Abstract
A Cameroonian patient with antibodies reacting simultaneously to human immunodeficiency virus type 1 (HIV-1) group O- and group M-specific V3-loop peptides was identified. In order to confirm that this patient was coinfected with both viruses, PCRs with O- and M-specific discriminating primers corresponding to different regions of the genome were carried out with both primary lymphocyte DNA and the corresponding viral strains isolated from three consecutive patient samples. The PCR data suggested that this patient is coinfected with a group M virus and a recombinant M/O virus. Indeed, only type M gag sequences could be amplified, while for the env region, both type M and O sequences were amplified, from plasma or from DNA extracted from primary lymphocytes. Sequence analysis of a complete recombinant genome isolated from the second sample (97CA-MP645 virus isolate) revealed two intergroup breakpoints, one in the vpr gene and the second in the long terminal repeat region around the TATA box. Comparison of the type M sequences shared by the group M and the recombinant M/O viruses showed that these sequences were closely related, with only 3% genetic distance, suggesting that the M virus was one of the parental viruses. In this report we describe for the first time a recombination event in vivo between viruses belonging to two different groups, leading to a replicative virus. Recombination between strains with such distant lineages (65% overall homology) may contribute substantially to the emergence of new HIV-1 variants. We documented that this virus replicates well and became predominant in vitro. At this time, group O viruses represent a minority of the strains responsible for the HIV-1 pandemic. If such recombinant intergroup viruses gained better fitness, inducing changes in their biological properties compared to the parental group O virus, the prevalences of group O sequences could increase rapidly. This will have important implications for diagnosis of HIV-1 infections by serological and molecular tests, as well as for antiviral treatment.
Phylogenetic analysis of human immunodeficiency type 1 (HIV-1) isolates from different geographic locales has revealed that HIV-1 can be divided into at least three distinctive groups, M, N, and O (41). Group M, the major group, comprises the majority of the HIV-1 strains responsible for the AIDS epidemic worldwide and can be further subdivided into at least 10 different genetic envelope subtypes (A to J) approximately equidistantly related, with intrasubtype divergence of up to 20% and intersubtype divergence of between 25 and 35% for the Env amino acid sequences (39). The genetic diversity results mainly from the error-prone nature of the viral reverse transcriptase and from the in vivo selection of variants (33). HIV-1 group O isolates represent a minority of the HIV-1 isolates and have only 50% homology with the HIV-1 group M isolates in the env gene (16, 45). HIV-1 group O seems to be endemic in West Central Africa, especially Cameroon, where the frequency of infection is estimated to be 2 to 5% of HIV-1-infected individuals (27, 31, 32). Group N viruses, recently reported for two Cameroonian patients only, are equidistant to group M and O strains (41).
Infections with HIV-1 isolates from group O present a public health problem because antibodies against them might not be detected (26, 36). Commercial viral load assays are still not able to detect and quantify viral RNA in plasma from patients infected with HIV-1 group O strains (25), and in vitro data showed that HIV-1 group O viruses are naturally resistant to nonnucleoside reverse transcriptase inhibitors, as is HIV-2 (7, 8).
Within group M, not all subtypes are pure or nonrecombinant. At least one full-length isolate characterized as a nonrecombinant has been sequenced for subtypes A, B, C, D, F, and H (21). Recombination events among sequences of different genetic subtypes of HIV-1 group M have been frequently identified. Indeed, up to 20% of the genomes that have been completely sequenced revealed a mosaic structure comprising fragments from two or more genetic subtypes (13). All representatives of subtypes E and G sequenced to date represent mosaic genomes, with parts of the genome clustering with subtype A viruses and other parts forming clearly distinguishable clades designated E and G (4, 5, 13, 14). In certain populations and regions where multiple HIV-1 subtypes cocirculate, many combinations of intersubtype recombinant viruses have been documented (A/C, A/D, B/F, A/G/H, and A/G/I) (12, 21, 34). Therefore, recombination may play a significant role in global HIV evolution, creating novel viral genotypes within given human populations.
Recombination requires the simultaneous infection of a cell with two different proviruses, allowing the encapsidation of one RNA transcript from each provirus into a heterozygous virion. After the subsequent infection of a new cell, the reverse transcriptase, by jumping back and forth between the two RNA templates, will generate a newly synthesized retroviral DNA sequence that is recombinant between the two parental genomes (17). Recombinant viruses may have some advantages over the parental strain, including eventual modifications of tropism and replication efficiency (fitness).
The first epidemiologic studies, conducted from 1988 to 1993, reported a low prevalence of HIV infection in Cameroon (29). However, since then, the reported incidence of AIDS cases increased, and this correlates well with the increased incidence and prevalence found in sentinel populations. For example, from 1985 to 1995, the seroprevalence rates of HIV infection in pregnant women and in students increased from 0.3 to 4.0%, with the highest rates in cities (28).
Recent reports have outlined a paradoxically great diversity of HIV-1 strains in Cameroon, including almost all of the group M subtypes, intersubtype recombinant viruses, and to a lesser extent the highly divergent HIV-1 group O viruses and also the recently discovered group N viruses (16, 30, 41, 42, 45).
Here we provide a comprehensive genetic analysis of an HIV-1 intergroup recombinant that arose in vivo. This intergroup M/O HIV-1 recombinant was isolated from a patient belonging to a cohort of HIV-1-infected patients initiated to study the impact of HIV-1 group M or group O viruses on disease development and pathogenicity. The characterization of this recombinant virus, which is highly replicative in vitro, illustrates the continuous emergence of new HIV-1 variants that are likely to be transmissible.
MATERIALS AND METHODS
Detection of antibodies against group O HIV-1.
