., or ), type I error rate is only slightly conservative (i.e., below the nominal ). Putting together, the type I error rate of the cosinor method can be correctly controlled against the heavy tail error distribution.
Existence of outliers To evaluate the impact of outliers on type I error control, we replace of the expression values with outliers, where . To be specific, for a gene , there is chance that the expression level is simulated from ; and chance that the expression level is simulated independently based on Equation 1 under (i.e., ). Figure. C1b shows that the cosinor method achieves accurate nominal type I error rate control (i.e., 5%), showing robustness to outliers.
Correlated gene structure Instead of assuming all genes are independent, we simulate every 50 genes as a gene module with correlation coefficient . The error term for each gene module where is a symmetric matrix with diagonal elements being and off-diagonal elements being . We simulate a grid of . Figure. C1c shows that when genes are correlated, the cosinor method maintains accurate nominal type I error rate control (i.e., 5%).
Since our goal is to evaluate the type I error rate control, which does not involve any multiple testing issue, we directly use 5% nominal level. Figure. C1 shows that the cosinor model only has slightly conservative type I error from cosinor model when error terms are drawn from very heavy tail distribution while maintains accurate type I error rate in all other scenarios suggesting the robustness against the three types of violation in general.
Figure C1.
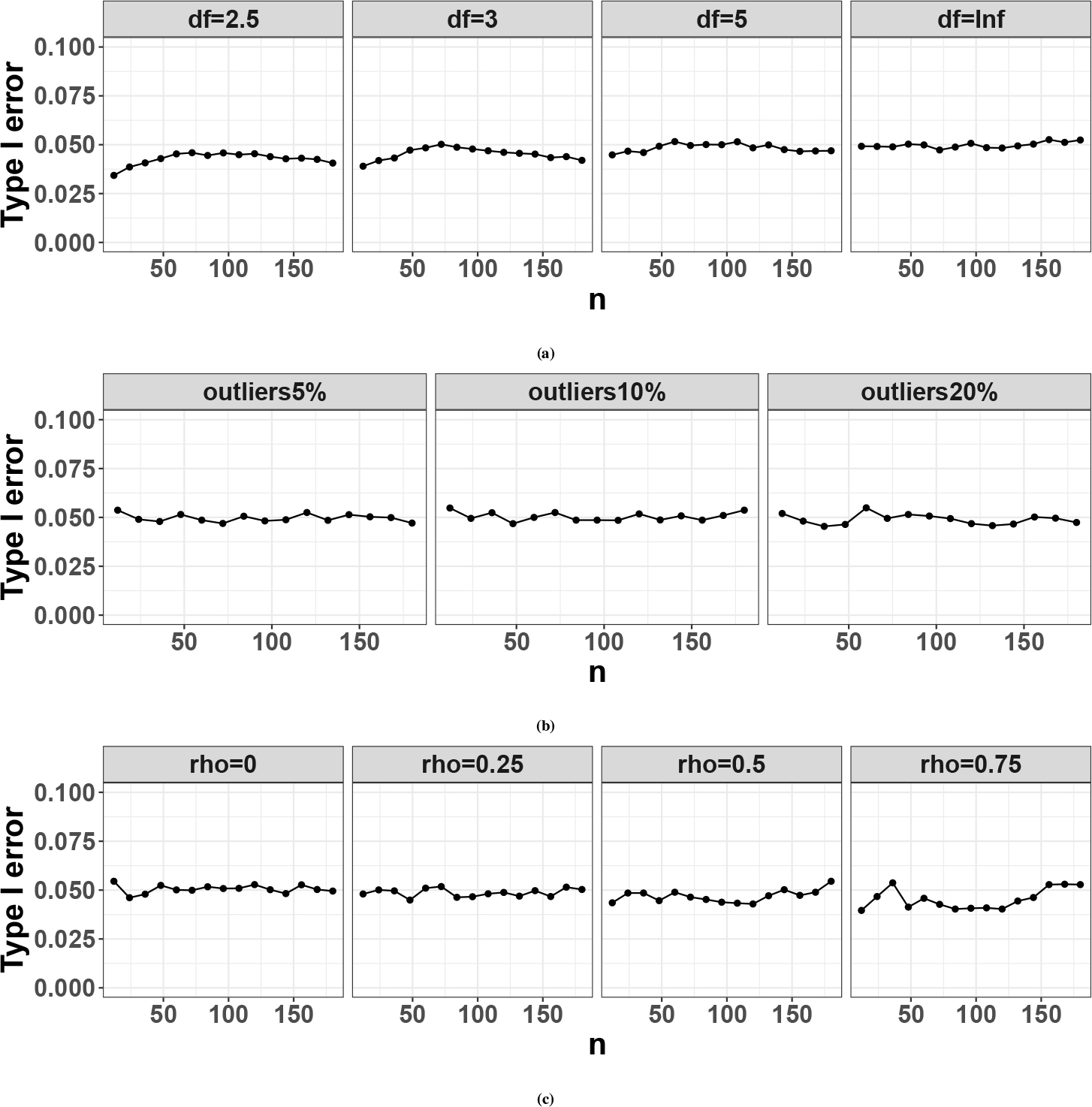
Actual type I error rate for the cosiner method in detecting circadian rhythmicity pattern at 5% nominal level when there exist violation of model assumptions (i.e., independent Gaussian errors.) (a) shows the case when the error term comes from a heavy tail distribution (i.e., , is the degree of freedom of the distribution); (b) shows the case when there exists outliers; (c) shows the case when the errors of multiple genes are correlated (i.e., , is number of correlated genes, and is the variance-covariance matrix for the multivariate normal distribution).
