Abstract
The evolution of influenza B viruses is poorly understood. Reassortment of influenza B viruses in nature as a means of genetic variation has not been considered to be a major contributor to their evolution. However, the current practice of assigning evolutionary relationships by antigenic analysis of the hemagglutinin of influenza B viruses would fail to detect reassortants. In this study, influenza B viruses isolated within the past 10 years from sites in the United States and China were studied by nucleotide sequencing of the hemagglutinin and neuraminidase genes and construction of phylogenetic trees to assess evolutionary relationships. A group of viruses represented by B/Houston/1/92 possess a hemagglutinin derived from a B/Yamagata/16/88-like strain and a neuraminidase derived from a B/Victoria/2/87-like strain. A second reassortment event between the hemagglutinin of a B/Yamagata/16/88-like virus closely related to the B/Beijing/184/93 strain and the neuraminidase of a B/Victoria/2/87-like strain is represented by a single virus, B/Memphis/3/93. The neuraminidase of the reassortant viruses is most closely related to that of B/Victoria/2/87-like viruses currently circulating in Nanchang, China. A pattern of insertions and deletions in the hemagglutinin and the neuraminidase of different strains of influenza B viruses is observed. Reassortment plays a role in the evolution of influenza B viruses and may necessitate a change in the methods used to assess and identify new influenza viruses.
There are fundamental differences in the evolution of influenza A and B viruses (25). Both types of influenza virus share a common means of antigenic variation termed antigenic drift, which allows these viruses to evade immune pressures from their hosts (22). However, while the evolution of influenza A viruses is driven by yearly selection of new variants through this mechanism (7, 9), it is uncertain at present how important antigenic drift is for the evolution of influenza B viruses (1, 6). It is well established that the evolutionary rates of the HA1 gene and protein of influenza B viruses are slower than those of influenza A viruses (1–3, 6, 11, 19, 20, 25), although the reason for this has yet to be satisfactorily explained. It is also well accepted that multiple strains of influenza B virus may cocirculate in a population at one time (14, 16, 19, 20), although how different this is from the differences observed within subtypes of influenza A virus has been challenged (6).
Influenza A viruses possess an additional means of antigenic variation: reassortment of gene segments between two viruses infecting the same host. Reassortment of avian influenza viruses with human influenza viruses has been responsible for at least two major pandemics in this century (21), and reassortment among human strains has been implicated in recent epidemics of influenza in Japan (12). Because influenza B viruses do not have an animal reservoir, antigenic shift of influenza B viruses has not been demonstrated, and recent publications on influenza B virus evolution fail to implicate reassortment among influenza B viruses as a factor in the evolution of these viruses (6, 15, 26). Preliminary evidence based on restriction endonuclease analysis of PCR-amplified gene segments NP and M indicates that reassortment does occur between circulating strains (24). However, evolutionary relationships among influenza B viruses have traditionally been assigned on the basis of antigenic or sequence data from the hemagglutinin (HA). This practice would fail to detect naturally occurring reassortant viruses.
The epidemiology of influenza A and B viruses appears to parallel their observed evolutionary behavior. Influenza A viruses cause yearly epidemics related to decreased immunity to antigenic sites on the surface glycoproteins HA and neuraminidase (NA) due to antigenic drift, punctuated by infrequent pandemics following antigenic shift. New variants succeed old viruses on a yearly basis at a fairly constant rate (21, 25). Influenza B viruses cause frequent epidemics worldwide, but with no established pattern. In some years influenza B viruses are the predominant influenza viruses isolated worldwide, and in others they are virtually absent from the human population. Reassortment between circulating strains of influenza B virus may play a role in this observed epidemiology.
In this report we describe evidence for reassortment of influenza B viruses in nature and characterize a pattern of insertions and deletions in both the HAs and the NAs of influenza B virus strains circulating in the past decade. We then describe the evolution of influenza B viruses over this period and speculate on the roles of various means of antigenic and genetic variation in the epidemiology of influenza B virus infections.
MATERIALS AND METHODS
Viruses.
Influenza viruses used in this study are listed in Table 1 with their abbreviations and sources of sequence data. Viruses whose genes were sequenced for this report were isolated in either embryonated chicken eggs or Madin-Darby canine kidney (MDCK) cells and then were propagated at least twice in MDCK cells to obtain stock viruses prior to RNA extraction and sequencing. Viruses were generously provided by Peter Wright (Nashville, Tenn.) and Paul Glezen (Houston, Tex.), were obtained by one of the authors, Shiqin He (Nanchang, China), or were from the influenza virus repository at St. Jude Children’s Research Hospital (Memphis, Tenn.).