The discrimination between group O and M viruses was done by serology, with all of the sera being tested for group O antibodies by an enzyme-linked immunosorbent assay with a combination of V3 peptides from HIV-1ANT70 and HIV-1MVP5180 (Research Product; Innogenetics, Antwerp, Belgium). Sera reactive in the enzyme-linked immunosorbent assay were retested in a line immunoassay, in which biotinylated V3 peptides from different group O and group M HIV-1 isolates (M-consensus, M-Mal, O-ANT70, O-VI686, and O-MVP5180) were applied as a streptavidin complex in parallel lines on nylon strips (Research Product; Innogenetics) (32).
Viral isolation and biological phenotype.
Peripheral blood mononuclear cells (PBMCs) from the HIV-positive patients were cocultivated with phytohemagglutinin (PHA)-stimulated lymphocytes from a healthy (HIV-negative) human donor in RPMI 1640 medium (Biowhittaker, Verviers, Belgium) supplemented with 15% heat-inactivated fetal calf serum (Gibco, Paisley, Scotland), 0.03% l-glutamine (Gibco), 2 μg of Polybrene (Murex, Dartford, England) per ml, antibiotics, and 20 U of recombinant interleukin-2 (Boehringer, Mannheim, Germany) per ml. The release of viral particles in the culture supernatant was examined by an HIV p24 antigen-capturing test (Innogenetics).
Syncytium formation by the HIV-1 isolates was determined with the MT-2 assay essentially as described by Koot et al. (20). HIV-1 strains obtained from the initial culture were propagated by short-term passage (7 to 10 days), and then 1 million infected PBMCs were cocultivated with 2 million MT-2 cells at a concentration of 500,000 cells per ml. HIV-1 cultures were considered to exhibit syncytia if one multinucleated giant cell per field of the light microscope was observed.
Biological cloning.
Biological clones were generated by means of a direct limiting-dilution technique from the cultured PBMCs at the third positive time point in the p24 HIV-1 antigen-capturing test. Briefly, in a 96-well microtiter plate, 100-μl volumes containing 100, 101, 102, 103, and 104 cocultured patient PBMCs were cocultivated with 100 μl containing 105 3-day PHA-stimulated donor PBMCs in RPMI 1640 medium (Biowhittaker) containing 15% inactivated fetal calf serum (Gibco), 20 U of recombinant interleukin-2 (Boehringer) per ml, 0.03% glutamine (Gibco), 2 μg of Polybrene (Murex) per ml, and antibiotics. The cultures were kept at 37°C in a 5% CO2 incubator for 4 weeks. Culture supernatants from each well were tested for the expression of p24 antigen once a week by an HIV-1 p24 antigen-capturing assay (Innogenetics). The cultures were refed with fresh medium twice a week, and fresh PHA-stimulated donor PBMCs (104) were added once a week. When fewer than 37% of the wells were positive for p24 antigen expression after 4 weeks of culture, the cells in these wells were assumed to be infected with a single virus according to the Poisson distribution (22, 37). HIV-1 clones were then expanded, and the PBMCs and supernatants were cryopreserved at −80°C until use.
Detection of group O or group M HIV-1.
Table 1 summarizes the different primer sets used for the different genomic regions. Nested PCR, specific for group O, was used to amplify a fragment of approximately 420 bp in the gp41 envelope region as previously described (2). Briefly, the outer primers (sense, 41-1; antisense, 41-4) allow amplification of HIV-1 from groups O and M, and the inner primers (sense, 41-6; antisense, 41-7) were specific for group O.
TABLE 1.
Primers used to amplify group O and group M sequences in different parts of the HIV-1 genome
| Amplified fragment | Primer | Sense | Sequence | Reference |
|---|---|---|---|---|
| Group M gag | G00 | Outer, sense | GACTAGCGGAGGCTAGAG | 35 |
| G01 | Outer, antisense | AGGGGTCGTTGCCAAAGA | 35 | |
| G60 | Inner, sense | CAGCCAAAATTACCCTATAGTGCAG | 35 | |
| G25 | Inner, antisense | ATTGCTTCAGCCAAAACTCTTGC | 35 | |
| Group O gag-pol | GAGCAM-EX5 | Outer, sense | GAGAATTCCAGGGACAAATGGTACATCA | 7 |
| Hpol 4481 | Outer, antisense | GCTGTCCCTGTAATAAACCCG | 10 | |
| GAGCAM-IN5 | Inner, sense | GAGAATTCTAAATGCATGGGTAAAGGCAGT | 7 | |
| POLCAM52 | Inner, antisense | AGGAATTCGATAGATTTGACTTGCCCAAT | 7 | |
| Group M accessory genes | VIF-1 | Outer, sense | GGGTTTATTACAGGGACAGCAGAG | 1a |
| VPU-1 | Outer, antisense | GGTTGGGGTCTGTGGGTACACAGG | 1a | |
| M-vif | Inner, sense | GGGGTCTGCATACAGGAGAA | ||
| M-vpu | Inner, antisense | TACTATRGTCCACACAACTATKGCT | ||
| Group O accessory genes | VIF 1 | Outer, sense | GGGTTTATTACAGGGACAGCAGAG | 1a |
| VPU1 | Outer, antisense | GGTTGGGGTCTGTGGGTACACAGG | 1a | |
| O-vif | Inner, sense | CATATTGGGGATTGATGCCAG | 1a | |
| O-vpu | Inner, antisense | GCATYAGCGTTACTTACTGC | 1a | |
| Group O env (gp41) | 41-1 | Outer, sense | GGGTTCTTGGGAGCAGCAGGAAGCACTATGGGCG | 2 |
| 41-4 | Outer, antisense | TCTGAAACGACAGAGGTGAGTATCCCTGCCTAA | 2 | |
| 41-6 | Inner, sense | TGGATCCCACAGTGTACTGAAGGGTATAGTGCA | 2 | |
| 41-7 | Inner, antisense | CATTTAGTTATGTCAAGCCAATTCCAAA | 2 | |
| Group M env | envA | Outer, sense | GGCTTAGGCATCTCCTATGGCAGGAAGAA | 11 |
| envN | Outer, antisense | CTGCCATTCAGGGAAGTAGCCTTGTGT | 11 | |
| ED3 | Inner, sense | TTAGGCATCTCCTATGGCAGGAAGAAGCGG | 6 | |
| ED14 | Inner, antisense | TCTTGCCTGGAGCTGTTTGATGCCCCAGAC | 6 |
A group M fragment of the envelope was amplified with a nested PCR by using as outer primers the A and N primers previously described (11) to amplify the entire gp160 region and as inner primers ED3-ED14, previously described for subsequent genetic subtyping by the heteroduplex mobility assay and to amplify specifically group M viruses (6). The amplification reaction was performed with the Expand high-fidelity PCR system according to the instructions of the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
A nested PCR was necessary to amplify the accessory gene region. The outer primers (sense, VIF-1; antisense, VPU-1) allow amplification of HIV-1 from groups O and M (1a). PCRs were carried out in a final volume of 100 μl containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase (Promega), and 0.4 μM each primer. The PCR rounds consisted of 35 cycles with a denaturing step at 94°C for 30 s, an annealing step at 50°C for 30 s, and an elongation step at 72°C for 2 min. The inner primers were specific for either group O (sense, O-vif; antisense, O-vpu) or group M (sense, M-vif; antisense, M-vpu). The PCR conditions were similar to those described for the first round.