D. ALTERNATIVE POWER CALCULATION METHOD BY MONTE-CARLO SIMULATION
Without the proposed analytical method CircaPower, a conventional method for circadian detection power calculation is by Monte-Carlo simulation (MC), which assumes known , , , and , in Equation 1. The detailed algorithm for MC is described as following:
Given the ZT for samples and key parameters (, , , and ), we simulate gene expression based on Equation 11 where is the gene index and is the total number of genes. genes is used in the simulation comparison between CircaPower and MC algorithm in Section 3.1 in the main text.
We apply the cosinor method31 to derive the rhythmic p-value for each gene . Given a pre-specified alpha level , the power of MC algorithm is calculated as .
Although both the CircaPower and the MC algorithm rely on the statistics for rhythmicity detection (see Figure. D2), the CircaPower has several obvious advantages over the MC algorithm. First of all, the explicit representation of total effect size in CircaPower provides insights on the three determining factors in circadian detection power calculation while it is hard for MC algorithm to determine selections and trends on the many parameters (, , , , and , ). In addition, our simulation shows the closed-form solution by CircaPower is at least 10,000 folds faster than the MC algorithm (see Section 3.1 in the main text). More importantly, even though both approaches can calculate power given sample size, only CircaPower can directly solve the inverse problem of deriving the smallest sample size meeting the desired detection power, while MC algorithm needs repeated interpolation to obtain an answer.
Figure D2.
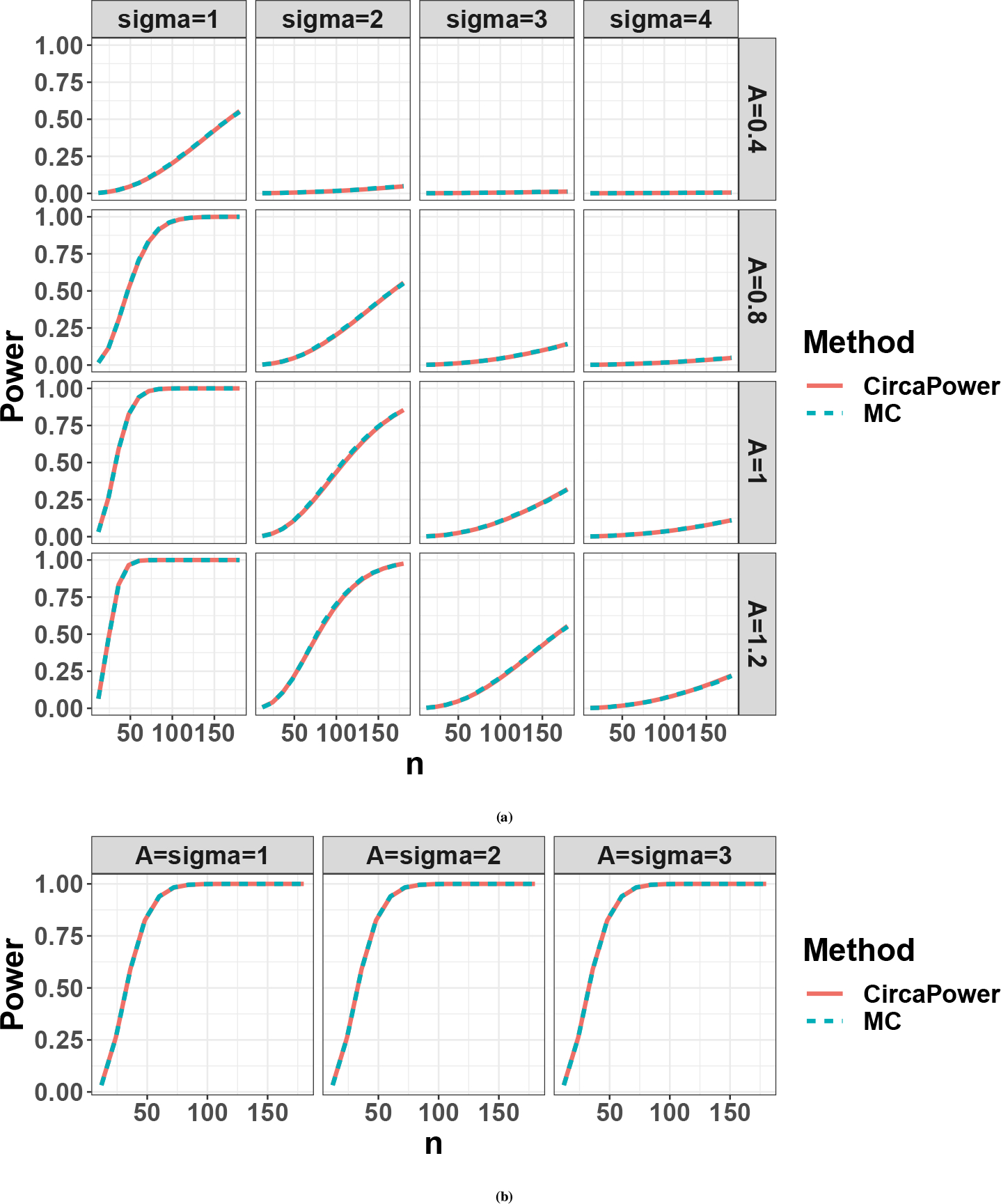
Compare power trajectories from the CircaPower with the Monte-Carlo (MC) algorithm. The red color curve denotes the CircaPower method, and the cyan color curve denotes the MC approach. (a) We vary the amplitude and . (b) We co-vary and simultaneously (i.e., ) while keeping their ratio as a constant (i.e., .