TABLE 1.
Sequence data and abbreviations for influenza B viruses used in this study
| Strain | Abbreviation | Sequence referencea
|
|
|---|---|---|---|
| HA | NA | ||
| B/Lee/40 | Lee40 | K00423 | J02095 |
| B/Maryland/59 | Mar59 | K00424 | M30633 |
| B/Hong Kong/8/73 | HK73 | K00425 | M30631 |
| B/Singapore/222/79 | Sin79 | K00038 | M30637 |
| B/Oregon/5/80 | Ore80 | K02713 | M30636 |
| B/USSR/100/83 | USR83 | X13552 | M30638 |
| B/Victoria/3/85 | Vic85 | X13553 | M30639 |
| B/Ann Arbor/1/86 | Ann86 | U70385 | |
| B/Leningrad/179/86 | Len86 | M30632 | |
| B/Memphis/6/86 | Mem86 | X13551 | M39634 |
| B/Beijing/1/87 | Bei87 | X53098 | M54967 |
| B/Victoria/2/87 | Vic87 | M58428 | |
| B/Finland/56/88 | Fin88 | L19647 | |
| B/Yamagata/16/88 | Yam88 | M36105 | X67013 |
| B/India/3/89 | Ind89 | M65168 | |
| B/Memphis/3/89 | Mem89 | AF129889* | M30635 |
| B/Nashville/6/89 | Nas89 | AD129895* | AF129907* |
| B/Panama/45/90 | Pan90 | M65171 | AF129908* |
| B/Paris/329/90 | Par90 | M65173 | |
| B/Houston/1/91 | Hou91 | AF129896* | AF129909* |
| B/Nashville/48/91 | Nas91 | AF129897* | AF129910* |
| B/Houston/1/92 | Hou92 | AF129899* | AF129912* |
| B/Sichuan/8/92 | Sic92 | AF129898* | AF129911* |
| B/Beijing/184/93 | Bei93 | AF050061 | |
| B/Finland/268/93 | Fin93 | L76320 | |
| B/Houston/2/93 | Hou93 | AF129900* | AF129914* |
| B/Memphis/3/93 | Mem393 | AF129890* | AF129915* |
| B/Memphis/4/93 | Mem493 | AF129901* | AF129916* |
| B/Memphis/5/93 | Mem593 | AF129902* | AF129917* |
| B/Mie/1/93 | Mie93 | D38643 | |
| B/Nanchang/26/93 | Nan93 | AF134911* | AF134906* |
| B/Nashville/107/93 | Nas93 | AF129903* | AF129913* |
| B/Osaka/c19/93 | Osa93 | D38644 | |
| B/Harbin/7/94 | Har94 | AF050065 | |
| B/Kobe/1/94 | Kob94 | D38646 | |
| B/Nanchang/480/94 | Nan94 | AF134912* | AF134907* |
| B/Memphis/18/95 | Mem95 | AF129891* | AF129918* |
| B/Nanchang/8/95 | Nan95 | AF134913* | AF134908* |
| B/Alaska/12/96 | Ala96 | AF050060 | |
| B/Houston/1/96 | Hou96 | AF129904* | AF129919* |
| B/Memphis/19/96 | Mem1996 | AF129905* | AF129920* |
| B/Memphis/20/96 | Mem2096 | AF129892* | AF129921* |
| B/Nanchang/6/96 | Nan96 | AF134914* | AF134909* |
| B/Nashville/3/96 | Nas96 | AF129906* | AF129922* |
| B/Tokyo/924/96 | Tok96 | AF050067 | |
| B/Beijing/243/97 | Bei97 | AF050062 | |
| B/Memphis/10/97 | Mem1097 | AF129893* | AF129923* |
| B/Memphis/12/97 | Mem1297 | AF129894* | AF129924* |
| B/Nanchang/5/97 | Nan97 | AF134915* | AF134910* |
| B/Osaka/491/97 | Osa97 | AF05006 | |
GenBank accession number. *, gene sequenced for this study.
RNA extraction and nucleotide sequencing.
RNA was extracted from virus-containing material according to the manufacturer’s instructions (RNAeasy kit; Qiagen, Chatsworth, Calif.). Reverse transcription (RT) and PCR amplification of genes of interest were performed by using standard methodologies. RT-PCR products were purified with the QIAquick PCR purification kit (Qiagen) and sequenced by Taq Dye Terminator chemistry according to the manufacturer’s instructions (Applied Biosystems, Inc.), then analyzed on an ABI 373 DNA sequencer.