To amplify a group M fragment of 700 bp in the gag gene, corresponding to the p24 region, a nested PCR was done with previously described primers G00 and G01 as outer primers and G60 and G25 as inner primers (35).
To amplify a group O fragment in the gag-pol region, a nested PCR was done with previously described primers GAGCAM-EX5 (7) and Hpol 4481 (10) as outer primers and GAGCAM-IN5 and POLCAM52 (7) as inner primers. The amplification reaction was performed with the Expand high-fidelity PCR system according to the instructions of the manufacturer (Boehringer Mannheim).
Cloning and sequencing.
Sequencing of the different amplified fragments was done either directly or after cloning. For direct sequencing the amplified DNA was purified by using a QiaQuick gel extraction kit (QIAGEN S.A., Courtaboeuf, France). Cycle sequencing was performed with fluorescent dye terminator technology (dye terminator cycle sequencing with AmpliTaq DNA polymerase FS [Perkin-Elmer, Roissy, France]) according to the instructions of the manufacturer. Electrophoresis and data collection were done on an Applied Biosystems 373A automatic DNA sequencer.
Some PCR fragments were purified with the QIAquick PCR purification kit (QIAGEN S.A.), cloned with the TOPO-XL-PCR cloning kit (Invitrogen, Leek, The Netherlands), and then sequenced.
Overlapping sequence fragments were assembled by using SeqEd (Applied Biosystem, Inc.) to generate the full-length sequence of the complete viral genome.
Phylogenetic analysis and recombination analysis.
Nucleotide sequences were aligned by using CLUSTAL W (43) with minor manual adjustments, considering the protein sequences. Regions that could not be aligned unambiguously, due to length or sequence variability, were omitted from the analysis. Phylogenetic trees obtained with the neighbor-joining method and reliability of the branching orders obtained with the bootstrap approach were implemented by using CLUSTAL W. Genetic distances were calculated with Kimura’s two-parameter method (19).
In order to analyze whether the viruses were recombinant in the sequenced regions, several additional analysis were performed: the Recombinant Identification Program (RIP) (40), available on-line through the Los Alamos Database (23a); diversity plotting (DIVERT), available on-line through the Agence National de Recherche sur le SIDA (ANRS) website (1); and Blast subtyping, available on-line through the National Center for Biotechnology Information website (28b). Informative site analysis (13) was done to estimate the locations and the significance of crossovers. The putative hybrid sequence was compared with a representative of each of the two HIV-1 groups inferred to have been involved in the recombination event and an appropriate outgroup. Phylogenetically informative sites in this context are those at which four taxa are divided equally into two groups, each of which has identity at that site. Each informative site supports one of the three possible phylogenetic relationships among the four taxa, and a cluster analysis maximizing the value of X2 is then used to select breakpoints among the clusters. P values for the resultant divisions were calculated by the Fisher exact test. These breakpoints were used to divide the alignment into segments for phylogenetic tree construction as described above. The positions of the breakpoints were confirmed by phylogenetic tree analysis of the corresponding regions.
Nucleotide sequence accession numbers.
The sequences of the intergroup M/O recombinant virus and the parental group M virus (vif to 5′env) have been submitted to GenBank under accession no. AJ239083 and AJ239084, respectively.
RESULTS
Patient, serology, and virus isolations.
Within the Cameroonian population studied, a patient with antibodies reacting simultaneously with group O and group M V3-loop peptides was identified. This patient was a 29-year-old unmarried woman with multiple heterosexual partners, who had received a blood transfusion. When the three samples were collected, in January 1997 (MP 575), March 1997 (MP 645), and March 1998 (MP 973), the patient was asymptomatic and belonged to stage A according to the Centers for Disease Control and Prevention clinical status classification. At a follow-up visit in October 1998, the patient was classified in stage B. Her CD4 counts decreased slightly, from 508 to 365, during the study period of about 2 years, and no weight loss was observed.
Virus was isolated from the three sequential blood samples taken from this patient. The biological phenotype on MT-2 cells was non-syncytium inducing for the three consecutive viruses isolated from this patient.
HIV-1 group O- and group M-specific PCRs.