E. NON-PARAMETERIC CURVE FITTING SIMULATION SETTING
From the perspective of circadian power calculation, Figure. 3a implies that the evenly-spaced sampling design is phase-invariant as long as , which is further corroborated by Corollary 1. However, in the perspective of curve fitting, smaller number of time points may not necessarily guarantee the goodness-of-fit for a sinusoidal curve, resulting in potentially false positive findings.
To explore the impact of number of on the goodness-of-fit for a sinusoidal wave, we simulate expression data from the sinusoidal model and perform non-parametric curve fitting to identify the minimum necessary to capture the sinusoidal wave curve. To be specific, we first choose the ZT points within a cycle to be , where starts from 3 as at least 3 sampling times are needed to characterize the three parameters in sine (or cosine) model. Then for each , we simulate samples, and evenly allocated them at time points across 2 full cycles (every hours, from - 12h to 36h), resulting in samples at each time point. The expression values of samples at each time point are simulated independently from Equation 1] where we set , , and . The LOESS regression is then used to fit a smooth curve through the data points. The LOESS regression is a nonparametric method using locally-weighted regression to fit a smooth curve over a scatter plot60. Such LOESS regression represents the smooth curve fitting without the sinusoidal assumption, which could reflect the minimum of that is necessary to capture the sinusoidal wave curve. The rationale for 2 full cycles is to improve the boundary behavior of the curve fitting within one cycle. In addition, to evaluate the effect of the phase shift, we set . By comparing the fitted non-parametric curves with the underlying sinusoidal wave, we can observe the minimum of NT that is necessary to capture the sinusoidal wave curve.
The data points and fitted smooth curves in one circadian cycle [0, 24] are shown in Figure. E3 When , it is uncertain whether the underlying curve fitting is a sinusoidal wave. Only when increases to 6 or more, the curve fitting is stable and almost identical to the underlying sinusoidal wave. To further justify this choice, we calculate the root mean square distance between the fitted non-parametric curve and the underlying sinusoidal curve at evenly spaced 1000 points between [0, 24] and plot against . The elbow plot (Figure. E4) suggests that is an inflection point after which the change of distance between the fitted curve and the underlying curve becomes stably small. Therefore, considering both circadian power calculation and smooth curve fitting, our results suggest to be the minimum number of ZT points to fully capture the circadian rhythmicity pattern, which is commonly adopted in the literature.
Figure E3.