Sequence analysis and phylogenetic analysis.
Sequence analysis was carried out by using the Wisconsin Package, version 9.1, of the Genetics Computer Group, Madison, Wis. Phylogenies were estimated by the parsimony method based on nucleic acid sequences by using PHYLIP (Phylogeny Inference Package), version 3.5c, Seattle, Wash.
Nucleotide sequence accession numbers.
The HA genes sequenced in this study have been assigned GenBank no. AF129889 through AF129906 (Mem89, Mem393, Mem85, Mem2096, Mem1097, Mem1297, Nas89, Hou91, Nas91, Sic92, Hou92, Hou93, Mem493, Mem593, Nas93, Hou96, Mem1996, and Nas96, respectively) and AF134911 through AF134915 (Nan93, Nan94, Nan95, Nan96, and Nan97, respectively). The NA genes sequenced in this study have been assigned GenBank no. AF129907 through AF129924 (Nas89, Pan90, Hou91, Nas91, Sic92, Hou92, Nas93, Hou93, Mem393, Mem493, Mem593, Mem85, Hou96, Mem1996, Mem2096, Nas96, Mem1097, and Mem1297, respectively) and AF134906 through AF134910 (Nan93, Nan94, Nan95, Nan96, and Nan97, respectively).
RESULTS
Reassortment between HAs of Yam88-like viruses and the NAs of Vic87-like viruses.
The HAs of Vic87-like viruses and Yam88-like viruses can be differentiated on the basis of a number of characteristic amino acid differences (19, 25). Examination of the deduced amino acid sequences of Vic87-like and Yam88-like viruses reveals strain-specific differences in approximately 5% of the HA1. Many of these differences are located within the proposed antigenic sites of the influenza B virus HA (2, 3) and are likely related to differences in antigenicity between the strains. Recent Yam88-like viruses, referred to below as Bei93-like viruses, have accumulated additional amino acid differences.
Analysis of the coding region of all full-length NAs available indicates that there are characteristic deduced amino acid differences between the Vic87-like and Yam88-like strains in this gene as well, representing approximately 2% of the NA. All are in the stalk or the first portion of the head region of the NA, and all are contained within the reading frame of the NB protein gene and account for six deduced amino acid changes in that protein (6% of the NB). A number of silent nucleotide changes are seen in both the HA and NA sequences, following this characteristic division between lineages.
The nucleotide sequences for portions of the HA and NA of influenza B viruses isolated during this decade were determined. An approximately 550 bp segment of the HA1 including amino acids 140 to 320 and an approximately 565 bp segment of the NA including amino acids 1 to 187 were sequenced for all viruses studied, and full-length sequences of the coding regions of the HA1 and the NA of Pan90, Hou92, Mem393, Mem95, Mem2096, Mem1097, and Mem1297 were determined. The selected regions studied contain the majority of the amino acid differences observed in the full-length sequences.
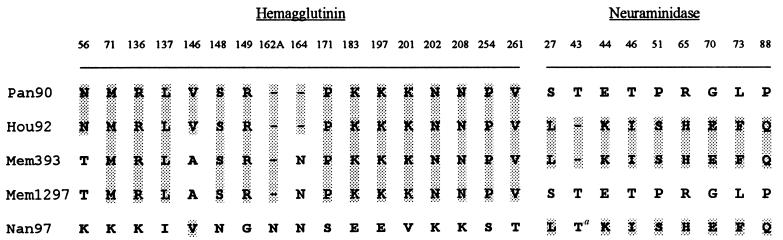
Examination of the nucleotide and deduced amino acid sequences of the HAs and NAs of influenza B viruses isolated from cities in the United States (Memphis, Nashville, and Houston) and China (Nanchang) between 1989 and 1997 reveals that a number of the viruses isolated from the United States are reassortants between the HA of a Yam88-like virus and the NA of a Vic87-like virus. One virus studied, Mem393, appears to be a reassortment between the HA of a Bei93-like virus and the NA of a Vic87-like virus. Figure 1 shows the amino acids found in selected viruses at characteristic positions that differentiate the HAs and NAs of Vic87-like viruses and Yam88-like viruses. Table 2 shows the derivations of the HA and NA genes of all viruses sequenced in this study.
FIG. 1.