In order to confirm whether the dual M/O seropositivity in this patient was due to a coinfection with HIV-1 group O and group M viruses, PCRs with group O- and group M-specific discriminating primers were performed for different regions of the genome. These PCRs were done with DNA extracted from primary lymphocytes and the corresponding viral strains of the three consecutive samples from this patient. Table 2 summarizes the results obtained with the different primer sets for the different genomic regions on primary and cultured PBMCs.
TABLE 2.
Results of the different group O- and group M-specific and discriminatory PCRs for different regions of the genome with primary and cultured PBMCs of the patient
| Amplified region and group | Reaction with samplea:
|
|||||
|---|---|---|---|---|---|---|
| MP 575
|
MP 645
|
MP 973
|
||||
| Primary PBMCs | Cultured virus | Primary PBMCs | Cultured virus | Primary PBMCs | Cultured virus | |
| gag, M | + | + | + | + | + | + |
| gag-pol, O | − | − | − | − | − | − |
| AGb | ||||||
| M | + | − | + | − | + | − |
| O | − | − | − | − | − | − |
| M/Oc | + | + | + | + | + | + |
| env | ||||||
| M | + | − | + | − | + | − |
| O | + | + | + | + | + | + |
Three sequential samples from the patient were tested: MP 575 (January 1997), MP 645 (March 1997), and MP 973 (March 1998).
AG, accessory genes.
A group M-specific primer was used for the 5′ end, and a group O-specific primer was used for the 3′ end.
In primary PBMCs, the gag, pol, and accessory gene regions could be amplified only with group M-specific primers, whereas the envelope region was simultaneously amplified with group O- and group M-specific primers. For the corresponding viral isolates, group M-specific primers also amplified only the gag-pol region, but only group O-specific primers could amplify the envelope region, and the accessory gene region remained negative with group O- and group M-specific primers.
These data showed that in primary PBMCs group O- and group M-type envelopes were simultaneously present, while in gag, pol, and the accessory genes only type M sequences could be detected. However, in the viral populations obtained after coculture, only group M gag and pol fragments and group O env sequences were detected. These data suggested that this patient is coinfected with a group M virus and a recombinant group M/group O virus. Since on cultured virus, neither group O- nor group M-specific primers could amplify the accessory gene region, we suspected that the recombination event occurred in this region. Indeed, a combination of M-vif and O-vpu inner primers could successfully amplify the accessory gene region in all of the samples (primary PBMCs and viral strains).
Sequence analysis of the complete genome of an intergroup M/O recombinant HIV-1 isolate.
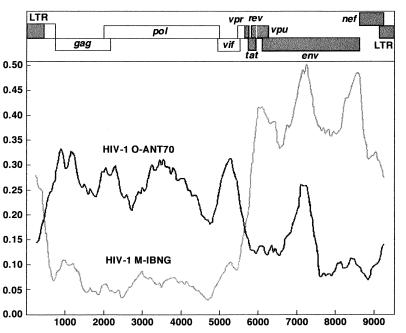
The complete genome for one sample (the 97CA-MP645 virus isolate) has been sequenced. Four different but overlapping fragments spanning the entire genome were amplified and subsequently sequenced: pol-gp41, gp41-long terminal repeat (LTR), LTR-gag, and gag-vpu. Diversity plotting, RIP analysis, and Blast subtyping clearly revealed a recombinant group M/O virus with two intergroup breakpoints, one in the vpr region, which confirms the PCR results, and a second in the LTR region. The phylogenetic analysis described below indicated that the group M sequence is close to the HIV-1IBNG sequence, an A/G intersubtype recombinant virus originating in Nigeria (4). Figure 1 shows the diversity plot analysis which indicates the two intergroup breakpoints of the 97CA-MP645 complete genome compared to HIV-1ANT-70 (group O) and HIV-1IBNG. Figure 2 shows the locations of the breakpoints in the vpr and LTR genes. In order to map more precisely the two possible breakpoints, we examined the distribution of phylogenetically informative sites along the vpr and LTR genes on an alignment including the HIV-1IBNG strain as a representative of group M, HIV-1ANT-70 as a representative of group O, and the simian immunodeficiency virus (SIV) SIVcpz-gab strain as outgroup. Each informative site supports one of three possible trees: (i) a tree in which M/O 97CA-MP645 clusters with HIV-1IBNG, (ii) a tree in which M/O 97CA-MP645 clusters with HIV-1ANT-70, and (iii) a tree in which M/O 97CA-MP645 clusters with the SIVcpz-gab outgroup strain. This analysis allowed us to locate the breakpoint between bp 201 and 209 in the vpr gene and between bp 489 and 496 in the LTR gene. The positions of the breakpoints were confirmed when other group M and group O strains were used for the analysis (data not shown). In the LTR gene the intergroup breakpoint is situated around the highly conserved TATAA box.
FIG. 1.
Diversity plot analysis of the intergroup M/O recombinant HIV-1 strain (97CA-MP645) versus HIV-1IBNG (an A/G intersubtype HIV-1 group M recombinant strain) and HIV-1ANT-70. By using the DIVERT program, which examines the extent of sequence divergence between a sequence and reference sequences, pairwise comparisons were done in an incremented window. A window size of 500 nucleotides with an increment of 50 was used. The vertical axis shows the genetic differences, expressed as percentages, between the 97CA-MP645 M/O recombinant strain and the two reference strains. The horizontal axis shows the genetic positions on the HIV-1 genome. This program is available online at the ANRS website (1).