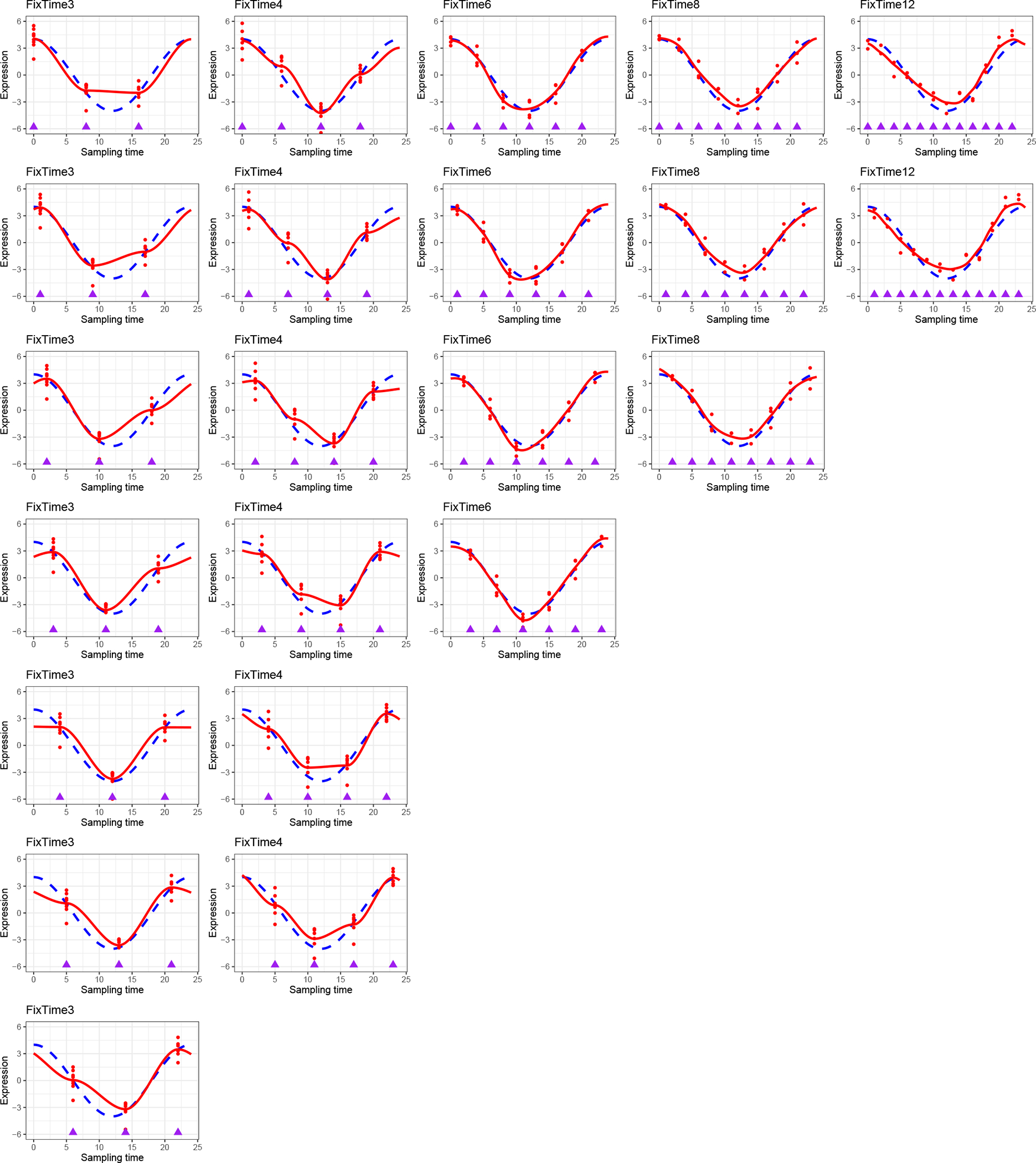
Non-parametric curve fitting to explore the impact of number of ZT points per cycle (i.e., ) on the goodness of fit for a sinusoidal wave. We choose the ZT points within a cycle to be (on columns). Then for each , we simulate samples evenly allocated at -time points across 2 full cycles (every hours, from −12h to 36h), resulting in samples at each time point. The rationale for 2 full cycles is to improve the boundary behaviour of the curve fitting within one cycle. The purple rectangles indicate sampling time points. The expression values of samples at each time point are simulated independently from Equation 1 with , , and . The LOESS regression is used to fit a smooth curve through the all data points. To evaluate the effect of the phase shift, we set (on rows). The data points and fitted smooth curve in one circadian cycle [0, 24] are shown in this figure, which represents the smooth curve fitting without the sinusoidal assumption.