Evidence for reassortment of influenza B viruses Hou92 and Mem93 by comparison of the amino acid sequences of their HAs and NAs with sequences from Pan90 (Yam88-like), Mem1297 (Yam88-like), and Nan97 (Vic87-like). Numbering is relative to that of Lee40. Amino acids contained in or identical to those in Hou92 are shaded to indicate conservation of the sequence. a, Vic87-like viruses isolated prior to 1990 have a deletion at this position.
TABLE 2.
Derivations of HA and NA genes for influenza B viruses isolated between 1989 and 1997
| Virus | Derivation of:
|
|
|---|---|---|
| HA | NA | |
| Mem89 | Vic87a | Vic87 |
| Nas89 | Vic87 | Vic87 |
| Hou91 | Yam88a | Vic87 |
| Nas91 | Yam88 | Vic87 |
| Hou92 | Yam88 | Vic87 |
| Hou93 | Yam88 | Vic87 |
| Mem393 | Yam88b | Vic87 |
| Mem493 | Yam88 | Vic87 |
| Mem593 | Yam88 | Vic87 |
| Nan93 | Yam88 | Yam88 |
| Nas93 | Yam88 | Vic87 |
| Nan94 | Yam88 | Yam88 |
| Mem95 | Yam88 | Vic87 |
| Nan95 | Vic87 | Vic87 |
| Hou96 | Yam88 | Yam88 |
| Mem1996 | Yam88 | Yam88 |
| Mem2096 | Yam88 | Vic87 |
| Nan96 | Vic87 | Vic87 |
| Nas96 | Yam88 | Yam88 |
| Mem1097 | Yam88 | Yam88 |
| Mem1297 | Yam88 | Yam88 |
| Nan97 | Vic87 | Vic87 |
Gene segment similar to those of Vic87-like or Yam88-like viruses.
Derived from a Bei93-like variant of the Yam88-like viruses.
Pattern of insertions and deletions.
A pattern of insertions and deletions in both the HAs and the NAs of influenza B viruses can be observed when the sequences of different lineages are compared (Table 3). The Vic87-like strains, which first appeared about 15 years ago, have 3 nucleotides inserted in the HA gene and 3 deleted in the NA gene relative to early influenza B viruses such as Lee40, resulting in changes in the lengths of these two genes and their deduced amino acid sequences; the change in the HA is the insertion of an Asn at position 162A by Lee40 numbering, and the change in the NA is the deletion of a Thr at position 43 by Lee40 numbering. Recent Vic87-like strains from Nanchang have regained a Thr at position 43 of the NA. The Yam88-like viruses have a deletion of an Asn in the HA relative to Lee40 at position 164, although more recent strains represented by Bei93 have regained this Asn and now have an HA and an NA identical in length to those of Lee40. Most of the reassortant viruses examined in this study have an HA deduced amino acid sequence with a deletion at position 164, as do Yam88-like viruses, but an NA sequence with a deletion at position 43 like Vic87-like viruses. The exception to this is Mem393, which appears to be a reassortant between the HA of a Bei93-like virus and the NA of a Vic87-like virus, with no change in the HA length relative to Lee40 but with a deletion at position 43 in the NA.
TABLE 3.