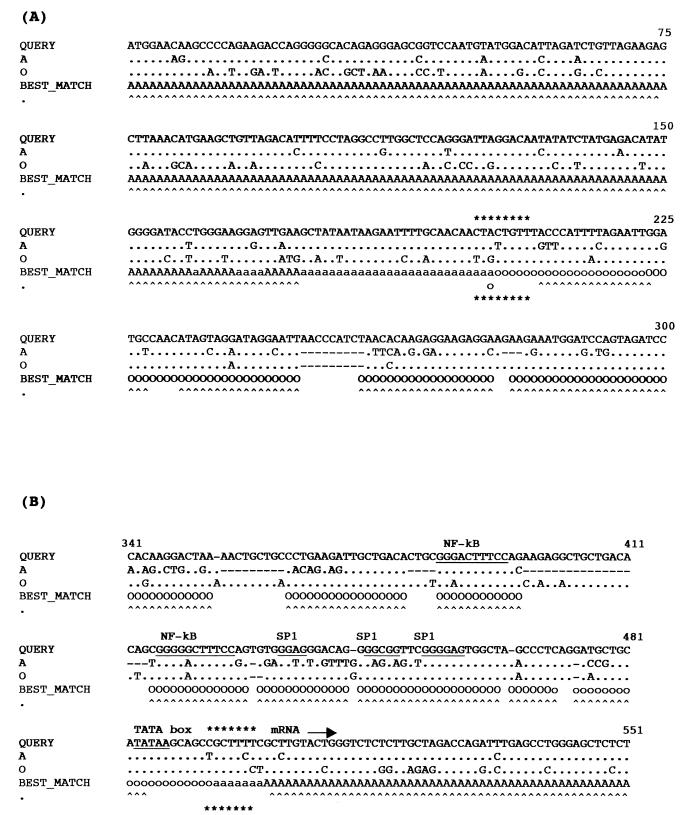
FIG. 2.
Mapping of the breakpoints of the intergroup recombinant M/O 97CA-MP645 isolate in the vpr (A) and LTR (B) genes by RIP and informative site analysis. For the RIP analysis, a window size of 100 nucleotides was used; capital letters indicate a threshold similarity of 0.9, lowercase letters indicate a 0.5 threshold similarity, and carets indicate matches with 90% certainty. The M/O 97CA-MP645 sequence was compared to HIV-1IBNG (A) (group M) and HIV-1ANT-70 (O) (group O) sequences. The putative recombination area calculated by informative site analysis is shown by asterisks above and below the RIP output alignment. The positions of the nucleotides in the genes are indicated by numbers at the right above the alignment.
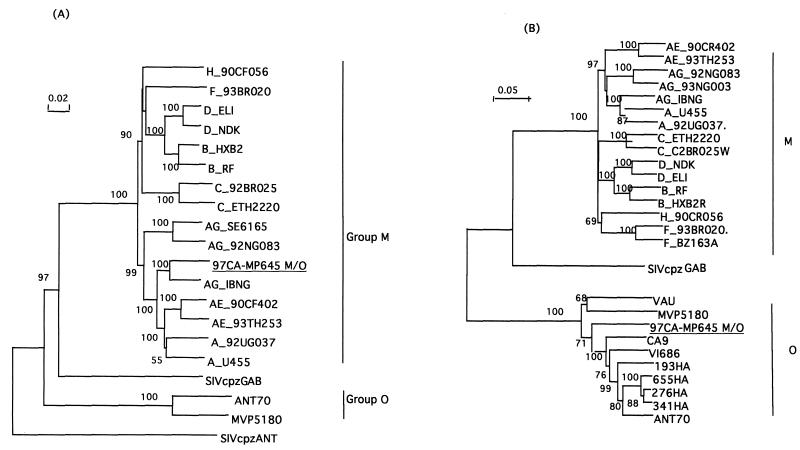
The positions of the breakpoints were confirmed by phylogenetic tree analysis: the gag-pol-vif fragment of the recombinant 97CA-MP645 strain clusters with the HIV-1IBNG strain (Fig. 3A), whereas the tat-vpu-env-nef fragment clusters with the group O viruses (data not shown). Figure 3B shows the phylogenetic analysis of the entire envelope region (gp160) of the intergroup M/O recombinant virus compared to reference strains of HIV-1 group M and group O. The gp160 sequence of our recombinant M/O 97CA-MP645 strain clusters with the ANT-70 virus. The A/G intersubtype breakpoints in the gag-pol region are similar to those observed for the HIV-1IBNG strain (data not shown). The intergroup M/O breakpoints in the vpr and LTR genes were confirmed for the primary PBMCs and the viral strains from the three consecutive samples by specific PCRs followed by sequence analysis of 800 and 1,700 bp, respectively.
FIG. 3.
Phylogenetic tree analysis of the gag-vif region (A) and the envelope gene (B) of the 97CA-MP645 intergroup M/O recombinant isolate. The nucleotide sequence of 97CA-MP645 was compared with analogous sequences from representative isolates from the Human Retroviruses and AIDS Database. The trees shown represent neighbor-joining consensus trees. (A) gag-vif region, including 4,772 aligned nucleotides after gap stripping. The tree was rooted, with the corresponding region of the chimpanzee SIVcpzANT isolate being used as an outgroup. (B) gp160, including 2,148 aligned nucleotides after gap stripping. The SIVcpzANT gp160 sequence was omitted due to the lack of C-terminal sequence information.
Partial sequence analysis of the parental HIV-1 group M strain.