Figure E4.
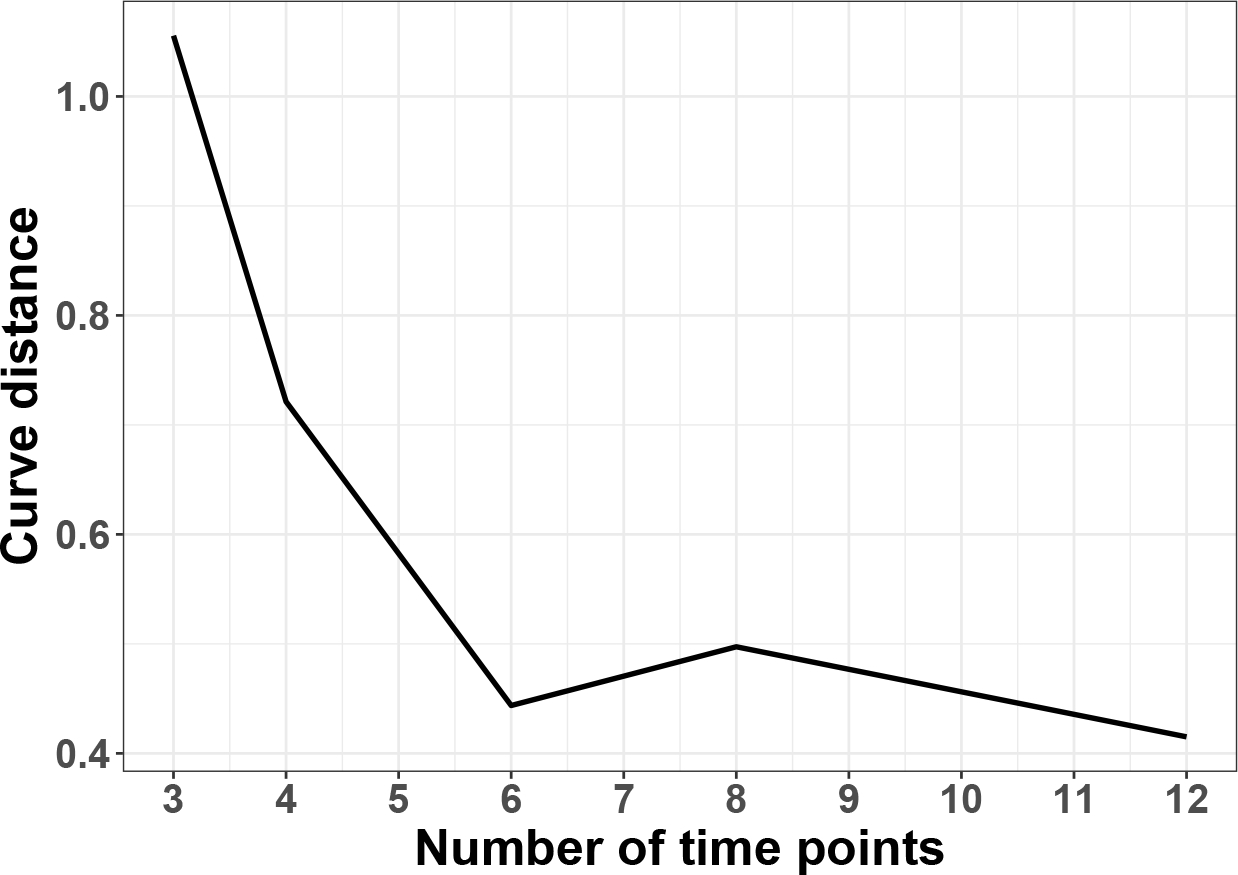
Elbow plot to evaluate the effect of number of ZT points per cycle (i.e., ) on the goodness of fit for a sinusoidal wave. The curve distance is calculated as where are 1000 points evenly spaced between 0 and 24, is the underlying sinusoidal curve evaluated at and is the fitted non-parametric curve evaluated at . To avoid randomness, for each , the non-parametric fitting is repeated 100 times and the mean curve distance is plot against the .
Figure E5.
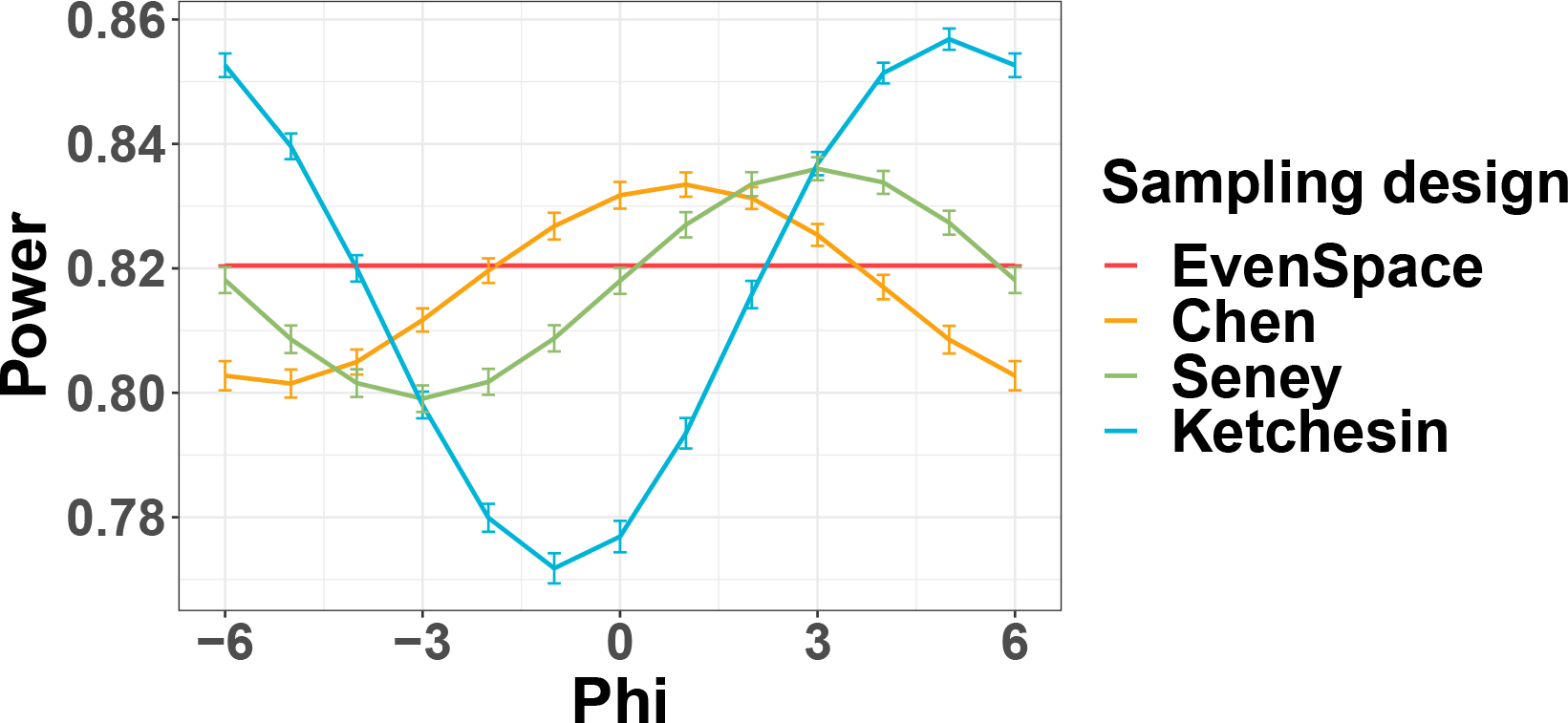
The mean power trajectories across different when and . For each and sampling distribution, we draw sampling times 1000 times and calculate corresponding power. Vertical bars indicate the 95% confidence interval of power estimates calculated form where and are mean and standard deviation of power estimates respectively. Maximum power drop ) is 3.8%, 4.4%, and 9.9% respectively.