Insertions and deletions in the HAs and NAs of influenza B viruses are related to strain differentiation
| Virus | HA sequence (aa 159–168)a | NA sequence (aa 39–47)a | Change inb:
|
|
|---|---|---|---|---|
| HA | NA | |||
| Early B | ||||
| Lee40 | VIPK-DNNKTA | KFSSTKTTA | 0 | 0 |
| Mar59 | AVPK-NKNKTA | KFSPTKRTA | 0 | 0 |
| HK73 | AVPK---NKTA | KFSSTKRTA | −2 | 0 |
| Vic87-like | ||||
| Vic85 | AVPKNNNNKTA | KFSP-KITA | +1 | −1 |
| Mem86 | AVPKNNNNKTA | KFSP-KITA | +1 | −1 |
| Bei87 | AVPKNDNNKTA | KFSS-KITA | +1 | −1 |
| Mem89 | AVPKNDNNKTA | RFSS-KITA | +1 | −1 |
| Nas89 | AVPKNDNNKTA | KFSS-KITA | +1 | −1 |
| Nan95 | AVPKNDNNKTA | KFSPTEITA | +1 | 0 |
| Nan96 | AVPKNDNNKTA | KFSPTKTIA | +1 | 0 |
| Nin97 | AVPKNDNNKTA | KFSPTEITA | +1 | 0 |
| Yam88-like | ||||
| Sin79 | AVPK-D-NKTA | KFSPTKRTA | −1 | 0 |
| Ore80 | AVPK-D-NKTA | KFSTTKITA | −1 | 0 |
| USR83 | AVPK-D-NKTA | KFSPTKRTA | −1 | 0 |
| Yam88 | AVPR-D-NKTA | KFSPTEITA | −1 | 0 |
| Pan90 | AVPR-D-NKTA | KFSPTEITA | −1 | 0 |
| Sic92 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Nan93 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Nan94 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Hou96 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Mem1996 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Nas96 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Mem1097 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Mem1297 | AVPR-DNNKTA | KFSPTEITA | 0 | 0 |
| Reassortants | ||||
| Hou91 | AVPR-D-NKTA | KFSP-KIIA | −1 | −1 |
| Nas91 | AVPR-D-NKTA | KFSP-KIIA | −1 | −1 |
| Hou92 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
| Hou93 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
| Mem393 | AVPR-DNNKTA | KFSP-KIIA | 0 | −1 |
| Mem493 | AVPR-D-NKTA | KFSP-KIIA | −1 | −1 |
| Mem593 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
| Nas93 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
| Mem95 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
| Mem2096 | AVPK-D-NKTA | KFSP-KIIA | −1 | −1 |
Numbering is for Lee40. aa, amino acid(s). Dashes were inserted for the purpose of alignment.
Change in the number of deduced amino acids for the gene due to insertion or deletion relative to Lee40.
Phylogenetic analysis.
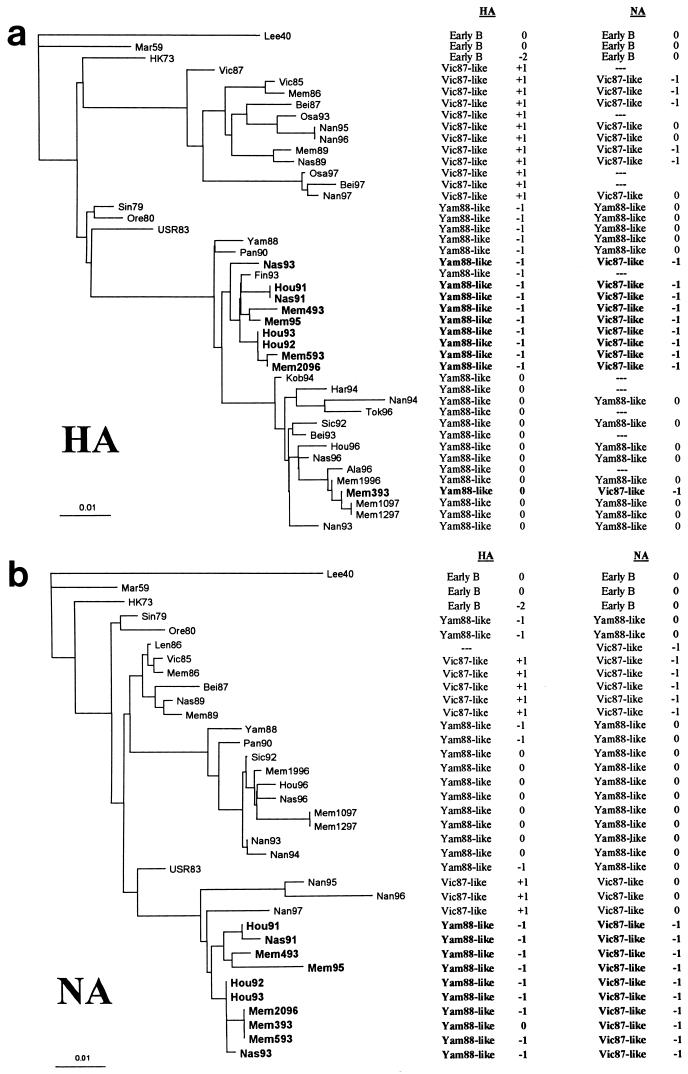
Phylogenetic trees were constructed by the distance method from the HA and NA nucleotide sequences of all viruses in this study and a number of influenza B virus sequences found in GenBank. All available influenza B virus NA sequences were used, while representative HA sequences were used in the analysis. Figure 2a shows the division of influenza B viruses into groups based on the HA1 sequence. Early B viruses, Vic87-like viruses, and Yam88-like viruses cluster separately in different branches of the tree. Yam88-like viruses are divided into one branch represented by Yam88 and Pan90 and a second branch represented by Bei93-like viruses, which have regained an Asn at position 164 relative to Lee40. The reassortant viruses have an HA1 most closely related to that of the former group of Yam88-like viruses, with the exception of Mem393. Figure 2b shows the tree generated by analysis of the NA sequences. The NAs of the reassortant viruses group with those of Vic87-like strains isolated in Nanchang within the past 4 years, indicating that the NAs of the reassortant viruses were derived from a common ancestor shared with the Vic87-like Nanchang viruses.