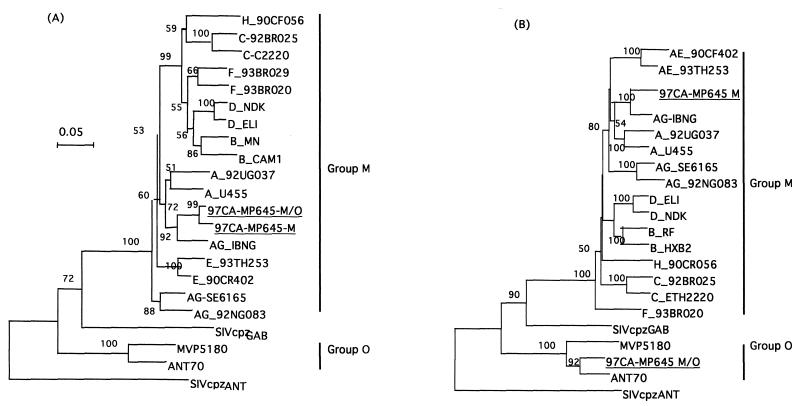
For the accessory gene region, a group M fragment could be amplified with the specific primers only for primary PBMCs. This group M fragment was amplified by a nested PCR with outer primers VIF-1 and VPU-1, which can amplify group O and M sequences in this region, followed by a second round with group M-specific primers M-vif and M-vpu. Sequence and phylogenetic analyses showed that this fragment clusters in this region of the genome with the HIV-1IBNG strain (Fig. 4A), as was previously seen for the group M gag-pol sequences from the M/O recombinant virus. Comparison of the common group M fragment (vif-vpr) between the group M and M/O viruses showed that these sequences were closely related, with a genetic distance of 3%, calculated by the Kimura two-parameter method (19) by using the same sequences as in the phylogenetic tree. A group M envelope fragment was also amplified in primary PBMCs; however, the signal was weaker than that obtained for the corresponding group O envelope sequences. A fragment of the group M envelope was amplified on primary PBMCs by a nested PCR with the group M-specific primers envA-envN as outer primers and ED3 and ED14 as inner primers. About 700 bp at the 5′ end was sequenced, and phylogenetic analysis of this group M fragment spanning the vpu gene and part of the 5′ end of the envelope gene showed that in this region the group M sequence also clusters with the HIV-1IBNG strains (Fig. 4B).
FIG. 4.
Phylogenetic tree analysis of the vif-vpr region (A) and the vpu-env region (B) of the HIV-1 97CA-MP645 intergroup M/O recombinant isolate and the 97CA-MP645 parental M virus. The trees shown represent the neighbor-joining consensus trees. The sequences were aligned against reference strains representing the different genetic subtypes and groups. The trees were rooted, with the corresponding region of the chimpanzee SIVcpzANT isolate being used as an outgroup. (A) Partial vif-vpr region, including 489 aligned nucleotides after gap stripping, of the type M fragment common to the parental group M virus and to the recombinant intergroup M/O HIV-1 isolate. (B) vpu-5′-env region, including 662 aligned nucleotides after gap stripping.
Identification of the predominant viral population.
In order to study which viral population was predominant, biological cloning was performed. Only limited amounts of primary PBMCs were available for the three sequential samples obtained from this patient; half of them were used for bulk virus isolation, and DNA was purified from the remaining primary patient material. Therefore, biological clones were prepared starting from the infected PBMCs at the third p24-positive time point from the viral culture, in order to see which viral populations were present in the viral supernatants from the three consecutive samples. More than 90 biological clones were obtained for the three sequential isolates by end point dilution cultures. The presence of the parental and recombinant viruses was checked in PCRs with the discriminating primers in the accessory gene region. Only the M/O recombinant virus was recovered in vitro. However, in the three consecutive plasma samples, group O and group M envelope sequences were amplified by PCR, although the signal was consistently weaker for the group M fragment.
DISCUSSION
In this report we describe for the first time a recombination event between viruses belonging to different groups that leads to a viable virus. We documented that this virus, which replicates well in vitro, also became predominant in vitro. The parental group M virus was detected in primary PBMCs and plasma, with a PCR signal weaker than that for the corresponding fragments of the recombinant M/O virus. On the other hand, the parental group O virus was undetectable in the patient, suggesting that the group O strain replicated even more poorly than the group M strain, which in turn replicated more poorly than the M/O recombinant. The present results demonstrate unequivocally that recombination between viruses with limited homology (65% overall homology) can occur in vivo. Most of the intersubtype mosaic genomes characterized to date originated from geographic regions where multiple subtypes cocirculate, and most of them have a complex genome structure with multiple crossover points (21, 34). The intergroup recombinant virus presented here has only two breakpoints, which induced three major changes, a chimeric vpr gene and a chimeric LTR gene leading to a heterologous TAR-M/Tat-O pair.
Recombinant viruses are already contributing substantially to the global pandemic, and the likelihood of generating recombinant viruses will continue to increase as the different HIV-1 subtypes spread to all continents (9). Virtually any isolate, including recombinant viruses, may compete more efficiently in a given region than the original genotype settled if it acquired some selective advantage. However, it remains striking to see how efficiently recombinant viruses have spread among different population groups in different geographic regions, suggesting that they could have a better viral fitness than the parental nonrecombinant strains. In Thailand, where subtypes E (an A/E recombinant) and B were initially introduced, subtype E became predominant (18). In West Africa subtype A is predominant, with prevalences ranging from 70 to >85% of the strains circulating (9); viruses similar to IBNG (an A/G recombinant) are highly represented among envelope subtype A viruses (28a). In China, subtype B and C viruses have been introduced, and a recombinant B/C virus is spreading now in different parts of the country (38). Several reports documented the introduction of subtypes A and B among intravenous-drug users in the former Soviet Union, and a recombinant A/B virus is actually rapidly spreading (3, 23).
Retrovirus recombination can generate viruses with altered biological properties. Experiments in vitro with feline and murine retroviruses have demonstrated that if appropriate selection pressures are applied, mixed infections can generate recombinant viruses with altered tissue tropism, pathogenicity, or host range or with changes in antigenic epitopes (15, 44). It is also likely that recombination can alter biological properties and pathogenesis among human retroviruses. It is important to monitor the impact of viral recombination on viral properties, since recombination may introduce genetic and biological consequences that are far greater than those resulting from the steady accumulation of single mutations.