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interests.
FINANCIAL DISCLOSURE
None reported.
DATA AVAILABILITY STATEMENT
To allow easy application by other researchers, our methods have been implemented in the R package CircaPower, which is publicly available in github (https://github.com/circaPower/CircaPower). Seneyl7 dataset is available in the Common Mind Consortium at https://www.nimhgenetics.org/available_data/commonmind/ through an approval process. All other datasets are publicly available on the NCBI GEO database with accession numbers shown in Table 1.
References
- 1.Badia P, Myers B, Boecker M, Culpepper J, Harsh J. Bright light effects on body temperature, alertness, EEG and behavior. Physiology & behavior 1991; 50(3): 583–588. [DOI] [PubMed] [Google Scholar]
- 2.Cagnacci A, Elliott J, Yen S. Melatonin: a major regulator of the circadian rhythm of core temperature in humans. The Journal of Clinical Endocrinology & Metabolism 1992; 75(2): 447–452. [DOI] [PubMed] [Google Scholar]
- 3.DIJK DJ, Duffy JF, Czeisler CA. Circadian and sleep/wake dependent aspects of subjective alertness and cognitive performance. Journal of sleep research 1992; 1(2): 112–117. [DOI] [PubMed] [Google Scholar]
- 4.Jung CM, Khalsa SBS, Scheer FA, et al. Acute effects of bright light exposure on cortisol levels. Journal of biological rhythms 2010; 25(3): 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nature Reviews Genetics 2017; 18(3): 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CY, Logan RW, Ma T, et al. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proceedings of the National Academy of Sciences 2016; 113(1): 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seney ML, Cahill K, Enwright JF, et al. Diurnal rhythms in gene expression in the prefrontal cortex in schizophrenia. Nature communications 2019; 10(1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodge BA, Wen Y, Riley LA, et al. The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skeletal muscle 2015; 5(1): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes ME, DiTacchio L, Hayes KR, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet 2009; 5(4): e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Möller-Levet CS, Archer SN, Bucca G, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proceedings of the National Academy of Sciences 2013; 110(12): E1132–E1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruben MD, Wu G, Smith DF, et al. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Science Translational Medicine 2018; 10(458). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proceedings of the National Academy of Sciences 2014; 111(45): 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mure LS, Le HD, Benegiamo G, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018; 359(6381). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim AS, Srivastava GP, Yu L, et al. 24-hour rhythms of DNA methylation and their relation with rhythms of RNA expression in the human dorsolateral prefrontal cortex. PLoS genetics 2014; 10(11): e1004792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Koike N, Yoo SH, Huang HC, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. science 2012; 338(6105): 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Song L, Liu M, et al. A proteomics landscape of circadian clock in mouse liver. Nature communications 2018; 9(1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proceedings of the National Academy of Sciences 2012; 109(7): 2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenvers DJ, Jongejan A, Atiqi S, et al. Diurnal rhythms in the white adipose tissue transcriptome are disturbed in obese individuals with type 2 diabetes compared with lean control individuals. Diabetologia 2019; 62(4): 704–716. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A, Van Gelder RN. Clocks, cancer, and chronochemotherapy. Science 2021; 371(6524). [DOI] [PubMed] [Google Scholar]
- 20.Ballesta A, Innominato PF, Dallmann R, Rand DA, Lévi FA. Systems chronotherapeutics. Pharmacological reviews 2017; 69(2): 161–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JZ, Bunney BG, Meng F, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proceedings of the National Academy of Sciences 2013; 110(24): 9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim AS, Klein HU, Yu L, et al. Diurnal and seasonal molecular rhythms in human neocortex and their relation to Alzheimer’s disease. Nature communications 2017; 8(1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G, Ruben MD, Lee Y, Li J, Hughes ME, Hogenesch JB. Genome-wide studies of time of day in the brain: Design and analysis. Brain Science Advances 2020; 6(2): 92–105. [Google Scholar]
- 24.Mei W, Jiang Z, Chen Y, Chen L, Sancar A, Jiang Y. Genome-wide circadian rhythm detection methods: systematic evaluations and practical guidelines. Briefings in Bioinformatics 2021; 22(3): bbaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundell LS, Parr EB, Devlin BL, et al. Time-restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nature communications 2020; 11(1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma D, Zhao M, Xie L, et al. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nature neuroscience 2020; 23(3): 456–467. [DOI] [PubMed] [Google Scholar]
- 27.Sato S, Basse AL, Schönke M, et al. Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell metabolism 2019; 30(1): 92–110. [DOI] [PubMed] [Google Scholar]