FIG. 2.
Phylogenetic tree of influenza B virus gene segments HA1 (a) and NA (b). Nucleotide sequences were analyzed for phylogenetic relationships by the distance method using the PHYLIP software package. Trees were constructed by the neighbor-joining method and are rooted to Lee40. The scale indicates the number of nucleotide substitutions per site. Strain abbreviations are listed in Table 1. Reassortment viruses are boldfaced. Columns adjacent to the phylogenetic tree give the derivations of the HA and NA gene segments for each strain and the lengths of the HA and NA gene segments relative to those of Lee40.
DISCUSSION
The data presented here demonstrate that at least two reassortment events occurred among influenza B viruses in the early 1990s. Similar viruses from Houston, Memphis, and Nashville contained an HA related to the Yam88-like strains which circulated in the years immediately prior to the event and an NA related to Vic87-like strains circulating in Nanchang, China. These reassortants circulated for several years and could be found as recently as 1996 in Memphis. No viruses isolated from these sites since 1996 that have been sequenced have been reassortants. The second reassortment event is presently represented by a single virus, Mem393, and differs from the former in that the HA is derived from a Bei93-like strain.
Evolutionary relationships among influenza viruses have traditionally been assigned by antigenic or sequence analysis of the HA1. This makes sense for influenza A viruses, as the antigenic domains on the HA are the most mutable parts of the virus, and selection of variants based on changes in these domains plays an important role in the evasion of the immune systems of hosts. Analysis of the ratio of amino acid substitutions to nucleotide substitutions in the HA1s of recent isolates compared to those of Yam88 (32.3 to 34.4%) and Vic87 (27.8 to 30.6%) and the evolutionary rates for the HA1s of Yam88 (3.2 × 10−3 to 3.3 × 10−3 nucleotides/site/year; 3.2 × 10−3 to 3.5 × 10−3 amino acids/site/year) and Vic87 (3.3 × 10−3 to 3.4 × 10−3 nucleotides/site/year; 2.9 × 10−3 to 3.2 × 10−3 amino acids/site/year) supports the contention by Air et al. that antibody selection does not dominate evolutionary change in influenza B viruses (1). Thus, the practice of assigning evolutionary relationships among influenza B viruses by differences in the HA alone may lead to misclassification of some viruses because it fails to account for other mechanisms of genetic variation such as reassortment.
While the HA is clearly the most important gene in terms of initiation of infection and is the most important target for the host’s immune system, the virulence of influenza viruses is a polygenic trait (10). With the increasing recognition of reassortment of influenza viruses in humans (12, 17, 18) and in animals (4, 8), knowledge of the derivations of all gene segments (or at least more than just the HA) may be desirable. In the present study, we show that the HAs and NAs of reassortants isolated in this decade are derived from different viruses. Xu et al. have offered evidence, based on analysis of restriction cleavage patterns, that the NP and M genes reassorted in some influenza B viruses isolated between 1987 and 1990 (24). Further work will be required to differentiate whether the reassortants described in this study are a homogenous population or a mixture of reassortants with multiple gene combinations.
The finding that most of the amino acid differences between the NAs of different strains are within the reading frame of the NB is interesting. Although it is believed that the NB protein plays a role in the life cycle of influenza B viruses similar to that of M2 in the influenza A virus life cycle, it is uncertain what changes to the sequence of the protein might do. The deduced changes in the NB protein would all occur in the hydrophobic region or the cytoplasmic tail of the molecule (23).