The highest diversity between group M and group O viruses is observed in the vpu, env, and nef genes, with 40 to 60% divergence at the nucleotide level, whereas for the other genes, including vpr and LTR, the divergence ranged from 25 to 30% (1a, 16, 45). Therefore, the level of divergence or similarity does not completely explain why the recombination occurred in these parts of the genome.
Importantly, recombination between group M and O occurred in vivo. Group O viruses represent a minority of the strains responsible for the HIV-1 pandemic, and the highest prevalences have been documented in Cameroon (32). Recombination between strains with such distant lineages may contribute substantially to the emergence of new HIV-1 variants. If these recombinant intergroup viruses have a better fitness than the parental group O viruses, the prevalence of group O sequences could increase rapidly. This will have important implications for diagnosis of HIV-1 infections by serological and molecular tests and for treatment, since differences among susceptibilities to certain antiretroviral drugs have been observed in vitro (7, 24, 26). This fact also has important implications for HIV vaccine strategies with live attenuated viruses, which could potentially form recombinants with wild-type strains even if the two viruses are only distantly related. Our finding also opens the hypothesis that distant SIVs and HIV can potentially recombine, particularly in individuals who are HIV positive and exposed to SIV by cross-species transmission or vice versa. Distant SIV sequences can thus spread more efficiently into the human population.
ACKNOWLEDGMENTS
This study was supported by grants from the European Union (contract IC18-CT97-0216), ANRS (AC12), and SIDACTION.
REFERENCES
- 1.Agence National de Recherche sur le SIDA. [Online.] DIVERT. http://193.50.234.246/beaudoin/anrs/Diversity.html. [30 June 1999, last date accessed.]
- 1a.Bibollet-Ruche F, Loussert-Ajaka I, Simon F, Mboup S, Ngole E M, Saman E, Delaporte E, Peeters M. Genetic characterization of accesory genes from human immunodeficiency virus type 1 group O strains. AIDS Res Hum Retroviruses. 1998;14:951–961. doi: 10.1089/aid.1998.14.951. [DOI] [PubMed] [Google Scholar]
- 2.Bibollet-Ruche F, Peeters M, Mboup S, Ekaza E, Gandji R, Torimiro J, Mpoudi E N, Amblard J, Dibanga G, Saidou M, Esu-Williams E, Vanden Haesevelde M, Saman E, Delaporte E. Molecular characterization of the envelope transmembrane glycoprotein of 13 new human immunodeficiency virus type 1 group O strains from six different African countries. AIDS Res Hum Retroviruses. 1998;14:1281–1285. doi: 10.1089/aid.1998.14.1281. [DOI] [PubMed] [Google Scholar]
- 3.Bobkov A, Kazennova E, Selimova L, Bobkova M, Khanina T, Ladnaya N, Kravchenko A, Pokrovsky V, Cheingsong-Popov R, Weber J. A sudden epidemic of HIV type 1 among injecting drug users in the former Soviet Union: identification of subtype A, subtype B, and novel gagA/envB recombinants. AIDS Res Hum Retroviruses. 1998;14:669–676. doi: 10.1089/aid.1998.14.669. [DOI] [PubMed] [Google Scholar]
- 4.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 5.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St. Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 7.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vezinet F. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1997;71:8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Descamps D, Collin G, Loussert-Ajaka I, Saragosti S, Simon F, Brun-Vezinet F. HIV-1 group O sensitivity to antiretroviral drugs. AIDS. 1995;9:977–978. . (Letter.) [PubMed] [Google Scholar]
- 9.European Commission and Joint United Nations Programme on HIV/AIDS. HIV-1 subtypes: implications for epidemiology, pathogenicity, vaccines and diagnostics. AIDS. 1997;11:UNAIDS17–UNAIDS36. [PubMed] [Google Scholar]
- 10.Fransen K, Zhong P, De Beenhouwer H, Carpels G, Peeters M, Louwagie J, Janssens W, Piot P, van der Groen G. Design and evaluation of new, highly sensitive and specific primers for polymerase chain reaction detection of HIV-1 infected primary lymphocytes. Mol Cell Probes. 1994;8:317–322. doi: 10.1006/mcpr.1994.1043. . (Erratum, 9:373, 1995.) [DOI] [PubMed] [Google Scholar]
- 11.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao F, Robertson D L, Carruthers C D, Li Y, Bailes E, Kostrikis L G, Salminen M O, Bibollet-Ruche F, Peeters M, Ho D D, Shaw G M, Sharp P M, Hahn B H. An isolate of human immunodeficiency virus type 1 originally classified as subtype I represents a complex mosaic comprising three different group M subtypes (A, G, and I) J Virol. 1998;72:10234–10241. doi: 10.1128/jvi.72.12.10234-10241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovkina T V, Jaffe A B, Ross S A R. Coexpression of exogenous and endogenous mouse mammary tumor viruses RNA in vivo results in viral recombination and broadens the virus host range. J Virol. 1994;68:5019–5026. doi: 10.1128/jvi.68.8.5019-5026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurtler L G, Hauser P H, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsague J M, Kaptue L. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 18.Kalish M L, Baldwin A, Raktham S, Wasi C, Luo C C, Schochetman G, Mastro T D, Young N, Vanichseni S, Rubsamen-Waigmann H, et al. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS. 1995;9:851–857. doi: 10.1097/00002030-199508000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 20.Koot M, Vos A H, Keet R P, de Goede R E, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C L. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 22.Lefkovits I, Waldmann H. Limiting dilution analysis of the cells from the immune system. The clonal basis of the immune response. Immunol Today. 1984;5:265–268. doi: 10.1016/0167-5699(84)90137-3. [DOI] [PubMed] [Google Scholar]
- 23.Liitsola K, Tashkinova I, Laukkanen T, Korovina G, Smolskaja T, Momot O, Mashkilleyson N, Chaplinskas S, Brummer-Korvenkontio H, Vanhatalo J, Leinikki P, Salminen M O. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS. 1998;12:1907–1919. doi: 10.1097/00002030-199814000-00023. [DOI] [PubMed] [Google Scholar]
- 23a.Los Alamos National Laboratory. [Online.] Recombinant identification program. http://linker.lanl.gov/RIP/RIPsubmit.html. [30 June 1999, last date accessed.]