- 28.Hughes ME, Abruzzi KC, Allada R, et al. Guidelines for genome-scale analysis of biological rhythms. Journal of biological rhythms 2017; 32(5): 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ness-Cohn E, Iwanaszko M, Kath WL, Allada R, Braun R. TimeTrial: An Interactive Application for Optimizing the Design and Analysis of Transcriptomic Time-Series Data in Circadian Biology Research. Journal of biological rhythms 2020; 35(5): 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes M, Deharo L, Pulivarthy S, et al. High-resolution time course analysis of gene expression from pituitary. In:. 72. Cold Spring Harbor Laboratory Press. ; 2007: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornelissen G Cosinor-based rhythmometry. Theoretical Biology and Medical Modelling 2014; 11(1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding H, Meng L, Liu AC, et al. Likelihood-based tests for detecting circadian rhythmicity and differential circadian patterns in transcriptomic applications. Briefings in Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheving LE, Halberg F. Chronobiology: Principles and applications to shifts in schedules. 3. Springer Science & Business Media. 1981. [Google Scholar]
- 34.Ketchesin KD, Zong W, Hildebrand MA, et al. Diurnal rhythms across the human dorsal and ventral striatum. Paroceedings of the National Academy of Sciences 2021; 118(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glynn EF, Chen J, Mushegian AR. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb–Scargle periodograms. Bioinformatics 2006; 22(3): 310–316. [DOI] [PubMed] [Google Scholar]
- 36.Straume M DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. In: 383. Elsevier. 2004. (pp. 149–166). [DOI] [PubMed] [Google Scholar]
- 37.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 2010; 26(12): i168–i174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. Journal of biological rhythms 2014; 29(6): 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of biological rhythms 2010; 25(5): 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding H, Meng L, Liu AC, et al. Likelihood-based tests for detecting circadian rhythmicity and differential circadian patterns in transcriptomic applications. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laloum D, Robinson-Rechavi M. Methods detecting rhythmic gene expression are biologically relevant only for strong signal. PLoS computational biology 2020; 16(3): e1007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nature structural & molecular biology 2013; 20(10): 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray MS, Shaw CA, Moore MW, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. American Journal of Physiology-Heart and Circulatory Physiology 2008; 294(2): H1036–H1047. [DOI] [PubMed] [Google Scholar]
- 44.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012; 485(7396): 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerstner JR, Koberstein JN, Watson AJ, et al. Removal of unwanted variation reveals novel patterns of gene expression linked to sleep homeostasis in murine cortex. BMC genomics 2016; 17(8): 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoogerwerf WA, Sinha M, Conesa A, et al. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology 2008; 135(6): 2019–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mari M, Tognini P, Liu Y, Kristin L, Baldi P, Paolo SC. Gut microbiota directs PPARgamma-driven reprogramming of the liver circadian clock by nutritional challenge. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masri S, Papagiannakopoulos T, Kinouchi K, et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 2016; 165(4): 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masri S, Rigor P, Cervantes M, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014; 158(3): 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Na YJ, Sung JH, Lee SC, et al. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Experimental & molecular medicine 2009; 41(9): 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolaeva S, Pradervand S, Centeno G, et al. The circadian clock modulates renal sodium handling. Journal of the American Society of Nephrology 2012; 23(6): 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paschos GK, Ibrahim S, Song WL, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nature medicine 2012; 18(12): 1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solanas G, Peixoto FO, Perdiguero E, et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 2017; 170(4): 678–692. [DOI] [PubMed] [Google Scholar]
- 54.Thaben PF, Westermark PO. Differential rhythmicity: detecting altered rhythmicity in biological data. Bioinformatics 2016; 32(18): 2800–2808. [DOI] [PubMed] [Google Scholar]
- 55.Singer JM, Hughey JJ. LimoRhyde: a flexible approach for differential analysis of rhythmic transcriptome data. Journal of biological rhythms 2019; 34(1): 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons R, Parsons R, Garner N, Oster H, Rawashdeh O. CircaCompare: a method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics 2020; 36(4): 1208–1212. [DOI] [PubMed] [Google Scholar]
- 57.Monahan JF. A primer on linear models. CRC Press. 2008. [Google Scholar]
- 58.Rohatgi VK, Saleh AME. An introduction to probability and statistics. John Wiley & Sons. 2015. [Google Scholar]