Nerome et al. have recently described a pattern of insertions and deletions in the HAs of influenza B viruses (15). The data presented here demonstrate that there is a matched pattern of insertions and deletions in both the HAs and the NAs of these viruses. Large deletions in the stalk of influenza A virus NA have been shown to impair or alter the function of the NA (5, 13). While the deletions observed in the stalk of the NA of influenza B viruses are deletions of only a single amino acid and are thus unlikely to significantly affect NA activity, it may be that altering the relative number of amino acids in the surface glycoproteins affects their interaction and that insertion or deletion in these areas is another means of generating genetic variability or of compensating for changes in the other surface glycoprotein. It is also interesting that the pattern of insertions and deletions seen in earlier Yam88-like and Vic87-like viruses appears not to have been preserved in more recently circulating viruses. Yam88-like viruses with a single deletion in the HA relative to Lee40 and an NA stalk of identical length to that of Lee40 have been replaced either by reassortants with the single deletion in the HA and a single deletion in the NA or by the Bei93-like strains, which have no deletions in the HA or NA relative to Lee40. No viruses with the original Yam88-like insertion-deletion pattern in the HA and NA can be found since 1990 in GenBank or among the more than 30 viruses sequenced for this report (data are shown in Table 3 only for viruses with sequence references for both HA and NA in Table 1), indicating that altering this pattern of insertions and deletions may play a role in strain variation in influenza B viruses. Similarly, the most recent Vic87-like viruses have regained a Thr at position 43 of the NA, bringing them within a single insertion in the HA of being identical to Lee40-like viruses in the lengths of the HA and NA.
Reassortant influenza B viruses appear to have emerged in the United States sometime during or before the 1991-to-1992 season, a season during which very little influenza B virus activity was recorded, and were replaced as the predominant circulating strains by the 1996-to-1997 season. Although reassortment might be expected to occur at a low frequency within the population and contribute to the mixed population of influenza B virus strains which is observed, it is noteworthy that the reassortant strain represented by Hou92 appears to be the dominant strain isolated in several areas of the United States (Memphis, Nashville, and Houston) and circulated for years in a fairly stable fashion, implying that these viruses possessed some sort of selective advantage in that population. During the 1992-to-1993 season, influenza B viruses with an HA antigenically similar to that of Yam88-like viruses were the predominant influenza viruses isolated and caused epidemic influenza in many parts of the world, despite the epidemics caused by Yam88-like viruses only a few years before, during the 1988-to-1989 season. It is possible that in the United States the resurgence of influenza B virus infections was due, at least in part, to this genetic reassortment event and this proposed selective advantage. Lindstrom et al. have offered similar arguments to explain an increase in epidemic influenza activity in Japan after the discovery that some influenza A viruses (H3N2) circulating in Japan between 1993 and 1997 were reassortants (12). The theoretical selective advantage that reassortant influenza B viruses might possess could be caused by an alteration in antigenic properties due to the presence of a new NA, or it could be a selective growth or replication advantage due to differing functional matches between the activities of the HA and the NA. One important caveat is that the viruses reported here were from only four sites, three in the United States and one in China. Thus, the importance of the event and the theoretical conclusions discussed here may be overestimated if this is a regional rather than a more widespread occurrence.
Reassortment appears to be a more important tool for genetic variability of influenza B viruses than was previously suspected and may help to explain the epidemiology of influenza B virus infection. The observed pattern of insertions and deletions in the surface glycoproteins deserves further study as a possible mechanism of genetic variability. This report highlights the importance of analysis of both the surface glycoproteins and perhaps the internal gene products in the evaluation of new or emerging strains of influenza B virus.
ACKNOWLEDGMENTS
This work was supported by Public Health research grant AI-08831 from the National Institute of Allergy and Infectious Diseases, Cancer Center Support (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC).
We thank Larisa V. Gubareva and Mikhail Matrosovich for helpful discussions and critical review.