- 24.Loussert-Ajaka I, Chaix M L, Korber B, Letourneur F, Gomas E, Allen E, Ly T D, Brun-Vezinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loussert-Ajaka I, Descamps D, Simon F, Brun-Vezinet F, Ekwalanga M, Saragosti S. Genetic diversity and HIV detection by polymerase chain reaction. Lancet. 1995;346:912–913. doi: 10.1016/s0140-6736(95)92762-x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 26.Loussert-Ajaka I, Ly T D, Chaix M L, Ingrand D, Saragosti S, Courouce A M, Brun-Vezinet F, Simon F. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet. 1994;343:1393–1394. doi: 10.1016/s0140-6736(94)92524-0. [DOI] [PubMed] [Google Scholar]
- 27.Mauclère P, Loussert-Ajaka I, Damond F, Fagot P, Souquières S, Monny Lobe M, Mbopi Keou F-X, Barré-Sinoussi F, Saragosti S, Brun-Vézinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Mbopi Keou F X, Mpoudi-Ngolle E, Nkengasong J, Zekeng L, Mbanya D, Affana G, Mauclere P, Monny Lobe M, Tapko J B, Ndumbe P, Salla R, Kaptue L, Belec L. Trends of AIDS epidemic in Cameroon, 1986 through 1995. J Acquired Immune Defic Syndr Hum Retrovirol. 1998;18:89–91. doi: 10.1097/00042560-199805010-00015. [DOI] [PubMed] [Google Scholar]
- 28a.Montavon, C., C. Toure-Kane, F. Liegeois, E. Mpoudi, A. Bourgeois, E. Esu-Williams, J.-L. Perret, A. Boumah, E. Saman, S. Mboup, E. Delaporte, M. Peeters. Unpublished data. [DOI] [PubMed]
- 28b.National Center for Biotechnology Information. [Online.] Blast subtyping program. http://www.ncbi.nlm.nih.gov/retroviruses/subtype/subtype.html. [30 June 1999, last date accessed.]
- 29.Ndumbe P M, Andela A, Ndoumou A, Yanga K, Befidi R, Kaptue L, Mbede J. HIV infection in selected populations in Cameroon. AIDS. 1991;5:465–466. doi: 10.1097/00002030-199104000-00024. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 30.Nkengasong J N, Janssens W, Heyndrickx L, Fransen K, Ndumbe P M, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, et al. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Nkengasong J N, Peeters M, vanden Haesevelde M, Musi S S, Willems B, Ndumbe P M, Delaporte E, Perret J L, Piot P, van den Groen G. Antigenic evidence of the presence of the aberrant HIV-1ant70 virus in Cameroon and Gabon. AIDS. 1993;7:1536–1538. . (Letter.) [PubMed] [Google Scholar]
- 32.Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, Ouedrago R, Gandji R, Mpele P, Dibanga G, Koumare B, Saidou M, Esu-Williams E, Lombart J P, Badombena W, Luo N, Vanden Haesevelde M, Delaporte E. Geographical distribution of HIV-1 group O viruses in Africa. AIDS. 1997;11:493–498. doi: 10.1097/00002030-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 33.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 34.Robertson D L, Gao F, Hahn B H, Sharp P M. Intersubtype recombinant HIV-1 sequences, p. III 25–30. In: Korber B, et al., editors. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 35.Sanders-Buell E, Salminen M O, McCutchan F E. Sequencing primers for HIV-1, p. III 15–21. In: Myers G, et al., editors. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 36.Schable C, Zekeng L, Pau C P, Hu D, Kaptue L, Gurtler L, Dondero T, Tsague J M, Schochetman G, Jaffe H, et al. Sensitivity of United States HIV antibody tests for detection of HIV-1 group O infections. Lancet. 1994;344:1333–1334. doi: 10.1016/s0140-6736(94)90695-5. [DOI] [PubMed] [Google Scholar]
- 37.Schuitemaker H, Kootstra N A, de Goede R E, de Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Y, Su L, Zhao F, Xing H, Zhang Y Z, Wolf H, Zhang L L. Genetic recombination of HIV-1 strains identified in China, abstr. 429/11179. 12th World AIDS Conference, Geneva. 1998. [Google Scholar]
- 39.Sharp P M, Robertson D L, Gao F, Hahn B H. Origins and diversity of human immunodeficiency viruses. AIDS. 1994;8:S27–S42. [Google Scholar]
- 40.Siepel A C, Halpern A L, Macken C, Korber B T. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 41.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin M C, Saragosti S, Georges-Courbot M C, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 42.Takehisa J, Zekeng L, Ido E, Mboudjeka I, Moriyama H, Miura T, Yamashita M, Gurtler L G, Hayami M, Kaptue L. Various types of HIV mixed infections in Cameroon. Virology. 1998;245:1–10. doi: 10.1006/viro.1998.9141. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumas K M, Poszgay J M, Avidan N, Ksiazek S J, Overmoyer B, Blank K J, Prystowsky M B. Loss of antigenic epitopes as the result of env gene recombination in retrovirus-induced leukemia in immunocompetent mice. Virology. 1993;192:587–595. doi: 10.1006/viro.1993.1075. [DOI] [PubMed] [Google Scholar]
- 45.Vanden Haesevelde M, Decourt J L, De Leys R J, Vanderborght B, van der Groen G, van Heuverswijn H, Saman E. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol. 1994;68:1586–1596. doi: 10.1128/jvi.68.3.1586-1596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]