- 59.Greitzer S Many cheerful facts. Arbelos 1986; 4(5): 14–17. [Google Scholar]
- 60.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. Journal of the American statistical association 1979; 74(368): 829–836. [Google Scholar]
- 61.Sahar S, Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends in Endocrinology & Metabolism 2012; 23(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000; 288(5466): 682–685. [DOI] [PubMed] [Google Scholar]
- 63.Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: Around the transcription-translation feedback loop and on to output. Trends in biochemical sciences 2016; 41(10): 834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annual review of physiology 2001; 63(1): 647–676. [DOI] [PubMed] [Google Scholar]
- 65.Hastings M, Brancaccio M, Maywood E. Circadian pacemaking in cells and circuits of the suprachiasmatic nucleus. Journal of neuroendocrinology 2014; 26(1): 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lück S, Thurley K, Thaben PF, Westermark PO. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell reports 2014; 9(2): 741–751. [DOI] [PubMed] [Google Scholar]
- 67.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 2016; 32(21): 3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mei W, Jiang Z, Chen Y, Chen L, Sancar A, Jiang Y. Genome-wide circadian rhythm detection methods: systematic evaluations and practical guidelines. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu PY, Harmer SL. Circadian phase has profound effects on differential expression analysis. PloS one 2012; 7(11): e49853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hughey JJ, Butte AJ. Differential phasing between circadian clocks in the brain and peripheral organs in humans. Journal of biological rhythms 2016; 31(6): 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parker T Finite-sample distributions of the Wald, likelihood ratio, and Lagrange multiplier test statistics in the classical linear model. Communications in Statistics - Theory and Methods 2017; 46(11): 5195–5202. [Google Scholar]
- 72.Elzhov TV, Mullen KM, Spiess AN, Bolker B. minpack.lm: R Interface to the Levenberg-Marquardt Nonlinear Least-Squares Algorithm Found in MINPACK, Plus Support for Bounds. 2016. R package version 1.2–1.
- 73.Nash JC, Varadhan R. Unifying Optimization Algorithms to Aid Software System Users: optimx for R. Journal of Statistical Software 2011; 43(9): 1–14. [Google Scholar]
- 74.Nash JC. On Best Practice Optimization Methods in R. Journal of Statistical Software 2014; 60(2): 1–14. [Google Scholar]
- 75.Huo Z, Ding Y, Liu S, Oesterreich S, Tseng G. Meta-Analytic Framework for Sparse K-Means to Identify Disease Subtypes in Multiple Transcriptomic Studies. Journal of the American Statistical Association 2016; 111(513): 27–42. doi: 10.1080/01621459.2015.1086354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kunieda T, Minamino T, Katsuno T, et al. Cellular senescence impairs circadian expression of clock genes in vitro and in vivo. Circulation research 2006; 98(4): 532–539. [DOI] [PubMed] [Google Scholar]
- 77.Crosby P, Hamnett R, Putker M, et al. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 2019; 177(4): 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris CJ, Aeschbach D, Scheer FA. Circadian system, sleep and endocrinology. Molecular and cellular endocrinology 2012; 349(1): 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pelikan A, Herzel H, Kramer A, Ananthasubramaniam B. Studies overestimate the extent of circadian rhythm reprogramming in response to dietary and genetic changes. bioRxiv 2020. [Google Scholar]
- 80.Done AJ, Traustadóttir T. Nrf2 mediates redox adaptations to exercise. Redox biology 2016; 10: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Box GEP, Cox DR. An Analysis of Transformations. Journal of the Royal Statistical Society. Series B (Methodological) 1964; 26(2): pp. 211–252. [Google Scholar]
- 82.Box GE, Cox DR. An analysis of transformations. Journal of the Royal Statistical Society: Series B (Methodological) 1964; 26(2): 211–243. [Google Scholar]
- 83.Wen S, Ma D, Zhao M, et al. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. 2020. [DOI] [PubMed]
- 84.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waltman L, Eck NJv. A smart local moving algorithm for large-scale modularity-based community detection. 2013. [Google Scholar]
- 86.Maaten v. dL, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research 2008; 9(86): 2579–2605. [Google Scholar]
- 87.Ypma J Introduction to nloptr: an R interface to NLopt. R Package 2014; 2. [Google Scholar]
- 88.Lonsdale J, Thomas J, Salvatore M, et al. The genotype-tissue expression (GTEx) project. Nature genetics 2013; 45(6): 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li J, Grant GR, Hogenesch JB, Hughes ME. Considerations for RNA-seq analysis of circadian rhythms. Methods in enzymology 2015; 551: 349–367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To allow easy application by other researchers, our methods have been implemented in the R package CircaPower, which is publicly available in github (https://github.com/circaPower/CircaPower). Seneyl7 dataset is available in the Common Mind Consortium at https://www.nimhgenetics.org/available_data/commonmind/ through an approval process. All other datasets are publicly available on the NCBI GEO database with accession numbers shown in Table 1.