REFERENCES
- 1.Air G M, Gibbs A J, Laver W G, Webster R G. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc Natl Acad Sci USA. 1990;87:3884–3888. doi: 10.1073/pnas.87.10.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berton M T, Naeve C W, Webster R G. Antigenic structure of the influenza B virus hemagglutinin: nucleotide sequence analysis of antigenic variants selected with monoclonal antibodies. J Virol. 1984;52:919–927. doi: 10.1128/jvi.52.3.919-927.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berton M T, Webster R G. The antigenic structure of the influenza B virus hemagglutinin: operational and topological mapping with monoclonal antibodies. Virology. 1985;143:583–594. doi: 10.1016/0042-6822(85)90396-4. [DOI] [PubMed] [Google Scholar]
- 4.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 5.Castrucci M R, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox N J, Bender C A. The molecular epidemiology of influenza viruses. Semin Virol. 1995;6:359–370. [Google Scholar]
- 7.Fitch W M, Leiter J M E, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinshaw V S, Bean W J, Webster R G, Sriram G. Genetic reassortment of influenza A viruses in the intestinal tract of ducks. Virology. 1980;102:412–419. doi: 10.1016/0042-6822(80)90108-7. [DOI] [PubMed] [Google Scholar]
- 9.Ina Y, Gojobori T. Statistical analysis of nucleotide sequences of the hemagglutinin gene of human influenza A viruses. Proc Natl Acad Sci USA. 1994;91:8388–8392. doi: 10.1073/pnas.91.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilbourne E D. Influenza pandemics in perspective. JAMA. 1977;237:1225–1228. [PubMed] [Google Scholar]
- 11.Krystal M, Young J F, Palese P, Wilson I A, Skehel J J, Wiley D C. Sequential mutations in hemagglutinins of influenza B virus isolates: definitions of antigenic domains. Proc Natl Acad Sci USA. 1983;80:4527–4531. doi: 10.1073/pnas.80.14.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindstrom S E, Hiromoto Y, Nerome R, Omoe K, Sugita S, Yamazaki Y, Takahashi T, Nerome K. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence of genetic reassortment of the six internal genes. J Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Air G M. Selection and characterization of a neuraminidase-minus mutant of influenza virus and its rescue by cloned neuraminidase genes. Virology. 1993;194:403–407. doi: 10.1006/viro.1993.1276. [DOI] [PubMed] [Google Scholar]
- 14.Lu B-L, Webster R G, Brown L E, Nerome K. Heterogeneity of influenza B viruses. Bull W H O. 1983;61:681–687. [PMC free article] [PubMed] [Google Scholar]
- 15.Nerome R, Hiromoto Y, Sugita S, Tanabe N, Ishida M, Matsumoto M, Lindstrom S E, Takahashi T, Nerome K. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch Virol. 1998;143:1569–1583. doi: 10.1007/s007050050399. [DOI] [PubMed] [Google Scholar]
- 16.Oxford J S, Klimov A I, Corcoran T, Ghendon Y Z, Schild G C. Biochemical and serological studies of influenza B viruses: comparisons of historical and recent isolates. Virus Res. 1984;1:241–258. doi: 10.1016/0168-1702(84)90042-x. [DOI] [PubMed] [Google Scholar]
- 17.Peng G, Hongo S, Kimura H, Muraki Y, Sugawara K, Kitame F, Numazaki Y, Suzuki H, Nakamura K. Frequent occurrence of genetic reassortment between influenza C virus strains in nature. J Gen Virol. 1996;77:1489–1492. doi: 10.1099/0022-1317-77-7-1489. [DOI] [PubMed] [Google Scholar]
- 18.Peng G, Hongo S, Muraki Y, Sugawara K, Nishimura H, Kitame F, Nakamura K. Genetic reassortment of influenza C viruses in man. J Gen Virol. 1994;75:3619–3622. doi: 10.1099/0022-1317-75-12-3619. [DOI] [PubMed] [Google Scholar]
- 19.Rota P A, Wallis T R, Harmon M W, Rota J S, Kendal A P, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 20.Rota P A, Hemphill M A, Whistler T, Regnery H L, Kendal A P. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J Gen Virol. 1992;73:2737–2742. doi: 10.1099/0022-1317-73-10-2737. [DOI] [PubMed] [Google Scholar]
- 21.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster R G, Laver W G, Air G M, Schild G C. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 23.Williams M A, Lamb R A. Determination of the orientation of an integral membrane protein and sites of glycosylation by oligonucleotide-directed mutagenesis: influenza B virus NB glycoprotein lacks a cleavable signal sequence and has an extracellular NH2-terminal region. Mol Cell Biol. 1986;6:4317–4328. doi: 10.1128/mcb.6.12.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Guo Y, Rota P A, Hemphill M, Kendal A P, Cox N. Genetic reassortment of human influenza virus in nature. In: Hannoun C, Kendal A P, Klenk H D, Rudenko A A, editors. Options for the control of influenza. II. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1993. pp. 203–207. [Google Scholar]
- 25.Yamashita M, Krystal M, Fitch W M, Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988;163:112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- 26.Zou S, Prud’homme I, Weber J M. Evolution of the hemagglutinin gene of influenza B virus was driven by both positive and negative selection pressures. Virus Genes. 1997;14:181–185. doi: 10.1023/a:1007927725332. [DOI] [PubMed] [Google Scholar]